Abstract
Sucrose (Suc) synthase (SUS) cleaves Suc to form UDP glucose and fructose, and exists in soluble and membrane-associated forms, with the latter proposed to channel UDP glucose to the cellulose-synthase complex on the plasma membrane of plant cells during synthesis of cellulose. However, the structural features responsible for membrane localization and the mechanisms regulating its dual intracellular localization are unknown. The maize (Zea mays) SUS1 isoform is likely to have the intrinsic ability to interact directly with membranes because we show: (1) partial membrane localization when expressed in Escherichia coli, and (2) binding to carbonate-stripped plant microsomes in vitro. We have undertaken mutational analyses (truncations and alanine substitutions) and in vitro microsome-binding assays with the SUS1 protein to define intrinsic membrane-binding regions and potential regulatory factors that could be provided by cellular microenvironment. The results suggest that two regions of SUS1 contribute to membrane affinity: (1) the amino-terminal noncatalytic domain, and (2) a region with sequence similarity to the C-terminal pleckstrin homology domain of human pleckstrin. Alanine substitutions within the pleckstrin homology-like domain of SUS1 reduced membrane association in E. coli and with plant microsomes in vitro without reducing enzymatic activity. Microsomal association of wild-type SUS1 displayed cooperativity with SUS1 protein concentration and was stimulated by both lowering the pH and adding Suc. These studies offer insight into the molecular level regulation of SUS1 localization and its participation in carbon partitioning in plants. Moreover, transgenics with active SUS mutants altered in membrane affinity may be of technological utility.
Glycosyltransferases (GTases) constitute a large group of enzymes involved in the biosynthesis of carbohydrates and glycoconjugates in prokaryotes and eukaryotes. The diversity of compounds synthesized by these enzymes and their varied intracellular locations implicates them in storage, structural, and signaling functions. Currently, 77 families of GTases are recognized (Coutinho et al., 2003; http://afmb.cnrs-mrs.fr/CAZY/). Although these families possess very little sequence homology, the enzymes within a given GTase family are expected to fold similarly into a GT-A- or GT-B-type structure. These enzymes are further classified as inverting or retaining GTases depending upon whether the products formed invert or maintain, respectively, the stereochemistry at the C1 position of the donor sugar.
In bacteria and fungi, numerous GTases have been implicated in the biosynthesis of extracellular or membrane-localized compounds. For instance, the Streptococcus pneumoniae enzyme WciS is a retaining GTase included in family GT4 that is involved in capsular polysaccharide biosynthesis. This multimeric protein is membrane associated, although it lacks any predicted transmembrane sequences (Saksouk et al., 2005). Similarly, the Escherichia coli MurG enzyme is an inverting GTase with a GT-B fold included in family GT28 that is involved in peptidoglycan (bacterial cell wall) biosynthesis. This GTase as well is only moderately hydrophobic and lacks a membrane-spanning domain, yet it is still associated with the cytoplasmic surface of the inner membrane (Ha et al., 2000). The Pichia pastoris Ugt51 enzyme is an inverting GTase included in family GT1 that produces sterol glucosides. This protein contains two lipid-binding modules, a GRAM (glucosyltransferases, Rab-like GTPase activators, and myotubularins) and a pleckstrin homology (PH) domain, and is associated with the micropexophagic membrane apparatus in vivo (Oku et al., 2003). Bacterial α-monoglucosyldiacylglycerol synthases are included in family GT4 and are membrane-associated GTases involved in modulating bilayer and lipid surface properties (Berg et al., 2001; Edman et al., 2003). These proteins also lack transmembrane domains but have predicted structural and sequence similarities to E. coli MurG that may impart membrane affinity.
Cellulose is one of the most abundant carbon reserves in nature, constituting a large portion of the dry matter content of grasses and wood. Consistent with bacterial extracellular matrix deposition, plants utilize numerous GTases during cell wall biosynthesis (Buckeridge et al., 1999; Charnock et al., 2001; Egelund et al., 2004). Cellulose synthase is itself an inverting GTase belonging to family GT2 that synthesizes the β-1,4-linked Glc polymer cellulose from UDP-Glc. Moreover, a membrane-bound GTase (UDP-Glc:sterol glucosyltransferase) has been implicated in the production of a sitosterol-β-glucoside primer that initiates cellulose polymerization (Peng et al., 2002). In developing cotton (Gossypium hirsutum) ovules during the peak of secondary cell wall formation, cellulose synthesis can consume roughly 80% of all imported carbon (Haigler et al., 2001). In most plant species, the cellulose biosynthetic pathway is supported by carbon skeletons obtained from imported Suc.
Suc synthase (SUS) enzymes are also classified as retaining GTases belonging to family GT4, and are thought to adopt a GT-B-type fold similar to E. coli MurG (MacGregor, 2002). SUS catalyzes a reaction that converts Suc and UDP into UDP-Glc and -Fru (Tsai, 1974), and can exist in association with the plasma membrane (Amor et al., 1995; Carlson and Chourey, 1996; Winter et al., 1997; Sturm et al., 1999; Haigler et al., 2001; Komina et al., 2002). Plasma membrane-associated SUS (m-SUS) is postulated to channel its product, UDP-Glc, toward the synthesis of cellulose and callose (Amor et al., 1995; Winter and Huber, 2000; Haigler et al., 2001; Koch, 2004). The involvement of SUS in cellulose deposition is supported by expression and localization of protein and activity (Amor et al., 1995; Ruan et al., 1997; Kordel and Kutschera, 2000; Salnikov et al., 2001, 2003; Uggla et al., 2001; Kutschera and Heiderich, 2002; Albrecht and Mustroph, 2003) in cotton, Zinnia elegans, wheat (Triticum aestivum), pea (Pisum sativum), sunflower (Helianthus annuus), and pine (Pinus sylvestris). Moreover, the importance of SUS in this process is evidenced by genetic overexpression (Konishi et al., 2004) and suppression or mutation (Chourey et al., 1998; Sturm and Tang, 1999; Ruan et al., 2003). It is therefore well established that SUS can exist on membranes and channel UDP-Glc to cellulose biosynthesis. However, it is unclear how this mainly soluble enzyme is redistributed to the plasma membrane to fulfill this function.
In maize (Zea mays), SUS is a tetrameric enzyme composed of subunits encoded by the Sh1, Sus1, and Sus2 loci, which encode the SUS-SH1, SUS1, and SUS2 proteins, respectively (http://www.maizegdb.org/cgi-bin/displaygprecord.cgi?id=13861). Both the SUS-SH1 and SUS1 proteins associate with the plasma membrane in maize endosperm (Carlson and Chourey, 1996), whereas the intracellular distribution of SUS2 (previously known as SUS3) in maize is currently being determined (Duncan et al., 2005). SUS is peripherally associated with maize leaf microsomes and the abundance of m-SUS is variable along the longitudinal axis of developing maize leaves (Hardin et al., 2004). Therefore, it is unlikely that m-SUS represents a distinct gene product but rather that individual SUS isoforms can exist in multiple intracellular locations. At maximum, m-SUS constituted about 9% to 10% of total cellular SUS in maize leaves (Winter et al., 1997; Hardin et al., 2004) and 3% to 14% in endosperms (Carlson and Chourey, 1996), whereas it represented 25% to 50% of total SUS in cotton fiber membranes and carrot (Daucus carota) protoplasts (Amor et al., 1995; Sturm et al., 1999). These observations strongly suggest that the association of SUS with the plasma membrane is a controlled and regulated process in plants.
In this manuscript, we identify a region of SUS that possesses significant sequence homology to the C-terminal PH domain of human pleckstrin. Since PH domains are known to impart polyphosphoinositide and/or membrane localization to about 30% of the proteins that contain them (Lemmon et al., 2002; Yu et al., 2004; Edlich et al., 2005), we investigated the involvement of this region in SUS1 membrane binding. We demonstrate that SUS1 has an intrinsic ability to interact directly with membranes, and that this ability is due, in part, to the identified PH-like domain. In addition, we demonstrate that the N-terminal noncatalytic region also contributes to membrane binding. The presence of Suc and low pH were identified as factors that promote partitioning of SUS1 to membranes. These experiments provide a foundation for guiding the rational creation of mutant SUS transgenics that possess altered membrane localization, and to potentially control carbon allocation to structural compounds in plants.
RESULTS
Structural Analyses of SUS Substantiate Its Assignment as a GTase and Reveal Regions Potentially Involved in Membrane Binding
Bioinformatic analyses of the SUS1 primary sequence revealed the presence of a region between amino acids 360 and 457 with striking sequence similarity to the C-terminal PH domain contained within human pleckstrin (Fig. 1A). The alignment of these regions in SUS1 and human pleckstrin required only a single amino acid gap over this 98-amino acid stretch and possessed 34% similarity. The C-terminal PH domain of human pleckstrin binds phosphatidylinositol-(3,4)-bisphosphate (Edlich et al., 2005), making the related region of SUS a possible contributor to membrane binding. This PH-like domain of SUS1 is also highly conserved in the two other known maize SUS isoforms (≥74% identity among the SUS isoforms; Fig. 1A). However, the secondary structures predicted by the Jpred consensus method (http://www.compbio.dundee.ac.uk/∼www-jpred/) differ significantly between SUS and the PH domain of human pleckstrin (Fig. 1A). SUS possesses an alternating β strand-α helix arrangement, whereas the PH domain of pleckstrin consists of sequential β strands bordered at both termini by α helices. Therefore, an identical tertiary structure of these two regions is unlikely.
Figure 1.
SUS proteins can be modeled onto a GT-B-type glucosyltransferase fold and contain a region with sequence similarity to a PH domain. A, Alignment of amino acid sequences of the maize SUS isoforms SUS1 (P49036), SUS-SH1 (P04712), and SUS2 (Q8L5H0), with the C-terminal PH domain of human pleckstrin (HsPlek; P08567) by CLUSTALW (v 1.82). Secondary structure elements for SUS1 identified by the Jpred method are shown above the sequences, and those of pleckstrin below the sequences. Conservation of similar amino acids between HsPlek and the SUS isoforms is indicated by gray shading. Consensus residues are conserved across all four proteins. h, Hydrophobic; a, acidic; b, basic residues. The locations of Ala-substituted mutants are indicated by “A” and truncation (stop) mutants with arrows above the SUS1 sequence, residues identified in this study as important for membrane/lipid binding are marked with asterisks. Asterisks below the HsPlek sequence are important for PH domain membrane binding (Isakoff et al., 1998; Edlich et al., 2005). B, Diagram of SUS1 sequence showing Jpred secondary structural elements (α helices as circles, β strands as arrows) for the putative catalytic domain on a GT-B-type fold (drawn after MacGregor, 2002). Bold lines represent the PH-like domain of SUS1, locations of truncation mutants are indicated by “stop”, and Ala-substituted mutants by an “A”. The putative catalytic Glu residues are shown in gray.
The secondary structure of SUS has recently been modeled onto a GTase GT-B-type fold (MacGregor, 2002). Indeed, our own analyses (Fig. 1B) demonstrated that the secondary structure of amino acids 281 to 769 of maize SUS1 predicted by Jpred was consistent with this putative catalytic structure, although the α2 and α5 helixes seemed to be replaced by β strands. Therefore, it is presumed that the amino-terminal 280 amino acids and the carboxyl-terminal 47 amino acids represent noncatalytic regions of SUS (Fig. 1B). Moreover, amino acids 281 to 780 of SUS1 could be modeled onto the structures of Agrobacterium tumafaciens glycogen synthase (structural classification of proteins [SCOP] code c1rzvA), and E. coli MurG (SCOP code d1f0ka) by the Protein Homology/Analogy Recognition Engine (phyre) server (http://www.sbg.bio.ic.ac.uk/∼phyre/) with 100% precision and E-values less than 1.4e-06 even though sequence identity was less than 13%. Notably, both of these enzymes are known to posses a GT-B-type fold (Ha et al., 2000; Buschiazzo et al., 2004). The predicted GT-B-type structure represents the entire catalytic domain of SUS1 (amino acids 281–780) and consists of a bilobed configuration with large amino and carboxyl domains joined by a helical linker (Fig. 1B). The N-terminal lobe is thought to possess the acceptor-binding sites, while the C-terminal lobe provides the donor-binding sites, as well as the putative catalytic Glu residues within the Ex7E motif (E678E686 in SUS1) characteristic of retaining GTases. The identified PH-like domain of SUS (Fig. 1A) falls within the N-terminal catalytic lobe between β2 and β6 (Fig. 1B). The SUS β3 segment and the β3/α3 loop sequence are enriched in hydrophobic residues and well conserved among SUS isoforms. Notably, it is amino acids within this region of the C-terminal human pleckstrin PH domain that contact phosphatidylinositol-(3,4)-bisphosphate and confer specificity (Isakoff et al., 1998; Edlich et al., 2005). The presence of a Pro (P371) and different secondary structural elements in this region of SUS may alter this property in its PH-like domain. Both termini of SUS polypeptides contain additional sequences outside of the catalytic domain, the largest (280 amino acids) being at the N terminus (NT; Fig. 1B). Acceptable models for these noncatalytic regions of SUS1 could not be identified through the Protein Homology/Analogy Recognition Engine (phyre) server. However, the predicted tertiary structure of the SUS catalytic domain allows for comparisons with other known GTase structures and membrane-binding regions. Significantly, the PH-like domain of SUS occupies an identical region of the GTase molecule (β3-α5) as does the proposed membrane-association site of MurG (β3-α5; Ha et al., 2000). This region of the MurG structure is occupied by a surface-exposed hydrophobic patch surrounded by basic residues; similar amino acid types are found in SUS between β3 and α3 (Fig. 1A).
Analyses of SUS1 Truncation Mutants Reveal That Membrane Binding Involves the Amino-Terminal Noncatalytic Domain
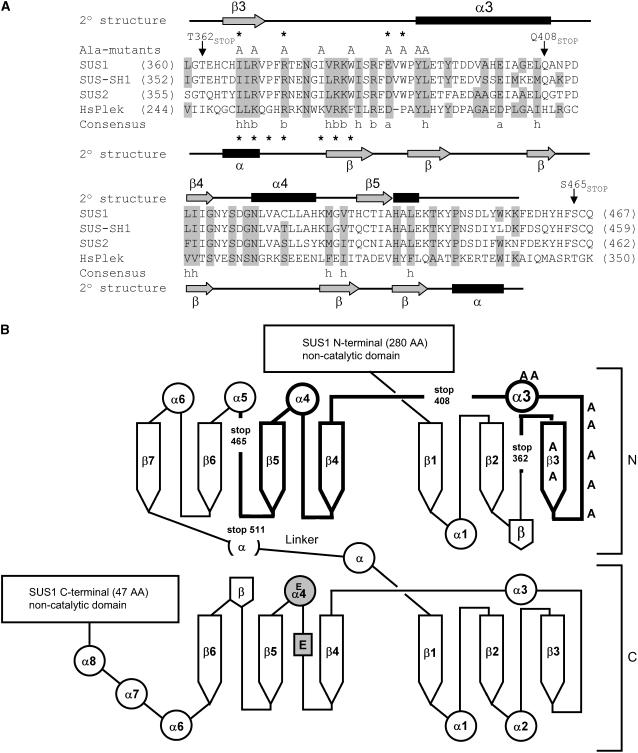
To address the involvement of the PH-like domain of SUS in membrane binding, we produced N-terminal maltose binding protein (MBP)-tagged truncation mutants of the SUS1 polypeptide (Figs. 1B and 2) to isolate this region. The large MBP tag was selected for its recognized ability to maintain recombinant protein solubility in E. coli. As reported earlier, we have found that a significant portion of 6His-tagged SUS1 recombinant protein was associated with bacterial membranes when expressed in E. coli (Hardin et al., 2004). When the various protein truncation mutants of MBP-SUS1 were expressed and the bacterial extracts fractionated into soluble (100 K supers) and insoluble (100 K pellets) components, even the smallest construct (T362stop) that contained the N-terminal noncatalytic domain but lacked the PH-like domain partitioned strongly to the membrane fraction (Fig. 2). However, both full-length MBP-SUS1 and the Y511stop proteins, which contain the entire N-terminal catalytic lobe and the PH-like region, displayed a slightly greater tendency to partition with the insoluble material.
Figure 2.
SUS1 truncation mutants partition to the membrane-containing insoluble phase in E. coli. Equal amounts of total protein (0.25 μg) isolated in E. coli 100,000g supernatants (100 K supers) and pellets (100 K pellets) of recombinants expressing full-length SUS1 or the truncation mutant listed above the sections were probed on immunoblots (IB) with an antibody against MBP (anti-MBP). The positions of PAGE molecular mass markers are shown in kilodaltons on the left of each section. Densitometry of these blots is displayed in the graph at the bottom.
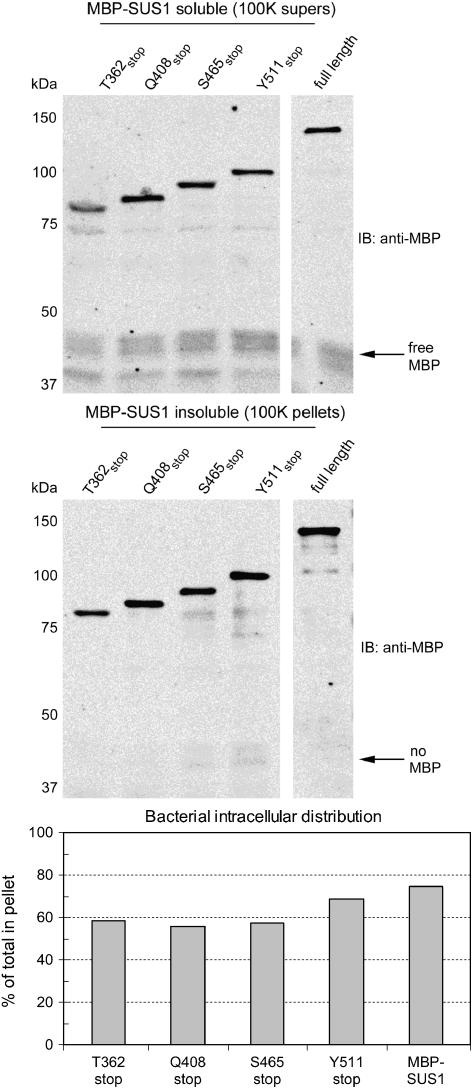
The 100 K soluble fraction of these MBP-SUS1 recombinant proteins was analyzed by size-exclusion chromatography to determine whether the oligomeric structure of SUS1 was affected by truncation, as this may be relevant to its observed partitioning and membrane-binding behavior. MBP itself eluted as a monomeric protein (Fig. 3), whereas all of the MBP-SUS1 proteins eluted as dimers. This observation differs from the typically tetrameric form usually observed for native SUS and may be related to the presence of an MBP tag. However, it clearly validated that all the MBP-SUS1 recombinant proteins were identical in this regard and therefore oligomeric structure would not be the source of any potential differences. It should also be stated that equivalency in analysis by size-exclusion chromatography (Fig. 3) does not imply that these proteins are equivalently folded or structured as predicted (Fig. 1B).
Figure 3.
Size-exclusion chromatography demonstrates that SUS1 truncation mutants are all dimers. Equal volumes of total protein isolated in E. coli 100,000g supernatants of recombinants expressing full-length SUS1 or the truncation mutant listed to the left of the sections were resolved by size-exclusion chromatography and fractions probed on immunoblots (IB) with an antibody against MBP. The elution positions of size-exclusion chromatography molecular mass markers are shown above the sections, the calculated monomer Mr (MW) of each protein and its deduced oligomeric type are listed to the left of the sections.
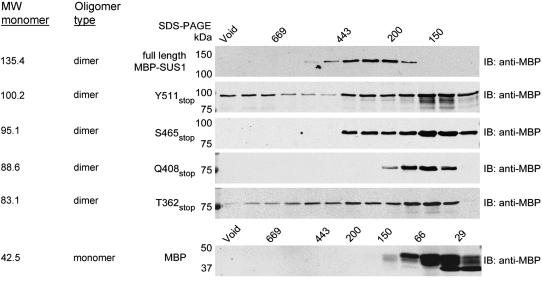
Although the relative intracellular partitioning of the MBP-SUS1 recombinant proteins in E. coli (Fig. 2) suggested that the full-length protein contained additional determinants for segregation into the 100 K pellet, it did not necessarily show that this partitioning behavior reflected membrane binding. Therefore, as a rigorous demonstration of membrane association that did not rely on cosedimentation, we fractionated the 100 K pellets by Suc gradient flotation (Fig. 4). Under these conditions membranes and their associated proteins floated to the top of the gradient (Ahola et al., 1999), while inclusion bodies and soluble protein remained in the bottom of the gradient. Full-length MBP-SUS1 protein obtained from the 100 K supernatant (soluble) fraction did not float in these assays (Fig. 4), whereas about 35% of the full-length protein recovered from the 100 K pellet was shown to be unequivocally membrane associated as it floated to the top of these gradients. Between 15% to 25% of the other MBP-SUS1 truncation proteins were also membrane associated (Fig. 4). These results demonstrated that even the smallest recombinant protein (T362stop), which includes the N-terminal noncatalytic region but not the PH-like domain, contained determinants that provided association with bacterial membranes.
Figure 4.
Suc gradient flotation reveals that the N-terminal 362 amino acids of SUS1 bind to bacterial membranes. Equal volumes of total protein isolated in E. coli 100,000g pellets of recombinants expressing full-length SUS1 or the truncation mutant listed to the left of the sections were subjected to Suc gradient flotation and fractions probed on immunoblots (IB) with an antibody against MBP. Densitometry of these blots is displayed in the graph at the bottom.
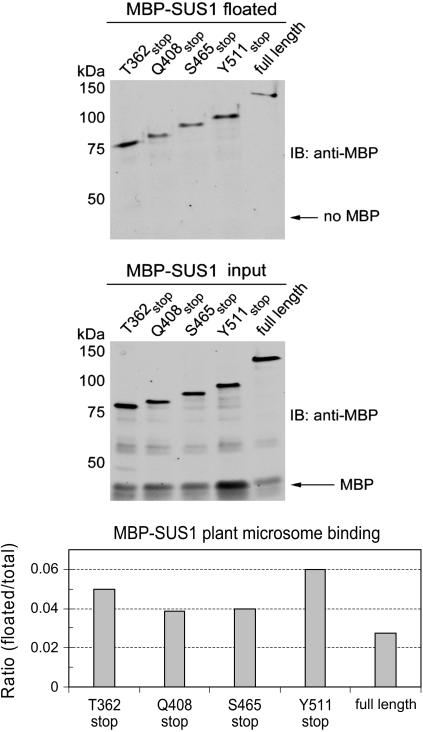
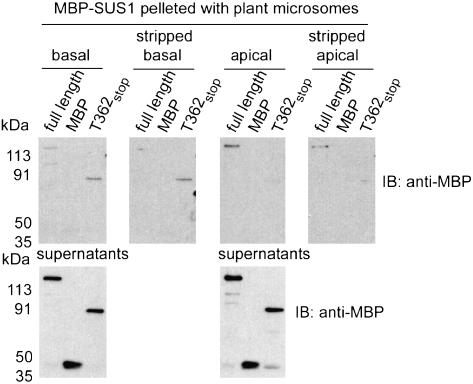
To test the involvement of these SUS1 regions in binding to plant-derived membranes, we mixed the various MBP-SUS1 recombinant proteins with microsomes obtained from the basal 4 to 16 cm region of elongating maize leaves and subsequently floated the membranes, and any protein that had bound to them, to the top of Suc gradients (Fig. 5). Since SUS is known to interact with a variety of different membrane types in maize (Carlson and Chourey, 1996; Winter et al., 1998; Buckeridge et al., 1999; Etxeberria and Gonzalez, 2003) we purposefully prepared unfractionated total microsomes containing all these membrane types for these studies. The fact that the free MBP in these MBP-SUS1 recombinant protein samples did not float demonstrated the selectivity that this assay provided for membrane association. Under these conditions, the Y511stop protein, which contains the N-terminal noncatalytic lobe and the N-terminal catalytic lobe with its PH-like domain, associated with plant microsomes slightly better than all other recombinant proteins (Fig. 5). Therefore, the PH-like domain of SUS1 might be contributing determinants that are of significance for binding to plant membranes, in addition to the determinants within the N-terminal noncatalytic region, but our experiments cannot discern the origin of the membrane being bound in these plant-derived microsomes. The most striking difference between bacterial (Fig. 4) and plant (Fig. 5) membrane association assays was observed for the full-length MBP-SUS1 protein. Its binding to plant microsomes was relatively weaker than to E. coli membranes, suggesting that differences in the source of the membranes were likely to contribute to SUS1 affinity. This assumption was supported by similar membrane-binding experiments in which microsomes derived from two different regions of the elongating maize leaf were used (Fig. 6). No binding of the full-length MBP-SUS1 protein to microsomes derived from the basal (4–16 cm) region was again observed, whereas it interacted well with microsomes derived from the apical (24–36 cm) region. Conversely, the T362stop protein interacted best with the basal microsome sample. These results suggested that the two regions of SUS1 implicated in membrane binding (i.e. the N-terminal noncatalytic domain and the PH-like domain) potentially provide affinity to membranes that may differ in lipid or lipid headgroup types. Moreover, treatment of these microsomes with carbonate, to remove peripherally associated proteins that SUS1 may have affinity for, eliminated >50% of the protein content (data not shown) but did not significantly effect the binding of either the T362stop or full-length MBP-SUS1 proteins (Fig. 6). Although integral membrane proteins remain in these plant microsomes, these carbonate washing results and the observation that SUS1 will bind to bacterial membranes (Fig. 4) that contain a membrane protein composition dissimilar to plant microsomes, supports the supposition that SUS1 can interact directly with the lipid bilayer rather than another protein to confer membrane localization.
Figure 5.
Suc gradient flotation reveals that the N-terminal 362 amino acids of SUS1 bind to plant membranes. Equal amounts (0.18 pmol μL−1) of full-length SUS1 or the truncation mutant listed above the sections were mixed with equal amounts of basal leaf microsomes at pH 5.5 and 150 mm Suc. After pelleting the microsomes and removing aliquots of the supernatants (input) the pellets were subjected to Suc gradient flotation and the top fraction probed on immunoblots (IB) with an antibody against MBP. Densitometry of these blots is displayed in the graph at the bottom. Note that free MBP did not bind to membranes.
Figure 6.
The source region of leaf microsomes affects membrane binding of SUS1, implicating multiple membrane-interacting protein regions. Equal amounts (0.28 pmol μL−1) of MBP, full-length MBP-SUS1, or the T362stop truncation mutant were mixed with equal amounts of basal or apical leaf microsomes (before or after being carbonate stripped) at pH 7.5 and 250 mm Suc. The microsomes were pelleted, aliquots of the supernatants removed before washing the pellets, and then both were probed on immunoblots (IB) with an antibody against MBP.
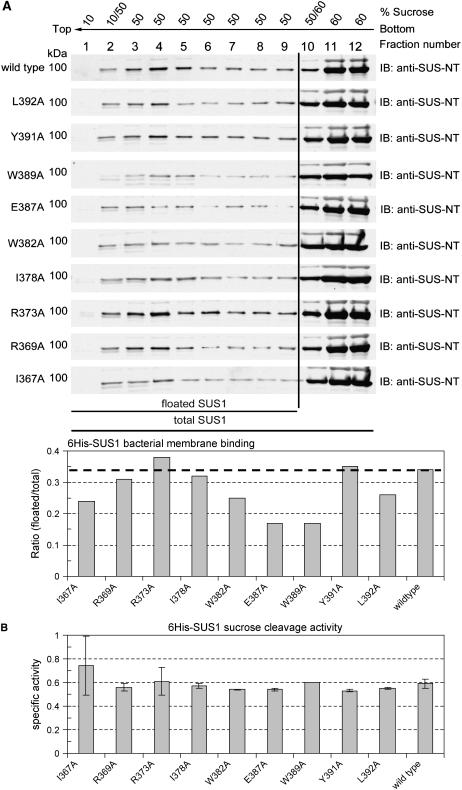
Site-Directed Mutants in the SUS1 PH-Like Domain Reveal Its Contribution to Membrane Binding
Individual amino acids within the identified PH-like domain of SUS1 were converted to Ala (Fig. 1) to determine their contribution to membrane affinity. All Ala-substituted mutant proteins were evaluated for intracellular distribution in E. coli but behaved very similar to wild type (approximately 70% partitioned to 100 K pellet; data not shown). However, when the amount of these recombinant proteins that bound to bacterial membranes was evaluated by Suc gradient flotation of the 100 K pellet, the substitution of certain hydrophobic and acidic residues with Ala significantly reduced the membrane-binding ability of SUS1 (Fig. 7A). In particular, binding to E. coli membranes was reduced by 30% to 50% in the I367A, E387A, and W389A recombinant mutant proteins. Conversely, substitution of the basic residue at position 373 with Ala (R373A) marginally (approximately 10%) increased SUS1 binding. These results not only enforced the conclusion that the SUS1 PH-like domain positively contributes to membrane affinity, but also suggested that binding may involve both hydrophobic and electrostatic interactions. These mutated residues occur primarily in a loop between β3 and α3 and do not alter the predicted secondary structure of the region (Fig. 1A; data not shown), implicating the importance of the individual amino acid itself to membrane binding. Significantly, these mutations did not interfere with the enzymatic (Suc cleavage) activity of SUS1 (Fig. 7B), as all recombinant proteins maintained essentially the same activity as wild type.
Figure 7.
Suc gradient flotation reveals that the PH-like domain of SUS1 influences binding to bacterial membranes. A, Equal volumes of total protein isolated in E. coli 100,000g pellets of recombinants expressing wild-type SUS1 or the Ala-substituted mutant listed to the left of the sections were subjected to Suc gradient flotation and fractions probed on immunoblots (IB) with an antibody against the SUS1 N terminus (anti-SUS-NT). Densitometry of these blots is displayed in the graph at the bottom. B, Suc cleavage activity (μmol UDP-Glc min−1 mg−1) of wild-type SUS1 and the Ala-substituted mutants. Y axis bars indicate ses.
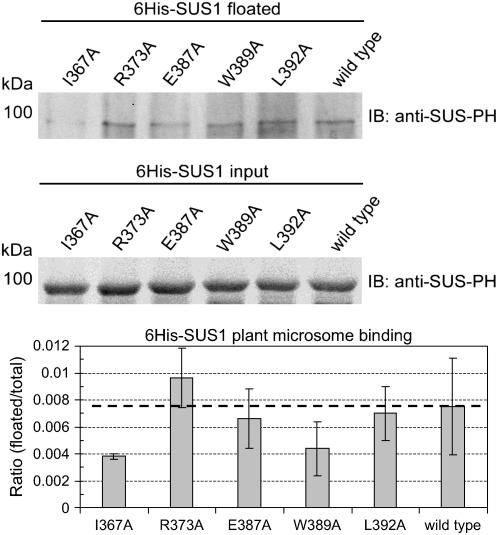
Selected Ala-substituted mutant proteins were tested for binding to plant membranes by mixing them with microsomes obtained from the apical 24 to 36 cm region of elongating maize leaves and flotation in Suc gradients. Similar to the binding observed for these proteins to bacterial membranes (Fig. 7A), mean values for binding to plant microsomes were reduced in the I367A and W389A mutants by about 50% (Fig. 8). Binding to plant microsomes was also marginally higher again in the R373A mutant protein in comparison to wild type.
Figure 8.
Suc gradient flotation reveals that the PH-like domain of SUS1 influences binding to plant microsomes. Equal amounts (0.30 pmol μL−1) of wild-type SUS1 or the Ala-substituted mutants listed above the sections were mixed with equal amounts of apical leaf microsomes at pH 7.5 and 150 mm Suc. After pelleting the microsomes and removing aliquots of the supernatants (input) the pellets were subjected to Suc gradient flotation and the top fraction probed on immunoblots (IB) with an antibody against SUS1 (anti-SUS-PH). A representative immunoblot is shown and densitometry of three separate blots is displayed in the graph at the bottom. Y axis bars indicate ses.
Sucrose and Low pH Are Identified as Potential Physiological Effectors of SUS1 Membrane Binding
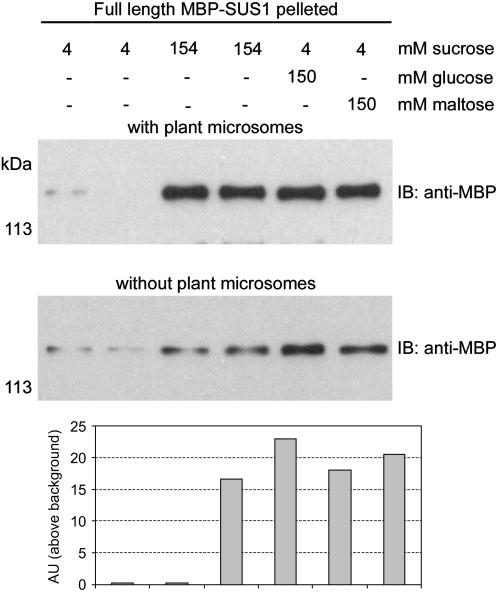
The effect of different sugars on the interaction of SUS1 with membranes was evaluated by mixing the full-length MBP-SUS1 protein with carbonate-washed microsomes obtained from the apical 24 to 36 cm region of elongating maize leaves followed by pelleting and washing to remove unbound protein (Fig. 9). This type of assay produced a higher level of background due to tube absorption but was necessitated by the need to perform the binding reactions under reduced sugar levels, which cannot be done in the Suc gradient-dependent flotation assays that completely lacked background. However, these experiments clearly revealed that Suc was important for the ability of SUS1 to bind plant microsomes (Fig. 9), as binding was reduced about 95% at low Suc concentrations. It is important to note that in all other experiments Suc was always maintained at 150 mm or greater. Glc and maltose at equimolar amounts to Suc mimicked this effect in these assays, suggesting the involvement of the glucosyl unit of Suc.
Figure 9.
Sugars stimulate binding of SUS1 to plant microsomes. Equal amounts (0.75 pmol μL−1) of full-length MBP-SUS1 were analyzed without added microsomes or mixed with equal amounts of carbonate-stripped apical leaf microsomes at pH 6.5 and residual (4 mm) Suc (duplicate reactions shown), or after addition of 150 mm Suc (duplicate reactions shown), Glc, or maltose. The controls and microsomes were pelleted, washed, and then probed on immunoblots (IB) with an antibody against MBP. Densitometry of these blots is displayed in the graph at the bottom.
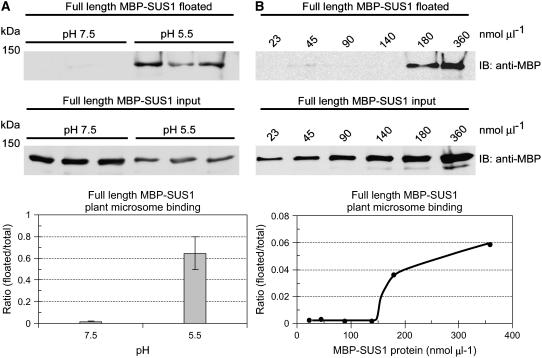
When the association of full-length MBP-SUS1 to apical (24–36 cm) microsomes was tested at pH 7.5 versus pH 5.5 by flotation (Fig. 10A), a dramatic stimulation (>6-fold) in membrane binding was observed at the lower pH. In similar flotation assays performed with increasing amounts of MBP-SUS1, the binding of SUS1 to carbonate-washed apical microsomes at pH 5.5 was sigmoidal (Fig. 10B). Binding was not apparent at MBP-SUS1 concentrations less than 140 nmol μL−1 and increased sharply at 180 nmol μL−1, suggestive of a cooperativity effect of SUS1 concentration on binding. This effect could be due to the presence of the MBP tag, but cannot be a source of variability in previous experiments because protein concentration within an experiment was constant and above the binding threshold. A further doubling of MBP-SUS1 concentration (360 nmol μL−1) did not double the amount bound, indicating that membrane binding is saturable (Fig. 10B).
Figure 10.
Low pH and high SUS1 protein concentration positively influence binding to plant microsomes. A, Equal amounts (0.18 pmol μL−1) of full-length MBP-SUS1 were mixed with equal amounts of apical leaf microsomes at pH 5.5 or pH 7.5 and 150 mm Suc. After pelleting the microsomes and removing aliquots of the supernatants (input) the pellets were subjected to Suc gradient flotation and the top fraction probed on immunoblots (IB) with an antibody against MBP. Densitometry of these blots is displayed in the graph at the bottom, y axis bars indicate ses. B, Increasing amounts (0.023–0.36 pmol μL−1) of full-length MBP-SUS1 were mixed with equal amounts of carbonate-stripped apical leaf microsomes at pH 5.5 and 150 mm Suc. After pelleting the microsomes and removing aliquots of the supernatants (input) the pellets were subjected to Suc gradient flotation and the top fraction probed on immunoblots (IB) with an antibody against MBP. Densitometry of these blots is displayed in the graph at the bottom.
DISCUSSION
The biosynthesis of cellulose is a universal aspect of plant growth that consumes a significant amount of photosynthetically fixed carbon. An emerging feature of carbon partitioning toward this extracellular sink involves the channeling of Suc-derived UDP-Glc to the cellulose-synthase complex by m-SUS (Amor et al., 1995; Winter and Huber, 2000; Haigler et al., 2001; Koch, 2004). However, we do not currently understand how m-SUS abundance is regulated, whether m-SUS interacts with the plasma membrane directly or complexes with other membrane proteins (e.g. CesA), and what regions of the SUS molecule are involved in membrane localization. In this study, we have addressed the protein structural requirements that allow the soluble enzyme SUS to interact with membranes. Specifically we focused on an approximately 100-amino acid region of SUS that we identified as having significant sequence homology to the C-terminal PH domain of human pleckstrin (Fig. 1A). We conclude that: (1) SUS has an inherent affinity for membranes (Figs. 4, 6, and 7) and therefore does not necessarily need to colocalize with another protein for membrane localization; (2) The interaction of SUS with membranes involves both the N-terminal noncatalytic region (residues 1–362) and the PH-like domain (residues 360–457; Figs. 5 and 8); and (3) Factors that influence the binding of SUS to membranes include pH, membrane source, and the concentrations of both SUS protein and Suc (Figs. 6, 9, and 10). We do not suggest that the intrinsic ability of SUS to bind membranes is exclusively responsible for dictating membrane localization in vivo, and cannot exclude the possibility that the interaction of SUS1 with plant microsomes in vitro reflects affinity for an integral membrane protein. Additional positive and negative influences on m-SUS may be supplied by localized protein:protein interactions that have been demonstrated or suggested to exist (Winter et al., 1998; Etxeberria and Gonzalez, 2003; Wachter et al., 2003; Wienkoop and Saalbach, 2003; Koch, 2004). However, an inherent affinity for membranes may partially explain the localization of SUS on membrane systems other than the plasma membrane, such as the tonoplast (Etxeberria and Gonzalez, 2003), golgi (Buckeridge et al., 1999), and peribacteroid membrane (Wienkoop and Saalbach, 2003), that are not involved in cellulose synthesis and are certain to differ in their membrane protein composition.
The Sequence Determinants Identified for SUS Membrane Affinity Have Functional Counterparts in Various Proteins
In GTases that obviously lack transmembrane domains but are nonetheless found in association with membranes, sparse experimental data exists to suggest the regions involved in this ability. Our analyses of the interaction of the SUS1 GTase with bacterial membranes in vivo (Figs. 4 and 7) and plant microsomes in vitro (Figs. 5, 6, 8, 9, and 10) identified two regions involved in binding ability, namely the N-terminal 362 amino acids and the PH-like domain (amino acids 360–457; Fig. 1A). The PH-like domain was the focus of this study, and the determinants of membrane binding within the N-terminal domain are currently under investigation. The demonstration that multiple regions are involved in membrane affinity is rather common, particularly for PH domain-containing proteins (Lemmon et al., 2002; Oku et al., 2003; Yu et al., 2004).
The N-terminal 362 amino acids of SUS1 include the entire noncatalytic domain (Fig. 1B) and a small portion of the catalytic domain. No structural templates for this region of SUS1 could be identified, and it lacks significant homology to other proteins known to interact with membranes. The entire SUS1 protein is not hydrophobic, possessing a grand average of hydrophobicity (GRAVY) score of −0.282, where higher positive values indicate greater hydrophobicity. Moreover, the N-terminal noncatalytic domain is also not hydrophobic (GRAVY score −0.353). However, as pointed out earlier, transmembrane prediction algorithms identified a region (FLGTIPMVFNVVILSPHGYFA) of the SUS1 sequence between amino acids 274 and 294 with high hydrophobicity (GRAVY score 1.324; Carlson and Chourey, 1996; Winter and Huber, 2000). This hydrophobic domain of SUS1 is conserved in the other two maize SUS isoforms and dicot SUS sequences, and is included within the smallest truncation mutant (T362stop) that we demonstrate to have membrane-binding affinity. It is clear that this hydrophobic domain does not function as a transmembrane helix because SUS is peripherally associated with both microsomal and tonoplast membranes (Etxeberria and Gonzalez, 2003; Hardin et al., 2004), and the core of this hydrophobic domain (VFNVVILS) constitutes the predicted β1 strand of the N-terminal catalytic domain (Fig. 1B). However, the preceding loop (FLGTIPM) is likely to be surface exposed and could contribute a hydrophobic determinant (GRAVY score 1.471) to membrane binding. This hydrophobic domain is only one possible contributor to membrane affinity within the N-terminal noncatalytic domain and does not rule out additional regions or modifications that influence m-SUS abundance. In particular, SUS1 is known to have a peptide S-thiolation site (Rohrig et al., 2004) and two sites of phosphorylation (Hardin et al., 2004) within the N-terminal noncatalytic domain, and phosphorylation has been shown to influence the binding of SUS to certain membrane types (Hardin et al., 2004).
The determinant of SUS1 membrane binding identified in this study is the PH-like domain (amino acids 360–457; Fig. 1A), which resides between the predicted N-terminal β3 strand and the α5 helix (Fig. 1B). Although this region of SUS has significant sequence homology to the C-terminal PH domain of human pleckstrin, it is unlikely to adopt a similar overall structural fold (Fig. 1A). Therefore, GTase structures may provide a better structural template to evaluate this region. Significantly, the MurG crystal structure (Ha et al., 2000) demonstrates that many of the hydrophobic and basic residues within the same region (β3-α5) occupied by the SUS PH-like domain fold to form a surface-exposed patch in MurG that has been suggested to constitute the membrane-association site (Ha et al., 2000). Since SUS1 can easily be threaded onto the atomic coordinates of the solved MurG structure by the Protein Homology/Analogy Recognition Engine (phyre) server, it is likely that the hydrophobic residues within the SUS1 PH-like domain form a similar hydrophobic patch. Further studies have demonstrated that the MurG protein has affinity for negatively charged lipids (i.e. cardiolipin; van den Brink-van der Laan et al., 2003). This suggests that the SUS1 PH-like domain may confer affinity to anionic lipids, and indeed the loop sequence between β3 and α3 (ILRVPFRTENGIVRKWISRFEVWPYL) has a pI of 10.83. In fact, effects on membrane binding were demonstrated by mutation of specific residues (underlined in the preceding sequence) within this region (Figs. 7 and 8). The predicted secondary structure of this region (Fig. 1A) is not altered by these Ala substitutions (data not shown), implicating a contribution of the individual amino acids themselves. Since the substitution of hydrophobic residues (I367 and W389) and an acidic residue (E387) with Ala decreased binding, whereas the basic residue mutation R373A slightly increased binding, presumably the position-specific contributions of hydrophobicity and negative charge influence SUS membrane affinity. Two of the residues (I367 and R373) we identified as contributing to SUS membrane binding are conserved (L251 and R257) in pleckstrin and have been identified as important for phosphatidylinositol-(3,4)-bisphosphate interaction (Isakoff et al., 1998; Edlich et al., 2005).
Interactions of Variables Is Likely to Control the Abundance of m-SUS
Despite the fact that certainly some of the regulation of SUS abundance occurs at the transcriptional level (Winter and Huber, 2000; Koch, 2004), we propose that much of the regulation of m-SUS abundance occurs at the protein level via the targeting of soluble SUS protein onto membranes. The relative abundance of m-SUS can vary dramatically (3%–50% of total) between plant species and organs (Amor et al., 1995; Carlson and Chourey, 1996; Winter et al., 1997; Sturm et al., 1999) and is developmentally or conditionally variable within a given organ (Amor et al., 1995; Winter et al., 1997; Subbaiah and Sachs, 2001). In maize leaves, the abundance of m-SUS is developmentally variable and not a fixed percentage of soluble SUS (Hardin et al., 2004), demonstrating that m-SUS protein abundance is regulated by factors other than SUS abundance within the soluble phase.
In this report, we identify several factors that potentially influence the abundance of m-SUS in vivo, including pH, membrane source, and the concentrations of both SUS protein and Suc (Figs. 6, 9, and 10). Effectors such as Ca2+ and phosphorylation also influence m-SUS abundance directly (Winter et al., 1997; Haigler et al., 2001; Hardin et al., 2004), and salt and inorganic N stresses preferentially reduce m-SUS abundance in nodules (Komina et al., 2002). Lowering the pH from 7.5 to 5.5 enhanced the binding of SUS1 to microsomes (Fig. 10A). The pH optimum for the Suc cleavage activity of SUS is acidic (pH 6.0–6.5), whereas Suc synthetic activity is maximal at alkaline pH (8.0–8.8; Tsai, 1974), suggesting that pH directly influences the conformation and enzymatic activities of SUS. Low pH (5.5) also reduces the helical conformation of at least the extreme NT of SUS1 (Hardin et al., 2004). Coincident with these low pH-promoted conformational changes could be the exposure of membrane-binding determinants.
The diversion of carbon toward cellulose biosynthesis rather than core metabolic requirements has been suggested to occur only when conditions are nearly optimal (Haigler et al., 2001). A component of establishing a near optimal condition for cellulose synthesis would be provision of a sufficient supply of Suc. In our experiments, the presence of higher levels (154 mm) of Suc was nearly essential for SUS1 to bind leaf microsomes (Fig. 9). These effects on m-SUS binding occurred within the reported range for physiological, cytosolic Suc concentrations (approximately 20–200 mm; Gerhardt et al., 1987; Winter et al., 1994), and was seemingly a direct effect on SUS itself. It has been shown that Suc effects the conformation of SUS directly, but curiously it decreased the overall surface hydrophobicity (Winter et al., 1997). Hypothetically, SUS could be part of a Suc-sensing mechanism for the carbohydrate level in sink tissues (Winter and Huber, 2000). These data also suggest the potential for m-SUS to localize in close proximity to Suc:proton cotransporters, as locally the concentration of Suc would be high and the pH low. Both of these conditions (Figs. 9 and 10A) would be expected to promote the membrane binding of SUS in the vicinity of these transporters. The existence of acidic pH microdomains has been experimentally demonstrated (Schwiening and Willoughby, 2002), and data to suggest that SUS:Suc transporter complexes exist has been provided (Etxeberria and Gonzalez, 2003).
SUS is known to interact with a variety of different membrane types and, while beyond the scope of this manuscript, a logical next level of evaluation involves determining the contribution of these sequence determinants and effectors on the binding of SUS to individual membrane types. Additionally, we are actively pursuing: (1) the creation of multiple and additional mutations within the N-terminal noncatalytic and PH-like domains, and (2) the production of transient and stable transgenic plants to assess the in vivo significance of altered SUS membrane affinity on cellulose deposition and carbon utilization.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Maize (Zea mays L. cv Pioneer 3183 or inbred line B73) plants were grown in a soil mixture in a greenhouse and fertilized three times weekly with a modified Hoagland solution. Temperature was controlled by an Argus control system (White Rock) and supplemental lighting was provided by 1,000 W metal halide lamps. Immature leaves from the top of 4- to 8-week-old plants were harvested, 12 cm segments were removed from the elongating basal (4–16 cm) or apical (24–36 cm) regions of the leaves, immediately frozen in liquid nitrogen, and stored at −80°C.
Preparation of Maize Leaf Microsomes
Frozen maize leaf was ground in 2 volumes of protein extraction buffer [100 mm MOPS, pH 7.5, 10 mm dithiothreitol (DTT), 5 mm EDTA, 5 mm EGTA, 20 mm NaF, 0.5 μm microcystin-LR, 1 mm 4-(2-Aminoethyl)benzenesulfonyl fluoride (AEBSF), 1 mm benzamidine, 5 mm caproic acid, 2 μm N-(trans-Epoxysuccinyl)-Leu-4-guanidinobutylamide (E64), 10 μm Carbobenzoxy-leucyl-leucyl-leucinal (MG132; CalBiochem), 0.05 g g−1 fresh weight polyvinylpolypyrrolidone, and 0.33 m Suc]. Clarified extracts were produced by filtration through Miracloth (CalBiochem) and two sequential centrifugations at 9,500g and 4°C for 20 min. The microsomal pellet was recovered at 100,000g for 1 h at 4°C, then salt and Brij58 washed to invert vesicles and release entrapped protein (Johansson et al., 1995) by resuspension in 50 mm MOPS pH 7.5, 5 mm DTT, 1 mm EDTA, 1 mm EGTA, 20 mm NaF, 0.1 μm microcystin-LR, 1 mm benzamidine, 5 mm caproic acid, 2 μm E64, 0.25 m Suc, 150 mm NaCl, and 0.05% (w/v) Brij 58. The membrane fraction was pelleted at 100,000g for 1 h at 4°C and resuspended on ice in 4% to 10% of the original volume in 50 mm MOPS pH 7.5, 1 mm DTT, 0.1 mm EDTA, 5 mm NaF, 0.5 mm benzamidine, 2.5 mm caproic acid, 1 μm E64, and 0.25 m Suc. Aliquots of membranes were treated with carbonate buffer (100 mm Na2CO3 pH 11.5, 1 mm DTT, 0.1 mm EDTA, 5 mm NaF, 0.5 mm benzamidine, 2.5 mm caproic acid, 1 μm E64, and 0.25 m Suc for 30 min on ice; Fujiki et al., 1982), then recovered at 145,000g for 1 h at 4°C and resuspended as above.
Production of Mutant SUS1 Recombinants and Proteins
A maize SUS1 cDNA (GI #514945) in the pRSETB vector was used to construct sequencing-verified, site-directed mutants (Ala substitutions) with the QuikChange XL kit as recommended by Stratagene. Subcloning of a T4 DNA polymerase-blunt ended PvuI, BamHI SUS1 fragment into a T4 DNA polymerase-blunt ended PstI, BamHI digested pMAL-c2x vector (New England Biolabs) was used to produce the full-length MBP-SUS1 construct. Sequencing-verified, site-directed mutants (stop codon insertions) were produced as above. All constructs were transformed into Escherichia coli BL21 (DE3) for protein expression.
Transformants expressing MBP-SUS1 proteins were induced with 0.5 mm isopropylthio-β-galactoside at 30°C, cell pellets were washed and resuspended in 20 mm Tris pH 7.4, 200 mm NaCl, 1 mm DTT, 1 mm EDTA, 10 μm leupeptin, 0.5 mm AEBSF, 1 mm benzamidine, 5 mm caproic acid, and 1 μm E64. Cells were lysed by sonication in an ice bath and clarified extracts were produced by two sequential centrifugations at 35,000g and 4°C for 15 min. Batch binding with amylose resin (New England Biolabs) equilibrated in column buffer (20 mm Tris pH 7.4, 200 mm NaCl, 1 mm DTT, and 1 mm EDTA) was performed on ice for 20 min, washing with 15 bed volumes of column buffer, and elution with 10 bed volumes of column buffer plus 20 mm maltose was performed in gravity-feed columns at 4°C. Dialyzed samples were loaded onto SOURCE 15Q (Amersham) anion-exchange resin in buffer A (50 mm MOPS pH 7.5 and 2 mm DTT) and eluted at 4°C with a 30-mL linear gradient of 0 to 500 mm NaCl in buffer A. Peak fractions were dialyzed against 10 mm MOPS pH 7.5 and 1 mm DTT at 4°C.
Transformants expressing 6His-SUS1 proteins were induced with 0.5 mm isopropylthio-β-galactoside at 30°C, cell pellets were washed and resuspended in 50 mm MOPS pH 7.5, 300 mm NaCl, 50 mm Suc, 250 μg mL−1 lysozyme, and incubated at room temperature for 30 min. Prior to sonication in an ice bath, 10 μm leupeptin, 1 mm AEBSF, 1 mm benzamidine, 5 mm caproic acid, 1 μm E64, and 0.2 m Suc were added. Clarified extracts were produced by two sequential centrifugations at 35,000g and 4°C for 15 min. Batch binding with Talon metal affinity resin (CLONTECH) equilibrated in column buffer (50 mm MOPS pH 7.5, 300 mm NaCl, and 50 mm Suc) was performed at 4°C for 30 min, washing with 26 bed volumes of column buffer plus 2.5 mm imidazole and elution with 10 bed volumes of column buffer plus 250 mm imidazole was performed in gravity-feed columns at 4°C. Samples were immediately adjusted to 1 mm DTT and 5 mm EDTA. Dialyzed samples were loaded onto SOURCE 15Q (Amersham) anion-exchange resin in buffer B (50 mm MOPS pH 7.5, 1 mm DTT, and 50 mm Suc) and eluted at 4°C with a 30-mL linear gradient of 0 to 500 mm NaCl in buffer B. Peak fractions were dialyzed against 10 mm MOPS pH 7.5, 1 mm DTT, and 10 mm Suc at 4°C. Protein concentrations were determined in duplicate by the dye-binding assay (Bio-Rad) with bovine serum albumin as the standard. Concentration of protein samples was performed by centrifugal filtration at 4°C in 10 K molecular weight cutoff Centricons (Millipore).
Size-exclusion chromatography was performed by loading 200 μL of recombinant MBP-SUS1 samples onto a Superdex 200 HR 10/30 column (Amersham) equilibrated in 10 mm MOPS pH 7.5, 1 mm DTT, and 100 mm NaCl. Isocratic elutions were at 4°C and a flow rate of 0.5 mL min−1 and 0.25 mL fractions were collected. The column was calibrated with thyroglobulin (669 kD), apoferritin (443 kD), β amylase (200 kD), alcohol dehydrogenase (150 kD), bovine serum albumin (66 kD), and carbonic anhydrase (29 kD).
Ultracentrifugal Fractionation and Suc Gradient Flotation of Membranes
Bacterial protein extracts from MBP-SUS1 transformants (in 50 mm MOPS pH 7.5, 1 mm DTT, 1 mm EDTA, 50 mm NaCl, 10 μm leupeptin, 1 mm AEBSF, 0.5 mm benzamidine, 1 μm E64, and 0.25 m Suc) and 6His-SUS1 transformants (in 50 mm MOPS pH 7.5, 1 mm DTT, 1 mm EDTA, 300 mm NaCl, 10 μm leupeptin, 1 mm AEBSF, 1 mm benzamidine, 5 mm caproic acid, 1 μm E64, and 0.25 m Suc) were clarified of any intact cells and inclusion bodies by filtration through Miracloth (Calbiochem) and two sequential centrifugations at 9,000 to 10,000g and 4°C for 30 min. Membrane vesicles within these extracts were recovered at 100,000g for 1.5 to 2.5 h at 4°C and the supernatants retained as the soluble phase extract (100 K supers). Membrane vesicles were washed in 50 mm MOPS pH 7.5, 1 mm DTT, 1 mm EDTA, 10 μm leupeptin, 1 mm AEBSF, 0.5 mm benzamidine, 1 μm E64, 0.25 m Suc, 150 mm NaCl, and 0.05% (w/v) Brij 58. The membrane fraction was pelleted at 100,000g for 1.5 h at 4°C and resuspended on ice in 10% of the original volume in a 60% (w/w) Suc solution made in 50 mm MOPS pH 7.5, 1 mm DTT, 0.1 mm EDTA, 0.5 mm benzamidine, 2.5 mm caproic acid, and 1 μm E64. This sample was used as the pelleted fraction (100 K pellets) and for flotation as previously described (Ahola et al., 1999; Hardin et al., 2004).
Microsome-Binding Experiments
The ability of various SUS1 constructs to bind plant leaf microsomes was determined by pelleting or adapting the Suc gradient flotation conditions to a microscale format. For the flotation assays, equimolar amounts of each recombinant SUS1 protein (10 or 15 pmol) was mixed with microsomes in 50 mm MOPS pH 7.5 or 50 mm MES pH 5.5, and 150 mm Suc on ice for 30 min. Membranes were pelleted in an airfuge (Beckman Coulter) at 100,000g for 15 min and a portion of the supernatant (input) removed. The pellets were resuspended in 50 μL of a 60% (w/w) solution of Suc made in 50 mm MOPS pH 7.5 or 50 mm MES pH 5.5, and then overlaid with 175 μL of 50% (w/w) Suc and 15 L of 10% (w/w) Suc both made in 50 mm MOPS pH 7.5 or 50 mm MES pH 5.5 and 100 mm NaCl. The gradients were centrifuged at 100,000g for 1 h and the top 17% to 30% of the gradient sampled. Some samples were delipidated using a methanol-chloroform-water procedure (Wessel and Flugge, 1984) prior to SDS-PAGE analysis. For the pelleting assays, recombinant SUS1 proteins were ensured to be soluble by prespinning at 145,000g for 15 min in an Airfuge, and on occasion the tubes used for the binding reactions were blocked with a 0.1% (w/v) solution of purified casein (I-block, Tropix) in 50 mm MOPS pH 7.5. An equimolar amount of each recombinant SUS1 protein (11–22 pmol) was mixed with microsomes in 50 mm MOPS pH 7.5, 250 mm Suc, or 50 mm MES pH 6.5, 150 mm Suc, Glc, or maltose on ice for 30 min. Membranes were pelleted in an airfuge (Beckman Coulter) at 145,000g for 15 min and a portion of the supernatant (input) removed. The pellets and tubes were washed with the appropriate buffered, sugar solution and then denatured for SDS-PAGE analysis.
Enzyme Activity Assays
SUS activity was assayed in the catabolic (cleavage) direction as described (Hardin et al., 2004) using a fixed-time assay in 50 mm MES pH 6.5 containing 1 mm UDP and 100 mm Suc. Production of UDP-Glc was determined enzymatically using UDP-Glc dehydrogenase (Calbiochem) coupled to the reduction of NAD.
Immunological Methods
The SUS1 peptides PH (CHILRVPFRTENGIVRKWISR) and NT (MGEGAGDRVLSRL) were purified by HPLC, verified by mass spectrometry, and used to generate immune serum in rabbits (Bethyl Laboratories). Immunosorbent columns coupled with PH or NT peptides were used to obtain the affinity-purified anti-SUS-PH and anti-SUS-NT antibodies. The anti-MBP polyclonal antiserum was from a commercial vendor (New England Biolabs). Proteins were denatured, separated on 7% to 10% polyacrylamide-0.1% SDS gels, transferred onto polyvinylidene fluoride membranes (Immobilon-FL, Millipore), and immunoblotted. Blocking was performed in 2% (w/v) fish gelatin (Sigma) in phosphate-buffered saline (PBS; 5 mm NaH2PO4 pH 7.4 and 150 mm NaCl); washes and antibody dilutions were done in PBS containing 0.1% (v/v) Tween 20 (PBST). The Alexa Fluor 680-conjugated secondary antibodies (Molecular Probes) were detected by scanning on an Odyssey infrared imager (LI-COR) and densitometry performed with the supplied software.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AAA68209.
Acknowledgments
Mention of a trademark or proprietary product does not constitute a guarantee or warranty by the U.S. Department of Agriculture-Agricultural Research Service and does not imply its approval to the exclusion of other products that might also be suitable.
This work was supported in part by funds from the U.S. Department of Energy (grant no. DE–AIOS–91ER20031 to S.C. Huber).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Steven C. Huber (schuber1@life.uiuc.edu).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.078006.
References
- Ahola T, Lampio A, Auvinen P, Kaariainen L (1999) Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J 18: 3164–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht G, Mustroph A (2003) Localization of sucrose synthase in wheat roots: increased in situ activity of sucrose synthase correlates with cell wall thickening by cellulose deposition under hypoxia. Planta 217: 252–260 [DOI] [PubMed] [Google Scholar]
- Amor Y, Haigler CH, Johnson S, Wainscott M, Delmer DP (1995) A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proc Natl Acad Sci USA 92: 9353–9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg S, Edman M, Li L, Wikstrom M, Wieslander A (2001) Sequence properties of the 1,2-diacylglycerol 3-glucosyltransferase from Acholeplasma laidlawii membranes. J Biol Chem 276: 22056–22063 [DOI] [PubMed] [Google Scholar]
- Buckeridge MS, Vergara CE, Carpita CC (1999) The mechanism of synthesis of a mixed-linkage (1-3),(1-4)β-d-glucan in maize: evidence for multiple sites of glucosyl transfer in the synthase complex. Plant Physiol 120: 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschiazzo A, Ugalde JE, Guerin ME, Shepard W, Ugalde RA, Alzari PM (2004) Crystal structure of glycogen synthase: homologous enzymes catalyze glycogen synthesis and degradation. EMBO J 23: 3196–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson SJ, Chourey PS (1996) Evidence for plasma membrane-associated forms of sucrose synthase in maize. Mol Gen Genet 252: 303–310 [DOI] [PubMed] [Google Scholar]
- Charnock SJ, Henrissat B, Davies GJ (2001) Three-dimensional structures of UDP-sugar glycosyltransferases illuminate the biosynthesis of plant polysaccharides. Plant Physiol 125: 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourey PS, Taliercio EW, Carlson SJ, Ruan Y-L (1998) Genetic evidence that the two isozymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Mol Gen Genet 259: 88–96 [DOI] [PubMed] [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B (2003) An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328: 307–317 [DOI] [PubMed] [Google Scholar]
- Duncan KA, Hardin SC, Huber SC (2005) The third sucrose synthase in Zea mays (SUS3) is a ubiquitous protein that is phosphorylated and membrane associated (abstract no. 22005). In Plant Biology 2005, July 16–20, 2005, Seattle. American Society of Plant Biologists, Rockville, MD, http://abstracts.aspb.org/pb2005/public/M11/9079.html
- Edlich C, Stier G, Simon B, Sattler M, Muhle-Goll C (2005) Structure and phosphatidylinositol-(3,4)-bisphosphate binding of the C-terminal PH domain of human pleckstrin. Structure 13: 277–286 [DOI] [PubMed] [Google Scholar]
- Edman M, Berg S, Storm P, Wikstrom M, Vikstrom S, Ohman A, Wieslander A (2003) Structural features of glycosyltransferases synthesizing major bilayer and nonbilayer-prone membrane lipids in Acholeplasma laidlawii and Streptococcus pneumoniae. J Biol Chem 278: 8420–8428 [DOI] [PubMed] [Google Scholar]
- Egelund J, Skjot M, Geshi N, Ulvskov P, Petersen BL (2004) A complementary bioinformatics approach to identify potential plant cell wall glycosyltransferase-encoding genes. Plant Physiol 136: 2609–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etxeberria E, Gonzalez P (2003) Evidence for a tonoplast-associated form of sucrose synthase and its potential involvement in sucrose mobilization from the vacuole. J Exp Bot 54: 1407–1414 [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB (1982) Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol 93: 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt R, Stitt M, Heldt HW (1987) Subcellular metabolite levels in spinach leaves. Plant Physiol 83: 399–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S, Walker D, Shi Y, Walker S (2000) The 1.9A crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis. Protein Sci 9: 1045–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler CH, Ivanova-Datcheva M, Hogan PS, Salnikov VV, Hwang S, Martin K, Delmer DP (2001) Carbon partitioning to cellulose synthesis. Plant Mol Biol 47: 29–51 [PubMed] [Google Scholar]
- Hardin SC, Winter H, Huber SC (2004) Phosphorylation of the amino terminus of maize sucrose synthase in relation to membrane association and enzyme activity. Plant Physiol 134: 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakoff SJ, Cardozo T, Andreev J, Li Z, Ferguson KM, Abagyan R, Lemmon MA, Aronheim A, Skolnik EY (1998) Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. EMBO J 17: 5374–5387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson F, Olbe M, Sommarin M, Larsson C (1995) Brij58, a polyoxy-ethylene acyl ether, creates membrane vesicles of uniform sidedness: a new tool to obtain inside-out (cytoplasmic side-out) plasma membrane vesicles. Plant J 7: 165–173 [DOI] [PubMed] [Google Scholar]
- Koch K (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7: 235–246 [DOI] [PubMed] [Google Scholar]
- Komina O, Zhou Y, Sarath G, Chollet R (2002) In vivo and in vitro phosphorylation of membrane and soluble forms of soybean nodule sucrose synthase. Plant Physiol 129: 1664–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T, Ohmiya Y, Hayashi T (2004) Evidence that sucrose loaded into the phloem of a poplar leaf is used directly by sucrose synthase associated with various β-glucan synthases in the stem. Plant Physiol 134: 1146–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordel B, Kutschera U (2000) Effects of gibberellin on cellulose biosynthesis and membrane-associated sucrose synthase activity in pea internodes. J Plant Physiol 156: 570–573 [Google Scholar]
- Kutschera U, Heiderich A (2002) Sucrose metabolism and cellulose biosynthesis in sunflower hypocotyls. Physiol Plant 114: 372–379 [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, Abrams CS (2002) Pleckstrin homology domains and the cytoskeleton. FEBS Lett 513: 71–76 [DOI] [PubMed] [Google Scholar]
- MacGregor EA (2002) Possible structure and active site residues of starch, glycogen and sucrose synthases. J Protein Chem 21: 297–306 [DOI] [PubMed] [Google Scholar]
- Oku M, Warnecke D, Noda T, Muller F, Heinz E, Mukaiyama H, Kato N, Sakai Y (2003) Peroxisome degradation requires catalytically active sterol glucosyltransferase with a GRAM domain. EMBO J 22: 3231–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Kawagoe Y, Hogan P, Delmer D (2002) Sitosterol-β-glucoside as primer for cellulose synthesis in plants. Science 295: 147–150 [DOI] [PubMed] [Google Scholar]
- Rohrig H, John M, Schmidt J (2004) Modification of soybean sucrose synthase by S-thiolation with ENOD40 peptide A. Biochem Biophys Res Commun 325: 864–870 [DOI] [PubMed] [Google Scholar]
- Ruan Y-L, Chourey PS, Delmer DP, Perez-Grau L (1997) The differential expression of sucrose synthase in relation to diverse patterns of carbon partitioning in developing cotton seed. Plant Physiol 115: 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y-L, Llewellyn DJ, Furbank RT (2003) Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 15: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksouk N, Pelosi L, Colin-Morel P, Boumedienne M, Abdian PL, Geremia RA (2005) The capsular polysaccharide biosynthesis of Streptococcus pneumoniae serotype 8: functional identification of the glycosyltransferase WciS (Cap8H). Biochem J 389: 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikov VV, Grimson MJ, Delmer DP, Haigler CH (2001) Sucrose synthase localizes to cellulose synthesis sites in tracheary elements. Phytochemistry 57: 823–833 [DOI] [PubMed] [Google Scholar]
- Salnikov VV, Grimson MJ, Seagull RW, Haigler CH (2003) Localization of sucrose synthase and callose in freeze-substituted secondary-wall-stage cotton fibers. Protoplasma 221: 175–184 [DOI] [PubMed] [Google Scholar]
- Schwiening CJ, Willoughby D (2002) Depolarization-induced pH microdomains and their relationship to calcium transients in isolated snail neurones. J Physiol 538: 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, Lienhard S, Schatt S, Hardegger M (1999) Tissue-specific expression of two genes for sucrose synthase in carrot (Daucus carota L.). Plant Mol Biol 39: 349–360 [DOI] [PubMed] [Google Scholar]
- Sturm A, Tang G-Q (1999) The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends Plant Sci 4: 401–407 [DOI] [PubMed] [Google Scholar]
- Subbaiah CC, Sachs MM (2001) Altered patterns of sucrose synthase phosphorylation and localization precede callose induction and root tip death in anoxic maize seedlings. Plant Physiol 125: 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C-Y (1974) Sucrose-UDP glucosyltransferase of Zea mays endosperm. Phytochemistry 13: 885–891 [Google Scholar]
- Uggla C, Magel E, Moritz T, Sundberg B (2001) Function and dynamics of auxin and carbohydrates during earlywood/latewood transition in scots pine. Plant Physiol 125: 2029–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink-van der Laan E, Boots J-WP, Spelbrink REJ, Kool GM, Breukink E, Killian JA, de Kruijff B (2003) Membrane interaction of the glycosyltransferase MurG: a special role for cardiolipin. J Bacteriol 185: 3773–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter R, Langhans M, Aloni R, Gotz S, Weilmunster A, Koops A, Temguia L, Mistrik I, Pavlovkin J, Rascher U, et al (2003) Vascularization, high-volume solution flow, and localized roles for enzymes of sucrose metabolism during tumorigenesis by Agrobacterium tumefaciens. Plant Physiol 133: 1024–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D, Flugge UI (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Wienkoop S, Saalbach G (2003) Proteome analysis: novel proteins identified at the peribacteroid membrane from Lotus japonicus root nodules. Plant Physiol 131: 1080–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter H, Huber JL, Huber SC (1997) Membrane association of sucrose synthase: changes during the graviresponse and possible control by protein phosphorylation. FEBS Lett 420: 151–155 [DOI] [PubMed] [Google Scholar]
- Winter H, Huber JL, Huber SC (1998) Identification of sucrose synthase as an actin-binding protein. FEBS Lett 430: 205–208 [DOI] [PubMed] [Google Scholar]
- Winter H, Huber SC (2000) Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. CRC Crit Rev Plant Sci 19: 31–67 [DOI] [PubMed] [Google Scholar]
- Winter H, Robinson DG, Heldt HW (1994) Subcellular volumes and metabolite concentrations in barley leaves. Planta 193: 532–555 [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA (2004) Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell 13: 677–688 [DOI] [PubMed] [Google Scholar]