Abstract
In order to probe the structure and function of the mycobacterial catalase-peroxidase enzyme (KatG), we employed a genetic approach using dominant-negative analysis of katG merodiploids. Transformation of Mycobacterium bovis BCG with various katG point mutants (expressed from low-copy-number plasmids) resulted in reductions in peroxidase and catalase activities as measured in cell extracts. These reductions in enzymatic activity usually correlated with increased resistance to the antituberculosis drug isoniazid (INH). However, for the N138S trans-dominant mutant, the catalase-peroxidase activity was significantly decreased while the sensitivity to INH was retained. trans-dominance required katG expression from multicopy plasmids and could not be demonstrated with katG mutants integrated elsewhere on the wild-type M. bovis BCG chromosome. Reversal of the mutant phenotype through plasmid exchange suggested the catalase-peroxidase deficiency occurred at the protein level and that INH resistance was not due to a second site mutation(s). Electrophoretic analysis of KatG proteins from the trans-dominant mutants showed a reduction in KatG dimers compared to WT and formation of heterodimers with reduced activity. The mutants responsible for these defects cluster around proposed active site residues: N138S, T275P, S315T, and D381G. In an attempt to identify mutants that might delimit the region(s) of KatG involved in subunit interactions, C-terminal truncations were constructed (with and without the D381G dominant-negative mutation). None of the C-terminal deletions were able to complement a ΔkatG strain, nor could they cause a dominant-negative effect on the WT. Taken together, these results suggest an intricate association between the amino- and carboxy-terminal regions of KatG and may be consistent with a domain-swapping mechanism for KatG dimer formation.
More than a century after Robert Koch's seminal studies, tuberculosis remains a devastating international public health problem. The World Health Organization has reported that there are more than 8 million new cases of active disease each year and that 2 million people die annually from tuberculosis (6). Moreover, the global prevalence of tuberculous infection has been estimated to be 32%, with approximately 2 billion individuals being infected worldwide. A recent international burden of disease study has determined that tuberculosis is currently the seventh leading cause of disability-adjusted life years and will remain among the top 10 causes of disability-adjusted life years through the year 2020 (22).
Among the most important components of tubercle bacilli is the KatG protein. This multifunctional heme-dependent enzyme belongs to an expanding group of proteins, referred to as hydroperoxidases I or catalase-peroxidases, which have been characterized from a variety of diverse bacteria, including Escherichia coli, Bacillus stearothermophilus, Halobacterium halobium, and Mycobacterium intracellulare (16, 20, 32). For the intracellular pathogen Mycobacterium tuberculosis, the KatG protein is a virulence factor that probably detoxifies antibacterial reactive oxygen compounds generated by host macrophages (18). Studies in mice and guinea pigs have demonstrated that the capacity of avirulent katG-deficient strains to persist in vivo can be greatly enhanced by complementing these strains with the wild-type (WT) katG gene (14). In addition to being a virulence determinant, the KatG protein also plays a critical role in antituberculosis therapy, because KatG activates the prodrug isoniazid (INH), the primary first-line antibiotic used against M. tuberculosis infections, to a yet-undefined active form (11, 12). Mutations in KatG that eliminate or decrease its catalase-peroxidase activity cause INH resistance by dramatically reducing the extent of INH activation. A number of alterations in the katG gene that have been mapped from patient isolates and in vitro site-directed mutants result in reduced enzymatic function and ultimately decreased sensitivity to INH (9, 26-30).
Although the primary enzymatic function of M. tuberculosis KatG is almost certainly as a catalase-peroxidase, the protein has been shown to have several other related enzymatic activities that may contribute to the ability of M. tuberculosis to persist in vivo. For example, an Mn(II)-dependent peroxidase activity has been described for the tuberculosis KatG protein (17). This protein also exhibits a significant peroxynitrase activity; decomposition of the ONOOH moiety at a rate comparable to those of other known peroxynitrases has been documented for the tuberculosis KatG enzyme (35). Additionally, the M. tuberculosis KatG protein enhances the rate of DNA repair when expressed in E. coli (21).
Despite the multifaceted nature and the overall importance of the M. tuberculosis KatG protein, knowledge of its physical structure is limited. It is known that M. tuberculosis KatG is a homodimer of identical 82-kDa subunits (23). Furthermore, the M. tuberculosis KatG protein and bacterial catalase-peroxidases, in general, consist of two related domains which likely evolved by gene duplication of an ancestral peroxidase (34). While sequence homology assessments have indicated that both the catalase and peroxidase activities are associated with the N-terminal domain, no definite function has been assigned to the carboxyl terminus of the protein. Interestingly, X-ray structural data have indicated that the cytochrome c peroxidase gene, which is homologous to the M. tuberculosis katG, does not have a C-terminal domain (8). However, deletion of the entire C terminus of the M. tuberculosis katG gene results in loss of catalase-peroxidase function and increased resistance to INH (39). Recent genetic evidence suggests that while the C-terminal domain does not play a role in protein dimerization, intramolecular interactions between the N- and C-terminal KatG domains may play an important functional role for the enzyme (38). In this study, we have used dominant-negative and deletion mapping approaches to further define the structural determinants of this multifunctional protein. We have shown that a deletion of only 41 amino acids eliminates the activity of the KatG protein. Furthermore, we have identified specific KatG mutations that alter dimer formation. Importantly, our studies suggest that while heterodimer formation between specific mutants and WT KatG can substantially decrease protein function, reductions in the catalase-peroxidase activity do not necessarily correlate with changes in sensitivity to INH.
MATERIALS AND METHODS
Bacterial strains.
E. coli DH5α and JM109 were used for plasmid manipulations. Mycobacterium bovis BCG (Montreal) and its ΔkatG derivative were obtained from the American Type Culture Collection in Manassas, Va. (ATCC no. 35735 and no. 35747, respectively). Determination of MICs of INH (Sigma, St. Louis) was performed according to NCCLS protocols (7).
Cloning vectors and plasmids.
The creation of plasmids expressing the various KatG point mutants have been described elsewhere (27). Briefly, various M. tuberculosis katG point mutants (generated by site-directed mutagenesis) were cloned into an E. coli-mycobacterium shuttle vector (pMD31) for subsequent introduction to any mycobacterial species. These plasmids contain the mycobacterium origin of replication from pAL5000 (five to eight copies per cell), and katG expression is driven from the katG promoter upstream of the coding region. Shuttle vectors pPE207 (conferring resistance to apramycin [50 μg/ml], a generous gift J. Davies, University of British Columbia) and pMV206 (conferring resistance to hygromycin [50 μg/ml], a generous gift W. Jacobs, Albert Einstein College of Medicine, Yeshiva University) both contain the pAL5000 origin as well (25). The single-copy integrative vector pYUB412.2 was a kind gift of W. Jacobs. Cloning of the katG expression cassettes into pYUB412.2 was accomplished by using T4 DNA polymerase (New England Biolabs, Beverly, Mass.) to fill in KpnI-XbaI fragments containing the WT, N138S, T275P, and D381G katG mutant cassettes. These fragments were used in a blunt-end ligation to pYUB412.2 previously treated with BclI and T4 DNA polymerase. All clones oriented katG transcription with the β-lactamase gene: pJD144 through pJD147 contained katG WT, N138S, T275P, and D381G, respectively.
C-terminal truncation mutants of katG were generated using unique restriction endonuclease sites present in the distal half of the gene. Briefly, WT katG plasmids were digested with the appropriate enzyme, blunted with mung bean nuclease (New England Biolabs), and religated in the presence of a frameshift oligonucleotide linker (5′-TGACTGAGCTCTAGACGTCACCTA-3′). Plasmid pJD150 contains the N-terminal 695 amino acids of KatG plus 4 amino acids from the frameshift oligonucleotide (Leu-Thr-Glu-Leu) to yield the Δ41 mutant. Plasmid pJD114 contains the N-terminal 598 amino acids of KatG plus 4 amino acids from the frameshift oligonucleotide (Val-Gly-Asp-Val) to yield the Δ119 mutant. Similarly, plasmids pJD115 and pJD116 have deletions of 221 and 344 amino acids from the C terminus of KatG, respectively. All constructs were confirmed by DNA sequencing (Lofstrand Laboratories, Gaithersburg, Md.). Double mutants, combining each of the C-terminal truncations with a trans-dominant point mutation, were constructed by substituting an internal restriction fragment in a deletion mutant with the same restriction fragment from the D381G mutant. Briefly, pJD150 was treated with MluI and KpnI, and the small fragment was replaced with an MluI-KpnI fragment from the pD381G mutant to yield pJD165. To construct double mutants of the Δ119 and Δ221 truncations, internal restriction fragments generated with NdeI-MluI were exchanged with the pD381G NdeI-MluI fragment generating pJD136 and pJD137, respectively. The double mutant of Δ344 truncation was constructed using the same strategy with BamHI-MluI restriction enzymes to generate pJD138.
Transformation of bacterial strains.
Plasmids were transformed to E. coli using TSS (4). For M. bovis BCG transformants, competent cells were prepared by growing stationary cultures in 50 ml of 7H9 broth (with oleic acid-albumin-dextrose-catalase [OADC] and 0.05% Tween 80) to approximately mid-log phase (10 to 14 days), and this was followed by a 1-h incubation on ice. Cells were pelleted by centrifugation at 5,000 × g at 4°C for 10 min, washed five times with ice-cold 10% glycerol (25 ml each), and concentrated to 1 ml. A minimum of 50 μl of concentrated cells was added to 200 ng of plasmid DNA, mixed, and electroporated at 25 μF, 1,000 Ω, and 2.5 kV in a 0.1-cm path length cuvette. One milliliter of fresh 7H9 broth containing 10% OADC (BBL, Sparks, Md.) and 0.05% Tween 80 was used to remove the transformation mix to a 15-ml tube for incubation at 37°C for 3 h with gentle shaking at 90 rpm. One-half milliliter was spread on 7H11 agar (supplemented with ADC [BBL] and appropriate antibiotics). Plates were incubated in gas-permeable plastic bags at 37°C in 5% CO2 for 14 to 21 days.
Preparation of cell extracts and enzyme assays.
Crude cellular extracts from all mycobacterial species were prepared using the following procedure. Single colonies were inoculated to 10 ml of 7H9-OADC-0.05% Tween 80 broth (with appropriate antibiotics) in a 25-cm2 flask and grown to mid-log phase at 37°C and 5% CO2 for 14 days. After a 5-min incubation on ice, the cells were transferred to a 15-ml conical tube and centrifuged at 4°C for 5 min at 5,000 × g in a tabletop centrifuge. Following aspiration of the growth medium, the cells were resuspended in 5 ml of ice-cold phosphate-buffered saline (PBS) by pipetting and were centrifuged again. Following a second rinse step, cell pellets were resuspended by pipetting in 125 μl of ice-cold PBS-5 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF protease inhibitor; ICN Biomedicals, Aurora, Ohio) and transferred to a chilled 1.5-ml microcentrifuge tube. Cells were lysed by four cycles of sonication in an ice-water bath (30-s pulse, 30-s rest per cycle). Lysates were centrifuged at 4°C for 10 min at 8,000 × g, and the soluble fraction was removed to fresh tubes. Total protein concentrations of the soluble fractions were determined using bicinchoninic acid (Pierce, Rockford, Ill.).
Total peroxidase activity present in the cell extracts was detected by measuring the oxidation of 2,2′-azino-bis-(3-ethylbenzthiazoline)6-sulfonic acid (ABTS) (Sigma) at room temperature. Thirty micrograms of cellular extract (or PBS for blank) was added to a 1.5-ml microcentrifuge tube, and this mixture was mixed with 1 ml of assay buffer (50 mM NaH2PO4, 25 mM citric acid [at pH 5.0], ABTS [100 μg/ml], 10 mM H2O2). The assay mixture was transferred immediately to a quartz cuvette, and the absorbance was monitored over time in a spectrophotometer at 405 nm. Peroxidase activity was calculated using the absorption coefficient (ɛ = 3.6 × 104 M−1 cm−1) defined by Childs and Bardsley (3) and is represented as units per milligram. For each cell extract, the background peroxidase activity (1.8 × 103 U/mg) of extracts from a katG-negative strain of BCG Montreal was subtracted.
Total catalase activity was detected by measuring the breakdown of hydrogen peroxide at room temperature. Briefly, 30 μg of cell extract was aliquoted to each of two 1.5-ml microcentrifuge tubes. For the reference tube, 1 ml of assay buffer (50 mM NaH2PO4, pH 7.0) was added to the extract, mixed, placed in a quartz cuvette, and used as a blank. For the assay tube, 1 ml of assay buffer (50 mM NaH2PO4 [pH 7.0] with 13 mM H2O2) was added to the extract, mixed, and transferred immediately to a quartz cuvette, and the absorbance was monitored over time in a spectrophotometer at 240 nm. Catalase activity is represented as units per milligram and was calculated using the absorption coefficient for hydrogen peroxide (ɛ = 43.6 M−1 cm−1) as defined by Beers and Sizer (2). For each cell extract, the activity of a katG-negative strain (1.8 U) was subtracted as background. The statistical analyses of the catalase and peroxidase results were completed using the In Stat program (Graph Pad Software, San Diego, Calif.).
Protein gel electrophoresis, immunoblotting, and activity staining.
Denaturing gel electrophoresis of cell extracts was performed according to the method of Laemmli (13) using minigels (resolving gel concentration of 10%). Nondenaturing electrophoresis was performed at 4°C under identical conditions, omitting sodium dodecyl sulfate and other denaturants from all buffers and gels. The separated proteins were electrotransferred to polyvinylidene difluoride (PVDF) membranes using standard protocols, but native gels were soaked for 10 min in running buffer supplemented with 0.1% sodium dodecyl sulfate prior to transfer. Following the transfer, membranes were washed three times in Tris-buffered saline plus 0.05% Tween 20 (TBS-T) and blocked in TBS-T supplemented with 2% casein and 1% fish gelatin (Norland Products, Cranbury, N.J.) for 16 to 18 h at 4°C. The membranes were probed for 60 min with a goat polyclonal antibody directed against mycobacterial catalase (27) or a mouse polyclonal antibody directed against the C-terminal 18 amino acids of the M. tuberculosis KatG (a generous gift of C. Barry, National Institutes of Health, Bethesda, Md.) in TBS-T plus 2% casein. Following washing and incubation with the appropriate horseradish peroxidase-conjugated secondary antibody (Sigma), the blots were developed using enhanced chemiluminescence reagents (Amersham, Piscataway, N.J.).
Peroxidase staining of native gels was performed as described by Wayne and Diaz (33). For peroxidase staining, the gels were rinsed three times for 10 min in ice-cold PBS following electrophoresis and incubated for approximately 20 min with 2.3 mM 3,3′′-diaminobenzidine (Sigma) and 5 mM H2O2 in PBS (room temperature with gentle shaking). The KatG protein appears as a brown band in the gel.
RESULTS
Coexpression of KatG mutants in BCG is dominant negative for enzyme activity.
In order to probe the structure and function of the M. tuberculosis KatG protein, various point mutants of the enzyme were episomally expressed from their native promoter in the attenuated strain of M. bovis (BCG) ATCC 35735. The cloned M. tuberculosis katG gene and the BCG chromosomal katG gene have identical sequences except for a codon 463 alteration found in the BCG gene. This R463L modification is a katG polymorphism that is also found in a significant proportion of M. tuberculosis strains (26). In both BCG and M. tuberculosis, the R463L enzyme is highly active as both a catalase and peroxidase and can efficiently convert INH from a prodrug to its activated form (27, 30).
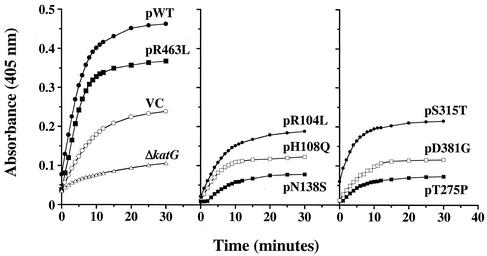
Coexpression of KatG mutant proteins (from a plasmid) with the active KatG enzyme (from the genome) revealed a trans-dominant effect on both catalase and peroxidase activity in repeated experiments. Figure 1 shows a representative kinetic analysis of the peroxidase activity for M. bovis BCG in one set of cellular extracts prepared from log-phase cells. When katG mutants at amino acids 138, 275, and 381 are coexpressed in BCG, a strong inhibition of peroxidase activity is seen (Fig. 1, center and right panels relative to controls in left panel). In Table 1, the average activity values taken at 5 min (linear range of reaction) for three separate sets of extracts are listed. Overall, coexpression of the pN138S mutant caused an 81% reduction in peroxidase activity, and a 70% reduction was detected with the pT275P mutant compared to the vector control (P < 0.05 for both mutants). For the mutant at amino acid 381, a 60% decrease in peroxidase activity relative to the vector control was calculated at 5 min (P < 0.05). While expression of the WT M. tuberculosis katG gene and the pR463L mutant significantly increased activity in the extracts (more than the BCG strain with the vector control plasmid), expression of the mutant at position 315 only modestly increased the total cellular peroxidase activity relative to controls.
FIG. 1.
Peroxidase kinetics of trans-dominant KatG mutants. The absorbance at 405 nm versus time for the reduction of ABTS substrate is plotted. Vector only and katG-expressing plasmids were transformed to M. bovis BCG strain ATCC 35735; katG-deficient M. bovis BCG strain ATCC 35747 (shown as ΔkatG) was transformed with vector only in this study. Abbreviations: WT, WT katG; VC, vector control (no katG on plasmid).
TABLE 1.
Enzymatic activity and the MIC of INH for trans-dominant KatG mutantsa
| Plasmid | Mean peroxidase activity ± SD (103 U/mg) | Mean catalase activity ± SD (U/mg) | INH MIC (μg/ml) |
|---|---|---|---|
| VC | 13.0 ± 0.9 | 9.7 ± 0.5 | 0.5 |
| pWT | 49.8 ± 4.7 | 24.6 ± 0.5 | 0.25 |
| pR104L | 12.7 ± 2.1 | 10.4 ± 0.5 | 0.5 |
| pH108Q | 11.6 ± 5.9 | NDc | 0.5 |
| pN138S | 2.5 ± 0.2b | 6.8 ± 0.6b | 0.5 |
| pT275P | 3.9 ± 0.5b | 7.1 ± 0.1b | 500 |
| pS315T | 19.3 ± 5.8 | 16.1 ± 0.4 | 10 |
| pD381G | 5.2 ± 2.2b | 8.2 ± 0.1 | 250 |
| pR463L | 50.3 ± 6.9 | 17.2 ± 2.6 | 0.25 |
All plasmids transformed to M. bovis (BCG) ATCC 35735. VC denotes vector control plasmid (with no katG). Peroxidase activity is based on results for three different cell extracts. Catalase activity is based on results for two different cell extracts.
The enzymatic activity is significantly less (P < 0.05) than that for VC.
ND, not determined.
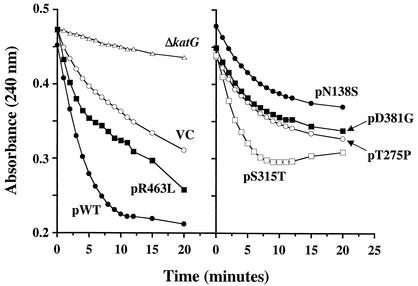
Testing the BCG extracts for catalase activity revealed a weaker but measurable effect from coexpression of the trans-dominant mutants. Similar to peroxidase studies, coexpression of the WT and R463L versions of M. tuberculosis katG increased the total cellular enzyme activity compared to the control strain (Fig. 2, left panel). The representative data shown in Fig. 2 illustrates the fact that coexpression of mutants pN138S and pT275P decreased catalase activity below levels detected in vector control extracts. The summary data of Table 1 show that 30 and 29% reductions in catalase activity, relative to the vector control, were detected for the pN138S and pT275Pmutants, respectively (P < 0.05). Moreover, the catalase activity seen after expression of these mutants is approximately 30% of the catalase levels detected in extracts from cells transfected with plasmid encoding the WT katG. Again, the pS315T mutant did not have a dominant-negative effect on enzymatic activity. In fact, a stimulatory effect on catalase activity was seen at 5 min.
FIG. 2.
Catalase kinetics of trans-dominant KatG mutants. The absorbance at 240 nm versus time for the reduction of H2O2 substrate is plotted. In this assay, the catalase activity is evaluated by measuring the breakdown of hydrogen peroxide (resulting in a reduction in the absorbance at 240 nm) during the 20-min period. Vector only and katG-expressing plasmids were transformed to strain M. bovis BCG strain ATCC 35735; M. bovis BCG strain ATCC 35747 (shown as ΔkatG) was transformed with vector only in this study. Abbreviations: WT, WT katG; VC, vector control (no katG on plasmid).
For specific dominant-negative mutants the MICs of INH are increased.
Since reduction of peroxidase and catalase activities normally correlates with resistance to the antituberculosis drug INH, the dominant-negative mutants were tested for their ability to grow in the presence of INH (Table 1). The MIC of INH for the WT BCG strain is 0.5 μg/ml, while the MIC for the corresponding katG deletion mutant of BCG is 1,000 μg/ml. When KatG mutants at positions 381 and 275 were expressed in BCG, the MICs for INH increased 500- to 1,000-fold, respectively, to values approaching that of the katG deletion mutant strain. Surprisingly, coexpression of the pN138S mutant which strongly inhibited the peroxidase activity did not increase the MIC for the BCG pN138S katG transformants. In contrast, while coexpression of the pS315T katG mutant had only minor effects on enzyme activity, 20-fold increases in the MIC (to 10 μg/ml) were detected for the BCG pS315T katG transformants.
trans-dominance is due to mutant enzyme expression from the plasmid and not secondary mutation.
In order to demonstrate that inhibition of enzyme activity and INH resistance were caused by the trans-dominant mutants acting at the protein level, plasmid exchange experiments were performed. These experiments were designed to prove that the BCG mutant phenotypes described above did not result from alterations of the chromosomal copy of katG or through second-site mutations. Previous reports have clearly shown that INH resistance can map to genetic loci other than katG (1, 19). For these exchange studies, plasmid vectors having the same origin of replication (approximately five to eight copies per cell) as the KatG plasmid panel, but containing a different selectable marker, were used to isolate cells that lost the KatG plasmid through incompatibility. Two plasmid exchange vectors were used: pPE207 harbors an aminoglycoside-modifying enzyme that allows resistance to apramycin and to kanamycin, and pMV206 encodes resistance to another aminoglycoside, hygromycin. Both of these plasmids contain only vector sequences.
The pPE207 plasmids were transformed into the strains carrying the pMD31 katG mutant plasmids (which are resistant to kanamycin but not to apramycin) and the resultant colonies were tested for peroxidase activity and INH resistance (Table 2). The phenotypes of the original trans-dominant mutants are shown for comparison in the top row of the table. For the katG mutants at amino acids 275 and 381, the whole-cell peroxidase activities were increased considerably by exchange with pPE207. Moreover, when the enzyme activity had nearly returned to normal levels through plasmid exchange, the INH sensitivity of the mutants was also restored. Since the pN138S dominant-negative mutant showed low-level resistance to apramycin, the pMD31 KatG mutant plasmid was exchanged with pMV206 by hygromycin selection. Upon removal of the N138S mutant plasmid, peroxidase activity was restored to WT levels (data not shown).
TABLE 2.
Removal of mutant katG plasmids from trans-dominant BCG strains
| Competing plasmid | Mean peroxidase activity ± SD (103 U/mg)
|
MIC of INH (μg/ml)
|
||||
|---|---|---|---|---|---|---|
| VCc | pT275P | pD381G | VC | pT275P | pD381G | |
| Nonea | 13.0 ± 0.9 | 3.9 ± 0.5 | 5.2 ± 2.2 | 0.5 | 500 | 250 |
| pPE207b | 18.3 ± 5.0 | 11.1 ± 1.1 | 11.6 ± 3.6 | 0.5 | 0.5 | 0.5 |
Peroxidase activity and MIC for the pT275P and pD381G KatG trans-dominant mutants in M. bovis (BCG) ATCC 35735 (mutant katG on Kanr plasmid).
Peroxidase activity and MIC for the same strains as in the top row except that their plasmids have been replaced by pPE207. (Aprr plasmid with no katG).
VC, vector control plasmid with no katG gene.
To further show that the dominant-negative effect of the plasmid-borne KatG mutants was due to expression of the genes from the multicopy plasmid, the entire mutant panel was cloned individually into the single-copy integrative mycobacterial plasmid pYUB412. When transformed into BCG, these single-copy constructs did not demonstrate a dominant-negative phenotype and showed no differences in enzyme activity or in the MIC (data not shown).
Mechanism of inhibition by the trans-dominant mutants.
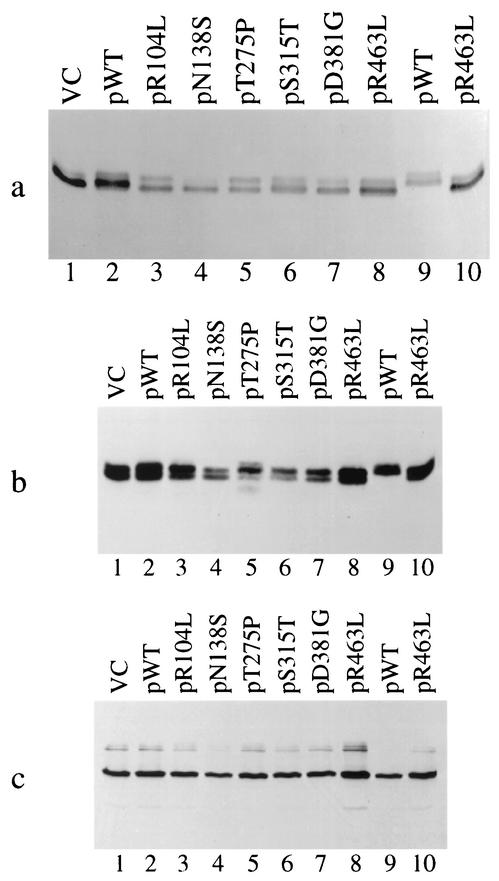
Since the KatG enzyme from M. tuberculosis complex organisms has been shown to function as a homodimer (23), the trans-dominant effect of the plasmid mutants could occur by disrupting the interaction of WT monomers in the cell. In order to investigate the mechanism of inhibition, KatG protein from the mutants was analyzed by electrophoresis of cell extracts. Following separation by nondenaturing electrophoresis, the activity of the native enzymes was monitored by staining with the peroxidase substrate 3,3′′-diaminobenzidine (Fig. 3a). The identity and relative amounts of the KatG bands in the nondenaturing gels were estimated by Western blotting with anti-KatG antibodies (Fig. 3b). Total KatG protein was monitored by Western blot analysis of the same extracts that had been separated by denaturing gel electrophoresis (Fig. 3c).
FIG. 3.
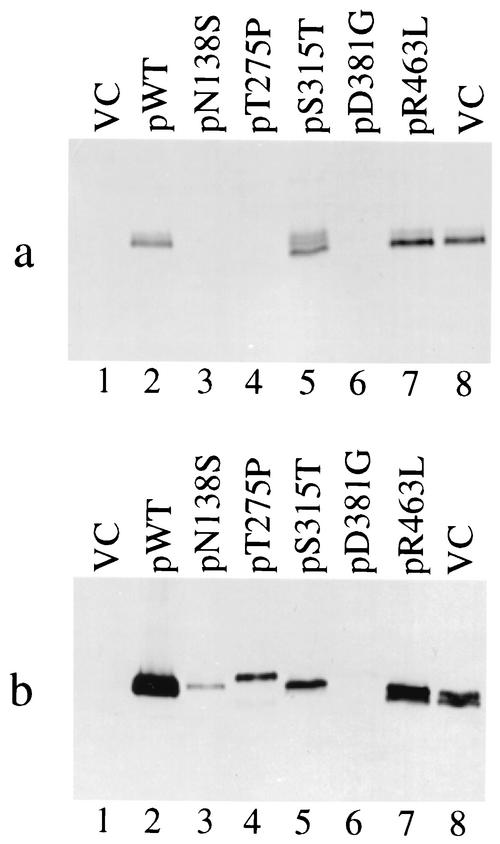
Analysis of trans-dominant mutants by gel electrophoresis. (a) Peroxidase activity gel of katG mutant plasmids in M. bovis BCG strain ATCC 35735 (lanes 1 to 8). Lanes 9 and 10 show vector control (VC) and the R463L mutant plasmid in the ΔkatG strain ATCC 35747. (b) Nondenaturing gel transferred to PVDF membrane and probed with anti-KatG polyclonal antibody. Lane assignments are identical to those for panel a. (c) Denaturing gel transferred to PVDF membrane and probed with anti-KatG polyclonal antibody. Lane assignments are identical to those in panel a.
As seen in Fig. 3b (VC, lane 1) KatG dimers from BCG separate into two bands of equal intensity following native gel electrophoresis. For these two bands, likely to be different forms of the enzyme, more than 90% of the peroxidase activity associates with the faster-migrating species (Fig. 3a, lane 1). A similar electrophoretic profile was seen for the plasmid-borne R463L katG protein expressed in a ΔkatG background (Fig. 3a and b, lanes 10). The WT M. tuberculosis KatG protein (expressed in a ΔkatG background) also migrates as a doublet on nondenaturing gels (Fig. 3a and b, lanes 9) but has an overall lower mobility, consistent with having an arginine instead of a leucine at residue 463. When WT M. tuberculosis katG (from the plasmid) is coexpressed with the BCG chromosomal allele, a mixture of dimeric forms is detected in the gel (Fig. 3b, lane 2). These forms are difficult to resolve, but the most-active band holds an intermediate position in relation to the controls, suggesting the formation of active heterodimers (Fig. 3a, lane 2).
When the M. tuberculosis katG point mutants are coexpressed in BCG, the amount of total KatG protein remains essentially unchanged (as shown by the Western blot in Fig. 3c) but the relative distribution of protein between the different forms is altered. For instance, the faster-migrating species is the most-active form for the KatG mutants at positions 315 and 381 (Fig. 3a, lanes 6 and 7, respectively). However, this oligomeric form of the pS315T and pD381G dominant-negative mutants is less prevalent than the slower-migrating species. Importantly, a new slower-migrating protein species having no enzyme activity is seen for the trans-dominant pT275P mutant (compare Fig. 3a and b, lanes 5). Overall, the prevalence of the less active species correlates with the dominant-negative phenotype exhibited by these mutants. These results suggest the mutants exert their effect by either disrupting the KatG dimer or through formation of less active (or inactive) heterodimers.
When expressed in the ΔkatG background, the katG point mutants at positions 138, 275, and 381 show no detectable peroxidase activity upon native gel electrophoresis (Fig. 4a, lanes 3, 4, and 6). When analyzed by Western blotting, the mutants are present in greatly reduced amounts compared to the WT and R463L mutant and do not demonstrate the characteristic KatG doublet (Fig. 4b). In fact, the pT275P and pD381G mutants run as single bands with a mobility even lower than that of the WT doublet, while pN138S and pS315T have intermediate mobilities. Interestingly, the pS315T mutant shows three different active species when expressed in the ΔkatG background (Fig. 4a, lane 5) but not all are detected in the Western blot (Fig. 4b, lane 5). In agreement with previously published results (27), analysis of total protein by denaturing gel and Western blotting confirms the reduction in KatG protein levels for the mutants at positions 138, 275, and 381.
FIG. 4.
Analysis of katG point mutants by native gel electrophoresis. (a) Peroxidase activity gel of katG mutant plasmids in the ΔkatG strain ATCC 35747 (lanes 1 to 7). Lane 8 shows vector control (VC) in the katG+ M. bovis (BCG) strain ATCC 35735. (b) Nondenaturing gel transferred to PVDF membrane and probed with anti-KatG polyclonal antibody. Lane assignments are identical to those in panel a. Although barely visible, the pD381G mutant does show a protein band with altered mobility (similar to the pT275P mutant).
Deletion mutants implicate the C terminus of KatG in protein stability and dimer formation.
The similarity of the amino- and carboxy-terminal sequences of KatG has been attributed to a gene duplication event that may have some catalytic or regulatory implications for the enzyme (34). Given that monomeric catalases show no evidence of gene duplication, a deletion analysis of the katG gene from M. tuberculosis was performed to assign a function to the duplicated C-terminal domain and test its potential role in dimerization. C-terminal deletions were constructed by introducing translational stop sites in the distal half of katG; the truncated genes expressed proteins lacking the C-terminal 41, 119, 221, and 334 amino acids, respectively. Plasmids containing the truncation mutants were transformed into a ΔkatG derivative of M. bovis BCG and tested for complementation. Interestingly, a deletion of only 41 amino acids from the C terminus was sufficient to inactivate the enzyme and prevent complementation. None of the deletion mutants were capable of restoring enzyme activity or INH sensitivity to the katG deletion mutant strains.
In order to test if these inactive truncation mutants retained the ability to dimerize, these truncated genes were introduced into BCG and analyzed for a dominant-negative phenotype. None of the C-terminal deletions were trans-dominant with respect to either enzyme activity or INH resistance. When analyzed by denaturing and nondenaturing gel electrophoresis, the deletion mutants were not detectable even after Western blotting, and the mutants did not effect the mobility of the WT enzyme when coexpressed in the cell (data not shown). To rule out the possibility that enzymatically active KatG heterodimers had formed between the WT and the truncation mutants but did not demonstrate mobility differences upon native gel analysis, double mutants were constructed. To each deletion mutant, the D381G point mutation was added by exchange of an internal restriction fragment. If any of the deletion mutants were capable of dimerization, the D381G dominant-negative mutation would inhibit enzyme activity and induce INH resistance. When tested for biochemical activity and drug resistance, none of the KatG C-terminal truncation mutants containing a D381G point mutation were trans-dominant.
DISCUSSION
Despite the pharmacological and pathogenic importance of the multifunctional M. tuberculosis KatG enzyme, the molecular definition of its physical structure has been limited. Primarily because the M. tuberculosis KatG crystal structure has not been solved, the details of specific subunit interactions are not well understood and the tertiary and quaternary structural determinants have not been defined. In this study, we have utilized dominant-negative genetic analyses to investigate KatG protein-protein interactions. We focused primarily on mutations that had been previously shown to eliminate or substantially reduce enzymatic activity. Dominant-negative effects were demonstrated for four mutant KatG proteins. The pT275P dominant-negative BCG mutants showed substantial reductions in catalase-peroxidase function and elevated MICs for INH. These data were not unexpected, because residue 275 is likely located near the KatG active site and substitution of a proline for the threonine at this residue should alter the physical structure of the heterodimer active site, resulting in reduced substrate binding and catalysis (30). Similarly, diminished peroxidase activity and decreased sensitivity to INH were demonstrated for the D381G dominant-negative BCG recombinant. This result was again not surprising since the KatG 381 aspartic acid residue has been predicted to hydrogen bond to a catalytic residue (10). The absence of this hydrogen bond may alter enzymatic function by affecting the active site structure.
It is of interest that a 20-fold-elevated MIC of INH was seen for the pS315T dominant-negative mutants, although nearly WT peroxidase and catalase levels were detected in the corresponding cellular extracts. The S315T substitution is the most-common mutation that has been associated with INH resistance in M. tuberculosis (26). In previous site-directed mutagenesis studies, we estimated that this KatG alteration reduced the catalase-peroxidase activity in cellular extracts by 50% and increased the MIC of INH by 90-fold relative to that for WT cells (27). Using purified enzymes, Wengenack et al. demonstrated that KatG S315T protein was a competent catalase-peroxidase (with only a sixfold and twofold relative reduction in catalase and peroxidase activity, respectively) but was much less efficient than WT KatG at metabolizing INH (37). Biophysical studies by the same group recently suggested that the addition of a single methyl group to KatG residue 315 confers resistance to INH through minor changes in the INH binding site (36). Our results support this hypothesis. The finding of higher MICs of INH for pS315T dominant-negative strains with essentially unaltered catalase and peroxidase activities strongly suggests that subtle alterations in the INH binding site are responsible for this phenotype.
Expression of the pN138S mutant in BCG dramatically reduced peroxidase function and also decreased catalase activity. Based on the Saccharomyces cerevisiae CCP structure, the M. tuberculosis KatG 138 residue is probably located adjacent to the peroxidase substrate binding channel and the N138S modification may effect substrate access to the active site (30). Paradoxically, although the peroxidase function of the N138S BCG transformants is reduced substantially, these cells retain their sensitivity to INH. This result is of interest because it suggests that the enzyme's capacity to activate INH can be uncoupled from its catalase-peroxidase activity. It should be emphasized that because of the potential importance of this finding, eight independent pN138S trans-dominant mutants were tested. All eight of these dominant-negative mutants had substantially reduced catalase-peroxidase activity yet remained highly sensitive to INH.
In addition to investigating protein-protein interactions via a dominant-negative approach, we examined the function of the C-terminal domain of KatG using deletion analysis. By comparing the sequences of bacterial catalase-peroxidases, Welinder has shown that the N-terminal region of KatG is closely related to the yeast CCP (34). This observation suggested that the N terminus is largely responsible for the catalytic function of the enzyme. Welinder also hypothesized that bacterial KatG C-terminal domains may have evolved from a gene duplication event because the N-terminal and C-terminal domains of these catalase-peroxidases exhibit considerable similarity at the primary sequence level. Comparisons of the sequences of the two M. tuberculosis KatG domains have supported this hypothesis, with considerable homology being shown between residues 55 to 422 (N terminus) and 423 to 735 (C terminus) of this protein (10). Although the sequence comparisons and the mutational analyses have indicated that the catalytic residues of KatG reside largely in the N terminus, the function of the related C-terminal region of KatG has remained undefined. Several studies have suggested that the C terminus of the M. tuberculosis KatG protein does play a role in enzymatic function. Zhang et al. demonstrated that expression of a truncated M. tuberculosis KatG (lacking approximately 300 C-terminal amino acids) yielded an inactive enzyme that was unable to restore INH sensitivity to katG-lacking mycobacterial cells (39). Based on the instability of an L587P KatG mutant, Saint-Joanis et al. speculated that the C-terminal domain was necessary for subunit-subunit interactions (30). Recently, using a yeast two-hybrid approach, Wilming and Johnsson have shown that intramolecular interactions between the amino and carboxy termini of M. tuberculosis KatG may be important for the enzymatic function of the protein (38). We have confirmed the importance of the KatG C terminus using a systematic deletion analysis. Our studies indicate that deletion of as few as 41 residues from the C terminus of M. tuberculosis KatG yields a nonfunctional enzyme that cannot activate INH. In addition, all of the C-terminal deletions that we evaluated were unable to exert a trans-dominant effect when expressed in WT cells. Although the precise role of the C terminus in KatG function is unknown, these data clearly demonstrate that sequences within the C-terminal region are required for optimal enzymatic function.
The phenotypes that result when singly mutated and truncated KatG proteins are coexpressed with WT KatG in BCG strongly suggest that heterodimers must form for the dominant-negative effects to be seen. However, the structure-related mechanisms by which specific point mutants can dramatically impact the enzymatic activity of the KatG heterodimers have not been elucidated. Clearly, the dominant-negative mutants may affect the stability of the KatG protein. Additionally, the effect of the mutant KatG on the physical structure of the heterodimer may be magnified because the dimer must swap domains to be productive. Three-dimensional domain swapping is a relatively common feature of oligomeric proteins and has been identified in dimeric proteins such as diphtheria toxin, RNase A, and nitric oxide synthase (5, 15, 24, 31). Monomers that swap domains usually contain two associated domains that are linked covalently by a hinge region. Opening of the hinge allows the two domains to disassociate from each other and reassociate with the equivalent domains from a different monomer. In an evolutionary sense, the replacement of intrasubunit domain interactions with intersubunit interactions reduces the evolutionary pressure on the maintenance of intrasubunit domains (24). A domain-swapping process may explain how single point mutants could substantially disrupt the KatG heterodimers. In this domain-swapping scenario, the impact of the point mutations would be amplified because of the physical linkage of the two subunits. Definitive proof of domain swapping is clearly dependent on completion of the crystal structure for KatG.
In conclusion, using genetic analyses, we have identified four M. tuberculosis KatG residues (138, 275, 315, and 381) that play prominent roles in subunit-subunit interactions and overall protein structure. Moreover, we have shown that the C-terminal region of KatG significantly contributes to the enzymatic function of the protein. Our study has also highlighted important questions such as the relative location of the substrate and INH binding sites within the KatG dimer and the mechanistic relationship between peroxidase-catalase activity and INH metabolism. Overall, these results increase our understanding of the structure and function of this clinically important protein and suggest that further KatG structural studies are needed.
Acknowledgments
We are grateful to Bill Jacobs, Lee Rosner, Clif Barry, and Julian Davies for bacterial strains, plasmids, and antibodies; to Frank Collins for helpful insights; and to Dave Rouse for assistance with the antibody binding and exposure steps for the Western blots.
J.A.D. was supported by a fellowship from the National Research Council.
REFERENCES
- 1.Banerjee, A., E. Dubnau, A. Quemard, V. Balasubramanian, K. S. Um, T. Wilson, D. Collins, G. de Lisle, and W. R. Jacobs, Jr. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227-230. [DOI] [PubMed] [Google Scholar]
- 2.Beers, R. F., and I. W. Sizer. 1952. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 195:133-140. [PubMed] [Google Scholar]
- 3.Childs, R. E., and W. G. Bardsley. 1975. The steady-state kinetics of peroxidase with 2′,2′-azino-di-(3-ethyl-benzathiazoline-6-sulphonic acid) as chromagen. Biochem. J. 145:93-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung, C. T., S. L. Niemela, and R. H. Miller. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. USA 86:2172-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crane, B. R., R. J. Rosenfeld, A. S. Arvai, D. K. Ghosh, S. Ghosh, J. A. Tainer, D. J. Stuehr, and E. D. Getzoff. 1999. N-terminal domain swapping and metal ion binding in nitric oxide synthase dimerization. EMBO J 18:6271-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dye, C., S. Scheele, P. Dolin, V. Pathania, M. C. Raviglion, et al.. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 7.Eliopoulos, G. M., and R. C. Moellering. 1996. Antimicrobial combinations, p. 330-396. In V. Lorian (ed.), Antibiotics in laboratory medicine. Williams and Wilkins, Baltimore, Md.
- 8.Finzel, B. C., T. L. Poulos, and J. Kraut. 1984. Crystal structure of yeast cytochrome C peroxidase refined at the 1.7 angstrom resolution. J. Biol. Chem. 259:13027-13036. [PubMed] [Google Scholar]
- 9.Heym, B., P. M. Alzari, N. Honore, and S. T. Cole. 1995. Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol. Microbiol 15:235-245. [DOI] [PubMed] [Google Scholar]
- 10.Heym, B., Y. Zhang, S. Poulet, D. Young, and S. T. Cole. 1993. Characterization of the katG gene encoding a catalase-peroxidase required for the isoniazid susceptibility of Mycobacterium tuberculosis. J. Bacteriol. 175:4255-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnsson, K., D. S. King, and P. Schultz. 1995. Studies on the mechanism of action of isoniazid and ethionamide in the chemotherapy of tuberculosis. J. Am. Chem. Soc. 117:5009-5010. [Google Scholar]
- 12.Johnsson, K., and P. Schultz. 1994. Mechanistic studies of the oxidation of isoniazid by the catase-peroxidase from Mycobacterium tuberculosis. J. Am. Chem. Soc. 116:7225-7226. [Google Scholar]
- 13.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.Li, Z., C. Kelley, F. Collins, D. Rouse, and S. Morris. 1998. Expression of katG in Mycobacterium tuberculosis is associated with its growth and persistence in mice and guinea pigs. J. Infect. Dis. 177:1030-1035. [DOI] [PubMed] [Google Scholar]
- 15.Liu, Y., G. Gotte, M. Libonati, and D. Eisenberg. 2001. A domain-swapped RNase A dimer with implications for amyloid formation. Nat. Struct. Biol. 8:211-214. [DOI] [PubMed] [Google Scholar]
- 16.Loprasert, S., S. Negoro, and H. Okada. 1989. Cloning, nucleotide sequence, and expression in Escherichia coli of the Bacillus stearothermophilus peroxidase gene (perA). J. Bacteriol. 171:4871-4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magliozzo, R. S., and J. A. Marcinkeviciene. 1997. The role of Mn(II)-peroxidase activity of mycobacterial catalase-peroxidase in activation of the antibiotic isoniazid. J. Biol. Chem. 272:8867-8870. [DOI] [PubMed] [Google Scholar]
- 18.Manca, C., S. Paul, C. E. Barry III, V. H. Freedman, and G. Kaplan. 1999. Mycobacterium tuberculosis catalase and peroxidase activities and resistance to oxidative killing in human monocytes in vitro. Infect. Immun. 67:74-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mdluli, K., R. A. Slayden, Y. Zhu, S. Ramaswamy, X. Pan, D. Mead, D. D. Crane, J. M. Musser, and C. E. Barry, III. 1998. Inhibition of a Mycobacterium tuberculosis beta-ketoacyl ACP synthase by isoniazid. Science 280:1607-1610. [DOI] [PubMed] [Google Scholar]
- 20.Morris, S. L., J. Nair, and D. A. Rouse. 1992. The catalase-peroxidase of Mycobacterium intracellulare: nucleotide sequence analysis and expression in Escherichia coli. J. Gen. Microbiol. 138:2363-2370. [DOI] [PubMed] [Google Scholar]
- 21.Mulder, M. A., S. Nair, V. R. Abratt, H. Zappe, and L. M. Steyn. 1999. Involvement of the N- and C-terminal domains of Mycobacterium tuberculosis KatG in the protection of mutant Escherichia coli against DNA-damaging agents. Microbiology 145:2011-2021. [DOI] [PubMed] [Google Scholar]
- 22.Murray, C. J., and A. D. Lopez. 1997. Global mortality, disability, and the contribution of risk factors: global burden of disease study. Lancet 349:1436-1442. [DOI] [PubMed] [Google Scholar]
- 23.Nagy, J. M., A. E. Cass, and K. A. Brown. 1997. Purification and characterization of recombinant catalase-peroxidase, which confers isoniazid sensitivity in Mycobacterium tuberculosis. J. Biol. Chem. 272:31265-31271. [DOI] [PubMed] [Google Scholar]
- 24.Ostermeier, M., and S. J. Benkovic. 2000. Evolution of protein function by domain swapping. Adv. Protein Chem. 55:29-77. [DOI] [PubMed] [Google Scholar]
- 25.Paget, E., and J. Davies. 1996. Apramycin resistance as a selective marker for gene transfer in mycobacteria. J. Bacteriol. 178:6357-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 27.Rouse, D. A., J. A. DeVito, Z. Li, H. Byer, and S. L. Morris. 1996. Site-directed mutagenesis of the katG gene of Mycobacterium tuberculosis: effects on catalase-peroxidase activities and isoniazid resistance. Mol. Microbiol. 22:583-592. [DOI] [PubMed] [Google Scholar]
- 28.Rouse, D. A., Z. Li, G. H. Bai, and S. L. Morris. 1995. Characterization of the katG and inhA genes of isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 39:2472-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rouse, D. A., and S. L. Morris. 1995. Molecular mechanisms of isoniazid resistance in Mycobacterium tuberculosis and Mycobacterium bovis. Infect. Immun. 63:1427-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saint-Joanis, B., H. Souchon, M. Wilming, K. Johnsson, P. M. Alzari, and S. T. Cole. 1999. Use of site-directed mutagenesis to probe the structure, function and isoniazid activation of the catalase/peroxidase, KatG, from Mycobacterium tuberculosis. Biochem. J. 338:753-760. [PMC free article] [PubMed] [Google Scholar]
- 31.Steere, B., and D. Eisenberg. 2000. Characterization of high-order diphtheria toxin oligomers. Biochemistry 39:15901-15909. [DOI] [PubMed] [Google Scholar]
- 32.Triggs-Raine, B. L., B. W. Doble, M. R. Mulvey, P. A. Sorby, and P. C. Loewen. 1988. Nucleotide sequence of katG, encoding catalase HPI of Escherichia coli. J. Bacteriol. 170:4415-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wayne, L. G., and G. A. Diaz. 1986. A double staining method for differentiating between two classes of mycobacterial catalase in polyacrylamide electrophoresis gels. Anal. Biochem. 157:89-92. [DOI] [PubMed] [Google Scholar]
- 34.Welinder, K. G. 1991. Bacterial catalase-peroxidases are gene duplicated members of the plant peroxidase superfamily. Biochim. Biophys. Acta 1080:215-220. [DOI] [PubMed] [Google Scholar]
- 35.Wengenack, N. L., M. P. Jensen, F. Rusnak, and M. K. Stern. 1999. Mycobacterium tuberculosis KatG is a peroxynitritase. Biochem. Biophys. Res. Commun. 256:485-487. [DOI] [PubMed] [Google Scholar]
- 36.Wengenack, N. L., S. Todorovic, L. Yu, and F. Rusnak. 1998. Evidence for differential binding of isoniazid by Mycobacterium tuberculosis KatG and the isoniazid-resistant mutant KatG(S315T). Biochemistry 37:15825-15834. [DOI] [PubMed] [Google Scholar]
- 37.Wengenack, N. L., J. R. Uhl, A. L. St. Amand, A. J. Tomlinson, L. M. Benson, S. Naylor, B. C. Kline, F. R. Cockerill III, and F. Rusnak. 1997. Recombinant Mycobacterium tuberculosis KatG(S315T) is a competent catalase-peroxidase with reduced activity toward isoniazid. J. Infect. Dis. 176:722-727. [DOI] [PubMed] [Google Scholar]
- 38.Wilming, M., and K. Johnsson. 2001. Inter- and intramolecular domain interactions of the catalase-peroxidase KatG from M. tuberculosis. FEBS Lett. 509:272-276. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, Y., B. Heym, B. Allen, D. Young, and S. Cole. 1992. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591-593. [DOI] [PubMed] [Google Scholar]