Abstract
Hic-5 (hydrogen peroxide–inducible clone-5) is a focal adhesion protein that is involved in cellular senescence. In the present study, a yeast two-hybrid screen identified Hic-5 as a protein that interacts with a region of the glucocorticoid receptor that includes a nuclear matrix–targeting signal and the τ2 transcriptional activation domain. In transiently transfected mammalian cells, overexpression of Hic-5 potentiated the activation of reporter genes by all steroid receptors, excluding the estrogen receptor. The activity of the estrogen receptor and the thyroid hormone receptor was stimulated by Hic-5 in the presence but not in the absence of coexpressed coactivator GRIP1. In biochemical fractionations and indirect immunofluorescence assays, a fraction of endogenous Hic-5 in REF-52 cells and transiently expressed Hic-5 in Cos-1 cells was associated with the nuclear matrix. The C-terminal region of Hic-5, which contains seven zinc fingers arranged in four LIM domains, was required for interaction with focal adhesions, the nuclear matrix, steroid receptors, and the τ2 domain of glucocorticoid receptor. The N-terminal region of Hic-5 possesses a transcriptional activation domain and was essential for the coactivator activity of Hic-5. Given the coexisting cytoplasmic and nuclear distributions of Hic-5 and its role in steroid receptor–mediated transcriptional activation, it is proposed that Hic-5 might transmit signals that emanate at cell attachment sites and regulate transcription factors, such as steroid receptors.
INTRODUCTION
The nuclear matrix provides a framework for organizing large macromolecular assemblies that carry out the fundamental nuclear processes of DNA replication, transcription, and splicing. Given the enormous variety of developmental, cell type–specific, and hormonal factors that influence these processes, it seems likely that the nuclear matrix maintains some degree of plasticity in its structure and composition. The composition of low-abundance nuclear matrix proteins of unknown function varies between different cell types (Fey and Penman, 1988; Getzenberg and Coffey, 1990). The development of tumors within particular tissues, such as the prostate, appears to be associated with alterations in the protein composition of the nuclear matrix (Partin et al., 1993). In some cases, the identity of specific nuclear matrix proteins has been established. The association of specific transcription factors with the nuclear matrix has also been shown to vary between different cell types (Van Wijnen et al., 1993). Because some transcription factors partition between the nuclear matrix and the soluble compartments of the nucleus (Van Wijnen et al., 1993; Sun et al., 1994), it seems likely that nuclear matrix binding of transcription factors is not a static process but includes the dynamic exchange between distinct nuclear compartments (Tang and DeFranco, 1996; Mancini et al., 1999).
What types of signaling pathways might regulate nuclear matrix composition or function? A number of studies have suggested that a solid-state pathway may link the nuclear matrix to signals that emanate from the extracellular matrix (ECM) (Bissell et al., 1999). In fact, changes in the organization of specific nuclear matrix proteins occur in response to ECM-directed changes in the architecture of human mammary epithelial cells (Lelievre et al., 1998). These ECM-dependent alterations in nuclear matrix protein compartmentalization may be associated with specific gene regulatory events that are influenced by the ECM (Myers et al., 1998). Although much is known about the signaling pathways that are mobilized upon the interactions of ECM components with cell surface receptors of the integrin family (Guan and Chen, 1996), there are large gaps in our understanding of how these changes are coupled with changes in gene expression or nuclear organization.
At focal adhesions, the ECM makes specialized contacts with the actin cytoskeleton and focal adhesion kinase (FAK) (Cary and Guan, 1999). Upon activation of integrins, FAK activity is increased, leading to its autophosphorylation and the mobilization of numerous downstream signaling pathways. Signaling molecules and regulators that are known to be associated with FAK include Src, phosphatidylinositol 3-kinase, p130Cas, Grb2, and Graf. In some cases, downstream targets of the signaling molecules activated by FAK in response to integrin activation are known (Cary and Guan, 1999). However, for some FAK-associated proteins, such as the LIM domain protein paxillin, downstream targets relevant for ECM-directed signaling have not been identified, and it is unclear how and where they function in the overall ECM–integrin signaling cascade.
Steroid receptors were the first transcription factors found to bind to the nuclear matrix (Barrack and Coffey, 1980), mainly because of the availability of high-specific-activity radiolabeled steroids. The interaction between steroid receptors and the nuclear matrix is hormone dependent and involves saturable, high-affinity interactions (Barrack, 1987). Discrete domains of steroid receptors required for nuclear matrix binding have been identified. For the androgen receptor (AR) and glucocorticoid receptor (GR), the DNA-binding domain (DBD) and hormone-binding domain (HBD) contribute to nuclear matrix binding, although the relative contributions of these domains differ between these two highly related proteins (Barrack, 1987; van Steensel et al., 1995; Tang et al., 1998). The relative proportion of steroid receptors associated with the nuclear matrix varies in different target tissues, particularly for sex steroid receptors (Barrack, 1987). It has been proposed that specific acceptor proteins may mediate this cell type–specific or tissue-specific binding of steroid receptors to the nuclear matrix (Barrett and Spelsberg, 1999). A candidate steroid receptor nuclear matrix acceptor protein has been isolated from chick oviduct (Schuchard et al., 1991), but its role in steroid receptor regulation of transcription has yet to be established.
We previously identified a minimal nuclear matrix targeting signal (NMTS) sequence within GR that includes both its DBD and its τ2 transcriptional activation domain (Tang et al., 1998). Using functional assays, we identified heterogeneous nuclear ribonucleoprotein U/scaffold attachment factor-A, which is known to be an RNA- and DNA-binding component of the nuclear matrix (Fackelmayer et al., 1994), as a potential GR NMTS-binding protein (Eggert et al., 1997; Tang et al., 1998). Overexpression of heterogeneous nuclear ribonucleoprotein U/scaffold attachment factor-A led to decreased activation of transiently transfected reporter genes by GR. To search for other nuclear matrix proteins that interact with steroid receptors, we performed a yeast two-hybrid screen with the DBD-τ2 NMTS of GR as bait. As reported here, with this screen we have identified a novel steroid receptor–binding protein and transcriptional coactivator, hydrogen peroxide–inducible clone-5 (Hic-5), which not only localizes to the nuclear matrix but also associates with focal adhesions.
MATERIALS AND METHODS
Plasmids
Yeast expression vectors for Gal4 DBD fusion proteins were constructed by inserting PCR-amplified cDNA fragments containing the following coding regions into pGBT9 (Clontech, Palo Alto, CA): GR τ2 (mouse GR amino acids 513–562, including the hinge region and τ2 region, followed by a stop codon) in SmaI–SalI sites; GR DBD-τ2 (mouse GR amino acids 395–562, including the DBD, hinge region, and τ2 region, followed by a stop codon) in SmaI–SalI sites; GR DBD (mouse GR amino acids 395–519) in SmaI–PstI sites; and Hic-5 full length (amino acids 1–444), Hic-5N (amino acids 1–200, including the leucine–aspartic acid [LD] domains), and Hic-5C (amino acids 201–444, including the LIM domains) into EcoRI–SalI sites. Yeast expression vectors for Gal4 activation domain (AD) fusion proteins with Hic-5 and its fragments were constructed by inserting PCR-amplified EcoRI–SalI cDNA fragments encoding Hic-5 full length, Hic-5N, and Hic-5C into pGAD424 (Clontech).
Mammalian expression vectors for Gal4 DBD fusion proteins with Hic-5 and its fragments were constructed by inserting PCR-amplified EcoRI–SalI cDNA fragments encoding Hic-5 full length, Hic-5N, and Hic-5C into pM (Clontech). The mammalian expression vector for full-length Hic-5 with an N-terminal hemagglutinin (HA) epitope tag was constructed by inserting a PCR-amplified EcoRI–XhoI cDNA fragment into pSG5.HA (Chen et al., 1999). pSG5.HA-GRIP1, encoding HA-tagged coactivator GRIP1, was described previously, as were the mammalian luciferase reporter gene plasmids GK1, controlled by Gal4 response elements, MMTV-LUC, controlled by glucocorticoid response elements, MMTV(ERE)-LUC, controlled by an estrogen response element, and MMTV(TRE)-LUC, controlled by a thyroid hormone response element (Chen et al., 1999). Mammalian expression vectors for nuclear receptors and their fragments were described previously as follows: pHE0 for human estrogen receptor (ER), PSVAR0 for human AR, and pCMX.hTRβ1 for human thyroid hormone receptor (TR) β1 (Chen et al., 1999); pKSX for mouse GR (Milhon et al., 1994); p6RMR for rat mineralocorticoid receptor (MR) (Pearce and Yamamoto, 1993); pCMV.hPR-B for human progesterone receptor (PR) B (Boonyaratanakornkit et al., 1998); and pC7mGR(395–533) for a mouse GR fragment including the DBD and hinge region and pC7mGR(395–562) for the mouse GR DBD, hinge region, and τ2 region (Milhon et al., 1997).
Yeast Two-Hybrid System
A mouse 17-d-old embryo cDNA library in the Gal4 AD fusion vector pGAD10 (Clontech) was screened as described previously (Hong et al., 1999), with Gal4 DBD fused to GR DBD-τ2 as bait, encoded by a derivative of plasmid pGBT9. A total of 3 million cDNA clones were screened in yeast strain Hf7c, which has Gal4-controlled reporter genes encoding His3 and β-galactosidase (β-gal). Thirty-two clones were confirmed positive by both growth on plates lacking histidine and β-gal assays. Four of these were identical clones encoding full-length Hic-5 (Shibanuma et al., 1994), including codons 1–444, 19 5′-flanking nucleotides, 219 3′-flanking nucleotides, and a poly(A) region. To measure protein–protein interactions, quantitative yeast two-hybrid assays were performed in yeast strain SFY526 as described previously (Ding et al., 1998).
Mammalian Cell Transfections
Transient transfection of CV-1 cells and luciferase reporter gene assays were performed as described previously (Chen et al., 1999). Where indicated, the following hormones at a final concentration of 100 nM were included during the last 48 h before harvesting the transfected cell cultures: for GR, dexamethasone; for ER, estradiol; for AR, dihydrotestosterone; for MR, corticosterone; for PR, progesterone; for TR, 3,5,5′-triiodo-l-thyronine.
Cos-1 cells and REF-52 cells were grown in DMEM (Life Technologies-BRL, Grand Island, NY) supplemented with 10% FBS (Irvine Scientific, Santa Ana, CA). Cellular ATP depletion was performed as described previously (Tang and DeFranco, 1996) by culturing cells in DMEM with 10 mM sodium azide and 6 mM deoxyglucose for 90 min. For in situ extractions, cells were grown on glass coverslips (22 × 22 mm) on 35-mm Petri plates. Cells were transfected by the calcium phosphate method (Tang et al., 1998) with the use of 2 μg of DNA per plate.
Indirect Immunofluorescence
Indirect immunofluorescence (IIF) assays were carried out as described previously (Matsuya et al., 1998). Briefly, Cos-1 monkey kidney fibroblast or REF-52 rat embryo fibroblast cell lines were fixed with 4% paraformaldehyde and then permeabilized with 0.1% Triton X-100. In transfected Cos-1 cells, an anti-HA mouse mAb (Roche, Indianapolis, IN, number 1583816) was used to recognize the HA-tagged Hic-5 for in situ assays. FITC-conjugated goat anti-mouse immunoglobulin G (Boehringer Mannheim, Indianapolis, IN) was used as a secondary antibody. For REF-52 cells, an anti-Hic-5 rabbit polyclonal antibody (kindly provided by Dr. K. Tachibana, Harvard University, Cambridge, MA) was used for in situ assays. Rhodamine Red–conjugated goat anti-rabbit immunoglobulin G (Jackson ImmunoResearch Laboratories, West Grove, PA) was the secondary antibody. DAPI (Sigma Chemical, St. Louis, MO) was used to visualize DNA in fixed cells.
Nuclear Matrix Preparation and Subcellular Fractionation
For in situ extractions, cells were grown on coverslips and treated as described previously (Tang et al., 1998). Briefly, Cos-1 or REF-52 cells were washed and treated with ice-cold CK buffer (10 mM piperazine-N,N′-bis[2-ethanesulfonic acid], pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 4 mM vanadyl riboside complex, 1.2 mM PMSF, and protease inhibitors). A nuclear matrix fraction was prepared by subjecting cells to DNAse I digestion and ammonium sulfate extraction. For Western blot analyses, analogous extractions were performed with cells in suspension.
Western Blots
Western blot analysis was used to detect endogenous Hic-5 in REF-52 cells and transiently expressed HA-tagged Hic-5 and Hic-5 fragments in various subcellular fractions (Tang and DeFranco, 1996). In each case, Hic-5 levels were compared in whole cell extracts or fractions obtained from equivalent amounts of cells with the use of the polyclonal rabbit HA.11 anti-HA antibody (BabCO, Richmond, CA) or the polyclonal rabbit anti-Hic-5 antibody. Separate blots were probed with a mouse monoclonal lamin B antibody (Oncogene Science, Uniondale, NY) to provide an internal control for nuclear matrix recovery. Primary antibodies were detected with the use of appropriate HRP-conjugated secondary antibodies (Bio-Rad Laboratories, Richmond, CA) and a chemiluminescence detection kit (New England Nuclear, Boston, MA). Where indicated, quantification of scanned Western blot images was performed with the use of NIH Image software version 1.62.
RESULTS
Identification of Hic-5 as a Protein That Binds to the τ2 Activation Domain of GR
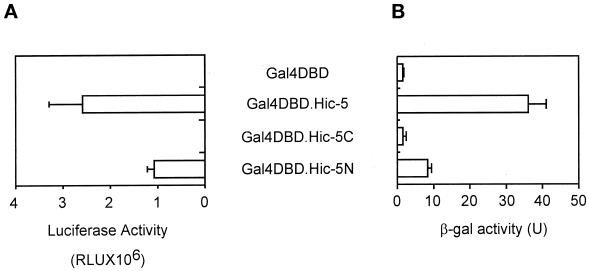
To investigate the mechanism of nuclear matrix targeting, we used the yeast two-hybrid system to screen a mouse embryo cDNA library for clones encoding proteins that interact with a minimum NMTS of GR (Tang et al., 1998), i.e., mouse GR amino acids 395–562, which include the DBD, the hinge region between the DBD and the HBD, and the τ2 region, which overlaps the boundary between the hinge region and the HBD. The clone that produced the strongest interaction encoded full-length Hic-5 (Shibanuma et al., 1994), a previously identified zinc finger protein that can bind DNA and has multiple cellular locations, including the nucleus and focal adhesion complexes (Shibanuma et al., 1997; Fujita et al., 1998; Matsuya et al., 1998). Hic-5 has multiple LD domains in its N-terminal region and four LIM domains (Schmeichel and Beckerle, 1994), each containing two zinc fingers, in its C-terminal region. In quantitative yeast two-hybrid assays, Hic-5 (fused to Gal4 AD) bound to mouse GR amino acids 395–562 (GR DBD, hinge, and τ2) and to mouse GR amino acids 513–562 (hinge and τ2), which were fused to Gal4 DBD (Figure 1A). However, no binding to Gal4 DBD or to GR DBD fused to Gal4 DBD was detected. While this work was in progress, Fujimoto et al. (1999) reported the identification of Hic-5 (designated ARA55) as a protein that bound to the AR HBD. We also observed binding of several full-length steroid receptors (GR, AR, ER, and TR) and/or their HBDs to Hic-5 in vitro (our unpublished results).
Figure 1.
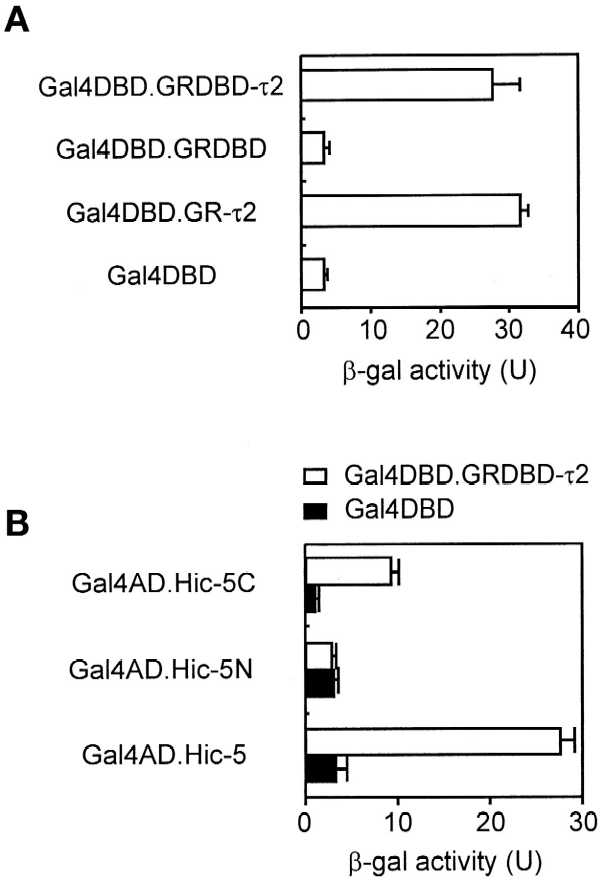
Binding of Hic-5 and its C-terminal fragment to GR τ2 region. (A) Yeast strain SFY526 was transformed with plasmids encoding a Gal4 AD–Hic-5 fusion protein and the indicated Gal4 DBD fusion protein. Cell extracts were assayed for β-gal activity produced by the integrated β-gal reporter gene, which was controlled by Gal4 response elements. Activity is given in units (U) as the mean and SD of triplicate samples. (B) Yeast two-hybrid analyses were conducted as in A with the use of yeast transformed with plasmids encoding the indicated Gal4 fusion proteins. Hic-5N and Hic-5C, the N-terminal (amino acids 1–200) and C-terminal (amino acids 201–444) fragments of Hic-5.
Functional Domains of Hic-5
To investigate which region of Hic-5 bound GR τ2, the N-terminal region containing the LD domains and the C-terminal region containing the LIM domains were separately fused to Gal4 AD. In yeast two-hybrid assays, the C-terminal Hic-5 fragment bound to GR DBD-τ2, but the N-terminal fragment did not (Figure 1B).
To test for a potential transcriptional activation domain, full-length Hic-5 and each of the two fragments of Hic-5 were expressed as Gal4 DBD fusion proteins in yeast and mammalian CV-1 cells. In both cell types, a Gal4 DBD fusion protein containing full-length Hic-5 or the N-terminal Hic-5 fragment activated a reporter gene containing Gal4 response elements, but a fusion protein with the C-terminal Hic-5 fragment had no activity (Figure 2).
Figure 2.
Transcriptional activation domain in Hic-5 and its N-terminal fragment. (A) CV-1 cells were transiently transfected with 0.5 μg of an expression vector for the indicated Gal4 DBD fusion protein and 0.5 μg of luciferase reporter gene GK1, controlled by Gal4 response elements. Luciferase activity of the transfected cell extracts is presented as the mean and SD of three transfected cultures. (B) Yeast were transformed with plasmids encoding the indicated Gal4 DBD fusion protein, and β-gal activity was determined as in Figure 1.
Enhancement of the Activity of a Subset of Nuclear Receptors by Hic-5
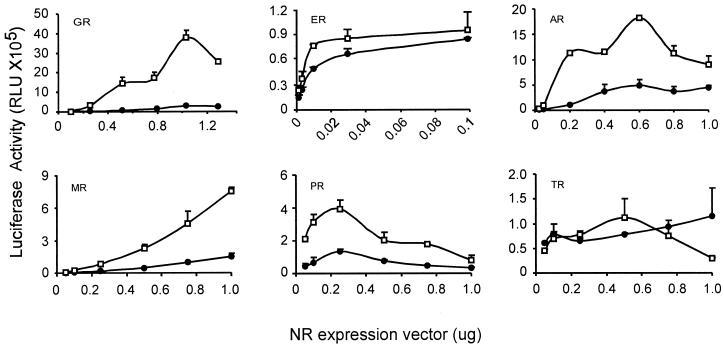
The ability of Hic-5 to bind steroid receptors and the presence of a potential transcriptional activation domain in Hic-5 suggested that it might function as a coactivator for steroid receptors or other nuclear receptors. To test this possibility, varying amounts of six different nuclear receptor expression vectors were transiently transfected in CV-1 cells with reporter genes containing appropriate nuclear receptor–binding elements in the presence and absence of an expression vector for full-length Hic-5. Hic-5 acted as a potent coactivator for GR, AR, MR, and PR but had little or no effect on ER or TR activity (Figure 3). The ability of Hic-5 to stimulate reporter gene activity was dependent on the presence of the steroid receptor. The coactivator effect of Hic-5 was observed over a wide range of steroid receptor expression vector amounts, including subsaturating and saturating amounts. Maximum enhancements observed were as follows: GR, 20-fold; AR, 11-fold; MR, 5-fold; PR, 5-fold; ER, 1.6-fold; TR, 1.4-fold. The failure of Hic-5 to enhance ER function was also observed with a different ER-activated reporter gene containing a thymidine kinase promoter and two estrogen response elements (our unpublished results).
Figure 3.
Hic-5 acts as a coactivator for a subset of nuclear receptors (NR). CV-1 cells were transiently transfected with varying amounts of the indicated nuclear receptor expression vector, 0.5 μg of a suitable reporter gene for each nuclear receptor, and, where indicated, 0.5 μg of expression vector for HA-tagged Hic-5. ●, no Hic-5; □, plus Hic-5. Reporter genes: for GR, AR, MR, and PR, MMTV-LUC; for ER, MMTV(ERE)-LUC; for TR, MMTV(TRE)-LUC.
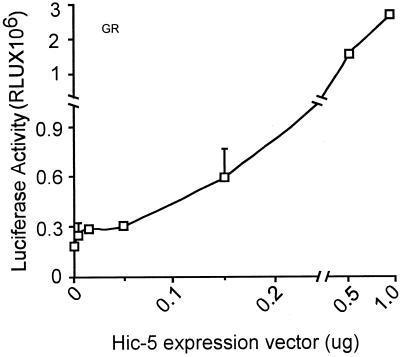
When different amounts of Hic-5 expression vector were cotransfected with a subsaturating amount of GR expression vector, the reporter gene expression increased in a roughly linear manner with the amount of Hic-5 vector used (Figure 4). Deletion of either the N-terminal activation domain of Hic-5 (amino acids 1–200) or the C-terminal steroid receptor–binding domain (amino acids 201–444) caused loss of the coactivator function of Hic-5 (our unpublished results).
Figure 4.
GR activity with varying amounts of Hic-5. CV-1 cells were transiently transfected with 0.25 μg of GR expression vector, 0.5 μg of MMTV-LUC reporter gene, and the indicated amount of expression vector for HA-tagged Hic-5.
Comparison of Hic-5 and GRIP1 Coactivator Effects for Nuclear Receptors
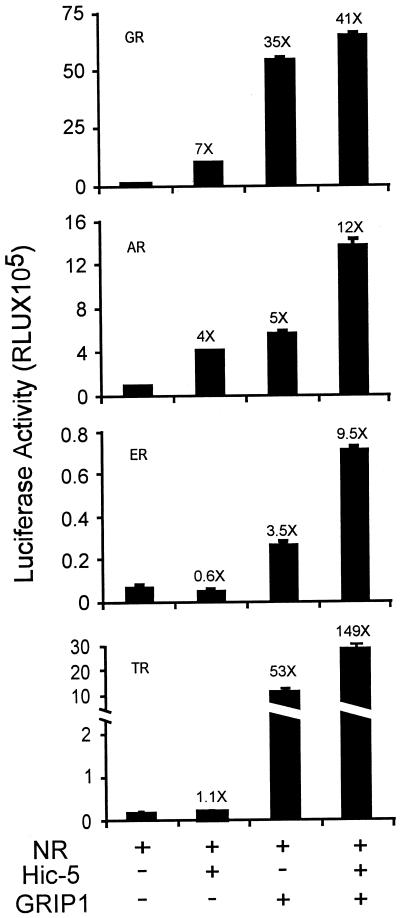
The coactivator effects of Hic-5 for steroid receptors were compared with those of GRIP1 (Hong et al., 1997), a member of the well-characterized 160-kDa family of nuclear receptor coactivators (Torchia et al., 1998; Xu et al., 1999). Hic-5 and GRIP1 each enhanced substantially the function of GR and AR (Figure 5). When both coactivators were used together, the effects were approximately additive (for AR) or less than additive (for GR). As reported previously, GRIP1 was also an effective coactivator for ER and TR (Ding et al., 1998; Voegel et al., 1998), but Hic-5 had no effect on the activity of these nuclear receptors (Figure 3). However, in the presence of GRIP1, Hic-5 caused a further threefold enhancement of ER and TR activity. Thus, the presence of GRIP1 in some way potentiated the effects of Hic-5 with TR and ER.
Figure 5.
Comparison of coactivator activity of GRIP1 and Hic-5. CV-1 cells were transfected with nuclear receptor expression vector as follows: GR, 250 ng; AR, 500 ng; ER, 7.5 ng; TR, 100 ng. Also included were 0.5 μg of suitable reporter gene for each nuclear receptor (as in Figure 3) and, where indicated, 0.5 μg of Hic-5 and/or GRIP1 expression vectors.
Enhancement of GR τ2 Activity by Hic-5
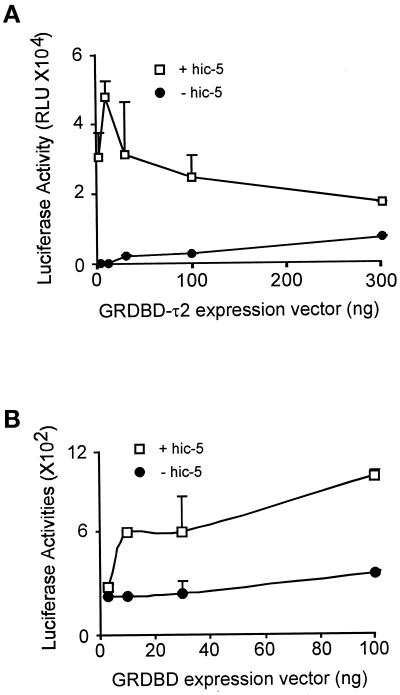
As described above, Hic-5 can bind the isolated GR τ2 domain as well as the intact GR HBD. The isolated GR τ2 region also functions as a weak transcriptional activation domain (Hollenberg and Evans, 1988; Milhon et al., 1997), and the GR DBD-τ2 fragment constitutes a NMTS (Tang et al., 1998). Therefore, we tested Hic-5 as a coactivator for τ2. In transiently transfected CV-1 cells, Hic-5 enhanced the activity of the GR DBD-τ2 fragment (mouse GR amino acids 395–562) by up to 500-fold (Figure 6A). The optimum enhancement by Hic-5 was observed at subsaturating levels of the GR DBD-τ2 expression vectors; the effect of Hic-5 was dramatically decreased at higher levels of GR DBD-τ2. In contrast, the activity of GR DBD alone was enhanced only approximately twofold by Hic-5 at all levels of GR DBD expression vector tested (Figure 6B). Thus, Hic-5 bound to and served as a coactivator for both the intact GR and the isolated τ2 domain of GR. Further studies will be required to determine whether these physical interactions mediate the enhancement of τ2 and GR activity by Hic-5.
Figure 6.
Hic-5 enhances transcriptional activation by the GR τ2 region. CV-1 cells were transiently transfected with the indicated amount of expression vector for GR DBD-τ2 (mouse GR amino acids 395–562) (A) or GR DBD (mouse GR amino acids 395–533) (B) and 0.5 μg of reporter gene MMTV-LUC in the presence (□) or absence (●) of 0.5 μg of Hic-5 expression vector.
Coexisting Cytoplasmic and Nuclear Distributions of Hic-5
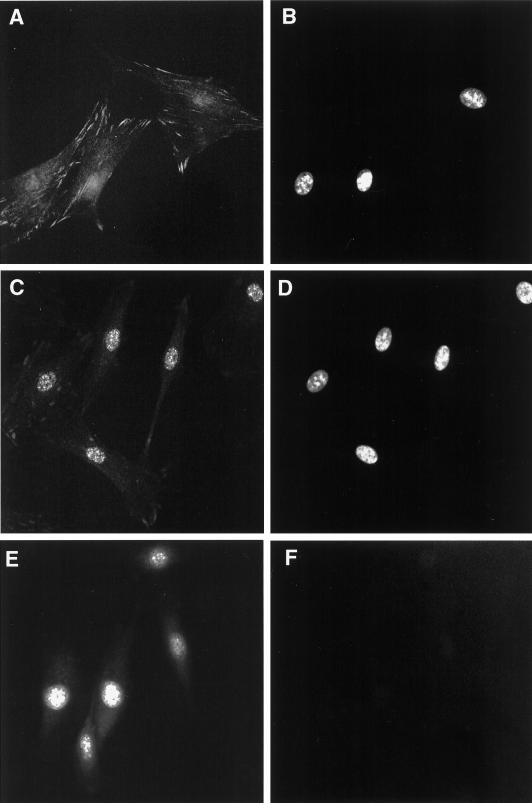
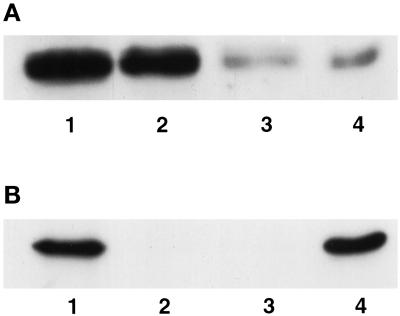
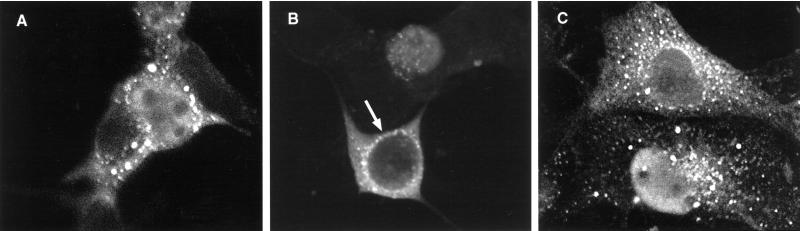
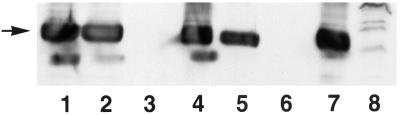
Although Hic-5 was initially identified as a protein associated with focal adhesions, there have been conflicting reports of its localization within the nucleus (Shibanuma et al., 1994; Matsuya et al., 1998; Thomas et al., 1999). In none of these cases was the subnuclear compartmentalization of Hic-5 assessed. With the use of IIF, endogenous Hic-5 was detected in both the cytoplasm and the nuclei of unextracted, fixed REF-52 cells (Figure 7A). The cytoplasmic staining pattern for Hic-5 appeared identical to that previously reported for Hic-5 in REF-52 and other rat fibroblast cell lines and was consistent with the location of Hic-5 in focal adhesions (Fujita et al., 1998; Matsuya et al., 1998; Thomas et al., 1999). The weak nuclear staining of Hic-5 was more readily observed after a high-salt and detergent extraction (i.e., with CK buffer), which eliminated cytoplasm- and focal adhesion–localized Hic-5 (Figure 7C). Further high-salt extraction and DNAse digestion of CK buffer–extracted cells generates a nuclear matrix fraction (Tang et al., 1998). When REF-52 cells were treated in this manner, some endogenous Hic-5 remained in the nuclear matrix fraction (Figure 7E). Western blot analysis (Figure 8) confirmed the IIF results and revealed that ∼20% of Hic-5 in REF-52 cells was associated with the nuclear matrix (Figure 8A, lane 4). The Western blot shown in Figure 8B demonstrates that lamin B, a nuclear matrix protein, was present only in whole cell extracts (lane 1) and in the nuclear matrix fraction (lane 4).
Figure 7.
IIF analysis of endogenous Hic-5 localization in REF-52 cells. REF-52 cells were fixed with paraformaldehyde and processed for IIF analysis with the use of an anti-Hic-5 antibody (A, C, and E), as described previously (Nishiya et al., 1999). DAPI staining of the fields in A, C, and E are shown in B, D, and F, respectively. Before fixation, cells were either permeabilized with low-salt and detergent buffer (C) or further digested with DNAse I and then extracted with ammonium sulfate buffer to generate a nuclear matrix (E).
Figure 8.
Western blot analysis of endogenous Hic-5 in subcellular fractions from REF-52 cells. Proteins present in whole cell extracts (lanes 1), CK buffer–extracted supernatant (lanes 2), DNAse I–digested supernatant (lanes 3), or nuclear matrix pellets (lanes 4) were separated by SDS-PAGE and subjected to Western blot analysis to detect Hic-5 (A) or the nuclear matrix protein lamin B (B).
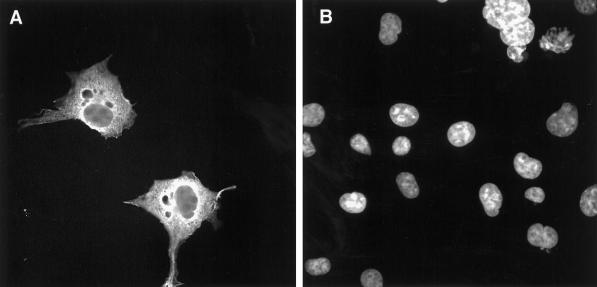
To map the domains of Hic-5 required for its nuclear matrix targeting, we expressed HA-tagged Hic-5 in Cos-1 cells by transient transfection. HA-tagged Hic-5 was localized throughout transfected Cos-1 cells (Figure 9A). Background staining in these experiments was minimal, as evident from the presumed nontransfected cells, which were stained with DAPI (Figure 9B) but exhibited no detectable staining by anti-HA antibody (Figure 9A). When nontransfected cultures were processed for IIF in the presence of anti-HA antibody, all cells showed uniform, low-level background staining (our unpublished results). Focal adhesions are not pronounced in Cos-1 cells plated in the absence of ECM proteins (Angers-Loustau et al., 1999), which most likely accounts for our inability to detect transfected HA-tagged Hic-5 in focal adhesions.
Figure 9.
IIF analysis of HA-tagged Hic-5 localization in transiently transfected Cos-1 cells. Cos-1 cells transiently transfected with HA-tagged Hic-5 were fixed with paraformaldehyde and processed for IIF analysis with an anti-HA antibody (A) with the use of standard procedures (Nishiya et al., 1999). DAPI staining of the field in A is shown in B.
In experiments analogous to those performed with endogenous Hic-5 in REF-52 cells, Cos-1 cells transiently expressing HA-tagged Hic-5 were extracted with detergent-containing CK buffer before fixation. In all positively stained transfected cells, HA-tagged Hic-5 was observed within a large number of cytoplasmic foci that had fairly uniform sizes and shapes (Figure 10A). These cytoplasmic foci of HA-tagged Hic-5 observed in extracted Cos-1 cells do not represent focal adhesions because they were not associated with actin filaments (our unpublished results). Furthermore, endogenous FAK staining was lost upon CK buffer extraction (our unpublished results), indicating that this extraction disrupted focal adhesion complexes. This pool of HA-tagged Hic-5 may either represent a transient intermediate in the intracellular trafficking of Hic-5 or be due to cell-type differences in the cytoplasmic localization of Hic-5. Cytoplasmic foci do not appear to result strictly from overexpression of HA-tagged Hic-5, because they were detected in Cos-1 cells transfected with low-input amounts of DNA (our unpublished results). Interestingly, in some positively staining Cos-1 cells transfected with HA-tagged Hic-5, the nucleus was devoid of any detectable signal (Figure 10B, arrow). However, in ∼80% of positively stained extracted Cos-1 cells, HA-tagged Hic-5 was localized within nuclei as well as cytoplasmic foci (Figure 10A).
Figure 10.
IIF analysis of HA-tagged Hic-5 localization in transiently transfected Cos-1 cells that were extracted before fixation. Cos-1 cells transiently transfected with HA-tagged Hic-5 were fixed with paraformaldehyde and processed for IIF analysis with an anti-HA antibody with the use of standard procedures (Nishiya et al., 1999). Before fixation, cells were permeabilized with a low-salt and detergent buffer. Representative cells are shown in A and B. C shows cells that were subsequently digested with DNAse I and extracted with ammonium sulfate. Arrow in B indicates an HA-tagged Hic-5–positive cell devoid of nuclear staining.
In addition to the CK buffer extraction, transfected Cos-1 cells were also treated with DNAse I and extracted with a high-salt buffer to prepare nuclear matrices (Tang et al., 1998). HA-tagged Hic-5 remained associated with both the nuclei and cytoplasmic foci after these harsh extractions (Figure 10C). The efficiency of the extraction and DNAse digestion was confirmed by the loss of DAPI staining (our unpublished results). Thus, a fraction of nuclear Hic-5 is associated with the nuclear matrix. In addition, the transiently expressed Hic-5 in these cytoplasmic foci is resistant to high-salt and detergent extraction.
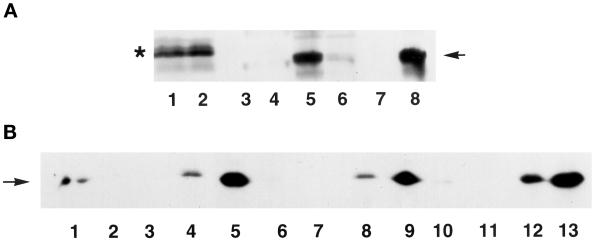
To confirm that nuclear matrix–associated HA-tagged Hic-5 represented intact protein, fractions obtained after various extractions were subjected to Western blot analysis. The amount of residual Hic-5 remaining after CK buffer extraction and DNAse digestion, unlike GR (Tang et al., 1998), was not altered by ATP depletion (Figure 11, compare lanes 2–4 with lanes 5–7). Thus, nuclear matrix binding by Hic-5 is unlikely to result strictly from its association with GR. This notion is supported by the findings that glucocorticoid agonists are required for nuclear matrix binding by GR (Barrack, 1987) but had no effect on the association of Hic-5 with the nuclear matrix (our unpublished results). Approximately 50% of total cellular HA-tagged Hic-5 was extracted with CK buffer (Figure 11, compare lanes 1 and 2). By reference to Figure 10C, the Hic-5 that remained insoluble after CK buffer extraction and DNAse digestion (Figure 11, lanes 4 and 7) represents nuclear matrix and cytoplasmic foci. As a control, the samples from Figure 11, lanes 1–4, were retested by Western blotting with antibodies against the nuclear matrix protein lamin B (Figure 12B, lanes 1–4); the nuclear matrix fraction (Figure 12B, lane 4), which resisted CK buffer extraction and DNAse I digestion, contained lamin B. Because cytoplasmic foci of HA-tagged Hic-5 in transiently transfected Cos-1 cells remained even after harsh extractions (Figure 10C), we are unable to estimate precisely the relative fraction of HA-tagged Hic-5 bound to the nuclear matrix.
Figure 11.
Western blot analysis of HA-tagged Hic-5 in subcellular fractions of aerobic and ATP-depleted Cos-1 cells. Cos-1 cells were either untransfected (lane 8) or transfected with full-length HA-tagged Hic-5 (lanes 1–7) and either grown under aerobic conditions (lanes 1–4 and 8) or subjected to ATP depletion (lanes 5–7). Proteins present in whole cell extracts (lanes 1 and 8), CK buffer–extracted supernatants (lanes 2 and 5), DNAse I–digested supernatants (lanes 3 and 6), or nuclear matrix pellets (lanes 4 and 7) were separated by SDS-PAGE and subjected to Western blot analysis to detect HA-tagged Hic-5 (arrow). Samples in lanes 1–4 were also probed on a separate blot to detect lamin B protein (see Figure 12B, lanes 1–4).
Figure 12.
Western blot analysis of HA-tagged Hic-5 and its fragments in subcellular fractions of Cos-1 cells. (A) Cos-1 cells were transfected with an expression vector for either the N-terminal (amino acids 1–200; lanes 1–4) or C-terminal (amino acids 201–444; lanes 5–8) fragment of HA-tagged Hic-5. Proteins present in whole cell extracts (lanes 1 and 5), CK buffer–extracted supernatants (lanes 2 and 6), DNAse I–digested supernatants (lanes 3 and 7), or nuclear matrix pellets (lanes 4 and 8) were separated by SDS-PAGE and subjected to Western blot analysis to detect HA-tagged Hic-5. The asterisk indicates the position of the N-terminal Hic-5 fragment, and the arrow points to the C-terminal Hic-5 fragment. The N-terminal fragment of Hic-5 migrates anomalously slower than the larger C-terminal fragment. (B) Samples from Figure 11 and Figure 12A were also probed on a separate blot to detect lamin B protein. Lanes 1–4 contain fractions from cells expressing full-length Hic-5 (from Figure 11, lanes 1–4); lanes 5–8 contain fractions expressing the N-terminal Hic-5 fragment (from panel A, lanes 1–4); and lanes 9–12 contain fractions expressing the C-terminal Hic-5 fragment (from panel A, lanes 5–8). Lane 13 contains whole cell extract from untransfected cells.
The specificity of Hic-5 binding to the nuclear matrix was confirmed by transfections with separate Hic-5 domains. The C-terminal fragment (Figure 12A, lanes 5–8), but not the N-terminal fragment (lanes 1–4), of Hic-5 localized to the nuclear matrix (nuclear matrix fractions are in lanes 4 and 8). As a control, lamin B was shown to associate only with the nuclear matrix fraction in each of these extractions (Figure 12B, lanes 5–12). Thus, lamin B served as a positive control, and the N-terminal fragment of Hic-5 served as a negative control, for the specificity of nuclear matrix binding in this assay. The C-terminal fragment of Hic-5 encodes its four LIM domains and has the capacity to interact with focal adhesions, the nucleus (Shibanuma et al., 1994; Matsuya et al., 1998; Thomas et al., 1999), and steroid receptors (Figure 1) (Fujimoto et al., 1999), and it possesses zinc-dependent DNA-binding activity (Nishiya et al., 1998).
DISCUSSION
The 30-amino acid τ2 domain of GR (amino acids 533–562 of mouse GR) possesses transcriptional activation (Hollenberg and Evans, 1988; Milhon et al., 1997) and nuclear matrix targeting (Tang et al., 1998) activity. In addition, we now show that the τ2 domain of rat GR is capable, on its own, of binding with a newly identified steroid receptor coactivator, Hic-5 (Figure 1), and that Hic-5 can potentiate transcriptional activation by the τ2 domain by up to 500-fold (Figure 6). Hic-5 also functioned as a coactivator when coexpressed with a subset of nuclear receptors (Figure 3) or their HBDs (our unpublished results), which in both cases included the τ2 domain. While this work was in progress, Fujimoto et al. (1999) reported the identification of Hic-5 (designated ARA55) as a protein that bound to AR and its HBD and served as a coactivator for AR, GR, and PR but not ER.
Although the isolated τ2 region exhibits a variety of activities, as described above, the precise roles of τ2 in the context of the full-length steroid receptors have been more difficult to assess because of the complex three-dimensional structure of the HBD (Wurtz et al., 1996). Many mutations in the τ2 region dramatically reduce or eliminate hormone binding (Milhon et al., 1997); loss of hormone binding precludes investigation of the effects of these mutations on transcriptional activation and nuclear matrix targeting. A few τ2-region mutations that do not severely affect hormone binding but reduce transactivation in the contexts of the full-length HBD and the isolated τ2 region were identified (Milhon et al., 1997). These and other mutations that do not severely affect hormone binding, tested in the context of the full-length HBD and the isolated τ2 domain, may be useful in determining the physiological role of this domain in the intact HBD and the relationships among the various activities currently ascribed to τ2, i.e., nuclear matrix targeting, Hic-5 binding, transactivation, and the ability to be functionally enhanced by Hic-5. In addition to the complex three-dimensional structure of the HBD, the preliminary analysis of some of the τ2 mutations also suggests that the spatial and functional relationships among these various activities will be complex. For example, in the context of the isolated τ2 domain, we have already found mutations that genetically separate transcriptional activation and nuclear matrix targeting (Milhon et al., 1997; Tang et al., 1998). The τ2 domain has high homology among most steroid receptors (less homology with ER), but there is very little sequence homology in this region between steroid receptors and other nuclear receptors (Milhon et al., 1997). The high homology in the τ2 region among GR, AR, MR, and PR and the divergence from this consensus in ER and TR may provide clues for further genetic tests to determine why Hic-5 can potentiate the former group of steroid receptors in the absence of GRIP1, whereas it can enhance the activity of ER and TR only in the presence of coexpressed GRIP1 (Figure 5).
We have not directly assessed whether τ2 is the only determinant within the GR HBD for Hic-5 interaction, but this should also become apparent by analyzing the ability of Hic-5 to bind and serve as a coactivator for GR HBD with mutations in the τ2 region. Given the nuclear matrix association of Hic-5 and its binding to the τ2-containing NMTS of GR (i.e., GR DBD-τ2), it is tempting to assign Hic-5 as a nuclear matrix acceptor protein for steroid receptors. A putative acceptor protein has been identified within chick oviduct tissue, but its impact on transcriptional activation by steroid receptors has not been assessed (Schuchard et al., 1991). If Hic-5 serves to recruit steroid receptors to the matrix, this activity may be distinct from its coactivator function, because the coactivator effect is observed even with transiently transfected reporter genes, which are not believed to be associated with the nuclear matrix. Alternatively, overexpression of Hic-5 might cause recruitment of the transiently transfected templates to the nuclear matrix.
Both the C-terminal steroid receptor/τ2-binding domain and the N-terminal activation domain of Hic-5 were required for its coactivator function with steroid receptors. The specific downstream target of the N-terminal activation domain of Hic-5 is currently unknown. However, the C-terminal domain binds specifically to a number of different cellular targets. Both the τ2-binding and the nuclear matrix–binding activities of Hic-5 localize within the C-terminal LIM domains of Hic-5 (Figures 1B and 12A). In addition, the LIM domains of Hic-5 are also responsible for its association with focal adhesions (Shibanuma et al., 1994; Matsuya et al., 1998), the protein tyrosine phosphatase–PEST (Nishiya et al., 1999), and DNA (Nishiya et al., 1998). The LIM domain is a unique cysteine zinc finger motif that has the consensus sequence CXXCX16–23HXX(H/C)XXCXXCX16–21CXX(D/H/C) and is found in a variety of proteins of diverse functions and localization (Schmeichel and Beckerle, 1994; Shibanuma et al., 1994). Although Hic-5 contains four LIM domains, they are not functionally equivalent. For example, the third LIM domain, LIM3, is required for protein tyrosine phosphatase–PEST interaction (Nishiya et al., 1999), whereas LIM4 alone, or LIM1 and LIM2 together, possess zinc-dependent DNA-binding activity (Nishiya et al., 1998). Thus, it is possible that separate LIM domains of Hic-5 contribute to its interaction with the nuclear matrix versus GR.
The identification of Hic-5, a focal adhesion protein (Fujita et al., 1998; Matsuya et al., 1998), as a steroid receptor coactivator (Figure 3) (Fujimoto et al.,1999) implies that if Hic-5 effects on transcriptional activation by steroid receptors are direct, it must have some capacity to localize within nuclei. Hic-5 had been observed to localize within nuclei in some reports (Shibanuma et al., 1997), but the significance of this alternative localization was unclear. We have not only confirmed the nuclear localization of Hic-5 but also shown that Hic-5 is targeted to the nuclear matrix (Figures 7, 8, 10, and 11). Zyxin, another LIM domain containing focal adhesion protein, has also been found to localize to nuclei, although its targeting to distinct subnuclear compartments (i.e., the nuclear matrix) has not been assessed. The resident time of zyxin within the nucleus was dramatically increased when its nuclear export signal sequence was deleted (Nix and Beckerle, 1997). The N-terminal domain of Hic-5 possesses five LD motifs that bear some resemblance to prototypical leucine-rich nuclear export signal sequences (Thomas et al., 1999). Thus, Hic-5 may share the nucleocytoplasmic shuttling properties of zyxin. Testing for possible physical and functional interactions between GR and zyxin should reveal whether this Hic-5–related protein, which also has LIM domains and binds to focal adhesion complexes, shares the steroid receptor coactivator activity of Hic-5.
Signals emanating from the ECM can affect gene expression. For example, a specific enhancer element (i.e., BCE-1) in the bovine casein gene responds to ECM-directed signaling (Myers et al., 1998). ECM activation of integrins can mobilize many signaling pathways (e.g., MAPKs; Guan and Chen, 1996) that ultimately target specific transcription factors. Although various second messenger systems may be used to transmit ECM-directed signals that regulate gene expression, there also appears to be a more direct physical link between the ECM and the nuclear matrix (Bissell et al., 1999). Given the fact that Hic-5 localizes to both focal adhesions and the nuclear matrix and participates in steroid hormone regulation of gene transcription, we propose that another pathway may exist that directly transmits ECM-directed signals to the nucleus, i.e., the shuttling of focal adhesion proteins to the nucleus. There is precedence for this paradigm, because the c-Abl protein has been found to shuttle between the nucleus and focal adhesions in response to cell adhesion (Lewis et al., 1996).
Hormone-regulated transcription of some target genes (e.g., casein, MMTV) is potentiated when cells are plated on particular ECMs (Schmidhauser et al., 1994; Myers et al., 1998). What mechanism accounts for these selective effects of the ECM on hormone-regulated gene transcription? ECM regulation of glucocorticoid-induced transcription could be due to the activation of various signaling pathways that are mobilized upon integrin stimulation. Alternatively, ECM effects on gene transcription might be transmitted directly to specific target genes in the nucleus, perhaps via a direct effect on shuttling focal adhesion proteins. Hic-5 could be one such shuttling protein because it has been found to associate with both focal adhesions and the nuclear matrix. All of the assays of Hic-5 effects on transcriptional activation by steroid receptors have used transient transfections in which nuclear matrix association seems unlikely to play a role in transcriptional regulation. Thus, it will be imperative to establish whether Hic-5 can regulate transcription from glucocorticoid-responsive target genes in native chromatin. Interestingly, ECM did not affect glucocorticoid induction mediated by the MMTV glucocorticoid response elements in CID-9 mammary epithelial cells in transient transfections but required stable integration of the hormone-regulated template (Myers et al., 1998). Recent results with Hic-5–overexpressing human fibroblast cell lines suggest that Hic-5 may participate in the regulation of ECM-related genes such as collagenase and cell cycle regulators such as Cip/WAF/sdi1 (Shibanuma et al., 1997). In this case as well, Hic-5 did not affect transcription from transiently transfected Cip and collagenase promoters.
Ultimately, we are interested in establishing whether proteins such as Hic-5, or other LIM domain–containing focal adhesion proteins, play a role in regulating ECM-mediated effects on glucocorticoid responsiveness. Cellular responses to ECM clearly play a role in physiologically relevant steroid hormone effects in specific tissues (Bissell et al., 1999). Furthermore, alterations in focal adhesion protein function have been detected in metastatic prostate cancer. It is premature to suggest that this change in focal adhesion protein function affects AR function during the progression to hormone independence of metastatic cancer cells (Tremblay et al., 1996a,b). Nonetheless, a detailed mechanistic analysis of LIM domain protein involvement in subnuclear trafficking and transcriptional activation by steroid receptors could increase our understanding of how cell attachment to the ECM influences gene expression both during normal development and in pathophysiological conditions.
ACKNOWLEDGMENTS
We thank Dr. D. Pearce (University of California, San Francisco) for providing the MR expression vector, Dr. D. Edwards (University of Colorado Health Sciences Center, Denver) for the PR expression vector, T. Harper and Dr. W. Saunders (University of Pittsburgh) for assistance with microscopy, Dr. D. Brautigan (University of Virginia, Charlottesville) for REF-52 cells, Dr. K. Tachibana (Harvard University Medical School) for the anti-Hic-5 antibody, and S.-M. Huang and Dr. H. Ma (University of Southern California, Los Angeles) for helpful technical consultations. This work was supported by U.S. Public Health Service grants DK43093 (to M.R.S.) and CA43037 (to D.B.D.) from the National Institutes of Health.
Abbreviations used:
- AD
activation domain
- AR
androgen receptor
- DBD
DNA-binding domain
- ECM
extracellular matrix
- ER
estrogen receptor
- FAK
focal adhesion kinase
- β-gal
β-galactosidase
- GR
glucocorticoid receptor
- HA
hemagglutinin
- HBD
hormone-binding domain
- IIF
indirect immunofluorescence
- LD
leucine–aspartic acid
- MR
mineralocorticoid receptor
- NMTS
nuclear matrix targeting signal
- PR
progesterone receptor
- TR
thyroid hormone receptor
REFERENCES
- Angers-Loustau A, Cote J-F, Charest A, Dowbenko D, Spencer S, Laskay LA, Tremblay ML. Protein tyrosine phosphatase-PEST regulates focal adhesion disassembly, migration, and cytokinesis in fibroblasts. J Cell Biol. 1999;144:1019–1031. doi: 10.1083/jcb.144.5.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrack ER. Localization of steroid receptors in the nuclear matrix. In: Clark CR, editor. Steroid Hormone Receptors: Their Intracellular Localization. Chichester, England: Ellis Horwood; 1987. pp. 86–127. [Google Scholar]
- Barrack ER, Coffey DS. The specific binding of estrogens and androgens to the nuclear matrix of sex hormone responsive tissues. J Biol Chem. 1980;255:7265–7275. [PubMed] [Google Scholar]
- Barrett TJ, Spelsberg TC. Nuclear matrix and steroid hormone action. Vitam Horm. 1999;55:127–163. doi: 10.1016/s0083-6729(08)60935-8. [DOI] [PubMed] [Google Scholar]
- Bissell MJ, Weaver VM, Lelievre SA, Wang F, Petersen OW, Schmeichel KL. Tissue structure, nuclear organization, and gene expression in normal and malignant breast. Cancer Res. 1999;59(suppl):1757s–1764s. [PubMed] [Google Scholar]
- Boonyaratanakornkit V, Melvin V, Prendergast P, Altmann M, Ronfani L, Bianchi ME, Taraseviciene L, Nordeen SK, Allegretto EA, Edwards DP. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol. 1998;18:4471–4487. doi: 10.1128/mcb.18.8.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary LA, Guan J-L. Focal adhesion kinase in integrin-mediated signaling. Front Biosci. 1999;4:102–113. doi: 10.2741/cary. [DOI] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang S-M, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- Ding XF, Anderson CM, Ma H, Hong H, Uht RM, Kushner PJ, Stallcup MR. Nuclear receptor-binding sites of coactivators Glucocorticoid Receptor Interacting Protein 1 (GRIP1) and Steroid Receptor Coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- Eggert M, Michel J, Schneider S, Bornfleth H, Baniahmad A, Fackelmayer FO, Schmidt S, Renkawitz R. The glucocorticoid receptor is associated with the RNA-binding nuclear matrix protein hnRNP U. J Biol Chem. 1997;272:28471–28478. doi: 10.1074/jbc.272.45.28471. [DOI] [PubMed] [Google Scholar]
- Fackelmayer FO, Dahm K, Renz A, Ramsperger U, Richter A. Nucleic acid-binding properties of hnRNP-U/SAF-A, a nuclear matrix protein which binds DNA and RNA in vivo and in vitro. Eur J Biochem. 1994;221:749–757. doi: 10.1111/j.1432-1033.1994.tb18788.x. [DOI] [PubMed] [Google Scholar]
- Fey EG, Penman S. Nuclear matrix proteins reflect cell type of origin in cultured human cells. Proc Natl Acad Sci USA. 1988;85:121–125. doi: 10.1073/pnas.85.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto N, Yeh S, Kang H-Y, Inui S, Chang H-C, Mizokami A, Chang C. Cloning and characterization of androgen receptor coactivator, ARA55, in human prostate. J Biol Chem. 1999;274:8316–8321. doi: 10.1074/jbc.274.12.8316. [DOI] [PubMed] [Google Scholar]
- Fujita H, Kamiguchi K, Cho D, Shibanuma M, Morimoto C, Tachibana K. Interaction of Hic-5, a senescence-related protein, with focal adhesion kinase. J Biol Chem. 1998;273:26516–26521. doi: 10.1074/jbc.273.41.26516. [DOI] [PubMed] [Google Scholar]
- Getzenberg RH, Coffey DS. Tissue specificity of the hormonal response in sex accessory tissues is associated with nuclear matrix protein patterns. Mol Endocrinol. 1990;4:1336–1342. doi: 10.1210/mend-4-9-1336. [DOI] [PubMed] [Google Scholar]
- Guan J-L, Chen H-C. Signal transduction in cell-matrix interactions. Int Rev Cytol. 1996;168:81–121. [PubMed] [Google Scholar]
- Hollenberg SM, Evans RM. Multiple and cooperative trans-activation domains of the human glucocorticoid receptor. Cell. 1988;55:899–906. doi: 10.1016/0092-8674(88)90145-6. [DOI] [PubMed] [Google Scholar]
- Hong H, Kohli K, Garabedian MJ, Stallcup MR. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Yang L, Stallcup MR. Hormone independent transcriptional activation and coactivator binding by novel orphan nuclear receptor ERR3. J Biol Chem. 1999;274:22618–22626. doi: 10.1074/jbc.274.32.22618. [DOI] [PubMed] [Google Scholar]
- Lelievre SA, Weave VM, Nickerson JA, Larabell CA, Bhaumik A, Petersen OW, Bissell MJ. Tissue phenotype depends on reciprocal interactions between the extracellular matrix and the structural organization of the nucleus. Proc Natl Acad Sci USA. 1998;95:14711–14716. doi: 10.1073/pnas.95.25.14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JM, Baskaran R, Taagepera S, Schwarta MA, Wang JY. Integrin regulation of c-Abl tyrosine kinase activity and cytoplasmic-nuclear transport. Proc Natl Acad Sci USA. 1996;93:15174–15179. doi: 10.1073/pnas.93.26.15174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini MG, Liu B, Sharp ZD, Mancini MA. Subnuclear partitioning and functional regulation of the Pit-1 transcription factor. J Cell Biochem. 1999;72:322–338. [PubMed] [Google Scholar]
- Matsuya M, Sasaki H, Aoto H, Mitaka T, Nagura K, Ohba T, Ishino M, Takahashi S, Suzuki R, Sasaki T. Cell adhesion kinase β forms a complex with a new member, Hic-5, of proteins localized at focal adhesions. J Biol Chem. 1998;273:1003–1014. doi: 10.1074/jbc.273.2.1003. [DOI] [PubMed] [Google Scholar]
- Milhon J, Kohli K, Stallcup MR. Genetic analysis of the N-terminal end of the glucocorticoid receptor hormone binding domain. J Steroid Biochem Mol Biol. 1994;51:11–19. doi: 10.1016/0960-0760(94)90110-4. [DOI] [PubMed] [Google Scholar]
- Milhon J, Lee S, Kohli K, Chen D, Hong H, Stallcup MR. Identification of amino acids in the τ2 region of the mouse glucocorticoid receptor that contribute to hormone binding and transcriptional activation. Mol Endocrinol. 1997;11:1795–1805. doi: 10.1210/mend.11.12.0018. [DOI] [PubMed] [Google Scholar]
- Myers CA, Schmidhauser C, Mellentin-Michelotti J, Fragoso G, Roskelley CD, Casperson G, Mossi R, Pujuguet P, Hager G, Bissell MJ. Characterization of BCE-1, a transcriptional enhancer regulated by prolactin and extracellular matrix and modulated by the state of histone acetylation. Mol Cell Biol. 1998;18:2184–2195. doi: 10.1128/mcb.18.4.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiya N, Iwabuchi Y, Shibanuma M, Cote J-F, Tremblay ML, Nose K. Hic-5, a paxillin homologue, binds to the protein-tyrosine phosphatase PEST (PTP-PEST) through its LIM3 domain. J Biol Chem. 1999;274:9847–9853. doi: 10.1074/jbc.274.14.9847. [DOI] [PubMed] [Google Scholar]
- Nishiya N, Sabe H, Nose K, Shibanuma M. The LIM domains of hic-5 protein recognize specific DNA fragments in a zinc-dependent manner in vitro. Nucleic Acids Res. 1998;26:4267–4273. doi: 10.1093/nar/26.18.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix DA, Beckerle MC. Nuclear-cytoplasmic shuttling of focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partin AW, Getzenberg RH, Carmichael MJ, Vindivich D, Yoo J, Epstein JI, Coffey DS. Nuclear matrix protein patterns in human benign prostatic hyperplasia and prostate cancer. Cancer Res. 1993;53:744–746. [PubMed] [Google Scholar]
- Pearce D, Yamamoto KR. Mineralocorticoid and glucocorticoid receptor activities distinguished by nonreceptor factors at a composite response element. Science. 1993;259:1161–1165. doi: 10.1126/science.8382376. [DOI] [PubMed] [Google Scholar]
- Schmeichel KL, Beckerle MC. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Schmidhauser C, Casperson GF, Bissell MJ. Transcriptional activation by viral enhancers: critical dependence on extracellular matrix-cell interactions in mammary epithelial cells. Mol Carcinog. 1994;10:66–71. doi: 10.1002/mc.2940100203. [DOI] [PubMed] [Google Scholar]
- Schuchard M, Subramaniam M, Ruesink T, Spelsberg TC. Nuclear matrix localization and specific matrix DNA binding by Receptor Binding Factor-1 of the avian progesterone receptor. Biochemistry. 1991;30:9516–9522. doi: 10.1021/bi00103a019. [DOI] [PubMed] [Google Scholar]
- Shibanuma M, Mashimo J-i, Kuroki T, Nose K. Characterization of the TGFβ1-inducible hic5 gene that encodes a putative novel zinc finger protein and its possible involvement in cellular senescence. J Biol Chem. 1994;269:26767–26774. [PubMed] [Google Scholar]
- Shibanuma M, Mochizuki E, Maniwa R, Mashimo J-i, Nishiya N, Imai S-I, Takano T, Oshimura M, Nose K. Induction of senescence-like phenotypes by forced expression of hic-5, which encodes a novel LIM motif protein, in immortalized human fibroblasts. Mol Cell Biol. 1997;17:1224–1235. doi: 10.1128/mcb.17.3.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J-M, Chen HY, Davie JR. Nuclear factor 1 is a component of the nuclear matrix. J Cell Biochem. 1994;55:252–263. doi: 10.1002/jcb.240550212. [DOI] [PubMed] [Google Scholar]
- Tang Y, DeFranco DB. ATP-dependent release of glucocorticoid receptors from the nuclear matrix. Mol Cell Biol. 1996;16:1989–2001. doi: 10.1128/mcb.16.5.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Getzenberg RH, Vietmeier BN, Stallcup MR, Eggert M, Renkawitz R, DeFranco DB. The DNA-binding and τ2 transactivation domains of the rat glucocorticoid receptor constitute a nuclear matrix-targeting signal. Mol Endocrinol. 1998;12:1420–1431. doi: 10.1210/mend.12.9.0169. [DOI] [PubMed] [Google Scholar]
- Thomas SM, Hagel M, Turner CE. Characterization of a focal adhesion protein, Hic-5, that shares extensive homology with paxillin. J Cell Sci. 1999;112:181–190. doi: 10.1242/jcs.112.2.181. [DOI] [PubMed] [Google Scholar]
- Torchia J, Glass C, Rosenfeld MG. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- Tremblay L, Hauck W, Aprikian AG, Begin LR, Chapdelaine A, Chevalier S. Focal adhesion kinase (pp125FAK) expression, activation and association with paxillin and p50CSK in human metastatic prostate carcinoma. Int J Cancer. 1996a;68:164–171. doi: 10.1002/(sici)1097-0215(19961009)68:2<169::aid-ijc4>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Tremblay L, Hauck W, Nguyen LT, Allard P, Landry F, Chapdelaine A, Chevalier S. Regulation and activation of focal adhesion kinase and paxillin during the adhesion, proliferation, and differentiation of prostatic epithelial cells in vitro and in vivo. Mol Endocrinol. 1996b;10:1010–1020. doi: 10.1210/mend.10.8.8843417. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Jenster G, Damm K, Brinkmann AO, van Driel R. Domains of the human androgen receptor and glucocorticoid receptor involved in binding to the nuclear matrix. J Cell Biochem. 1995;57:465–478. doi: 10.1002/jcb.240570312. [DOI] [PubMed] [Google Scholar]
- Van Wijnen AJ, Bidwell JP, Fey EG, Penman S, Lian JB, Stein JL, Stein GS. Nuclear matrix association of multiple sequence-specific DNA binding activities related to SP1, ATF, CCAAT, C/EBP, OCT-1, and AP-1. Biochemistry. 1993;32:8397–8402. doi: 10.1021/bi00084a003. [DOI] [PubMed] [Google Scholar]
- Voegel JJ, Heine MJ, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz J-M, Bourguet W, Renaud J-P, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]