Abstract
Inorganic phosphate (Pi)-signaling pathways in plants are still largely unknown. The Arabidopsis (Arabidopsis thaliana) pho2 mutant overaccumulates Pi in leaves in Pi-replete conditions. Micrografting revealed that a pho2 root genotype is sufficient to yield leaf Pi accumulation. In pho2 mutants, Pi does not repress a set of Pi starvation-induced genes, including AtIPS1, AT4, and Pi transporters Pht1;8 and Pht1;9. Map-based cloning identified PHO2 as At2g33770, an unusual E2 conjugase gene. It was recently shown that Pi deprivation induces mature microRNA (miRNA [miR399]) and that overexpression of miR399 in Pi-replete conditions represses E2 conjugase expression and leads to high leaf Pi concentrations, thus phenocopying pho2. We show here that miR399 primary transcripts are also strongly induced by low Pi and rapidly repressed after addition of Pi. PHO2 transcripts change reciprocally to miR399 transcripts in Pi-deprived plants and in miR399 overexpressers. However, responses after Pi readdition and in β-glucuronidase reporter lines suggest that PHO2 expression is also regulated by Pi in a manner unrelated to miR399-mediated transcript cleavage. Expression of miR399 was strongly reduced in Pi-deprived Arabidopsis phr1 mutants, and a subset of Pi-responsive genes repressed in Pi-deprived phr1 mutants was up-regulated in Pi-replete pho2 mutants. This places miR399 and PHO2 in a branch of the Pi-signaling network downstream of PHR1. Finally, putative PHO2 orthologs containing five miR399-binding sites in their 5′-untranslated regions were identified in other higher plants, and Pi-dependent miR399 expression was demonstrated in rice (Oryza sativa), suggesting a conserved regulatory mechanism.
Inorganic phosphate (Pi) is frequently limiting for plant growth (Marschner, 1995). Plants adapt to low Pi in several ways: Root growth and architecture are altered to access a larger soil volume, organic acids, phosphatases, and nucleases are exuded to solubilize or release Pi from organic sources, the capacity for Pi uptake is increased, and internal Pi is recycled (Raghothama, 1999; Abel et al., 2002; Poirier and Bucher, 2002). Although some components of Pi starvation signaling in plants have been identified, the overall pathways are still poorly understood.
Arabidopsis (Arabidopsis thaliana) mutants have been isolated that are impaired in the induction of Pi-responsive genes (pdr2, pho3, and phr1; Chen et al., 2000; Rubio et al., 2001; Zakhleniuk et al., 2001; Ticconi et al., 2004). PHO3 was recently shown to encode the Suc transporter SUC2 (Lloyd and Zakhleniuk, 2004), indicating it acts rather indirectly. PHR1 has homology to PSR1, a MYB-like transcription factor that is involved in Pi responses in Chlamydomonas reinhardtii (Wykoff et al., 1999), and contributes to downstream Pi signaling by regulating the expression of a few Pi starvation-induced genes (Rubio et al., 2001). Genetic evidence and the presence of sumoylation sites in PHR1 indicate that the SUMO E3 ligase SIZ1 acts upstream of PHR1 (Miura et al., 2005). Very recently, it was shown that Pi modulates the level of a microRNA (miRNA [miR399]), which targets an E2 conjugase (Fujii et al., 2005; Chiou et al., 2006).
A further set of Arabidopsis mutants have been identified with altered levels of Pi (pup1, pho1, and pho2; Poirier et al., 1991; Delhaize and Randall, 1995; Trull and Deikman, 1998). PUP1 encodes a phosphatase (Poirier and Bucher, 2002). The pho1 mutant has low shoot, but normal root, Pi levels (Poirier et al., 1991). Molecular cloning identified PHO1 as the founder member of a novel EXS transporter family, indicating a role in xylem loading and possibly also in signaling (Hamburger et al., 2002; Wang et al., 2004). The pho2 mutant has 2- to 4-fold increased levels of Pi in leaves and unaltered levels in the roots (Delhaize and Randall, 1995; Dong et al., 1998). The molecular function of PHO2 was previously unknown.
We show here by map-based cloning that PHO2 encodes the E2 conjugase that is modulated by the Pi-dependent miR399. By exploiting the pho2 mutant, we show that a discrete set of Pi-responsive genes lies downstream of PHO2 and that PHO2 expression in the roots is sufficient to restore wild-type levels of Pi in the shoot. We also provide data to suggest that PHO2 expression is also regulated at a level beyond miR399-mediated transcript decay. We then demonstrate that the phr1 mutant has altered expression of a far wider range of Pi-responsive genes than previously reported and that the low-Pi-dependent increase of miR399 is substantially suppressed in phr1. This defines the miR399/PHO2 pathway as a subcomponent of the Pi-signaling network that operates downstream of PHR1 in Arabidopsis and regulates a subset of Pi-dependent responses, including Pi allocation, between the shoot and the root. Finally, we show that Arabidopsis PHO2 has close homologs in higher plant species and that phosphate-dependent expression of miR399 is conserved in rice (Oryza sativa).
RESULTS
Identification of PHO2
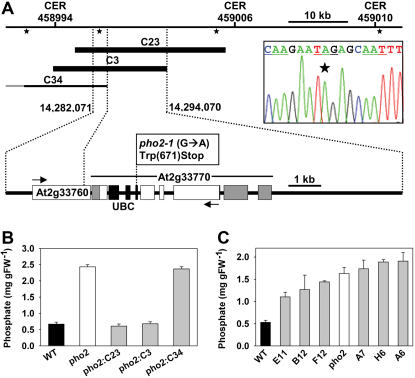
We identified PHO2 by map-based cloning based on the high-Pi phenotype (Supplemental Fig. 1, A and B) as described in “Materials and Methods.” A set of 13 microsatellite markers placed PHO2 in an approximately 50-kb region between CER458994 and CER459010 (Supplemental Fig. 1, A and B). Genomic cosmid clones covering this region (see Fig. 1A) were introduced into pho2 mutants. Transformed pho2 mutants containing clones C23 and C3 had wild-type-like leaf Pi levels (Fig. 1B), indicating these clones contain PHO2. End sequencing showed that clones C23 and C3 contain two annotated genes (At2g33760 and At2g33770), which are absent or truncated in C34. Sequencing of both reported mutant alleles (pho2-1 and pho2-2; Delhaize and Randall, 1995) did not reveal any mutations in At2g33760. Both mutant alleles contained the same G→A mutation in At2g33770, resulting in replacement of W671 by a stop codon (Fig. 1A). A derived cleaved-amplified polymorphic sequence (dCAPS) marker was developed, based on the single base change found in At2g33770 in the pho2 mutants (Supplemental Fig. 1C). Cosegregation of the low-shoot Pi phenotype and a heterozygous dCAPS genotype was found for the pho2 mutants transformed with clones C23 and C3, as expected (data not shown). To provide further and independent confirmation of the identity of PHO2, we produced an RNA interference (RNAi) construct to At2g33770 and introduced it into the wild type. Many of the resulting lines contained pho2-like high leaf Pi (Fig. 1C).
Figure 1.
Identification of PHO2. A, View of the approximately 50-kb mapping interval determined by CER458994 and CER459010 (compare with Supplemental Fig. 1). The positions of hybridization probes for cosmid library screening are shown as asterisks and three clones (C23, C3, and C34) are depicted as black bars, with the complementing clones C23 and C3 in bold. An expanded blow-up shows the structure of the two annotated genes (At2g33760 and At2g33770) within a 12-kb segment harboring PHO2. Arrows indicate the direction of transcription, exons are depicted as white boxes, and UTRs of PHO2 (At2g33770) as gray boxes. The UBC domain of At2g33770 is shaded in black. The point mutation in the pho2-1 and pho2-2 alleles (G→A; electropherogram) causes an early amber stop codon at the beginning of UBC. B, Leaf Pi levels in wild type (black), pho2 mutant (white), and complemented (C23, C3) and noncomplemented (C34) pho2 mutants (gray; mean ± sd; n = 5). C, Leaf Pi levels in wild type (black bar), pho2 mutant (white bar), and wild type transformed with a PHO2 RNAi construct (gray bars; mean ± sd; n = 3).
The PHO2 gene spans approximately 5.5 kb, contains nine exons, and encodes a 4.1-kb transcript (Fig. 1A). It includes a 1.1-kb 5′-untranslated region (UTR) and encodes a predicted 907-amino acid protein. A region toward the C terminus corresponds to the canonical ubiquitin-conjugating (UBC) domain of E2 enzymes (Smalle and Vierstra, 2004). This domain catalyzes the ATP-dependent transfer of a thioester-linked ubiquitin molecule via an active-site Cys residue onto a substrate protein, either directly or via an E3 ligase, targeting it for degradation via the 26S proteosome pathway (Smalle and Vierstra, 2004). The introduced stop codon in the pho2 mutant deletes this UBC domain (Fig. 1A).
Micrografting
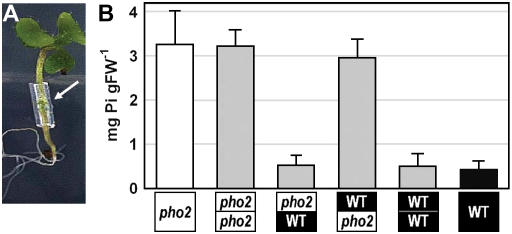
Reciprocal micrografting experiments were performed with wild-type and pho2 mutant seedlings (Fig. 2). High-shoot Pi was found when the roots derived from the pho2 mutant, but not when a pho2 mutant shoot was grafted onto a wild-type root, demonstrating that a pho2 root genotype is sufficient and necessary for Pi accumulation in the shoot. This, and the possibility that high Pi levels in the shoot could lead to secondary changes that might complicate interpretation of the results, led us to concentrate on the roots for molecular characterization of the pho2 mutant.
Figure 2.
Reciprocal micrografting of a pho2 mutant and wild type. A, Aspect of a 14-d-old micrografted Arabidopsis seedling. B, Leaf Pi levels in wild type (black), pho2 mutant (white), and reciprocally grafted chimeric seedlings (gray; mean value ± sd; n = 6).
Molecular Phenotypes of pho2 Mutants
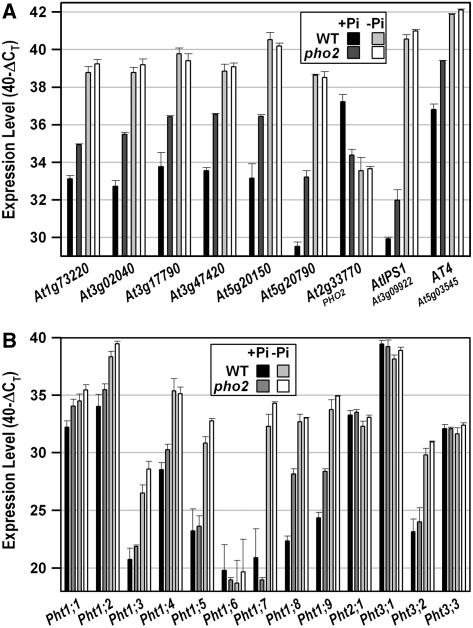
Root material from the wild type and the pho2 mutant grown in +Pi and −Pi conditions was analyzed using ATH1 genechips (Supplemental Table I). Table I shows the 12 genes that reproducibly exhibited >2-fold altered expression in +Pi-grown pho2, compared to +Pi-grown wild type. Remarkably, many of the genes induced in pho2 belong to those that are most strongly induced by Pi deprivation in Arabidopsis wild type (Table I; R. Morcuende, R. Bari, and W.-R. Scheible, unpublished data). These genes are also rapidly repressed after Pi readdition (Table I). The ATH1 results were confirmed for a subset of the genes, using real-time quantitative reverse transcription (qRT)-PCR (Fig. 3A). Two Pi starvation-induced genes (AtIPS1, AT4), which are not represented on the ATH1 array and are down-regulated in phr1 mutants (Rubio et al., 2001), were also included in the qRT-PCR analysis (Fig. 3A). Their expression was also increased in Pi-replete pho2 mutant roots (Fig. 3A). The changes in gene expression demonstrate that part of the Pi-deprived signaling network remains activated in Pi-replete conditions in the pho2 mutant. The annotations of the genes and encoded proteins/domains (e.g. SPX) suggest involvement in Pi transport, acquisition, and possibly signaling during Pi starvation. These changes were absent when pho2 and the wild type were compared in −Pi conditions (Table I; Fig. 3A). This is consistent with the observation that pho2 shows increased expression of genes that are normally expressed in Pi-deficient wild type.
Table I.
Pi starvation-inducible genes that remain induced in Pi-replete pho2 mutants
Genes that displayed >2-fold higher or lower expression in Pi-replete pho2 mutant roots in two independent Affymetrix ATH1 genechip experiments. +Pi, Pi-replete conditions; −Pi, Pi deprivation; 3-h Pi, 3 h after readdition of 0.5 mm Pi to previously Pi-deprived seedlings.
| Probe ID | Gene Identifier | Annotation | pho2 versus Wild Type (+Pi) | pho2 versus Wild Type (−Pi) | −Pi versus +Pia (Fold Induced) | 3-h Pi versus −Pib (%) |
|---|---|---|---|---|---|---|
| 258158_at | At3g17790 | Acid phosphatase 5 (ACP5) | 5.90 | 1.09 | 13.8 | 16.2 |
| 246001_at | At5g20790 | Expressed protein | 5.55 | 1.18 | 192 | 4.9 |
| 252414_at | At3g47420 | Glc-3-P permease | 5.45 | 1.15 | 13.9 | 15.5 |
| 246071_at | At5g20150 | SPX domain protein | 4.56 | 1.21 | 52 | 8.7 |
| 256597_at | At3g28500 | 60S ribosomal protein P2 | 2.45 | 1.42 | 0.42 | 107 |
| 266184_s_at | At2g38940 | Pi transporter Pht1;4 | 2.43 | 1.12 | 27.9 | 12.1 |
| 258856_at | At3g02040 | Glycerophosphodiester phosphodiesterase (SRG3) | 2.41 | 1.23 | 34.3 | 5.5 |
| 260097_at | At1g73220 | Sugar Pi transporter | 2.21 | 1.23 | 144 | 8.3 |
| 248770_at | At5g47740 | Expressed protein | 2.20 | 1.19 | 6.3 | 32.6 |
| 245928_s_at | At5g24780 | Acid phosphatase (VSP1) | 2.08 | 1.69 | 7.3 | 88.1 |
| 248970_at | At5g45380 | Sodium:solute symporter | 0.46 | 1.20 | 0.32 | 191 |
| 267456_at | At2g33770 | E2 conjugase (PHO2) | 0.17 | 0.86 | 0.43 | 45.2 |
Expressed as fold induction.
Expressed as the percentage of transcript remaining after 3-h readdition.
Figure 3.
Molecular phenotypes of Pi-replete pho2 mutant roots. Transcript levels of nine selected P-responsive genes (A) and of 13 genes encoding phosphate transporters (B) are shown. Both images compare transcript levels in pho2 mutant roots and wild-type roots grown in Pi-replete or Pi-deprived conditions. Expression levels are given on a log scale expressed as 40 − ΔCT, where ΔCT is the difference in qRT-PCR threshold cycle number between the respective gene and the reference gene (UBQ10; At4g05320); 40 therefore equals the expression level of UBQ10; the number 40 was chosen because the PCR run stops after 40 cycles. The fold difference in expression is 2ΔΔCT when PCR efficiency is 2 (e.g. an ordinate value of 34 represents 16-fold lower expression than a value of 38). The results are the mean ± sd of four biological replicates.
To establish a link between pho2 and the high-shoot Pi phenotype, we used qRT-PCR to analyze the expression of the nine members of the Arabidopsis Pht1 Pi transporter gene family (Mudge et al., 2002) and four additional Pi transporters (Pht2;1, Pht3;1–Pht3;3). Transcripts for some of these genes cannot be reliably measured using the Affymetrix ATH1 genechip due to low expression, nonspecific probe sets (Usadel et al., 2005), or absence from the array. As expected, Pi strongly represses several Pi transporters in the wild type (Fig. 3B). The major difference in pho2 roots was that this repression was considerably attenuated for Pht1;8 and Pht1;9, which encode two closely related family members (Poirier and Bucher, 2002). Threshold fluorescence during qRT-PCR was reached approximately six and four cycles, respectively, earlier for Pht1;8 and Pht1;9 in cDNA prepared from Pi-replete pho2 roots (Fig. 3B). This equals an approximately 60-fold and 15-fold, respectively, higher expression when primer efficiencies are taken into account, indicating that these transporters contribute to the establishment of the pho2 leaf Pi accumulation phenotype. The expression and Pi responsiveness of the other Pi transporters were very similar in pho2 and wild-type plants, except that expression of Pht1;1 and Pht1;4 was still significantly higher in phosphorus (P)-replete pho2 compared to P-replete wild type. These results again highlight that a subcomponent of the Pi-dependent changes in gene expression is suppressed in the pho2 mutant.
Expression of PHO2
Inspection of the expression profile of PHO2 in data files from >720 ATH1 genechips (Supplemental Fig. 2) revealed that PHO2 is expressed ubiquitously and at considerable levels. The only tissues or developmental stages to show a marked increase in the PHO2 transcript were senescing leaves (no. 25) and maturating seeds (nos. 79–84; Supplemental Fig. 2A). Nitrogen deficiency and an extended night (Bläsing et al., 2005; Supplemental Fig. 2, E and F) also caused the PHO2 transcript to rise, possibly because these treatments trigger senescence. The ubiquitous distribution of the PHO2 transcript was further confirmed by qRT-PCR transcript measurements (data not shown) and by analyzing GUS reporter lines in which expression is driven by the 1.8-kb sequence upstream of the PHO2 start codon (Supplemental Fig. 3). Pi deprivation led to a significant, but small (approximately 2-fold), decrease of PHO2 transcripts (Supplemental Fig. 2E), confirming the results in Figure 3A. In pho2 mutants, the PHO2 (i.e. At2g33770) transcript level was low regardless of Pi status (Fig. 3A). This indicates that wild-type PHO2 exerts feed-forward regulation to increase its own expression in Pi-replete conditions.
Regulation of PHO2 Expression by Phosphate-Dependent miR399
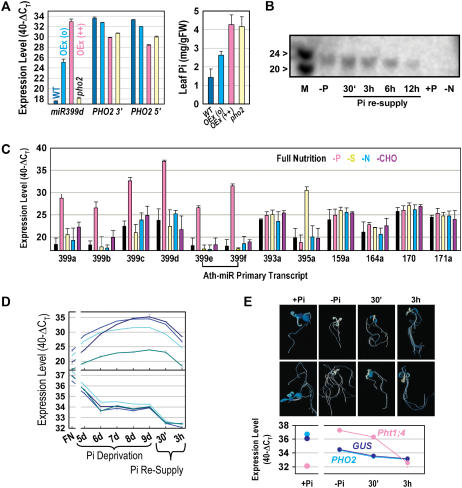
The annotated 1.1-kb 5′-UTR of PHO2 contains five, almost identical, approximately 21-nucleotide (nt) motifs that are complementary to Arabidopsis miR399 species (Supplemental Fig. 4A), miR399a to f, which are encoded by five loci in the Arabidopsis genome. RACE analysis was previously used to show that At2g33770 (i.e. PHO2) is a target for miR399-directed transcript degradation (Allen et al., 2005), and overexpression of miR399b, c, and f was recently shown to lead to reduced At2g33770 (i.e. PHO2) transcripts and increased leaf Pi levels (Chiou et al., 2006). We overexpressed miR399d and found that moderate overexpression of miR399d led to moderately reduced PHO2 transcripts and moderately increased leaf Pi levels, whereas in strong overexpressers PHO2 transcript levels were even lower and leaf Pi levels were as high as in pho2 mutants (Fig. 4A). This provides additional genetic evidence that miR399 species regulate the expression of PHO2 in planta.
Figure 4.
Regulation of PHO2 via Pi deprivation induced miR399. A, Impact of overexpression of miR399d on PHO2 leaf transcript level (left) and leaf Pi (right). Results are shown for wild type, pho2 mutant, moderate (o), and strong (++) overexpressers. The results are from leaves of 4-week-old soil-grown plants and show the mean ± sd of five biological replicates or five transgenic lines. PHO2 3′ and PHO2 5′ are designators for qRT-PCR products amplified from the 3′ and 5′ regions of the cDNA (compare with Supplemental Table III, primer pairs At2g33770_4 and At2g33770_5). B, Northern blot of the response of mature miR399 species in whole seedlings in full nutrition (+P), after Pi deprivation (−P) and 30-min to 12-h Pi resupply, and in nitrogen limitation (−N). The marker (M) is a mixture of 20- and 24-mer ribonucleotides. C, Levels of miRNA PTs in whole seedlings grown in full nutrition or Pi, sulfur, nitrogen, or carbohydrate limitation (P, S, N, and CHO, respectively). Results are shown for miR399a, miR399b, miR399c, miR399d, miR399e, and miR399f (the latter two are bracketed as they are encoded by the same gene), miR393a (previously shown to be stress responsive; see text), miR395a (previously shown to be S responsive; see text), and four miRNAs with developmental functions (159a, 164a, 170, and 171a). D, Time course of miR399 PT expression levels (top; miR399d, c, a, and e from the top at day 9) and PHO2 transcript level (bottom; analyzed with four different primer pairs as shown in Supplemental Fig. 4B) in wild-type seedlings during the development of Pi deprivation and after Pi resupply. E, Analysis of expression in reporter gene lines expressing GUS under control of an approximately 1.8-kb PHO2 upstream sequence. Two representative seedlings are shown for each condition. The transcript levels of PHO2, GUS, and Pht1;4 were determined by qRT-PCR in the same materials. The results in B to E are from seedlings grown in liquid culture in full nutrient (FN) medium for 9 d or were transferred after 7 d from FN to conditions in which a particular nutrient is missing, kept there for 2 d, and then resupplied with the respective nutrient (Scheible et al., 2004). Seedlings were harvested from full nutrition conditions, 2 d of P deprivation (−P), and 30 min (30′) and 3 h (3 h) after resupply of 0.5 mm KPi, or after 2 d of S, N, or CHO limitation. In D, 5-d-old seedlings were transferred to Pi depletion and Pi was added back 4 d later. For all qRT-PCR analyses, expression levels are given on a log scale as described in the legend to Figure 3.
Regulation of miR399 Expression by Pi
Fujii et al. (2005) and Chiou et al. (2006) recently reported that expression of miR399 is regulated by Pi availability. Using northern blotting, these authors showed that miR399 is readily detectable in Pi-deprived plants but undetectable in Pi-replete plants. The results in Figure 4 extend these findings in four ways. First, the 21-nt-long mature miR399s decrease within 6 to 12 h after readdition of 0.5 mm KPi to Pi-deprived wild type (Fig. 4B). Second, use of qRT-PCR reveals that all five miR399 primary transcripts (PTs; e and f are likely to be encoded by the same precursor) are very strongly induced by Pi deprivation (ΔΔCT = 11–14; Fig. 4, C and D). When primer efficiencies are taken into account, this represents a 1,000- to 10,000-fold increase in abundance during Pi deprivation (to a level similar to the ones of the most highly expressed Arabidopsis genes [e.g. the ubiquitin-10 reference transcript]). From these results and other data (R. Morcuende, R. Bari, and W.-R. Scheible, unpublished data), we conclude that miR399 PTs are among the most Pi-responsive transcripts in Arabidopsis. Third, these PTs decrease strongly and rapidly (already after 30 min) after supplying Pi to Pi-deprived seedlings (Fig. 4D, top). Fourth, the response of miR399 to Pi is highly specific. As shown in Figure 4B, nitrogen deprivation does not affect miR399 abundance, and none of the miR399 PTs responded significantly to carbohydrate, nitrogen, or sulfur depletion (Fig. 4C). None of these nutrient treatments affected the level of the PT for miR393a, which was reported to respond to abiotic stresses (Sunkar and Zhu, 2004), or the levels of four PTs for miRNAs with developmental functions (Dugas and Bartel, 2004). miR395a is induced by sulfur starvation (Jones-Rhoades and Bartel, 2004). The corresponding PT was specifically and strongly induced by sulfur deprivation, providing a second example of nutrient stress altering the level of the PT and the mature miRNA species (compare with Schmittgen et al., 2004).
PHO2 Transcripts Change Reciprocally to miR399 during Pi Deprivation and in miR399-Overexpressing Lines, But Not during Pi Readdition
As might be expected, PHO2 transcripts and miR399 PT levels show reciprocal behavior in (1) a comparison of wild-type plants grown in Pi-replete and Pi-deprived conditions (Supplemental Fig. 5); (2) miR399d overexpressers (Fig. 4A); and (3) a time course when wild-type seedlings are subjected to increasing Pi deprivation (Fig. 4D). However, closer inspection of these data reveals some discrepancies. Although the PHO2 transcript drops about 8-fold during the early phase of Pi deprivation when miR399 PT levels are rising (Fig. 4D), it remains easily detectable and stable during the later phase when miR399 PT levels have reached their maximum. Further, PHO2 transcripts decrease after readdition of Pi even though miR399 PTs are decreasing.
The miR399-binding sites in the PHO2 transcript are located in the 5′-UTR region. It is therefore possible that miR399 acts to generate a truncated PHO2 transcript with altered biological activity. To exclude this possibility, PHO2 transcript was measured using four different qRT-PCR primer pairs amplifying in the reverse-transcribed 5′-UTR (Fig. 4D; compare with Supplemental Fig. 4B and Supplemental Table III) and a fifth primer pair amplifying approximately 2.5 kb downstream of the 3′ region of the transcript (Fig. 4D, data not shown; compare with Supplemental Fig. 4B and Supplemental Table III). Even though primer pairs 1, 2, and 3 were designed to amplify upstream or around the miR399 cleavage sites (Allen et al., 2005; compare with Supplemental Fig. 4B), they yielded very similar amplification to that from primer pairs 4 and 5, which are located downstream of the miR399-binding sites (Supplemental Fig. 4B). This result indicates that all detectable PHO2 transcripts are intact (i.e. miR399-cleaved PHO2 transcript fragments do not persist during increasing and long-term Pi deprivation).
Is There Regulation of PHO2 Expression Independently of miR399-Mediated Transcript Decay?
The observation that PHO2 transcript levels remain relatively high in Pi-deficient plants when miR399 levels are high (Fig. 4D) could indicate that miR399-directed transcript cleavage is not the only mechanism that prevents PHO2 expression. We used the above-mentioned reporter lines, in which GUS expression is driven by the 1.8-kb sequence upstream of the PHO2 start codon, to visualize GUS staining in response to Pi availability and readdition. GUS staining was detected in all major organs and tissues of Pi-replete seedlings, which is consistent with the results described above (Supplemental Fig. 3). GUS staining was, however, strongly dependent on Pi availability (Fig. 4E). Seedlings grown with Pi showed strong GUS staining, whereas long-term Pi-deprived seedlings showed virtually no GUS staining. Interestingly, GUS staining appeared within 30 min after Pi readdition to long-term Pi-deprived seedlings and was marked by 3 h. This qualitative conclusion was checked by preparing extracts and quantitatively assaying GUS activity. GUS activity rose 2-fold on a protein basis (data not shown). The increase in GUS activity after Pi readdition occurred even though GUS and endogenous PHO2 transcript levels decreased in these conditions (Fig. 4E). These results and those mentioned above could indicate that PHO2 expression is subject to another level of posttranscriptional regulation.
PHR1 Acts Upstream of PHO2 in Pi Signaling
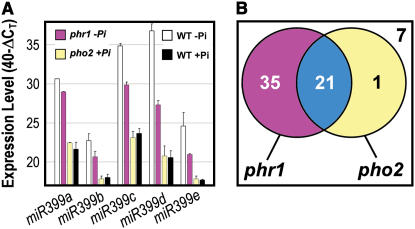
As outlined in the introduction, the MYB factor PHR1 has previously been shown to be required for induction of a small number of genes under Pi starvation, including AtIPS1 and AT4 (Rubio et al., 2001). These two genes are up-regulated in Pi-replete pho2 mutants (Fig. 3A), indicating that PHR1 and PHO2 share some common downstream targets. To clarify the relationship between PHO2 and PHR1, we extended the molecular phenotyping analysis by investigating the expression levels of 59 highly P-responsive genes (ΔΔCT > 3 between −Pi and +Pi conditions) and the five miR399 PTs in Pi-deprived phr1 and in Pi-replete pho2 seedlings (Fig. 5; Supplemental Fig. 6). The high expression of all five miR399 PTs in P deprivation was clearly impaired in phr1 mutants. For example, the PT of miR399d was several hundred-fold (ΔΔCT > 9) less abundant in P-deprived phr1 mutants compared to P-deprived wild type (Fig. 5A). These results indicate that PHR1 is required for miR399 expression, placing PHR1 upstream of PHO2 in plant Pi signaling. Expression of 56 (88%) of the 64 transcripts examined was significantly reduced (ΔΔCT < −1) in phr1 mutants (Fig. 5B; Supplemental Fig. 6), indicating a prominent, but not exclusive, role for PHR1 in Pi signaling. In contrast, expression of only 22 genes (34%) was altered in pho2 mutants. Importantly, all but one of the genes affected in pho2 were also affected in phr1. This analysis again suggests that PHR1 acts upstream of PHO2 in Pi signaling.
Figure 5.
PHR1 acts upstream of PHO2. A, qRT-PCR expression levels of miR399 PTs in Pi-deprived phr1 mutant seedlings, in Pi-deprived and Pi-replete wild-type seedlings, and in Pi-replete pho2 mutant seedlings. Expression levels are given as described in the legend to Figure 3. B, Venn diagram showing the overlap in the response of 64 Pi-responsive gene transcripts in phr1 mutants and pho2 mutants. Detailed results for all transcripts are shown in Supplemental Figure 6.
Conservation of Pi-Regulated miR399 Expression and PHO2 Gene Structure
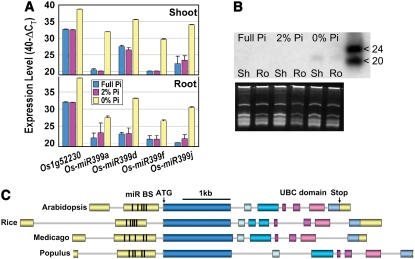
We investigated whether miR399 and PHO2 represent a conserved mechanism for Pi sensing in plants. Northern analysis and qRT-PCR both revealed induction of miR399 PTs and mature 21-nt miR399 in 3-week-old rice plants grown in the absence of external Pi (Fig. 6, A and B). Using public genomic DNA and expressed sequence tag (EST) data, we assembled potential orthologs of PHO2 from rice, Medicago truncatula, and poplar (Populus trichocarpa; Fig. 6C). The gene structure was fully conserved in these diverse species, except that the fifth exon was split into two in rice. In all cases, five miR399-binding sites were present in the second untranslated exons (Supplemental Fig. 7). Comparison of the protein sequences showed conserved stretches along the entire length, suggesting that the proteins are orthologs (Supplemental Fig. 7). Transcribed sequences encoding miR399, miR399-binding sites, or protein sequences homologous to the N- or C-terminal extensions of PHO2 were also found for wheat (Triticum aestivum), soybean (Glycine max), cotton (Gossypium hirsutum), orange (Citrus aurantium), or apple (Malus domestica), but not in the Physcomitrella patens EST database or the shotgun genome sequences of P. patens and C. reinhardtii. These results show that the regulatory mechanism is conserved across angiosperms and indicate that it may have emerged during the evolution of higher plants.
Figure 6.
Conservation of PHO2 genes and induction of miR399 by Pi starvation in higher plants. A, Expression levels of Pi starvation marker gene Os1g52230 (putative acid phosphatase homologous to the strongly Pi starvation-induced At1g17710), and Os-miR399a, d, f, and j PTs in shoots and roots of rice. The results are given as described in the legend to Figure 3. The reference gene was β-tubulin (Os1g59150). Colors represent the Pi concentrations in the external medium (full Pi, 320 μm, 2% Pi, 6.4 μm). B, Northern blot (top) showing the induction of mature Os-miR399 in shoots (Sh) and roots (Ro) of rice grown in the absence of external Pi. The bottom image shows the RNA gel used for blotting. C, Gene structures of AtPHO2 and potential orthologs from rice (Os5g48390), M. truncatula, and poplar. Gene structures are drawn to scale. UTRs are shaded yellow and coding regions are shown in shades of blue and red. The black ticks in the second exon depict the position of the five miR399-binding sites (miR BS).
DISCUSSION
Little is known about the components involved in signal transduction during Pi limitation in plants. A step forward was recently made by the discovery of phosphate-regulated miR399, which regulates expression of an E2 conjugase gene (Fujii et al., 2005; Chiou et al., 2006; this work). Chiou et al. (2006) showed that a T-DNA insertion in this E2 conjugase gene leads to a phosphate accumulation phenotype. The same authors claim the identification of a nucleotide mutation causing a stop codon in this E2 gene of the pho2 mutant. However, the claim was made without presentation of the results or complementation of the pho2 mutant. In this work, we identify Arabidopsis PHO2 by recombinational mapping as this E2 conjugase gene. We show that a G→A point mutation in pho2 predicts the replacement of W671 by a stop codon and the loss of the UBC domain. A PHO2 RNAi approach phenocopies pho2 and a genomic fragment harboring the entire E2 conjugase gene complements the mutant, providing definite proof for the correct identification of PHO2.
PHO2 is implicated in Pi signaling based on the (1) shoot Pi accumulation phenotype in Pi-replete pho2 mutants (Delhaize and Randall, 1995; Dong et al., 1998) and (2) continuous expression of a subset of Pi starvation-induced genes in Pi-replete conditions (Fig. 3). Some of these genes are probably involved in mobilization of external Pi and recycling from cellular components. However, the pho2 phenotype also points to a role for PHO2 in non-cell-autonomous responses, including regulation of Pi allocation between roots and shoots. Long-distance signaling is probably required to explain why the pho2 mutant shows a shoot-specific accumulation of Pi. In particular, we have shown in grafted plants that a pho2 root is necessary and sufficient to confer the high-shoot Pi phenotype. These results indicate that the pho2 mutant is either insensitive to feedback signals when Pi accumulates to high levels in the shoot, or the impact of a shoot-derived low-Pi signal is continuously present in pho2 roots.
In the roots of wild-type plants, the phosphate transporters Pht1;8 and Pht1;9 are highly expressed when Pi is limited and repressed when Pi is abundant. These two Pht1 genes represent segmentally duplicated twins and can be phylogenetically clearly separated from the other seven family members by the presence of an unusually large first intron (Poirier and Bucher, 2002). In the pho2 mutant, expression of Pht1;8 and Pht1;9 remains high in Pi-replete conditions. The possibility that this might contribute to the establishment of high Pi in pho2 leaves is supported by the observation that RNAi inhibition of Pht1;8 in pho2 decreases leaf Pi levels to the range found in the wild type (R. Bari and W.-R. Scheible, unpublished data). Expression of Pht1;1 and Pht1;4 also appears to remain higher in pho2 mutants, possibly facilitating uptake of Pi from the soil (Shin et al., 2004). This contrasts with other members of the Pht1 family, which are repressed in pho2 by Pi in a similar manner to the response in wild-type plants.
PHO2 is the first Arabidopsis UBC gene to be identified based on a mutant phenotype. Arabidopsis contains at least 37 genes annotated as E2 conjugases (Smalle and Vierstra, 2004). This reflects a general expansion of the number of genes encoding components of ubiquitin-dependent protein degradation and the key role that this pathway plays in plant responses to hormonal and environmental signals (Smalle and Vierstra, 2004). The PHO2 gene has two unusual features. First, UBC genes usually encode small (approximately 20-kD) proteins. The predicted PHO2 protein is much larger (approximately 100 kD) and contains N- and C-terminal extensions to the UBC domain (Fig. 1A; see below). These may assist in target recognition, association with appropriate E3 ligases, and/or localization (Smalle and Vierstra, 2004). Interestingly, the PHO2 E2 conjugase has high homology to the human E2 APOLLON protein (Kraft et al., 2005), which was shown to function as a chimeric E2 conjugase-E3 ligase (Hao et al., 2004). Second, PHO2 transcripts contain five binding sites for miR399 species in the 5′-UTR. The presence of miRNA-binding sites in a 5′-UTR is unusual, and the presence of five sites is unique (Sunkar and Zhu, 2004; Allen et al., 2005).
miR399 rises in Pi-deprived plants (Fujii et al., 2005; Chiou et al., 2006; Fig. 4) and falls rapidly after addition of Pi (Fig. 4). This provides a mechanism to inhibit PHO2 and hence stimulate Pi transfer to the shoot during Pi deprivation, and to derepress PHO2 in response to Pi resupply. The importance of these binding sites for transcriptional regulation, the regulation of miR399 by Pi availability, and its role in regulating transcript levels of the E2 conjugase encoded by At2g33770 were recently reported (Fujii et al., 2005). Chiou et al. (2006) showed that overexpression of miR399c, d, or f leads to Pi accumulation in plants. We have extended these results by showing that (1) the targeted E2 conjugase encodes PHO2, which has a well-defined whole-plant phenotype; (2) overexpressing a further and probably the major miR399 species (i.e. miR399d) also leads to transgenic plants with a pho2-like shoot Pi accumulation phenotype; (3) Pi specifically and strongly affects the expression of all five known miR399 precursors and this is rapidly reversed by Pi readdition; and (4) expression of miR399 and PHO2 are usually, but not always, anticorrelated, suggesting that miR399 mediates PHO2 transcript decay. This anticorrelation is apparent in miR399d overexpresser plants as well as in a comparison of wild-type plants grown in Pi-replete and Pi-deprived conditions.
However, further features of our data indicate that there are additional levels of regulation, in addition to miR399-mediated decay of the PHO2 transcript. First, the anticorrelation of miR399 and PHO2 transcripts sometimes breaks down. For example, miR399 PT and PHO2 transcript levels both decrease after adding Pi to Pi-deprived seedlings. In contrast to the results obtained with less-sensitive northern blotting (Fujii et al., 2005), our qRT-PCR measurements also reveal that, although PHO2 transcripts drop by 5- to 8-fold, they remain easily detectable in Pi-deprived plants, even though miR399 is highly abundant. These observations indicate that miR399-mediated degradation of the PHO2 transcript can be inhibited by further unknown mechanisms or counteracted by rapid continued transcription of PHO2. Second, some features of our data point to the possibility of translational regulation of the PHO2 transcript by Pi. In PHO2-GUS lines, PHO2 and GUS transcripts remain relatively high in Pi deprivation, but GUS reporter activity is almost undetectable. After readdition of Pi, GUS activity/staining rapidly emerges even though GUS and endogenous PHO2 transcripts decrease in a congruent manner. Whether such potential translational repression is mediated by miR399 binding to the PHO2 5′-UTR and whether rapid GUS translation after Pi readdition is due to fast dissociation and decrease of miR399s will require further study (e.g. mutating the miR399 target sites in the GUS reporter construct). It is interesting to note that the maximal levels of miR399 PTs in Pi deprivation are high enough to yield sufficient miR399s for saturation of the target sites on the PHO2 transcript (Supplemental Fig. 5).
The initial observation that transcript levels of two Pi starvation-induced genes (AtIPS1 and AT4) are both changed in pho2 mutants (Fig. 3A) and phr1 mutants (Rubio et al., 2001) led us to ask whether PHR1 and PHO2 might belong to the same signaling pathway. To this effect, we investigated the transcriptional changes in phr1 mutants in more detail. Crucially, induction of all miR399 PTs is clearly attenuated in Pi-deprived phr1 mutants but unaffected in pho2 mutants (Fig. 6). In addition, PHR1 GNATATNC cis-elements (Rubio et al., 2001) are usually found 160 to 270 nt upstream of the miR399s (data not shown). These results place PHR1 upstream of PHO2 and miR399 in Pi signaling. They also suggest that the MYB-like transcription factor PHR1 is an important, although not the only, factor required for full miR399 induction. Suppression of At4g20400, the segmental duplication of PHR1, in the phr1 background, or a phr1 enhancer screen to identify mutants in which the loss of induction is more pronounced could contribute to identifying the additionally required factors.
The levels of hundreds of transcripts change in abundance when plants are subjected to Pi deprivation (Misson et al., 2005; R. Morcuende, R. Bari, and W.-R. Scheible, unpublished data). However, only a small subset of these transcripts is highly responsive to Pi availability and shows a significant reversal of the response 3 h after Pi readdition to Pi-deprived plants (R. Morcuende, R. Bari, and W.-R. Scheible, unpublished data), indicating that they are involved in primary responses to Pi availability. Most of the 64 strongly Pi-responsive transcripts we analyzed in Figure 5 belong to this subset. Because approximately 90% of these transcripts showed an attenuated induction in phr1 mutants (Fig. 6B; Supplemental Fig. 7), it seems that PHR1 has an important role in the signaling of primary Pi responses. A genome-wide analysis of the transcriptional changes in Pi-deprived phr1 mutants and the wild type will be required to cement this hypothesis. The finding that only a smaller subset of these genes shows altered expression in the pho2 mutant provides independent evidence that PHO2 operates in a branch of the Pi-signaling pathways, which is located downstream of PHR1.
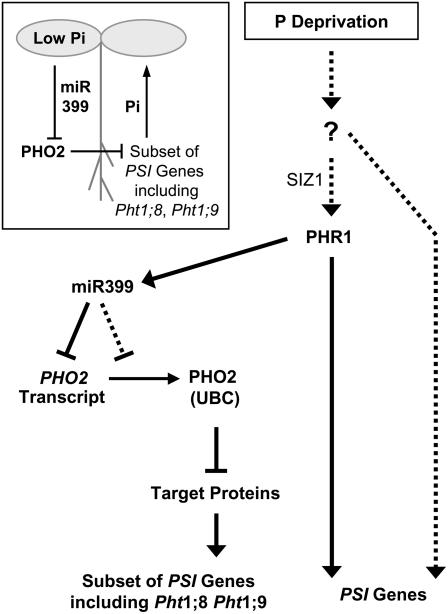
Figure 7 presents a working model for the mechanism of Pi sensing in higher plants. In this model, PHR1 takes a central role and the miR399/PHO2 pathway regulates the expression of a subset of the Pi starvation-induced genes. The phenotype of the pho2 mutant indicates that this branch is important for regulation of the allocation of Pi within the plant. It is intriguing that a miRNA plays a key role in this part of the Pi-signaling network, raising the possibility (see also Fujii et al., 2005) that transport of miR399 (Fig. 7, inset) may allow coordinated responses between different parts of the plant. The high-shoot Pi phenotype and the results of our grafting experiments could be explained if miR399 moves from the shoot to the root where it is perceived via PHO2, leading to changes in the expression of transporters and other genes that are involved in the movement of Pi from the root to the shoot. In this context, it is interesting that the levels of the miR399 PTs in Arabidopsis (Supplemental Fig. 5) and rice (Fig. 5A) are much higher in shoots than in roots during Pi deprivation. The possibility of additional levels of regulation in addition to PHO2 transcript turnover and translation and miR399-mediated transcript decay might contribute to a highly dynamic system for tuning the throughput of Pi from the root to the shoot.
Figure 7.
A model for plant Pi signaling involving PHO2, miR399, and PHR1. Expression of a subset of phosphate starvation-induced (PSI) genes, including the Pi transporter genes Pht1;8 and Pht1;9, is regulated through the UBC protein PHO2. Expression of PHO2 is regulated by miR399 species, which in turn require PHR1 for induction. PHR1 is also important for PHO2-independent expression of other PSI genes, and SIZ1-dependent sumoylation might be required for PHR1 activity. Other parts of the transcriptional response of plants to Pi deprivation might be PHR1 independent (dashed arrows). The inset illustrates the hypothesis that miR399 might serve as a mobile signal allowing coordinated responses to Pi stress between different parts of the plant.
Many open questions remain in the proposed signaling pathway. The immediate targets of PHO2 remain obscure. A pho2 suppressor/enhancer screen could contribute to identifying these components. Evidence is still required that miR399 actually moves between the shoot and the root. Further questions relate to the primary mechanisms by which Pi is sensed and to the mechanism by which PHR1 activity is regulated. The abundance of PHR1 transcripts and PHR1 protein is not Pi responsive (Rubio et al., 2001; R. Morcuende, R. Bari, and W.-R. Scheible, unpublished data), suggesting posttranslational regulation of PHR1 activity. The SUMO E3 ligase SIZ1 was recently shown to sumoylate PHR1 in vitro, and siz1 mutants were shown to cause alterations in Pi deprivation signaling (Miura et al., 2005). However, the few molecular phenotypes investigated to date in siz1 mutants are not identical to those found in phr1 mutants (compare with Miura et al., 2005; Supplemental Fig. 7), which is in contrast to what is expected if SIZ1-dependent sumoylation is a mechanism activating PHR1 during Pi deprivation. To explain this difference, it was necessary to postulate that SIZ1 has a complex dual role as a repressor as well as a positive regulator of low Pi-induced responses that also lie downstream of PHR1 (Miura et al., 2005). Clearly, more experiments, including a broader analysis of Pi starvation-related gene expression in siz1 mutants, are required to clarify the role of SIZ1 in controlling Pi deficiency responses.
MATERIALS AND METHODS
Plant Growth
Arabidopsis (Arabidopsis thaliana) plants for mapping were grown under normal greenhouse conditions (at Max Planck Institute-Molecular Plant Physiology [MPI-MPP]) on GS90 soil (Gebr. Patzer). PHO2 RNAi lines, miR399d overexpressers, and their respective wild-type and pho2 mutant controls were grown on GS90 soil in Percival AR 36-L growth chambers (Percival Scientific) in a 12-h light (approximately 120 μE, 25°C)/12-h dark (20°C) cycle at approximately 70% relative humidity. Arabidopsis seedlings (wild type, pho2, phr1) were also grown in a sterile liquid culture (Scheible et al., 2004; R. Morcuende, R. Bari, and W.-R. Scheible, unpublished data) either for 9 d in full nutrition conditions (+Pi) or first during 7 d in Pi-sufficient conditions (150 μm KPi) and then another 2 d in Pi-deprived conditions (−Pi). On day 9, some batches of Pi-starved seedlings were resupplied with 0.5 mm KPi for 30 min or 3 h. Materials were washed in demineralized water, quickly dissected into shoots and roots, and harvested into liquid nitrogen before storage at −80°C. Rice (Oryza sativa spp. japonicus cv Nipponbare) grains were germinated during 5 d in the dark at 28°C, then transferred into a growth chamber in a 12-h light (approximately 500 μE, 26°C)/12-h dark (20°C) cycle at 75% relative humidity for 3 d before they were mounted on a hydroponic culture system (24 plantlets/10-L box) filled with a standard nutrient solution (Yang et al., 1994) of varying Pi concentrations (320 μm, 6.4 μm, 0 μm). After 2 weeks, the plants were harvested.
Pi Measurements
Pi levels were measured using a colorimetric micromethod (Itaya and Ui, 1966).
PHO2 Mapping
The recessive pho2 mutant was isolated from an ethylmethane sulfonate-mutagenized population of Arabidopsis ecotype Columbia (Col-0) and the mutation was shown to be linked to visible marker as1 and RFLP/CAPS marker m429 on chromosome 2 (Delhaize and Randall, 1995). To obtain a high-resolution map position for the PHO2 locus, the pho2 mutant was crossed to the Arabidopsis Landsberg erecta wild type, and 96 F2 plants displaying high leaf Pi content were initially selected. For each plant, a DNA sample was prepared from inflorescence tissue using a quick alkaline lysis protocol (Lukowitz et al., 2000) and PCR genotyped with five microsatellite (simple sequence-length polymorphism [SSLP]) markers (nga1126, nga361, mpi14, nga168, and mpi17) equally spaced between 11.7 and 17.6 Mb on chromosome 2 (Supplemental Table II). The markers were identified from the genetic maps provided by The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org) or designed using the CEREON genomics database of Columbia-Landsberg polymorphisms (Jander et al., 2002). PCR conditions for SSLP markers were 50 mm KCl; 10 mm Tris-HCl (pH 9.0 at 25°C); 0.1% Triton X-100; 200 μm each of dATP, dGTP, dTTP, and dCTP; 10 pmol of each primer; 1.5 to 2.5 mm MgCl2, 1 unit Taq polymerase (Promega); and 10 to 50 ng of genomic DNA to a final volume of 22 μL. PCR program was as follows: 1 min 94°C; 40 cycles (20 s 94°C, 20 s 50°C–55°C, 30 s 72°C), and 2 min 72°C (Supplemental Table II); 4% agarose gels (3:1 HR agarose; Amresco) were used to resolve SSLP markers. The analysis placed the pho2 gene between nga361 and nga168 and close to mpi14 for which no recombinant chromosome was initially detected. Additional 3,840 F2 individuals were PCR genotyped with the flanking markers to reveal plants carrying a genome recombination in between. These recombinant F2 plants and their F3 progeny were analyzed for leaf Pi content to determine whether the recombination resides between pho2 and nga361 or between pho2 and nga168. This resulted in a total of 120 and 119 recombinants for the former and the latter, respectively (Supplemental Fig. 1A). The genotype of each recombinant F2 plant was further analyzed with a series of additional chromosome 2 SSLP markers (mpi11-mpi16, mpi18-mpi20; Supplemental Table II), finally placing the PHO2 locus in an approximately 50-kb region between polymorphisms CER458994 and CER459010 (Supplemental Fig. 1B; Fig. 1A).
Cosmid Complementation of pho2 Mutants
Cosmid clones covering the region (Fig. 1A) were isolated by colony hybridization (Scheible et al., 2003) from a genomic Col-0 library constructed in the binary vector pBIC20 (Meyer et al., 1994). Primers used for amplification of sequence-specific, digoxygenin-11-dUTP-labeled DNA probes are available on request. Clones were individually electroporated into Agrobacterium tumefaciens GV3101 and transformed into pho2 mutants by floral dipping (Clough and Bent, 1998). T1 transformants were selected on one-half-strength Murashige and Skoog agar plates containing kanamycin (50 μg mL−1), grown to the rosette stage in soil, and tested for leaf phosphate content. Sequences of the primers used for cosmid end sequencing are available on request.
DNA Sequencing
Genomic DNA was prepared by using a cetyl-trimethylammonium bromide detergent extraction method (Lukowitz et al., 2000) from Col-0 wild type and pho2-1 and pho2-2 mutants. Two (2,098 and 1,977 bp) fragments spanning the entire PHO2 (At2g33770) coding sequence were independently PCR amplified two times from each genotype using the primers 5′-CTCAGCATTTCTTGTATCATTTG-3′ and 5′-CAACTTTCACCAACACTGTAGAG-3′ or 5′-GCTCTTAGATGTTTGGGTGTC-3′ and 5′-CCTATTTTACAGCGGTTTTATTC-3′ and a mixture of Taq and proofreading Pfu polymerases (Promega). Cycle sequencing of both strands of the PCR products was performed with a set of five additional primers and BigDye dideoxy terminator reaction mix (Applied Biosystems), and products were resolved on an ABI310 sequencer. Likewise, a 1,992-bp genomic fragment containing At2g33760 was PCR amplified with primers 5′-GTATTTCCCCATGAATTTTACAAGT-3′ and 5′-AAACATCTCCCTAACATCCAATAACT-3′ and sequenced with three additional primers. Primer sequences are available on request.
dCAPS Marker Analysis
The Web-based program dCAPS finder 2.0 (Neff et al., 2002) was used for selection of a suitable restriction enzyme (MslI) that can distinguish between the wild-type and the pho2-1 allele after PCR fragment amplification with a mismatch and a match primer. See Supplemental Table II for primer sequences, PCR conditions, and PCR product lengths after MslI cleavage.
RNAi PHO2
A PHO2 gene-specific 874-bp-long PCR fragment was amplified using primers 5′-GAAGAAACCATTGGCAAAAT-3′ and 5′-CATTAACAGCACCAACAGGA-3′ and cloned into GATEWAY entry vector pENTR/SD/D-TOPO (Invitrogen) before recombining into destination vector pK7GWIWG2(II) (Karimi et al., 2002) and transformation into wild-type Col-0 via A. tumefaciens GV3101.
Overexpression of AthmiR399d
A 289-bp-long PCR fragment containing the entire annotated AthmiR399d precursor sequence was amplified using primers 5′-CACCAGGATTCGATCTATAAAACCTGA-3′ and 5′-TTGCTAGTCCAACTTCCAATAA-3′ and directionally cloned into GATEWAY entry vector pENTR/SD/D-TOPO (Invitrogen) before recombining into destination vector pMDC32 (Curtis and Grossniklaus, 2003) and transformation into wild-type Col-0 via A. tumefaciens GV3101.
Micrografting
Micrografting of 6-d-old pho2 mutant and wild-type Col-0 seedlings grown on sterile vertical agar plates was performed as described (Turnbull et al., 2002). Chimeric plants were maintained for 2 to 4 d at low light intensity (60 μE) on sterile one-half-strength Murashige and Skoog agar (1.5%) plates supplemented with 1% (w/v) Suc and then placed at higher light intensity (120 μE) for 4 to 6 d. Successfully grafted 14-d-old plantlets were transferred to soil and grown under greenhouse conditions for another 2 weeks before leaf Pi content was determined.
RNA Isolation, cDNA Preparation, and qRT-PCR
RNA isolation, cDNA preparation, and qRT-PCR were performed according to Czechowski et al. (2005). Rice total RNA for cDNA synthesis was isolated using the RNeasy plant mini kit (Qiagen). All RT-PCR primer sequences used in this study are given in Supplemental Table III. PCR primer efficiencies (E) were determined using the LinRegPCR program (Ramakers et al., 2003), did not change significantly between different cDNA samples, and were always higher than (1 + E) = 1.84.
Affymetrix ATH1 Genechip Data Analysis
Two independent experiments were carried out to obtain root materials from liquid culture-grown Pi-replete and Pi-deprived wild type and pho2 mutants. Expert Affymetrix array service, including all steps from total RNA to data acquisition, was provided by the German Resource Center for Genome Research (RZPD), Berlin. Affymetrix CEL files were processed using Robust Multi-Array Average normalization software (Bolstad et al., 2003). The normalized data were delogged, ratios were built on samples from the same experiment, and mean ratios for each probe set were calculated for the independent experiments.
Northern Analysis of miR399 Species
Northern analysis of miR399 species was performed according to the small RNA protocol available for download from the J. Carrington lab home page (http://jcclab.science.oregonstate.edu). Forty micrograms of total RNA from Pi-replete seedlings, Pi-starved seedlings during a Pi-readdition time course, nitrogen-starved seedlings, and a mix of 20- and 24-mer oligo-ribonucleotides (5′-UGCCAAAGGAGAUUUGCCCU-3′ and 5′-UGCCAAAGGAGAUUUGCCCUGUAA-3′; Ambion) were resolved on 17% polyacrylamide gels containing 7 m urea in 0.5 × Tris-borate/EDTA, and transferred to a nylon membrane (Biodyne B; Pall Europe Ltd.) using a semidry transfer cell (Bio-Rad). After UV cross-linking, the membrane was hybridized with a 32P end-labeled probe (5′-TTACAGGGCAAATCTCCTTTGGCA-3′) complementary to miR399 at 42°C.
Promoter 5′-UTR GUS Construct
A 1.833-kb sequence upstream of the PHO2 start codon was PCR amplified using primers 5′-CACCCGCTTTGTGTCATCATTTAGAG-3′ and 5′-CATTTCCATAGTTTAGACGCCT-3′ and cloned into GATEWAY entry vector pENTR/SD/D-TOPO (Invitrogen) before recombining into destination vector pMDC163 (Curtis and Grossniklaus, 2003) and transformation into wild-type Col-0 via A. tumefaciens GV3101. T2 progeny of kanamycin-resistant transformants were GUS stained at various ages and developmental stages (Jefferson et al., 1987).
Sequence Extraction and Alignments
Using the PHO2 protein sequence and the PHO2 5′-UTR sequence as queries, TBLASTN and BLAST searches were performed on the Web pages of the National Center for Biotechnology Information (NCBI), the Department of Energy Joint Genome Institute (JGI), TAIR, and The Institute for Genomic Research (TIGR) to identify genomic sequences containing putative PHO2 orthologs. This resulted in the identification of the rice chromosome 5 clone OJ1214_E03 (GenBank AC104709.2), the Medicago truncatula chromosome 2 clone mth2-154m16 (GenBank AC159143.1), and the poplar (Populus trichocarpa) scaffold_124 contig 26, encoding PHO2 homologs preceded by five miR399-binding sites. The gene structure, transcript, and protein sequence for the rice homolog (LOC_Os05g49380.1) are available at the Gramene Mapping Resource (http://www.gramene.org). Gene structures for Medicago truncatula and poplar were assembled by using cDNA and EST sequences and manual annotation. Multiple sequence alignments were created with the PileUp program of Accelrys Seqweb version 2 (Accelrys), processed with Boxshade 3.21 (http://www.ch.embnet.org), and manually arranged into the final layout.
Supplementary Material
Acknowledgments
We thank D. Schindelasch, B. Sundaram, and P. Matt (MPI-MPP) for technical assistance, the Arabidopsis Biological Resource Center (ABRC; Ohio State University, Columbus, OH,) and E. Delhaize (Commonwealth Scientific and Industrial Research Organization [CSIRO], Canberra, Australia) for pho2-1 and pho2-2 mutant seeds, and E. Grill (Technical University Munich) for the Col-0 cosmid library. N. Gaude and P. Dörmann (MPI-MPP) provided homozygous seeds of the phr1 T-DNA insertion mutant (SALK line 067629). Affymetrix Service was provided by the RZPD (Berlin). We also thank our colleagues R. Schmidt, M. Udvardi, and J. Lunn (MPI-MPP) for excellent suggestions on the manuscript.
This work was supported by the Max Planck Society and the German Federal Ministry of Education and Research.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Wolf-Rüdiger Scheible (scheible@mpimp-golm.mpg.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.079707.
References
- Abel S, Ticconi CA, Delatorre CA (2002) Phosphate sensing in higher plants. Physiol Plant 115: 1–8 [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Bläsing OE, Gibon Y, Günther M, Hohne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, Stitt M (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Chen DL, Delatorre CA, Bakker A, Abel S (2000) Conditional identification of phosphate-starvation-response mutants in Arabidopsis thaliana. Planta 211: 13–22 [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL (2006) Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ (1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Rengel Z, Delhaize E (1998) Uptake and translocation of phosphate by pho2 mutant and wild-type seedlings of Arabidopsis thaliana. Plant Physiol 205: 251–256 [DOI] [PubMed] [Google Scholar]
- Dugas DV, Bartel B (2004) MicroRNA regulation of gene expression in plants. Curr Opin Plant Biol 7: 512–520 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15: 2038–2043 [DOI] [PubMed] [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber Petétot J, Somerville C, Poirier Y (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Sekine K, Kawabata A, Nakamura H, Ishioka T, Ohata H, Katayama R, Hashimoto C, Zhang X, Noda T, et al (2004) Apollon ubiquitinates SMAC and caspase-9, and has an essential cytoprotection function. Nat Cell Biol 6: 849–860 [DOI] [PubMed] [Google Scholar]
- Itaya K, Ui M (1966) A new micromethod for the colorimetric determination of inorganic phosphate. Clin Chim Acta 14: 361–366 [DOI] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MV, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau OS, Deng XW, Callis J (2005) Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol 139: 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JC, Zakhleniuk OV (2004) Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. J Exp Bot 55: 1221–1230 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H (1995) Mineral Nutrition of Higher Plants, Ed 2. Academic Press, London
- Meyer K, Leube MP, Grill E (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW (2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Neff MM, Turk E, Kalishman M (2002) Web-based primer design for single nucleotide polymorphism analysis. Trends Genet 18: 613–615 [DOI] [PubMed] [Google Scholar]
- Poirier Y, Bucher M (2002) Phosphate transport and homeostasis in Arabidopsis. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0024, www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J (1991) Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol 97: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 13: 62–66 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Fry B, Kochevenko A, Schindelasch D, Zimmerli L, Somerville S, Loria R, Somerville CR (2003) An Arabidopsis mutant resistant to thaxtomin A, a cellulose synthesis inhibitor from Streptomyces species. Plant Cell 15: 1781–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136: 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Jiang J, Liu Q, Yang L (2004) A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res 32: e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16: 2001–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Lahner B, Salt DE, Abel S (2004) Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant J 37: 801–814 [DOI] [PubMed] [Google Scholar]
- Trull MC, Deikman J (1998) An Arabidopsis mutant missing one acid phosphatase isoform. Planta 206: 544–550 [DOI] [PubMed] [Google Scholar]
- Turnbull CG, Booker JP, Leyser OHM (2002) Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J 32: 255–262 [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ribot C, Rezzonico E, Poirier Y (2004) Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol 135: 400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff DD, Grossman AR, Weeks DP, Usuda H, Shimogawara K (1999) Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc Natl Acad Sci USA 96: 15336–15341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Roemheld V, Marschner H (1994) Uptake of iron, zinc, manganese, and copper by seedlings of hybrid and traditional rice cultivars. J Plant Nutr 17: 319–331 [Google Scholar]
- Zakhleniuk OV, Raines CA, Lloyd JC (2001) pho3: a phosphorus-deficient mutant of Arabidopsis thaliana (L.) Heynh. Planta 212: 529–534 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.