Abstract
Expression of the C4-specific phosphoenolpyruvate carboxylase (C4-PEPC) gene in maize (Zea mays) is regulated in a tissue-specific manner, but affected by light and nutrient availability. We manipulated these stimuli in a combinatorial manner and analyzed concomitant changes in histone acetylation of the nucleosomes associated with the C4-PEPC gene in relation to transcriptional activity and steady-state mRNA levels. Whereas the transition from the lowest activity to an intermediate activity was observed in the absence of histone acetylation, the light-induced boost to full activity was associated with strong enhancement of the acetylation of both histones H3 and H4 limited to the gene region. Once activated by light, prolonged darkness was necessary to reduce both transcription and, in parallel, histone acetylation. Unexpectedly, histone acetylation was also induced in bundle sheath cells, although the transcriptional activity did not respond to illumination in this tissue. Furthermore, we were able to down-regulate the promoter by nitrogen depletion in the light without any decrease in the hyperacetylation of histone H4. When plants kept in prolonged darkness were nitrogen depleted and then exposed to light, transcription was not induced, but the promoter chromatin became hyperacetylated. We suggest a model where inhibition of a histone deacetylase in the light triggers H4 hyperacetylation at the C4-PEPC gene promoter regardless of the transcriptional activity of the gene. Our data indicate that an understanding of the interplay between histone modification and transcription requires analysis of signal integration on promoters in vivo.
Gene expression is controlled in response to both external stimuli and internal information (e.g. the position of a cell within a tissue or the developmental stage). Multiple stimuli have to be integrated at the level of transcription initiation to adapt the transcriptome of the cell to the requirements of the specific environment. An interesting model for such an integrative function is the C4-specific phosphoenolpyruvate carboxylase (C4-PEPC) promoter in maize (Zea mays). Transcriptional activity is strongly induced by light (Sheen, 1999), but modulated by additional stimuli. First, maize is a C4 plant where primary and secondary CO2 fixation are separated in two photosynthetic leaf cell types, the mesophyll and the bundle sheath, respectively (Nelson and Langdale, 1992). C4-PEPC catalyzes the initial fixation of inorganic carbon in the mesophyll and, accordingly, the corresponding mRNA does not accumulate in bundle sheath cells (Kausch et al., 2001). Therefore, gene transcription differs in two tissues that receive very similar external stimuli, in particular, light stimuli. Second, C4-PEPC expression in leaf tissues is regulated at the transcriptional level by the availability of nitrogen. This stimulus is seemingly transported from the root to the leaf by the cytokinin Zeatin (Sugiharto et al., 1992). Transcription of the C4-PEPC gene rapidly decreases in detached leaves and can be rescued by exogenous application of the hormone (Suzuki et al., 1994). Moreover, the metabolic state of a cell has an impact on the expression of the C4-PEPC gene. Most prominently, high concentrations of hexoses act negatively on transcriptional activity (Sheen, 1990). It has been suggested that hexokinase acts as the major sugar sensor in this pathway (Jang and Sheen, 1994).
In eukaryotic cells, DNA, including the cis-acting control elements of transcription, is packed into chromatin. The fundamental repeat unit of chromatin is the nucleosome, a particle of around 10 nm in size that is made up from two molecules each of the core histones H2A, H2B, H3, and H4, and approximately 160 bp of DNA. The nucleosomes are interconnected by short stretches of linker DNA (Kornberg and Lorch, 1999). In this form, the structure is usually not permissive for transcription initiation (Struhl, 1999). There are two main mechanisms that are tightly correlated with the transcriptional activation of a promoter in the chromatin context. First, chromatin remodeling complexes alter the interaction of the histone core with DNA. This may induce the repositioning of nucleosomes, exposure of previously occluded binding sites on DNA, or loss or replacement of histone dimers within the nucleosome core, respectively (Flaus and Owen-Hughes, 2004). Second, the N-terminal tails, mainly of histones H3 and H4, are covalently modified by acetylation, methylation, phosphorylation, ubiquitinylation, and ADP ribosylation (Berger, 2002). Among these modifications, acetylation of Lys residues in histones associated with promoters and coding regions shows a strong positive correlation with gene transcription in both plant and animal systems (Lusser et al., 2001; Schubeler et al., 2004).
In this study, we investigated the individual and combined effects of different stimuli on transcription and steady-state mRNA levels of the C4-PEPC gene and measured the degree of histone acetylation at different points in the associated chromatin. We provide conclusive evidence that histone acetylation is controlled by illumination independent from the transcriptional state. Furthermore, histone acetylation is seemingly only required for the highest transcriptional activity, but not for an intermediate activity state. We provide a model for the role of histone acetylation in signal integration on the C4-PEPC promoter.
RESULTS
Light-Dependent Histone Acetylation at the C4-PEPC Promoter
Transcripts from the C4-PEPC gene only accumulate to low levels in etiolated plants that were never exposed to light, but to high levels in illuminated green plants (Sheen, 1999). We used real-time, reverse transcription PCR to quantify this effect and to verify whether this is due to changes in the transcriptional activity of the gene. The accumulation of nonspliced heterogeneous nuclear RNA (hnRNA) and spliced mRNA transcripts was therefore compared. Primary transcripts are rapidly spliced even during transcription, and the abundance of hnRNAs is therefore a good indicator of transcriptional activity, whereas mRNA abundance is also affected by posttranscriptional mechanisms (Elferink and Reiners, 1996; Delany, 2001; Reed, 2003). The primer pair for the detection of nonspliced hnRNA spans an intron at the 3′ end of the gene (see Fig. 1A, amplified region C3). The accumulation of mRNA was monitored in parallel with a primer system specific for spliced transcripts (Hahnen et al., 2003). As shown in Figure 1B, the abundance of hnRNA as well as mRNA was clearly higher in green plants compared to etiolated plants. In both situations, the ratio of mRNA to hnRNA abundance was similar (roughly 500-fold more mRNA than hnRNA). This indicates that the light-induced increase in the amount of transcripts is mainly controlled on the level of transcriptional activity.
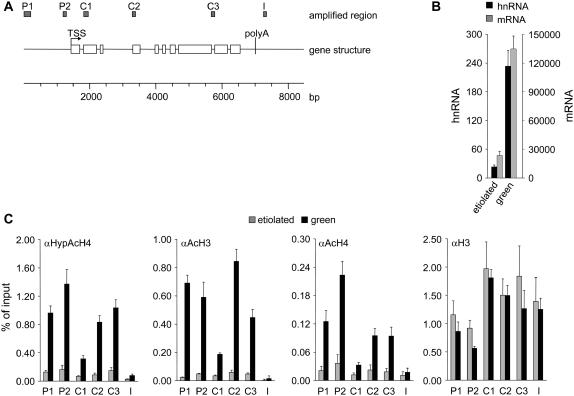
Figure 1.
Transcription of the C4-PEPC gene and histone acetylation in etiolated and green maize seedlings. A, Survey of the amplified regions on the C4-PEPC gene used in this study. Primer systems P1, P2, C1, C2, C3, and I were used to quantify the amount of precipitated DNA after ChIP. The gene structure is shown as an intron-line, exon-block diagram. Positions of the transcription start site (TSS) and the putative polyadenylation site (poly A) are indicated. Numbers are given in base pair. The corresponding primer systems are listed in Table I. Primer system C3 was also used to quantify the amount of C4-PEPC hnRNA in cDNA preparations. B, Quantification of C4-PEPC hnRNA synthesis and mRNA accumulation in etiolated and green maize seedlings. Etiolated = seedlings were grown for 10 to 12 d in complete darkness and harvested in the dark; Green = seedlings were grown in a diurnal rhythm of 16-h light and 8-h darkness for 10 to 12 d and were harvested 4 h after the onset of illumination. Numbers are given as molecules hnRNA (black columns) or mRNA (gray columns) per 50 μg of leaf material. hnRNA was amplified from cDNA with a primer system specific for an intron (C3). mRNA was amplified with a primer system that spans an intron (see Table I). A dilution series of PCR products of known concentrations was used as a standard. C, Histone acetylation on the C4-PEPC gene. Amounts of chromatin precipitated with antibodies specific for hyperacetylated histone H4 (αHypAcH4), acetylated histone H3 (αAcH3), acetylated histone H4 (αAcH4), and an invariant C-terminal epitope of histone H3 (αH3) from green (black columns) and etiolated (gray columns) leaf material. Numbers are given as the percentage of input. Input is the amount of chromatin subjected to immunoprecipitation (see also “Materials and Methods”). The data points are based on at least three independent experiments performed as technical duplicates. Vertical lines indicate ses.
To test whether this is accompanied by changes in the acetylation state of histones on the promoter or in the transcribed region, we used chromatin immunoprecipitation (ChIP). At the time of sampling, DNA and protein were cross-linked with formaldehyde and chromatin was isolated and immunoprecipitated with antibodies against acetylated H3 and H4, as well as an antibody that specifically recognizes hyperacetylated H4 where at least four of the five N-terminal Lys residues are acetylated (HypAcH4). The coprecipitated DNA was isolated and quantified by real-time PCR. Figure 1A shows an overview of the C4-PEPC locus and the amplified regions. We tested two positions in the distal and proximal promoter region (P1 and P2), three positions within the coding region (C1–C3, where C3 is used both for the determination of hnRNA levels and for the acetylation of chromatin), and one probably intergenic position about two nucleosomes behind the predicted polyadenylation site of the gene (I). Signals were corrected for the amount of DNA precipitated with the negative control serum. This background was never more than 5% of a positive signal. Furthermore, signals were standardized for the amount of chromatin input at the beginning of the immunoprecipitation (see also “Materials and Methods”). Thus, any variability during chromatin preparation was integrated into the calculation.
Figure 1C shows a comparison of immunoprecipitates derived from green and etiolated plants. The acetylation-specific antibodies differ in their efficiency of precipitation, but the observed relative patterns were very similar in all cases. In etiolated plants, the degree of histone acetylation was at the limit of detection at the tested positions. In green plants, a clear increase in histone acetylation was observed within the promoter and coding sequence, but not in the downstream intergenic region. Compared to the promoter or the residual coding sequence, a clearly weaker degree of acetylation was observed in the 5′ part of the coding region (system C1). This result was independently reproduced with a second primer system, thus excluding possible PCR artifacts (data not shown).
Identical experiments were performed with an antibody directed against the invariant C-terminal domain of histone H3. This antigen is not subject to covalent modifications and the amount of precipitated DNA is therefore an indicator of nucleosome density in the investigated genomic region (Pokholok et al., 2005). As shown in Figure 1C, the number of nucleosomes was slightly reduced at the promoter compared to the coding and intergenic regions. However, this effect was observed in both etiolated and green plants and therefore did not correlate with transcriptional activity.
We conclude that the C4-PEPC chromatin becomes highly acetylated at the N-terminal tails of histones H3 and H4 during light-induced activation of transcription. Histone acetylation appeared to be limited to the gene region and absent from the downstream intergenic region. We also conclude that the low transcriptional activity observed in etiolated plants does not require histone acetylation.
PEPC Transcriptional Activity and Histone Acetylation under Different Light Regimes
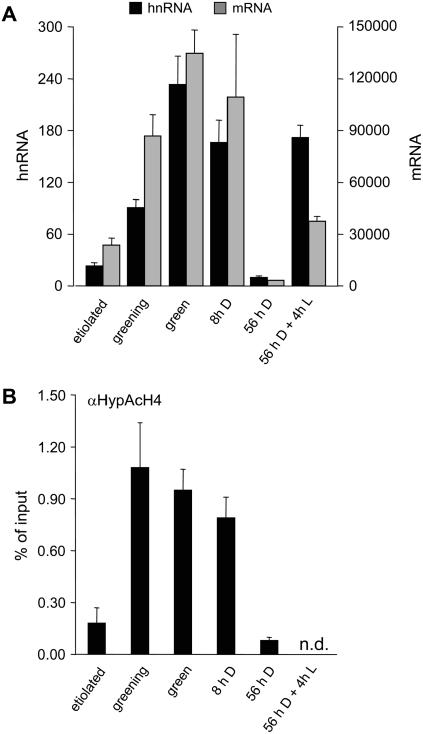
We wanted to see how closely histone acetylation correlates with the light regime given to the plant. Figure 2A shows the accumulation of hnRNA and mRNA under the different light conditions tested and Figure 2B shows the corresponding degree of H4 hyperacetylation at the proximal promoter position P2. Because of the almost identical patterns obtained with all antibodies directed to acetylated histones, we used H4 hyperacetylation at this position as a measure of the acetylation state of the gene. As shown before, transcription and H4 hyperacetylation were low in etiolated plants. When these plants were illuminated for only 12 h (greening plants), the transcriptional activity increased to one-half the level observed in fully green illuminated plants, whereas H4 hyperacetylation had already reached maximal levels. In parallel experiments, green plants were shifted to darkness for two time periods. At the end of the diurnal dark (D) period (8-h D), almost no decrease in both transcriptional activity and steady-state mRNA levels was observed and H4 hyperacetylation was still comparable to the illuminated state. The promoter was therefore not inactivated within the normal diurnal light (L) regime. By 56 h of darkness (56-h D), transcriptional activity was decreased and finally reached levels that were less than one-half those observed in etiolated plants. This was accompanied by a decrease in H4 hyperacetylation to background levels. These plants still had a green leaf blade with a pale base and only 4 h of reillumination (56-h D + 4-h L) were sufficient to fully restore transcriptional activity, whereas steady-state mRNA levels accumulated to only one-fourth the maximal value within this period. The data indicate that both gene transcription and histone acetylation were turned on rapidly after illumination, but were turned off slowly in the absence of light.
Figure 2.
Histone acetylation correlates with light-induced transcription at the C4-PEPC gene. Etiolated = grown in complete darkness and harvested in the dark; greening = etiolated plants that were illuminated for 12 h; green = seedlings were grown in the normal diurnal rhythm and harvested 4 h after the onset of illumination; 8-h D = green plants that were harvested at the end of the dark period; 56-h D = green plants that were darkened for 56 h; 56-h D + 4-h L = 56-h of darkness + 4 h of illumination. A, Quantification of C4-PEPC hnRNA synthesis and transcript accumulation. Numbers are given as molecules hnRNA (black columns) or mRNA (gray columns) per 50 μg of leaf material. hnRNA was amplified from cDNA with a primer system specific for an intron (C3). mRNA was amplified with a primer system that spans an intron (see Table I). A dilution series of PCR products of known concentrations was used as a standard. Transcription data for etiolated and green plants were taken from Figure 1. B, Histone H4 hyperacetylation at the C4-PEPC promoter position P2. Numbers are given as the percentage of input. Input is the amount of chromatin subjected to immunoprecipitation (see also “Materials and Methods”). n.d., Not determined. The data points are based on at least three independent experiments performed as technical duplicates. Vertical lines indicate ses.
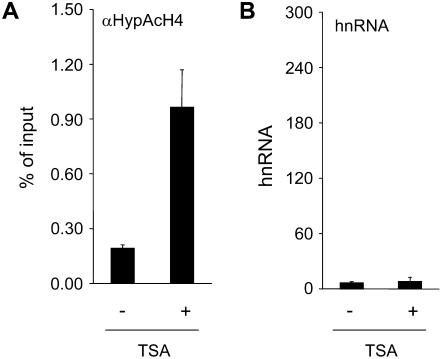
To test whether histone acetylation is sufficient to induce activity of the C4-PEPC promoter, we darkened plants for 56 h and, afterward, fed detached leaves the potent histone deacetylase (HD) inhibitor Trichostatin A (TSA). This treatment induced an increase in H4 hyperacetylation that was similar to levels found in green illuminated plants (Fig. 3A). However, very low levels of transcription were measured for both treated and untreated plants (Fig. 3B), indicating that histone hyperacetylation does not suffice for transcriptional activation in this system.
Figure 3.
Histone H4 hyperacetylation and transcription of the C4-PEPC gene after TSA treatment in the dark. Seedlings were grown in a diurnal rhythm of 16-h light and 8-h darkness for 10 to 12 d and afterward darkened for 56 h. At the end of the last dark period, leaves were detached in the dark and subjected to TSA treatment (+) or incubated in tap water (−) for an additional 12 h in the dark. A, Histone H4 hyperacetylation at the C4-PEPC promoter position P2. Numbers are given as the percentage of input. Input is the amount of chromatin subjected to immunoprecipitation (see also “Materials and Methods”). B, C4-PEPC hnRNA accumulation. Numbers are given as molecules of hnRNA per 50 μg of leaf material. hnRNA was amplified from cDNA with a primer system specific for an intron (C3). A dilution series of PCR products of known concentrations was used as a standard. The data points are based on at least three independent experiments performed as technical duplicates. Vertical lines indicate ses.
Histone H4 Hyperacetylation in Nitrogen-Depleted Leaves
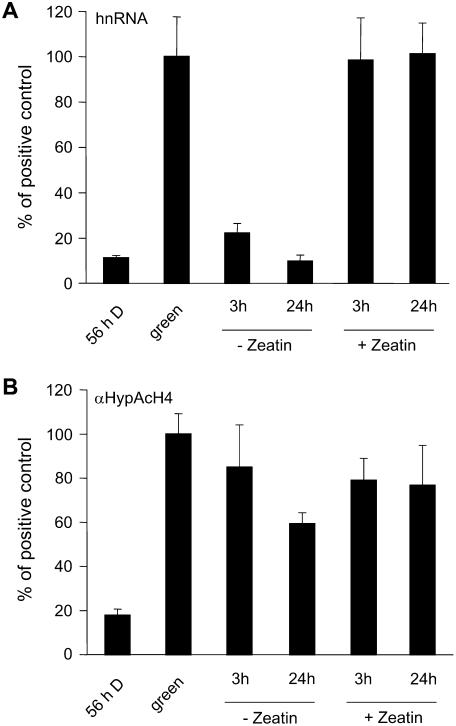
When leaves are detached and incubated in water, the C4-PEPC promoter is rapidly inactivated. Presumably, this is due to the depletion of a nitrogen signal from the root that is transduced by the hormone Zeatin and, consequently, addition of a mixture of Zeatin and nitrate negates the reduction in C4-PEPC transcription (see introduction). We used this effect to further test the connection between histone acetylation and transcription. Leaves were detached from green illuminated plants and incubated for two different time periods in the light in solutions with and without Zeatin/nitrate, respectively (Fig. 4). Detached leaves supplied with Zeatin/nitrate showed high transcriptional activity and high H4 hyperacetylation at both tested time points. In untreated leaves, the C4-PEPC transcriptional activity was reduced by a factor of 5 after only 3 h and, by 24 h, was further reduced to levels comparable to the fully inactivated state in control plants (not detached, but darkened for 56 h). Unexpectedly, the hyperacetylation of histone H4 on the promoter remained high in these plants. This indicates that histone deacetylation is not a prerequisite for transcriptional inactivation of the C4-PEPC gene.
Figure 4.
Inactivation of transcription without loss of histone H4 hyperacetylation. Green = seedlings were grown in the normal diurnal rhythm and harvested 4 h after onset of illumination; 56-h D = green plants that were darkened for 56 h; ±Zeatin = leaves from green plants were detached 4 h after the onset of illumination and incubated in a solution containing Zeatin/KNO3 or in tap water for 3 h in the light or for 24 h in the normal diurnal rhythm (12-h light, 8-h dark, 4-h light), respectively. A, C4-PEPC hnRNA synthesis. hnRNA was amplified from cDNA with a primer system specific for an intron (C3). A dilution series of PCR products of known concentrations was used as a standard. B, Histone H4 hyperacetylation at the C4-PEPC promoter position P2. Values were normalized for the amount of input. Input is the amount of chromatin subjected to immunoprecipitation (see also “Materials and Methods”). All values were further normalized for the amount of hnRNA or precipitated chromatin, respectively, in green leaves (positive control). Each data point is based on four independent experiments. Vertical lines indicate ses.
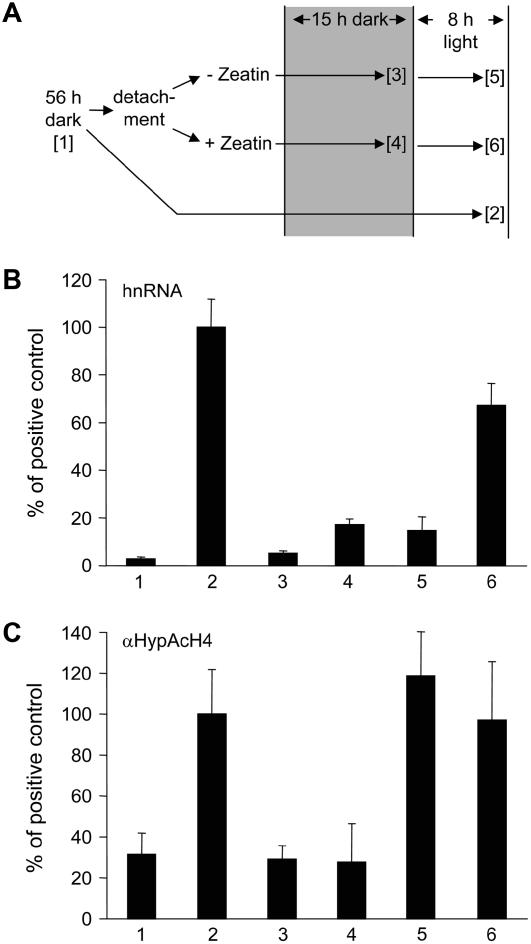
In a second series of experiments, green plants were grown in the dark for 56 h to reduce the transcriptional activity of the C4-PEPC gene to minimal levels. Leaves were then detached in the dark and incubated for a further 15 h in water or a mixture of Zeatin and nitrate. After this period, some of the leaves were additionally illuminated for 8 h (see Fig. 5A). The resulting transcriptional activities are shown in Figure 5B and the corresponding levels of H4 hyperacetylation are shown in Figure 5C. When comparing Zeatin-/nitrate-treated and untreated plants in the dark (columns 3 and 4), a more than 3-fold increase in hnRNA levels was observed in the treated plants. This increase in transcriptional activity was not reflected at the level of histone acetylation. After illumination, plants without Zeatin/nitrate treatment showed a weak light response, resulting in a low transcriptional activity (column 5), whereas transcription was further increased by a factor of 4 after illumination of treated plants (column 6). Surprisingly, H4 hyperacetylation at the promoter was induced to high levels in all illuminated plants regardless of Zeatin/nitrate treatment and therefore regardless of transcriptional activity. We conclude that light induces H4 hyperacetylation at the C4-PEPC promoter regardless of additional signals modulating transcriptional activity. An intermediate activity of the promoter is induced in the absence of the light signal and does not require induction of histone acetylation.
Figure 5.
Induction of H4 hyperacetylation without promoter activation. A, Schematic overview of the experimental design. Seedlings were grown in a diurnal rhythm of 16-h light and 8-h darkness for 10 to 12 d and the last dark period was extended to 56 h ([1]). The leaves were detached in the dark and incubated in tap water ([3]) or Zeatin/KNO3 ([4]) for a further 15 h. After this, one-half of the leaves were additionally illuminated for 8 h ([5] and [6]). Positive control plants were kept in the dark for 71 h and illuminated for 8 h ([2]). B, C4-PEPC hnRNA synthesis. hnRNA was amplified from cDNA with a primer system specific for an intron (C3). A dilution series of PCR products of known concentrations was used as a standard. C, Histone H4 hyperacetylation at the C4-PEPC promoter position P2. Values were normalized for the amount of input. Input is the amount of chromatin subjected to immunoprecipitation (see also “Materials and Methods”). All values were further normalized for the amount of hnRNA or precipitated chromatin, respectively, in the positive control ([2]). Each data point is based on four independent experiments. Vertical lines indicate ses.
Histone H4 Hyperacetylation in Bundle Sheath Cells
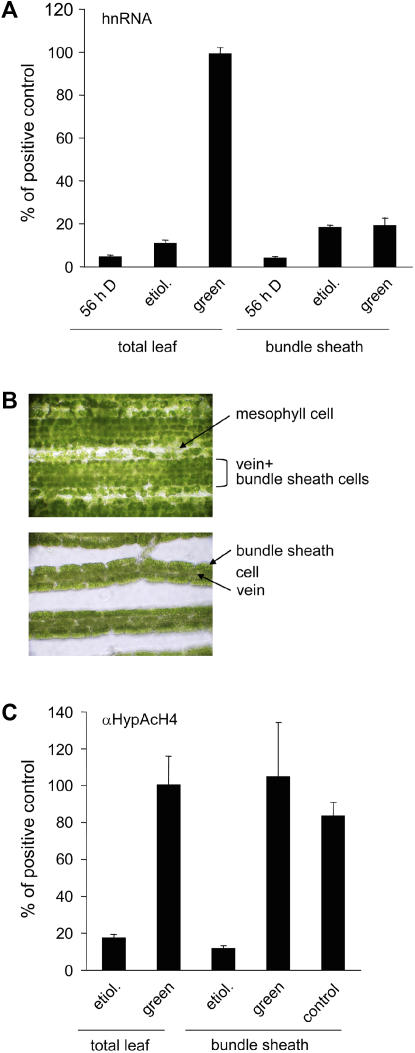
The maize leaf blade contains two photosynthetically active tissues; the mesophyll and the bundle sheath. Available data indicate that the C4-PEPC gene is preferentially transcribed in mesophyll cells and that transcript accumulation is very low in bundle sheath cells (Sheen, 1999; Hahnen et al., 2003). We used the established hnRNA system to verify these results (Fig. 6A). The total leaf samples showed the typical pattern with three different levels of activity. In green plants grown for 56 h in the dark (56-h D), transcription was at the limit of detection. Etiolated plants that were never exposed to light showed intermediate transcription and illuminated green plants showed high transcription. In isolated bundle sheath cells, the difference between the dark-treated green plants and the etiolated plants could be reproduced. However, the transcription in green bundle sheath cells remained on the level observed in the bundle sheath of etiolated plants and thus no light induction of transcription could be observed in this tissue.
Figure 6.
Histone H4 hyperacetylation at the C4-PEPC promoter in bundle sheaths is triggered by light independent of transcription. Etiolated = grown in complete darkness and harvested in the dark; green = seedlings were grown in the normal diurnal rhythm and harvested 4 h after onset of illumination; 56-h D = green plants that were darkened for 56 h; leaf = total leaves; bundle sheath = isolated bundle sheath strands (see Fig. 6B). A, Quantification of C4-PEPC hnRNA synthesis. hnRNA was amplified from cDNA with a primer system specific for an intron (C3). A dilution series of PCR products of known concentrations was used as a standard. The data for etiolated total leaves were taken from Figure 1. B, Photographic representation of leaves after cross-linking and limited enzymatic digestion. The top photograph shows the preparation before manual sorting of bundle sheath strands and the bottom photograph shows isolated bundle sheath strands (magnification is 200×). The different tissues are labeled in the figure. C, Histone H4 hyperacetylation at the C4-PEPC promoter position P2. Values were normalized for the amount of input. Input is the amount of chromatin subjected to immunoprecipitation (see also “Materials and Methods”). Control = leaves were enzymatically degraded but the different leaf tissues were not separated manually. All values were further normalized for the amount of hnRNA or precipitated chromatin, respectively, in green total leaves. The data points are based on four independent experiments. Vertical lines indicate ses.
For analysis of the hyperacetylation of histone H4 in bundle sheath cells, we established a different preparation method. Total leaves were first treated with a cross-linking reagent to fix the current acetylation state. Afterward, bundle sheath strands were isolated by limited enzymatic digestion and manual sorting. Photographs of cross-linked and digested leaves before (top image) and after (bottom image) tissue separation are shown in Figure 6B. Unseparated leaves contained veins surrounded by bundle sheath strands and interveining mesophyll cells. After separating the strands, the mesophyll cells were completely removed and only veins and bundle sheath cells remained. Each of the preparations was examined microscopically for the absence of mesophyll cells. The pattern of histone H4 hyperacetylation was unexpectedly identical in isolated bundle sheath strands and total leaves (Fig. 6C). Low levels were observed in etiolated tissues and high levels were found in illuminated green tissues. This result was not an artifact of the preparation method because control samples from total leaves that had been subjected to the same enzymatic digestion, but where the cell types were not separated, showed a similar degree of acetylation (Fig. 6C, control). We conclude that histone acetylation at the C4-PEPC promoter in bundle sheath cells was induced by light even though transcription was not.
DISCUSSION
Three Activity Levels of the C4-PEPC Promoter
We have manipulated the transcriptional activity of the C4-PEPC gene by three different means in a combinatorial manner. The gene responds to the illumination state of the plant, the nitrogen supply, and is expressed in a cell type-specific manner (see also introduction). Importantly, the two photosynthetic cell types investigated are both part of the leaf blade and therefore receive identical light stimuli. Based on these studies, we can discriminate three different levels of transcription of the C4-PEPC gene. The first level is an almost complete inactivation in green plants that were kept in the dark for extended periods. A clearly higher intermediate transcriptional activity can be observed in etiolated leaves. A similar level of C4-PEPC transcription was also measured in detached leaves that were treated with the cytokinin Zeatin and nitrate in the absence of light (Fig. 5), and conversely in illuminated leaves in the absence of Zeatin/nitrate (Fig. 4). Furthermore, bundle sheath cells isolated from both green and etiolated leaves show intermediate C4-PEPC transcriptional activity (Fig. 6). Thus, the intermediate state is induced when only some, but not all, of the stimuli necessary for full transcription are available. Full transcriptional activity is only observed in mesophyll cells under optimal nitrogen supply and light availability. The pattern with three activity levels is reminiscent of the situation described for the light-induced plastocyanin gene in pea (Pisum sativum), where the lowest activity is found in roots, a basal activity in etiolated plants, and activated transcription in green plants (Chua et al., 2001).
Induction of the highest activity level by light is a fast process, whereas the reciprocal loss of transcriptional activity in the dark is very slow (Fig. 2; Markelz et al., 2003). This is not due to high transcript stability because the mRNA levels correlate well with the amounts of hnRNA, indicating that RNA synthesis is the major determinant of the total amount of C4-PEPC transcripts. Thus, the gene is not diurnally regulated by light. This is in accordance with previous nuclear run-on analyses also indicating high transcriptional activity of the C4-PEPC gene after 12 h of darkness (Suzuki et al., 1994).
Histone Acetylation at the C4-PEPC Promoter
We have shown that the difference in transcriptional activity between etiolated plants (intermediate activity) and green plants (highest activity) is correlated with an increase in the acetylation of both histones H3 and H4 at the promoter and coding regions of the C4-PEPC gene (Fig. 1). However, at a position only two nucleosomes downstream of the transcription unit, no changes in histone acetylation were found (Fig. 1). Thus, histone acetylation is seemingly limited to the gene region. The limit of acetylation upstream of the promoter could not be determined because no further sequence information was available. A different pattern was described for the pea plastocyanin gene (Chua et al., 2001). Here, acetylation of H4 was found in both the promoter and the transcribed region, whereas H3 was mainly acetylated at the promoter. However, a genome-wide study in Drosophila indicated an almost perfect correlation between H3 and H4 acetylation in coding regions (Schubeler et al., 2004).
Interestingly, the degree of acetylation of histones H3 and H4 was clearly reduced in the 5′ part of the coding sequence (C1) compared to the promoter-proximal position P2 and position C2, which are three and seven nucleosomes upstream or downstream of the C1 position, respectively (Fig. 1A). By immunoprecipitation with an antibody to the C-terminal domain of H3, we were able to exclude that this was due to removal of nucleosomes from DNA as has been shown before (e.g. for the activated pho5 promoter in yeast [Saccharomyces cerevisiae]; Svaren and Horz, 1997). The nucleosome density at C1 is identical or even higher compared to the surrounding positions. Two interpretations are compatible with the available data: The acetylation level could in fact be reduced at this position. This interpretation would be in accordance with data from Liu et al. (2005), who have shown that nucleosomes adjacent to the transcription start site of active genes are hypoacetylated compared to the surrounding regions. Or, alternatively, modified histone epitopes at position C1 could have been masked by the presence of cross-linked chromatin binding factors, as has been speculated before for the induction of histone methylation on flowering locus C in Arabidopsis (Arabidopsis thaliana) during vernalization (Bastow et al., 2004). We favor the latter scenario because treatment of green leaves with the strong HD inhibitor butyrate induced a massive increase in overall histone acetylation, but did not alter the relative signal distribution when comparing positions P2 and C1 (data not shown).
The nucleosome density on the C4-PEPC gene is not clearly affected by illumination, although there is a tendency for a decrease in green plants compared to etiolated plants (Fig. 1C). However, in both etiolated and green plants, we measured a reduction in nucleosome density at the promoter compared to the transcribed region. We have previously shown that accessibility of the promoter chromatin for restriction endonucleases is enhanced by illumination of etiolated plants (Kalamajka et al., 2003). We can now add that this effect is due to changes in the interaction of histones and DNA or the precise repositioning of nucleosomes rather than to a complete removal of nucleosomes from the promoter.
Histone acetylation is rapidly induced in illuminated plants concomitant with an increase in transcription. In the opposite experiment, gene transcription and histone acetylation are maintained in green plants over extended periods of darkness (Fig. 2). The time course is much longer than the turnover rate of histone acetylation that has been estimated to be in the range of a few minutes to 1 h with H4 hyperacetylation having the fastest turnover rate (Waterborg, 2002). This means that histones at the promoter have to be actively acetylated in the dark. Furthermore, pharmacological inhibition of HD activity is sufficient to induce a degree of H4 hyperacetylation at the promoter in the dark that is almost identical to the physiologically active state (Fig. 3). We therefore conclude that histone acetyltransferases remain active at the C4-PEPC promoter in darkened plants over extended time periods. Accordingly, light-induced histone acetylation should be brought about by repression of HD activity.
Histone acetylation accompanies the light-activated boost of transcription. We cannot see such changes when investigating the transition from the lowest activity state to the intermediate activity state (e.g. Zeatin activation in the dark; Fig. 5, columns 3 and 4). These studies were limited to the hyperacetylation of H4 at the proximal promoter. However, in etiolated plants, a second situation where the promoter also shows intermediate activity, histone acetylation was investigated in more detail. The additional use of antibodies that recognize acetylated H3 and H4 in a more unspecific manner (i.e. independent of the specific position of the acetylated Lys residue on the N-terminal histone tail) did not reveal evidence for any acetylation of the C4-PEPC gene above background and all measurements were close to the detection limit (Fig. 1). In contrast, Ng et al. (2005) have recently shown that different patterns of histone acetylation can be detected during different phases of promoter activation. Specific Lys residues are acetylated on the N-terminal tail of H4 during potentiation and activation of the phaseolin promoter, respectively. Future studies will reveal whether any histone modification at the C4-PEPC gene is associated with the induction of the intermediate activity level.
Illumination Is Necessary and Sufficient for H4 Hyperacetylation
Under all light regimes tested, histone acetylation correlates with transcriptional activity at the C4-PEPC locus. However, this is only true for total leaves with optimal nitrogen supply and we have provided several examples of loss of concurrence of both effects: In illuminated bundle sheath cells, steady-state transcriptional activity is much lower than in total leaves, but histone acetylation is high and indistinguishable from total leaves (Fig. 6). Furthermore, nitrogen depletion of detached leaves represses transcription of the C4-PEPC gene, but not H4 hyperacetylation in the light (Fig. 4). This could still be explained by different rates of promoter inactivation and histone deacetylation. For instance, Kouskouti and Talianidis (2005) have shown that histone modifications defining active genes can persist after transcriptional inhibition by α-Amanitin treatment for at least 5 h and even through mitosis. We therefore additionally tested the opposite setup where darkened leaves are first depleted for the nitrogen signal and afterward illuminated. In this experiment, transcription was only slightly enhanced by the light signal, but H4 hyperacetylation was fully induced (Fig. 5, column 5). Combination of these experiments conclusively shows that illumination is necessary and sufficient to induce hyperacetylation of histone H4 at the C4-PEPC promoter regardless of the input from additional regulatory stimuli and therefore independent of the transcriptional state of the gene.
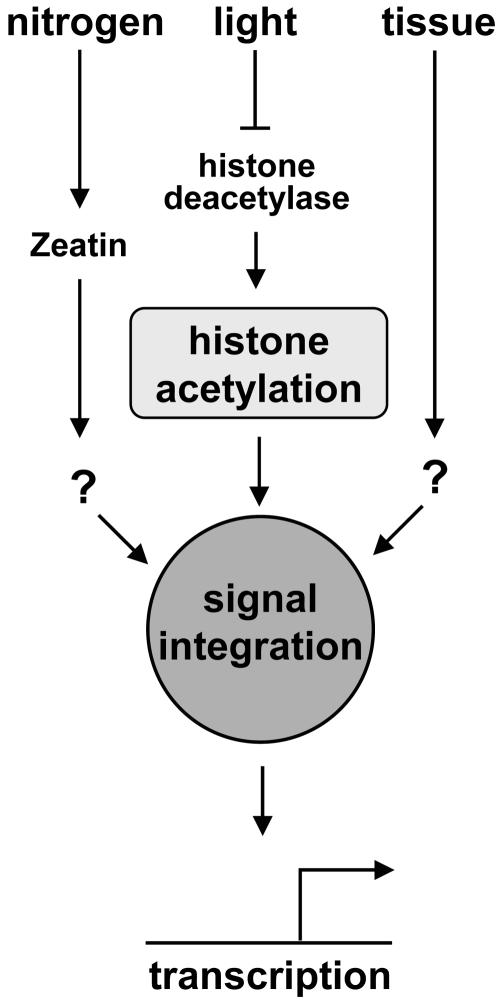
Figure 7 recapitulates our current working model for the activation of the C4-PEPC promoter. Illumination acts negatively on HD and the promoter becomes hyperacetylated. This is an early event in gene activation and independent from other tested stimuli that are transduced to the promoter chromatin by so far unknown mechanisms. Nitrogen, light, and the developmental program act additively and not in a linear cascade because intermediate transcriptional activity is possible in the absence of some of the positive stimuli. Nevertheless, histone acetylation is tightly linked to the highest activation state because it transduces the important light stimulus. Only downstream from histone acetylation all signals are integrated into one transcriptional response. When plants are shifted to darkness, but the other positive stimuli remain, the promoter stays highly active until the deacetylase activity is sufficient to remove the hyperacetylation signal. Because this process is slow, the response of the gene to darkness is retarded. However, the slow response is not an inherent property of the signal integrator because illumination rapidly induces transcriptional activity and because depletion of the nitrogen signal is capable of repressing transcription within only 3 h.
Figure 7.
A model of the impact of histone acetylation on the activation of the C4-PEPC gene in maize. Nitrogen availability, tissue specificity, and illumination act independently on the C4-PEPC promoter. Only illumination is necessary and sufficient to induce histone acetylation. This is probably brought about by inhibition of HD activity. The integration of all signals is downstream of the control of histone acetylation.
This scenario is unexpected because very good correlation between histone acetylation and transcription has been described in many systems. For instance, Chua et al. (2004) have shown that the degree of H4 acetylation correlates well with transcription in tobacco (Nicotiana tabacum). An analysis of the complete Drosophila genome also revealed that the degree of histone H4 acetylation on a gene is strongly correlated to gene transcription. Furthermore, all histone modifications linked to activity, including the trimethylation of Lys 4 on histone H3, show an almost identical distribution (Schubeler et al., 2004). Similar results have also been obtained with human and mouse cells (Bernstein et al., 2005). A recent study by Dion et al. (2005) in yeast suggests simply that the number of Lys residues acetylated on the N-terminal tail of H4 controls the activity of a gene without any impact of a specific pattern of acetylation as would be predicted by the histone code hypothesis (Strahl and Allis, 2000).
All of these experiments have been performed with single cell types under optimal growth conditions. If our experiments would have been designed similarly, the result would have been identical with a perfect correlation of histone acetylation and transcription at the C4-PEPC gene. By using entire leaves with different tissues that are exposed to different external stimuli, we can identify light as the stimulus triggering histone acetylation at the investigated locus. We conclude that an understanding of the histone code requires analysis of signal integration on promoters. Further experiments will show whether this example describes a common pattern for C4-specific genes or for light-activated transcription in plants in general.
MATERIALS AND METHODS
Sequence Assembly
The sequence of the C4-PEPC gene from maize (Zea mays) was deduced from GenBank accession X15642. The overlapping genome survey sequence tuc11-12-04.2773.1 was added to the 5′ end and tuc11-12-04.181719.1 to the 3′ end, respectively (http://www.plantgdb.org).
Plant Material and Growth Conditions
Maize cv Montello was cultivated similar to as described in Markelz et al. (2003), with a diurnal rhythm of 16 h of illumination at 25°C and 8 h of darkness at 20°C. The plants were illuminated with Osram Superstar HQI-T 400W/DH lamps. The photon flux density was between 120 and 180 μmol m−2 s−1. Seedlings were grown in soil (ED 73; Werkverband) containing 30% sand for 10 to 12 d with varying light-dark regimes (as indicated in the figures) or in complete darkness (etiolated plants).
Preparation of Bundle Sheath Strands
For ChIP analysis from isolated bundle sheath strands, leaves were treated with formaldehyde as described below and afterward incubated in SMC buffer (0.5 m sorbitol, 5 mm MES, and 10 mm CaCl2, pH 5.8) containing 15% w/v Rohament CL (AB Enzymes), 10% w/v Rohament PL (AB Enzymes), and 0.6% w/v Macerozyme R-10 (Serva) for 2.5 h at 25°C. Mesophyll protoplasts and remaining epidermal strips were sorted out manually. The quality of each preparation was evaluated microscopically. For gene expression analysis, bundle sheath strands were isolated mechanically as described before (Hahnen et al., 2003).
RNA Preparation and Reverse Transcription
RNA was prepared from tissues following the TRIzol protocol (Chomczynski, 1993). RNA amounts were estimated from dilution series on a nondenaturing agarose gel. One unit of DNAseI (Roche Applied Science) per microgram of RNA and MgCl2 to a final concentration of 2 mm were added and reactions were incubated for 15 min at 37°C, followed by a denaturation step of 15 min at 70°C to remove traces of contaminating DNA. cDNA synthesis was performed with approximately 1 μg of total RNA and 50 pmol of random nonamer primer. Reactions were incubated for 5 min at 70°C and cooled down on ice before adding 200 units of Moloney murine leukemia virus reverse transcriptase (Promega) and 1 mm dNTP in reaction buffer as specified by the manufacturer. The size of the amplified regions was confirmed by gel electrophoresis and products were sequenced to ensure specificity of the assay.
Quantitative Real-Time PCR
Quantitative PCR was performed on an ABI PRISM 7000 sequence detection system (Applied Biosystems) using SYBR Green fluorescence (qPCR core kit for SYBR Green I; Eurogentec) for detection. Oligonucleotides were purchased from Metabion. Sequences are as given in Table I. Dilutions of template DNA were stabilized with a final concentration of 100 ng μL−1 bovine serum albumin. Amplification conditions were 10 min of initial denaturation at 95°C, followed by 40 cycles of 15 s at 95°C, 30 s at 59°C, and 1 min at 72°C. General amplification conditions were 3 mm MgCl2 and 300 nm of each oligonucleotide. For the amplification system P1, C2, and C3 (see Table I; Fig. 2), and 5 mm MgCl2 were used. For the amplification system P1, the primer concentration was 250 nm. For the amplification system I, the MgCl2 concentration was 6 mm and the primer concentration was 600 nm. Systems P2, C1, and C2 were supplemented with 1 m Betain and the C3 system with 1.5 m Betain. The size of the amplified regions was confirmed by gel electrophoresis and products were sequenced to ensure specificity of the assay.
Table I.
Oligonucleotides used in this study
| Name/Function | Sequence (5′ → 3′) |
|---|---|
| P1 f | gta caa atg agg tgc cgg att gat g |
| P1 r | cgg cca tgg cat gat aca att ctc a |
| P2 f | cga ttg ccg cca gca gt |
| P2 r | gaa ccg gct gtg gct gag |
| C1 f | ctg taa tgc atg cag gtc cag gag t |
| C1 r | atg gag ctc gcc acg agg atg g |
| C2 f | ttc aca gat cca agc agc ctt cag |
| C2 r | gat gta gct cat ccc ata gcg cat |
| C3 f/C4-PEPC hnRNA f | gta tgc tgc cat tgc cca ttg c |
| C3 r/C4-PEPC hnRNA r | gtc tcc ggt gta gcc tga tag tga |
| I f | gga agg gga cac tag aga tgt cag |
| I r | ctc ctc aga aac aag tgt gat gtc ca |
| C4-PEPC mRNA f | aga act caa gcc ctt tgg gaa gc |
| C4-PEPC mRNA r | gtc ggc gaa ctc ctt gga cag c |
Zeatin and TSA Treatment
For nitrogen depletion experiments, 10- to 12-d-old leaves were detached 1 cm above the laminar joint under water and placed into tap water or a solution containing 5 μm trans-Zeatin (Sigma) and 16 mm KNO3 (Sugiharto et al., 1992). For inhibition of HDs, detached leaves were treated with a 300 μm TSA solution (ICS) for 12 h in the dark. This is the lowest concentration of TSA that induces the maximal level of global histone hyperacetylation as determined by western analysis with antibodies specific for acetylated histones (data not shown).
ChIP
ChIP was performed according to Bowler et al. (2004) with the following modifications: 3 g of leaf material from 10- to 12-d-old maize seedlings were harvested and cross-linked by vacuum infiltration in extraction buffer 1 supplemented with 3% v/v formaldehyde. The material was ground, resuspended in extraction buffer 2, and incubated for 15 min at 4°C. After purification, the nuclei were resuspended in nuclei lysis buffer containing 25 mm Tris-HCl, pH 8.0, 5 mm EDTA, 0.5% (w/v) SDS, 0.1 mm phenylmethylsulfonyl fluoride, and 1× Complete (Roche Applied Science). The purified chromatin was sheared by sonication using a G 70 sonifier (Bandelin Electronic), 6 × 20 bursts, 25% power, cycle = 50. The average fragment size was approximately 700 bp. The sheared chromatin solution was diluted 10-fold with ChIP dilution buffer (50 mm Tris-HCl, pH 8.0, 1 mm EDTA, 150 mm NaCl, 0.1% [w/v] Triton X-100) and precleared with 40 μL protein A agarose (Roche Applied Science). Precleared chromatin was split into five aliquots of 400 μL for immunoprecipitation and one aliquot of 40 μL for determination of the amount of input. The chromatin aliquots were added to 30 μL protein A agarose and 5 μL antihyperacetylated histone H4 (Penta; 06–946, antigen sequence AGGAcKGGAcKGGAcKGGAcKGGAcKGGC), 11 μL antiacetyl-histone H3 (06–599, antigen sequence ARTKQTARAcKSTGGAcKAPRKQLC), 11 μL antiacetyl-histone H4 (06–866, antigen sequence AGGAcKGGAcKGMGAcKVGAAcKRHS; all Upstate), or 1 μL antihistone H3 (ab1791; Abcam; antigen sequence not available), respectively, and incubated for 2.5 h at 4°C on an overhead centrifuge. The control serum was derived from rabbits immunized with an unrelated protein from potato (Solanum tuberosum). After washing, the bead-bound complexes were released and de-cross-linked by incubation in elution buffer (62.5 mm Tris-HCl, pH 6.8, 200 mm NaCl, 2% [w/v] SDS, and 10 mm dithiothreitol) at 65°C overnight. The coprecipitated DNA was purified using the MSB Spin PCRapace kit (Invitek). Typically, 5 μL of eluted DNA were used as a template for quantitative PCR analysis. All results were normalized for the amount of chromatin used for immunoprecipitation (% of input).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers X15642, CG252268, and CC426077.
Acknowledgments
We are grateful to Fritz Kreuzaler for continuous support and the opportunity to perform these experiments at his institute. We would like to thank Maike Stam and Max Haring for collaboration during the establishment of the ChIP technique and Alan Slusarenko and our coworkers at the Institute for Biology I for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. Pe819 to C.P.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Christoph Peterhänsel (cp@bio1.rwth-aachen.de).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.080457.
References
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167 [DOI] [PubMed] [Google Scholar]
- Berger SL (2002) Histone modifications in transcriptional regulation. Curr Opin Genet Dev 12: 142–148 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ III, Gingeras TR, et al (2005) Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181 [DOI] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J (2004) Chromatin techniques for plant cells. Plant J 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Chomczynski P (1993) A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 15: 532–534 [PubMed] [Google Scholar]
- Chua YL, Brown AP, Gray JC (2001) Targeted histone acetylation and altered nuclease accessibility over short regions of the pea plastocyanin gene. Plant Cell 13: 599–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua YL, Mott E, Brown APC, MacLean D, Gray JC (2004) Microarray analysis of chromatin-immunoprecipitated DNA identifies specific regions of tobacco genes associated with acetylated histones. Plant J 37: 789–800 [DOI] [PubMed] [Google Scholar]
- Delany AM (2001) Measuring transcription of metalloproteinase genes: nuclear run-off assay vs analysis of hnRNA. Methods Mol Biol 151: 321–333 [DOI] [PubMed] [Google Scholar]
- Dion MF, Altschuler SJ, Wu LF, Rando OJ (2005) Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci USA 102: 5501–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elferink CJ, Reiners JJ Jr (1996) Quantitative RT-PCR on CYP1A1 heterogeneous nuclear RNA: a surrogate for the in vitro transcription run-on assay. Biotechniques 20: 470–477 [DOI] [PubMed] [Google Scholar]
- Flaus A, Owen-Hughes T (2004) Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr Opin Genet Dev 14: 165–173 [DOI] [PubMed] [Google Scholar]
- Hahnen S, Joeris T, Kreuzaler F, Peterhänsel C (2003) Quantification of photosynthetic gene expression in maize C3 and C4 tissues by real-time PCR. Photosynth Res 75: 183–192 [DOI] [PubMed] [Google Scholar]
- Jang JC, Sheen J (1994) Sugar sensing in higher plants. Plant Cell 6: 1665–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamajka R, Hahnen S, Cavalar M, Töpsch S, Weier D, Peterhänsel C (2003) Restriction accessibility in isolated nuclei reveals light-induced chromatin reorganization at the PEPC promoter in maize. Plant Mol Biol 52: 669–678 [DOI] [PubMed] [Google Scholar]
- Kausch AP, Owen TP, Zachwieja SJ, Flynn AR, Sheen J (2001) Mesophyll-specific, light and metabolic regulation of the C4 PPCZm1 promoter in transgenic maize. Plant Mol Biol 45: 1–15 [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98: 285–294 [DOI] [PubMed] [Google Scholar]
- Kouskouti A, Talianidis I (2005) Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J 24: 347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ (2005) Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol 3: e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusser A, Kolle D, Loidl P (2001) Histone acetylation: lessons from the plant kingdom. Trends Plant Sci 6: 59–65 [DOI] [PubMed] [Google Scholar]
- Markelz NH, Costich DE, Brutnell TP (2003) Photomorphogenic responses in maize seedling development. Plant Physiol 133: 1578–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T, Langdale JA (1992) Developmental genetics of C4 photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 43: 25–47 [Google Scholar]
- Ng DWK, Chandrasekharan MB, Hall TC (2005) Ordered histone modifications are associated with transcriptional poising and activation of the phaseolin promoter. Plant Cell 18: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [DOI] [PubMed] [Google Scholar]
- Reed R (2003) Coupling transcription, splicing and mRNA export. Curr Opin Cell Biol 15: 326–331 [DOI] [PubMed] [Google Scholar]
- Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, et al (2004) The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev 18: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J (1990) Metabolic repression of transcription in higher plants. Plant Cell 2: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J (1999) C4 gene expression. Annu Rev Plant Physiol Plant Mol Biol 50: 187–217 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Struhl K (1999) Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98: 1–4 [DOI] [PubMed] [Google Scholar]
- Sugiharto B, Burnell JN, Sugiyama T (1992) Cytokinin is required to induce the nitrogen-dependent accumulation of mRNAs for phosphoenolpyruvate carboxylase and carbonic anhydrase in detached maize leaves. Plant Physiol 100: 153–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki I, Cretin C, Omata T, Sugiyama T (1994) Transcriptional and posttranscriptional regulation of nitrogen-responding expression of phosphoenolpyruvate carboxylase gene in maize. Plant Physiol 105: 1223–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svaren J, Horz W (1997) Transcription factors vs. nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem Sci 22: 93–97 [DOI] [PubMed] [Google Scholar]
- Waterborg JH (2002) Dynamics of histone acetylation in vivo: a function for acetylation turnover? Biochem Cell Biol 80: 363–378 [DOI] [PubMed] [Google Scholar]