Abstract
The resistance profile and its correlation with mobile genetic elements were investigated in 11 Vibrio cholerae O1 and 2 Vibrio parahaemolyticus clinical isolates, as well as in 1 V. cholerae O1 and 1 V. cholerae non-O1 environmental isolate, isolated between 1991 and 1996 in different provinces of Angola. All clinical isolates of V. cholerae O1 were resistant to ampicillin, chloramphenicol, trimethoprim, sulfamethoxazole, and tetracycline. They also contained a large conjugative plasmid (p3iANG) with a set of three class 1 integrons harboring dfrA15, blaP1, and qacH-aadA8 cassettes, which code for resistance to trimethoprim, beta-lactams, quaternary ammonium compounds, and aminoglycosides, clustered in a 19-kb region. Chloramphenicol (cat1), kanamycin (aph), sulfonamide (sul2), and tetracycline (tetG) resistance genes were also carried on the plasmid within the same 19-kb region. A chromosomal integron containing the dfrA15 cassette was also revealed in V. parahaemolyticus strains. SXT integrase genes were present in six V. cholerae isolates but apparently were not associated with known SXT-associated resistance genes. This study indicates that plasmids and integrons contributed mainly to the circulation of multiple-drug resistance determinants in Vibrio strains from Angola.
The genus Vibrio includes harmless aquatic strains as well as strains capable of causing diseases and epidemics, mainly in developing countries. Vibrio cholerae O1 and O139, the causative agents of cholera epidemics, and Vibrio parahaemolyticus, a cause of acute gastroenteritis in humans, can be isolated from infected patients as well as from the aquatic environment.
Besides morbidity and mortality, multiple-drug resistance expressed by pathogenic bacteria is a major public health problem, and both V. cholerae- and V. parahaemolyticus-resistant strains may contribute to the spread of resistance through mobile genetic elements. Genetic horizontal transfer alone may explain the rapid acquisition of resistance determinants seen in epidemic strains and their high variability among different outbreaks in the same area. It is well known that in V. cholerae as well as in other enteric pathogens, large plasmids are able to confer resistance to drugs and to be naturally conjugative (7, 14).
Integrons are an important mechanism for the acquisition of antibiotic resistance genes in many bacteria (16, 17), including V. cholerae. These elements are not autonomously mobile but are able to capture, integrate, and express resistance cassettes in their variable regions (28). So far, several classes of integrons have been identified according to integrase sequence located in the 5′ conserved sequence (CS). Class 1 integrons have been found to be present among clinical V. cholerae isolates and are usually characterized by the presence of qacEΔ1 and sul1 genes, which provide resistance to quaternary ammonium compounds and sulfonamide, respectively, in the 3′ CS. To our knowledge, no resistance integrons have been found in Vibrio spp. other than V. cholerae, with the exception of V. fluvialis (1).
Integrative and conjugative elements (ICEs) form a large class of mobile genetic elements able to encode many properties, such as drug resistance (4). The SXT element is an ICE that contributes to horizontal transmission and rearrangement of resistance genes in V. cholerae. It was originally found in the chromosome of V. cholerae O139 MO10 from India (SXTMO10) in 1992 (2, 30). This element is able to mobilize plasmids and chromosomal DNA from strain to strain (20) and codes for resistance to chloramphenicol (coded by floR), streptomycin (strA and strB), sulfamethoxazole (sul2), and trimethoprim (dfrA18).
In relation to resistance profiles and cholera epidemics, many authors have described resistance integrons and SXT in V. cholerae (2, 10). Chromosomally located class 1 integrons, containing the aadA1 gene cassette, were found among V. cholerae O1 strains isolated during a cholera outbreak in Albania and Italy in 1994 (12). The presence of integrons of class 1 was demonstrated in V. cholerae non-O1 and in V. cholerae O139 in India (29).
In Africa, Dalsgaard et al. (9) showed that an epidemic strain of V. cholerae O1 isolated in 1996-1997 in Guinea-Bissau contained a 150-kb conjugative multiple-resistance plasmid with class 1 integron-borne gene cassettes encoding resistance to trimethoprim (dfrA12) and aminoglycosides (aadA1). An outbreak of cholera in 1998 in South Africa (8) was characterized by chromosomally located integrons containing an aadA2 gene cassette for resistance to spectinomycin and the SXT element. Nevertheless, the information about integrons and ICE circulation and the analysis of their role in drug resistance emergence are limited, at least in Africa.
In Angola, after a lapse of 12 years, a new cholera epidemic started in 1987 (13) and recurred in a seasonal pattern, spreading throughout the country. In a large microbiological study of acute diarrheal diseases (5, 6) and drug resistance conducted in 1991 to 1994 in Luanda province (Angola), we isolated V. cholerae O1 and other Vibrio spp. from patients and from the aquatic environment. During the epidemic period under study, Angola was affected by civil war; the circulation of people was very limited. In spite of this, cholera spread to at least seven provinces. V. cholerae O1 isolates from different provinces and from the environment were characterized by the presence of a large conjugative multiresistant plasmid and gave identical enterobacterial repetitive intergenic consensus profiles and PCR ribotypes, suggesting the same clonal origin (6).
In the present study, 15 representative strains of Vibrio species isolated from the environment and from patients in five different provinces of Angola, during the period from 1991 to 1996, were analyzed for the presence, location, and content of plasmids and class 1 integrons. ICE presence was also investigated to better define the mobile genetic elements borne by the strains.
MATERIALS AND METHODS
Bacterial strains and susceptibility testing.
This is a retrospective study of 15 Vibrio strains, isolated during the cholera epidemic that occurred between 1991 and 1996 in five different representative provinces of Angola. Five V. cholerae O1 isolates and two V. parahaemolyticus isolates from Luanda province were selected from a collection previously described (6). Six additional V. cholerae O1 isolates were further collected in different representative provinces (Bengo, Benguela, Cabinda, and Cuando Cubango).
The environmental V. cholerae O1 and non-O1 V. cholerae strains were isolated from Bengo River (Luanda province) surface water as described previously (6). All of the strains under study are described in Table 1.
TABLE 1.
Vibrio strains under study
| Strain | Date of isolation (day/mo/yr) | Place of isolation (Angolan province) | Resistances in addition to common profilea |
|---|---|---|---|
| V. cholerae O1 582 | 04/03/92 | Luanda | KAN, SUL, TMP, TET |
| V. cholerae O1 583 | 09/03/92 | Luanda | KAN, SUL, TMP, TET |
| V. cholerae O1 588 | 10/03/92 | Luanda | ERY, KAN, SUL, TMP, TET |
| V. cholerae O1 611 | 26/03/92 | Luanda | KAN, SUL, TMP, TET |
| V. cholerae O1 647 | 21/04/92 | Luanda | KAN, SUL, TMP, TET |
| V. cholerae O1 817 | 28/06/94 | Bengo (Caxito) | KAN, SUL, TMP, TET |
| V. cholerae O1 819 | 28/06/94 | Bengo (Dondo) | KAN, SUL, TMP, TET |
| V. cholerae O1 1383 | 09/11/94 | Benguela | KAN, SUL, TMP, TET |
| V. cholerae O1 1356 | 23/11/94 | Cuando Cubango | KAN, SUL, TMP, TET |
| V. cholerae O1 1361 | 03/05/95 | Cabinda | KAN, SUL, TMP, TET |
| V. cholerae O1 908 | 16/04/96 | Cabinda | ERY, KAN, SUL, TMP, TET |
| V. parahaemolyticus 570 | 06/12/91 | Luanda | SUL, TMP |
| V. parahaemolyticus 586 | 17/03/92 | Luanda | SUL, TMP |
| V. cholerae non-O1 698b | 02/03/93 | Bengo River | ERY, SUL, TET |
| V. cholerae O1 547b | 31/09/94 | Bengo River | KAN, SUL, TMP, TET |
Abbreviations: AMP, ampicillin; CHL, chloramphenicol; ERY, erythromycin; KAN, kanamycin; PEN, penicillin; STR, streptomycin; SPT, spectinomycin; SUL, sulfamethoxazole; TET, tetracycline; TMP, trimethoprim. The common profile is resistance to AMP, CHL, PEN, STR, and SPT.
Environmental isolate.
All of the clinical strains were isolated from stool samples and/or rectal swabs from patients. After isolation on thiosulfate citrate bile sucrose agar and identification as previously described, bacterial strains were routinely grown in Luria-Bertani (LB) and/or nutrient broth (NB) or agar plates at 37°C and maintained at −80° in LB broth containing 30% (vol/vol) glycerol. The MICs of the antimicrobial agents for the isolates under study were already determined in a previous report (6).
V. cholerae O1 N16961 (18), V. cholerae O139 MO10 (kindly provided by D. Mazel, Institute Pasteur, Paris, France), and Salmonella enterica serotype Typhimurium containing In-t1 class 1 integron (kindly provided by B. Colonna, University la Sapienza, Rome, Italy) were used appropriately as negative and positive controls in PCR experiments for ICE and for class 1 integron detection.
Transfer of drug resistance.
The following concentrations of drugs were used for selection in conjugation, transformation, and electroporation experiments according to their MICs: ampicillin (30 μg/ml), chloramphenicol (2 and 20 μg/ml), erythromycin (10 μg/ml), kanamycin (30 μg/ml), nalidixic acid (40 μg/ml), penicillin (20 μg/ml), rifampin (100 μg/ml), streptomycin (10 and 100 μg/ml), spectinomycin (20 μg/ml), sulfamethoxazole (160 and 600 μg/ml), tetracycline (2 and 10 μg/ml), and trimethoprim (10 μg/ml) (6). Conjugation assays were performed by mixing donor and recipient strains at a ratio of 3:1, filtering through Millipore 0.45-μm-pore-size filters, and incubating the filter on NB agar plates overnight at 37°C. After mating, bacteria were collected, washed, and plated on selecting plates containing appropriate drug concentrations. The frequency of transfer was expressed as the number of resistant recipient cells per donor cell in the mating mixture at the time of plating.
Rifampin- and nalidixic acid-resistant derivatives of V. cholerae O1 N16961, Escherichia coli 803 (rifampin resistant), and E. coli CSH26 (nalidixic acid resistant) were used as recipients in conjugation experiments.
Transformation of E. coli DH10b (streptomycin resistant) and electroporation of E. coli JM109 (nalidixic acid resistant) with plasmid DNA were performed as previously described (3). An Eppendorf Electroporator 2510 was used for transferring plasmids eventually unable to conjugate.
Generally at least three exconjugants or transformants from each donor were further examined by molecular procedures.
Molecular biology procedures.
Bacterial DNA for PCR testing was extracted according to the technique described by Ausubel et al. (3). The amplicons to be sequenced were directly purified from PCR by a Nucleospin extract kit (Macherey-Nagel, Oensingen, Switzerland), cloned into pGEM-T Easy vector system I (Promega, Madison, WI), or extracted from agarose gel by a Rapid Gel extraction system (Marligen BioSciences, Ijamsville, MD) according to the manufacturer's instructions.
Plasmid DNA preparation was obtained as described by Holmes and Quigley (21) or by a Nucleobond PC500 purification kit (Macherey-Nagel, Oensingen, Switzerland) according to the manufacturer's instructions.
Specific primer pairs and PCR conditions for the detection of class 1 integrons, the blaP1 β-lactamase cassette, and SXT int, floR, strA, strB, sul2, dfrA18, and dfrA1 were already described and are listed in Table 2. We designed original primer pairs for PCR detection of aadA1 resistance to the spectinomycin cassette, dfrA15 resistance to the trimethoprim cassette (Fig. 1), and the internal sequence of the SXT int gene, as well as primers 3′CSrev, 5′CSrev, dfr-F, and bla-F, to map the relative displacement of integrons (Fig. 1). We also designed int1A (to be paired with int1B), sul1A (to be paired with 5′CSrev), and dfrF2-sul1B to sequence one of the resulting bond regions. All primers are listed in Table 2.
TABLE 2.
PCR primers used in this study
| Primer and purpose | Sequence (5′ to 3′) | Target gene or description | Amplicon size (bp) | Reference, accession no., and/or chromosomal locationd |
|---|---|---|---|---|
| Class 1 integron and cassette detection | ||||
| In-F | GGCATCCAAGCAGCAAG | Class 1 integron | Variable | 23 |
| In-B | AAGCAGACTTGACCTGA | |||
| Dfr-Ba | TCACCTTCTGGCTCAATGTCG | dfrA15 | 490 | Z83311, bp 374-354 |
| Aad-Ba | GCCGACTACCTTGGTGATCTCG | aadA1 | 1,476 | AY103457, bp 786-765 |
| blaP1-Ba | CTGGTTCATTTCAGATAGCG | blaP1 | 874 | 11 |
| Integron cluster mapping and characterization | ||||
| 3′CSrev | AGTCCAGTTCAGACGAA | Integron spacers | Variable | U12338, bp 1433-1416 |
| 5′CSrev | GAACGACGAACCTACGG | U12338, bp 4814-4831 | ||
| Dfr-Fb | GGAGTTATCGGAAATGGCCCAG | Integron spacers | Variable | Z83311, bp 37-58 |
| Bla-Fb | GGCAATCAGCGCTTCCCGTT | AF221899, bp 334-353 | ||
| Dfr-F2 | TTAGCCAAGACTTCGTGT | dfrA15-sul1 | AB113114, bp 515-532 | |
| Sul1B | GCAAGGCGGAAACCCGCGCC | X12869, bp 1360-1341 | ||
| Int1A | AAAACCGCCACTGCGCCGTTA | intI1 | 1,200 | 12 |
| Int1B | GAAGACGGCTGCACTGAACG | |||
| Sul1Ac | GCACGTGCTGTCGAACCTTCA | AY0462763, bp 38844-38865 | ||
| Aph F | GGCAATCAGGTGCGACAAT | kan | 484 | AY090559, bp 12978-12997 |
| Aph R | GTGACGACTGAATCCGGTGA | AY090559, bp 13461-13441 | ||
| CAT1-F | GGCATTTCAGTCAGTTG | cat1 | 584 | V00622, bp 312-328 |
| CAT1-B | CCGCCCTGCCACTCATC | V00622, bp 880-896 | ||
| TET(G) 1 | GCTCGGTGGTATCTCTGCTC | tetG | 468 | 25 |
| TET(G) 2 | AGCAACAGAATCGGGAACAC | |||
| ICE detection and characterization | ||||
| Int1-F | GCTGGATAGGTTAAGGGCGG | SXT int | 592 | 8 |
| Int1-B | CTCTATGGGCACTGTCCACATTG | |||
| Nint-F | TACCTACAGCAGGAACGGGC | SXT int | 258 | AF099172, bp 255-274 |
| Nint-B | GCAGCACAGACACCAGACGT | AF099172, bp 513-494 | ||
| FLOR-F | TTATCTCCCTGTCGTTCCAGCG | floR | 526 | 22 |
| FLOR-2 | CCTATGAGCACACGGGGAGC | |||
| TMP-F | TGGGTAAGACACTCGTCATGGG | dfr18 | 389 | 19 |
| TMP-B | ACTGCCGTTTTCGATAATGTGG | |||
| SUL2-F | AGGGGGCAGATGTGATCGC | sul2 | 625 | AY034138, bp 16726-16706 |
| SUL2-B | TGTGCGGATGAAGTCAGCTCC | 19 | ||
| STRA-F | TTGATGTGGTGTCCCGCAATGC | strA | 383 | 19 |
| STRA-B | CCAATCGCAGATAGAAGGCAA | |||
| strB-F | GGCACCCATAAGCGTACGCC | strB | 470 | 10 |
| strB-R | TGCCGAGCACGGCGACTACC | |||
| dfr1-F | CGAAGAATGGAGTTATCGGG | dfrA1 | 372 | 22 |
| dfr1-B | TGCTGGGGATTTCAGGAAAG |
Paired with In-F.
Paired by different primer pair combination for mapping the integron cluster (see the text).
To be paired with 5′CSrev.
Primers without references were newly designed.
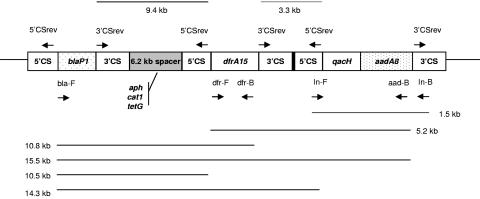
FIG. 1.
Map of the 19-kb region on the p3iANG plasmid, clustering together the three integrons and chloramphenicol, kanamycin, and tetracycline resistance genes (approximate sizes; not to scale). The upper and lower lines represent the amplicons obtained by the primer pairs as indicated and oriented by arrows. Primer pairs and resulting amplicons for sequencing the 3.30-kb amplicon are described in the text; the 140-bp junction between the two integrons is indicated by the solid black rectangle. Sequence DQ227350 corresponds to the 3.3-kb 3′CSrev/5′CSrev amplicon. Sequence DQ149925 corresponds to the contiguous 1.5-kb In-F/In-B amplicon.
All PCRs were set in a 50-μl volume of reaction buffer containing 1 U of Taq polymerase as directed by the manufacturer (Promega).
Integron mapping required the amplification of large templates (>3,000 bp). Therefore, we utilized a specific Taq polymerase. PCR amplification of large fragments was set in a 50-μl volume of reaction buffer containing 2.5 U of La TaqTM′ polymerase, as directed by the manufacturer (TaKaRa BIO Inc.). Cycling conditions were the following: initial denaturation at 94°C for 1 min was followed by 30 cycles of amplification at 98°C for 10 s, annealing at 55 to 65°C (according to primer pair melting temperature) for 1 min, extension at 68°C for 15 min, and a last cycle of 72°C for 10 min. All amplifications were carried out in an MJ Research, Inc. DNA thermal cycler. Optimal conditions were set in an Eppendorf Mastergradient thermal cycler.
Electrophoresis of amplicons was performed with 1 or 0.8% agarose (Bio-Rad) gels in 40 mM Tris-acetate, 1 mM EDTA. Nucleotides sequences were determined by the Istituto Dermopatico dell’Immacolato-Istituto di Ricovero e Cura a Carattere Scientifico (Nucleic Acid Facility, Rome, Italy) by using the Sanger method and an ABI Prism 377-96 genetic analyzer as well as a Servizio Sequenziamento Automatico (Progetto Camilla, Pomezia, Italy). DNAMAN software was used to design DNA primers and to analyze DNA sequences against GenBank.
Nucleotide sequence accession numbers.
The accession numbers of the integron sequences described herein were submitted to GenBank and have been assigned accession numbers DQ149925 (qacH-aadA8) and DQ227350 (qacE, sul1, orf5, and int1).
RESULTS
Antimicrobial susceptibility and plasmid-mediated resistance transfer.
Testing of the drug resistance patterns was in accordance with our previous report and revealed that, irrespective of species, all 15 Vibrio strains were susceptible to nalidixic acid and rifampin and showed a common profile of multiresistance to ampicillin, chloramphenicol, penicillin, streptomycin, and spectinomycin (6).
Detailed resistance profiles of each strain are listed in Table 1. Among clinical V. cholerae O1 strains, besides the common resistance pattern, all showed resistance to kanamycin, trimethoprim-sulfamethoxazole, and tetracycline. Additional resistance to erythromycin was also found in V. cholerae O1 588 and 908. Both V. parahaemolyticus 570 and 586 showed resistance to trimethoprim-sulfamethoxazole in addition to having the common profile. Among the environmental strains, the toxigenic V. cholerae O1 547 was resistant to kanamycin and trimethoprim-sulfamethoxazole, similar to the clinical strains.
As drug resistance is often associated with genetic mobile elements, we preliminarily submitted all the strains to conjugation experiments and monitored resistance transfer to recipient strains. Ampicillin, spectinomycin, and trimethoprim resistances were used as selective donor markers according to resistance profiles, and nalidixic acid or rifampin was used as a selective recipient marker. The environmental V. cholerae non-O1 698 and V. cholerae O1 547 did not show any detectable transfer in repeated mating with different selective plates. All of the clinical V. cholerae O1 isolates were able to transfer into both V. cholerae O1 and E. coli (and back into V. cholerae O1) a common multiple resistance to ampicillin, chloramphenicol, kanamycin, streptomycin, spectinomycin, sulfamethoxazole, tetracycline, and trimethoprim (the selective transferable marker). Erythromycin resistance was cotransferred with the common resistance profile by V. cholerae O1 588 and 908 donors (Table 3). Conjugative transfer of the resistance traits occurred at a frequency of 4 × 10−3 to 1 × 10−4 transconjugants per donor. These findings suggested the presence of a common conjugative plasmid. DNA extraction from V. cholerae O1 582 and from the respective E. coli recipients confirmed the presence of a plasmid population of about 100 kb in size associated with multiresistance. We named this conjugative plasmid present in clinical V. cholerae O1 strain p3iANG.
TABLE 3.
Resistances transferred and class 1 integron cassettes identified in Vibrio strains under study
| Strain | Transferred resistances in addition to common profilea | Class 1 integron cassette(s) |
|---|---|---|
| V. cholerae O1 582d | KAN, STR, SPT, TET | dfrA15, blaP1, qacH-aadA8b,g |
| V. cholerae O1 583d | KAN, STR, SPT, TET | dfrA15, blaP1, qacH-aadA8b |
| V. cholerae O1 611d | KAN, STR, SPT, TET | dfrA15, blaP1, qacH-aadA8b |
| V. cholerae O1 1383d | KAN, STR, SPT, TET | dfrA15, blaP1, qacH-aadA8b |
| V. cholerae O1 647 | KAN, STR, SPT, TET | dfrA15, blaP1, qacH-aadA8b |
| V. cholerae O1 817 | KAN, STR, SPT, TET | dfrA15, blaP1, qacH-aadA8b |
| V. cholerae O1 819 | KAN, STR, SPT, TET | dfrA15, blaP1, qacH-aadA8b |
| V. cholerae O1 1356 | KAN, STR, SPT, TET | dfrA15, blaP1, qacH-aadA8b |
| V. cholerae O1 1361 | KAN, STR, SPT, TET | dfrA15, blaP1, qacH-aadA8b |
| V. cholerae O1 588d | ERY, KAN, STR, SPT, TET | dfrA15, blaP1, qacH-aadA8b |
| V. cholerae O1 908 | ERY, KAN, STR, SPT, TET | dfrA15, blaP1, qacH-aadA8b |
| V. parahaemolyticus 570 | dfrA15c,g | |
| V. parahaemolyticus 586 | dfrA15c,g | |
| V. cholerae non-O1 698d | NCe | |
| V. cholerae O1 547 | NCf | dfrA15, blaP1, qacH-aadA8b |
Abbreviations: AMP, ampicillin; CHL, chloramphenicol; ERY, erythromycin; KAN, kanamycin; PEN, penicillin; STR, streptomycin; SPT, spectinomycin; SUL, sulfamethoxazole; TET, tetracycline; TMP, trimethoprim. The transferred common profile is resistance to AMP, CHL, SUL, and TMP.
Plasmid located.
Chromosome located.
SXT integrase positive.
NC, not conjugative.
AMP, CHL, and TMP resistance transferred by electroporation.
Confirmed by amplicon sequencing.
Since the environmental V. cholerae O1 547 showed a resistance profile similar to that of the clinical V. cholerae O1 isolates but was unable to conjugate under any condition, we tried to monitor resistance transfer by electroporation. Eventually, we succeeded in transferring the plasmid into E. coli JM109. This transformed strain was still unable to conjugate. The plasmid size was apparently similar to that of p3iANG.
Regarding V. parahaemolyticus, both the 570 and 586 isolates were submitted to mating experiments with E. coli as the recipient, and the comobilization of ampicillin, chloramphenicol, trimethoprim, and sulfamethoxazole resistance was demonstrated, evidence of the presence of a conjugative plasmid. DNA extraction both from donors and from exconjugants confirmed the presence of a plasmid approximately 80 kb in size.
Occurrence of class 1 integrons and cassette characterization in V. cholerae.
In-F and In-B primers, located in the 5′ and 3′ CS regions, respectively, were used to identify class 1 integron-bearing strains among Vibrio isolates under study.
All clinical and environmental V. cholerae O1 isolates yielded a common profile of three different amplicons: A, B, and C. Their approximate sizes were 0.75 kb, 1.2 kb, and 1.5 kb, respectively, and they are evidence of three independent integrons. V. cholerae non-O1 698 did not show any amplification; thus, this isolate did not seem to contain class 1 integrons.
The amplicons obtained from V. cholerae O1 582 as a representative strain were sequenced for characterization of their cassette content. Three different cassettes were identified: the dfrA15 (accession no. AF221900) dihydrofolate reductase gene for trimethoprim resistance in amplicon A, the blaP1 β-lactamase gene (accession no. AF221899) in amplicon B, and the qacH (accession no. AF205943) gene for quaternary ammonium compound resistance in amplicon C. The last cassette was followed by an aadA8 aminoglycoside 3′ adenyl transferase resistance cassette (accession no. AY139603); (Fig. 1).
In order to confirm the presence of these integron cassettes in all the other isolates, we performed PCR testing with specific internal primer pairs (Fig. 1). In-F as 5′ primer and dfr-B, blaP1-B, and aad-B as 3′ primers were used to detect, respectively, dfrA15 and blaP1 cassettes and the qacH-aadA8 sequence. All V. cholerae O1 isolates conserved the identity of the common integron profile, including the environmental isolate.
Since all the integron-bearing isolates contain transferable plasmids associated with multiple resistance, we tracked the integron mobilization into E. coli recipients using In-F and In-B primers. All of the E. coli exconjugants were positive for the triple integron profile, indicating its location on the p3iANG plasmid.
Mapping of p3iANG plasmid integrons.
To define the relative distances between the three integrons on the plasmid, we performed a PCR test utilizing the 3′CSrev/5′CSrev primer pair (in an orientation opposite that of In-F and In-B) (Fig. 1). This amplification yielded two amplicons of approximately 9,400 bp and 3,300 bp (Fig. 1), suggesting that the three integrons were clustered at a relatively short distance from each other. The latter PCR product was cloned in pGEM-T, resulting in plasmid pSPA12. According to the primer location and the integron structure, the amplicons were expected to include the terminal 3′ CS (about 1.9 kb) of the integron at one end and the 5′ CS (about 1.3 kb) of the following integron at the other end. Thus, the first amplicon of 9.4 kb apparently contained a spacer of about 6.2 kb between the two integrons, while the second amplicon of 3.3 kb was apparently lacking any spacer between the two integrons (Fig. 1). To determine the correct order of the three integrons, we performed amplification with the following primer combinations: bla-F/aad-B, bla-F/dfr-B, and dfr-F/aad-B. These primer pairs gave the following amplicon sizes, respectively: 15.5 kb, 10.8 kb, and 5.2 kb. The combination of bla-F/5′CSrev primer pair gave two amplicons, 10.5 and 14.3 kb, an indication that the blaP1 integron cassette was followed by two 5′ CS regions corresponding to two integrons. The combination of 3′CSrev/blaP1-B gave no amplicons, confirming the upstream displacement of the blaP1 integron cassette (Fig. 1).
To characterize the joining region of the dfrA15 and qacH-aadA8 integrons (3.3-kb amplicon), we amplified three overlapping amplicons (dfrF2-sul1B with p3iANG DNA as template as well as int1A-int1B and sul1A-5′CSrev with pSPA12 DNA as template, with sizes 1.3, 1.2, and 2.3 kb, respectively) spanning from the dfrA15 cassette and including the 3.3-kb 3′CSrev/5′CSrev region. This sequence includes, as expected, the following genes: the remnants of qacE, sul1, and orf5 and the downstream portion of int1 (in opposite orientation as that found in the 5′ CS); the two integrons, in a head-to-tail string, were tied by a 140-bp noncoding junction (Fig. 1).
To characterize the 6.2-kb spacer, the 9.4-kb amplicon was purified from agarose gel and used as template for specific PCR detection of the aph, cat1, and tetG resistance genes (in accordance with the transferable resistance profile). They were positive and therefore localized in the 6.2-kb spacer.
Analyzing the results obtained by the above-described matching and sequencing, we succeeded in mapping the hypothetical order of the integron region: blaP1 integron and 6.2-kb spacer (containing cat1, aph, and tetG), followed by dfrA15 integron joined head to tail with the qacH-aadA8 integron, all clustered with the same orientation in a 19-kb region of the p3iANG plasmid (Fig. 1).
Integrons in V. parahaemolyticus.
Both V. parahaemolyticus 570 and 586 were submitted to the integron investigation. Their DNA yielded one amplicon of 0.75 kb. An internal specific amplification by the In-F/dfr1-B primer pair revealed the presence of the dfrA15 cassette, confirmed by amplicon sequencing. On the contrary, exconjugants from V. parahaemolyticus 570 and 586 into E. coli gave a negative result. This finding suggested no association of the integron with the transferable resistance plasmid.
ICE detection and mobility.
Since SXT contributes to multiple antibiotic resistance, is known to be conjugative, and is involved in plasmid mobilization in V. cholerae (20), we investigated its presence both in V. cholerae isolates and in their E. coli exconjugants.
PCR analysis was conducted to look for the SXT integrase gene with int1-F- and int1-B-specific primers (Table 2), and amplicon identity was confirmed for each isolate either by nested PCR with nint-F- and nint-B-specific primers or by sequencing (94% of homology to SXT integrase; accession no. AF099172). An SXT integrase gene was found in five clinical V. cholerae O1 isolates (four from Luanda and one from Benguela) and in the environmental non-O1 isolate. Some of the exconjugants (6 out 10) obtained from V. cholerae O1 isolate 582 were also positive for the presence of the SXT integrase gene, suggesting its cotransfer into E. coli with p3iANG in conjugation experiments. V. cholerae O1 583, 588, and 611 from Luanda and isolate 1383 from Benguela were integrase positive, but their exconjugants were negative.
Since both positive and negative integrase strains showed the ability to transfer the p3iANG plasmid, we deduced that there was no direct correlation between its presence and plasmid mobility.
ICE characterization.
Isolates positive for STX integrase genes were tested for the presence of the ICE-associated resistance genes floR, strA, strB, sul2, dfrA18, and dfrA1 (22). All of them were negative for floR, strA, strB, dfrA18, and dfrA1. The specific sul2 primer pair gave the expected 625-bp amplicon (and its sequence was fully homologous to sul2) only in those isolates bearing the p3iANG plasmid. Since isolates and exconjugants characterized by integrase-negative, p3iANG-positive profiles were sul2 positive and the integrase-positive, p3iANG-negative isolate (V. cholerae non-O1 698) was sul2 negative, we deduced that the resistance gene sul2 should not be associated with integrase presence but should be located on p3iANG only, though not located on the 6.2-kb spacer, as indicated by specific PCR testing of the 9.4-kb amplicon.
DISCUSSION
The last cholera epidemic in Angola started in 1987 and persisted. Resistance to drugs soon became an important public health problem (5, 6, 13). This is shown by the evidence that all of the Vibrio isolates under study showed resistance to at least seven drugs. We found the ubiquitous presence (from the Cabinda enclave in the north to Cuando Cubango province in the south) of the conjugative p3iANG plasmid, the main factor of resistance spread among clinical and environmental V. cholerae O1 in Angola, mainly because of integron presence. These integrons contained resistance cassettes not found in Africa before. Both dfrA15 and blaP1 integron cassettes showed complete identity to the cassettes in Thailand described previously (11). The qacH cassette was previously found in E. coli in a class 1 integron carried by the plasmid-located In53 (24); our integron also bears qacEΔ1 in the 3′ CS and therefore contains two determinants for ammonium quaternary compound resistance. However, it was established that qacEΔ1 does not provide resistance to ammonium compounds (26), and we did not find any enhanced phenotypic resistance to disinfectants in these strains (data not shown). This new resistance cluster, flanked by the blaP1 integron on the left and the tail-to-head dfrA15-qacH-aadA8 integron tandem on the right, and separated by the 6.2-kb spacer containing the resistance genes to chloramphenicol, kanamycin, and tetracycline (this internal resistance assembly also was not described before), constitutes about 20% of the plasmid and suggests that it is an autonomous mobile element itself that merits further investigation.
A large conjugative resistance plasmid with class 1 integron-borne gene cassettes (encoding resistance to trimethoprim, dfr12, and aminoglycosides, aadA1) was also found in 1996 in Guinea-Bissau (9), but no evidence relates it to the Angolan p3iANG plasmid. This comparison, nevertheless, confirms the high variability of plasmids and integrons in epidemic V. cholerae O1.
V. parahaemolyticus isolates contain the dfrA15 class 1 integron not associated with its conjugative resistance plasmid. We have no evidence to hypothesize the identity or common origin with the analogous integron found on the V. cholerae O1 p3iANG plasmid. Its presence, however, may indicate a strong pressure on Vibrio strains to select resistance to trimethoprim and sulfonamides, which are widely used in drug therapy.
Although superintegrons have already been described for many Vibrio spp., including V. parahaemolyticus (27), to our knowledge this is the first report of the presence of class 1 integrons in V. parahaemolyticus isolates. This reinforces the idea that integrons represent a major factor for setting the drug resistance profile in bacterial strains. Our findings are further evidence of the main role played by these elements in genome plasticity and variability in the Vibrio genus.
SXT integrase interestingly characterized most of the clinical V. cholerae O1 strains isolated in 1992, the same year of isolation in India of V. cholerae O139 MO10 containing SXT. We have evidence that integrase was transferred into an E. coli recipient from at least one of these isolates and that it was not associated with resistance genes. Apparently this element made no contribution to resistance performance in our isolates. Hochhut et al. (19) also described an SXT (SXTS) unable to confer a phenotypic pattern, because the resistance cluster was entirely deleted. A preliminary molecular characterization (data not reported) of the ICE scaffold associated with Angolan integrase and its chromosomal integration site showed a high degree of divergence to that of SXTMO10. Its molecular structure and genetic implications are the topics of a manuscript in preparation.
Acknowledgments
We are very grateful to M. Waldor and V. Burrus for valuable advice, G. Prosseda and S. Bani for technical help, M. L. Bernardini and B. Colonna for encouragements and critical suggestions, and J. Cliff and C. Grim for paper revision.
This work was supported by MURST and by MAE-DGCS, Italy. D. Ceccarelli was supported by a “Cellular and Developmental Biology” doctorate fellowship, Dip. Biologia Cellulare e Sviluppo, Univ. Roma, La Sapienza, Italy.
REFERENCES
- 1.Ahmed, A. M., T. Nakagawa, E. Arakawa, T. Ramamurthy, S. Shinoda, and T. Shimamoto. 2004. New aminoglycoside acetyltransferase gene, aac(3)-Id, in a class 1 integron from a multiresistant strain of Vibrio fluvialis isolated from an infant aged 6 months. J. Antimicrob. Chemother. 53:947-951. [DOI] [PubMed] [Google Scholar]
- 2.Amita, S. R. Chowdhury, M. Thungapathra, T. Ramamurthy, G. B. Nair, and A. Ghosh. 2003. Class I integrons and SXT elements in El Tor strains isolated before and after 1992 Vibrio cholerae O139 outbreak, Calcutta, India. Emerg. Infect. Dis. 9:500-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A Smith, and K. Struhl. 1990. Current protocols in molecular biology. Greene Publishing Associates and Wiley Interscience, New York, N.Y.
- 4.Burrus, V., and M. K. Waldor. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155:376-386. [DOI] [PubMed] [Google Scholar]
- 5.Colombo, M. M., M. Francisco, B. D. Ferreira, S. Rubino, and P. Cappuccinelli. 1993. The early stage of the recurrent cholera epidemic in Luanda, Angola. Eur. J. Epidemiol. 9:563-565. [DOI] [PubMed] [Google Scholar]
- 6.Colombo, M. M., S. Mastrandrea, F. Leite, A. Santona, S. Uzzau, P. Rappelli, M. Pisano, S. Rubino, and P. Cappuccinelli. 1997. Tracking of clinical and environmental Vibrio cholerae O1 strains by combined analysis of the presence of toxin cassette, plasmid content and ERIC PCR. FEMS Immunol. Med. Microbiol. 19:33-45. [DOI] [PubMed] [Google Scholar]
- 7.Coppo, A., M. M. Colombo, C. Pazzani, R. Bruni, K. A. Mohamud, K. H. Omar, S. Mastrandrea, A. M. Salvia, G. Rotigliano, and F. Maimone. 1995. Vibrio cholerae in the Horn of Africa: epidemiology, plasmids, tetracycline resistance gene amplification, and comparison between O1 and non-O1 strains. Am. J. Trop. Med. Hyg. 53:351-359. [DOI] [PubMed] [Google Scholar]
- 8.Dalsgaard, A., A. Forslund, D. Sandvang, L. Arntzen, and K. Keddy. 2001. Vibrio cholerae O1 outbreak isolates in Mozambique and South Africa in 1998 are multiple-drug resistant, contain the STX element and the aadA2 gene located on class 1 integrons. J. Antimicrob. Chemother. 48:827-838. [DOI] [PubMed] [Google Scholar]
- 9.Dalsgaard, A., A. Forslund, A. Petersen, D. J. Brown, F. Dias, S. Monteiro, K. Molbak, P. Aaby, A. Rodrigues, and A. Sandstrom. 2000. Class 1 integron-borne, multiple-antibiotic resistance encoded by a 150-kilobase conjugative plasmid in epidemic Vibrio cholerae O1 strains isolated in Guinea-Bissau. J. Clin. Microbiol. 38:3774-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalsgaard, A., A. Forslund, N. Tam, D. X. Vinh, and P. D. Cam. 1999. Cholera in Vietnam: changes in genotypes and emergence of class I integrons containing aminoglycoside resistance gene cassettes in Vibrio cholerae O1 strains isolated from 1979 to 1996. J. Clin. Microbiol. 37:734-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalsgaard, A., A. Forslund, O. Serichantalergs, and D. Sandvang. 2000. Distribution and content of class 1 integrons in different Vibrio cholerae O-serotype strains isolated in Thailand. Antimicrob. Agents Chemother. 44:1315-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falbo, V., A. Carattoli, F. Tosini, C. Pezzella, A. M. Dionisi, and I. Luzzi. 1999. Antibiotic resistance conferred by a conjugative plasmid and a class I integron in Vibrio cholerae O1 El Tor strains isolated in Albania and Italy. Antimicrob. Agents Chemother. 43:693-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreira, E., M. Costa, and M. V. Pato. 1992. Résistance aux antibiotiques de souches de Vibrio cholerae isolée en Angola. Pathol. Biol. 40:561-565. [PubMed] [Google Scholar]
- 14.Finch, M. J., J. G. Morris, Jr., J. Kaviti, W. Kagwanja, and M. M. Levine. 1988. Epidemiology of antimicrobial resistant cholera in Kenya and East Africa. Am. J. Trop. Med. Hyg. 39:484-490. [DOI] [PubMed] [Google Scholar]
- 15.Hall, R. M., H. I. Brown, D. E. Brookes, and H. W. Stokes. 1994. Integrons found in different locations have identical 5′ ends but variable 3′ ends. J. Bacteriol. 176:6286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall, R. M., and C. M. Collis. 1995. Mobile gene cassettes and integrons: capture and spread of genes by site specific recombination. Mol. Microbiol. 15:593-600. [DOI] [PubMed] [Google Scholar]
- 17.Hall, R. M., and H. W. Stokes. 1993. Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica 90:115-132. [DOI] [PubMed] [Google Scholar]
- 18.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, and O. White. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hochhut, B., Y. Lotfi, D. Mazel, S. M. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochhut, B., J. Marrero, and M. K. Waldor. 2000. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J. Bacteriol. 182:2034-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes, D. S., and M. Quigley. 1981. A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem. 114:193-197. [DOI] [PubMed] [Google Scholar]
- 22.Iwanaga, M., C. Toma, T. Miyazato, S. Insisiengmay, N. Nakasone, and M. Ehara. 2004. Antibiotic resistance conferred by a class I integron and SXT constin in Vibrio cholerae O1 strains isolated in Laos. Antimicrob. Agents Chemother. 48:2364-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naas, T., Y. Mikami, T. Imai, L. Poirel, and P. Nordmann. 2001. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183:235-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng, L. K., I. Martin, M. Alfa, and M. Mulvey. 2001. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15:209-215. [DOI] [PubMed] [Google Scholar]
- 26.Paulsen, I. T., T. G. Littlejohn, P. Radstrom, L. Sundstrom, O. Skold, G. Swedberg, and R. A. Skurray. 1993. The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob. Agents Chemother. 37:761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rowe-Magnus, D. A., A. M. Guerout, L. Biskri, P. Bouige, and D. Mazel. 2003. Comparative analysis of superintegrons: engineering extensive genetic diversity in the vibrionaceae. Genome Res. 13:428-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowe-Magnus, D. A., and D. Mazel. 2001. Integrons: natural tools for bacterial genome evolution. Curr. Opin. Microbiol. 4:565-569. [DOI] [PubMed] [Google Scholar]
- 29.Thungapathra, M., K. Amita, K. Sinha, S. R. Chaudhuri, P. Garg, T. Ramamurthy, G. B. Nair, and A. Ghosh. 2002. Occurrence of antibiotic resistance gene cassettes aac(6′)-Ib, dfrA5, dfrA12, and ereA2 in class I integrons in non-O1, non-O139 Vibrio cholerae strains in India. Antimicrob. Agents Chemother. 46:2948-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waldor, M. K., H. Tschäpe, and J. J. Mekalanos. 1996. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J. Bacteriol. 178:4157-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]