Abstract
The influence of salicylic acid (SA) on elicitation of defense mechanisms in Arabidopsis (Arabidopsis thaliana) seeds and seedlings was assessed by physiological measurements combined with global expression profiling (proteomics). Parallel experiments were carried out using the NahG transgenic plants expressing the bacterial gene encoding SA hydroxylase, which cannot accumulate the active form of this plant defense elicitor. SA markedly improved germination under salt stress. Proteomic analyses disclosed a specific accumulation of protein spots regulated by SA as inferred by silver-nitrate staining of two-dimensional gels, detection of carbonylated (oxidized) proteins, and neosynthesized proteins with [35S]-methionine. The combined results revealed several processes potentially affected by SA. This molecule enhanced the reinduction of the late maturation program during early stages of germination, thereby allowing the germinating seeds to reinforce their capacity to mount adaptive responses in environmental water stress. Other processes affected by SA concerned the quality of protein translation, the priming of seed metabolism, the synthesis of antioxidant enzymes, and the mobilization of seed storage proteins. All the observed effects are likely to improve seed vigor. Another aspect revealed by this study concerned the oxidative stress entailed by SA in germinating seeds, as inferred from a characterization of the carbonylated (oxidized) proteome. Finally, the proteomic data revealed a close interplay between abscisic signaling and SA elicitation of seed vigor.
As sessile organisms, plants have devised sophisticated mechanisms to cope with biotic and abiotic stresses imposed by their environment. Among these mechanisms, those collectively referred to as defense reactions induced by endogenous elicitors such as salicylic acid (SA), jasmonic acid, or ethylene have received considerable attention (Gaffney et al., 1993; Clarke et al., 2000). This is of agronomical importance since a better knowledge of the involved genes and mechanisms can offer novel means for crop protection, for example helping characterize novel chemical elicitors of plant defense toward the development of sustainable agriculture. In the plant life cycle, the seed and seedling stages are key developmental stages conditioning the final yield of crops. Both are very sensitive to environmental stresses (Bewley and Black, 1994; Koornneef et al., 2002). Consequently, most commercialized crop seeds are submitted to various treatments in which fungicides, for example, are included via seed pelleting and/or coating (Barratt et al., 1995). Paradoxically, plant defense mechanisms are principally studied on adult plant organs and there is only very scarce information about the onset of defense mechanisms at the level of seed germination.
To address this question we have in this work investigated the effect of a model elicitor compound, SA, on seed germination of the reference plant Arabidopsis (Arabidopsis thaliana), notably under salt stress conditions. There are some conflicting reports on the effect of SA on seed germination and seedling establishment, suggesting that this molecule can either inhibit germination (e.g. as in maize [Zea mays]; Guan and Scandalios, 1995) or enhance seed vigor (e.g. as in wheat [Triticum aestivum; Shakirova et al., 2003] and pea [Pisum sativum; McCue et al., 2000]). In Arabidopsis, SA has been proposed to increase the oxidative damage generated by NaCl and osmotic stresses, thus causing seedling lethality under these conditions (Borsani et al., 2001).
Based on previous work (for review, see Rajjou et al., 2006), we decided to investigate this question by proteomics. This type of approach brings robust information about the biological functions affected in physiological changes (Pandey and Mann, 2000). In particular, we used a dynamic proteomics approach to characterize the effect of SA on the de novo synthesized proteome during germination. The combined results obtained by the classical proteomics approach based upon two-dimensional (2D) gel staining by silver nitrate and the dynamic proteomics approach revealed several processes potentially affected by SA. They are discussed in relation to the observed improvement of seed vigor brought about by this elicitor molecule.
RESULTS AND DISCUSSION
Sensitivity to SA of Wild-Type Arabidopsis Seed Germination and Seedling Establishment
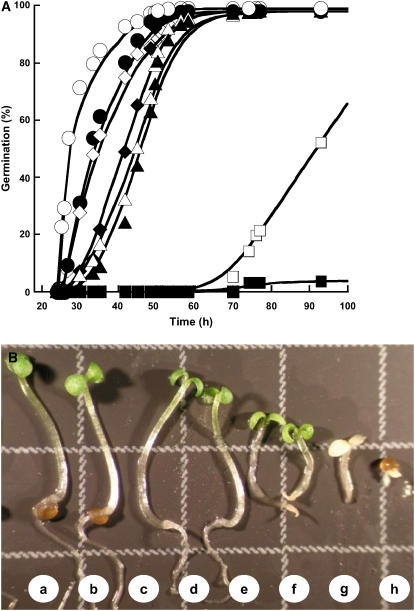
SA induced several effects depending on the concentration applied and quite high doses were required to observe symptoms (Metraux et al., 1990). In all cases, the first effect observed was a delay in seed germination. Above a concentration of 1 mm, the seed germination process was dramatically impeded (Fig. 1A). Similar results have been reported for maize embryo germination, for which high doses of SA, in the range of 3 to 5 mm, completely inhibited germination (Guan and Scandalios, 1995). However, it is noted that at concentrations lower than 1 mm, SA did not affect the speed, the homogeneity, and the final extent of Arabidopsis germination (Fig. 1). Above a concentration of 0.5 mm, this compound entailed a strong retardation of growth and plants appeared bleached, most presumably because at these high concentrations SA induced an oxidative stress (Rao et al., 1997; Fig. 1B). Therefore, all subsequent experiments were carried out by using SA at 0.5 mm, a concentration that appeared harmless for both seed germination and establishment of the seedling (Fig. 1).
Figure 1.
Influence of SA on seed germination and seedling establishment of Arabidopsis Ler ecotype. A, Influence of SA on the germination profiles. Seeds were germinated as described in “Materials and Methods,” in the absence (○) or presence of SA (•, 0.1 mm; ⋄, 0.25 mm; ♦, 0.5 mm; ▵,0.75 mm; ▴, 1 mm; □, 2.5 mm; ▪, 5 mm). The graph shows a representative experiment carried out three times in triplicate. B, Phenotypes observed 7 d after sowing on water without SA (a) and with SA (b, 0.1 mm; c, 0.25 mm; d, 0.5 mm; e, 0.75 mm; f, 1 mm; g, 2.5 mm; h, 5 mm). Square, 3 mm × 3 mm.
Influence of SA on Stress Tolerance during Seed Germination
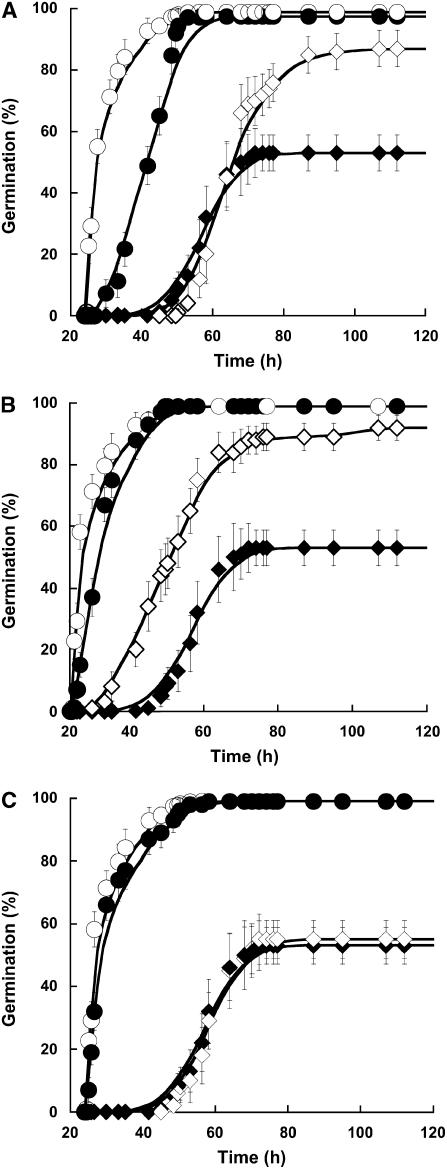
Previous studies demonstrated that SA plays an important role in determining the sensitivity of plants to various abiotic stresses (Dat et al., 1998; Rao and Davis, 1999), notably at the seedling stage (Borsani et al., 2001). To further document the influence of this molecule at the level of seed germination, Arabidopsis seeds were germinated under salt stress conditions with or without SA addition (Fig. 2). At 100 mm NaCl, seeds were only able to achieve 53% (±5.7%) germination. The inclusion of SA at 0.5 mm in the germination medium was associated with a marked increase in maximum germination percentage, which reached 87% (±5.9%; Fig. 2A). These results show that SA substantially improved germination vigor under salt stress conditions. To address the question of the specificity of this molecule, we assessed the influence of several SA derivatives on Arabidopsis seed germination, both under control and salt stress conditions. The influence of two of them, sulfo-SA (SSA) and acetyl-SA (ASA), is documented here. Contrary to what was observed with SA, these two molecules did not delay Arabidopsis seed germination. SSA strongly improved seed germination in the presence of 100 mm NaCl, both enhancing the rate and extent of germination compared to SA (Fig. 2B). In marked contrast, ASA had no effect on Arabidopsis seed germination under salt stress conditions (Fig. 2C). These results suggested the importance of structure/activity relationships for enhancement of Arabidopsis seed vigor.
Figure 2.
Analysis of the effect of SA, SSA, and ASA on the time courses of seed germination in optimal conditions or under salt stress. A, Influence of SA on the germination profiles. Seeds were germinated as described in “Materials and Methods” in optimal conditions without SA (○) or with 0.5 mm SA (•), and under salt stress (100 mm NaCl) without SA (♦) or with 0.5 mm SA (⋄). B, Influence of SSA on the germination profiles. Seeds were germinated as described in “Materials and Methods” in optimal conditions without SSA (○) or with 1 mm SSA (•), and under salt stress (100 mm NaCl) without SSA (♦) or with 1 mm SSA (⋄). C, Influence of ASA on the germination profiles. Seeds were germinated as described in “Materials and Methods” in optimal conditions without ASA (○) or with 0.5 mm ASA (•), and under salt stress (100 mm NaCl) without ASA (♦) or with 0.5 mm ASA (⋄). The figure shows a representative experiment carried out three times in triplicate.
Proteomic Analyses Based upon Silver-Stained 2D Gels (Classical Proteomics)
The effect of SA on the seed germination process was assessed by proteomics. Total soluble proteins prepared from seeds during germination on water or in the presence of 0.5 mm SA were separated by 2D gel electrophoresis and, following silver-nitrate staining, protein patterns were characterized by image analysis. In the following, these experiments are referred to as classical proteomics.
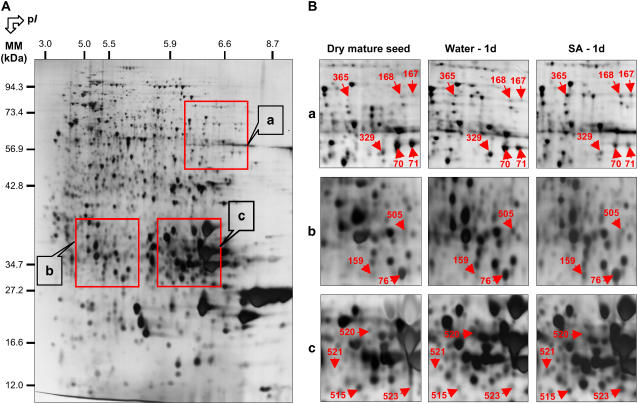
Out of 1,332 protein spots detected on reproducible 2D gels, 27 showed reproducible variations in their accumulation level upon incubation in the presence of SA compared to incubation in water (Fig. 3; Table I). The abundance of six spots (nos. 70, 71, 329, 504, 505, and 520) decreased upon SA treatment. The 21 other proteins spots (nos. 5, 11, 33, 76, 159, 167, 168, 254, 255, 365, 420, 509, 510, 512, 513, 515, 516, 518, 519, 521, and 523) showed a higher abundance in the presence of SA compared to control seeds (Table I).
Figure 3.
Influence of SA on the proteome of Arabidopsis seeds after 1 d imbibition. An equal amount (150 μg) of total protein extracts was loaded in each gel. A, A silver-stained 2D gel of total proteins from wild-type Arabidopsis seeds incubated for 24 h in water. The indicated portions of the gel, a, b, and c, are reproduced in B. B, Enlarged windows (a–c) of 2D gels as shown in A for dry mature seeds (left), seeds incubated in water for 24 h (middle), and seeds incubated in 0.5 mm SA for 24 h (right). The 13 labeled protein spots (protein nos. 70, 71, 76, 159, 167, 168, 329, 365, 505, 515, 520, 521, and 523) were identified by comparison with Arabidopsis seed protein reference maps (Gallardo et al., 2001, 2002a; http://seed.proteome.free.fr; Table I) or by mass spectrometry. Protein spot quantification was carried out as described in “Materials and Methods,” from at least three gels for each seed sample.
Table I.
Arabidopsis proteins whose abundance was significantly different in wild-type germinating Ler seeds in the presence or absence of 0.5 mm of SA 24 h post imbibition
DS, Dry seeds; H2O, seeds imbibed for 24 h on water; SA, seeds imbibed for 24 h in the presence of 0.5 mm of SA; % Cov., coverage; Exp., experimental; MM, molecular mass; Th., theoretical; AGI, Arabidopsis Genome Initiative; H2O/DS, normalized spot volume in the Arabidopsis seeds incubated for 24 h in water divided by the normalized spot volume in the dry mature seeds, from three different gels and independent extractions; SA/DS, normalized spot volume in the Arabidopsis seeds incubated for 24 h in the presence of 0.5 mm SA divided by the normalized spot volume in the dry mature seeds, from three different gels and independent extractions; SA/H2O, normalized spot volume in the Arabidopsis seeds incubated for 24 h in the presence of 0.5 mm SA divided by the normalized spot volume in the Arabidopsis seeds incubated for 24 h in water, from three different gels and independent extractions.
| No. | Exp. MM | Exp. pI | Arabidopsis Protein Name | % Cov. | Th. MM | Th. pI | AGI No. | Ratioa H2O/DS | Ratioa SA/DS | Ratioa SA/H2O |
|---|---|---|---|---|---|---|---|---|---|---|
| kD | kD | |||||||||
| 70b | 49.43 | 7.19 | 12S seed storage protein precursor | 22% | 52.49 | 7.68 | At5g44120 | 0.96 | 0.31 | 0.32 |
| 71b | 50.44 | 7.67 | 12S seed storage protein precursor | 34% | 52.49 | 7.68 | At5g44120 | 0.67 | 0.18 | 0.27 |
| 329b | 50.44 | 7.67 | 12S Seed storage protein precursor | 35% | 52.49 | 7.68 | At5g44120 | 3.98 | 1.17 | 0.29 |
| 504c | 60.87 | 6.20 | 12S seed storage protein precursor | 7% | 55.86 | 6.36 | At4g28520 | 2.15 | 1.08 | 0.50 |
| 505c | 28.87 | 5.30 | 20S proteasome α subunit A1 (PAA1) | 23% | 27.28 | 5.66 | At5g35590 | 2.98 | 1.01 | 0.34 |
| 510c | 25.98 | 4.00 | 20S proteasome α subunit E2 or E1 (PAE2 or PAE1) | 9% | 25.96 | 4.41 | At3g14290 or At1g53850 | ≤0.001 | 1.22 | ≥1,000 |
| 520c | 32.81 | 5.99 | α-Cruciferin 12S (seed storage protein fragment) | 12% | 31.63 | 9.08 | At5g44120 | 45.21 | 2.02 | 0.04 |
| 159b | 23.54 | 5.33 | α-Cruciferin 12S (seed storage protein fragment) | 15% | 27.24 | 6.34 | At4g28520 | 1.01 | 2.66 | 2.63 |
| 76b | 23.94 | 5.58 | α-Cruciferin 12S (seed storage protein fragment) | 16% | 34.68 | 6.42 | At4g28520 | 0.85 | 2.06 | 2.42 |
| 521c | 25.34 | 5.81 | α-Cruciferin 12S (seed storage protein fragment) | 40% | 34.68 | 6.42 | At5g44120 | 1.00 | 2.9 | 2.90 |
| 518c | 15.82 | 8.66 | α-Cruciferin 12S (seed storage protein fragment) | 36% | 20.84 | 9.06 | At5g44120 | 1.00 | 31.95 | 31.95 |
| 519c | 15.16 | 8.80 | β-Cruciferin 12S (seed storage protein fragment) | 53% | 20.84 | 9.06 | At5g44120 | 1.00 | 27.79 | 27.79 |
| 5b | 57.35 | 4.89 | Tubulin-α 3.5 | 27% | 50.73 | 4.70 | At5g19770 | ≥1,000 | ≥1,000 | 1.51 |
| 33b | 14.86 | 5.87 | β-Cruciferin 12S (seed storage protein fragment) | 18% | 20.84 | 9.06 | At5g44120 | 0.96 | 2.71 | 2.82 |
| 420c | 15.05 | 4.81 | Cytochrome b5 | 49% | 15.07 | 4.93 | At5g53560 | 1.00 | ≥1,000 | ≥1,000 |
| 254b | 22.10 | 4.96 | Dehydrin | 6% | 18.46 | 7.10 | At5g66400 | 0.32 | 1.16 | 3.63 |
| 255b | 21.65 | 5.18 | Dehydrin | 6% | 18.46 | 7.10 | At5g66400 | 0.29 | 1.21 | 4.17 |
| 11b | 19.61 | 5.75 | EM-like protein | 3% | 16.61 | 5.75 | At3g51810 | 0.59 | 1.09 | 1.85 |
| 361b | 47.75 | 5.22 | 4-Methyl-5(b-hydroxyethyl)-thiazole monophosphate biosynthesis protein | 6% | 41.84 | 5.08 | At3g14990 | 2.18 | 3.23 | 1.48 |
| 365b | 62.77 | 6.30 | Isocitrate lyase | 16% | 50.42 | 6.29 | At3g21720 | 240.34 | 634.52 | 2.64 |
| 167b | 62.96 | 8.00 | Malate synthase | 25% | 63.89 | 8.02 | At5g03860 | 1.28 | 3.54 | 2.77 |
| 168b | 62.91 | 7.08 | Malate synthase | 10% | 63.89 | 8.02 | At5g03860 | 1.46 | 2.17 | 1.49 |
| 509c | 34.11 | 4.01 | NAC domain-containing protein | 24% | 17.87 | 4.85 | At5g13850 | 64.94 | 183.04 | 2.82 |
| 513c | 18.67 | 5.50 | NAC domain-containing protein | 6% | 17.99 | 6.00 | At1g73230 | 51.15 | 102.31 | 2.00 |
| 516c | 16.11 | 6.30 | Pathogenesis-related protein | 23% | 19.12 | 6.34 | At1g70840 | 0.18 | 1.89 | 10.50 |
| 515c | 23.94 | 5.89 | Superoxide dismutase | 21% | 25.44 | 8.83 | At3g10920 | 0.44 | 1.27 | 2.89 |
| 523c | 23.94 | 6.42 | Superoxide dismutase | 16% | 26.87 | 6.76 | At3g56350 | 4.48 | 18.93 | 4.23 |
| 512c | 19.41 | 5.50 | Temperature stress induced lipocalin | 12% | 21.42 | 6.29 | At5g58070 | 3.13 | 7.38 | 2.36 |
Data obtained from densitometric analysis of individual spots from proteins in 2D gels stained with silver nitrate.
Listed proteins correspond to previously identified proteins (Gallardo et al., 2001, 2002a; Rajjou et al., 2004; Job et al., 2005).
Listed proteins correspond to proteins identified during this work; the peptide sequences determined are available in Supplemental Table V.
Three spots corresponding to 12S cruciferin seed storage precursor (nos. 70, 71, and 329) showed decreased abundance following SA treatment compared to control seeds (Fig. 3; Table I). Furthermore, several polypeptides (nos. 520, 521, 518, and 519) corresponding to fragments of the A- and B-subunits of 12S cruciferins showed a higher abundance in seeds germinated in the presence of SA compared to control seeds (Fig. 3; Table I). Thus, SA appeared to accelerate seed storage protein mobilization during germination.
Two spots (nos. 509 and 513; Table I; Supplemental Table I) whose accumulation levels were up-regulated by SA during seed germination corresponded to the nascent polypeptide-associated complex (NAC). NAC is a heterodimeric complex that can reversibly bind to eukaryotic ribosomes (Rospert et al., 2002). It is presumed to prevent ribosome-associated nascent polypeptide from inappropriate interaction with proteins in the cytosol (Wiedmann et al., 1994). A recent study demonstrated that the NAC was down-regulated in rice (Oryza sativa) roots submitted to a salt stress in the presence of 150 mm NaCl (Yan et al., 2005). Also, a proteomic study of sugar beet (Beta vulgaris) leaves identified the α-chain NAC as being down-regulated in response to drought (Hajheidari et al., 2005). It is therefore tempting to correlate the induction of this protein by SA in germinating Arabidopsis seeds (Table I; Supplemental Table I) with the observed enhancement of germination under salt stress elicited by this molecule (Fig. 2). This data also agrees with the proteomic results of Campo et al. (2004) showing the induction in fungal-infected maize embryos of several proteins involved in initiation of protein synthesis or proteins that participate in the protein folding process.
As shown in Figure 3 and Table I, the accumulation levels of two superoxide dismutases, spots numbers 515 and 523, were more abundant in seed germinated in the presence of SA that in its absence. The accumulation of spot number 515 encoded by the At3g10920 gene decreased during germination on water but this decrease was inhibited by SA. For spot number 523 encoded by the At3g56350 gene the situation was different. In this case, the accumulation level of this protein increased during normal germination while SA strongly enhanced further this accumulation. This behavior is in agreement with previous reports showing that exogenous SA treatment leads to increased antioxidant capacity in barley (Hordeum vulgare) leaves (Ananieva et al., 2004) and stimulates peroxidase/catalase activities in plant cells (Dixon et al., 1995) because of an enhanced accumulation of hydrogen peroxide under such conditions (Chen et al., 1993; Fauth et al., 1996; Rao et al., 1997). It is known that salt stress induces the generation of reactive oxygen species in plants (Polle, 1997; Borsani et al., 2001). Thereby, it is possible that the presently observed induction of such enzymes by SA can provide an explanation for the improvement of Arabidopsis seed germination under salt stress brought about by this elicitor.
Effect of SA on Protein Carbonylation during Germination
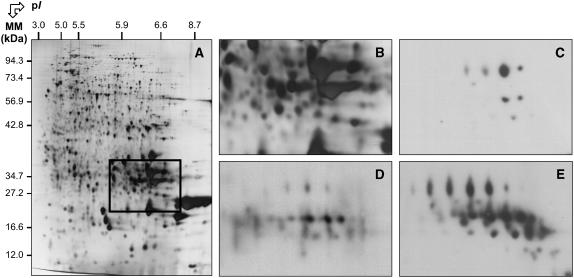
The above data suggest that SA can induce an oxidative stress in germinating Arabidopsis seeds. To address more directly this question we determined the influence of this molecule on the extent of protein carbonylation during germination. Protein carbonylation is a widely used marker of protein oxidation (Johansson et al., 2004; for review, see Nyström, 2005) and sensitive methods for its detection have been developed (Levine et al., 1994). Protein extracts were prepared from control and SA-treated germinating seeds collected after 24 h imbibition. They were separated by 2D electrophoresis, transferred onto nitrocellulose membrane, and oxidatively modified proteins were detected as described under “Materials and Methods.” There was a massive increase in the carbonylation of 12S cruciferin α (acidic) subunits in the presence of SA (Fig. 4). It is known that in 12S globulins (cruciferins), the α subunits are more exposed to the solvent, whereas the β (basic) chains are buried inside the hexameric protein complexes (Adachi et al., 2003). The observed increased carbonylation of the α chains, but not of the β chains, is in agreement with these structural features. These results demonstrate that SA entailed an oxidative stress during germination. Since carbonylation of proteins increases their susceptibility toward proteolytic cleavage (Nyström, 2005), this enhanced carbonylation of 12S cruciferin α subunits brought about by SA might provide a means to facilitate seed storage protein mobilization during germination, a feature that we do observe experimentally (Table I).
Figure 4.
Increased protein carbonyl levels in α subunits of 12S cruciferins following 1 d imbibition of Arabidopsis seeds in the presence of 0.5 mm SA. A, A silver-stained 2D gel of total proteins from wild-type Arabidopsis seeds incubated for 24 h in water. The indicated portion of the gel showing the migration of the α subunits of 12S Arabidopsis cruciferins (Job et al., 2005) is reproduced in B. C to E, Revelation of carbonylated proteins with the anti-DNP immunoassay in same gel window as shown in B. C, Dry mature seeds. D, Seeds incubated in water for 24 h. E, Seeds incubated in the presence of 0.5 mm SA for 24 h.
Dynamic Proteomic Analyses during Germination
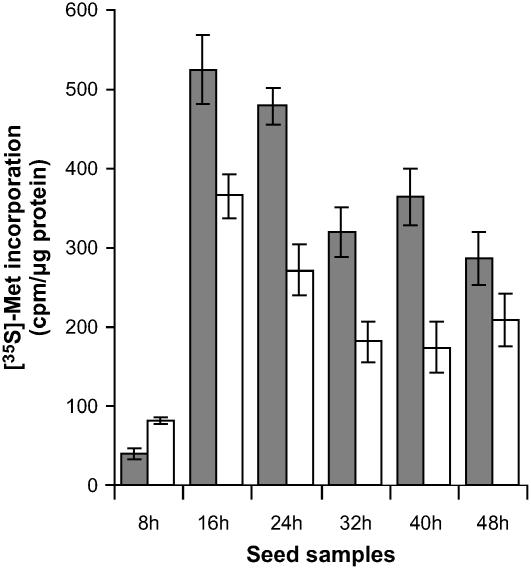
To investigate more closely the influence of SA on germinating Arabidopsis seeds, we wished to characterize de novo protein synthesis during this process. To this end, pulses of [35S]-Met were provided to the seeds at various times during germination. In this dynamic proteomics approach, [35S]-Met was added to the germination medium with or without SA for a period of 8 h, namely from 0 to 8 h, 8 to16 h, 16 to 24 h, 24 to 32 h, 32 to 40 h, or 40 to 48 h after imbibition. Total incorporation of [35S]-Met into proteins was quantitated by trichloroacetic acid precipitation as described under “Materials and Methods.” For the seeds germinated on water only, translational activity was low but detectable during the first 8 h after imbibition, then strongly increased to reach a maximum during the labeling periods of 8 to16 h and 16 to 24 h, and finally steadily declined for longer times of germination (Fig. 5). The same profile was observed in the presence of SA although the translational activity of the seeds was lower, except for the first labeling period of 0 to 8 h (Fig. 5). Following 2D electrophoresis, the radiolabeled proteomes (Fig. 6) were compared with the total proteome obtained after silver staining of the same gels and also with the reference maps established for the Arabidopsis seed proteome (Gallardo et al., 2001, 2002a, 2002b; Rajjou et al., 2004; Job et al., 2005). These comparisons can provide a means to assign some of the [35S]-Met labeled protein spots to proteins previously characterized by mass spectrometry analysis. Interestingly, during the first 8-h imbibition SA increased de novo synthesis of proteins normally accumulated during seed maturation and that are under abscisic acid (ABA) control, such as 12S globulin subunits, late embryogenesis abundant (LEA) proteins, dehydrins, and heat shock proteins (Finkelstein, 1993; Bewley and Black, 1994; Cuming, 1999; Fig. 6; Supplemental Table II). These results are in keeping with those of Lopez-Molina et al. (2002) showing that Arabidopsis seed germination comprises a short window of time after the start of imbibition during which the seed can still recruit late maturation programs. It is worth noting that SA can induce a transient accumulation of ABA in wheat seedlings (Shakirova et al., 2003). If the same situation holds during Arabidopsis seed germination, it could provide an explanation to our results showing that the synthesis of maturation proteins was up-regulated by SA. Since ABA is a potent inhibitor of seed germination (Garciarrubio et al., 1997), it would also account for the delay in Arabidopsis seed germination observed in the presence of SA (Fig. 1).
Figure 5.
Incorporation of [35S]-Met in proteins synthesized de novo during seed germination of wild-type Arabidopsis Ler ecotype in the absence (gray bars) or presence of 0.5 mm SA (white bars). Seed samples: 8 h, proteins radiolabeled between 0 and 8 h after imbibition; 16 h, proteins radiolabeled between 8 and 16 h after imbibition; 24 h, proteins radiolabeled between 16 and 24 h after imbibition; 32 h, proteins radiolabeled between 24 and 32 h after imbibition; 40 h, proteins radiolabeled between 32 and 40 h after imbibition; 48 h, proteins radiolabeled between 40 and 48 h after imbibition. The figure shows a representative experiment carried out three times.
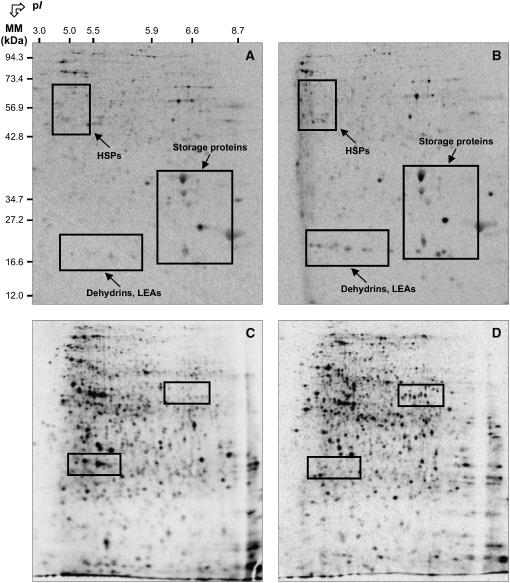
Figure 6.
Comparison of de novo protein synthesis patterns during germination of wild-type Arabidopsis Ler ecotype seeds in the presence or absence of 0.5 mm SA. A and B, Protein profiles of de novo synthesized proteins in the time period between 0 and 8 h of imbibition in water (A) or 0.5 mm SA (B). C and D, Protein profiles of de novo synthesized proteins in the time period between 16 and 24 h of imbibition in water (C) or 0.5 mm SA (D). Radiolabeling of proteins was carried out by introducing [35S]-Met in the germination assays, as described in “Materials and Methods.” Proteins were extracted, submitted to 2D gel electrophoresis, and the radiolabeled proteins revealed as described in “Materials and Methods.” The labeled protein spots in indicated windows were identified either by mass spectrometry or by comparison with Arabidopsis seed protein reference maps (Gallardo et al., 2001, 2002a; http://seed.proteome.free.fr). Several labeled spots exhibiting contrasting specific accumulation in the presence or absence of SA could not be identified because they had no match with proteins listed in Arabidopsis seed protein reference maps. Protein spot quantification was carried out as described in “Materials and Methods” and from at least three gels for each seed sample.
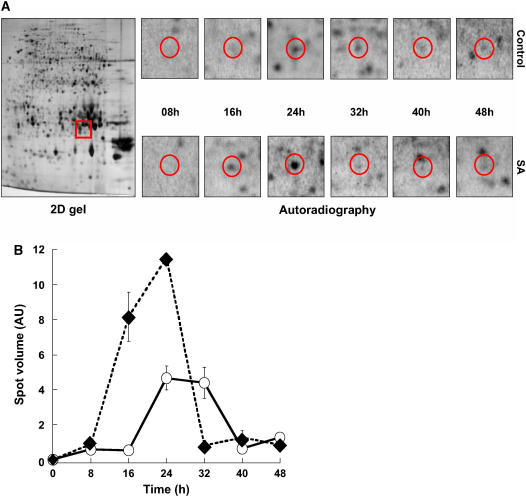
From a methodological point of view, dynamic proteomics proved much more potent than classical proteomics to reveal differential accumulation of proteins. However, it must be stressed that there was overall good agreement between the results obtained with these two approaches. For example, by monitoring the kinetics of de novo synthesis of a superoxide dismutase (spot no. 265) during germination, it appeared clearly that this protein was synthesized more precociously and more abundantly in response to SA than in its absence (Fig. 7, A and B), a result that is fully consistent with that obtained by classical proteomics (Table I).
Figure 7.
Kinetics of superoxide dismutase (At3g10920) de novo synthesis during germination of wild-type Arabidopsis Ler seeds in the presence or absence of 0.5 mm SA. Radiolabeling of de novo synthesized proteins was effected as described in “Materials and Methods.” Protein extracts were prepared, submitted to 2D gel electrophoresis, and de novo synthesized proteins were revealed by autoradiography as indicated in “Materials and Methods.” A, 2D gel analysis. Left, 2D gel of dry mature seeds stained with silver nitrate (2D gel); the red window marks the migration of the superoxide dismutase spot (no. 265). Right, radiolabeling (autoradiography) of this protein during seed germination (red circles) in water (top) or in the presence of 0.5 mm SA (bottom). B, Quantitation of de novo synthesis of superoxide dismutase (spot 265). De novo synthesis of superoxide dismutase was monitored by measuring the volume of its corresponding spot on the autoradiograms shown in A. AU, Arbitrary units; ○, imbibition of the seeds on water; ♦, imbibition of the seeds on 0.5 mm SA.
An Overview of Seed Proteins Differentially Expressed during Germination upon SA Treatment
Consistent with the data obtained by classical proteomics, the dynamic proteomics approach revealed that several initiation and elongation factors were up-regulated by SA during germination (Supplemental Table II). Thus, at 24-h seed imbibition seven of such factors were up-regulated by at least 2-fold upon SA treatment, with the translation initiation factor eIF-6 (spot no. 1,129) being up-regulated by a factor of 70 (Supplemental Table II). Concomitantly, five proteases and two subunits of the 20S proteasome were strongly up-regulated by SA (Supplemental Table II). The results are consistent with highly active protein metabolism in seed germination (Rajjou et al., 2004) and strengthen the hypothesis that SA can enhance seed germination by supporting the synthesis of proteins that are essential for germination and by favoring the mobilization/degradation of seed proteins accumulated during seed maturation.
By both classical and dynamic proteomics, isocitrate lyase and malate synthase were found to be more abundant in seeds germinated in the presence of SA than in its absence (Table I; Supplemental Tables I and II). Isocitrate lyase and malate synthase are two key enzymes of the glyoxylate cycle that play a crucial role in the synthesis of carbohydrates from storage lipids during seed germination and seedling establishment (Eastmond and Graham, 2001). Also, it has been proposed that these two enzyme activities are good indicators of seedling emergence potential and seed vigor in sugar beet, notably under stress conditions (de los Reyes et al., 2003). This suggests that a rapid set up of the glyoxylate pathway during germination can help mobilize the lipid storage reserves, enabling the exit from quiescence and the establishment of a vigorous plantlet. It is noted that the biosynthesis of several enzymes involved in related metabolic pathways (citric acid cycle, pentose phosphate pathway, glycolysis, and gluconeogenesis) was strongly activated by SA during germination. For example, at 24-h germination the biosynthesis of transketolase, enolase, malate dehydrogenase, phosphoglycerate kinase, cytosolic glyceraldehyde 3-P dehydrogenase, Fru 1,6-bisphosphatase, aconitase, phosphoglyceromutase, phosphoglucomutase, Fru bisphosphate aldolase, and pyruvate decarboxylase was up-regulated by a factor of 2 to 20 (Supplemental Table II). An elicitation of these metabolic pathways by SA would favor the transition from a metabolically quiescent state to a metabolically active state during germination, thereby accounting for the observed improvement in seed vigor in the presence of this molecule (Fig. 2).
An examination of the data in Supplemental Table II indicated that SA induced an increased synthesis of several enzymes involved in Met metabolism, namely Met synthase (spots nos. 160 and 206), S-adenosyl-Met (AdoMet) synthetase (spots nos. 49 and 62), and S-adenosylhomo-Cys hydrolase (spots nos. 359 and 729). There are several possibilities to account for the observed effect of SA on this pathway. The first is that SA helped reactivate cellular activity in germinating seeds owing to the general importance of Met and AdoMet in plant metabolism (Ravanel et al., 1998). This finding is in agreement with previous work demonstrating a requirement for Met biosynthesis in Arabidopsis seedling establishment (Gallardo et al., 2002b). A second hypothesis could rely on the possibility for the germinating seeds to set up a detoxification mechanism in response to the toxicity of exogenously applied SA (Fig. 1). It is known that in plants SA can be methylated to form methyl SA by the action of an AdoMet-dependent methyl transferase. This volatile compound is biologically inactive and considered to be an airborne signal that induces the defense responses in nondamaged organs of the same plant or in adjacent ones, being absorbed by the tissue and then converted back into free active SA by an esterase activity (Forouhar et al., 2005). If such formation of methyl SA occurred in Arabidopsis seeds to counteract the toxicity of applied SA (Fig. 1), it would require a very active synthesis of AdoMet during seed germination and seedling establishment.
Germination Behavior of the NahG Seeds
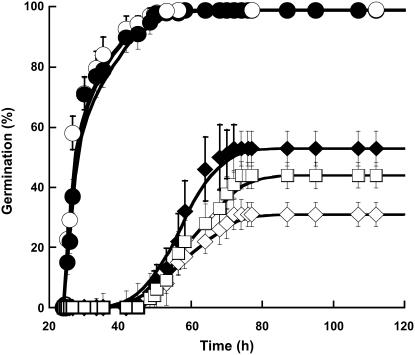
Another way to assess the influence of SA on seed germination is to use seeds descended from transgenic plants expressing the SA-metabolizing enzyme salicylate hydroxylase (SAH), which is encoded by the NahG gene of Pseudomonas putida. Owing to this transformation, NahG plants are unable to accumulate SA and thereby they have been widely used to demonstrate the essential role of this elicitor in the induction of systemic acquired resistance (Gaffney et al., 1993; Friedrich et al., 1995).
Germination parameters were not significantly different for NahG and wild-type seeds (Fig. 8). However, the NahG seeds appeared much more sensitive to salt stress than wild-type seeds (Fig. 8), a finding that is in agreement with the observed protection of SA against salt toxicity during wild-type Arabidopsis seed germination (Fig. 2). Also, as for the wild-type seeds (Fig. 2), SA added to the germination medium of NahG seeds substantially improved the extent of germination in the presence of NaCl (Fig. 8). However, this improvement was lower than with the wild-type seeds, presumably because of an efficient degradation of exogenously applied SA by the SAH enzyme in the transgenic seeds.
Figure 8.
Effect of SA on the time courses of germination of the wild-type Arabidopsis and NahG seeds under optimal conditions or under salt stress (100 mm NaCl). Seeds were germinated as described in “Materials and Methods.” ○, Wild-type seeds germinated on water; ♦, wild-type seeds germinated on 100 mm NaCl; •, NahG seeds germinated on water; ⋄, NahG seeds germinated on 100 mm NaCl; □, NahG seeds germinated on 100 mm NaCl and 0.5 mm SA. The figure shows a representative experiment carried out three times in triplicate.
Proteome Analyses of the NahG Seeds during Germination
We characterized the proteome of dry mature NahG seeds and its evolution during germination (Fig. 9). In particular, this analysis disclosed an accumulation of the SAH hydroxylase in the dry mature seeds and during seed germination (Table II; Supplemental Tables III and IV).
Figure 9.
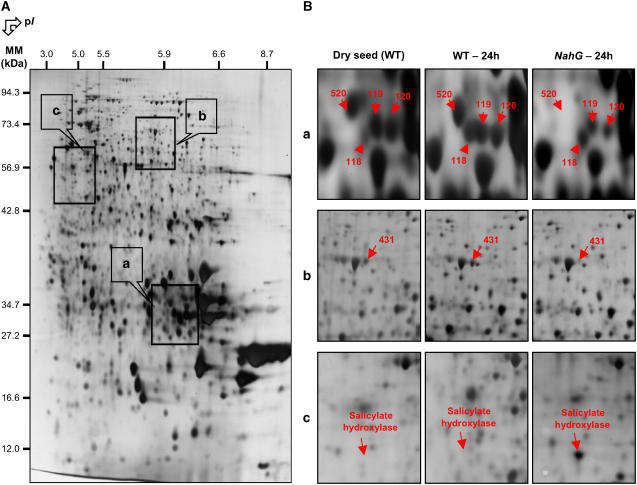
Comparison of the seed proteome of wild-type Arabidopsis and NahG mutant after 1 d imbibition. A, A silver-stained 2D gel of total proteins from Arabidopsis wild-type seeds incubated for 24 h in water. The indicated portions of the gel (a, b, and c) are reproduced in B. B, Enlarged windows (a–c) of 2D gels as shown in A for wild-type mature dry seeds (left), wild-type seeds incubated in water for 24 h (middle), and NahG seeds incubated in water for 24 h (right). The six labeled protein spots (protein nos. 118, 119, 120, 431, 520, and “Salicylate hydroxylase”) were identified by mass spectrometry or comparison with Arabidopsis seed protein reference maps (Gallardo et al., 2001, 2002a; http://seed.proteome.free.fr). Protein spot quantification was carried out as described in “Materials and Methods,” from at least three gels for each seed sample.
Table II.
Arabidopsis proteins whose abundance was significantly different in wild-type dry mature Ler seeds compared to NahG dry mature Ler seeds
% Cov., coverage; Exp., experimental; MM, molecular mass; Th., theoretical; WT, wild-type; AGI, Arabidopsis Genome Initiative; NahG/WT, normalized spot volume in the NahG Arabidopsis dry seeds divided by the normalized spot volume in the wild-type Arabidopsis dry seeds, from three different gels and independent extractions.
| No. | Exp. MM | Exp. pI | Arabidopsis Protein Name | % Cov. | Th. MM | Th. pI | AGI No. | Ratio NahG/WTa |
|---|---|---|---|---|---|---|---|---|
| kD | kD | |||||||
| 70b | 49.43 | 7.19 | 12S seed storage protein precursor | 22% | 52.49 | 7.68 | At5g44120 | 0.28 |
| 71b | 50.44 | 7.67 | 12S seed storage protein precursor | 34% | 52.49 | 7.68 | At5g44120 | 0.37 |
| 520c | 32.81 | 5.99 | α-Cruciferin 12S (seed storage protein fragment) | 12% | 31.63 | 9.08 | At5g44120 | <0.01 |
| 119b | 30.27 | 6.10 | α-Cruciferin 12S (seed storage protein fragment) | 36% | 29.23 | 6.49 | At5g44120 | 0.44 |
| 120b | 30.40 | 6.17 | α-Cruciferin 12S (seed storage protein fragment) | 28% | 29.23 | 6.49 | At5g44120 | 0.31 |
| 437c | 27.84 | 4.97 | α-Cruciferin 12S (seed storage protein fragment) | 21% | 22.81 | 9.30 | At1g03890 | 0.11 |
| 25b | 23.32 | 6.89 | β-Cruciferin 12S (seed storage protein fragment) | 27% | 21.20 | 6.19 | At4g28520 | 0.17 |
| 439c | 25.90 | 5.88 | β-Cruciferin 12S (seed storage protein fragment) | 21% | 23.36 | 6.96 | At5g44120 | 0.28 |
| 236c | 26.94 | 5.74 | Cupin family protein α subunit | 11% | 23.52 | 5.83 | At2g28490 | 2.55 |
| 140b | 60.26 | 4.15 | Calreticulin | 15% | 48.53 | 4.46 | At1g56340 | 0.53 |
| 438c | 25.92 | 5.39 | 20S proteasome α-subunit B (PAB1; PRC3) | 8% | 25.68 | 5.39 | At1g16470 | 0.19 |
| 440c | 27.27 | 5.88 | Expressed protein | 26% | 27.28 | 6.22 | At1g05510 | 5.38 |
| 431c | 66.68 | 5.84 | LEA | 32% | 67.18 | 5.97 | At2g42560 | 0.44 |
| 435c | 55.34 | 5.64 | Lectin family protein | 19% | 51.18 | 5.50 | At2g33070 | 0.38 |
| 436c | 52.15 | 5.93 | Lectin family protein | 27% | 49.69 | 6.03 | At3g21380 | 0.29 |
| SAHc | 46.95 | 4.80 | SAH (salicylate 1-monooxygenase) | 76% | 46.94 | 5.00 | P23262 | >100 |
Data obtained from densitometric analysis of individual spots from proteins in 2D gels stained with silver nitrate.
Listed proteins correspond to previously identified proteins (Gallardo et al., 2001, 2002a; Rajjou et al., 2004; Job et al., 2005).
Listed proteins correspond to proteins identified during this work; the peptide sequences determined are available in Supplemental Table V.
A comparison of the proteome for the dry mature NahG and wild-type seeds revealed 16 polypeptides showing reproducible differential variations in their accumulation levels (Table II). A 20S proteasome subunit (PAB1; spot no. 438; At1g16470) was less abundant in dry mature NahG seeds (Table II) as well as in germinating NahG seeds (Supplemental Tables III and IV) than in the corresponding wild-type seeds. This behavior therefore is fully consistent with the observation that the 20S proteasome α-subunits A1 (PAA1; spot no. 505; At5g35590) and E2 or E1 (PAE2 or PAE1; spot no. 510; At3g14290 or At1g53850) were more abundant in wild-type seeds germinated in the presence of SA than in its absence (Table I). Moreover, the dynamic proteomics approach disclosed that the 20S proteasome α-subunits B (PAB1; spot no. 438; At1g16470) and E2 or E1 (PAE2 or PAE1; spot no. 510; At3g14290 or At1g53850) and the 26S proteasome regulatory subunit S5A (RPN10; spot no. 557; At4g38630) were up-regulated during seed germination in the presence of SA (Supplemental Table II). These combined results suggest a link between SA and proteasome, as proposed in previous studies (Etienne et al., 2000; Kim et al., 2003). Also, an Arabidopsis mutant expressing an altered RPN10 proteasome subunit displayed a decreased germination phenotype, presumably because this mutant proved hypersensitive to ABA (Smalle et al., 2003). More work is needed to understand the role of the proteasome during seed germination and its possible modulation by ABA and SA.
An important result revealed by this proteomic analysis was that the NahG seeds had substantially lower contents of storage proteins than wild-type seeds. Thus, 12S cruciferin precursor (spots nos. 70 and 71) and 12S α- or β-cruciferin subunits (spots nos. 119, 120, 520, 437, 25, and 439) were much less abundant in dry mature NahG seeds than in corresponding wild-type seeds (Table II). Since the accumulation of the storage proteins is under ABA control during seed development (Nambara et al., 1992; Kroj et al., 2003), this result reinforces the finding that there exists a link between SA content in seeds and ABA signaling.
During germination, the proteome of the NahG seeds also differed from that of wild-type seeds (Supplemental Tables III and IV). Besides the variations in the accumulation levels of the seed storage proteins mentioned above, proteins associated to stress response and embryo protection exhibited different abundance in germinating NahG seeds compared to germinating wild-type seeds. Thus, spot number 431 corresponding to a LEA protein and spot number 436 corresponding to a lectin family protein showed decreased accumulation in germinating NahG seeds compared to wild-type seeds (Supplemental Tables III and IV). Likewise, spots numbers 56 and 61 corresponding to LEA proteins were more abundant during NahG seed germination than during wild-type seed germination (Supplemental Tables III and IV). Similarly, spots numbers 58 and 92 corresponding to the lectin family protein were more abundant in germinating NahG seeds than in germinating wild-type seeds (Supplemental Tables III and IV). This lectin protein family belongs to the myrosinase-binding protein family, which is known to be regulated by methyl jasmonate or jasmonic acid (Taipalensuu et al., 1997; Geshi and Brandt, 1998; Hass et al., 2004). The jasmonic acid content has been shown to be strongly increased under stress conditions in NahG Arabidopsis plants, consistent with cross talk operating between different defense pathways (Heck et al., 2003). Our data suggest that such a cross talk functions in seeds.
CONCLUSION
These data document that a defense response can be elicited in Arabidopsis seeds and seedlings by SA. In particular, this confers to the germinating seeds a marked tolerance toward salt stress. Proteomics have been used to describe the effect of SA on accumulation of specific spots during germination as well as to describe the behavior of NahG seeds during this process. In particular, the dynamic proteomics approach allowed characterizing the de novo synthesized proteome during germination in greater detail than the classical proteomics approach based upon 2D gel staining by silver nitrate. The combined results revealed several processes potentially affected by SA. This molecule was shown to enhance the reinduction of the late maturation program during early stages of germination, thereby allowing the germinating seeds to reinforce their capacity to mount adaptive responses in environmental water stress. Other processes affected by SA concerned the quality of protein translation (e.g. enhanced accumulation of NAC subunits), the priming of seed metabolism (e.g. glyoxylate and Met metabolisms), the synthesis of antioxidant enzymes, and the mobilization of seed storage proteins. All the observed effects are likely to improve seed vigor. Another aspect revealed by this study concerned the oxidative stress entailed by SA in germinating seeds, as inferred from a characterization of the carbonylated (oxidized) proteome. Finally, the proteomic data revealed a close interplay between ABA signaling and SA elicitation of seed vigor.
On a practical point of view, this study enlightens the possibility of developing new seed treatments based upon the use of chemical elicitors of plant defense that can be included in seed coating and pelleting.
MATERIALS AND METHODS
Plant Material and Germination Experiments
Wild-type or transgenic NahG nondormant seeds of Arabidopsis (Arabidopsis thaliana) ecotype Landsberg erecta (Ler) were used in all experiments. NahG seeds were a generous gift of Dr. Xinnian Dong (Duke University). Germination assays were carried out on three replicates of 100 seeds and independent experiments. Seeds were incubated at 25°C, with 8-h light daily, on three sheets of absorbent paper (Roundfilter paper circles, Ø 45 mm, Schleicher & Schuell) and a black membrane filter with a white grid (ME 25/31, Ø 45 mm, Schleicher & Schuell) wetted with 1.3 mL of distilled water in covered plastic boxes (Ø 50 mm). Assays were carried out in the presence or absence of various concentrations of SA (Sigma), SSA (Sigma), or ASA (Sigma), and/or 100 mm NaCl. A seed was regarded as germinated when the radicle protruded through the seed coat. The Seed Calculator software (Plant Research International B.V.) was used in curve-fitting analyses to estimate the germination parameters from the germination curves.
Preparation of SA and Derivative Solutions
Since the pH of a saturated aqueous solution of SA is 2.4 (Raskin, 1992), all experiments presently described have been carried out with 2 mm SA solutions prepared in water, neutralized to pH 7.0, and kept frozen (−80°C) in aluminum foil-wrapped flasks. Stock solutions of SSA and ASA were prepared in the same way.
Preparation of Total Protein Extracts
Total protein extracts were prepared from dry mature seeds and from seeds at different stages of germination. Following grinding of seeds using mortar and pestle (150 mg representing approximately 8,400 seeds) in liquid nitrogen, total proteins were extracted at 2°C in 1.2 mL of thiourea/urea lysis buffer (Harder et al., 1999) containing 7 m urea, 2 m thiourea, 4% (w/v) CHAPS, and 1% (v/v) Pharmalyte pH 3 to 10 carrier ampholytes (Amersham Biosciences). This extraction buffer also contained 18 mm Tris-HCl (Trizma HCl; Sigma), 14 mm Trizma base (Sigma), the protease inhibitor cocktail complete Mini from Roche Diagnostics GmbH, 53 units/mL DNAse I (Roche Diagnostics), 4.9 Kunitz units/mL RNAse A (Sigma), and 0.2% (v/v) Triton X-100. After 10 min at 4°C, 14 mm dithiothreitol (DTT) was added and the protein extracts were stirred for 20 min at 4°C, then centrifuged (35,000g, 10 min) at 4°C. The supernatant was submitted to a second clarifying centrifugation as above. The final supernatant corresponded to the total protein extract. Protein concentrations in the various extracts were measured according to Bradford (1976). Bovine serum albumin was used as a standard.
2D Electrophoresis
Proteins were analyzed by 2D gel electrophoresis as described (Görg et al., 1987; Job et al., 2005). Isoelectrofocusing was carried out with protein samples with an equivalent to an extract of 105 seeds, corresponding to about 150 μg protein for all samples. Proteins from the various extracts were separated using gel strips forming an immobilized nonlinear pH gradient from 3 to 10 (Immobiline DryStrip pH 3–10 NL, 18 cm; Amersham Biosciences). The second dimension was carried out in 10% SDS-polyacrylamide gels (Job et al., 2005). Ten gels (200 × 250 × 1.0 mm) were run in parallel (Isodalt system from Amersham Biosciences). For each condition analyzed, 2D gels were made in triplicate and from two independent protein extractions.
Protein Staining and Analysis of 2D Gels
2D gels were stained with silver nitrate according to either Blum et al. (1987) for densitometric analyses or to Shevchenko et al. (1996) for the mass spectrometry analyses. Stained gels were scanned with the UMAX Powerlook III scanner equipped with the MagicScan version 4.5 from UMAX Data Systems. Image analysis was carried out with the ImageMaster 2-D Elite version 4.01 software (Amersham Biosciences), according to the instruction booklet ImageMaster 2D Elite from Amersham Biosciences. After spot detection and background subtraction (mode: lowest on boundary) 2D gels were aligned, matched, and the quantitative determination of the spot volumes was performed (mode: total spot volume normalization).
Detection of Carbonylated Proteins and Western Blotting
The appearance of carbonyl groups in proteins was analyzed by immunodetection of protein-bound 2,4-dinitrophenylhydrazone (DNP) after derivitization with the corresponding hydrazine (Korolainen et al., 2002; Job et al., 2005). SDS was added to the protein extract (100 μL; 10 μg/μL) at a final concentration of 0.8%. Following dialysis, four volumes of 10 mm DNPH (Sigma)/2 m HCl were added. Samples were agitated for 30 min at room temperature, and five volumes of 20/80 ice-cold TCA acetone containing 1 mm DTT were added to each sample. The samples were centrifuged for 15 min at 15,000g at 4°C. The precipitated protein was then washed three times with 1 mL of 1:1 (v/v) ethanol:ethyl acetate and resolubilized in the thiourea/urea lysis buffer containing 2% (v/v) Triton X-100 and 20 mm DTT. Proteins were separated by 2D PAGE as above and transferred to nitrocellulose sheets (Bio-Rad) using standard procedures. Blots were rinsed twice for 5 min in 50 mm Tris-HCl, 150 mm NaCl, pH 7.5 (Tris-buffered saline [TBS]), then incubated for 1 h at 25°C in Blocking solution (Roche Diagnostics GmbH). After incubation for 1 h with rabbit anti-DNP antibodies (Serologicals) in TBS, blots were washed four times in TBS containing 0.05% Tween 20 (v/v), and incubated for 1 h in anti-rabbit secondary antibodies conjugated to horseradish peroxidase (Sigma). Bound antibodies were visualized by using the ECL kit (Roche Diagnostics). Relative protein carbonyl levels were quantitated by densitometric analyses of the blots as described above.
Protein Identification by Mass Spectrometry
Identification of the new proteins characterized in this work was obtained by mass spectrometry. Silver-stained protein spots of interest were obtained from at least three different 2D gels. The spots were excised from 2D SDS-PAGE gels with sterile tips and put in 1.5 mL sterile tubes. Each spot was rinsed then reduced with 10 mm DTT, alkylated with 55 mm iodoacetamide, and incubated overnight at 37°C with 12.5 ng/μL trypsin (sequencing grade, Roche) in 25 mm NH4HCO3 as described (Shevchenko et al., 1996). The tryptic fragments were extracted, dried, reconstituted with 2% acetonitrile (v/v), 0.1% formic acid, and sonicated for 10 min. Analysis of tryptic peptides by tandem mass spectrometry was performed on a nanoelectrospray ionization quadrupole time-of-flight hybrid mass spectrometer (Q-TOF Ultima Global, Waters Micromass) coupled with a nano-HPLC (Cap-LC, Waters). The samples were loaded and desalted on a C18 precolumn (LC-Packings PepMap C18, 5 μm, 100 Å, 300 μm × 5 mm) at a flow rate of 20 μL/min isocratically with 0.1% formic acid. The peptides were separated on a C18 column (Atlantis dC18, 3 μm, 75 μm × 150 mm Nano Ease, Waters). After a wash with solvent A (water/acetonitrile 98/2 [v/v], 0.1% formic acid), a linear gradient from 5% to 60% of solvent B (water/acetonitrile 20/80 [v/v], 0.1% formic acid) was developed over 80 min at a flow rate of 180 nL/min. The Q-TOF spectrometer was operated in the Data Dependent Analysis mode using a 1 s mass spectrometry survey scan on three different precursor ions. The peptide masses and sequences obtained were either matched automatically to proteins in a nonredundant database (National Center for Biotechnology Information) using the Mascot MS/MS Ions Search algorithm (http://www.matrixscience.com) or blasted manually against the current databases.
De Novo Protein Synthesis
Labeled proteins were synthesized in vivo by wild-type seeds imbibed on water for 8, 16, 24, 32, 40, and 48 h as above in the presence of [35S]-Met (1.85 MBq; ICN Biomedicals SARL) under continuous light and in the presence or absence of 0.5 mm SA. Following incubation, protein extracts were prepared and protein synthesis was measured by TCA precipitation of aliquots of reaction mixtures spotted on Whatmann GF/C filters; after eight washing steps in cold 5% TCA and 0.04 m sodium pyrophosphate and two washing steps in absolute methanol, filters were dried and counted for radioactivity in a liquid scintillation counter (Dietrich et al., 1985).
Protein extracts were also submitted to 2D gel electrophoresis as described above. Proteins on the 2D gels were stained by silver nitrate (see above). Then, stained 2D gels were dried for 2 d at room temperature in a sandwich composed of, from bottom to top: one sheet of cellophane model Gel Dryer (Bio-Rad), 2D gel, one sheet of Saran wrap (VWR international SAS), and one sheet of cellophane model Gel Dyer (Bio-Rad). After drying, the upper sheet of cellophane and the Saran wrap sheet were peeled and gels were submitted to Phosphorimager analysis (Molecular Dynamics Storm 840 phosphorimager, Amersham Biosciences). Labeled 2D protein patterns were scanned as described above for the silver-nitrate-stained gels. Relative protein neosynthesis levels were quantitated by densitometric analyses of the spots on the autoradiography as described above. Proteins of interest were identified by mass spectrometry as above or by comparison with the reference protein maps for Arabidopsis seed proteome available at http://seed.proteome.free.fr.
Sequence data from this article can be found in Supplemental Table V.
Supplementary Material
This work was supported by the French Ministry of Industry and Bayer CropScience (Ph.D. thesis support to L.R.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Dominique Job (dominique.job@bayercropscience.com).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.082057.
References
- Adachi M, Kanamori J, Masuda T, Yagasaki K, Kitamura K, Mikami B, Utsumi S (2003) Crystal structure of soybean 11S globulin: glycinin A3B4 homohexamer. Proc Natl Acad Sci USA 100: 7395–7400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananieva EA, Christov KN, Popova LP (2004) Exogenous treatment with salicylic acid leads to increased antioxidant capacity in leaves of barley plants exposed to paraquat. J Plant Physiol 161: 319–328 [DOI] [PubMed] [Google Scholar]
- Barratt BIP, Lowther WL, Ferguson CM (1995) Seed coating with insecticide to improve oversown white clover (Trifolium repens L.) establishment in tussock grassland. N Z J Agric Res 38: 511–518 [Google Scholar]
- Bewley JD, Black M (1994) Seeds. Physiology of development and germination. Plenum Press, New York
- Blum H, Beier H, Gross HJ (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8: 93–99 [Google Scholar]
- Borsani O, Valpuesta V, Botella MA (2001) Evidence for a role of salicylic acid in the oxidative damage generated by NaCl and osmotic stress in Arabidopsis seedlings. Plant Physiol 126: 1024–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Campo S, Carrascal M, Coca M, Abián J, San Segundo B (2004) The defense response of germinating maize embryos against fungal infection: a proteomics approach. Proteomics 4: 383–396 [DOI] [PubMed] [Google Scholar]
- Chen Z, Silva H, Klessig DF (1993) Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science 262: 1883–1886 [DOI] [PubMed] [Google Scholar]
- Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X (2000) Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuming AC (1999) LEA proteins. In PR Shewry, R Casey, eds, Seed proteins. Kluwer Academic Press, Dordrecht, The Netherlands, pp 753–780
- Dat JF, Lopez-Delgado H, Foyer CH, Scott IM (1998) Parallel changes in H2O2 and catalase during thermotolerance induced by salicylic acid or heat acclimation in mustard seedlings. Plant Physiol 116: 1351–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Reyes BG, Myers SJ, McGrath JM (2003) Differential induction of glyoxylate cycle enzymes by stress as a marker for seedling vigor in sugar beet (Beta vulgaris). Mol Genet Genomics 269: 692–698 [DOI] [PubMed] [Google Scholar]
- Dietrich J, Teissère M, Job C, Job D (1985) Polyd(AT) dependent trinucleotide synthesis catalysed by wheat-germ RNA polymerase II: effects of nucleotide substrates and cordycepin triphosphate. Nucleic Acids Res 13: 6155–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL, Bhattacharyya MK (1995) Engineering disease resistance in plants: an overview. In Singh RP, Singh US, eds, Molecular Methods in Plant Pathology. CRC Press, Boca Raton, FL, pp 249–270
- Eastmond PJ, Graham IA (2001) Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci 6: 72–78 [DOI] [PubMed] [Google Scholar]
- Etienne P, Petitot AS, Houot V, Blein JP, Suty L (2000) Induction of tcI 7, a gene encoding a beta-subunit of proteasome, in tobacco plants treated with elicitins, salicylic acid or hydrogen peroxide. FEBS Lett 466: 213–218 [DOI] [PubMed] [Google Scholar]
- Fauth M, Merten A, Hahn MG, Jeblick W, Kauss H (1996) Competence for elicitation of H2O2 in hypocotyls of cucumber is induced by breaching the cuticle and is enhanced by salicylic acid. Plant Physiol 110: 347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR (1993) Abscisic acid-insensitive mutations provide evidence for stage-specific signal pathways regulating expression of an Arabidopsis late embryogenesis-abundant (lea) gene. Mol Gen Genet 238: 401–408 [DOI] [PubMed] [Google Scholar]
- Forouhar F, Yang Y, Kumar D, Chen Y, Fridman E, Park SW, Chiang Y, Acton TB, Montelione GT, Pichersky E, et al (2005) Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. Proc Natl Acad Sci USA 102: 1773–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich L, Vernooij B, Gaffney T, Morse A, Ryals J (1995) Characterization of tobacco plants expressing a bacterial salicylate hydroxylase gene. Plant Mol Biol 29: 959–968 [DOI] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–756 [DOI] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D (2001) Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol 126: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D (2002. a) Proteomics of Arabidopsis seed germination: a comparative study of wild-type and gibberellin-deficient seeds. Plant Physiol 129: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D (2002. b) Importance of methionine biosynthesis for Arabidopsis seed germination and seedling growth. Physiol Plant 116: 238–247 [DOI] [PubMed] [Google Scholar]
- Garciarrubio A, Legaria JP, Covarrubias AA (1997) Abscisic acid inhibits germination of mature Arabidopsis seeds by limiting the availability of energy and nutrients. Planta 203: 182–187 [DOI] [PubMed] [Google Scholar]
- Geshi N, Brandt A (1998) Two jasmonate-inducible myrosinase-binding proteins from Brassica napus L. seedlings with homology to jacalin. Planta 204: 295–304 [DOI] [PubMed] [Google Scholar]
- Görg A, Postel W, Weser J, Günther S, Strahler JR, Hanash SM, Somerlot L (1987) Elimination of point streaking on silver stained two-dimensional gels by addition of iodoacetamide to the equilibration buffer. Electrophoresis 8: 122–124 [Google Scholar]
- Guan L, Scandalios JG (1995) Developmentally related responses of maize catalase genes to salicylic acid. Proc Natl Acad Sci USA 92: 5930–5934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajheidari M, Abdollahian-Noghabi M, Askari H, Heidari M, Sadeghian SY, Ober ES, Salekdeh GH (2005) Proteome analysis of sugar beet leaves under drought stress. Proteomics 5: 950–960 [DOI] [PubMed] [Google Scholar]
- Harder A, Wildgruber R, Nawrocki A, Fey SJ, Larsen PM, Görg A (1999) Comparison of yeast cell protein solubilization procedures for two-dimensional electrophoresis. Electrophoresis 20: 826–829 [DOI] [PubMed] [Google Scholar]
- Hass C, Lohrmann J, Albrecht V, Sweere U, Hummel F, Yoo SD, Hwang I, Zhu T, Schafer E, Kudla J, et al (2004) The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J 23: 3290–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck S, Grau T, Buchala A, Metraux JP, Nawrath C (2003) Genetic evidence that expression of NahG modifies defence pathways independent of salicylic acid biosynthesis in the Arabidopsis-Pseudomonas syringae pv. tomato interaction. Plant J 36: 342–352 [DOI] [PubMed] [Google Scholar]
- Job C, Rajjou L, Lovigny Y, Belghazi M, Job D (2005) Patterns of protein oxidation in Arabidopsis seeds and during germination. Plant Physiol 138: 790–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson E, Olsson O, Nyström T (2004) Progression and specificity of protein oxidation in the life cycle of Arabidopsis thaliana. J Biol Chem 279: 22204–22208 [DOI] [PubMed] [Google Scholar]
- Kim M, Yang KS, Kim YK, Paek KH, Pai HS (2003) Molecular characterization of NbPAF encoding the alpha6 subunit of the 20S proteasome in Nicotiana benthamiana. Mol Cells 15: 127–132 [PubMed] [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H (2002) Seed dormancy and germination. Curr Opin Plant Biol 5: 33–36 [DOI] [PubMed] [Google Scholar]
- Korolainen MA, Goldsteins G, Alafuzoff I, Koistinaho J, Pirttila T (2002) Proteomic analysis of protein oxidation in Alzheimer's disease brain. Electrophoresis 23: 3428–3433 [DOI] [PubMed] [Google Scholar]
- Kroj T, Savino G, Valon C, Giraudat J, Parcy F (2003) Regulation of storage protein gene expression in Arabidopsis. Development 130: 6065–6073 [DOI] [PubMed] [Google Scholar]
- Levine RL, Williams JA, Stadtman ER, Shacter E (1994) Carbonyl assays for determination of oxidatively modified proteins. Methods Enzymol 233: 346–357 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua N-H (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32: 317–328 [DOI] [PubMed] [Google Scholar]
- McCue P, Zheng Z, Pinkham JL, Shetty K (2000) A model for enhanced pea seedling vigor following low pH and salicylic acid treatments. Process Biochem 35: 603–613 [Google Scholar]
- Metraux JP, Signer H, Ryals J, Ward E, Wyss-Benz M, Gaudin J, Raschdorf K, Schmid E, Blum W, Inverardi B (1990) Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250: 1004–1006 [DOI] [PubMed] [Google Scholar]
- Nambara E, Naito S, McCourt P (1992) A mutant of Arabidopsis which is defective in seed development and storage protein accumulation is new abi3 allele. Plant J 11: 435–441 [Google Scholar]
- Nyström T (2005) Role of oxidative carbonylation in protein quality control and senescence. EMBO J 24: 1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A, Mann M (2000) Proteomics to study genes and genomes. Nature 405: 837–846 [DOI] [PubMed] [Google Scholar]
- Polle A (1997) Defence against photo-oxidative damage in plants. In JG Scandalios, ed, Oxidative Stress and Molecular Biology of Antioxidant Defences. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 623–666
- Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D (2004) The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol 134: 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Gallardo K, Job C, Job D (2006) Proteome analysis for the study of developmental processes in plants. In C Finnie, ed, Plant proteomics. Blackwell Publishing, Oxford, pp 151–184
- Rao MV, Davis RD (1999) Ozone-induced cell death occurs via two distinct mechanisms in Arabidopsis: the role of salicylic acid. Plant J 17: 603–614 [DOI] [PubMed] [Google Scholar]
- Rao MV, Paliyath G, Ormrod DP, Murr DP, Watkins CB (1997) Influence of salicylic acid on H2O2 production, oxidative stress, and H2O2-metabolizing enzymes: salicylic acid-mediated oxidative damage requires H2O2. Plant Physiol 115: 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin I (1992) Role of salicylic acid in plants. Annu Rev Plant Physiol Plant Mol Biol 43: 439–463 [Google Scholar]
- Ravanel S, Gakière B, Job D, Douce R (1998) The specific features of methionine biosynthesis and metabolism in plants. Proc Natl Acad Sci USA 95: 7805–7812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rospert S, Dubaquie Y, Gautschi M (2002) Nascent-polypeptide-associated complex. Cell Mol Life Sci 59: 1632–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirova FM, Sakhabutdinova AR, Bezrukova MV, Fatkhutdinova RA, Fatkhutdinova DR (2003) Changes in the hormonal status of wheat seedlings induced by salicylic acid and salinity. Plant Sci 164: 317–322 [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Emborg TJ, Babiychuk E, Kushnir S, Vierstra RD (2003) The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15: 965–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipalensuu J, Eriksson S, Rask L (1997) The myrosinase-binding protein from Brassica napus seeds possesses lectin activity and has a highly similar vegetatively expressed wound-inducible counterpart. Eur J Biochem 250: 680–688 [DOI] [PubMed] [Google Scholar]
- Wiedmann B, Sakai H, Davis TA, Wiedmann M (1994) A protein complex required for signal-sequence-specific sorting and translocation. Nature 370: 434–440 [DOI] [PubMed] [Google Scholar]
- Yan S, Tang Z, Su W, Sun W (2005) Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics 5: 235–244 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.