Abstract
Nine genes of Arabidopsis (Arabidopsis thaliana) encode for β-amylase isozymes. Six members of the family are predicted to be extrachloroplastic isozymes and three contain predicted plastid transit peptides. Among the latter, chloroplast-targeted β-amylase (At4g17090) and thioredoxin-regulated β-amylase (TR-BAMY; At3g23920; this work) are experimentally demonstrated to be targeted to plastids. Recombinant TR-BAMY was catalytically active only when expressed as a mature protein, i.e. with no transit peptide. Mature TR-BAMY was a monomer of 60 kD, hydrolyzing soluble starch with optimal activity between pH 6.0 and 8.0. The activity of recombinant TR-BAMY was strictly dependent on redox potential with an Em,7.0 of −302 ± 14 mV. Thioredoxins f1, m1, and y1 of Arabidopsis were all able to mediate the reductive activation of oxidized TR-BAMY. Site-specific mutants showed that TR-BAMY oxidative inhibition depended on the formation of a disulfide bridge between Cys-32 and Cys-470. Consistent with TR-BAMY redox dependency, total β-amylase activity in Arabidopsis chloroplasts was partially redox regulated and required reducing conditions for full activation. In Arabidopsis, TR-BAMY transcripts were detected in leaves, roots, flowers, pollen, and seeds. TR-BAMY may be the only β-amylase of nonphotosynthetic plastids suggesting a redox regulation of starch metabolism in these organelles. In leaves, where chloroplast-targeted β-amylase is involved in physiological degradation of starch in the dark, TR-BAMY is proposed to participate to a redox-regulated pathway of starch degradation under specific stress conditions.
Plants store primary starch in chloroplasts under active photosynthetic conditions and degrade it during the night to obtain energy and organic matter for growth. Unlike leaf primary starch, long-term starch deposits are found in storage organs such as seeds, roots, or tubers. Degradation of long-term starch does not follow a diurnal rhythm as in leaves and is triggered during sink-to-source conversion of storage organs (e.g. seed germination or tuber sprouting).
Starch granules are semicrystalline particles composed of long polymers of Glc, chiefly amylopectin, which is an α-1,4 glucan with a distinct pattern of α-1,6 branches (Zeeman et al., 2004). Plants contain a host of starch-degrading enzymes and degradation pathways are believed to vary in different organs (Smith et al., 2005). In leaves, much of our knowledge on starch-degradation pathways relies on studies of Arabidopsis (Arabidopsis thaliana). Leaf starch in Arabidopsis is phosphorylated at either C-3 or C-6 position of one out of 2,000 Glc monomers (Yu et al., 2001). Two types of amylopectin phosphorylating enzymes have been identified, namely α-glucan water dikinase (GWD; Ritte et al., 2002) and phosphoglucan water dikinase (Baunsgaard et al., 2005; Kötting et al., 2005). Although phosphorylation probably occurs during both starch synthesis and degradation (Nielsen et al., 1994; Ritte et al., 2004), the activity of starch phosphorylating enzymes is required for normal starch degradation, suggesting that partially phosphorylated amylopectin is more accessible to degradative enzymes (Smith et al., 2005).
While α-amylases do not play a crucial role in leaf starch degradation (Yu et al., 2005), hydrolysis of α,1-4 bonds by β-amylases seems to be essential (Scheidig et al., 2002). Debranching enzymes like isoamylase may also be required to hydrolyze α,1-6 bonds, although distinct debranching enzymes are involved in normal starch synthesis as well (Smith et al., 2005). β-Amylases remove β-maltose units from the nonreducing end of polyglucans, and because β-maltose is not further metabolized in plastids (Lu and Sharkey, 2004), the activity of β-amylases is functionally coupled to the maltose transporter MALTOSE EXCESS1 (MEX1), allowing maltose to reach the cytoplasm (Niittylä et al., 2004).
Nine β-amylases are encoded by the Arabidopsis genome, and three of them are predicted to be chloroplast targeted (At3g23920, At4g00490, and At4g17090). Among these, the product of gene At4g17090 was experimentally confirmed to be a chloroplastic β-amylase, chloroplast-targeted β-amylase (CT-BMY; Lao et al., 1999). The involvement of CT-BMY in leaf starch degradation was supported by Arabidopsis RNAi lines (Kaplan and Guy, 2005) and transgenic potato (Solanum tuberosum) plants with reduced levels of CT-BMY ortholog (Scheidig et al., 2002), which accumulated high levels of starch in leaves.
The pathway involving GWD, phosphoglucan water dikinase, isoamylase, β-amylase, and MEX1 is now believed to be the major route for starch degradation and sugar export from Arabidopsis chloroplasts (Lloyd et al., 2005; Smith et al., 2005). Consistently, mutants or transgenic plants with decreased contents of any of these proteins accumulate high levels of leaf starch during night (starch excess or sex mutants). Other chloroplast enzymes such as d-enzyme (Critchley et al., 2001) and Glc transporter (Weber et al., 2000) also participate in starch degradation, although their contribution in Arabidopsis may be smaller.
Regulatory mechanisms controlling the diurnal rhythm of leaf starch degradation are largely unknown. Interestingly, an Arabidopsis GWD has recently been shown to be redox regulated (Mikkelsen et al., 2005). This plastidic GWD isozyme was bound to starch granules in the dark in the oxidized/inactive conformation and was released/activated in the light by reduced thioredoxin (Trx).
In this work, we show that gene At3g23920 encodes a newly described plastid-targeted β-amylase. Similarly to GWD, this β-amylase isozyme is redox regulated and activated by reduced plastidial Trxs f1, m1, and y1 (Trx-regulated β-amylase [TR-BAMY]). Cys responsible for Trx-mediated regulation of TR-BAMY are absent in any other β-amylase of Arabidopsis. Specific roles for TR-BAMY may include starch and/or polyglucan degradation both in nonphotosynthetic tissues and in leaves. Although chloroplast TR-BAMY would be inactivated in the dark, when Trxs are oxidized and leaf starch is usually degraded, TR-BAMY may play a major role in leaf starch degradation under certain stress conditions when TR-BAMY gene expression is strongly up-regulated.
RESULTS
Arabidopsis Gene At3g23920 Encodes a Ubiquitous Plastid-Targeted β-Amylase Regulated by Trxs
In silico analysis of gene At3g23920 belonging to the family of putative β-amylases in Arabidopsis predicted a transit peptide of 41 amino acids for chloroplast targeting (Fig. 1). Both the precursor protein of 575 amino acids and the putative mature protein without transit peptide (534 residues) were obtained by heterologous expression in Escherichia coli. Recombinant proteins were expressed as fusion proteins with a cleavable His tag at the N terminus, allowing effective purification by metal affinity chromatography. The His tag was then cleaved off by thrombin protease and separated from untagged recombinant proteins by a second metal affinity column. Consistent with the amino acid sequence, single bands of 64 kD (precursor) and 60 kD (mature protein) showed up in SDS-PAGE (Fig. 2).
Figure 1.
Multiple alignment between plastidial β-amylase precursors of Arabidopsis CT-BMY (At4g17090), TR-BAMY (At3g23920), and At4g00490. Transit peptides as predicted by ChloroP (Emanuelsson et al., 1999) are in italics on top of the figure. The conserved region corresponding to the typical (α/β)8 barrel of β-amylases is shaded gray. The eight Cys of TR-BAMY, all of which have been individually mutated into Ser, are on a black field. Numeration of amino acids starts from the first predicted residue of mature proteins. The sequence of TR-BAMY precursor and mature TR-BAMY recombinant proteins, after cleavage of the His tag, started with GSHMALN… and GSHMNRN…, respectively (underlined residues derive from the thrombin cleavage site).
Figure 2.
SDS-PAGE of purified recombinant TR-BAMY. Lane A, Mature TR-BAMY (60 kD). Lane B, TR-BAMY precursor that includes the transit peptide of 41 amino acids (64 kD). Both recombinant proteins were purified from transformed E. coli cells as fusion proteins containing an N-terminal His tag, which was removed by thrombin protease before electrophoresis. On the right, molecular mass markers (in kilodaltons).
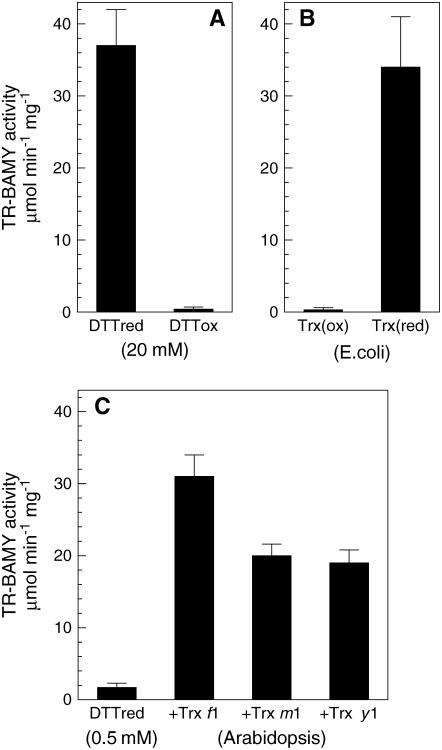
The purified mature protein was little active with β-amylase-specific substrate p-nitrophenyl maltopentaoside (PNPG5) unless reduced dithiothreitol (DTT) was added to the assay mixture, suggesting a redox control of enzyme activity. Indeed, when the mature protein was incubated with either reduced or oxidized DTT (20 mm), only the reduced enzyme was able to hydrolyze PNPG5 (Fig. 3A). To check the reversibility of the effect, both reduced and oxidized enzymes were desalted to eliminate the excess of DTT and incubated with either preoxidized or prereduced Trx from E. coli. Again, only the sample treated with reduced Trx was catalytically active, demonstrating the reversibility of the redox regulation (Fig. 3B).
Figure 3.
Redox regulation of TR-BAMY. A, TR-BAMY was incubated for 1 h with either 20 mm reduced DTT (DTTred) or 20 mm oxidized DTT (DTTox) before assaying the activity with PNPG5. B, After incubation with either DTTred or DTTox, excess DTT was removed by ultrafiltration, and oxidized TR-BAMY was treated with prereduced Trx from E. coli, while reduced TR-BAMY was treated with preoxidized Trx. C, TR-BAMY (oxidized) was incubated for 10 min with 0.5 mm reduced DTT alone or with the addition of 1 μm Trx f1, m1, or y1 from Arabidopsis (Collin et al., 2003, 2004).
The potential physiological meaning of TR-BAMY redox regulation was investigated by testing the role of plastidic Trxs as reductants. A low concentration of reduced DTT (0.5 mm) was unable to activate oxidized TR-BAMY, but a high activation state was attained if Trx f1 from Arabidopsis was present as a redox mediator (Fig. 3C). Albeit less effective than Trx f1, other plastidial Trxs of Arabidopsis (Collin et al., 2003, 2004) such as Trx m1 and Trx y1 were also activating (Fig. 3C).
Mature TR-BAMY (60 kD) was totally unable to hydrolyze α-amylase-specific substrate BPNPG7 (PNPG5 blocked at its nonreducing end), confirming that gene At3g23920 encodes a β-amylase (TR-BAMY). The activity of TR-BAMY was also tested with physiological carbohydrates and found to be similarly active with either soluble (boiled) starch or amylopectin provided that TR-BAMY was in the reduced state. Although soluble starch was a good substrate of TR-BAMY, the enzyme was unable to directly digest insoluble starch granules from potato tubers at any detectable rate (Supplemental Table I), and no activity with any substrate was detected when TR-BAMY was in the oxidized state.
The pH dependency of TR-BAMY activity was influenced by the substrate used. TR-BAMY hydrolytic activity with the artificial substrate PNPG5 was maximal at pH 6.5 to 6.0 and steeply decreased below pH 5.5 and above pH 6.5. However, maximum activity with soluble starch was detected at pH 7.0 with little change between pH 6.0 and 8.0 (Fig. 4), suggesting that TR-BAMY would not be limited in vivo by stromal pH values in the physiological range.
Figure 4.
pH-dependency of TR-BAMY activity. Purified recombinant TR-BAMY was assayed in Britton-Robinson buffer (Hirata et al., 2004) with either PNPG5 (white circles) or soluble starch (black circles). Data points are means ± sd of triplicate determinations.
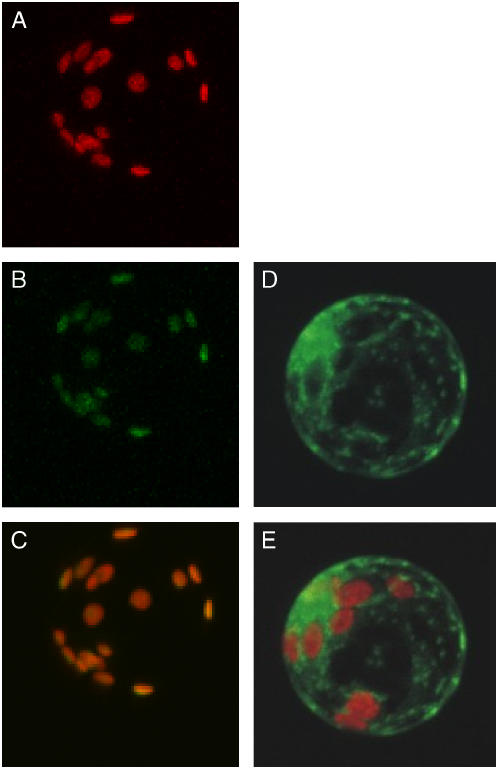
The TR-BAMY precursor (64 kD; Fig. 2) was inactive under any redox condition, indicating that the putative transit peptide fully inhibited enzyme activity. To investigate the localization of TR-BAMY in vivo, a C-terminal fusion between the first 119 residues of the precursor sequence and the green fluorescent protein (GFP) was transiently expressed in mesophyll tobacco (Nicotiana tabacum) protoplasts. Induced GFP fluorescence was exclusively associated with chloroplasts, demonstrating the targeting of TR-BAMY to these organelles (Fig. 5). In control protoplasts transformed with GFP alone the distribution of green fluorescence was clearly extrachloroplastic.
Figure 5.
Localization of GFP in transformed tobacco mesophyll protoplasts. In A, B, and C, protoplasts were transformed with a construct containing the first 119 amino acids of TR-BAMY precursor fused at the N terminus of GFP. A, Chlorophyll fluorescence. B, GFP fluorescence. C, Merge of image A and B. As a control, in D and E, protoplasts were transformed with a construct containing GFP alone. D, GFP fluorescence. E, Merge of chloroplast and GFP fluorescence.
Northern-blot analysis on Arabidopsis leaves and roots (data not shown) confirmed published microarray data (Schmid et al., 2005) showing that gene At3g23920 (TR-BAMY) is similarly expressed in photosynthetic and nonphotosynthetic tissues (Supplemental Fig. 1). It is therefore plausible that in vivo location of TR-BAMY is not restricted to chloroplasts but also includes nonphotosynthetic plastids.
Characterization of TR-BAMY Redox Regulation: Thermodynamics of Disulfide Formation and Identification of Regulatory Cys
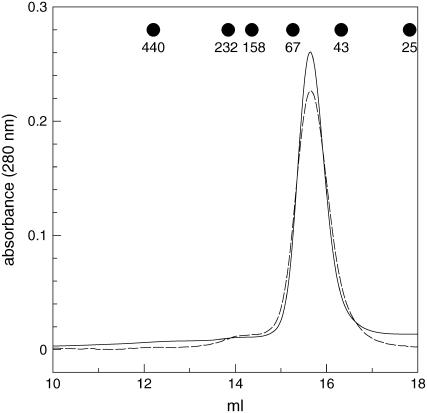
The sequence of TR-BAMY includes eight Cys (Fig. 1). Regulatory disulfides could either involve two Cys of the same subunit or two Cys belonging to different subunits. In the latter case, the polymerization state of TR-BAMY might be affected. The estimated molecular mass of TR-BAMY, as analyzed by gel filtration chromatography, was 60 kD under both reduced and oxidized conditions (Fig. 6). Hence, TR-BAMY was invariably monomeric and redox regulation depended on intramolecular disulfides.
Figure 6.
Size-exclusion chromatography on Superdex 200 of reduced versus oxidized TR-BAMY. Pure TR-BAMY was incubated for 1 h with 20 mm reduced (solid line) or oxidized DTT (broken line) prior to loading on the column. The column was equilibrated with 50 mm Tris-HCl, pH 7.5, 1 mm EDTA, and 150 mm KCl. The column was calibrated as described in Sparla et al. (2002), and elution volumes of standard proteins are indicated. The apparent molecular mass of either reduced or oxidized TR-BAMY was 60 kD.
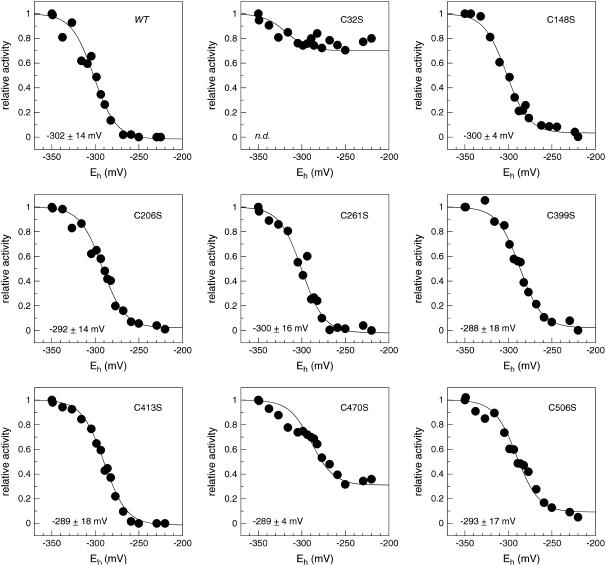
Thermodynamics of TR-BAMY redox regulation were investigated by redox titration analysis (Fig. 7). Enzyme activity with PNPG5 was assayed after 30 min of equilibration at 37°C with mixtures of reduced and oxidized DTT at defined molar ratios and total concentration of 20 mm. At pH 7.0, TR-BAMY shifted between an active form at −350 mV and an inactive form at −270 mV, and activity data nicely fit a Nernst equation for a two-electron oxidoreduction of a single disulfide with a midpoint redox potential (Em,7.0) of −302 ± 14 mV. Similar experiments performed at different pH gave Em values of −332 mV at pH 7.5 and −350 mV at pH 7.9 (data not shown). These values are in good agreement with the theoretical shift of −59 mV/(pH unit) expected for a redox reaction involving one proton for each electron exchanged, as for dithiol-disulfide equilibria (Hirasawa et al., 1999).
Figure 7.
Redox titrations at pH 7.0 of wild-type TR-BAMY and eight site-specific mutants of TR-BAMY. Recombinant proteins were incubated in Tricine-NaOH, pH 7.0, with 20 mm DTT in different dithiol/disulfide ratios. Samples were incubated for 30 min at 37°C before assaying the activity. Experimental data were fitted to the Nernst equation with n = 2 (CoHort software). Midpoint redox potentials (Em,7.0, means ± sd of two experiments for each protein) are indicated in the figure.
To identify which Cys were involved in the formation of the regulatory disulfide, eight site-specific mutants were analyzed, one for each Cys present in the sequence. Each mutant carried a single Cys to Ser substitution, and all of them were expressed in E. coli and purified to homogeneity before characterization. Mutants were all active under reducing conditions. Redox titrations of six out of eight mutants gave similar responses to wild-type TR-BAMY (C148S, C206S, C261C, C399S, C413S, and C506S; numeration starting from the first residue of the mature protein; Fig. 1). The Em,7.0 was between −288 and −300 mV for all these mutants and 95% to 100% inhibition was always achieved under oxidizing conditions (Fig. 7). At variance from all these TR-BAMY mutants, redox sensitivity of mutant C32S was very poor (the activity of the oxidized form was only 30% inhibited in respect to the reduced form) and redox sensitivity of mutant C470S was also impaired (70% inhibition). The relative insensitivity of both mutants to the redox poise suggests that Cys-32 and Cys-470 may form, in the wild-type enzyme, an inhibitory disulfide under oxidizing conditions.
Redox-Sensitive β-Amylase Activity in Arabidopsis Chloroplasts
TR-BAMY is the first β-amylase reported to be redox regulated. Because Arabidopsis chloroplasts contain other β-amylase isozymes, namely CT-BMY (Lao et al., 1999) and possibly At4g00490 as predicted by ChloroP (Emanuelsson et al., 1999) and PSORT (Nakai and Horton, 1999), the question arose as to whether redox-sensitive β-amylase activity could be detected in vivo. Indeed, under oxidizing conditions the β-amylase activity measured in Arabidopsis chloroplasts amounted to 55% ± 1% of the activity measured under reducing conditions (with PNPG5), confirming the existence of a measurable redox-sensitive β-amylase activity in Arabidopsis chloroplasts.
DISCUSSION
β-Amylases hydrolyze α-1,4 glucosidic bonds releasing β-anomeric maltose units from the nonreducing end of polyglucans like amylopectin. The Arabidopsis genome contains nine genes annotated as β-amylases (Lloyd et al., 2005; Smith et al., 2005). Among these, CT-BMY (At4g17090) was the first to be experimentally shown to be a chloroplast β-amylase (Lao et al., 1999). A potato ortholog of CT-BMY was also shown to be imported by chloroplasts and was found involved in leaf starch degradation (Scheidig et al., 2002). Here, we demonstrate that TR-BAMY, encoded by gene At3g23920, is a functional β-amylase also targeted to chloroplasts by means of its N-terminal transit peptide of 41 amino acids. TR-BAMY displays the typical activity of plant β-amylases that can release maltose units from the artificial substrate PNPG5 (a maltopentaoside derivative) as well as physiological polyglucans like amylopectin and soluble starch. A third member of the β-amylase family in Arabidopsis (At4g00490) is predicted to contain a chloroplast transit peptide by both ChloroP (Emanuelsson et al., 1999) and PSORT (Nakai and Horton, 1999). The remaining six members of the family are not predicted to contain a plastid transit peptide and their precise location and role remain obscure (Laby et al., 2001; Zeeman et al., 2004).
Strong pH dependency and acidic pH optimum are common features among β-amylases (Hirata et al., 2004). Indeed, TR-BAMY, when assayed with the artificial substrate PNPG5, showed a narrow pH-activity curve with a definite maximum at pH 6.0 to 6.5 and dramatically reduced activity above neutrality. However, the TR-BAMY was found to be active over a wider range of pH values with soluble starch as a substrate, >90% of maximal activity being maintained between pH 6.0 and 8.0. It seems, therefore, that TR-BAMY activity would not be limited in vivo by the pH status of the stroma, either under photosynthetic conditions (pH 7.5–8.0) or in the dark (pH approximately 7; Kramer et al., 1999).
The activity of TR-BAMY is strongly regulated by the redox potential. Under oxidizing conditions, TR-BAMY activity was negligible as compared to the fully reduced enzyme. Midpoint redox potential of TR-BAMY activity was −302 mV at pH 7.0 (−350 mV at pH 7.9), similar to Calvin cycle enzymes such as glyceraldehyde-3-P dehydrogenase (Sparla et al., 2002), Fru bisP phosphatase, or sedoheptulose bisP phosphatase (Hirasawa et al., 1999; Hutchinson et al., 2000). However, most Calvin cycle enzymes are specifically activated by Trx f (Schürmann and Jacquot, 2000), whereas TR-BAMY was also sensitive to Trx m1 and y1, similar to NADP-malate dehydrogenase in this property (Collin et al., 2003, 2004).
Among the eight Cys of TR-BAMY, only Cys-32 and Cys-470 are eligible to form a regulatory disulfide. However, while mutant C32S was basically redox insensitive, the redox regulation of mutant C470S was only partially abolished, suggesting that Cys-32 was able to form an alternative disulfide with another Cys partner. A similar situation with more than two Cys involved in redox regulation was previously observed in Fru bisP phosphatase (Jacquot et al., 1997; Rodriguez-Suarez et al., 1997). Multiple alignments of plant and bacterial β-amylases show a highly conserved, central region of about 430 amino acids (Fig. 1), folding into a single (α/β)8 barrel domain according to structural studies (Mikami et al., 1993). Differences among isozymes principally reside in N- and C-terminal extensions that are variable in sequence and length. TR-BAMY mature protein (without transit peptide) consists of 533 amino acids with a typical β-amylase conserved domain between residues 64 and 496. Redox-sensitive Cys-32 is thus included in the nonconserved N-terminal extension, while Cys-470 resides in the conserved region. Neither Cys-32 nor Cys-470 is individually conserved in any other β-amylase of Arabidopsis, suggesting that TR-BAMY may be the only redox-sensitive isoform of the family. Interestingly, TR-BAMY orthologs do not appear to be universal among higher plants and are noticeably absent in cereals (The Institute for Genomic Research [TIGR] databases, www.tigr.org), while present in dicotyledons: Brassica napus (GenBank AAL37169), tomato (Lycopersicon esculentum; TIGR TC163705), and potato (TIGR TC118938).
Published microarray data provide an indication of the relative abundance of three plastid β-amylases (CT-BMY, TR-BAMY, and At4g00490) in different tissues of Arabidopsis (Schmid et al., 2005; Supplemental Fig. 1). Within this group, At4g00490 is the least expressed gene with no peaks of expression in any tissue. CT-BMY is much more expressed than TR-BAMY in leaves, but the latter isozyme is strongly induced during senescence. In flowers and seeds, CT-BMY and At4g00490 transcripts decrease during development to eventually become undetectable in differentiated seeds, mature pollen, and roots. TR-BAMY shows an opposite trend during flower and seed development, constituting the only plastid β-amylase occurring in mature pollen, differentiated seeds, and roots (Supplemental Fig. 1).
Although little is known about starch degradation in nonphotosynthetic tissues of Arabidopsis, high levels of starch in nonphotosynthetic tissues of sex mutants (Caspar et al., 1991) do suggest a pathway of starch degradation similar to that of leaves (Smith et al., 2005). TR-BAMY is apparently the only β-amylase present in amyloplasts and might be involved in this process, possibly acting in concert with GWD, which is also activated by reduced Trxs and ubiquitously expressed (Mikkelsen et al., 2005). TR-BAMY activation in amyloplasts could involve Trx y1, a Trx isoform of nonphotosynthetic plastids, which can reduce TR-BAMY (Fig. 3) and is expressed in different Arabidopsis tissues similarly to TR-BAMY (Collin et al., 2004). The redox state of Trx y1 in amyloplasts might be modulated by a recently described ferredoxin/Trx system linked to NADPH as an electron source (Balmer et al., 2006).
However, redox regulation of starch degradation in amyloplasts is complicated by the concomitant regulation of starch biosynthesis via reductive activation of ADP-Glc pyrophosphorylase (Ballicora et al., 2000; Tiessen et al., 2002). These apparently contrasting results may be perhaps reconciled if TR-BAMY played a role in net starch biosynthesis by degrading water soluble polyglucans. This activity would prevent the accumulation of phytoglycogen that competes with the synthesis of semicrystalline starch granules, as proposed for other catabolic enzymes like debranching enzymes and d-enzyme (Ball and Morell, 2003). Interestingly, limit dextrinase, a particular debranching enzyme, was also reported to be redox regulated in both photosynthetic and nonphotosynthetic tissues (Ball and Morell, 2003). This assumption might support a role of TR-BAMY in net starch biosynthesis also in chloroplasts in the light, when Trxs are photosynthetically reduced.
Starch degradation in leaves normally occurs during the night (Zeeman et al., 2002), and the role of CT-BMY in this process has been established (Scheidig et al., 2002). Due to its strict redox dependency, TR-BAMY would not contribute to starch degradation in the dark, when chloroplast Trxs are oxidized (Buchanan and Balmer, 2005). Consistently, TR-BAMY knockout Arabidopsis plants do not accumulate starch in leaves under normal conditions or cold stress (Kaplan and Guy, 2005).
Activity and expression of β-amylases are known to be affected by abiotic stresses (Dreier et al., 1995; Nielsen et al., 1997; Datta et al., 1999; Kaplan and Guy, 2004), and published microarray data show that the high CT-BMY/TR-BAMY transcript ratio in normal leaves can even be reversed under stress (Zimmermann et al., 2004). In particular, osmotic and salt stress, potassium deficiency, and hydrogen peroxide treatments (Davletova et al., 2005; Supplemental Figs. 2 and 3) cause strong up-regulation of TR-BAMY and symmetrical down-regulation of CT-BMY in Arabidopsis leaves. Similarly, up-regulation of TR-BAMY and down-regulation of CT-BMY were observed in plants treated with abscisic acid, senescing cell cultures, and plants inoculated with Pseudomonas syringae (Supplemental Figs. 3–5). TR-BAMY was also up-regulated by heat shock in Arabidopsis leaves, while CT-BMY expression is specifically induced by cold stress (Kaplan and Guy, 2004). It is therefore apparent that in some particular stress conditions or senescence, the role of CT-BMY in chloroplasts is replaced by TR-BAMY, its redox-regulated counterpart. Interestingly, some elements of the Trx system in chloroplasts are also induced by stress, including a NADPH:Trx reductase involved in abiotic stress responses (Serrato et al., 2004) and Trx CDSP32 induced by drought stress (Rey et al., 2005).
In general, stimulation of starch degradation in leaves under stress is necessary both to directly support respiration under conditions of low photosynthesis and to protect cell structures by raising the levels of maltose as an osmoprotectant (Kaplan and Guy, 2004; Rizhsky et al., 2004). In fact, maltose accumulates in Arabidopsis leaves subjected to either cold stress (via induction of CT-BMY) or heat shock (via induction of TR-BAMY; Kaplan and Guy, 2004). The rise of TR-BAMY in leaves under defined stress conditions suggests its involvement in leaf starch degradation in the light under such conditions. Although oxidative stress is often considered as a general component of biotic and abiotic stresses, reducing conditions may prevail in cells responding to certain stresses. For instance, photosynthetic electron transport and NADPH levels in leaves are elevated under water stress (Lawlor, 2002), and expression of some pathogenesis-related genes requires a reducing cellular status to activate redox-sensitive factors (like NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 and TGA1; Fobert and Despres, 2005).
Light degradation of leaf starch has been firmly established in Arabidopsis mutants lacking the triose-phosphate/phosphate translocator (Walters et al., 2004). Under high-light regime, these mutants synthesize and degrade starch simultaneously, thereby providing a continued supply of substrate (maltose) for Suc synthesis in the cytosol. Interestingly, Trx-regulated malate dehydrogenase is fully activated, and CT-BMY transcripts decrease under these conditions, whereas expression levels of other chloroplast β-amylases (TR-BAMY and At4g00490) are kept constant (Walters et al., 2004). As speculated by the authors, redox and metabolic signals (possibly related to low Suc levels in the cytosol) might be involved in switching on starch degradation in the light (Walters et al., 2004). There is growing evidence that metabolic signals derived from Suc and Glc have a regulatory effect on starch metabolism (Tiessen et al., 2002), but it is still unclear how cytosolic sugar levels might influence the redox state in the plastid. It is therefore puzzling how the reductive activation of starch-degrading and starch synthesis enzymes might be physiologically coordinated by the Trx system (Ball and Morell, 2003; Smith et al., 2005; Balmer et al., 2006). Recently, it has been shown that trehalose-6-P could play a role in such metabolic crosstalk between cytosol and plastid (Kolbe et al., 2005).
In conclusion, we have described a novel β-amylase of Arabidopsis plastids, the activity of which is tightly regulated by the redox potential. TR-BAMY is probably the only β-amylase of nonphotosynthetic plastids in Arabidopsis, where it may play a role in starch degradation under the control of Trx y1 (Collin et al., 2004) or, alternatively, participate to net starch biosynthesis by degrading water soluble polyglucans. In Arabidopsis leaves, TR-BAMY gene expression is strongly induced by certain abiotic stresses. Conversely, these same conditions inhibit the expression of the CT-BMY, which is involved in physiological degradation of leaf starch in the dark (Scheidig et al., 2002). Chloroplast Trx CDSP32 (Rey et al., 2005) and NADPH:Trx reductase (Serrato et al., 2004) are also involved in abiotic stress responses. Interestingly, the reductive activation of TR-BAMY has much in common with the recently described redox regulation of GWD (Mikkelsen et al., 2005), also involved in leaf starch degradation. Overall, these findings concur to suggest the existence of a redox-regulated, stress-induced pathway of starch degradation in leaves.
MATERIALS AND METHODS
Identification and in Silico Analysis of Arabidopsis β-Amylases
The amino acid sequence of CT-BMY (NP567523) has been used as a query in a BLASTP search using the Arabidopsis Genome Initiative proteins database (www.arabidopsis.org). Nine putative Arabidopsis (Arabidopsis thaliana) proteins with E values lower than 2 × 10−87 were identified (The Arabidopsis Information Resource codes: At4g17090 [CT-BMY], At3g23920, At2g32290, At4g15210, At4g00490, At5g55700, At2g45880, At5g45300, and At5g18670, in order of similarity; see also Lloyd et al., 2005; Smith et al., 2005).
To predict the presence of a putative transit peptide for localization into organelles, amino acid sequences were analyzed by ChloroP (Emanuelsson et al., 1999) and PSORT (Nakai and Horton, 1999). Besides At4g17090 (CT-BMY; Lao et al., 1999), only At3g23920 and At4g00490 were predicted to be chloroplast targeted by both programs. In this work, we studied the putative CT-BMY At3g23920 (TR-BAMY).
Northern-blot analysis to check the expression of At3g23920 in both roots and leaves of Arabidopsis was performed as in Marri et al. (2005a).
Cloning, Expression, and Purification of Recombinant β-Amylase TR-BAMY
The cDNAs coding for β-amylase TR-BAMY, with or without the predicted N-terminal transit peptide of 41 amino acids (precursor and mature protein, respectively), were amplified by PCR using primers containing NdeI and SalI restriction sites (forward primer for mature TR-BAMY: 5′-CACCTAAACATATGAATCG-3′; forward primer for entire TR-BAMY: 5′-CATATGGCGCTTAATTTATCGC-3′; reverse primer for both recombinant proteins: 5′-ACCAGTCGACAATCTATC-3′; restriction sites are underlined). The plasmid template for PCR reactions was extracted from clone U18509, kindly provided by The Arabidopsis Biological Research Center at the Ohio State University. PCR cycles consisted of denaturation at 93°C for 2 min, annealing at 52°C for 1 min, and extension at 72°C for 2 min. After 30 cycles, products were checked on 0.8% (w/v) agarose gel. Amplified fragments were cut with NdeI and SalI and inserted into NdeI-SalI predigested expression vector pET28a(+) (Novagen). Both cDNAs were in frame with a His tag and a cleavable thrombin site at the 5′. The recombinant plasmids were amplified into Escherichia coli HB101 cells and sequenced before the transfer into BL21(DE3) cells for expression. The in silico translated amino acid sequence of both recombinant proteins was identical to the sequence of At3g23920.1 in the Arabidopsis Genome Initiative proteins database (except for the N-terminal His tag and thrombin cleavage site). Heterologous expression, purification of recombinant proteins, and removal of the His tag were performed as in Sparla et al. (1999). The purification grade of recombinant proteins was checked by SDS-PAGE and Coomassie staining.
Site-Specific Mutants
Site-specific mutagenesis was performed as previously described (Sparla et al., 2002). The following pairs of primers were used for each mutant: C32S: 5′-ctgtcggtggcgtctaaagctttcgcggtgg-3′, 5′-ccaccgcgaaagctttagacgccaccgacag-3′; C148S: 5′-cgtttcatcagagtggtggcaacgttggtgac-3′, 5′-tcaccaacgttgccaccactctgatgaaacg-3′; C206S: 5′-gaacacctgtgcaatcctatgcggatttcatgcg-3′, 5′-gcatgaaatccgcataggattgcacaggtgttc-3′; C261S: 5′-ggattggagccttccagtcctatgataagtactcg-3′, 5′-cgagtacttatcataggactggaaggctccaatcc-3′; C399S: 5′-gccatattcaacttcacaagtatcgagatgaggga-3′, 5′-tccctcatctcgatacttgtgaagttgaatatggc-3′; C413S: 5′-cctcaagacgcacttagtgcaccagagaagc-3′, 5′-gcttctctggtgcactaagtgcgtcttgagg-3′; C470S: 5′-gggagaacctcgagagatgtctgcatttacttacc-3′, 5′-ggtaagtaaatgcagacatctctcgaggttctccc-3′; and C506S: 5′-gagagactcacatagatctcgggaagaagttgagc-3′, 5′-gctcaacttcttcccgagatctatgtgagtctctc-3′.
Enzyme Activity Assays
Activities of α- and β-amylase were measured at 37°C using assay kits from Megazyme (Ceralpha Method and Betamyl Method for α- and β-amylase activity, respectively) according to the manufacturer's instructions. The activity of TR-BAMY with potato (Solanum tuberosum) starch granules (Sigma S-4251), soluble (boiled) starch (Sigma S-4251), and amylopectin (Sigma A-8515) was performed as described in Wang et al. (1995). If not otherwise stated, all assays were performed at pH 6.0. The pH dependency of purified TR-BAMY was determined in Britton-Robinson buffer (Hirata et al., 2004).
Redox modulation of TR-BAMY activity was assayed after 1 h incubation at 37°C of pure recombinant enzyme with 20 mm reduced DTT or 20 mm oxidized DTT (trans-4,5-dihydroxy-1,2-dithiane; Sigma D3511). Reversibility of redox modulation was tested as follows: Samples of oxidized and reduced TR-BAMY, obtained by incubation with DTT as above, were desalted by ultrafiltration (Centricon YM10) to remove DTT in excess, then incubated with reduced or oxidized E. coli Trx (Sigma), respectively, at Trx to TR-BAMY molar ratio of 3:1. Reduced and oxidized Trx were prepared as in Marri et al. (2005b). After 1 h incubation at 37°C with Trx, TR-BAMY activity was measured with PNPG5.
Trxs f1, m1, and y1 were kindly provided by Emmanuelle Issakidis-Bourguet (Collin et al., 2003, 2004). To test the specificity of TR-BAMY activation by Trxs, 0.2 μg of TR-BAMY was incubated for 10 min at 37°C with 0.5 mm reduced DTT with or without 1 μm Trxs, before measuring the activity with PNPG5 at pH 7.9.
Redox Titrations
For redox titrations at pH 7.9, 7.5, and 7.0, purified TR-BAMY was dissolved in 80 μL of 100 mm Tricine-NaOH at appropriate pH in the presence of 20 mm DTT in different dithiol/disulfide ratios. Samples were incubated for 30 min at 37°C before assaying the activity with PNPG5. Control experiments were performed to check that after 30 min incubation the activity of TR-BAMY was constant over time, indicating equilibration between protein and redox buffer.
Experimental data were fitted to the Nernst equation with n = 2 (CoHort software). Midpoint redox potentials of DTT at different pH values were calculated as in Hirasawa et al. (1999) based on a value of −330 mV for the Em of DTT at pH 7.0 (Hutchinson and Ort, 1995).
Size-Exclusion Chromatography
Native Mr of recombinant TR-BAMY was estimated by size-exclusion chromatography on a Superdex 200 HR10/30 column, connected to a SMART system (Amersham Biosciences). The column was equilibrated with 50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 150 mm KCl, and calibrated as described in Sparla et al. (2002). To determine the polymerization state under different redox conditions, TR-BAMY was incubated for 1 h with 20 mm reduced or oxidized DTT prior to loading on the column.
Subcellular Localization of TR-BAMY in Tobacco Protoplasts
The nucleotide sequence corresponding to the first 119 residues at the N terminus of TR-BAMY precursor was amplified by PCR using specific primers (5′-AGATCTATGGCGCTTAATTTATCG-3′ and 5′-CATGGTACCACTATCAAGC-3′) to generate BglII site and KpnI site at the 5′ and 3′ end, respectively (restriction sites are underlined). The resulting fragment was digested and fused in frame to the 5′ end of a GFP-encoded gene, inserted in a plant transient expression vector derived from pTZ-19U (Stratagene) and kindly provided by Geoffrey Duby (Duby et al., 2001). The resulting construct was sequenced before further use. Expression of the fusion protein was under the control of plant-enhanced transcription promoter of the plasma membrane H+-ATPase isoform 4 (Zhao et al., 1999) and the nopaline synthase terminator.
In vitro sterile shoot culture of tobacco (Nicotiana tabacum) cv SR1 was maintained on one-half Murashige and Skoog agar medium (Murashige and Skoog, 1962) containing 15 g/L Suc and grown at 25°C, with a light/dark cycle of 16/8 h in a growth chamber. Mesophyll protoplasts were isolated and transformed as described in Nagy and Maliga (1976) and Locatelli et al. (2003), respectively. Polyethylene-glycol-mediated transformation was obtained by adding 15 μg of plasmid DNA to 300,000 protoplasts. Transformation solution was gently mixed and dark incubated for 15 to 20 min at room temperature. After incubation, the polyethylene-glycol solution was washed out with 5 mL of W5 buffer (154 mm NaCl, 5 mm KCl, 125 mm CaCl2, and 5 mm Glc, pH 5.8). Protoplasts were collected by centrifugation at 70g for 5 min and suspended in 2 mL of K3 solution (0.4 m Suc; 1.67 mm Xyl; 5 mm CaCl2; 3 mm NH4NO3; 1× Gamborg B5 salts; 1× Kao and Michayluk organic acids; 5 μm α-naphthalene-acetic acid; and 4.4 μm 6-benzyl-aminopurine, pH 5.8). Before microscopy analysis, protoplasts were incubated at 20°C in the dark for at least 16 h.
Subcellular localization of the GFP fusion construct was performed using a Nikon PCM2000 (Bio-Rad) laser scanning confocal imaging system. For GFP detection, excitation was at 488 nm and detection between 515 and 530 nm. For chlorophyll detection, excitation was at 488 nm and detection over 570 nm. The images were processed using the software Corel Photo-Paint (Corel).
Chloroplast Purification
Arabidopsis plants were grown on sterile soil for 1 month at 22°C under a 15-h dark/9-h light cycle in a growth chamber. Chloroplast isolation from 15-h dark-adapted plants was performed as described (Marri et al., 2005b). To measure total β-amylase activity under reducing or oxidizing conditions, chloroplasts were made permeable by resuspension in hypoosmotic buffer (50 mm MES, pH 6.0) supplemented with 20 mm reduced or oxidized DTT and incubated for 1 h before measuring the activity.
Supplementary Material
Acknowledgments
We thank Emmanuelle Issakidis-Bourguet and Valérie Collin for the generous gift of Arabidopsis Trxs f1, m1, and y1.
This work was supported by the Ministero dell'Istruzione, dell'Università e della Ricerca (PRIN 2003 and 2005).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Paolo Trost (trost@alma.unibo.it).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.079186.
References
- Ball SG, Morell MK (2003) From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu Rev Plant Biol 54: 207–233 [DOI] [PubMed] [Google Scholar]
- Ballicora MA, Frueauf JB, Fu Y, Schurmann P, Preiss J (2000) Activation of the potato tuber ADP-glucose pyrophosphorylase by thioredoxin. J Biol Chem 275: 1315–1320 [DOI] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Cai N, Manieri W, Schurmann P, Hurkman WJ, Buchanan BB (2006) A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proc Natl Acad Sci USA 103: 2988–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baunsgaard L, Lutken H, Mikkelsen R, Glaring MA, Pham TT, Blennow A (2005) A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated alpha-glucans and is involved in starch degradation in Arabidopsis. Plant J 41: 595–605 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56: 187–220 [DOI] [PubMed] [Google Scholar]
- Caspar T, Lin TP, Kakefuda G, Benbow L, Preiss J, Somerville C (1991) Mutants of Arabidopsis with altered regulation of starch degradation. Plant Physiol 95: 1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin V, Issakidis-Bourguet E, Marchand C, Hirasawa M, Lancelin J-M, Knaff DB, Miginiac-Maslow M (2003) The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. J Biol Chem 278: 23747–23752 [DOI] [PubMed] [Google Scholar]
- Collin V, Lamkemeyer P, Miginiac-Maslow M, Hirasawa M, Knaff DB, Dietz KJ, Issakidis-Bourguet E (2004) Characterization of plastidial thioredoxins from Arabidopsis belonging to the new y-type. Plant Physiol 136: 4088–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley JH, Zeeman SC, Takaha T, Smith AM, Smith SM (2001) A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutant in Arabidopsis. Plant J 26: 89–100 [DOI] [PubMed] [Google Scholar]
- Datta R, Selvi MT, Seetharama N, Sharma R (1999) Stress-mediated enhancement of beta-amylase activity in pearl millet and maize leaves is dependent on light. J Plant Physiol 154: 657–664 [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreier W, Schnarrenberger C, Börner T (1995) Light- and stress-dependent enhancement of amylolytic activities in white and green barley leaves: beta-amylases are stress-induced proteins. J Plant Physiol 145: 342–348 [Google Scholar]
- Duby G, Oufattole M, Boutry M (2001) Hydrophobic residues within the predicted N-terminal amphiphilic alpha-helix of a plant mitochondrial targeting presequence play a major role in in vivo import. Plant J 27: 539–549 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fobert PR, Despres C (2005) Redox control of systemic acquired resistance. Curr Opin Plant Biol 8: 378–382 [DOI] [PubMed] [Google Scholar]
- Hirasawa M, Schürmann P, Jacquot JP, Manieri W, Jacquot P, Keryer E, Hartman FC, Knaff DB (1999) Oxidation-reduction properties of chloroplast thioredoxins, ferredoxin: thioredoxin reductase and thioredoxin f-regulated enzymes. Biochemistry 38: 5200–5205 [DOI] [PubMed] [Google Scholar]
- Hirata A, Adachi M, Sekine A, Kang YN, Utsumi S, Mikami B (2004) Structural and enzymatic analysis of soybean β-amylase mutants with increased pH optimum. J Biol Chem 279: 7287–7295 [DOI] [PubMed] [Google Scholar]
- Hutchinson RS, Groom Q, Ort DR (2000) Differential effects of chilling-induced photooxidation on the redox regulation of photosynthetic enzymes. Biochemistry 39: 6679–6688 [DOI] [PubMed] [Google Scholar]
- Hutchinson RS, Ort DR (1995) Measurement of equilibrium midpoint potentials of thiol/disulfide regulatory groups on thioredoxin-activated chloroplast enzymes. Methods Enzymol 252: 220–228 [DOI] [PubMed] [Google Scholar]
- Jacquot JP, Lopez-Jaramillo J, Miginiac-Maslow M, Lemaire S, Cherfils J, Chueca A, Lopez-Gorge J (1997) Cysteine-153 is required for redox regulation of pea chloroplast fructose-1,6-bisphosphatase. FEBS Lett 401: 143–147 [DOI] [PubMed] [Google Scholar]
- Kaplan F, Guy CL (2004) β-Amylase induction and the protective role of maltose during temperature shock. Plant Physiol 135: 1674–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Guy CL (2005) RNA interference of Arabidopsis beta-amylase8 prevents maltose accumulation upon cold shock and increases sensitivity of PSII photochemical efficiency to freezing stress. Plant J 44: 730–743 [DOI] [PubMed] [Google Scholar]
- Kolbe A, Tiessen A, Schluepmann H, Paul M, Ulrich S, Geigenberger P (2005) Trehalose 6-phosphate regulates starch synthesis via posttranslational redox activation of ADP-glucose pyrophosphorylase. Proc Natl Acad Sci USA 102: 11118–11123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kötting O, Pusch K, Tiessen A, Geigenberger P, Steup M, Ritte G (2005) Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves: the phosphoglucan, water dikinase. Plant Physiol 137: 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer DM, Sacksteder CA, Cruz JA (1999) How acidic is the lumen? Photosynth Res 60: 151–163 [Google Scholar]
- Laby RJ, Kim D, Gibson SI (2001) The ram1 mutant of Arabidopsis exhibits severely decreased β-amylase activity. Plant Physiol 127: 1798–1807 [PMC free article] [PubMed] [Google Scholar]
- Lao NT, Schoneveld O, Mould RM, Hibberd JM, Gray JC, Kavanagh TA (1999) An Arabidopsis gene encoding a chloroplast targeted β-amylase. Plant J 20: 519–527 [DOI] [PubMed] [Google Scholar]
- Lawlor DW (2002) Limitation to photosynthesis in water-stressed leaves: stomata vs. metabolism and the role of ATP. Ann Bot (Lond) 89: 871–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JR, Lossmann J, Ritte G (2005) Leaf starch degradation comes out of the shadows. Trends Plant Sci 10: 130–137 [DOI] [PubMed] [Google Scholar]
- Locatelli F, Vannini C, Magnani E, Coraggio I, Bracale M (2003) Efficiency of transient transformation in tobacco protoplasts is independent of plasmid amount. Plant Cell Rep 21: 865–871 [DOI] [PubMed] [Google Scholar]
- Lu Y, Sharkey T (2004) The role of amylomaltase in maltose metabolism in the cytosol of photosynthetic cells. Planta 218: 466–473 [DOI] [PubMed] [Google Scholar]
- Marri L, Sparla F, Pupillo P, Trost P (2005. a) Coordinated gene expression of photosynthetic glyceraldehyde-3-phosphate dehydrogenase, phosphoribulokinase and CP12 in Arabidopsis thaliana. J Exp Bot 56: 73–80 [DOI] [PubMed] [Google Scholar]
- Marri L, Trost P, Pupillo P, Sparla F (2005. b) Reconstitution and properties of the recombinant GAPDH/CP12/PRK supramolecular complex of Arabidopsis thaliana. Plant Physiol 139: 1433–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami B, Hehre EJ, Sato M, Katsube Y, Hirose M, Morita Y, Sacchettini JC (1993) The 2.0-A resolution structure of soybean beta-amylase complexed with alpha-cyclodextrin. Biochemistry 32: 6836–6845 [DOI] [PubMed] [Google Scholar]
- Mikkelsen R, Mutenda KE, Mant A, Schurmann P, Blennow A (2005) Alpha-glucan, water dikinase (GWD): a plastidic enzyme with redox-regulated and coordinated catalytic activity and binding affinity. Proc Natl Acad Sci USA 102: 1785–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays in tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nagy JI, Maliga P (1976) Callus induction and plant regeneration from mesophyll protoplasts of Nicotiana sylvestrys. Z Pflanzenphysiol 78: 453–544 [Google Scholar]
- Nakai K, Horton P (1999) PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem Sci 24: 34–35 [DOI] [PubMed] [Google Scholar]
- Nielsen TH, Wischmann B, Enevoldsen K, Moller BL (1994) Starch phosphorylation in potato tubers proceeds concurrently with de novo biosynthesis of starch. Plant Physiol 105: 111–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TM, Deiting U, Stitt M (1997) A beta-amylase in potato tubers is induced by storage at low temperature. Plant Physiol 113: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niittylä T, Messerli G, Trevisan M, Chen J, Smith AM, Zeeman SC (2004) A previously unknown maltose transporter essential for starch degradation in leaves. Science 303: 87–89 [DOI] [PubMed] [Google Scholar]
- Rey P, Cuine S, Eymery F, Garin J, Court M, Jacquot JP, Rouhier N, Broin M (2005) Analysis of the proteins targeted by CDSP32, a plastidic thioredoxin participating in oxidative stress responses. Plant J 41: 31–42 [DOI] [PubMed] [Google Scholar]
- Ritte G, Lloyd JR, Eckermann N, Rottmann A, Kossmann J, Steup M (2002) The starch-related protein R1 is an α-glucan, water dikinase. Proc Natl Acad Sci USA 99: 7166–7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritte G, Scharf A, Eckermann N, Haebel S, Steup M (2004) Phosphorylation of transitory starch is increased during degradation. Plant Physiol 135: 2068–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R (2004) When defense pathways collide: the response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Suarez RJ, Mora-Garcia S, Wolosiuk RA (1997) Characterization of cysteine residues involvement in the reductive activation and the structural stability of rapeseed (Brassica napus) chloroplast fructose-1,6-bisphosphatase. Biochem Biophys Res Commun 232: 388–393 [DOI] [PubMed] [Google Scholar]
- Scheidig A, Fröhlich A, Schulze S, Lloyd JR, Kossmann J (2002) Downregulation of chloroplast-targeted β-amylase leads to a starch excess phenotype in leaves. Plant J 30: 581–591 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann J (2005) A gene expression map of Arabidopsis development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schürmann P, Jacquot J-P (2000) Plant thioredoxin systems revisited. Annu Rev Plant Physiol Plant Mol Biol 51: 371–400 [DOI] [PubMed] [Google Scholar]
- Serrato AJ, Perez-Ruiz JM, Spinola MC, Cejudo FJ (2004) A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J Biol Chem 15: 43821–43827 [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC, Smith SM (2005) Starch degradation. Annu Rev Plant Biol 56: 73–97 [DOI] [PubMed] [Google Scholar]
- Sparla F, Pupillo P, Trost P (2002) The C-terminal extension of glyceraldehyde-3-phosphate dehydrogenase subunit B acts as an autoinhibitory domain regulated by thioredoxins and nicotinamide adenine dinucleotide. J Biol Chem 277: 44946–44952 [DOI] [PubMed] [Google Scholar]
- Sparla F, Tedeschi G, Pupillo P, Trost P (1999) Cloning and heterologous expression of NAD(P)H:quinone reductase of Arabidopsis thaliana, a functional homologue of animal DT-diaphorase. FEBS Lett 463: 382–386 [DOI] [PubMed] [Google Scholar]
- Tiessen A, Hendriks JHM, Stitt M, Branscheid A, Gibon Y, Farré EM, Geigenberger P (2002) Starch Synthesis in potato tubers is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase: a novel regulatory mechanism linking starch synthesis to the sucrose supply. Plant Cell 14: 2191–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RG, Ibrahim DG, Horton P, Kruger NJ (2004) A mutant of Arabidopsis lacking the triose-phosphate/phosphate translocator reveals metabolic regulation of starch breakdown in the light. Plant Physiol 135: 891–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Monroe J, Sjölund RD (1995) Identification and characterization of a phloem-specific β-amylase. Plant Physiol 109: 743–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Servaites JC, Geiger DR, Kofler H, Hille D, Groner F, Hebbeker U, Flugge UI (2000) Identification, purification, and molecular cloning of a putative plastidic glucose translocator. Plant Cell 12: 787–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TS, Kofler H, Hausler RE, Hille D, Flugge UI, Zeeman SC, Smith AM, Kossmann J, Lloyd J, Ritte G, et al (2001) The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in e chloroplast hexose transporter. Plant Cell 13: 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T-S, Zeeman SC, Thorneycroft D, Fulton DC, Dunstan H, Lue W-L, Hegemann B, Tung S-Y, Umemoto T, Chapple A, et al (2005) α-Amylase is not required for breakdown of transitory starch in Arabidopsis leaves. J Biol Chem 280: 9773–9779 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Smith SM, Smith AM (2004) The breakdown of starch in leaves. New Phytol 163: 247–261 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Tiessen A, Pilling E, Kato KL, Donald AM, Smith AM (2002) Starch synthesis in Arabidopsis: granule synthesis, composition, and structure. Plant Physiol 129: 516–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Moriau L, Boutry M (1999) Expression analysis of the plasma membrane H+-ATPase pma4 transcription promoter from Nicotiana plumbaginifolia activated by the CaMV 35S promoter enhancer. Plant Sci 149: 157–165 [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.