Abstract
An α-l-fucosidase (EC 3.2.1.51) able to release the t-fucosyl residue from the side chain of xyloglucan oligosaccharides has been detected in the leaves of Arabidopsis plants. Moreover, an α-l-fucosidase with similar substrate specificity was purified from cabbage (Brassica oleracea) leaves to render a single band on SDS-PAGE. Two peptide sequences were obtained from this protein band, and they were used to identify an Arabidopsis gene coding for an α-fucosidase that we propose to call AtFXG1. In addition, an Arabidopsis gene with homology with known α-l-fucosidases has been also found, and we proposed to name it as AtFUC1. Both AtFXG1 and ATFUC1 were heterologously expressed in Pichia pastoris cells and the α-l-fucosidase activities secreted to the culture medium. The α-l-fucosidase encoded by AtFXG1 was active against the oligosaccharides from xyloglucan XXFG as well as against 2′-fucosyl-lactitol but not against p-nitrophenyl-α-l-fucopyranoside. However, the AtFUC1 heterologously expressed was active only against 2′-fucosyl-lactitol. Thus, the former must be related to xyloglucan metabolism.
Plant cell walls are built by two independent networks, cellulose microfibers cross-linked by xyloglucan chains and cross-linked pectins, both networks contributing to their mechanical and functional properties (Roberts, 2001). The xyloglucan-cellulose complex has been considered as the network responsible for controlling the rate of cell expansion, xyloglucan being the load-bearing component in the primary cell walls because of its proposed cross-linking of the cellulose microfibers (Fry, 1989). It consists of a linear β-(1-4)-linked d-glucan backbone that carries α-d-xylosyl, β-d-galactosyl-(1-2)-α-d-xylosyl, and α-l-fuc-osyl-(1-2)-β-d-galactosyl-(1-2)-α-d-xylosyl side chains attached to the OH-6 of β-glucosyl residues. In addition to its structural role, xyloglucan may act as a source of signaling molecules. Oligosaccharides derived from xyloglucan have been found to be formed in vivo (Fry, 1986), and they have been shown to regulate auxin-induced (McDougall and Fry, 1988, 1990) and acid pH-induced (Lorences et al., 1990) growth, the t-Fuc being necessary for the regulatory effect of the XXFG, a nonasaccharide derived from xyloglucan (Fry et al., 1993). The Fuc-deficient mur mutants of Arabidopsis (Reiter et al., 1993) showed dwarf growth habit and fragile cell walls.
Enzymes that modify xyloglucan oligosaccharides have been detected in plant cell walls (Fry, 1995). An α-fucosidase that removes the α-l-fucosyl residue from XXFG has been purified from pea (Pisum sativum) epicotyls (Farkas et al., 1991; Augur et al., 1993) and has been cloned (Augur et al., 1995). Furthermore, cDNAs encoding α-l-fucosidase have been also isolated from human (Fukushima et al., 1985; Occhiodoro et al., 1989) and rat (Fisher and Aronson, 1989) livers. However, no α-l-fucosidase has yet been identified in Arabidopsis. Although some sequences homologous to pea α-l-fucosidase have been found in leguminous plants, there is no clear candidate for this activity in the whole genome of Arabidopsis.
Thus, our aim in the present paper has been to look for an Arabidopsis gene(s) encoding for α-fucosidase activity. Two different experimental approaches were used: (a) α-fucosidase purification from cabbage (Brassica oleracea), microsequencing, and search for homologous sequences in Arabidopsis; and (b) search for sequences homologous to known α-fucosidases from different sources. Finally, those sequences were expressed in Pichia pastoris cells.
RESULTS
α-Fucosidase Activity of Arabidopsis
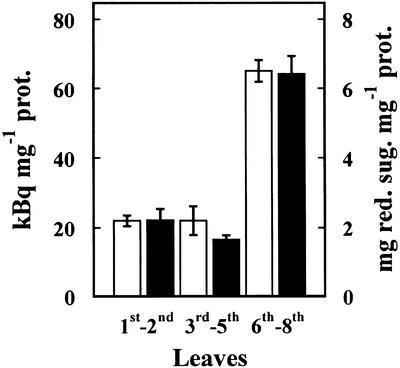
The presence of α-fucosidase activity in Arabidopsis was examined using leaves from 21-d-old plants (Fig. 1). These leaves showed α-fucosidase activity against [3H]Fuc-labeled XXFG as well as against 2′-fucosyl-lactitol, but not against p-nitrophenyl-fucoside. α-Fucosidase activity was higher in the younger leaves (sixth–eighth), being about three times that of the older ones (first–second). The pattern of the α-fucosidase changes with the developmental stage of leaves was similar for both substrates.
Figure 1.
α-Fucosidase activity extracted from Arabidopsis leaves. The leaves from 21-d-old plants were sorted according to their appearance order. α-Fucosidase activity was measured against [3H]Fuc-labeled XXFG (white bars) and 2′-fucosyl-lactitol (black bars). Mean values with ses are given (n = 3).
The apoplastic fluid from Arabidopsis seedlings grown under water with shaking was extracted as described in “Materials and Methods.” The α-fuco-sidase activity detected in the apoplastic fluid accounted for 80% of the activity in the whole plant. However, Glc-6-phosphate dehydrogenase accounted for less than 0.4% of the total activity, proving the absence of cytoplasmic contamination in the apoplastic preparation. Thus, most of the α-fucosidase activity in Arabidopsis plants was located in the apoplast.
α-Fucosidase Purification from Cabbage Leaves
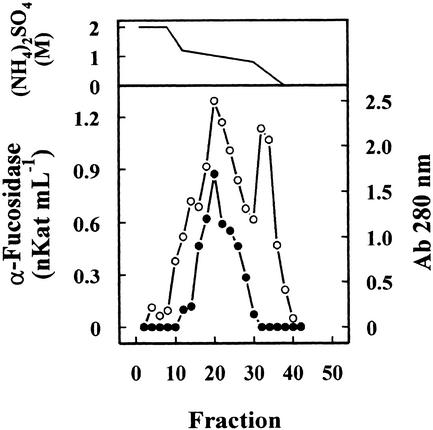
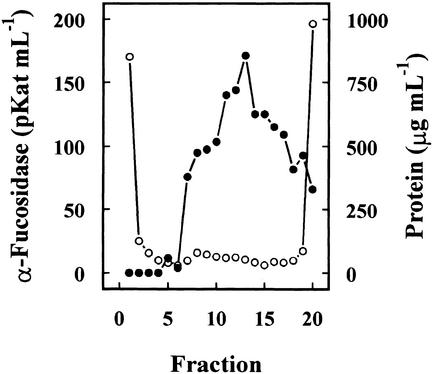
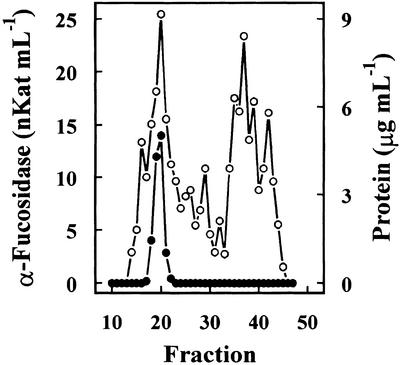
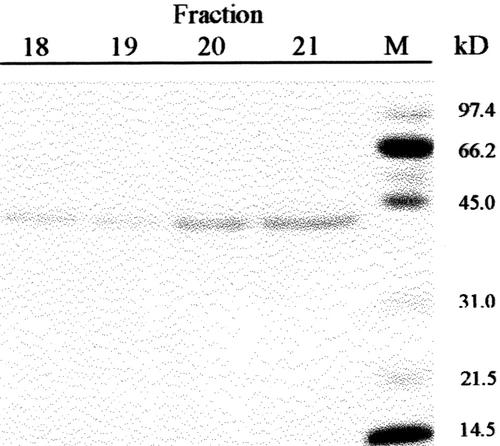
Previous results had shown the presence in cabbage leaves of an α-fucosidase with substrate specificity similar to that found in Arabidopsis (data not shown). Table I summarizes the purification of the α-fucosidase from cabbage leaves, its activity being measured against 2′-fucosyl-lactitol. The protein precipitated between the 40% to 80% of (NH4)2SO4 saturation apparently contained all the α-fucosidase activity present in the crude extract. However, an under-estimation of the activity in the crude extract cannot be excluded because of the interference of the reducing sugars in the plant extract. The protein extract was applied on a SP-Sepharose column and the α-fucosidase activity was recovered in the retained fraction (data not shown). This fraction was loaded onto a hydrophobic-interaction column that rendered a broad peak, with α-fucosidase activity eluting between 1.4 and 0.4 m (NH4)2SO4 (Fig. 2). The fractions containing α-fucosidase activity were pooled and applied on a Concana-Val-A Sepharose column. The α-fucosidase activity was retained on the column suggesting that the protein might be glycosylated. After affinity chromatography, the retained fraction was further fractionated by preparative isoelectric focusing (IEF; Fig. 3). α-Fucosidase activity was present from fraction 7 through 20 corresponding with a pH range between 7.1 and 7.9. These fractions obtained from the IEF were subjected to SDS-PAGE (data not shown). All the active fractions showed a 37-kD protein band, but some minor contaminant proteins were also present. Thus, fractions 7 through 19 were pooled and subjected to a further purification step using gel permeation chromatography. The chromatography on Sephacryl S-200 HR rendered a single peak of α-fucosidase activity (Fig. 4) eluting in high molecular mass fractions (approximately 200 kD). Aliquots from active fractions 18 to 21 were analyzed by SDS-PAGE, and a protein band of 37 kD was present in all fractions (Fig. 5). No other band was visible in fractions 19 and 20, which had α-fucosidase activity.
Table I.
Purification of α-l-fucosidase from cabbage leaves
| Step | Activity | Protein | Specific Activity | Recovery | Purification |
|---|---|---|---|---|---|
| nkat | mg | nkat mg−1 | % | ||
| Crude extract | 45 | 1,896 | 0.023 | 100 | 1 |
| Ammonium | 45 | 327 | 0.137 | 100 | 6 |
| Sulfate ppt SP-Sepharose | 41 | 177 | 0.231 | 91 | 10 |
| Phenyl-Sepharose | 35.2 | 86 | 0.409 | 78 | 17 |
| Con-A Sepharose | 31.7 | 21 | 1.509 | 70 | 65 |
| Isoelectric focusing | 1.5 | 0.798 | 1.879 | 3.3 | 81 |
| Sephacryl S-200 HR | 0.66 | 0.137 | 4.817 | 1.46 | 209 |
Purification procedures are described in “Materials and Methods.” Activity was assayed against 2′-fucosyl-lactitol.
Figure 2.
Hydrophobic-interaction chromatography. The cabbage protein extract partially purified after ammonium sulfate precipitation and cation-exchange chromatography was chromatographed on a Phenyl-Sepharose HR column. The column was eluted with a gradient between 2 and 0 m of (NH4)2SO4 (upper panel) dissolved in 0.1 m sodium acetate pH 5.5 containing 1 mm dithiotreitol (DTT). Four-milliliter fractions were collected and the α-fucosidase activity (●) was measured against 2′-fucosyl-lactitol. Protein was measured by A280 (○).
Figure 3.
Preparative IEF. The partially purified extract from cabbage leaves after affinity chromatography was subjected to a preparative IEF using a Mini-Rotofor equipment with an ampholytes pH range between 6.7 and 8.0. One-milliliter fractions were collected and the α-fucosidase activity (●) was measured against 2′-fucosyl-lactitol. Protein was measured by the Coomassie protein assay reagent (○).
Figure 4.
Gel permeation chromatography. Fractions 7 to 19 from IEF were pooled and chromatographed on a Sephacryl S-200 HR column. The column was eluted with 0.1 m sodium acetate, pH 5.5, containing 1 mm DTT. Four-milliliter fractions were collected. α-Fucosidase activity (●) was measured against 2′-fucosyl-lactitol, and protein was measured by the Coomassie protein assay reagent.
Figure 5.
SDS-PAGE. Selected fractions from the gel permeation chromatography were analyzed by SDS-PAGE on 12% (w/v) polyacrylamide gels. M, Molecular markers.
Thus, a protein with α-fucosidase activity was purified after six consecutive steps. The optimum pH for this enzyme was shown to be 5.5. The specific activity at the end of the purification process rose to 4.81 nkat mg−1 (Table I). This low activity might be caused by some inactivation during the purification procedure or high ionic concentration extraction.
Arabidopsis α-Fucosidase Genes
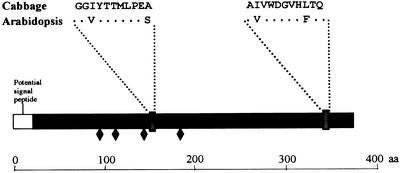
The purified protein was subjected to SDS-PAGE, and the 37-kD protein band was extracted and subjected to tryptic digestion as indicated in “Materials and Methods.” Two internal peptide sequences of 11 amino acids each were obtained (Fig. 6), and they were used as a query to identify homologous sequences in the databases of Arabidopsis. The BLAST 2.0 program found in BAC F12A21 (GenBank accession no. AC008113) a putative gene that could be translated to a protein (GenBank accession no. AC008113.4) that comprises two amino acid sequences with an identity of 73% with each of the cabbage peptides (Fig. 6). Thus, we expected this gene to encode an α-fucosidase in Arabidopsis and propose AtFXG1 as its name. As predicted by GENSCAN (Burge and Karlin, 1997), there appear to be two introns in this sequence. The expected length of the coding region is 1,116 bp, corresponding to an expected molecular mass of 38 kD for the mature protein, with a theoretical pI of 8.31. Both data fit with our findings on cabbage. Further analysis of this sequence using PSORT (Nakai and Kanehisa, 1992) revealed a potential signal peptide, suggesting a possible secretion pathway as well as four potential N-glycosylation sites (Fig. 6).
Figure 6.
Schematic representation of the AtFXG1 protein. Amino acid sequence of the two peptides obtained from purified cabbage α-fucosidase are given. Identical amino acids are noted as asterisks. ♦, Putative N-glycosylation sites.
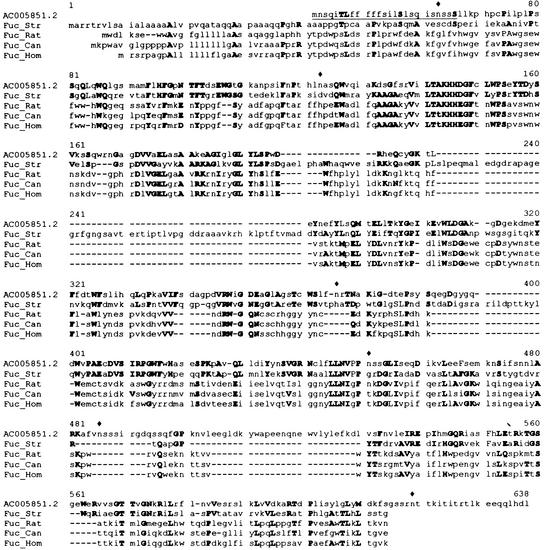
A new search based on homology with known α-fucosidases from animals and microorganisms of family 29 rendered a putative gene contained in BAC F24D13 from Arabidopsis (GenBank accession no. AC005851). This gene encodes a protein that exhibits 44% positive amino acids when compared with the α-fucosidase from Streptomyces sp. (Fig. 7). A translation of this DNA sequence can be obtained by GenBank accession number AC005851.2, although our prediction of the mature protein differs in length because of the existence of two introns as predicted by GENSCAN (Burge and Karlin, 1997). The expected coding region is 1,518 bp long. We propose AtFUC1 as its name. The protein encoded by this gene has a molecular mass of 54 kD and a theoretical pI of 5.2. Again, analysis of this sequence revealed a potential signal peptide and six potential N-gly-cosylation sites.
Figure 7.
Comparison of a putative gene from Arabidopsis (accession no. AC005851) with known fucosidases from different sources. Bold capital letters mean identical or conservative substitutions, lowercase letters mean non-conserved residues. Fuc_Rat, α-l-Fucosidase from Rattus norvegicus (S10235); Fuc_Hom, α-l-fucosidase from Homo sapiens (NP 000138); Fuc_Can, α-l-fucosidase from Canis familiaris (P48300); and Fuc_Str, α-l-fucosidase from Streptomyces sp. (AAD10477). ♦, Putative N-glycosylation sites.
Heterologous Expression
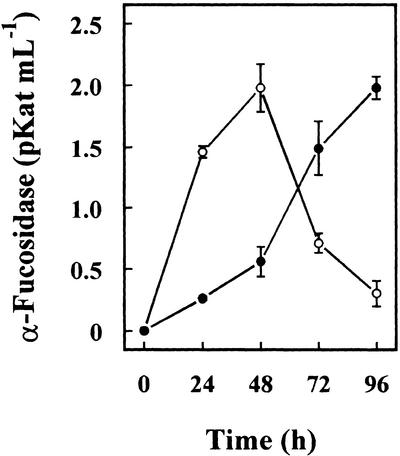
To confirm the putative α-fucosidase activity of both Arabidopsis genes, they were used to transform P. pastoris cells, and the heterologous proteins were purified from the culture media. α-Fucosidase activity against 2′-fucosyl-lactitol was detected in the culture medium of cells transformed with both sequences from Arabidopsis (Fig. 8). Cells transformed with AtFUC1 achieved the highest activity at 48 h, whereas cells transformed with AtFXG1 continued to increase for 96 h. The cells transformed with a control plasmid without insert did not show any activity. Because the recombinant protein should have incorporated a poly-His tag, both proteins were purified from the culture media by affinity chromatography and their α-fucosidase activity measured (Table II). When the activity of both heterologous proteins was assayed against XXFG, only AtFXG1 was able to release the t-Fuc from XXFG, as demonstrated by analysis of the reaction products by paper chromatography (Table II). Neither of expressed proteins showed activity against p-nitrophenyl-fucoside. Thus, although both Arabidopsis genes encode proteins with α-fucosidase activity, they differ in their substrate specificity.
Figure 8.
α-Fucosidase activity measured against 2′-fucosyl-lactitol in the culture medium of P. pastoris cells transformed with Arabidopsis α-fucosidase genes. ○, AtFUC1; ●, AtFXG1.
Table II.
α-Fucosidase activity of the affinity-purified AtFXG1 and AtFUC1 expression products from Pichia pastoris-transformed cells
| Gene | Substrate
|
||
|---|---|---|---|
| p-Nitrophenyl-α-fucoside | XXFG | 2′-Fucosyl-lactitol | |
| pkat mg−1 | |||
| AtFXG1 | N.D. | 21.1 ± 3.4 | 146.6 ± 6.5 |
| AtFUC1 | N.D. | N.D. | 455.0 ± 20.2 |
N.D., Not detected.
DISCUSSION
The presence of an enzymatic activity able to release t-Fuc from the oligosaccharide XXFG and from 2′-fucosyl-lactitol but not from p-nitrophenyl-α-l-fucopyranoside have been shown in Arabidopsis plants (Fig. 1). Previous results have shown the presence in cabbage of a similar enzymatic activity.
In the complete Arabidopsis genome, a putative gene (GenBank accession no. AC034106.4) showed 34% identities with pea α-fucosidase, the only plant fucosidase cloned at the moment. However, it seem to be more related to miraculins (47% identities). That Arabidopsis gene and all those closely related do not seem good candidates to code for the detected α-fucosidase activity. Thus, the homology to α-fucosidase of pea, a Leguminosae plant, is not sufficient to identify apoplastic fucosidases in relatively distant Brassicaceae species (e.g. Arabidopsis and cabbage).
Thus, we looked for Arabidopsis gene/s encoding α-fucosidase activity using two different experimental approaches: searching for sequences homologous to α-fucosidase peptides from cabbage, a very closely related taxon, and searching for sequences homologous to known α-fucosidases from different sources. An α-fucosidase was purified from cabbage leaves to render a single protein band on SDS-PAGE (Table I; Fig. 5). The molecular mass of α-fucosidase under denaturing conditions was 37 kD. Different molecular mass values for α-fucosidases purified from plant under denaturing conditions have been reported: 20 kD for pea (Augur et al., 1993) and 54 kD for almond (Scudder et al., 1990). The different behavior of α-fucosidase on gel permeation chromatography and SDS-PAGE might be explained in terms of a protein complex composed by a number of identical subunits. When the pH was raised to 8.5 during GPC, an additional peak of activity was observed at lower molecular mass (approximately 100 kD, data not shown), suggesting that cabbage α-fucosidase forms a non-covalently bound complex. An α-fuco-sidase of 220 kD from Pomacea canaliculata has been found to be tetramer (Hirata et al., 1996).
Important differences between cabbage and pea α-fucosidases were found for pI, their values being 7.5 (Fig. 4) and 5.5 (Augur et al., 1993), respectively. Thus, the α-fucosidase from cabbage showed different characteristics as compared with the α-fucosidase purified from other plants. Furthermore, if the peptide sequences obtained from the purified α-fucosidase from cabbage are compared with the protein product translated from the cDNA of pea α-fucosidase (Augur et al., 1995; accession no. CAA57931), no significant sequence homology is found.
The search based on both peptide sequences obtained from cabbage rendered a putative gene from Arabidopsis that could be translated to a protein (accession no. AC008113.4) that showed a high identity with both peptide sequences (Fig. 6). Thus, that protein, which does not yet have an assigned function, might be considered as a putative α-fucosidase. As expected for an apoplastic α-fucosidase, it showed a secretion signal peptide. Its molecular mass of 38 kD was very close to the molecular mass found for the α-fucosidase from cabbage. Finally, when that putative α-fucosidase sequence was heterologously expressed in P. pastoris cells, they secreted to the culture medium an α-fucosidase active against 2′-fucosyl-lactitol as well as against XXFG but not against p-nitrophenyl-fucoside (Table II). The α-fucosidase activity of the recombinant enzyme from P. pastoris was lower than the activity of the cabbage purified enzyme (Table I). The low activity is possibly caused by adverse post-translational modification or misfolding. These data clearly demonstrated that this Arabidopsis sequence (accession no. AC008113) encoded a α-fucosidase with similar substrate specificity to that detected in the leaves of Arabidopsis (Fig. 1). We propose AtFXG1 as the name for this Arabidopsis gene. This sequence shares some homology with a previously reported Arabidopsis gene with reported hydrolase activity against lipids (Brick et al., 1995), which belongs to a family that also comprises nodulins, acetyl-transferases, and several putative proteins with unassigned functions. However, when activity against lipids was checked we did not find detectable activity.
Furthermore, an Arabidopsis gene (accession no. AC005851; Fig. 7) with homology with known α-fucosidases from animals and microorganisms, also encoded a α-fucosidase active against 2′-fucosyl-lactose but not against XXFG (Table II). The translation product also showed a potential secretion signal peptide, but its theoretical molecular mass was 54 kD (as reported for the fucosidase purified from almond), higher than that of cabbage α-fucosidase. We propose AtFUC1 for this Arabidopsis gene. Although both Arabidopsis genes (AtFXG1 and AtFUC1) had α-fucosidase activity, the protein sequences showed no homology (Figs. 6 and 7).
AtFXG1, which is able to remove the t-fucosyl residues from xyloglucan oligosaccharides, might have a key role in the regulation of the XXFG levels as proposed for pea fucosidase by Augur et al. (1993). An inhibitory effect of auxin-induced (York et al., 1984; McDougall and Fry, 1988, 1989) and acid pH-induced (Lorences et al., 1990) growth has been found for XXFG when applied at nanomolar range. The action of AtFXG1 removing the t-Fuc would convert XXFG into XXLG, a growth inducer oligosaccharide that in turn would be further degraded by other glycanases as β-galactosidase (Edwards et al., 1988), α-xylosidase (Sampedro et al., 2001), and β-glucosidase (Cline and Albersheim, 1981). Thus, the α-fucosidase may inactivate the growth-inhibitor XXFG, leading to a growth-inducer product that will be further degraded, rendering small inactive oligosaccharides. Finally, the identification of the AtFXG1 gene encoding for an α-fucosidase able to remove t-Fuc from xyloglucan oligosaccharides opens new ways to elucidate the potential functions of that oligosaccharide in developing plant tissues.
The other identified α-fucosidase, AtFUC1, was not able to act on XXFG, at least when it was heterologously expressed in P. pastoris, suggesting that its function in vivo should be different from the function of AtFXG1. AtFUC1 probably acts on fucosylated substrates other than xyloglucan oligosaccharides. t-Fuc is also present on glycoproteins and on pectic rhamnogalacturonan II, which could be the in vivo substrate for AtFUC1 (Stevenson et al., 1988).
MATERIALS AND METHODS
Plant Material
Cabbages (Brassica oleracea L. var capitata) were purchased at a local market and processed in the same day. Arabidopsis ecotype Columbia plants were grown at 22°C under a 16-h light/8-h dark photoperiod. The leaves were harvested after 21 d and sorted in three groups depending on their developmental stage.
To obtain the apoplastic fraction, Arabidopsis seeds were superficially sterilized with 1% (w/v) NaClO for 10 min, thoroughly rinsed with sterile water, and grown for 3 d in 100-mL flasks containing 20 mL of sterile distilled water at 25°C, with orbital shaking at 120 rpm under continuous light.
Preparation of α-Fucosidase Substrates
[3H]Fuc-labeled xyloglucan was custom prepared as described by Fry (1988). Rapidly growing cell suspension cultures of spinach (Spinacia oleracea) were incubated aseptically with 30 MBq of [3H]Fuc (Amersham International, Buckinghamshire, UK). The cells were harvested after 7 d and the ethanol-insoluble residue prepared as described elsewhere. Hemicelluloses were extracted overnight with 6 m NaOH supplemented with 1% (w/v) NaBH4. The extract was neutralized, dialyzed, and freeze-dried. The lyophilized hemicelluloses were digested with cellulase (Megazyme, Megazyme International Ireland Limited, Wicklow, Ireland), and the oligosaccharides released were chromatographed on a Bio-Gel P-2 (Bio-Rad, Hercules, CA) column. The fractions corresponding to the main peak of kav ∼ 0.37 were pooled together and analyzed by paper chromatography in butan-1-ol/pyridine/water (4:3:4, v/v). The chromatogram resulted in a single peak of radioactivity with Rmaltoheptaose = 1 as expected for XXFG. Further analysis included a hydrolysis with trifluoroacetic acid of the [3H]oligosaccharide. The paper chromatography performed with the product of this hydrolysis showed a single peak of radioactivity that co-chromatographed with Fuc.
To prepare 2′-fucosyl-lactitol, 2 mg of 2′-fucosyl-lactose (Sigma, St. Louis) were incubated for 1 h at room temperature in 1 m NH4(OH) containing 2% (w/v) NaBH4, neutralized with acetic acid and vacuum-dried in a Speed-Vac (Savant Instruments, Inc., Holbrook, NY). After three washes with acetic acid:methanol (1:9) and four with methanol, the 2′-fucosyl-lactitol was diluted in the required amount of H2O.
α-Fucosidase Assay
2′-Fucosyl-lactitol was usually used as substrate for α-fucosidase determination. The activity was determined as reducing sugars release, according to the method of Lever (1972). Aliquots of extract containing α-fucosidase were incubated with 20 μg of 2′-fucosyl-lactitol in 100 mm sodium acetate buffer, pH 5.5, at 35°C for a variable period according to the extract activity. When [3H]XXFG was used as substrate, 1.6 kBq of radiolabeled substrate were incubated with the fucosidase-containing extract for 14 h. The amount of radioactivity released as free Fuc was determined by paper chromatography followed by scintillation counting.
The activity against p-nitrophenyl-α-l-fucopyranoside was measured with 10 mm substrate concentration in 0.1 m sodium acetate buffer pH 5.5 at 35°C for 6 h. After incubation, the reaction was stopped by adding 0.2 m Na2CO3 and the A400 measured.
Arabidopsis Protein Extract Preparation
Leaves from 21-d-old Arabidopsis were sorted according to their developmental stage. The leaves (100-mg fresh weight) were frozen in liquid N2 and then homogenized with sodium 1 m acetate buffer, pH 5.5. After centrifugation (15,000g; 20 min), the crude extract was dialyzed and concentrated using a Centricon-10 (Millipore Corporation, Bedford, MA). Aliquots of this extract were incubated separately with 2′-fucosyl-lactitol, [3H]radiolabeled XXFG, or p-nitrophenyl-α-l-fucopyranoside, and the α-fucosidase activity measured as previously described.
Apoplastic Fraction Extraction
The apoplastic fluid from Arabidopsis seedlings was obtained as described by Monroe et al. (1999) and Sampedro et al. (2001). Seedlings grown on water for 3 d were transferred to 10 mL of 0.1 m MES buffer pH 6.0 containing 1 m LiCl and shaken for 6 h at 120 rpm. The saline extract was considered to be the apoplastic fraction.
Seedlings were then homogenized in 5 mL of 0.1 m MES buffer, pH 6.0, containing 1 m LiCl. The homogenate was shaken for 2 h and centrifuged at 15,000g for 20 min. The supernatant was considered to be the cytoplasmic fraction. Both extracts were concentrated with a Centricon-10 (Millipore) to approximately 200 μL and then diluted to 2 mL with 100 mm MES, pH 6.0. This procedure was repeated twice. Finally, the extract was concentrated, and α-fucosidase activity was then measured.
The absence of cytoplasmic contamination was assessed according to Sánchez et al. (1997) by measuring Glc-6-phosphate dehydrogenase activity. To measure this activity extracts were obtained as stated above except for the extraction buffer that was 0.1 m MES, pH 6.0, containing 1 m NaCl. Glc-6-phosphate dehydrogenase activity was determined measuring the absorption increase at 340 nm, in a reaction mixture (1 mL) containing 100 μL of protein extract, 1 mm Glc-6-phosphate, 0.2 mm NADP+, 1 mm MgCl2, and 0.1 m Tricine, pH 8.0 (Takahama, 1993). α-Fucosidase activity was determined in these extracts by hydrolysis of 2′-fucosyl-lactitol hydrolysis, as described previously.
Protein Determination
Protein concentration was determined by the Coomassie protein assay reagent (Pierce, Rockford, IL) using bovine serum albumin as a standard (Bradford, 1976).
α-Fucosidase Purification
The inner leaves of cabbage (3.5-kg fresh weight) were cut in small pieces after removal of the main veins and homogenized in 0.1 m sodium acetate buffer, pH 5.5 (5 L), with a Polytron (Kinematica, Luzern, Switzerland). The homogenate was filtered through muslin cheesecloth and the residue suspended again in 1 m sodium acetate buffer, pH 5.5, containing 1 mm DTT and 1.2% (w/v) polyvinylpolypirrolidone (Sigma). The suspension was maintained under magnetic stirring for 2 h, filtered again and the filtrate centrifuged at 15,000g for 40 min. The supernatant was considered as the crude extract. Unless otherwise stated all further purification steps were carried out at 4°C.
The crude extract was brought to 40% saturation with (NH4)2SO4, the precipitate discarded and the supernatant was brought to 80% saturation. The new precipitate was dissolved in 0.1 m sodium acetate pH 5.5 containing 1 mm DTT and dialyzed against the same buffer.
After the ammonium sulfate fractionation, the partially purified extract was chromatographed on a Sepharose-SP column (2.5 × 10 cm; flow rate, 2 mL min−1; Amersham Pharmacia Biotech AB, Uppsala). The column was washed with 0.1 m sodium acetate, pH 5.5, containing 1 mm DTT (200 mL). The retained fraction was eluted with 45 mL of the same buffer supplemented with 2 m (NH4)2SO4 and 1 mm DTT.
The fraction retained in the cationic-exchange column was concentrated by ultrafiltration (AMICON YM10, Millipore) and applied on a Phenyl Sepharose HR (Amersham Pharmacia Biotech) column (1 × 30 cm; flow rate, 1 mL min−1). The column was washed with 0.1 m sodium acetate, pH 5.5, containing 2 m (NH4)2SO4 and 1 mm DTT. The column was eluted with 200 mL of 0.1 m sodium acetate, pH 5.5, containing 1 mm DTT with a linear gradient of (NH4)2SO4 from 2 to 0 m, and 4-mL fractions were collected.
The active fractions after the hydrophobic chromatography step were pooled and dialyzed against binding buffer (50 mm sodium acetate buffer, pH 5.5, containing 0.2 m NaCl, 1 mm CaCl2, and 1 mm Mn2Cl). The partially purified extract was then chromatographed on a ConA-Sepharose (Amersham Pharmacia Biotech) column (1.5 × 2.8 cm; flow rate, 0.8 mL min−1). The column was equilibrated and washed with the same buffer. The retained fraction was eluted with 20 mL of binding buffer supplemented with 100 mm methyl-O-d-glucopyranoside.
The extract was then applied to a Sephadex G25 column (Amersham Pharmacia Biotech) and eluted with 10 mm sodium acetate buffer, pH 5.5. Afterward, the sample was concentrated by ultrafiltration (Amicon YM 10, Millipore) to a final volume of 10 mL. Preparative IEF was performed using a Mini-Rotofor chamber (Bio-Rad), following the manufacturer's directions. Narrow range ampholytes (Biolyte 6.7–8.0, Bio-Rad) were used.
Fractions 7 to 19 from the IEF were pooled, and ampholytes were removed by dialysis-ultrafiltration against 0.1 m sodium acetate buffer, pH 5.5, containing 1 m NaCl and 1 mm DTT, and then concentrated to a final volume of 1.5 mL. The sample was chromatographed in a Sephacryl S-200 HR column (1.5 × 90 cm; flow rate, 1 mL min−1; Amersham Pharmacia Biotech), and 4-mL fractions were collected.
Analytical electrophoresis was performed using a Mini-Protean II electrophoresis cell (Bio-Rad) on 12% (w/v) polyacrylamide gels following the manufacturer's directions. Silver Stain Kit, Protein (Amersham Pharmacia Biotech) was used. Silver stain SDS-PAGE standards (low range, Bio-Rad) were employed to determine apparent molecular weights.
Gel permeation selected fractions containing α-fuco-sidase activity (40 μg of protein) were pooled, concentrated with a Centricon-10 (Millipore), vacuum-dried with a Speed Vac (Savant Instruments), and electrophoresed as described above. After a brief staining with Coomassie blue, the only visible band was sliced. Eurosequence (Groningen, The Netherlands) performed tryptic peptide sequencing of two peptides.
Sequence Analysis
Similarity searches were done with the National Center for Biotechnology Information BLAST 2.0 (Altschul et al., 1997). Signal peptide analysis was performed with PSORT (Nakai and Kanehisa, 1992). Multiple alignments were done with MultAlin (Corpet, 1988). For prediction of splice sites, GENSCAN (Burge et al., 1997) was used.
Heterologous Expression
AtFUC1 and AtFXG1 (accession nos. AC005851.2 and AC008113.4) were amplified by PCR from an Arabidopsis cDNA bank (Minet et al., 1992). We designed primers Ex5851-1 (5′-GAATTCTCATCACTACTAAAACCACACC-3′) and Ex5851-2 (5′-GCGGCCGCCAAATCATGTAGTTG- CTGCTCT-3′) for amplifying AtFUC1, whereas Ex8113-1 (5′-GAATTCCATCAATGCCATTTCCCAGCA-3′) and Ex8113-2 (5′-GCGGCCGCCTGCCTTTTACAGGCCTTGCT-3′) were used for AtFXG1. In both cases, the potential signal peptide was excluded. These four primers include restriction sites for EcoRI (Ex5851-1 and Ex8113-1) and NotI (Ex5851-2 and Ex8113-2). PCR conditions were as follows: 94°C for 2 min followed by 30 cycles of 94°C for 45 s, 50°C for 45 s, and 72°C for 90 s, followed by 72°C for 5 min. Pfu DNA polymerase (Promega, Madison, WI) was used according to the manufacturer's recommendations. The PCR reaction included 25 pmol of each primer and 100 ng of template.
For both sequences, a unique PCR product of the expected size was observed on agarose electrophoresis. The DNA was extracted and purified from the gel with QIAEX II gel extraction kit (QIAGEN GmbH, Hilden, Germany), and ligated to pGAPZαA Pichia pastoris expression vector (Invitrogen Corporation, Carlsbad, CA) for constitutive expression in Pichia spp., using T4DNA ligase (Promega). XL1-Blue Escherichia coli was transformed with the recombinant plasmid (Sambrook et al., 1989). The X-33 strain of P. pastoris was transformed by the lithium chloride method with plasmid purified from E. coli following the manufacturer's directions.
For analysis of both sequences, six cultures (10 mL of yeast peptone dextrose [1% {w/v} yeast extract, 2% {w/v} peptone, and 2% {w/v} Glc]) of independently transformed Zeocin-resistant colonies were used to inoculate 2-L flasks containing 500 mL of 1.34% (w/v) yeast nitrogen base, and 0.5% (w/v) Glc. Cultures were grown for 96 h at 30°C at 200 rpm. At 0, 24, 48, 72, and 96 h, aliquots of the culture were transferred to a microcentrifuge tube to determine the optimal time to harvest. Cells were harvested by centrifugation (4,500g for 10 min), disrupted with glass beads and extracted in 5 mL of 1 m sodium acetate buffer, pH 5.5, for 3 h. Control transformations were performed as indicated by the manufacturer, and cells were grown and treated as above.
Purification of the Expressed Protein
TALON Purification Kit (CLONTECH Laboratories Inc., Palo Alto, CA) was employed to purify the expressed proteins from the culture media, expecting the poly-His-tagged proteins to be retained by the cobalt-based resin. The supernatant obtained after centrifugation of the cultures was concentrated to 20 mL by ultrafiltration (AMICON YM10, Millipore), and the pH was raised to 7.5 by addition of 50 mm sodium phosphate buffer, pH 8, supplemented with 300 mm NaCl and 1 mm DTT. The sample was then chromatographed on a column containing 1 mL of TALON affinity resin. A hybrid batch/gravity-flow column purification following the manufacturer's directions was performed. Ten bed volumes of the same buffer were used to wash the column. The protein was eluted by lowering the pH using Elution buffer and 50 mm sodium acetate buffer, pH 5.0, containing 0.3 m NaCl.
ACKNOWLEDGMENTS
We are grateful to Dr. Ester P. Lorences for her helpful comments and to Duncan A. Lindsay for correcting the English version.
Footnotes
This work was supported by the Dirección General de Investigación (Ministerio de Ciencia y Tecnología, Spain; grant no. PB98–0640) and the Xunta de Galicia (grant no. PGIDT00PXI20002PN).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010508.
LITERATURE CITED
- Altschul SF, Madden TL, Schaffer TL, Zhang J, Zhang Z, Miller W, Lipman AJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augur C, Benhamou N, Darvill A, Albersheim P. Purification, characterization, and cell wall localization of an α-fucosidase that inactivates a xyloglucan oligosaccharin. Plant J. 1993;3:415–426. doi: 10.1046/j.1365-313x.1993.t01-24-00999.x. [DOI] [PubMed] [Google Scholar]
- Augur C, Stiefel V, Darvill A, Albersheim P, Puigdomenech P. Molecular cloning and pattern of expression of an α-l-fucosidase gene from pea seedlings. J Biol Chem. 1995;270:24839–24843. doi: 10.1074/jbc.270.42.24839. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brick DJ, Brumlik MJ, Buckley T, Cao JX, Davies PC, Misra S, Tranbarger TJ, Upton C. A new family of lipolytic plant enzymes with members in rice, Arabidopsis and maize. FEBS Lett. 1995;377:475–480. doi: 10.1016/0014-5793(95)01405-5. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Cline K, Albersheim P. Host-pathogen interactions: XVII. Purification and characterization of a β-glucosyl hydrolase/transferase present in the walls of soybean cells. Plant Physiol. 1981;68:207–220. doi: 10.1104/pp.68.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M, Bowman YJL, Dea ICM, Reid JSG. A β-d-galactosidase from nasturtium (Tropaeolum majusL.) cotyledons. J Biol Chem. 1988;263:4333–4337. [PubMed] [Google Scholar]
- Farkas V, Hanna R, Maclachlan G. Xyloglucan oligosaccharide α-l-fucosidase activity from growing pea stems and germinating nasturtium seeds. Phytochemistry. 1991;30:3203–3207. doi: 10.1016/0031-9422(91)83176-l. [DOI] [PubMed] [Google Scholar]
- Fisher K, Aronson N., Jr Isolation and sequence analysis of a cDNA encoding rat liver alpha-l-fucosidase. Biochem J. 1989;15:695–701. doi: 10.1042/bj2640695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC. In-vivo formation of xyloglucan nonasaccharide: a possible biologically active cell wall fragment. Planta. 1986;169:443–453. doi: 10.1007/BF00392143. [DOI] [PubMed] [Google Scholar]
- Fry SC. The Growing Plant Cell Wall: Chemical and Metabolic Analysis. Essex, UK: Longman Scientific & Technical; 1988. [Google Scholar]
- Fry SC. The structure and function of xyloglucan. J Exp Bot. 1989;40:1–11. [Google Scholar]
- Fry SC. Polysaccharide-modifying enzymes in the plant cell wall. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:497–520. [Google Scholar]
- Fry SC, Aldington SP, Hetherington PR, Aitken J. Oligosaccharides as signals and substrates in the plant cell wall. Plant Physiol. 1993;103:1–5. doi: 10.1104/pp.103.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima H, DeWet J, O'Brien J. Molecular cloning of a cDNA for human alpha-l-fucosidase. Proc Natl Acad Sci USA. 1985;82:1262–1265. doi: 10.1073/pnas.82.4.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata K, Nakahara Y, Kimura Y, Funatsu G. Purification and some properties of a β-xylosidase and an α-fucosidase from apple snails (Pomacea canaliculata) Biosci Biotechnol Biochem. 1996;60:249–254. doi: 10.1271/bbb.60.249. [DOI] [PubMed] [Google Scholar]
- Lever M. A new reaction for colorimetric determination of carbohydrates. Anal Biochem. 1972;47:273–279. doi: 10.1016/0003-2697(72)90301-6. [DOI] [PubMed] [Google Scholar]
- Lorences EP, McDougall GJ, Fry SC. Xyloglucan- and cello-oligosaccharides: antagonists of the growth promoting effect of H+ Physiol Plant. 1990;80:109–113. [Google Scholar]
- McDougall G, Fry SC. Inhibition of auxin-stimulated growth of pea stem segments by a specific nonasaccharide of xyloglucan. Planta. 1988;175:412–416. doi: 10.1007/BF00396348. [DOI] [PubMed] [Google Scholar]
- McDougall G, Fry SC. Structure-activity relationships for xyloglucan oligosaccharides with anti-auxin activity. Plant Physiol. 1989;89:883–887. doi: 10.1104/pp.89.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougall G, Fry SC. Xyloglucan oligosaccharides promote growth and activate cellulase: evidence for a role of cellulase in cell expansion. Plant Physiol. 1990;93:1042–1048. doi: 10.1104/pp.93.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet M, Dufour M-E, Lacroute F. Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thalianacDNAs. Plant J. 1992;2:417–422. doi: 10.1111/j.1365-313x.1992.00417.x. [DOI] [PubMed] [Google Scholar]
- Monroe JD, Gough CM, Chandler LE, Loch CM, Ferrante JE, Wright PW. Structure, properties, and tissue localization of apoplastic α-glucosidase in crucifers. Plant Physiol. 1999;119:385–397. doi: 10.1104/pp.119.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Occhiodoro T, Beckmann K, Morris CP, Hopwood J. Human α-l-fucosidase: complete coding sequence from cDNA clones. Biochem Biophys Res Commun. 1989;164:439–445. doi: 10.1016/0006-291x(89)91739-7. [DOI] [PubMed] [Google Scholar]
- Reiter WD, Chapple CCS, Somerville CR. Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science. 1993;261:1032–1035. doi: 10.1126/science.261.5124.1032. [DOI] [PubMed] [Google Scholar]
- Roberts K. How the cell acquired a cellular context. Plant Physiol. 2001;125:127–130. doi: 10.1104/pp.125.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sampedro J, Sieiro C, Revilla G, González-Villa T, Zarra I. Cloning and expression pattern of a gene encoding an α-xylosidase active against xyloglucan oligosaccharides from Arabidopsis. Plant Physiol. 2001;126:910–920. doi: 10.1104/pp.126.2.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez M, Queijeiro E, Revilla G, Zarra I. Changes in ascorbic acid levels in apoplastic fluid during growth of pine hypocotyls: effect on peroxidase activities associated with cell walls. Physiol Plant. 1997;101:815–820. [Google Scholar]
- Scudder P, Neville DCA, Butters TD, Fleet GWJ, Dwek RA, Rademacher TW, Jacob GS. The isolation by ligand affinity chromatography of a novel form of α-l-fucosidase from almond. J Biol Chem. 1990;265:16472–16477. [PubMed] [Google Scholar]
- Stevenson TT, Darvill AG, Albersheim P. Structural features of the plant cell-wall polysaccharide rhamnogalacturonan II. Carbohydr Res. 1988;182:207. [Google Scholar]
- Takahama U. Regulation of peroxidase dependent oxidation of phenolics by ascorbic acid: different effects of ascorbic acid on the oxidation of coniferyl alcohol by the apoplastic soluble and cell wall-bound peroxidases from epicotyls of Vigna angularis. Plant Cell Physiol. 1993;39:809–817. [Google Scholar]
- York W, Darvill A, Albersheim P. Inhibition of 2,4-dichlorophenoxyacetic acid stimulated elongation of pea stem segments by a xyloglucan oligosaccharide. Plant Physiol. 1984;75:295–297. doi: 10.1104/pp.75.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]