Abstract
A full-length cDNA clone (LeCDPK1) from tomato (Lycopersicon esculentum) encoding a calcium-dependent protein kinase (CDPK) was isolated by screening a cDNA library from tomato cell cultures exposed to Cladosporium fulvum elicitor preparations. The predicted amino acid sequence of the cDNA reveals a high degree of similarity with other members of the CDPK family. LeCDPK1 has a putative N-terminal myristoylation sequence and presents a possible palmitoylation site. The in vitro translated protein conserves the biochemical properties of a member of the CDPK family. In addition, CDPK activity was detected in soluble and particulate extracts of tomato leaves. Basal levels of LeCDPK1 mRNA were detected by northern-blot analysis in roots, stems, leaves, and flowers of tomato plants. The expression of LeCDPK1 was rapidly and transiently enhanced in detached tomato leaves treated with pathogen elicitors and H2O2. Moreover, when tomato greenhouse plants were subjected to mechanical wounding, a transient increase of LeCDPK1 steady-state mRNA levels was detected locally at the site of the injury and systemically in distant non-wounded leaves. The increase observed in LeCDPK1 mRNA upon wounding correlates with an increase in the amount and in the activity of a soluble CDPK detected in extracts of tomato leaves, suggesting that this kinase is part of physiological plant defense mechanisms against biotic or abiotic attacks.
Plants live in intimate contact with different microorganisms and insects and have evolved several mechanisms including physical barriers and inducible defenses to avoid or resist invasion by a pathogen or insect foraging. This capacity depends on early warning followed by the activation of wound/defense-response genes.
In gene-for-gene interactions, plants harboring specific disease resistance (R) genes avoid the infection of pathogens that carry the corresponding avirulence (Avr) genes (Flor, 1971). The R gene products act as receptors that recognize the matching Avr protein of potentially pathogenic microbes (Staskawicz et al., 1995). This recognition event triggers an hypersensitive response, which inhibits further infection of tissue by the pathogen. It was expected that R genes might encode components involved in signal recognition or signal transduction pathways. Four classes of R genes are now known: cytoplasmic protein kinases, protein kinases with an extracellular domain, cytoplasmic proteins with a region of Leu-rich repeats and a nucleotide binding site, and proteins with a region of Leu-rich repeats that appear to encode extracellular proteins (Halterman and Martin, 1997).
In addition to the hypersensitive response, plants use other defense mechanisms such as oxidative burst, strengthening of the cell wall, and expression of defense-related proteins to restrict the growth of pathogens. Changes in ion fluxes, protein phosphorylation/dephosphorylation, and generation of fatty-acid derivatives have been reported to occur in response to non-specific bacterial or fungal elicitors (for review, see Yang et al., 1997; Rushton and Somssich, 1998; Scheel, 1998).
Reactive oxygen species are common components of the defense responses of plants against pathogen and herbivore attacks. The oxidative burst is characterized by the rapid generation of hydrogen peroxide (H2O2). Wound-induced H2O2 accumulation is observed both locally and systemically in leaves of several plant species, apparently caused by oligogalacturonides (OGAs) released by a systemically wound-induced polygalacturonase (Bergey et al., 1999; Orozco-Cárdenas and Ryan, 1999).
Plants are also exposed to injuries caused by insect, pathogen attack, or mechanical wounding and respond by producing protective compounds, either at the site of the injury or systemically in distant unwounded tissues (Bowles, 1990; Ryan, 1990). Among the best studied defense-related genes are proteinase inhibitors, chitinase, Leu aminopeptidase, Phe ammonia lyase, and chalcone synthase, all of which accumulate systemically. The wound-induced response is regulated both by chemicals such as the phytohormones abscisic acid and JA (Peña-Cortés et al., 1989; Farmer and Ryan, 1990, 1992; Hildmann et al., 1992; Peña-Cortés et al., 1993), the octadecapeptide systemin (Pearce et al., 1991) and oligosaccharides (Ryan, 1987), and by physical signals such as hydraulic variation potentials and electrical activation potentials (Wildon et al., 1992; Herde et al., 1995).
Most of the genes involved in wounding are also activated in the plant defense mechanisms against pathogen invasion. It is now apparent that, regardless of the origin of the attack, the plant activates its defense against a variety of pathogens or wounding stresses by combining a limited number of common mechanisms. An increase in cytosolic calcium concentration, which occurs immediately after elicitation, appears to be a key regulator of the defense pathways triggered. Elevation of intracellular levels of calcium and changes in the pattern of protein phosphorylation are part of the responses to wounding in tomato (Lycopersicon esculentum) plant cells. Recent reports suggest that chelation of the ion affects the production of active oxygen species, phytoalexin production, mitogen-activated protein kinase activation, and defense gene activation (Scheel, 1998; Schaller and Oecking; 1999; Blume et al., 2000).
Typically, changes in calcium concentration are transduced via calcium-binding modulator proteins that affect directly or indirectly the activity of a protein kinase enzyme, protein phosphorylation being a major mechanism involved in transducing various external stimuli (Sopory and Munshi, 1998). A crucial role for protein phosphorylation has already been suggested by the isolation of the disease resistance Pto and Xa21 genes from tomato and rice (Oryza sativa; Martin et al., 1993; Song et al., 1995). In addition, Yang et al. (1997), Rojo et al. (1998), and Menke et al. (1999) have demonstrated that protein kinases and phosphatases are required for the activation of early defense responses and numerous reports suggest an important role of mitogen-activated protein kinases after race-specific and non-specific elicitation (Ligterink et al., 1997; Zhang et al., 1998; Romeis et al., 1999). Romeis et al. (2000) recently identified a calcium-dependent protein kinase (CDPK) in transgenic tobacco cell cultures expressing the Cf-9 gene, that underwent an Avr9/Cf-9-dependent transition from a nonelicited to an elicited form as result of reversible phosphorylation.
CDPKs are a unique class of Ser/Thr protein kinases very conserved in structure that consist of an N-terminal variable domain, a kinase catalytic domain, a junction domain, and a calmodulin-like domain (CLD) with conserved calcium binding motifs (Roberts and Harmon, 1992). These kinases are present only in plants and in some protists and have been reported to be involved in multiple signaling pathways and in the response to several environmental stresses.
In this work, we have cloned the first CDPK from tomato plants using a cDNA library from tomato cell cultures exposed to Cladosporium fulvum elicitor preparations. The expression of LeCDPK1 was studied both in detached leaves exposed to different defense signals and in greenhouse plants subjected to mechanical wounding. We report a rapid and transient increase of LeCDPK1 steady-state mRNA levels in response to the different treatments and in response to wounding. In addition, an increase in the amount and in the activity of CDPK occurs with a similar timing in extracts of wounded leaves. The data reported suggest that LeCDPK1 could play a role in the defense response and that the signal transduction pathways triggered in response to non-specific elicitation and wounding are interlinked by this kinase.
RESULTS
Molecular Characterization of LeCDPK1
The cloning of a CDPK from tomato was carried out by probing a cDNA library from tomato with a RT-PCR amplified fragment that corresponded to the catalytic domain of this protein kinase. The library was made from mRNA obtained from wild-type tomato cell cultures (L. esculentum cv Money maker) exposed during 1.5 h to C. fulvum (race 2.3) elicitor preparations. After three rounds of screening, a clone designated LeCDPK1 was purified and sequenced.
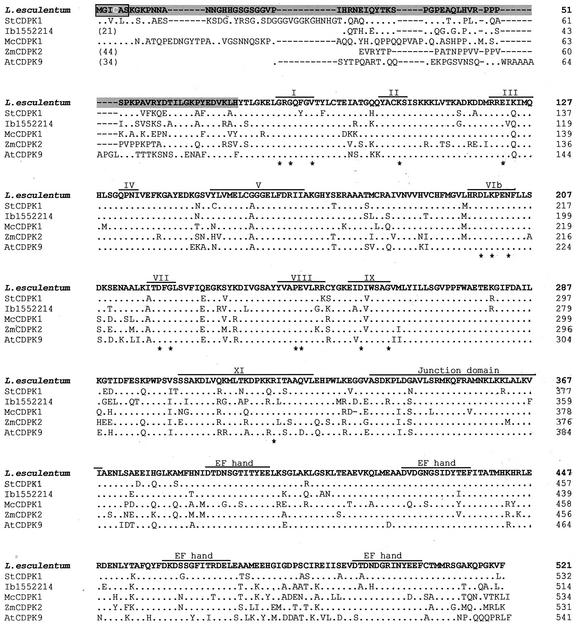
LeCDPK1 was a full-length clone with an open reading frame, 1,566 nucleotides long from the ATG to the stop codon, encoding a protein of 57,819 D (GenBank accession no. AF363784). The predicted amino acid sequence contains an N-terminal variable region, a kinase catalytic domain with subdomains I to XI from Ser/Thr kinases, a junction domain, and a CLD with four conserved calcium binding motifs characteristic of the CDPK family. Comparison of the predicted LeCDPK1 amino acid sequence with other CDPKs, shows a very strong homology that extends over all the conserved domains (Fig. 1). The highest identity and similarity (80% and 87%, respectively) is shared with the potato (StCDPK1) and sweet potato (Ib1552214) CDPKs. The alignment of LeCDPK1 with common ice plant (McCDPK1); maize (ZmCDPK2) and Arabidopsis (AtCDPK9) CDPKs also displayed high identity.
Figure 1.
Comparison of the amino acid sequence of LeCDPK1 with other CDPKs. Conserved residues are indicated with a dot. Dashes indicate absent residues. Asterisks indicate conserved residues in all Ser/Thr kinases. The N-terminal variable domain is shaded in gray, the myristoylation consensus is indicated with a box, and the putative palmitoylation site is written in white. Catalytic subdomains, the junction domain, and the EF-hands are indicated in the figure. The CDPKs aligned with LeCDPK1 are: potato (Solanum tuberosum; StCDPK1), sweet potato (Ipomoea batatas; Ib1552214), common ice plant (Mesembryanthemum crystallinum; McCDPK1), maize (Zea mays; ZmCDPK2), and Arabidopsis isoform 9 (AtCDPK9).
The less-conserved region of CDPKs is the N-terminal end. The first 26 amino acids of the N-terminal region of LeCDPK1 (Fig. 1) only share some homology with the N-terminal region of a potato CDPK (Raíces et al., 2001) and with a stress responsive CDPK from M. crystallinum (Patharkar and Cushman, 2000).
Biochemical Properties
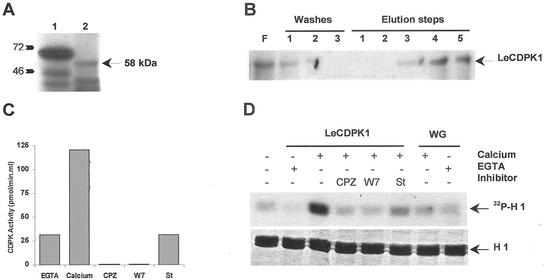
In vitro translation assays were carried out using a coupled transcription/translation system from wheat germ. As shown in Figure 2A, a protein of the predicted size of 58 kD was translated (lane 2). The positive control reaction (lane 1) done using T7 luciferase control DNA rendered a protein of approximately 61 kD.
Figure 2.
A, In vitro translation of LeCDPK1 (lane 2) was performed in the presence of [35S]Met. A control was carried out in parallel (lane 1). B, Purification of the in vitro translated [35S]LeCDPK1 using a Phenyl Sepharose column. Flowthrough (F), washes (1, 2, and 3), and elution steps (0.3 m NaCl [1 and 2], 0.4 m NaCl plus 5 mm EGTA [3 and 4], and 4 m urea [5]) were analyzed on 12% (w/v) SDS-PAGE. C, CDPK activity was determined in the urea fraction. A standard assay was performed in the presence of 1 mm EGTA or 1 mm CaCl2 using syntide-2 as substrate. Kinase inhibitors or calmodulin antagonists 0.5 mm chlorpromazine (CPZ), 1 mm W7, and 1 μm staurosporine (St) were tested in the presence of 1 mm CaCl2. D, Purified LeCDPK1 was incubated with 0.1 mg mL−1 histone H1 and 10 μm [γ-32P]ATP in the presence of 1 mm EGTA or 1 mm CaCl2. The same reaction was carried out in the presence of CPZ, W7, or staurosporine. An equivalent amount of a purified wheat germ translation reaction performed in the absence of pGEMT-LeCDPK1 (WG) was used as negative control. Histone loading (H 1), stained with Coomassie Brilliant Blue, is indicated.
The in vitro-translated [35S]LeCDPK1 was purified using a Phenyl Sepharose column, as described. The Phenyl Sepharose fractions were concentrated and loaded on an SDS-polyacrylamide gel. A band of the expected size could be observed in the percolate and in the fractions eluted with EGTA and urea (Fig. 2B). CDPK activity was determined in the urea eluted fraction, using syntide-2 or histone H1 as phosphate acceptors. LeCDPK1 activity was clearly dependent on the presence of calcium, whereas the addition of EGTA, CPZ, and W7 inhibited the enzyme (Fig. 2, C and D). Both CPZ and W7 are calmodulin antagonists that can also inhibit the CLD characteristic of CDPKs. The protein kinase inhibitor, staurosporine, also inhibited the enzyme's activity (Fig. 2, C and D). These results indicate that LeCDPK1 encodes an active CDPK.
The addition of 4 m urea was necessary to completely elute the enzyme suggesting that this protein displays a high affinity for a hydrophobic support. LeCDPK1 N-terminal region (shaded in gray in Fig. 1) is 72 amino acids long and contains a putative consensus sequence for N-myristoylation (MG2XXXS) as do other CDPKs (Hrabak et al., 1996; Ellard-Ivey et al., 1999; Martín and Busconi, 2000). A potential palmitoylation site, the Cys in position four is also present in this protein kinase. This suggests that LeCDPK1 could belong to a subgroup of plant CDPKs that have an SH4 domain containing sites for myristoylation and palmitoylation. These post-translational modifications are potentially involved in the protein interaction with the membrane fraction.
Southern-Blot Analysis of LeCDPK1
A Southern-blot assay was carried out using tomato genomic DNA digested with the restriction endonucleases EcoRI, EcoRV, and HindIII and hybridized with the full-length clone LeCDPK1 (Fig. 3). Two bands were revealed in the DNA digested with EcoRI (lane EI), four prominent bands were observed in the DNA digested with EcoRV (lane EV) whereas a prominent band and a fainter one were identified in the DNA digested with HindIII (lane H). These endonuclease sites are present in the sequence used to probe the blot; EcoRI has two restriction sites close to one another whereas EcoRV and HindIII have one site each. The four bands that appear in the EcoRV digestion may indicate the presence of other restriction sites in intronic sequences. The low number of hybridizing fragments revealed in each case suggests that LeCDPK1 is a single copy gene.
Figure 3.
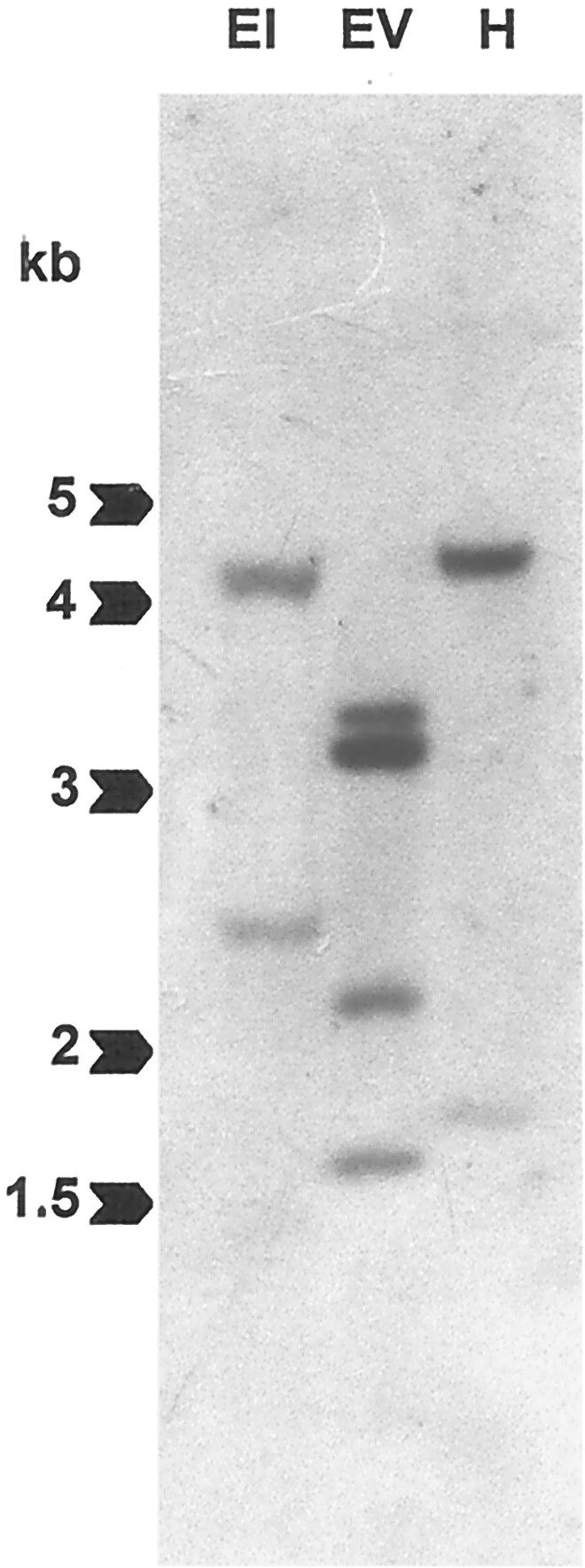
Southern-blot hybridization. Genomic DNA (12 μg) was digested with EcoRI (EI), EcoRV (EV), or HindIII (H), separated on 0.7% (w/v) agarose gels, blotted onto a nylon membrane, and hybridized with the full-length LeCDPK1 probe. Mr markers (in kb) are indicated with arrows.
General Expression Patterns of LeCDPK1
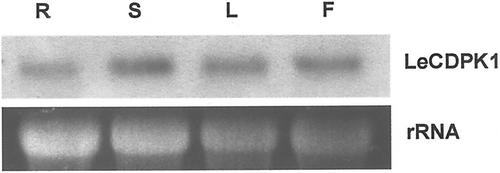
A band of about 2.5 kb corresponding to LeCDPK1 mRNA was detected in roots, stems, leaves, and flowers under high stringency conditions (Fig. 4). Though LeCDPK1 mRNA was present in all these organs, the steady-state levels of the transcript were lower in roots and higher in leaves and flowers. These experiments were carried out with flowering and non-flowering plants and, in both cases, the relative steady-state levels of LeCDPK1 mRNA were constant in the different vegetative organs.
Figure 4.
Northern-blot analysis of LeCDPK1 in roots (R), stems (S), leaves (L), and flowers (F) of tomato plants. RNA loading was checked with the ethidium bromide (EtBr) staining.
LeCDPK1 Is Involved in the Plant's Defense Response
A study with detached leaves was carried out to analyze whether signals involved in the induction of an active plant defense strategy could modify the basal levels of LeCDPK1 mRNA. The expression of LeCDPK1 was analyzed in response to the elicitors chitosan and polygalacturonide (PGA), to jasmonic acid (JA), and to H2O2.
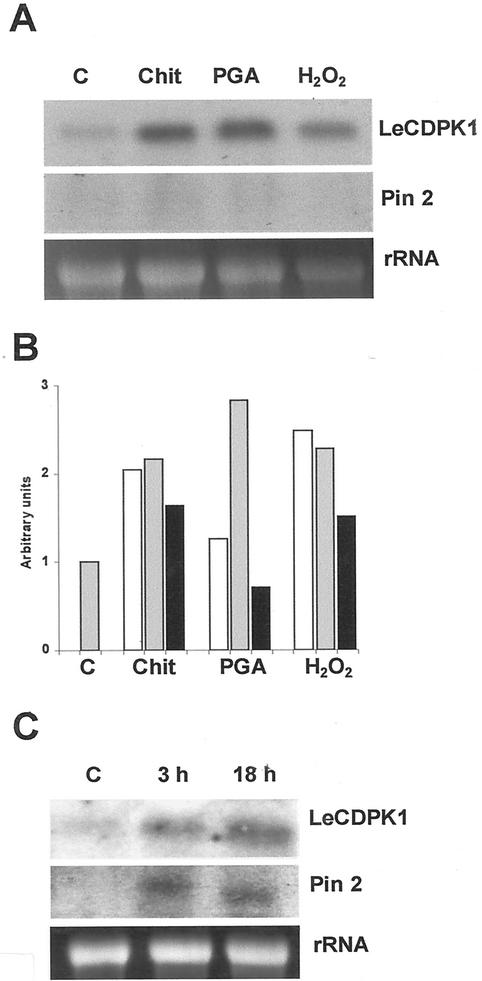
Detached leaflets were transferred to Murashige and Skoog medium for 24 h. Then, the different defense signals were added to the media; and 1, 4, or 8 h later, the leaflets were collected for total RNA extraction. In addition, expression analysis of LeCDPK1 was performed using RNA extracted from leaves incubated with conidiospores from the fungus Colletotrichum coccodes for 3 and 18 h. The expression of pin2 was analyzed in all cases as a pathogen-inducible control.
Chitosan, derived from pathogen cell walls, rapidly enhanced the transcription of LeCDPK1 mRNA, which remained high between 1 and 4 h and slowly declined thereafter. PGA, derived from the plant cell walls, was also able to enhance the transcription of LeCDPK1. Transcript levels reached a maximum at 4 h and rapidly declined to basal levels at 8 h. In the times analyzed, the elicitors were unable to induce the pin2 mRNA (Fig. 5A).
Figure 5.
Northern-blot analysis of LeCDPK1 and pin2 expression in leaves subjected to different treatments. A, Leaves were treated with 100 μg mL−1 chitosan (Chit), 50 μg mL−1 PGA, or 4 mm H2O2 for 4 h. Total RNA (10 μg in each lane) was isolated and hybridized. B, The accumulation of LeCDPK1 mRNA transcripts in control plants (C) and in response to chitosan (Chit), PGA, and H2O2 after 1 h (white bars), 4 h (gray bars), and 8 h (black bars) is plotted in the figure. C, Detached leaves were transferred to a conidial suspension of C. coccodes (6 × 104 conidia mL−1). Total RNA (15 μg in each lane) was extracted after 3 and 18 h, and membranes were hybridized with the full-length LeCDPK1 or with the pin2 probes. Equal RNA loading was checked with the EtBr staining.
An increase of basal levels of LeCDPK1 mRNA could be observed when leaves were incubated with a conidial suspension (6 × 104 conidia mL−1) from the fungus C. coccodes, which causes anthracnose of tomato. As shown in Figure 5C, LeCDPK1 and pin2 transcripts accumulated at 3 h and remained high for at least 18 h. Lazarovits et al. (1979) reported that germinating conidia from C. fulvum can produce non-specific elicitors, mostly glycopeptides, probably of cell wall origin, which are released in culture. Presumably, C. coccodes germinating conidia (20% of the conidial suspension as observed under light microscopy) also release non-specific elicitors to the media. This result and those obtained with chitosan and PGA suggest that the induction of LeCDPK1 is an early event in the signaling cascade triggered in response to pathogen elicitors.
The addition of H2O2 to the excised leaves enhanced the expression of LeCDPK1 rapidly (1 h) and the transcript levels remained high for at least 4 h and slowly declined thereafter (Fig. 5B). No induction of pin2 mRNA was detected at the times analyzed (Fig. 5A). In contrast, the steady-state levels of LeCDPK1 mRNA were only slightly enhanced at 1 h in leaves treated with JA and declined at 4 h, whereas pin2 mRNA was strongly induced at 4 h as reported in the literature (data not shown).
LeCDPK1 Is Systemically Induced upon Wounding
In response to herbivory or pathogen attack, tomato plants activate a signal transduction cascade that leads to the synthesis of more than 15 proteins, including the well-characterized proteinase inhibitors. Most of the genes involved in the plant defense mechanism are also induced in response to wounding, so we analyzed the expression of LeCDPK1 upon injury using a whole-plant system.
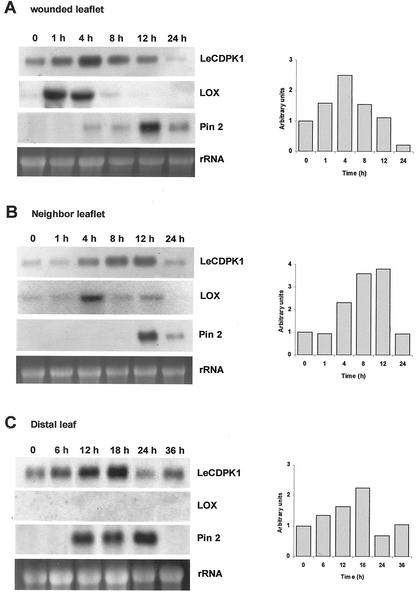
Tomato plants were subjected to mechanical wounding as described in experimental procedures. Total RNA was extracted from the directly wounded or from unwounded neighbor leaflets at 1, 2, 4, 8, 12, and 24 h after the injury. Noninjured plants were used as controls. Membranes were probed with LeCDPK1, TomLoxD, and pin2 clones.
The expression of LeCDPK1 was enhanced in a rapid and transient manner in directly wounded leaves peaking within 4 h after the damage, the mRNA remained high for 4 more h and declined thereafter (Fig. 6A). If a second cut was performed to a second leaflet 20 h after the first injury and total RNA was extracted 4 h later from the directly wounded leaves, the expression of LeCDPK1 was again enhanced to maximum levels (data not shown).
Figure 6.
Time course induction of LeCDPK1 and pin2 transcripts in response to wounding. Total RNA was extracted from wounded leaflets (A) non-wounded neighbor leaflets (B) or distal leaves (C) at the times indicated in the figure. Fifteen micrograms of total RNA was loaded in each lane and hybridized with radioactive LeCDPK1, TomLoxD (LOX), and pin2 probes. Equal RNA loading was checked with the EtBr staining. Relative expression levels of LeCDPK1 are plotted on the right.
In non-wounded neighbor leaflets, the mRNA for LeCDPK1 followed the same pattern of expression as in directly wounded leaves but with a temporal 4-h delay relative to the wounded leaflets. LeCDPK1 transcripts accumulated in neighbor leaflets between 4 and 12 h post-injury peaking at 12 h, followed by decay to control levels afterward (Fig. 6B).
Both membranes were stripped and probed first with a TomLoxD clone that encodes a chloroplast lipoxygenase up-regulated in tomato leaves in response to wounding (Heitz et al., 1997) and then with a pin2 clone used as marker for the wound response. The expression of TomLoxD was rapidly enhanced 1 h after wounding and remained high in wounded leaflets until 4 h post-injury (Fig. 6A); whereas in non-wounded neighbor leaflets the increase was observed only at 4 h (Fig. 6B). This is in agreement with data reported in the literature (Bell and Mullet, 1993; Heitz et al., 1997). Pin2 expression peaked at 12 h in both wounded and neighbor leaflets (Fig. 6, A and B).
To analyze the expression of LeCDPK1 in distal leaves, tomato plants with two compound leaves were used for the experiment. The terminal leaflet of the lower leaf was wounded, and 6, 12, 18, 24, and 36 h afterward the non-wounded upper leaf was collected. LeCDPK1 transcripts accumulated in distal leaves between 6 and 18 h after wounding reaching the highest levels at 18 h post-wounding (Fig. 6C). The decline to basal transcript levels was reached at 24 h. The same membrane was probed with the TomLoxD and pin2 clones. The systemic induction of the proteinase inhibitor (Green and Ryan, 1972) was observed at 12 h. No expression of TomLoxD was detected in distal leaves at the times analyzed (Fig. 6C) however, a systemic wound response has been reported for a lipoxygenase from soybean leaves (Saravitz and Siedow, 1996).
This is the first report of a systemic induction of a CDPK gene in response to wounding. Our results indicate that the transcriptional enhancement of LeCDPK1 observed locally, in the wounded leaflet, also occurs systemically, in distal leaves. However, it is interesting to notice that the delay in the induction of the kinase's mRNA in neighbor leaflets or distal leaves is proportional to the distance from the wounding site. Clearly, LeCDPK1 follows a different expression pattern than pin2, it is rapidly induced at the site of the injury and appears later in distal leaves. When comparing LeCDPK1 expression in the wounded or neighbor leaflets with that of TomLoxD, it can be observed that the kinase is up-regulated later than the lipoxygenase, and its induction lasts longer.
A CDPK Activity Is Induced upon Wounding
It was interesting to determine whether the induction observed in LeCDPK1 mRNA in response to wounding was paralleled by an increase in CDPK activity. Tomato plants were subjected to mechanical wounding as described, and soluble and particulate extracts were obtained from the directly wounded leaflets 20 min and 4.5, 6, 12, and 18.5 h after the injury. CDPK activity was also determined in distal leaves at 12 and 18.5 h after wounding. Noninjured plants were used as controls.
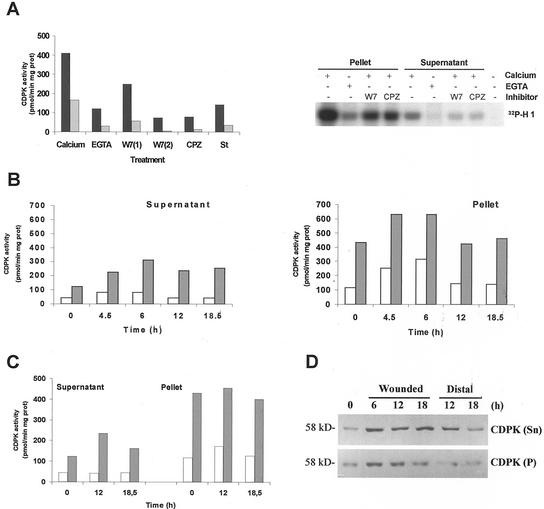
CDPK activity was detected in soluble and particulate fractions from leaves of control plants using syntide-2 or histone H1 as phosphate acceptors. Both enzymatic activities were clearly dependent on the presence of calcium. As shown in Figure 7A, the addition of CPZ or W7 reverted the 4-fold activation observed with calcium, whereas staurosporine also inhibited the kinase's activity. The particulate enzyme shows a higher specific activity when compared with the soluble one, but it corresponds to less than 7% of the total CDPK activity detected (Table I). The behavior of the CDPK activities detected in soluble or particulate extracts was similar to that of the in vitro translated LeCDPK1 (Fig. 2, C and D).
Figure 7.
A, CDPK activity was determined in soluble (gray bars) and particulate (black bars) fractions. A standard assay was performed using syntide-2 as substrate in the presence of 1 mm EGTA or 1 mm CaCl2. CPZ (0.5 mm), 0.25 (1) or 1 mm (2) W7, and 1 μm staurosporine (St) were tested in the presence of 1 mm CaCl2. Alternatively, extracts were incubated with 0.1 mg mL−1 histone H1 and 10 μm [γ-32P]ATP in the presence of 1 mm EGTA or 1 mm CaCl2. The same reaction was carried out in the presence of CPZ, W7, or staurosporine. Phosphorylated samples were analyzed on 12% (w/v) SDS-PAGE. Specific CDPK activity was expressed as picomoles of 32P incorporated per minute per milligram of protein. Time course induction of CDPK activity in response to wounding. Soluble (Supernatant) and particulate (Pellet) fractions were obtained from control and wounded leaflets (B) or distal leaves (C) at the times indicated in the figure. CDPK activity was assayed using syntide-2 as substrate in the presence of 1 mm EGTA (white bars) or 1 mm CaCl2 (gray bars). Specific CDPK activity was expressed as above. D, Western-blot analysis of soluble (50 μg) and particulate (100 μg) protein extracts from control, wounded, or distal leaves. Membranes were incubated with a polyclonal antibody (1:4,000) against the CLD domain of soybean CDPK.
Table I.
CDPK activity in supernatant and pellet fractions
| Time after Wounding | Supernatant
|
Induction* | Pellet
|
Induction* | |||
|---|---|---|---|---|---|---|---|
| Specific activity | %Total activity | Specific activity | %Total activity | ||||
| Wounded leaf | 0 | 79 | 93.6 | 1.00 | 314 | 6.4 | 1.00 |
| 20 min | 98 | 87.4 | 1.24 | 445 | 12.6 | 1.42 | |
| 4.5 h | 143 | 95.7 | 1.81 | 376 | 4.3 | 1.20 | |
| 6 h | 232 | 96 | 2.94 | 312 | 4 | 1.01 | |
| 12 h | 192 | 96.2 | 2.43 | 278 | 3.8 | 0.89 | |
| 18.5 h | 212 | 96.6 | 2.68 | 320 | 3.4 | 1.02 | |
| Distal leaf | 0 | 79 | 93.6 | 1.00 | 314 | 6.4 | 1.00 |
| 12 h | 192 | 96.2 | 2.43 | 283 | 3.8 | 0.90 | |
| 18.5 h | 117 | 96.2 | 1.48 | 274 | 3.8 | 0.87 | |
Soluble (Supernatant) and particulate (Pellet) fractions were obtained from wounded leaflets or distal leaves at the times indicated in the Table. CDPK activity was assayed using syntide-2 as substrate. Specific CDPK activity, expressed as pmol of 32P incorporated min−1 mg−1 protein, was the difference between the activity detected in the presence of 1 mM CaCl2 and 1 mM EGTA. Total CDPK activity was calculated in each sample and relative soluble and particulate CDPK activity was expressed as %Total Activity. Induction was calculated for soluble and particulate fractions as the ratio between CDPK activity of each sample and control (time after wounding = 0 min). The values reported are of one experiment representative of three independent ones.
Considering the pattern of LeCDPK1 expression upon wounding, we analyzed CDPK activity on injured leaves at the moment the mRNA level was highest (4.5 and 6 h) and once the mRNA had begun to decline (12 and 18.5 h). The soluble CDPK activity increased almost three times in response to wounding reaching a maximum at 6 h and remained high for at least 18.5 h (Fig. 7B; Table I). In contrast, the membrane-associated CDPK activity remained relatively constant; the enhancement observed at 4.5 and 6 h was not calcium dependent because a 3-fold increase of membrane associated protein kinase activity was also detected in the presence of EGTA (Fig. 7B). This increase returned to control levels 12 h after wounding.
To analyze the systemic response, CDPK activity was determined in distal leaves of injured plants at 12 and 18.5 h after wounding. A soluble CDPK activity increased 2.4-fold over the control at 12 h after wounding, whereas the particulate activity remained constant (Fig. 7C; Table I). At 18.5 h the soluble CDPK activity was still higher than control levels.
Soluble and particulate protein fractions of control, directly wounded, or distal leaves were analyzed by western blot using a polyclonal antibody against the CLD domain of soybean CDPK (Bachmann et al., 1996). A band of 58 kD that corresponds to the expected molecular mass of LeCDPK1 was revealed in both the soluble and particulate fractions of control leaves (Fig. 7D). The amount of soluble protein was considerably higher in injured leaves 6 h after wounding and remained high for at least 18 h. Moreover, when extracts from distal leaves were analyzed, an increase of the soluble CDPK protein was observed at 12 h declining at 18 h. These results suggest that de novo synthesis of a soluble CDPK occurs, both in the directly wounded and in distal leaves, in response to mechanical wounding.
When the particulate fractions were analyzed by western blot, the band corresponding to CDPK increased at 6 h post-injury in wounded leaves but no increase was observed in distal leaves. It can be noticed that CDPK activity determined in the particulate fraction at 6 h post-injury (Fig. 7B) does not reflect the increase of CDPK detected by western blot. It is then possible to suggest that others proteins present in this fraction might modulate CDPK activity.
To analyze if pre-existing CDPK could also play a role in an earlier step of the wound response, CDPK activity was determined in wounded leaflets collected 20 min post-injury. As shown in Table I, the membrane-associated CDPK activity increased 42% compared with that of control leaves, whereas the soluble one remained relatively constant. The ratio of membrane bound CDPK activity relative to total CDPK activity increased 2-fold (from 6.4% to 12.6%) 20 min post-injury (Table I). This is in agreement with data reported by Romeis et al. (2000) who identified a membrane-bound CDPK that was rapidly activated when transgenic Cf9 tobacco cell cultures were challenged with Avr9 elicitors.
DISCUSSION
In this paper we report the cloning of LeCDPK1, a CDPK from tomato plants that is up-regulated in response to wounding and in the presence of elicitors. LeCDPK1 is transcriptionally enhanced in a transient way both locally at the site of the injury and systemically in distal, non-wounded leaves.
LeCDPK1 encodes an active CDPK that presents features characteristic of this family of protein kinases. The enzyme's activity is dependent on the presence of free calcium; it can phosphorylate substrates such as histone H1 or syntide-2, and it can bind to hydrophobic matrixes such as Phenyl Sepharose in a calcium-dependent manner (Roberts and Harmon, 1992). In addition, calmodulin antagonists such as W7 and CPZ, that inhibit the CLD of CDPKs, and the kinase inhibitor staurosporine, were able to inhibit phosphorylation by LeCDPK1.
The predicted sequence of LeCDPK1 shares a high degree of similarity with the conserved domains of other CDPKs cloned so far. The N-terminal variable region only shares some homology with the N-terminal region of a potato CDPK induced transiently during tuber development (Raíces et al., 2001) and with a stress responsive CDPK from M. crystallinum induced in response to drought and salinity (Patharkar and Cushman, 2000). LeCDPK1 as well as these CDPKs seems to be involved in signaling pathways triggered in response to different environmental stresses.
Southern analysis suggests that LeCDPK1 is a single copy gene. However, this should be taken with caution considering the numerous isoforms of CDPKs present in the Arabidopsis genome and in other plant species. The expression of LeCDPK1 was analyzed in the different tissues of tomato plants, and though basal levels of the transcript could be observed in all tissues, it was more abundant in leaves and flowers.
The Cf9 R gene from tomato confers resistance to the fungus C. fulvum expressing the corresponding Avr protein. LeCDPK1 was purified from a cDNA library prepared with RNA obtained from tomato cell cultures exposed to C. fulvum elicitor preparations. As most of the genes involved in the plant's defense mechanisms against pathogen invasion are also activated upon wounding, it was interesting to analyze the expression of LeCDPK1 in whole tomato plants subjected to mechanical injury and in excised tomato leaves treated with different defense signals. A transient induction of LeCDPK1 mRNA was observed in excised leaves treated with different elicitors or with H2O2.
The analysis of LeCDPK1 mRNA, in whole plants subjected to wounding, showed that the kinase had a maximum expression at 4 h in injured leaves but its mRNA appeared later in neighboring leaflets (8–12 h) or in distal leaves (18 h). In contrast, TomLoxD was expressed earlier and only in wounded and neighbor leaflets, whereas pin2 was transcribed almost simultaneously (12 h) at all sites. According to Moura et al. (2001), there are two classes of wound-inducible genes that are differentially regulated in a temporal manner. Tomato LeCDPK1 could be a member of the “late wound-inducible genes” whose mRNAs increase 4 to 12 h after wounding, in contrast to several “early wound-inducible genes” that are transiently induced within 30 min. TomLoxD belongs to this latter class (Heitz et al., 1997).
One of the central issues of the wound response is the probability of many signals emanating from the wound challenge, changes in the injury site will take place sequentially leading to the likely release of molecular species capable of acting as signals over an extended time course (Bowles, 1998). It has been established by numerous reports that JA is rapidly biosynthesized from α-linolenic acid in plants upon contact with pathogens or wounding and triggers gene activation, leading to the synthesis of defensive secondary metabolites and proteins. Elicitors of the wound response, such as oligosaccharides and systemin, trigger the synthesis of lipid-derived second messengers via the octadecanoid pathway (Farmer and Ryan, 1992). A transient increase in JA levels, with a maximum 1 h after wounding, was reported in tomato excised leaves (Conconi et al., 1996; Parchmann et al., 1997). The local accumulation of LeCDPK1 transcripts occurred within the first 4 h after wounding, after a temporal pattern that could correlate with the rise in JA reported by other authors in tomato wounded leaves. Moreover, when TomLoxD expression was analyzed in wounded and neighbor leaflets, its induction preceded that of the kinase suggesting that the expression of LeCDPK1 might require of the presence of wound-induced JA. However, when excised leaves were treated with JA, only a slight increase in the steady-state levels of LeCDPK1 mRNA was observed (data not shown) indicating that although the timing suggests a causal relationship, JA itself is not the triggering signal of LeCDPK1 induction.
H2O2 directly regulates the expression of numerous genes, some of which are involved in plant defense and in the hypersensitive response that is accompanied by the development of systemic acquired resistance (Levine et al., 1994; Korsmeyer et al., 1995; Alvarez et al., 1998; Desikan et al., 2000; Kovtun et al., 2000). The finding that H2O2 is produced by cell wall-derived OGAs and acts as a second messenger for the activation of defense genes (Orozco-Cárdenas et al., 2001) brought a new perspective of the temporal, spatial, and functional relationships among systemic wound signals, which include systemin, JA, oligosaccharides, and H2O2. Reactive oxygen species, generated near the walls of vascular bundle cells by OGAs, result in the accumulation of H2O2 within 4 h after wounding, but not earlier (Orozco-Cárdenas et al., 2001). A substantial increase of LeCDPK1 mRNA in response to H2O2 was observed in excised treated leaves suggesting that this diffusible signal might modulate the kinase at the transcription level. In addition, the lag (4 h) observed between the induction of LeCDPK1 mRNA at the site of the injury and in a neighbor leaflet correlates with the time elapsed until H2O2 is reported to be detected after wounding. Though tentative, it can be suggested that H2O2 could be involved in the systemic induction of LeCDPK1 in planta.
Chitosan and PGA are currently used to elicit a plant defense response and were capable of enhancing a rapid and transient 2- or 3-fold accumulation of LeCDPK1 transcripts. Both pectic fragments are effective signals in eliciting defense gene induction in tissues adjacent to sites of pathogen attack (Darvill and Albersheim, 1984). Several years ago, OGAs were found to signal the induction of proteinase inhibitors (Ryan, 1988), however, their inability to be transported away from wound sites suggested that they were not involved in the long-distance induction of genes. At that time, OGAs were thought to be generated only by pathogen-derived pectin-degrading enzymes, because no polygalacturonase activities had been reported in leaves. However, Bergey et al. (1999) reported that herbivore attacks can produce OGAs, elicitors synthesized by an endogenous polygalacturonase, that are induced locally and systemically in tomato leaves in response to wounding. These molecules could be involved in the initial steps of LeCDPK1 induction in local and distant leaves.
Another possibility concerning the systemic induction of LeCDPK1 should be considered. There is a clear correlation between the time of induction of LeCDPK1 mRNA in distant tissues and the distance to the site of the injury that may suggest a direct delivery of presynthesized LeCDPK1 mRNA via the phloem. Phloem has recently been considered as a long distance signal pathway used by higher plants to integrate developmental and physiological processes on a whole-plant basis. The discovery of endogenous plant RNA molecules in the phloem (Sasaki et al., 1998; Ruiz-Medrano et al., 1999; Xoconostle-Cázares et al., 1999) suggests that mRNAs may be transported between cells as components of a systemic signaling system. Among the RNA molecules detected in rice phloem sap, a sequence identical to one of a known rice CDPK was found (Hanaoka et al., 1999). This data allows us to speculate on the systemic traffic of LeCDPK1 mRNA upon wounding, even though more experimental data are required to support this hypothesis.
Protein phosphorylation and dephosphorylation are essential events along the signaling pathways that lead to plant defense responses (for review, see León et al., 2001). Alkalinization of the growth medium has been described as an early response of suspension-cultured cells to a variety of elicitor preparations and to systemin (Felix et al., 1993; Schaller and Oecking, 1999). Previous reports establish a correlation between the alkalinization, which was found to depend on the influx of Ca2+, and changes in the pattern of protein phosphorylation. Schaller and Oecking (1999) propose that systemin exerts its effect via Ca2+-dependent protein phosphorylation and suggest the plasma membrane H+-ATPase as a possible target of a Ca2+-activated protein kinase. In a recent work, Romeis et al. (2000) identified a membrane-bound CDPK that was activated in a gene-to-gene-dependent manner in transgenic Cf9 tobacco cell cultures challenged with Avr9 elicitors. High CDPK activity was maintained between 10 and 60 min after elicitation suggesting that a CDPK participated in the Avr9/Cf9-mediated signaling to activate the plant's defense.
A CDPK activity with biochemical properties characteristic of this family was determined in soluble and particulate extracts of tomato leaves. An antibody against the soybean CDPK was able to recognize, both in soluble and particulate extracts of tomato leaves, a polypeptide of 58 kD that corresponds to the expected molecular mass of LeCDPK1. The 2.5-fold increase in LeCDPK1 mRNA observed after mechanical wounding in the wounded leaflet or in distal leaves was paralleled by a 2.4- to 2.9-fold increase in a soluble CDPK activity. This increase in activity correlates with an increase in the amount of soluble protein. However, a slighter increase (42%) in a membrane associated-CDPK activity could be observed 20 min after wounding. It might be suggested that this particulate activity could be involved in an early response to wounding, similar to the one detected by Romeis et al. (2000) in elicited tobacco cells.
The correlation between the induction of LeCDPK1 mRNA, the increase in soluble protein according to the western-blot analysis, and the increase in CDPK activity, together with similarities in timing between these increases, suggest that de novo synthesis of the enzyme occurs in response to wounding. However, the rapid increase in membrane-associated CDPK activity, possibly involved in early signaling events could be due to an activation of pre-existing protein by post-translational modifications rather than de novo transcription. At present we cannot establish whether both soluble and particulate CDPK activities are encoded by LeCDPK1 and their localization depends on the post-translational modifications or whether they are isoforms encoded by different mRNAs. Nevertheless, it is tempting to speculate a dynamic control of the localization of LeCDPK1 by palmitoylation.
Much of the recent progress in understanding the molecular basis of plant disease resistance has come from using cultured plant cells as model systems. The up-regulation of LeCDPK1 mRNA, the increase of a soluble CDPK protein and the enhancement of its activity in planta upon wounding, together with the messenger's induction in detached leaves in response to pathogen elicitors, strengthens the hypothesis that this kinase could be part of physiological plant defense mechanisms against biotic or abiotic attacks.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Tomato (Lycopersicon esculentum VF36) plants were cultivated in a greenhouse under a regime of 16 h of light (25°C) and 8 h of dark (20°C). All experiments were carried out with 4- to 5-week-old plants.
Isolation of cDNA Clones for LeCDPK1
Primers homologous to a potato CDPK (5′-GCCAAGGATGCTCAAGAACTT-3′ and 5′-GATTTTGGGCTGTCA/CG/ATA/GTTCATT-3′) were used to amplify a 340-bp fragment in a PCR reaction (35 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min). The amplified fragment was sequenced using fmol DNA Sequencing System (Promega, Madison, WI). The partial CDPK clone was labeled with [γ-32P]dCTP (109 cpm pmol−1) using the RadPrime DNA labeling system kit (Gibco-BRL, Cleveland) and used to probe a cDNA library from tomato (L. esculentum cv Moneymaker) that had been cloned 5′ EcoRI-3′ XhoI in the phagemid vector pBK-CMV. The library, a gift from Dr. Eduardo Blumwald (University of California, Davis), was made from mRNA obtained from cell cultures that had been exposed to Cladosporium fulvum (race 2.3) elicitor preparations for 1.5 h. Hybridization (Church and Gilbert, 1984) and sequential stringent washes were performed at 65°C. A CDPK clone designated LeCDPK1 was isolated. The plasmids, excised in vivo from hybridizing phages into E. coli JM109 strain using helper phage R408, were purified using Qiagen Plasmid Midi Kit. Automated sequencing was performed by BioResource Center, DNA Sequencing Facility, Cornell University (Ithaca, NY). The nucleotide sequence data reported (LeCDPK1) appears in the GenBank Database under the accession number AF363784.
In Vitro Translation Assays
LeCDPK1 was amplified using primers from the 5′- and 3′-untranslated region, and the product was cloned in a pGEMT-easy plasmid, such that the production of a sense transcript was under the control of T7 promoter. Transcription was carried out using the TNT Coupled Transcription/Translation Wheat Germ Extract System (Promega). Products were radiolabeled with [35S]Met (1,000 Ci mmol−1 at 10 mCi mL−1) from New England Nuclear (Boston) and analyzed by 12% (w/v) SDS-PAGE according to Laemmli, as described in MacIntosh et al. (1996). Gels were fixed, fluorographed (Bonner and Laskey, 1974), dried, and exposed. In parallel, a luciferase T7 control DNA, was used as positive control.
Purification of the in Vitro-Translated LeCDPK1
In vitro translation of LeCDPK1 was carried out as described. Translated LeCDPK1 was purified using a Phenyl Sepharose column equilibrated with 10 mm Tris-HCl, pH 7.4, and 0.5 mm CaCl2. Elution was performed stepwise using 0.3 m NaCl, 0.4 m NaCl plus 5 mm EGTA, and 4 m urea, in 10 mm Tris-HCl, pH 7.4. A mock in vitro translation reaction carried out in the absence of pGEMT-easy LeCDPK1 was purified in parallel. All fractions were concentrated and analyzed on 12% (w/v) SDS-polyacrylamide gels. In vitro translation and purification of LeCDPK1 was carried out as described but in the absence of labeled Met. CDPK activity was determined of the urea eluted fraction. A negative control was carried out using the purified wheat germ extract.
DNA Isolation and Southern-Blot Hybridization
Twelve micrograms of genomic DNA from tomato leaves (Murray and Thompson, 1980) were digested with restriction enzymes (EcoRI, EcoRV, and HindIII) (New England Biolabs, Beverly, MA), separated on 0.7% (w/v) agarose gels, and blotted onto a nylon membrane (Hybond N+, Amersham, Buckinghamshire, UK). Membranes hybridized with [32P]LeCDPK1 at 65°C were exposed after sequential stringent washes.
Treatments with Different Defense Signals
Detached leaflets (3–5 per treatment) with the corresponding petiole, were transferred to liquid Murashige and Skoog medium. After 24 h when LeCDPK1 mRNA was restored to basal levels, the following defense signals were added: 100 μg mL−1 of chitosan prepared in 85% (w/w) orto-phosphoric acid, 50 μg mL−1 of PGA, 10 or 50 μm JA prepared in dimethylformamide, or 4 mm H2O2. Chitosan, PGA, and JA were from Sigma (St. Louis). Leaflets were frozen in liquid nitrogen and total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen USA, Valencia, CA).
Treatment with the Fungus Colletotrichum coccodes
Spores of C. coccodes were cultured in yeast peptone dextrose medium in the presence of 0.3 μg mL−1 ampicillin during 2 weeks at 25°C (16-h photoperiod). Conidia from the grown fungus were transferred to liquid Murashige and Skoog medium for 24 h. Detached tomato leaves were first transferred to liquid Murashige and Skoog medium for 24 h and then to the conidial suspension in Murashige and Skoog medium (6 × 104 conidia mL−1 counted under light microscopy). Leaves were collected 3 or 18 h later and total RNA was extracted. During the experiments, leaves were kept in covered glass flasks to maintain conditions of high humidity. A control in Murashige and Skoog medium was done in parallel.
Wounding Treatments
Mechanical wounding was performed according to Carrera and Prat (1998) with modifications. The main veins of apical or subapical leaflets of compound leaves were cut with a dented forceps, and the directly wounded, the non-wounded neighbor leaflets, or the distal leaves were collected at the times indicated in the figures and immediately frozen in liquid nitrogen. Leaves from non-wounded plants were used as control. For each point, two tomato plants were used.
RNA Isolation and Northern-Blot Hybridization
Total RNA was isolated from the different plant tissues (roots, stems, leaves, and flowers) and from leaves subjected to wounding experiments or exposed to the different defense signals. Samples (0.1–1 g) were collected and ground in liquid nitrogen, and total RNA was extracted using the TRIzol Reagent (Gibco-BRL) or the RNeasy Plant Mini Kit (Qiagen USA). Total RNA (10–20 μg) was separated on 1.4% (w/v) formaldehyde agarose gels and blotted onto nylon membranes (Hybond N+, Amersham). Northern blots (Alwine et al., 1977) were hybridized with LeCDPK1, TomLoxD, and a proteinase inhibitor 2 (pin2) probes labeled with RadPrime DNA labeling system kit (Gibco-BRL). After sequential stringent washes at 65°C, blots were exposed. Equal RNA loading was checked in the EtBr staining of the electrophoresis. LeCDPK1 mRNA was quantified relative to the RNA loading using Scion Image software.
Preparation of Plant Extracts
Control, wounded, or distal leaves from tomato plants were harvested at the times indicated in the figure, rinsed with distilled water, ground in a mortar cooled with liquid nitrogen, and extracted with 50 mm Tris-HCl, pH 7.5, containing 2 mm β-mercaptoethanol, 1 mm EDTA, 1 mm EGTA, 20% (v/v) glycerol, 20 mm β-glycerophosphate, 1 mm Na3VO4, and protease inhibitors (0.5 mm phenylmethylsulfonyl fluoride, 1 mm benzamidine, 2 μg mL−1 soybean trypsin inhibitor, and 25 units mL−1 aprotinin). The suspensions (1 mL buffer g−1 wet tissue) were centrifuged 10 min at 1,000g (twice), and the pellet was discarded. The supernatants were centrifuged 1 h at 100,000g and the resulting supernatant and pellet fractions were analyzed.
Protein Kinase Activity Assays
CDPK activity was determined in aliquots of soluble and particulate fractions or in the in vitro translated LeCDPK1 according to MacIntosh et al. (1996) using syntide-2 as substrate with the addition of 1 mm EGTA or 1 mm CaCl2. Assays were performed at 30°C for 10 min. Specific CDPK activity of the different extracts was expressed as picomoles of 32P incorporated per minute per milligram of protein.
Alternatively, fractions were incubated 5 min at 30°C with 0.1 mg mL−1 histone H1 and 10 μm [γ-32P]ATP (specific activity 500 cpm pmol−1) in the presence of 1 mm EGTA or 1 mm CaCl2. Reactions were stopped with the addition of cracking buffer and analyzed on 12% (w/v) SDS-PAGE. Assays were also performed in the presence of 0.5 mm chloropromazine, 0.25 and 1 mm W7, or 1 μm staurosporine using both phosphate acceptors.
Western-Blot Analysis
Soluble (50 μg) and particulate (100 μg) protein extracts were resolved in 12% (w/v) SDS-PAGE and blotted onto nitrocellulose membranes. Blots were incubated overnight at 4°C in blocking solution and then 2 h with affinity purified polyclonal antibodies (1:4,000) directed against the CLD of soybean αCDPK (Bachmann et al., 1996) at room temperature. The blot was developed with Renaissance, Western-Blot Chemiluminescence reagent from NEN according to the manufacturer's procedure.
ACKNOWLEDGMENTS
We thank Dr. Eduardo Blumwald for kindly providing the cDNA library from tomato cell cultures and Dr. Salomé Prat Monguio for the pin2 clone. We thank Dr. Alice Harmon for the antibody against the soybean CDPK. We are grateful to Dr. Verna Higgings for helpful discussion of our results with C. coccodes and for providing the spores.
Footnotes
This work was supported by grants from Agencia de Promoción Científica y Tecnológica, Consejo Nacional de Investigaciones Científicas y Técnicas, and the University of Buenos Aires, Argentina.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010649.
LITERATURE CITED
- Alvarez ME, Pennell RI, Meijer PJ, Ishikawa A, Dixon RA, Lamb C. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell. 1998;92:773–784. doi: 10.1016/s0092-8674(00)81405-1. [DOI] [PubMed] [Google Scholar]
- Alwine JC, Kemp DJ, Stark GR. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci USA. 1977;74:5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann M, Shiraishi N, Campbell WH, Yoo BC, Harmon AC, Huber SC. Identification of Ser-543 as the major regulatory phosphorylation site in spinach leaf nitrate reductase. Plant Cell. 1996;8:505–517. doi: 10.1105/tpc.8.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell E, Mullet JE. Characterization of an Arabidopsis lipoxygenase gene responsive to methyl jasmonate and wounding. Plant Physiol. 1993;103:1133–1137. doi: 10.1104/pp.103.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergey DR, Orozco-Cárdenas M, de Moura DS, Ryan CA. A wound- and systemin-inducible polygalacturonase in tomato. Proc Natl Acad Sci USA. 1999;96:1756–1760. doi: 10.1073/pnas.96.4.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume B, Nurnberger T, Nass N, Scheel D. Receptor-mediated increase in cytoplasmic free calcium required for activation of pathogen defense in parsley. Plant Cell. 2000;12:1425–1440. doi: 10.1105/tpc.12.8.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner WM, Laskey RA. A film detection method for tritium-labeled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974;1:83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Bowles DJ. Defense-related proteins in higher plants. Annu Rev Biochem. 1990;59:873–907. doi: 10.1146/annurev.bi.59.070190.004301. [DOI] [PubMed] [Google Scholar]
- Bowles DJ. Signal transduction in the wound response of tomato plants. Philos Trans R Soc Lond B Biol Sci. 1998;353:1495–1510. doi: 10.1098/rstb.1998.0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E, Prat S. Expression of the Arabidopsis abi1-1 mutant allele inhibits proteinase inhibitor wound-induction in tomato. Plant J. 1998;15:765–771. doi: 10.1046/j.1365-313x.1998.00261.x. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conconi A, Smerdon MJ, Howe GA, Ryan CA. The octadecanoid signalling pathway in plants mediates a response to ultraviolet radiation. Nature. 1996;383:826–829. doi: 10.1038/383826a0. [DOI] [PubMed] [Google Scholar]
- Darvill A, Albersheim P. Phytoalexins and their elicitors: a defense against insects and pathogens. Annu Rev Plant Physiol. 1984;53:243–275. [Google Scholar]
- Desikan R, Neill SJ, Hancock JT. Hydrogen peroxide-induced gene expression in Arabidopsis thaliana. Free Radic Biol Med. 2000;28:773–778. doi: 10.1016/s0891-5849(00)00157-x. [DOI] [PubMed] [Google Scholar]
- Ellard-Ivey M, Hopkins RB, White TJ, Lomax TL. Cloning, expression and N-terminal myristoylation of CpCPK1, a calcium-dependent protein kinase from zucchini (Cucurbita pepo L.) Plant Mol Biol. 1999;39:199–208. doi: 10.1023/a:1006125918023. [DOI] [PubMed] [Google Scholar]
- Farmer EE, Ryan CA. Interplant communication: Airborne methyl jasmonate induces synthesis of proteinase inhibitor in plant leaves. Proc Natl Acad Sci USA. 1990;96:12947–12952. doi: 10.1073/pnas.87.19.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Ryan CA. Octadenoic precursors of jasmonic acid activate the synthesis of wound-inducible proteinase inhibitors. Plant Cell. 1992;4:129–134. doi: 10.1105/tpc.4.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Regenass M, Boller T. Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J. 1993;4:307–316. [Google Scholar]
- Flor HH. Current status of the gene-for-gene concept. Annu Rev Phytopathol. 1971;9:275–296. [Google Scholar]
- Green TR, Ryan CA. Wound-induced proteinase inhibitors in plant leaves: a possible defense against insects. Science. 1972;175:776–777. doi: 10.1126/science.175.4023.776. [DOI] [PubMed] [Google Scholar]
- Halterman DA, Martin GB. Signal recognition and transduction involved in plant disease resistance. Essays Biochem. 1997;32:87–99. [PubMed] [Google Scholar]
- Hanaoka H, Fukuda A, Sasaki T, Nemoto K, Fujiwara T, Hayashi H. CDPK in rice phloem sap (abstract no. 408). Interactions and Intersections in Plant Signaling Pathways–Keystone Symposia. 1999. p. 57. [Google Scholar]
- Heitz T, Bergey DR, Ryan CA. A gene encoding a chloroplast-targeted lipoxygenase in tomato leaves is transiently induced by wounding, systemin, and methyl jasmonate. Plant Physiol. 1997;114:1085–1093. doi: 10.1104/pp.114.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herde O, Fuss H, Peña-Cortés H, Fisahn J. Proteinase inhibitor II gene expression induced by electrical stimulation and control of photosynthetic activity in tomato plants. Plant Cell Physiol. 1995;36:737–742. [Google Scholar]
- Hildmann T, Ebneth M, Peña-Cortés H, Sánchez-Serrano JJ, Willmitzer L, Prat S. General roles of abscisic and jasmonic acid in gene activation as a result of mechanical wounding. Plant Cell. 1992;4:1157–1170. doi: 10.1105/tpc.4.9.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabak EM, Dickmann LJ, Satterlee JS, Sussman MR. Characterization of eight new members of the calmodulin-like domain protein kinase gene family from Arabidopsis thaliana. Plant Mol Biol. 1996;31:405–412. doi: 10.1007/BF00021802. [DOI] [PubMed] [Google Scholar]
- Korsmeyer SJ, Yin XM, Oltvai ZN, Veis-Novack DJ, Linette GP. Reactive oxygen species and the regulation of cell death by the Bcl-2 gene family. Biochem Biophys Acta. 1995;1271:63–66. doi: 10.1016/0925-4439(95)00011-r. [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Tena G, Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA. 2000;97:2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarovits G, Bhullar BS, Sugiyama HJ, Higgins VJ. Purification and partial characterization of a glycoprotein toxin produced by Cladosporium fulvum. Phytopathology. 1979;69:1062–1068. [Google Scholar]
- León J, Rojo E, Sánchez-Serrano JJ. Wound signalling in plants. J Exp Bot. 2001;52:1–9. doi: 10.1093/jexbot/52.354.1. [DOI] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- Ligterink W, Kroj T, Zur Nieden U, Hirt H, Scheel D. Receptor-mediated activation of a MAP kinase in pathogen defense of plants. Science. 1997;276:2054–2057. doi: 10.1126/science.276.5321.2054. [DOI] [PubMed] [Google Scholar]
- MacIntosh GC, Ulloa RM, Raíces M, Téllez-Iñón MT. Changes in calcium-dependent protein kinase activity during in vitro tuberization in potato. Plant Physiol. 1996;112:1541–1550. doi: 10.1104/pp.112.4.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD. MAP-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- Martín ML, Busconi L. Membrane localization of a rice calcium-dependent protein kinase (CDPK) is mediated by myristoylation and palmitoylation. Plant J. 2000;24:429–435. doi: 10.1046/j.1365-313x.2000.00889.x. [DOI] [PubMed] [Google Scholar]
- Menke FL, Parchmann S, Mueller MJ, Kijne JW, Memelink J. Involvement of the octadecanoid pathway and protein phosphorylation in fungal elicitor-induced expression of terpenoid indole alkaloid biosynthetic genes in Catharanthus roseus. Plant Physiol. 1999;119:1289–1296. doi: 10.1104/pp.119.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura DS, Bergey DR, Ryan CA. Characterization and localization of a wound-inducible type I serine-carboxypeptidase from leaves of tomato plants (Lycopersicon esculentum Mill.) Planta. 2001;212:222–230. doi: 10.1007/s004250000380. [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Cárdenas M, Narvaez-Vasquez J, Ryan CA. Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. Plant Cell. 2001;13:179–191. [PMC free article] [PubMed] [Google Scholar]
- Orozco-Cárdenas M, Ryan CA. Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc Natl Acad Sci USA. 1999;96:6553–6557. doi: 10.1073/pnas.96.11.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchmann S, Gundlach H, Mueller MJ. Induction of 12-oxo-phytodienoic acid in wounded plants and elicited plant cell cultures. Plant Physiol. 1997;115:1057–1064. doi: 10.1104/pp.115.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patharkar OR, Cushman JC. A stress-induced calcium-dependent protein kinase from Mesembryanthemum crystallinum phosphorylates a two-component pseudo-response regulator. Plant J. 2000;24:679–691. doi: 10.1046/j.1365-313x.2000.00912.x. [DOI] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Jonhnson S, Ryan CA. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science. 1991;253:895–898. doi: 10.1126/science.253.5022.895. [DOI] [PubMed] [Google Scholar]
- Peña-Cortés H, Albrecht T, Prat S, Weiler EW, Willmitzer L. Aspirin prevents wound-induced gene expression in tomato leaves by blocking jasmonic acid biosynthesis. Planta. 1993;191:123–128. [Google Scholar]
- Peña-Cortés H, Sánchez-Serrano JJ, Mertens R, Willmitzer L. Abscisic acid is involved in the wound-induced expression of th proteinase inhibitor II gene in potato and tomato. Proc Natl Acad Sci USA. 1989;86:9851–9855. doi: 10.1073/pnas.86.24.9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raíces M, Chico JM, Téllez-Iñón MT, Ulloa RM. Molecular characterization of StCDPK1, a calcium-dependent protein kinase from Solanum tuberosum that is induced at the onset of tuber development. Plant Mol Biol. 2001;46:591–601. doi: 10.1023/a:1010661304980. [DOI] [PubMed] [Google Scholar]
- Roberts DM, Harmon AC. Calcium-modulated proteins: targets of intracellular calcium signals in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:375–414. [Google Scholar]
- Rojo E, Titarenko E, León J, Berger S, Vancanneyt G, Sánchez-Serrano JJ. Reversible protein phosphorylation regulates jasmonic acid-dependent and independent wound signal transduction pathways in Arabidopsis thaliana. Plant J. 1998;13:153–165. doi: 10.1046/j.1365-313x.1998.00020.x. [DOI] [PubMed] [Google Scholar]
- Romeis T, Piedras P, Jones JD. Resistance gene-dependent activation of a calcium-dependent protein kinase in the plant defense response. Plant Cell. 2000;12:803–816. doi: 10.1105/tpc.12.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T, Piedras P, Zang S, Klessig DF, Hirt H, Jones JD. Avr9- and Cf-9-dependent activation of MAP kinase in tobacco cell cultures and leaves: convergence of resistance genes, elicitor, wound and salicylate responses. Plant Cell. 1999;11:273–287. doi: 10.1105/tpc.11.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano R, Xoconostle-Cázares B, Lucas WJ. Phloem long-distance transport of CmNACP mRNA: implications for supracellular regulation in plants. Development. 1999;126:4405–4419. doi: 10.1242/dev.126.20.4405. [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE. Transcriptional control of plant genes responsive to pathogens. Curr Opin Plant Biol. 1998;1:311–315. doi: 10.1016/1369-5266(88)80052-9. [DOI] [PubMed] [Google Scholar]
- Ryan CA. Oligosaccharide signalling in plants. Annu Rev Cell Biol. 1987;3:295–317. doi: 10.1146/annurev.cb.03.110187.001455. [DOI] [PubMed] [Google Scholar]
- Ryan CA. Oligosaccharides as recognition signals for the expression of defensive genes in plants. Biochemistry. 1988;27:8879–8883. [Google Scholar]
- Ryan CA. Protease inhibitors in plants: genes for improving defenses against insects and pathogens. Annu Rev Phytopathol. 1990;28:425–449. [Google Scholar]
- Saravitz DM, Siedow JN. The differential expression of wound-inducible lipoxygenase genes in soybean leaves. Plant Physiol. 1996;110:287–299. doi: 10.1104/pp.110.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Chino M, Hayashi H, Fujiwara T. Detection of several mRNA species in rice phloem sap. Plant Cell Physiol. 1998;39:895–897. doi: 10.1093/oxfordjournals.pcp.a029451. [DOI] [PubMed] [Google Scholar]
- Schaller A, Oecking C. Modulation of plasma membrane H+-ATPase activity differentially activates wound and pathogen defense responses in tomato plants. Plant Cell. 1999;11:263–272. doi: 10.1105/tpc.11.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel D. Resistance response physiology and signal transduction. Curr Opin Plant Biol. 1998;1:305–310. doi: 10.1016/1369-5266(88)80051-7. [DOI] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald P. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Sopory SK, Munshi M. Protein kinases and phosphatases and their role in cellular signaling in plants. Crit Rev Plant Sci. 1998;17:245–318. [Google Scholar]
- Staskawicz BJ, Ausubel FM, Baker BJ, Ellis JG, Jones JD. Molecular genetics of plant disease resistance. Science. 1995;268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- Wildon DC, Thain JF, Minchin PEH, Gubb IR, Reilly AJ, Skipper YD, Doherty HM, O'Donnell PJ, Bowles DJ. Electrical signalling and systemic proteinase inhibitor induction in the wounded plant. Nature. 1992;360:62–65. [Google Scholar]
- Xoconostle-Cázares B, Xiang Y, Ruiz-Medrano R, Wang HL, Monzer J, Yoo BC, McFarland KC, Franceschi VR, Lucas WJ. Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science. 1999;283:94–98. doi: 10.1126/science.283.5398.94. [DOI] [PubMed] [Google Scholar]
- Yang Y, Shah J, Klessig DF. Signal perception and transduction in plant defense responses. Genes Dev. 1997;11:1621–1639. doi: 10.1101/gad.11.13.1621. [DOI] [PubMed] [Google Scholar]
- Zhang S, Du H, Klessig DF. Activation of the tobacco SIP kinase by both a cell wall-derived carbohydrate elicitor and purified proteinaceous elicitins from Phytophthora spp. Plant Cell. 1998;10:435–449. doi: 10.1105/tpc.10.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]