Abstract
Lateral organs in flowering plants display polarity along their adaxial-abaxial axis with distinct cell types forming at different positions along this axis. Members of three classes of transcription factors in Arabidopsis (Arabidopsis thaliana; the Class III homeodomain/leucine zipper [HD-ZIP] proteins, KANADI proteins, and YABBY proteins) are expressed in either the adaxial or abaxial domain of organ primordia where they confer these respective identities. Little is known about the factors that act upstream of these polarity-determining genes to regulate their expression. We have investigated the relationship between AINTEGUMENTA (ANT), a gene that promotes initiation and growth of lateral organ primordia, and polarity genes. Although ant single mutants do not display any obvious defects in organ polarity, loss of ANT activity in combination with mutations in one or more YABBY genes results in polarity defects greater than those observed in the yabby mutants alone. Our results suggest that ANT acts in combination with the YABBY gene FILAMENTOUS FLOWER (FIL) to promote organ polarity by up-regulating the expression of the adaxial-specifying HD-ZIP gene PHABULOSA. Furthermore, we show that ANT acts with FIL to up-regulate expression of the floral homeotic gene APETALA3. Our work defines new roles for ANT in the development of lateral organs.
In flowering plants, leaves and floral organs are produced on the periphery of apical meristems. These lateral organs possess an inherent asymmetry with regard to the meristem in that their adaxial side is adjacent and close to the meristem, while their abaxial side is located further from the meristem. This asymmetry gives rise to a polarity that is readily apparent at the cellular and whole organ level and which can have important functional consequences. For example, cells within the adaxial region of a leaf are specialized for light capture, while those in the abaxial region are specialized for gas exchange. In addition, outgrowth of the leaf lamina is dependent on the juxtaposition of cells with adaxial and abaxial identities (Waites and Hudson, 1995). A similar mechanism may be responsible for the outgrowth of flattened floral organs such as sepals and petals.
Members of three classes of transcription factors contribute to the establishment of adaxial and abaxial cell fates in lateral organs of Arabidopsis (Arabidopsis thaliana; for review, see Engstrom et al., 2004). Class III homeodomain/Leu zipper (HD-ZIP) proteins specify adaxial identity, while KANADI and YABBY proteins specify abaxial identity. Three HD-ZIP genes, PHABULOSA (PHB), PHAVOLUTA (PHV), and REVOLUTA (REV), are expressed in the adaxial domain of lateral organ primordia (McConnell et al., 2001; Emery et al., 2003). Dominant mutations in PHB or PHV result in transformation of abaxial cells to adaxial fates and the production of radially symmetric lateral organs (McConnell and Barton, 1998; McConnell et al., 2001). phb phv rev triple mutants produce just a single abaxialized and radialized cotyledon in the most severe case (Emery et al., 2003).
At least three members of the KANADI gene family (KAN1, KAN2, and KAN3) redundantly specify abaxial identity (Eshed et al., 2001; Kerstetter et al., 2001). Transformation of abaxial cell types into adaxial cell types is observed with increasing severity in kan double and triple mutants, while ectopic expression of KAN genes results in the development of abaxial tissues in adaxial regions and the radialization of lateral organs (Eshed et al., 2001, 2004; Kerstetter et al., 2001). KAN1 is expressed in the abaxial domain of developing lateral organs, complementary to the expression of PHB-like genes in the adaxial domain (Kerstetter et al., 2001). Members of the YABBY gene family also contribute to the specification of abaxial identity. Three YABBY genes, FILAMENTOUS FLOWER (FIL), YABBY2 (YAB2), and YABBY3 (YAB3), are expressed in the abaxial half of all lateral organ primordia (Siegfried et al., 1999). Ectopic expression of these YABBY genes can convert some adaxial cell types into abaxial cells (Sawa et al., 1999; Siegfried et al., 1999). Although loss of both FIL and YAB3 activity does not result in conversion of abaxial cells into adaxial cells, increased adaxialization of lateral organs occurs in kan1/+ kan2 plants upon loss of FIL and YAB3 activity (Eshed et al., 2004). KANADI proteins are members of the GARP family of transcriptional regulators, while YABBY proteins are zinc finger-containing proteins with an HMG box-like domain (Bowman and Smyth, 1999; Sawa et al., 1999; Eshed et al., 2001; Kerstetter et al., 2001).
It has been proposed that a meristem-derived signal is responsible for the establishment of adaxial-abaxial polarity in lateral organs (for review, see Bowman et al., 2002). Cells closest to the meristem, those in the adaxial domain, perceive this signal, while cells further away do not. In response, PHB-like proteins are activated in cells of the adaxial domain. Because of the antagonism between PHB-like and KANADI genes, KANADI gene expression is subsequently inhibited in the adaxial region and becomes restricted to the abaxial domain (Eshed et al., 2004). Thus, an initial asymmetry due to the meristem-derived signal is thought to be maintained by antagonism between the PHB-like and KANADI genes and results in restriction of PHB-like gene expression to the adaxial domain and KANADI gene expression to the abaxial domain. Additionally, the mutual repression between the PHB-like and KANADI genes leads to the abaxial specific expression of the YABBY genes (Eshed et al., 2004).
We are interested in the relationship between factors that promote the initiation and growth of lateral organ primordia, such as AINTEGUMENTA (ANT), and factors that act within lateral organ primordia to establish their polarity, such as PHB-like, KANADI, and YABBY proteins. ANT expression is up-regulated in leaf and flower founder cells in apical meristems and is one of the earliest markers of lateral organ specification (Elliott et al., 1996; Long and Barton, 2000). It has been suggested that YABBY genes could initially be activated by proteins that promote primordia initiation (such as ANT), while other factors act later to restrict YABBY gene expression to abaxial regions (Bowman, 2000). ANT encodes a transcription factor of the APETALA2/ethylene-responsive element binding factor family that binds to 5′-gCAC(A/G)N(A/T)TcCC(a/g)ANG(c/t)-3′ DNA sequences (Nole-Wilson and Krizek, 2000). Although we find that ANT can bind in vitro to such a sequence within the FIL and YAB3 promoters, ANT is not required for normal levels of FIL or YAB3 expression. Characterization of fil-8 ant-4 and fil-8 yab3-2 ant-4 double and triple mutants does suggest a role for ANT in the establishment of adaxial-abaxial polarity in leaves and floral organs.
RESULTS
ANT Binds to a Conserved Element in the FIL and YAB3 Promoters
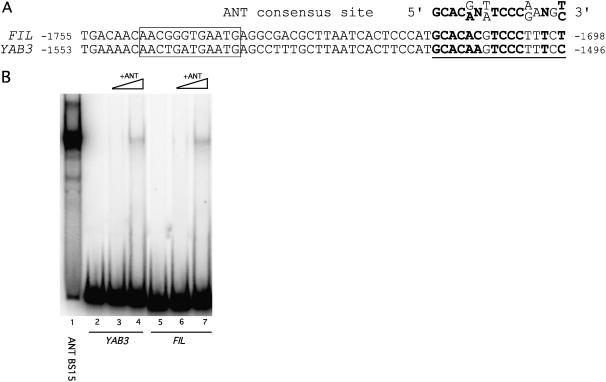
FIL and YAB3 are expressed in largely overlapping domains and share sequence similarity within an approximately 300-bp 5′ regulatory region, part of which is shown in Figure 1A (Siegfried et al., 1999; Watanabe and Okada, 2003). Deletion analysis of the FIL promoter identified two cis-acting regulatory elements required for proper FIL expression (Watanabe and Okada, 2003). A region proximal to the FIL coding sequence (−1,742 to −1,547) is required for expression in both adaxial and abaxial domains, while a 12-bp (−1,748 to −1,737) sequence is required for the abaxial-specific expression of the FIL gene (Fig. 1A). A sequence with similarity to the ANT consensus binding site is present within the former FIL region and within the YAB3 promoter. The putative ANT binding sites in the FIL and YAB3 promoters match the in vitro-determined ANT consensus binding site in 10 of 14 conserved positions (Fig. 1A). Gel mobility shifts revealed that ANT binds in vitro to fragments of both promoters that contain these sequences (Fig. 1B). The observed binding to either of these sites is weaker than that of ANT to the consensus binding site (BS15; Fig. 1B). Binding to the FIL and YAB3 promoter sites was competed by unlabeled BS15 but not by a mutated version of this binding site (data not shown).
Figure 1.
ANT binds to a sequence within the FIL and YAB3 promoters. A, Alignment of a conserved element within the 5′ regulatory region of FIL and YAB3 to the ANT consensus site. The putative ANT binding site is underlined. Nucleotides shared between the ANT consensus binding site and the FIL and YAB3 promoters are shown in bold. A putative Kruppel binding site (−1,748 to −1,737), required for repression of FIL in the adaxial domain, is boxed. Numbers indicate positions relative to the start codons. B, Gel shift showing binding of ANT to the consensus binding site (BS15), the YAB3 promoter site, and the FIL promoter site. The YAB3 promoter fragment is a 106-bp sequence corresponding to nucleotides −1,561 to −1,456 and the FIL promoter fragment is a 131-bp sequence corresponding to nucleotides −1,763 to −1,633. Lanes 2 and 5 contain no protein. Increasing amounts of ANT protein are shown in lanes 3 and 4 (and 6 and 7). The same amount of ANT protein was used in lanes 1, 4, and 7.
FIL and YAB3 Expression Is Normal in ant Flowers
To probe the potential role of ANT in FIL and YAB3 regulation, we examined the expression of FIL and YAB3 in an ant mutant background. If ANT is a positive regulator of FIL and/or YAB3, we might expect FIL and YAB3 expression to be reduced in ant mutants. FIL expression was examined in ant-4 flowers by in situ hybridization.
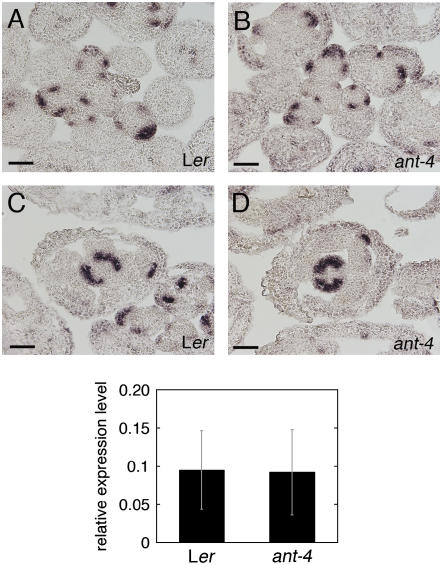
A similar level and pattern of FIL expression was observed in Landsberg erecta (Ler) and ant-4 flowers (Fig. 2, A–D). YAB3 mRNA was examined in Ler and ant-4 inflorescences by real-time reverse transcription (RT)-PCR. Similar levels of YAB3 mRNA were present in both genotypes (Fig. 2E). These results suggest that ANT activity is not required for activation of YAB3 or FIL in flowers.
Figure 2.
FIL and YAB3 expression in wild-type and ant-4 plants. A, FIL mRNA is present in floral primordia and sepal primordia in this transverse Ler inflorescence section. B, FIL mRNA in an ant-4 inflorescence. C, FIL expression in a stage 8 Ler flower. D, FIL expression in a stage 8 ant-4 flower. Size bars correspond to 50 μm in A to D. Bottom section, Relative expression levels (compared to ACTIN2) of YAB3 in Ler and ant-4 inflorescences. The average of two experiments is shown. The bars show sd.
fil ant and fil yab3 ant Plants Are Reduced in Size
To gain insight into the relationship between ANT and the two YABBY genes, we generated fil-8 ant-4 double mutants and fil-8 yab3-2 ant-4 triple mutants. The double and triple mutant plants were dwarfed and exhibited severe alterations in organ development during both vegetative and reproductive development (Figs. 3, A–D, and 5, A and B). While the leaves of yab3-2, fil-8, and ant-4 single mutants were not dramatically different in size from those of wild type (Kumaran et al., 2002), the leaves of fil-8 yab3-2, fil-8 ant-4, and fil-8 yab3-2 ant-4 were considerably smaller than those of wild type (Fig. 3, E–H; Table I). The decreased leaf area of the double and triple mutants resulted from reductions in both length and width (Table I). In the triple fil-8 yab3-2 ant-4 mutant, there was a dramatic reduction in lamina expansion such that the petioles of fil-8 yab3-2 ant-4 leaves were often not clearly distinguishable from the lamina (Fig. 3, H and L).
Figure 3.
Wild-type, fil-8 yab3-2, fil-8 ant-4, and fil-8 yab3-2 ant-4 leaves. A to D, Mature rosettes just prior to bolting: Ler (A), fil-8 yab3-2 (B), fil-8 ant-4 (C), and fil-8 yab3-2 ant-4 (D). The pictures in A to D are taken at the same magnification. Insets in B and D show closer views. E to H, Fully expanded leaves from Ler (E), fil-8 yab3-2 (F), fil-8 ant-4 (G), and fil-8 yab3-2 ant-4 (H) plants. The pictures in E to H are taken at the same magnification. I to L, Vascular patterns of Ler (I), fil-8 yab3-2 (J), fil-8 ant-4 (K), and fil-8 yab3-2 ant-4 (L) fully expanded leaves. The pictures in I to L are taken at the same magnification. Leaves shown in E to L were from position 5 or 6 of the rosette. Size bars correspond to 1 mm in I to L.
Figure 5.
Flowers from wild-type, ant-4, fil-8, fil-8 yab3-2, fil-8 ant-4, and fil-8 yab3-2 ant-4 plants. A, Side view of fil-8, fil-8 yab3-2, fil-8 ant-4, and fil-8 yab3-2 ant-4 plants of approximately 3 weeks of age. B, A single image showing the side views of fil-8, fil-8 yab3-2, fil-8 ant-4, and fil-8 yab3-2 ant-4 flowers (from left to right) and the relative sizes of the flowers. C, Ler flower. D, ant-4 flower. E, fil-8 flower. F, fil-8 yab3-2 flower. G, fil-8 ant-4 flower. H, Early arising fil-8 ant-4 flower. Arrow points to a staminoid organ and arrowhead points to a white petaloid organ. I, fil-8 yab3-2 ant-4 flower. J, fil-8 yab3-2 ant-4 inflorescence meristem that has initiated filament production. Arrow points to the inflorescence meristem.
Table I.
Leaf size in wild-type and mutant plants
The data are indicated as averages ± sd. *, Values that are significantly different from wild type (Student's t test, P < 0.01).
| Leaf Area | Leaf Width | Leaf Length | |
|---|---|---|---|
| mm2 | mm | mm | |
| Ler | 55 ± 8.4 | 6.8 ± 0.52 | 12.5 ± 1.1 |
| ant-4 | 48 ± 5.7* | 7.0 ± 0.49 | 9.3 ± 0.64* |
| fil-8 | 54 ± 14 | 7.3 ± 0.94 | 11.5 ± 2.0 |
| fil-8 yab3-2 | 10 ± 2.3* | 3.2 ± 0.60* | 5.3 ± 0.85* |
| fil-8 ant-4 | 27 ± 13* | 5.0 ± 1.1* | 8.9 ± 1.9* |
| fil-8 yab3-2 ant-4 | 4.1 ± 1.1* | 1.6 ± 0.48* | 3.7 ± 1.9* |
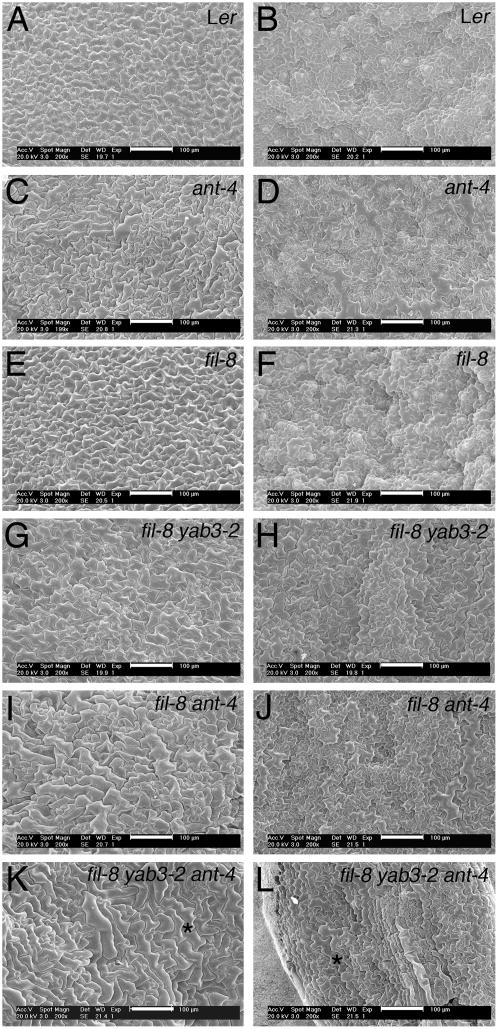
To determine the basis for the smaller leaf size in fil-8 ant-4 and fil-8 yab3-2 ant-4 plants, the size of mature leaf epidermal cells was examined using scanning electron microscopy (SEM). Epidermal cells were larger in fil-8 ant-4 and fil-8 yab3-2 ant-4 plants compared to Ler (Fig. 4, A, B, and I–L). This indicates that the smaller leaf blades of the double and triple mutants are due to the presence of fewer cells. Similarly, the reduced height of the double and triple mutant plants (Fig. 5A; Table II) was due to fewer cells in the stem (data not shown).
Figure 4.
Adaxial and abaxial surfaces of leaves from wild-type, ant-4, fil-8, fil-8 yab3-2, fil-8 ant-4, and fil-8 yab3-2 ant-4 plants. Shown are adaxial leaf surfaces of Ler (A), ant-4 (C), fil-8 (E), fil-8 yab3-2 (G), fil-8 ant-4 (I), and fil-8 yab3-2 ant-4 (K). Also shown are abaxial leaf surfaces of Ler (B), ant-4 (D), fil-8 (F), fil-8 yab3-2 (H), fil-8 ant-4 (J), and fil-8 yab3-2 ant-4 (L). Two cells with similar morphologies in K and L are noted with *. Size bars correspond to 100 μm.
Table II.
Height of wild-type and mutant plants
The data are indicated as averages ± sd. *, Values that are significantly different from wild type (Student's t test, P < 0.01).
| Height | |
|---|---|
| cm | |
| Ler | 14.2 ± 2.52 |
| ant-4 | 11.9 ± 1.87 |
| fil-8 | 10.4 ± 1.85 |
| fil-8 yab3-2 | 1.8 ± 0.492* |
| fil-8 ant-4 | 3.8 ± 0.898* |
| fil-8 yab3-2 ant-4 | 1.1 ± 0.084* |
fil ant and fil yab ant Mutants Show Disruptions in Leaf Polarity
A juxtaposition of adaxial and abaxial cell types is thought to be required for leaf blade expansion (for review, see Bowman, 2000; Bowman et al., 2002). To determine whether the reduced expansion of leaf blades in fil-8 ant-4 and fil-8 yab3-2 ant-4 plants might be a consequence of reduced leaf polarity, epidermal cell morphologies were examined. The adaxial and abaxial surfaces of wild-type leaves are distinct. The adaxial epidermis is flat, while the abaxial epidermis is undulating (Fig. 4, A and B). In addition, adaxial epidermal cells are fairly uniform in size, while abaxial epidermal cells are variably sized and puzzle shaped (Fig. 4, A and B). The adaxial and abaxial surfaces of ant-4 and fil-8 leaves are normal (Fig. 4, C–F). The adaxial surface of fil-8 yab3-2 leaves is normal (Fig. 4G). However, the abaxial surface of fil-8 yab3-2 leaves was altered slightly from wild type, indicating a partial loss of abaxial identity (Fig. 4H; Siegfried et al., 1999).
More dramatic changes in adaxial and abaxial identities were observed in fil-8 ant-4 and fil-8 yab3-2 ant-4 plants. Adaxial epidermal cells of fil-8 ant-4 leaves were variable in size and sometimes puzzle shaped, slightly resembling abaxial epidermal cells (Fig. 4I). In addition, the abaxial surface was flatter than wild type and the cells larger than wild type (Fig. 4J). This suggests a partial loss of both adaxial and abaxial identities in fil-8 ant-4 plants. Thus, at least some of the reduced growth of fil-8 ant-4 leaves may result from a loss of polarity. The more dramatic reduction in lamina expansion in the triple mutant was correlated with a more complete loss of polarity. In fil-8 yab3-2 ant-4 leaves, adaxial and abaxial epidermal cells closely resembled each other with neither the adaxial or abaxial surface displaying its characteristic appearance (Fig. 4, K and L). Loss of both adaxial and abaxial identities distinguishes the fil ant and fil yab ant mutants from mutations in either the KAN or PHB-like genes, where there is replacement of one identity with the other identity (i.e. replacement of abaxial cell fates with adaxial identities in kan mutants).
fil-8 ant-4 and fil-8 yab3-2 ant-4 leaves also exhibit alterations in their vascular patterning. Vascular tissue in wild-type leaves exhibits a reticulate pattern with minor veins branching from the major vein (Fig. 3I). There was a marked decrease in vascular branching in the leaves of fil-8 ant-4 plants (Fig. 3K). This phenotype is similar to that reported previously for yab3-1 fil-5 leaves (Siegfried et al., 1999) and fil-8 yab3-2 (Fig. 3J). fil-8 yab3-2 ant-4 leaves often have just a single vein running the length of the leaf (Fig. 3L). In some cases, one to several shorter veins branch from this central vein.
fil ant and fil yab ant Mutants Show Disruptions in Floral Organ Identity and Polarity
fil-8 ant-4 and fil-8 yab3-2 ant-4 plants exhibit inflorescence defects similar to those observed in fil-8 plants with the inflorescence meristem switching between the production of flowers and filaments (Sawa et al., 1999). In fil-8 ant-4, around 12 flowers were produced before the inflorescence meristem started to produce filaments (Fig. 6A). This is similar to the number of individual flowers initiated by the inflorescence meristem of fil-8 plants prior to filament production. Fewer flowers were produced prior to filament production in fil-8 yab3-2 ant-4 plants (Fig. 5J). After producing some filaments, fil-8 ant-4 and fil-8 yab3-2 ant-4 inflorescence meristems switched to producing a mixture of flower-like structures and filaments (Fig. 6). This was subsequently followed by termination of the inflorescence meristem. After termination of the primary inflorescence, secondary and axillary inflorescences grew out, resulting in the production of short and bushy fil-8 ant-4 and fil-8 yab3-2 ant-4 plants (Fig. 5).
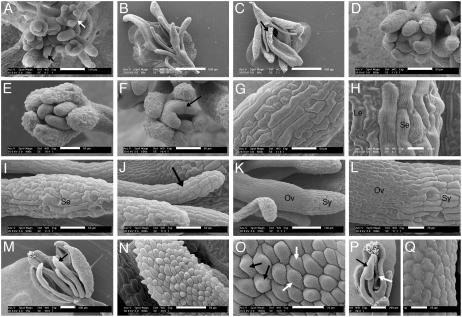
Figure 6.
SEM analyses of fil-8 ant-4 flowers. A, fil-8 ant-4 inflorescence meristem producing a mixture of flowers (white arrow) and filamentous structures (black arrow). B, fil-8 ant-4 flower. C, fil-8 ant-4 flower. Stigmatic papillae are indicated with an arrow. D, Stage 7 fil-8 ant-4 flower showing three whorls of organ primordia. E, fil-8 ant-4 flower with two visible whorls of organs. The inner whorl contains three unfused organ primordia. F, Stage 8 fil-8 ant-4 flower with two visible whorls of organs. A black arrow points to a region of fusion between the inner whorl organ primordia. G, An outer whorl organ with sepal-like cells. H, An outer whorl organs with leaf-like (Le) and sepal-like (Se) cells. I, An outer whorl filamentous organ with sepal-like (Se) cells. J, An inner whorl filamentous organ (arrow). K, An inner whorl carpel-like organ with ovary valve-like (Ov) cells at the base and style-like (Sy) cells at the top. L, Close-up of organ shown in K. M, An early arising fil-8 ant-4 flower. A black arrow indicates a petal-like organ. N, Close-up of the petal-like organ in M showing cells with petal morphologies. O, Close-up of the organ in M. Petal cells with adaxial (black arrows) and abaxial (white arrows) morphologies are present on the abaxial surface of this fil-8 ant-4 organ. The adaxial petal epidermal cells are conical in shape with epicuticular thickenings oriented along the cone axis. The abaxial epidermal petal cells are flatter with more zigzagged epicuticular thickenings. P, Staminoid organ (black arrow) present in an early arising fil-8 ant-4 flower. The white arrow points to adaxial carpel tissue. Q, Close-up of the anther-like region of the staminoid organ in P. Size bars correspond to 20 μm in H, L, O, and Q; 50 μm in D to G, I, J, and N; 100 μm in A and K; 200 μm in M and P; and 500 μm in B and C.
The flowers produced by fil-8 ant-4 and fil-8 yab3-2 ant-4 plants were much smaller than flowers of wild type, fil-8, or fil-8 yab3-2 (Fig. 5B). In addition, they exhibited loss of floral identity as demonstrated by the presence of flowers with subtending leaves and by a loss of floral organ identity. fil-8 ant-4 and fil-8 yab3-2 ant-4 flowers typically consisted of narrow, flat, green organs; filaments; and carpelloid organs (Figs. 5, G and I, and 6, B and C). Because these organs lack most recognizable features of floral organs, we examined their development and cell types by SEM to better characterize them. Because fil-8 ant-4 and fil-8 yab3-2 ant-4 flowers were quite similar, we present a detailed SEM analysis of just fil-8 ant-4 flowers.
fil-8 ant-4 flowers typically produced two or three whorls of floral organs with variable numbers and positions of organs within each whorl (Fig. 6, D–F). Flowers arising later on the inflorescence typically produced a fewer number of organs. The flat, outermost organs from early arising fil-8 ant-4 flowers had epidermal cells resembling those of sepals (Fig. 6G). In later-arising flowers, these organs became thinner and more pointed. SEM analysis showed that these later-arising outer whorl organs were mosaics containing both leaf-like and sepal-like cells (Fig. 6H). Filamentous organs present in the outer two whorls of fil-8 ant-4 flowers were variable in appearance. Those in the outermost whorl were typically dark green and had epidermal cells resembling those of sepals (Fig. 6I), while filaments in the second whorl were light green or white in color with more regular cells in files (Fig. 6J). In addition, filaments in the outer whorl tended to be thicker than those in the second whorl. The flat innermost organs of fil-8 ant-4 flowers resembled unfused or partially fused carpels. Style-like cells were found at the top of these organs (Fig. 6, K and L), and valve-like ovary cells were present along the rest of their length. Stigmatic papillae were sometimes present, although not necessarily at the apex of these organs (Fig. 6C). The carpel-like organs arose as distinct primordia rather than the fused ring of tissue that develops in wild-type flowers (Fig. 6D). These primordia gave rise to either distinct organs (Fig. 6E) or partially fused organs (Fig. 6F). Internal tissues present within a normal gynoecium (septum, transmitting tract, placenta, and ovules) were almost completely lacking in fil-8 ant-4 flowers (Fig. 6, B and C).
In a few early arising fil-8 ant-4 flowers, narrow white organs, and/or yellow stamen-like organs were present (Figs. 5H and 6M). SEM examination indicated that petal epidermal cells were present on the surface of some of these white organs (Fig. 6N). In some cases, these organs exhibited polarity defects, as petals cells with both adaxial and abaxial morphologies were present on the abaxial surface of these organs (Fig. 6O). Epidermal cells characteristic of stamens were present on the stamen-like organs (Fig. 6, P and Q). A small amount of internal carpel tissue was occasionally present in early arising fil-8 ant-4 flowers (Fig. 6P). fil-8 yab3-2 ant-4 flowers exhibit a slightly more severe phenotype than fil-8 ant-4 flowers in that organs with petal and stamen-like cells were never observed in fil-8 yab3-2 ant-4 flowers.
fil-8 ant-4 and fil-8 yab3-2 ant-4 flowers have a more severe phenotype than fil-8 yab3-2 flowers (Fig. 5, F, G, and I). fil-8 yab3-2 flowers consist of radialized or flat sepal-like organs, no petals, small stamen filaments lacking anthers, and a carpel with a larger style and no replum (Siegfried et al., 1999; Kumaran et al., 2002). In comparison, fil-8 ant-4 and fil-8 yab3-2 ant-4 flowers consisted of fewer floral organs and show increased radialization of outer whorl organs and loss of carpel adaxial tissue.
fil ant Flowers Show Altered Expression of Floral Organ Identity Genes
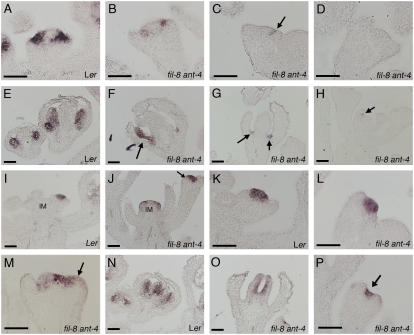
Because of the greatly reduced floral organ identity in fil-8 ant-4 flowers, we examined the expression patterns of the floral homeotic genes APETALA3 (AP3) and AGAMOUS (AG) by in situ hybridization. AP3 is a B class floral homeotic gene involved in the specification of petal and stamen identities, and AG is a C function gene involved in the specification of stamen and carpel identities (Yanofsky et al., 1990; Jack et al., 1992). AP3 expression is greatly reduced or absent in young fil-8 ant-4 flowers as compared with wild type (Fig. 7, A–D). When AP3 mRNA was detected in young fil-8 ant-4 flowers, it was present in a normal spatial pattern (Fig. 7B). AP3 mRNA was only rarely observed in organs of older fil-8 ant-4 flowers (Fig. 7, E and F). AP3 mRNA was often observed in a few cells located between the inner and outer whorls of fil-8 ant-4 flowers (Fig. 7G). Weak patches of AP3 expression were rarely observed in filaments produced in place of flowers (Fig. 7H).
Figure 7.
AP3 and AG expression in wild-type and fil-8 ant-4 flowers. AP3 expression is shown in A to H, and AG expression is shown in I to P. Size bars correspond to 50 μm in A to P. A, AP3 mRNA in a Ler stage 3 flower. B, AP3 mRNA in a stage 3 fil-8 ant-4 flower. C, Arrow points to region of AP3 expression in a stage 3 fil-8 ant-4 flower. D, No AP3 mRNA was detected in this fil-8 ant-4 flower. E, AP3 mRNA is detected in the second and third whorls of stage 5 (left) and stage 8 (right) flowers. F, AP3 mRNA is detected in a stamen-like organ of a fil-8 ant-4 flower. G, AP3 mRNA is present in a few cells located between the outer and inner whorl in older fil-8 ant-4 flowers. H, AP3 mRNA was rarely detected in filaments (arrow) initiated by the inflorescence meristem of fil-8 ant-4 plants. I, AG expression is first detected in a stage 3 flower in wild type. J, AG is expressed in the inflorescence meristem of this fil-8 ant-4 plant. K, AG mRNA is detected in the central part of the floral meristem in a stage 3 Ler flower. L, AG expression in a stage 3 fil-8 ant-4 flower. M, AG expression in the outer organ primordia (arrow) of a stage 4 fil-8 ant-4 flower. N, AG is expressed in the stamens and carpels of a stage 8 Ler flower. O, AG mRNA is detected on the adaxial surface of the central carpel-like organs in a fil-8 ant-4 flower. P, AG mRNA is detected in the central region of filamentous structures (arrow) produced by a fil-8 ant-4 inflorescence meristem.
In wild-type plants, AG mRNA was first detected in the center of the floral meristem of stage 3 flowers (Fig. 7I). In fil-8 ant-4 plants, AG was misexpressed in the inflorescence meristem (Fig. 7J). AG mRNA was also detected in stage one and two floral meristems, earlier than the first appearance of AG mRNA in Ler flowers (data not shown). AG was expressed at high levels in the center of young fil-8 ant-4 floral meristems, similar to the pattern seen in young Ler stage 3 floral meristems (Fig. 7, K and L). In some cases, this AG expression domain was broader in stage 4 fil-8 ant-4 flowers than stage 4 Ler flowers and extended into the outermost organ primordia (Fig. 7M). In Ler flowers, AG is expressed throughout developing stamens and carpels until late stages of flower development (Fig. 7N). In older fil-8 ant-4 flowers, AG mRNA was detected on the inner surface of carpel-like organs (Fig. 7O). AG mRNA was also present in the center of filamentous structures produced by the inflorescence meristem (Fig. 7P).
fil ant Flowers Show Reduced Floral Expression of the Adaxial Cell Fate Regulator PHB
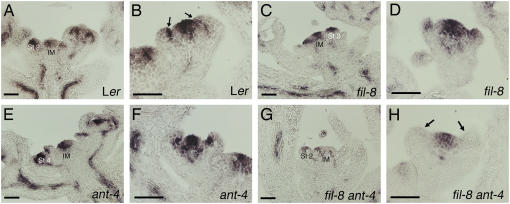
The radialization of fil-8 ant-4 and fil-8 yab3-2 ant-4 floral organs suggests that these organs have lost adaxial-abaxial polarity. To further investigate this possibility, we examined the expression of the adaxial cell fate regulator PHB in fil-8 ant-4 flowers. PHB mRNA is present in the inflorescence meristem and throughout young floral meristems of Ler flowers (Fig. 8A). In stage 4 flowers, PHB mRNA was detected in the center of the floral meristem and in the adaxial half of developing sepal primordia (Fig. 8B). The expression pattern and levels of PHB mRNA were similar in fil-8 and ant-4 single mutants as compared with Ler (Fig. 8, C–F). In fil-8 ant-4 plants, PHB mRNA was typically present at lower levels in the inflorescence meristem and stage 1 and 2 floral meristems as compared with wild type (Fig. 8G). PHB mRNA was usually absent from the outer whorl organs of fil-8 ant-4 flowers and was present in reduced amounts in the floral meristem of young stage 3 and 4 fil-8 ant-4 flowers (Fig. 8H).
Figure 8.
PHB expression in wild-type, fil-8, ant-4, and fil-8 ant-4 flowers. Size bars correspond to 50 μm in A to H. A, PHB expression in a wild-type inflorescence. B, PHB is expressed in the adaxial region of sepal primordia (arrows) and in the floral meristem dome in a stage 4 Ler flower. C, PHB expression in a fil-8 inflorescence. D, PHB expression in a stage 3 fil-8 flower. E, PHB expression in an ant-4 inflorescence. F, PHB expression in a stage 4 ant-4 flower. G, PHB mRNA is present at reduced levels in fil-8 ant-4 inflorescences. H, PHB is expressed in the floral meristem dome but not in the outer whorl organ primordia of this stage 3 fil-8 ant-4 flower. St 2/4, Stage 2 and stage 4 flowers; IM, inflorescence stem.
DISCUSSION
ANT is an important regulator of lateral organ development. ANT expression marks cells that will leave the meristem to form lateral organs, and it is required for proper initiation and growth of lateral organs. Mutations in ANT result in the production of fewer and smaller floral organs (Elliott et al., 1996; Klucher et al., 1996). Despite its importance in lateral organ development, little is known about the genes regulated by this transcription factor. Our results here suggest that ANT acts with FIL to regulate organogenesis and to up-regulate genes establishing organ polarity and those specifying organ identity.
ANT Regulates Organ Polarity
While lateral organs in ant single mutants exhibit normal polarity, defects along the adaxial/abaxial axis were visible at the whole organ, cellular, and molecular level in fil ant and fil yab ant plants. fil yab ant leaves showed dramatic reductions in lamina growth and loss of both adaxial and abaxial epidermal cell identities. fil ant and fil yab ant floral organs were severely radialized with many floral organs replaced by filaments or very narrow organs. In some floral organs, adaxial cell types were found in abaxial positions. These defects are more severe than those observed in either fil or fil yab mutants.
To investigate whether the role of ANT in polarity establishment involves regulation of known adaxial and/or abaxial identity factors, we examined the expression of FIL, YAB3, and PHB in ant mutants. While the expression of FIL, YAB3, and PHB are normal in ant-4 flowers, PHB expression was reduced in fil ant double mutants. This suggests that ANT and FIL together are required for normal levels of PHB mRNA. Examination of the PHB promoter revealed a sequence matching the ANT consensus binding site in 12 of 14 conserved positions including one gap (S. Nole-Wilson and R. Franks, personal communication). We were unable to detect binding of ANT to this site in vitro, suggesting that ANT is not a direct regulator of PHB expression, its role in PHB regulation involves additional factors, and/or that ANT binds to a different DNA sequence within the context of the PHB promoter.
It is curious that an ANT binding site is present within a conserved region of the FIL and YAB3 promoters but that ANT is not required for FIL or YAB3 expression. Seven AINTEGUMENTA-like (AIL) genes are present within the Arabidopsis genome, and several of these genes are expressed in young floral primordia in overlapping domains with ANT (Nole-Wilson et al., 2005). Sequence conservation within the DNA-binding APETALA2 repeat regions of AIL proteins suggest that they may have similar DNA-binding specificities. It is possible that redundancy within the AIL family masks any effect of loss of ANT activity on FIL and YAB3 expression. It will be necessary to generate double, triple, and perhaps higher order mutants within members of the AIL gene family to investigate this possibility.
As both adaxial and abaxial identities are partially lost in fil yab ant mutants and ANT is expressed throughout lateral organs, our results suggest that ANT is a positive regulator of both genes specifying adaxial fates and those specifying abaxial fates. ANT may function as a general activator of PHB-like and YABBY genes throughout organ primordia with their region-specific expression resulting from mutually repressive interactions between the PHB-like and KAN genes. Supporting our view that ANT is a positive regulator of genes specifying either adaxial or abaxial fates, preliminary examination of ant rev double mutants revealed enhanced carpel phenotypes including enhanced loss of some adaxial tissues relative to either single mutant (S. Nole-Wilson and R. Franks, personal communication).
ANT May Act with SEUSS and LEUNIG to Regulate Expression of PHB
SEUSS (SEU) and LEUNIG (LUG) have been proposed to promote polarity along the adaxial/abaxial axis in petals by positively regulating PHB and FIL expression (Franks et al., 2006). Similar effects on leaf lamina/petal blade expansion and vascular development are observed in ag seu lug petals as reported here for fil yab ant leaves (Franks et al., 2006). These similarities are particularly intriguing as SEU, LUG, and ANT share other functions during flower development. All three proteins act as negative regulators of the floral homeotic gene AG (Liu and Meyerowitz, 1995; Krizek et al., 2000; Franks et al., 2002). Interestingly, FIL also acts as an AG repressor in whorls one and two (Chen et al., 1999). Similar carpel growth defects result from the combined loss of LUG and ANT or the combined loss of FIL and ANT. In both fil ant and lug ant flowers, the inner whorl consists of unfused or partially fused valve-like structures with style cells at their tips and an almost complete loss of adaxial tissues (placenta, ovules, and septa; Krizek et al., 2000; Liu et al., 2000).
The similarities in these phenotypes suggest that ANT, FIL, SEU, and LUG have overlapping and partially redundant functions. These proteins might form a complex in which the SEU-LUG transcriptional corepressor (Sridhar et al., 2004) is recruited to promoter sequences via interaction with either of the DNA-binding proteins ANT or FIL (Nole-Wilson and Krizek, 2000; Kanaya et al., 2002). The Antirrhinum LUG ortholog STYLOSA has been shown to physically interact with YABBY proteins (Navarro et al., 2004). ANT and FIL could share some functions such that severe phenotypes only result in the absence of both ANT and FIL.
ANT and YABBY Genes Promote Lamina Expansion and Floral Organ Identity
Besides contributing to the specification of abaxial identity, YABBY genes are important regulators of lamina expansion. Polar expression of YABBY genes has been proposed to regulate signaling events between the adaxial and abaxial domains that control cell division in each domain and expansion of the leaf lamina (Eshed et al., 2004). Although ant mutants show only slight reductions in leaf area, the narrower lamina of fil yab ant leaves as compared with fil yab leaves indicates that ANT plays an important growth promotion role in leaves as well as flowers. Consistent with a role in lamina expansion, ANT expression in developing leaves becomes refined to the central and marginal regions in a pattern that is quite similar to the Solanum tuberosum YABBY gene StYABBY1 (Long and Barton, 2000; Eshed et al., 2004).
While petals and stamens are present in fil and ant single mutants, organs with petal or stamen characteristics are rare in fil ant double mutants. The loss of floral organ identify in fil ant double mutants was correlated with altered floral homeotic gene expression. AP3 expression was reduced in fil ant flowers while the pattern of AG expression was altered. Thus, ANT acts as a positive regulator of the class B gene AP3 and acts to prevent AG expression in inflorescences and flowers prior to stage 3. A role for ANT in AG repression has been noted previously (Krizek et al., 2000; Liu et al., 2000). Similar losses in floral organ identity have been observed in other genotypes with polarity defects. For example, kan1 kan2 fil yab flowers consist of carpels and radialized organs that lack cell types characteristic of sepals, petals, or stamens (Eshed et al., 2004). These results suggest a connection between the establishment of polarity and the specification of organ identity during flower development. Further studies will be needed to probe this relationship and to better understand the diverse processes that ANT regulates during lateral organ development.
MATERIALS AND METHODS
Protein Expression
Full-length ANT lacking a stop codon was cloned into pQE12 (Qiagen) and expressed by induction with 1 mm isopropyl-β-d-thiogalactoside in XL1-Blue MRF′ Tet cells (Stratagene) at 30°C. Cells were harvested between 6 and 8 h after induction. ANT was purified using Ni-NTA (Qiagen) under denaturing conditions according to the manufacturer's instructions. ANT refolded upon dilution in the DNA-binding reactions.
Gel Mobility Shift Assays
Gel mobility shift assays were carried out as described previously (Nole-Wilson and Krizek, 2000), except that binding reactions were incubated for 4 h at room temperature or overnight at 4°C. The YAB3 and FIL binding sites were created by PCR amplification of Columbia genomic sequence using YAB-12 (5′-CTCGAGATTAAGTGTGAAAACAACTGAT-3′) and YAB-13 (5′-GAATTCAAAGGACGCAAAGTTCGATG-3′) or FIL-3 (5′-TACACTCGAGTTAAGGAATGACAACAACGGG-3′) and YAB-9 (5′-TACCGGATCCGAATTCGCAGTTCCCAATGGA-3′), respectively. These fragments correspond to nucleotides at positions 1,561 to 1,456 upstream of the YAB3 start codon and nucleotides at positions 1,763 to 1,633 upstream of the FIL start codon. The PCR products were cloned into pCRScript (Stratagene) and the probes were prepared as described previously (Nole-Wilson and Krizek, 2000).
Plant Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Ler was used as the wild type. Plants were grown in a soil mixture of Fafard 4P:perlite:vermiculite in a ratio of 4:1:1 under continuous light (70–100 μmol m−2 s−1) at a temperature of 22°C. Plants were fertilized once at 1 to 2 weeks postgermination.
Real-Time RT-PCR
Inflorescences were collected from 3- to 4-week-old Ler and ant-4 plants grown on soil under continuous light at 22°C. Total RNA was extracted and DNase treated as described previously (Nole-Wilson et al., 2005). Approximately 5 μg of total RNA was reverse transcribed using the Superscript First-Strand Synthesis system for RT-PCR (Invitrogen). Real-time RT-PCR was performed as described previously except that ACTIN2 (ACT2) was used for normalization purposes (Nole-Wilson et al., 2005). The ACT2 primers were ACT-3 (5′-CCTTTGTTGCTGTTGACT-3′) and ACT-4 (5′-GAACAAGACTTCTGGGCATCT-3′). The YAB3 primers were YAB3-10 (5′-GCGGAGGGCAGAATATAAAC-3′) and YAB3-11 (5′-CACTGATCTTCCGTTGCGA-3′).
In Situ Hybridization
Inflorescences were fixed, embedded, sectioned, hybridized, and washed as described previously (Krizek, 1999). Digoxigenin-labeled RNA probes were synthesized by in vitro transcription using T7 RNA polymerase (AP3, AG, and FIL probes) or T3 RNA polymerase (PHB probe) and the appropriate linearized plasmids. The AP3, AG, and FIL plasmids used for probe production have been described before (Yanofsky et al., 1990; Jack et al., 1992; Siegfried et al., 1999). The PHB probe corresponds to nucleotides 609 to 2,559 of PHB and was made after linearization of PHB/pBSKS with XbaI.
Generation of fil-8 ant-4 and fil-8 yab3-2 ant-4 Plants
yab3-2 fil-8/+ seeds were obtained from John Bowman. These alleles have been described previously (Kumaran et al., 1999, 2002). Both yab3-2 and fil-8 mutants are Ds insertion lines in the Ler background. YAB3 transcripts are not detectable in yab3-2 (Kumaran et al., 2002). ant-4 seeds were obtained from Charles Gasser (Baker et al., 1997). ant-4 is in the Ler background and contains a T-to-A transversion at nucleotide 1,335, altering the donor splice site of the fourth intron. These alleles were chosen as they are all strong alleles in the Ler background. The double and triple mutants were generated by pollinating putative fil-8 yab3-2/+ plants with ant-4 pollen. F2 and subsequent generations were observed for segregation of plants with novel phenotypes. fil-8 ant-4 double mutants and fil-8 yab3-2 ant-4 triple mutants were confirmed by PCR genotyping.
PCR Genotyping
yab3-2
Green leaf tissue was prepared as described previously (Klimyuk et al., 1993) and subjected to PCR. PCR reactions using YAB35I-F (5′-GCCCTCCTCTCTCTCTTACTC-3′) and YAB35I-R2 (5′-TCTGACCGTCACCGTCTTGA-3′) verified the absence of the wild-type YAB3 allele and PCR reactions using Ds5I-1 (5′-CCGTTTACCGTTTTGTATATCCCG-3′) and YAB35I-R2 verified the presence of the yab3-2 mutant allele.
fil-8
Green leaf tissue was prepared as described above and subjected to PCR. PCR reactions using AFO-FW3 (5′-AGATTCCTAAAGCACCACCC-3′) and YAB1R (5′-GATACGTTGGATCTCCTCCC-3′) verified the absence of the wild-type FIL allele and PCR reactions using AFO-FW3 and Ds5I-1 verified the presence of the fil-8 allele.
ant-4
DNA was isolated from green leaf tissue by grinding in 200 mm Tris, pH 7.5, 250 mm NaCl, 25 mm EDTA, 0.5% SDS, and precipitation with isopropanol. The DNA was then PCR amplified using ANT-6 (5′-TCAAGGATCCACTTTTGGACAACGAACTTCT-3′) and ANT-33 (5′-TCTTGGATCCTGCAACATATTCTTGTCTAGT-3′). The PCR product was gel purified and cloned into the BamHI site of pGEM3Z (Promega). Sequencing of multiple clones confirmed the presence of only the ant-4 allele in putative fil-8 ant-4 and fil-8 yab3-2 ant-4 plants.
Leaf Area and Plant Height Measurements
For each genotype, the largest rosette leaf was removed from six different plants at the time of bolting. Leaf surface area was measured with a LI-COR LI-3000 portable area meter. The length and width of rosette leaves were measured using an ocular micrometer or a ruler. Plant heights were determined for six different plants of each genotype at the time when their primary inflorescences were starting to senesce. Bolting time was approximately the same for all genotypes.
Leaf Vascular Staining
Leaves were fixed overnight at room temperature in a 3:1 solution of ethanol:acetic acid. The tissue was mounted in 70% ethanol and examined using a dissecting microscope with illumination from below.
SEM
Tissue for SEM was fixed, dried, dissected, and coated as described previously (Krizek, 1999). SEM analysis was performed on a FEI XL30 ESEM (Hillsboro).
Acknowledgments
We thank John Bowman for the fil-8 yab3-2/+ seeds and the FIL in situ plasmid, Charles Gasser for the ant-4 seeds, David Lincoln for help with leaf area measurements, John Herr for assistance with the xylem staining, and Mike Prigge and Steve Clark for the PHB in situ plasmid. We also thank Bob Franks for sharing unpublished data and providing valuable comments on the manuscript.
This work was supported by the U.S. Department of Energy (grant no. 98ER20312).
The author responsible for the distribution of materials integral to the finding presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Beth A. Krizek (krizek@sc.edu).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.076604.
References
- Baker SC, Robinson-Beers K, Villanueva JM, Gaiser JC, Gasser CS (1997) Interactions among genes regulating ovule development in Arabidopsis thaliana. Genetics 145: 1109–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL (2000) The YABBY gene family and abaxial cell fate. Curr Opin Plant Biol 3: 17–22 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Eshed Y, Baum SF (2002) Establishment of polarity in angiosperm lateral organs. Trends Genet 18: 134–141 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR (1999) CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126: 2387–2396 [DOI] [PubMed] [Google Scholar]
- Chen Q, Atkinson A, Otsuga D, Christensen T, Reynolds L, Drews GN (1999) The Arabidopsis FILAMENTOUS FLOWER gene is required for flower formation. Development 126: 2715–2726 [DOI] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQJ, Gerentes D, Perez P, Smyth DR (1996) AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Engstrom EM, Izhaki A, Bowman JL (2004) Promoter bashing, microRNAs, and KNOX genes. New insights, regulators, and targets-of-regulation in the establishment of lateral organ polarity in Arabidopsis. Plant Physiol 135: 685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL (2001) Establishment of polarity in lateral organs of plants. Curr Biol 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Izhaki A, Baum SF, Floyd SK, Bowman JL (2004) Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131: 2997–3006 [DOI] [PubMed] [Google Scholar]
- Franks RG, Liu Z, Fischer RL (2006) SEUSS and LEUNIG regulate cell proliferation, vascular development and organ polarity in Arabidopsis petals. Planta (in press) [DOI] [PubMed]
- Franks RG, Wang C, Levin JZ, Liu Z (2002) SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development 129: 253–263 [DOI] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM (1992) The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68: 683–687 [DOI] [PubMed] [Google Scholar]
- Kanaya E, Nakajima N, Okada K (2002) Non-sequence-specific DNA binding by the FILAMENTOUS FLOWER protein from Arabidopsis thaliana in reduced by EDTA. J Biol Chem 227: 11957–11964 [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS (2001) KANADI regulates organ polarity in Arabidopsis. Nature 411: 706–709 [DOI] [PubMed] [Google Scholar]
- Klimyuk VI, Carroll BJ, Thomas CM, Jones JDG (1993) Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J 3: 493–494 [DOI] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL (1996) The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8: 137–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA (1999) Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev Genet 25: 224–236 [DOI] [PubMed] [Google Scholar]
- Krizek BA, Prost V, Macias A (2000) AINTEGUMENTA promotes petal identity and acts as a negative regulator of AGAMOUS. Plant Cell 12: 1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran MK, Bowman JL, Sundaresan V (2002) YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 14: 2761–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran MK, Ye D, Yang W-C, Griffith ME, Chaudhury AM, Sundaresan V (1999) Molecular cloning of ABNORMAL FLORAL ORGANS: a gene required for flower development in Arabidopsis. Sex Plant Reprod 12: 118–122 [Google Scholar]
- Liu Z, Franks RG, Klink VP (2000) Regulation of gynoecium marginal tissue formation by LEUNIG and AINTEGUMENTA. Plant Cell 12: 1879–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Meyerowitz EM (1995) LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121: 975–991 [DOI] [PubMed] [Google Scholar]
- Long J, Barton MK (2000) Initiation of axillary and floral meristems in Arabidopsis. Dev Biol 218: 341–353 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Barton MK (1998) Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- Navarro C, Efremova N, Golz JF, Rubiera R, Kuckenberg M, Castillo R, Tietz O, Saedler H, Schwarz-Sommer Z (2004) Molecular and genetic interactions between STYLOSA and GRAMINIFOLIA in the control of Antirrhinum vegetative and reproductive development. Development 131: 3649–3659 [DOI] [PubMed] [Google Scholar]
- Nole-Wilson S, Krizek BA (2000) DNA binding properties of the Arabidopsis floral development protein AINTEGUMENTA. Nucleic Acids Res 28: 4076–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole-Wilson S, Tranby T, Krizek BA (2005) AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Mol Biol 57: 613–628 [DOI] [PubMed] [Google Scholar]
- Sawa S, Watanabe K, Goto K, Kanaya E, Morita EM, Okada K (1999) FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev 13: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL (1999) Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126: 4117–4128 [DOI] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Gonzalez D, Conlan RS, Liu Z (2004) Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc Natl Acad Sci USA 101: 11494–11499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites R, Hudson A (1995) phantastica: a gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121: 2143–2154 [Google Scholar]
- Watanabe K, Okada K (2003) Two discrete cis elements control the abaxial side-specific expression of the FILAMENTOUS FLOWER gene in Arabidopsis. Plant Cell 15: 2592–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldman KA, Meyerowitz EM (1990) The protein encoded by the Arabidopsis homeotic gene AGAMOUS resembles transcription factors. Nature 346: 35–39 [DOI] [PubMed] [Google Scholar]