Abstract
Quantitative trait loci (QTL) analysis was used to identify genes underlying natural variation in primary cell wall composition in Arabidopsis (Arabidopsis thaliana). The cell walls of dark-grown seedlings of a Bay-0 × Shahdara recombinant inbred line population were analyzed using three miniaturized global cell wall fingerprinting techniques: monosaccharide composition analysis by gas chromatography, xyloglucan oligosaccharide mass profiling, and whole-wall Fourier-transform infrared microspectroscopy. Heritable variation and transgression were observed for the arabinose-rhamnose ratio, xyloglucan side-chain composition (including O-acetylation levels), and absorbance for a subset of Fourier-transform infrared wavenumbers. In total, 33 QTL, corresponding to at least 11 different loci controlling dark-grown hypocotyl length, pectin composition, and levels of xyloglucan fucosylation and O-acetylation, were identified. One major QTL, accounting for 51% of the variation in the arabinose-rhamnose ratio, affected the number of arabinan side chains presumably attached to the pectic polysaccharide rhamnogalacturonan I, paving the way to positional cloning of the first gene underlying natural variation in pectin structure. Several QTL were found to be colocalized, which may have implications for the regulation of xyloglucan metabolism. These results demonstrate the feasibility of combining fingerprinting techniques, natural variation, and quantitative genetics to gain original insight into the molecular mechanisms underlying the structure and metabolism of cell wall polysaccharides.
Elucidation of the biogenesis and function of cell wall components remains a major challenge in plant biology. The use of functional genomics has led to progress in identification of the actors involved in the synthesis of nucleotide sugars, cellulose, xyloglucan, pectin, and galactomannan (Perrin et al., 1999; Madson et al., 2003; Robert et al., 2004; Scheible and Pauly, 2004), but most of the molecular mechanisms underlying the biosynthesis, deposition, assembly, and turnover of polysaccharides remain unknown. The analysis of natural variation, complementary to mutant analysis, provides a powerful tool for the dissection of complex processes. Natural variation in cell wall composition underlies various inheritable agronomic properties, such as digestibility in maize (Zea mays; Mechin et al., 2000), bread-making quality in wheat (Triticum aestivum; Courtin and Delcour, 1998), and brewing quality in barley (Hordeum vulgare; Han et al., 1995). Quantitative trait loci (QTL) have been identified for β-(1 > 3),(1 > 4)-linked glucan content in oat (Avena sativa; Kianian et al., 2000) and the sugar composition of pericarp cell walls (Hazen et al., 2003), and fiber content and silage digestibility (Lubberstedt et al., 1997, 1998; Mechin et al., 2000; Guillet-Claude et al., 2004) in maize. Despite the availability of genomics tools for some of these species, it remains difficult to identify the specific genes associated with QTL. The cloning of QTL is much more efficient in Arabidopsis (Arabidopsis thaliana). In addition to all the sophisticated molecular genomics tools available for this species, numerous natural accessions have been collected from all over the world, and there is a growing number of recombinant inbred line (RIL) populations and efficient single nucleotide polymorphism-based genotyping methods. The identification and cloning of cell wall-related QTL from Arabidopsis is, of course, relevant to our understanding of cell wall biogenesis and function in this species. However, orthologs of Arabidopsis QTL may also underlie natural variation in similar traits in crop species (Borevitz and Chory, 2004). This method could therefore speed up the identification of agronomically important cell wall-related QTL. Cell wall structure is difficult to determine due to the hierarchical organization of the wall polymers. Detailed information is required concerning the structure of particular polymers, including their side chains and substitutions, and the abundance of each polymer and the orientation and interactions of the various polymers. In this study, we used three analytical techniques, global monosaccharide composition analysis, xyloglucan oligosaccharide (XyGO) mass profiling (Lerouxel et al., 2002), and Fourier transform infrared (FTIR) microspectroscopy (Mouille et al., 2003), to analyze cell wall composition in dark-grown seedlings. We also measured hypocotyl length because differences in growth may reflect differences in cell wall composition. All methods were robust, reproducible, and allowed the medium- to high-throughput analysis of samples. The plant material studied was a well-established RIL population from a cross between the Arabidopsis accessions Bay-0 and Shahdara (Loudet et al., 2002). We identified 33 QTL. The colocalization of some QTL may have implications for the regulation of xyloglucan modifications. One major locus affected the number of arabinan side chains, which were presumably attached to the pectic polysaccharide rhamnogalacturonan I (RGI), paving the way for positional cloning of the first gene underlying natural variation in pectin structure.
RESULTS
Monosaccharide Composition
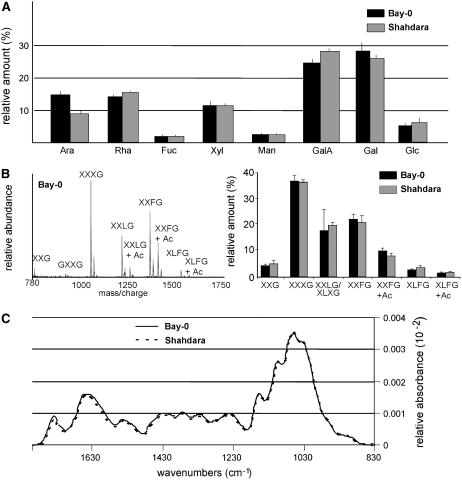
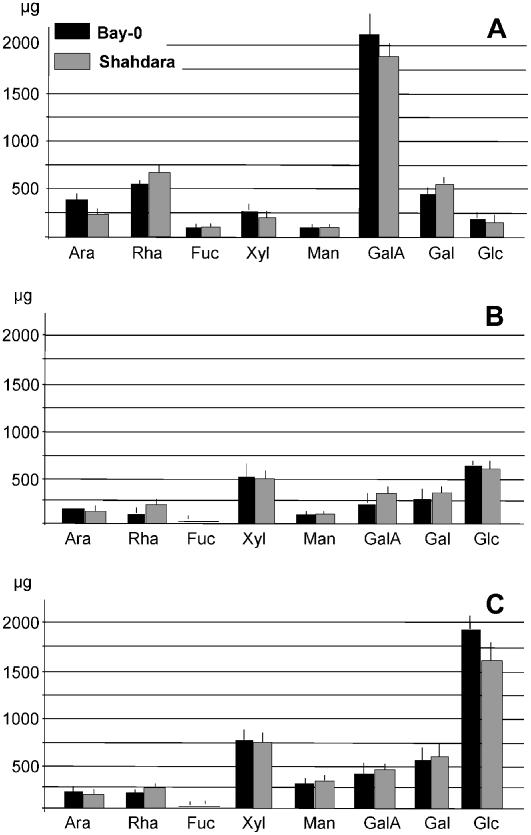
The relative amounts of individual monosaccharides in Bay-0 and Shahdara (Fig. 1A) were determined, together with the ratios of the principal monosaccharides: Ara-Rha, Gal-Rha, Glc-Xyl, Ara-Xyl, and GalA-Rha. Significant differences were found in the abundance of wall-derived Ara and GalA, and the largest difference was that for the Ara-Rha ratio (t test; P < 0.05). In RIL analysis, a significant genotype effect was found for the Ara-Rha ratio (Table I), but not for any of the other monosaccharide contents or ratios (data not shown).
Figure 1.
Cell wall-related traits in 4-d-old dark-grown seedlings of Bay-0 and Shahdara. A, Noncellulose monosaccharide composition, obtained by hydrolysis and gas chromatography analysis of ethanol-insoluble material. B, MALDI-TOF spectrum of XyGOs released by endoxyloglucanase treatment of ethanol-insoluble wall material from hypocotyls. Left, Example of a representative mass spectrum obtained from Bay-0 seedlings; for explanation of the one-letter code, see Figure 2. Right, Relative quantities of the various XyGOs. C, Average infrared spectrum.
Table I.
Mean and range values of traits measured in the Bay-0 × Shahdara RIL population and parents
| Bay-0 | Shahdara | RIL Minimum | RIL Maximum | Heritability | |
|---|---|---|---|---|---|
| Ara-Rha ratio | 1.03 ± 0.08 | 0.68 ± 0.05 | 0.53 | 1.17 | 0.68 |
| XXG | 0.043 ± 0.005 | 0.049 ± 0.013 | 0.03 | 0.08 | 0.10 |
| XXXG | 0.361 ± 0.019 | 0.356 ± 0.010 | 0.30 | 0.40 | 0.07 |
| XXLG | 0.174 ± 0.080 | 0.195 ± 0.011 | 0.11 | 0.22 | 0.20 |
| XXFG | 0.217 ± 0.019 | 0.203 ± 0.029 | 0.16 | 0.29 | 0.21 |
| XXFG Ac | 0.099 ± 0.010 | 0.080 ± 0.009 | 0.08 | 0.14 | 0.13 |
| XLFG | 0.028 ± 0.002 | 0.035 ± 0.008 | 0.02 | 0.06 | 0.16 |
| XLFG Ac | 0.015 ± 0.002 | 0.017 ± 0.004 | 0.01 | 0.04 | 0.09 |
| XXLG/XXFG | 0.805 ± 0.048 | 0.982 ± 0.181 | 0.37 | 1.17 | 0.14 |
| Aca | 0.148 ± 0.011 | 0.117 ± 0.023 | 0.11 | 0.24 | 0.14 |
| XLFG ± Acb | 0.039 ± 0.008 | 0.047 ± 0.009 | 0.03 | 0.1 | 0.17 |
| FTIR 1,041 | 0.0032 ± 0.00032 | 0.0033 ± 0.00026 | 0.0029 | 0.0035 | 0.25 |
| FTIR 1,049 | 0.0033 ± 0.00034 | 0.0034 ± 0.00026 | 0.0030 | 0.0037 | 0.25 |
| FTIR 1,060 | 0.0034 ± 0.00037 | 0.0036 ± 0.00027 | 0.0032 | 0.0038 | 0.24 |
| FTIR 1,157 | 0.0017 ± 0.00010 | 0.0016 ± 0.000062 | 0.0015 | 0.0017 | 0.14 |
| FTIR 1434 | 0.00097 ± 0.00005 | 0.00095 ± 0.000035 | 0.0009 | 0.0010 | 0.16 |
| FTIR 1,473 | 0.00049 ± 0.00009 | 0.00046 ± 0.000069 | 0.0004 | 0.0006 | 0.21 |
| FTIR 1,496 | 0.00041 ± 0.00008 | 0.00042 ± 0.000068 | 0.0003 | 0.0005 | 0.21 |
| FTIR 1,554 | 0.00070 ± 0.00013 | 0.00068 ± 0.000085 | 0.0006 | 0.0008 | 0.22 |
| FTIR 1,639 | 0.0016 ± 0.00020 | 0.0016 ± 0.000085 | 0.0015 | 0.0018 | 0.20 |
| FTIR 1,678 | 0.00091 ± 0.0012 | 0.0009 ± 0.000071 | 0.0008 | 0.0011 | 0.21 |
| Hypocotyl length (mm) | 11.79 ± 1.32 | 10.5 ± 1.15 | 8.7 | 12.7 | 0.37 |
Cumulative relative abundance of all Ac XyGOs (XXLG Ac, XXFG Ac, XLFG Ac).
Cumulative relative abundance of XLFG and XLFG Ac values of XyGO content corresponds to relative abundance of each oligosaccharide. Values of FTIR correspond to relative absorbance at the indicated wavenumber (cm−1).
XyGO Mass Profiling
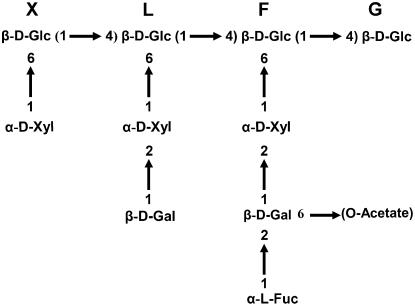
The oligosaccharide profile of xyloglucan was determined by digesting cell wall material with a xyloglucan-specific endoglucanase (XEG; EC 3.2.1.151; Pauly et al., 1999) and carrying out matrix-assisted laser-desorption time-of-flight (MALDI-TOF) analysis of the solubilized XyGOs. Arabidopsis walls contain XyGOs of various sizes, ranging from the pentasaccharide XXG to the decasaccharide XLFG, with various degrees of O-acetylation (Ac; Fig. 2; Pauly et al., 2001). The oligosaccharide-profiling method provides a fingerprint of the native xyloglucan polysaccharide structure, including Ac patterns (Lerouxel et al., 2002). Unfortunately, the structural isomers XXLG and XLXG, which are present in a 2:1 ratio in wild-type Arabidopsis leaves, cannot be distinguished with this technique. Hereafter, for the sake of simplicity, we refer to these two structures as XXLG. The various XyGO ion signals can be integrated to generate a semiquantitative relative abundance profile. XyGOs were quantified using a PERL-based oligosaccharide mass profiling (OLIMP) program (Lerouxel et al., 2002). The reproducibility of the method was demonstrated by the XyGO profiles obtained in three independent growth experiments with the parental accessions (Fig. 1B; Table I). Oligosaccharide profiling of the parents, Bay-0 and Shahdara (Fig. 1B), indicated that only the overall degree of Ac and the XXLG-XXFG ratio were significantly different between the two parents (P < 0.05; data not shown).
Figure 2.
Example of a XyGO structure present in Arabidopsis and the established one-letter abbreviation code (Fry et al., 1993).
FTIR Microspectroscopy
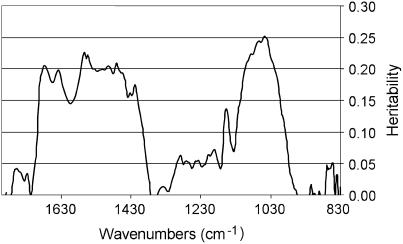
FTIR spectra were collected in transmission mode from a 50-μm × 50-μm window halfway up the hypocotyl, avoiding the central cylinder. Hypocotyls are anatomically very simple and the spectra obtained therefore correspond to the absorbance of epidermal and cortical cell layers only. We selected a window between 829 and 1,801 cm−1, represented by 253 data points, containing information characteristic for polysaccharides. Representative baseline-corrected and area-normalized spectra for Bay-0 and Shahdara are shown in Figure 1C. With the exception of a few clearly resolved frequencies, such as the band at 1,740 cm−1 corresponding to carboxylic esters (Kacurakova et al., 2002), most of the spectrum is too complex to be interpreted by the eye. Student's t tests were used to determine the significance of differences in the mean values for Bay-0 and Shahdara for each individual wavenumber. None of the t values obtained was significant, indicating that the hypocotyl cell walls of the two parental accessions are very similar. Nevertheless, as shown below, significant transgressions were observed for several wavenumbers in the RIL population, making it possible to calculate heritability (Fig. 3).
Figure 3.
Heritability of FTIR absorbance values. Each wavenumber was considered as a single trait and the heritability of the absorbance values was calculated by ANOVA for an average of 140 RIL data (three to four replicates).
Genetic Variation in RILs and Parents
Significant genetic variation was observed for 22 traits P < 0.05 (Table I), but heritability was highest for the Ara-Rha ratio (ARH; h2 = 0.68) and hypocotyl length (h2 = 0.37). Lower heritability values were obtained for several FTIR wavenumbers and various xyloglucan fragments (Table I). Considering each wavenumber as a single trait, an ANOVA of phenotypic variation across the 140 RILs showed that heritability was heterogeneous across the spectrum, with regions of significant genetic variation (h2 between 0.07 and 0.25) and regions of no genetic variation (Fig. 3). The wavenumbers corresponding to the 10 most significant local heritability peaks (h2 from 0.14–0.25) were considered as single traits and included in subsequent QTL mapping. All trait values for the core population of 140 RILs were normally distributed, consistent with quantitative genetic variation (data not shown). Transgressions were considered significant if extreme RIL trait values differed from the parental trait values by more than twice the se. Significant transgression was observed for all traits listed in Table I.
QTL Mapping
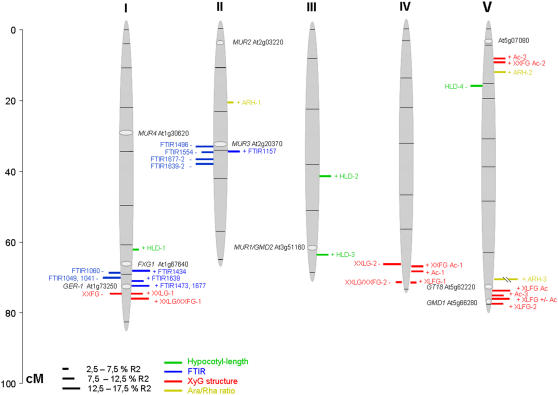
The results of QTL mapping are presented in Table II and Figure 4. We detected 33 QTL for the entire set of traits. Each trait was significantly controlled by one to four QTL dispersed among the chromosomes. Individual QTL explained between 5% and 51% of the total phenotypic variation (R2) of the given trait, and as many as 12 QTL had an R2 exceeding 10%. QTL with positive and negative allelic effects were identified, a positive effect implying a higher value for the trait conferred by the Bay-0 allele. QTL were considered to be potentially colocalized when their 1-log of the odds (LOD) support intervals (estimated by decreasing the QTL peak by 1 LOD) overlapped.
Table II.
Summary of QTL affecting the traits measured in the Bay-0 × Shahdara RIL population
| QTL Cluster | QTLa | Chromosome Markerb | Positionc | LOD Score | R2d | Additivee |
|---|---|---|---|---|---|---|
| HLD-1 | 1-F5I14 | 65.0 | 2.7 | 7 | + | |
| IRA-1 | FTIR 1,434 | 1-F5I14 | 70.1 | 3.3 | 13 | + |
| FTIR 1,639 | 1-MSAT1.13 | 72.3 | 2.5 | 9 | + | |
| FTIR 1,060 | 1-MSAT1.13 | 74.5 | 2.4 | 12 | − | |
| FTIR 1,049 | 1-MSAT1.13 | 74.9 | 2.4 | 13 | − | |
| FTIR 1,041 | 1-MSAT1.13 | 74.9 | 2.3 | 12 | − | |
| FTIR 1,473 | 1-MSAT1.13 | 77.2 | 2.4 | 13 | + | |
| FTIR1 677 | 1-MSAT1.13 | 77.2 | 3.0 | 13 | + | |
| XGM-1 | XXFG | 1-MSAT1.13 | 78.0 | 2.7 | 11 | − |
| XXLG-1 | 1-MSAT1.13 | 78.3 | 3.1 | 11 | + | |
| XXLG/XXFG-1 | 1-MSAT1.13 | 78.7 | 4.5 | 15 | + | |
| ARH-1 | 2-MSAT2.38 | 25.8 | 3.0 | 5 | + | |
| IRA-2 | FTIR 1,496 | 2-MSAT2.36 | 34.3 | 3.6 | 13 | − |
| FTIR 1,157 | 2-MSAT2.41 | 35.3 | 2.3 | 9 | + | |
| FTIR 1,554 | 2-MSAT2.41 | 36.8 | 3.1 | 12 | − | |
| FTIR 1,677-2 | 2-MSAT2.41 | 37.3 | 3.5 | 14 | − | |
| FTIR 1,639-2 | 2-MSAT2.41 | 38.5 | 3.2 | 13 | − | |
| HLD-2 | 3-MSAT3.32 | 45.7 | 4.2 | 12 | + | |
| HLD-3 | 3-MSAT3.18 | 62.5 | 2.7 | 9 | + | |
| XGM-2 | XXLG-2 | 4-MSAT4.9 | 65.6 | 4.3 | 16 | − |
| XXFG Ac-1 | 4-MSAT4.9 | 65.9 | 3.2 | 12 | + | |
| Ac-1 | 4-MSAT4.9 | 68.1 | 3.3 | 10 | + | |
| XXLG/XXFG-2 | 4-MSAT4.9 | 69.7 | 2.7 | 7 | − | |
| XLFG-1 | 4-MSAT4.9 | 69.7 | 2.3 | 6 | + | |
| XGM-4 | Ac-2 | 5-NGA249 | 8.2 | 2.9 | 10 | + |
| XXFG Ac-2 | 5-NGA249 | 8.6 | 2.6 | 10 | + | |
| ARH-2 | 5-NGA249 | 12.4 | 4.6 | 12 | + | |
| HLD-4 | 5-MSAT5.14 | 18.2 | 3.5 | 9 | − | |
| ARH-3 | 5-MSAT5.12 | 70.9 | 18.5 | 51 | + | |
| XGM-3 | XLFG Ac | 5-MSAT5.12 | 73.9 | 5.0 | 16 | + |
| Ac-3 | 5-MSAT5.12 | 74.9 | 4.0 | 11 | + | |
| XLFG ± Ac | 5-MSAT5.12 | 75.1 | 4.2 | 13 | + | |
| XLFG-2 | 5-MSAT5.12 | 76.4 | 2.5 | 8 | + |
The name given to a local LOD-score peak includes the trait name and a suffix, with an order number, if necessary.
The corresponding marker was used in CIM detection and for ANOVA.
The position of the QTL is expressed in centiMorgans with respect to the first marker of the chromosome.
Percentage of variance accounted for by the QTL.
The sign of the allelic effect is calculated as the mean effect of replacing both Shahdara alleles by Bay-0 alleles at the QTL. Thus, + indicates that the Bay-0 allele increases the trait value.
Figure 4.
Map positions of wall-related QTL. Each QTL (for nomenclature, see Tables I and II) is represented by a bar located at its most probable position. The length of the bar is proportional to the contribution of the QTL (R2). Bars on the left side of the chromosome correspond to positive additive-effect QTL (Bay-0 allele increasing the trait value) and those on the right side to negative additive-effect QTL. The position of some candidate genes is also indicated. The framework genetic map (indicating marker positions) is from Loudet et al. (2002).
Dark-Grown Hypocotyl Length
Hypocotyl length was measured because variation in cell wall composition may lead to variation in cell elongation. Four QTL of moderate effect (R2 from 7%–12%) affected etiolated hypocotyl length: three had positive (HLD-1, 2, and 3) and one had negative (HLD-4; Fig. 4; Table II) effects. None of these QTL colocalized with the dark-grown hypocotyl-length QTL previously identified in RIL populations of crosses between Columbia-0 and Kashmir-1 (Wolyn et al., 2004) and between Landsberg erecta and Cape Verde Island (Borevitz et al., 2002). None of the three hypocotyl-length QTL identified colocalized with any of the cell wall-related QTL described below.
Xyloglucan Structure
In total, 10 xyloglucan-structure QTL were identified. Seven corresponded to the relative abundance of single typical XyGOs, one corresponded to the cumulative abundance of O-acetylated XyGOs (Ac), and another to the cumulative abundance of XLFG with and without Ac (XLFG ± Ac). The remaining QTL (XXLG/XXFG) corresponded to the ratio of the relative areas for ions with mass-to-charge ratios of 1,247 and 1,393 (XXLG and XXFG, respectively). We identified QTL for all types of combinations of galactosyl-, fucosyl-, and O-acetyl-substituted oligosaccharides, but none of the QTL was linked to XyGOs containing only xylosyl substitutions, not even the highly abundant XXXG.
A QTL for the XXLG-XXFG ratio (positive effect) was identified on chromosome 1 at about 78 cM. This QTL colocalized with two other QTL, corresponding to the relative amounts of XXFG and XXLG, with negative and positive effects, respectively. These three QTL probably correspond to a single locus influencing the level of fucosylation of XXLG. We have named this locus XYLOGLUCAN MODIFICATION-1 (XGM-1).
The second QTL, XXLG/XXFG-2 (chromosome 4, around 67 cM), colocalized with XXLG-2, both QTL having a negative effect. In contrast to what was observed for XGM-1, no QTL for XXFG levels was found to colocalize with XXLG/XXFG-2. Instead, XXFG + Ac and Ac-1, both of which had a positive effect, mapped to this locus. The most likely explanation for this association of QTL is that a single QTL (referred to as XGM-2) at this locus affects XXLG fucosylation levels, with an effect opposite to that of XGM-1, and that Ac may compensate for changes in XXFG levels. Interestingly, the positive-effect QTL XLFG-1 also mapped to this locus, suggesting that XGM-2, unlike XGM-1, affects the fucosylation levels of both XLLG and XXLG.
A third QTL influencing xyloglucan fucosylation, XLFG-2 (chromosome 5, around 75 cM), was also identified. This positive-effect QTL colocalized with XLFG + Ac, Ac-3, and XLFG ± Ac, all three with positive effects. Theoretically, these four QTL may all correspond to a single locus (XGM-3) affecting the degree of XLLG fucosylation or XXFG galactosylation.
The QTL Ac-2 and XXFG + Ac-2 had positive effects and colocalized to the top of chromosome 5. They presumably correspond to a single locus (XGM-4) specifically affecting Ac of XXFG in particular.
FTIR Wavenumbers
All the QTL related to FTIR wavenumbers mapped to only two regions: one on chromosome 1 and the other on chromosome 2. On chromosome 1, QTL for seven wavenumber values colocalized within the 70- to 77-cM interval: 1,041, 1,049, and 1,060 cm−1 with negative effects, and 1,434, 1,473, 1,639, and 1,677 cm−1 with positive effects. Wavenumbers 1,041, 1,049, and 1,060 cm−1 are part of the carbohydrate fingerprint region and may correspond to the absorbances of cellulose, xyloglucan, or even RGI side-chain bonds (McCann et al., 1992). Wavenumbers 1,639 and 1,677 cm−1 correspond to the absorbances of the COOH groups of pectic polysaccharides. The simplest explanation for this pattern is that the seven QTL correspond to a single locus (IR-ABSORBANCE-1 or IRA-1), with opposing effects on cellulose and nonesterified pectin in the cell walls.
Within the 34- to 38-cM interval on chromosome 2, we identified an association of positive-effect QTL for the absorbance values of wavenumbers 1,496, 1,554, 1,677, and 1,639 cm−1 and a negative-effect QTL for 1,157 cm−1. Wavenumber 1,157 cm−1 corresponds to the absorbance of polysaccharide glycoside bonds, 1,496 cm−1 to amide band II, and 1,554 cm−1 to carboxylates. Again, it seems likely that there is a single locus, IRA-2, coordinately influencing all five wavenumber values. The results obtained suggest that IRA-1 and IRA-2 have opposite effects on the amount of acidic pectin (COOH wavenumbers 1,639 and 1,677 cm−1) in the wall.
Variation in the Ara-Rha Ratio Reflects Differences in RGI Structure
Three QTL (ARH-1 to 3) with positive allelic effects on the Ara-Rha ratio were identified, with ARH-3 having the strongest effect (R2 = 51%). The molecular basis of this variation was investigated by fractionating cell wall polysaccharides from the parental accessions by means of successive chemical extractions and determining the sugar composition of the resulting fractions. Variation in the Ara-Rha ratio may reflect differences in the arabinan side chains of RGI or arabinogalactan proteins (AGPs). RGII and glucuronoarabinoxylan also contain Ara, but are unlikely to have a major effect given their low abundance in Arabidopsis primary cell walls (Zablackis et al., 1995). We distinguished between RGI and AGP by extracting cell wall material from both parental accessions successively with ammonium oxalate, 0.1% KOH, and 1% KOH, and analyzing the monosaccharide composition of each fraction. Most of the pectins were extracted with ammonium oxalate, as indicated by the amount of GalA (Fig. 5A). The alkali-extracted cell wall fractions were enriched in hemicelluloses (Fig. 5, B and C) and contained only small amounts of pectin. Differences in the Ara-Rha ratio were found between the ammonium oxalate fractions of the two accessions (Bay-0, Ara-Rha = 0.63 ± 0.10; Shahdara, Ara-Rha = 0.37 ± 0.08). Because this fraction may contain AGPs, we then used Yariv reagent to prepare fractions enriched in AGPs from the parental accessions. As expected, no significant differences between accessions were observed for the Gal or Ara content of this fraction (45% ± 2% and 12% ± 3%, respectively, of total monosaccharide content for both accessions). Thus, the difference in the Ara-Rha ratio between the parental accessions did not reflect differences in the composition of AGPs. Instead, it was primarily, if not entirely, due to variation in the amount of RGI-linked arabinan. Decreases in the amount of arabinan may result from defects in the metabolism of Ara or UDP-Ara. We investigated this possibility by growing seedlings of both accessions in the presence of 20 and 100 mm Ara. Cell wall sugar analysis showed that the Ara-Rha ratio was similar to that observed for seedlings grown in the absence of Ara (data not shown). The difference in the Ara-Rha ratio between Shahdara and Bay-0, therefore, probably results from differences in Ara incorporation into pectic material or pectin degradation rather than limited synthesis of Ara or UDP-Ara.
Figure 5.
Monosaccharide composition of cell wall fractions of Bay-0 and Shahdara. Cell wall fractions were extracted from 20 mg of alcohol-insoluble cell wall material with ammonium oxalate (A), 1 m KOH (B), and 4 m KOH (C).
DISCUSSION
This study demonstrates the feasibility of exploiting natural variation in Arabidopsis for identification of the processes underlying cell wall metabolism. We first showed that the miniaturized analytical techniques were sufficiently quantitative and reproducible for the detection and dissection of natural variation in cell wall composition. We then showed that the homeostasis of a number of cell wall-related features, including previously unsuspected polysaccharide decorations, such as xyloglucan acetylation and fucosylation, was genetically controlled in developing dark-grown seedlings.
The Ara-Rha ratio was the most heritable trait identified in this RIL population (h2 = 0.68). Analysis of cell wall fractions from the two parental lines showed that this trait probably reflects differences primarily, if not exclusively, in the number and/or length of arabinan side chains on the pectic polysaccharide RGI. No difference was observed between the parental lines in terms of the arabinan content of AGPs precipitated with Yariv reagent. Three QTL were identified for this trait, including one major locus accounting for more than one-half of the total phenotypic variation. All three QTL are thought to affect arabinan side chains on RGI, rather than other polysaccharides. Indeed, we observed no difference between the parental lines for other polysaccharides, with only a weak transgression for the Ara-Rha ratio in the RIL population, and all three QTL acted in the same direction. However, we cannot exclude the possibility of a hidden transgression for another polysaccharide contributing to this character.
Several mutants with low (mur4, 5, 6, and 7) or high (mur10) Ara content have been described (Reiter et al., 1997). Three mutant alleles of MUR4, which encodes a UDP-d-Xyl 4-epimerase, have been shown to be associated with decreases in Ara content of up to 75%, affecting all Ara-containing polymers (Burget et al., 2003). MUR4 maps to the bottom of chromosome 1 and is therefore different from ARH1, 2, or 3. ARH-3 is also unlikely to be related to UDP-Ara synthesis because supplying the plant with Ara had no effect on the Ara-Rha ratio in the parental lines. ARH-3 is also different from mur10, which does not map to the bottom of chromosome 5 (G. Seifert, personal communication). The map positions of the poorly characterized mutants mur5, 6, and 7 have not yet been reported. The cloning of ARH-3 will therefore provide insight into the metabolism of Ara and presumably of arabinan side chains on RGI in the cell wall.
Ara content varies more strongly than the content of any other neutral sugar during development and is higher in growing than in nongrowing tissues (Reiter et al., 1997; Burget and Reiter, 1999). The amount of arabinan epitopes, detected with the arabinan-specific LM6 antibody, also varies considerably during development, as shown in Arabidopsis roots (Willats et al., 2001) and potato (Solanum tuberosum) tubers (Bush et al., 2001), in which LM6 labeling is restricted to meristematic cells, being entirely absent from more highly differentiated cells. Pectic arabinan side chains may play a key role in cell wall architecture because they interact directly with cellulose and may therefore be involved in the cross-linking of cellulose microfibrils (Zykwinska et al., 2005). Jones et al. (2003) showed, with various glycan hydrolases, that arabinan side chains in guard cell walls play a crucial role in the reversible opening and closing of the stomata. These observations suggest that arabinan side chains make a major contribution to the physicochemical properties of the cell wall, growth, development, and key physiological processes. They also suggest that selection pressure might maintain heritable variation for this trait.
Natural Variation, Leading to Differences between Bay-0 and Shahdara, Occurs Primarily in Late Steps of Xyloglucan Modification
Whereas many mutants have strong, easily identifiable phenotypes, natural variation typically gives rise to a whole range of highly subtle phenotypes. These subtle phenotypic differences make molecular analysis more difficult but provide a genetic basis for correlations between traits, potentially providing original insight into the regulation of the underlying processes. Although XXXG accounts for one-third of all ions, no QTL corresponding to this XyGO was identified. In contrast, high heritability was observed for later biosynthetic steps, relating to the fucosylation and Ac of XyGOs, in particular. Interestingly, the composition of xyloglucan side chains has been shown to play a critical role in growth and maintenance of the mechanical properties of the cell wall (Pena et al., 2004). This character may therefore also be under selection pressure.
Complex Control of Xyloglucan Side-Chain Modification
At least three loci influence xyloglucan fucosylation, each acting in a different way. XGM-1 specifically affects fucosylation of XXLG, having no effect on that of XLLG. XGM-2 affects fucosylation of both XXLG and XLLG. XGM-3 affects galactosylation of XXFG or, alternatively, fucosylation of XLLG, but not of XXLG. Any change in the side-chain structures of xyloglucan may result primarily from a net change in biosynthesis, production of the substrate (GDP-Fuc) by fucosyltransferases, or catabolism by fucosidases. A number of enzymes affecting xyloglucan side chains have been identified: MUR1/GMD1, GMD2, and GER are involved in the synthesis of GDP-Fuc (Bonin and Reiter, 2000); MUR2 is a xyloglucan-specific fucosyltransferase (Perrin et al., 1999); MUR3 is a galactosyl transferase, catalyzing the addition of Gal to the third position of XXXG but not to the equivalent position of XLXG (Madson et al., 2003); and FXG1 is a fucosidase that removes fucosyl moieties from XyGOs (De La Torre et al., 2002). Neither MUR2 nor any of the other nine members of the fucosyltransferase gene family map to any of the three QTL. However, GER1, which has been implicated in the synthesis of GDP-Fuc, maps to XGM-1. It is therefore feasible that changes in the size of the substrate pool lead to a decrease in xyloglucan fucosylation, affecting XXFG, the most abundant fucosylated XyGO, most severely. Other genes mapping to the XGM-3 region include GMD1, another gene encoding a protein implicated in GDP-Fuc biosynthesis. GT18, a gene encoding an enzyme catalyzing the transfer of Gal to the second position of various XyGOs and producing XLFG from XXFG (Li et al., 2004; W.-D. Reiter, personal communication), also maps to XGM-3, consistent with the structural data for this QTL.
All four XGM loci also affect xyloglucan Ac in different ways. However, nothing is yet known about the enzymes controlling polysaccharide Ac in plants. A gene encoding a putative O-linked acetyl transferase (At5g07080; see Fig. 4) maps within the confidence interval of XGM-4. For the other QTLs, no obvious candidate genes can be identified. However, recent studies strongly suggest interdependence between the Ac and fucosylation of xyloglucan. For example, decreases in fucosylation, but not in galactosylation, lead to the almost total absence of O-acetyl-substituents, as shown for the mur1, Atfut1, and mur2 mutants (Pauly et al., 2001; Perrin et al., 2003). The ectopic expression of MUR2 (fucosyltransferase) leads to an increase in the number of O-acetyl substituents on galactosyl-residues without increasing fucosylation levels. So, XGM-2 may influence the activity of a combination of xyloglucan-modifying enzymes. Alternatively, the confidence interval of XGM-1 and/or XGM-2 may contain two different genes, one affecting fucosylation and the other acetylation.
The different behaviors of XGM1 to 4 may reflect differences in the specificity of the modifying enzymes or their production in different cell types. Alternatively, the observed effects on xyloglucan modifications may reflect pleiotropic effects of the developmental changes influenced by the QTL. For example, the amounts of XLFG and XLFG + Ac, unlike those for other XyGOs, vary during development, with low levels in young seedlings and higher levels in older tissues (Lerouxel et al., 2002). Thus, XGM-1 to 4 may therefore have a primarily heterochronic effect on seedling development.
Thus, whereas the effects of XGM-1 and XGM-3 may be accounted for by known genes present in these chromosome areas, XGM-2 and XGM-4 are new loci, not previously identified as involved in xyloglucan metabolism.
FTIR microspectroscopy, unlike the other two techniques used, provides information about the cell wall composition of only two cell types (epidermis and cortex) in a 50-μm × 50-μm area of the hypocotyl. The validity and sensitivity of the technique have been demonstrated in Arabidopsis mutants. FTIR fingerprints can be used to distinguish between a series of mutants with defects in cellulose, pectins, xyloglucans, or the cytoskeleton (Mouille et al., 2003). We detected no significant differences between the parental lines. The differences between parental lines observed with the other two techniques may reflect variation in other parts of the seedling or may simply be undetectable with the FTIR technique. Nevertheless, the transgression observed within the RIL population made it possible to identify QTL for 10 of the 253 wavenumbers. These QTL corresponded to a minimum of two loci. IRA-1 has opposite effects on cellulose and pectin content. Increases in pectin content have also been observed in cellulose-deficient mutants and wild-type plants treated with cellulose inhibitors (His et al., 2001; Pilling and Hofte, 2003), indicating the existence of feedback mechanisms coordinating the deposition of pectin and of cellulose. IRA-2 also has an effect on acidic pectin content that is opposite to and independent of that of IRA-1.
In conclusion, this study demonstrates the feasibility of combining multivariate datasets obtained using high-throughput miniaturized cell wall analysis techniques, natural variation, and quantitative genetics to obtain new insights into cell wall metabolism and to identify key genes underlying the observed natural variation. At least one locus with a strong effect can now be cloned and will provide new information about the metabolism and function of RGI-linked arabinans. The observed colocalizations of QTL suggest complex regulation of steps late in the xyloglucan biosynthesis pathway and lead to the identification of new loci influencing xyloglucan fucosylation and Ac.
MATERIALS AND METHODS
Plant Material
We used the Bay-0 × Shahdara RIL population (Loudet et al., 2002). The Core-Pop set of 165 RILs (N57921) was selected from the original population of 411 RILs as an optimal set for QTL mapping experiments (see www.inra.fr/qtlat for details). Seeds were plated on medium (Estelle and Somerville, 1987) without Suc, incubated for 2 d at 4°C, and then exposed to continuous white light (200 μmol m−2 s−1) for 4 h. They were then incubated in the dark (petri dishes wrapped in three layers of aluminum foil) for 4 d in a growth chamber at 20°C with 75% relative humidity. Seedlings originating from three or four independent plant growth experiments were harvested separately and split into three batches. One batch of seedlings was used for hypocotyl-length measurements and FTIR analysis. The other two batches of around 50 seedlings each were used for the analysis of monosaccharide composition and oligosaccharide mass profiling. Seedlings were collected in Eppendorf tubes containing 100% ethanol (1 mL) and stored at 4°C. For nutritional experiments, the culture medium was supplemented with 20 to 100 mm Ara.
Measurement of Hypocotyl Length
Seedling growth was arrested by treatment with an aqueous solution of 0.4% (w/v) formaldehyde. Seedlings were spread on agar plates and an image was captured with a digital camera. Hypocotyl length was measured with image analysis software (Optimas 5.2; IMASYS), as described previously (Gendreau et al., 1997).
Monosaccharide Composition of Noncellulose Cell Wall Material
Four-day-old dark-grown seedlings were used for cell wall analyses because they are easy to grow in highly controlled conditions and do not produce starch, thereby simplifying sugar analysis. Neutral sugars and uronic acids were quantified in the noncellulose cell wall fraction. About 10 seedlings, corresponding to a fresh weight of approximately 0.2 mg, were heated at 70°C for 15 min in 70% ethanol and ground in a Potter homogenizer. The homogenate was washed twice with hot 70% ethanol. The resulting ethanol-insoluble residue was hydrolyzed by incubation with trifluoroacetic acid (2 m, 2 h at 110°C) and then for 18 h at 80°C with dry 2 m methanolic-HCl (Lerouxel et al., 2002). The resulting methyl glycosides were then converted into their trimethylsilylated derivatives (York et al., 1985) and separated by gas chromatography, with helium as the carrier gas. The gas chromatograph was equipped with a flame ionization detector and a WCOT-fused silica capillary column (length 25 m, i.d. 0.25 mm, and film thickness 0.4 μm) with a CP-Sil 5 CP stationary phase. AGPs were extracted from the parent accessions by incubation of the ethanol-insoluble residue with 17 mm sodium-MOPS buffer, pH 5.4, followed by precipitation in Yariv reagent (Schultz et al., 2000). Pectins and hemicelluloses were isolated from 20 mg of cell wall material by successive extractions in 0.5% ammonium oxalate at 100°C for 1 h and 1 m KOH and 4 m KOH at 4°C for 2 h. The sugar composition of AGPs and cell wall fractions was determined by gas chromatography, as described for the ethanol-insoluble fractions.
XyGO Mass Profiling
About 10 seedlings were placed in a 2-mL Eppendorf tube to which 1 mL 100% methanol was added. The tissue was homogenized, using a Ball-Retchmill (model MM200; F. Kurt Retch GmbH), at 25 Hz for 1 min. The macerated tissue was centrifuged at 14,000g for 15 min. The supernatant was decanted and the pellet was centrifuged in a methanol-chloroform mixture (1:1 [v/v]) for 5 min. The resulting suspension was centrifuged, the supernatant was decanted, and the cell wall material pellet was dried under vacuum. The cell wall residue was then treated with 0.2 units of a purified XEG (EC 3.2.1.151; Pauly et al., 1999; 1 unit releases 1 μmol of reducing XyGOs/min) in 50 μL of 50 mm ammonium formate, pH 5.0, for 18 h at 37°C. The undigested wall material was removed by centrifugation at 14,000g for 5 min. Some of the supernatant (10 μL), containing solubilized XyGOs, was transferred to a fresh tube (0.5-mL volume) and treated for 5 to 7 min with approximately 10 washed Biorex MSZ 501 resin beads (Bio-Rad) for desalting. The solution was then spotted onto dried 2,5-dihydroxybenzoic acid (1 μL, 10 mg/mL) on a target plate. The sample was analyzed by MALDI-TOF mass spectrometry (Voyager DE-Pro; Applied Biosystems) using an acceleration voltage of 20,000 V and a delay of 350 ns in reflector mode.
The quality of the mass spectra obtained was evaluated with software provided by the manufacturer (Applied Biosystems). The relative abundance of XyGO ions was determined using an in-house PERL- and mass spectrometry-based tool (Lerouxel et al., 2002; http://www.mpimp-golm.mpg.de/pauly).
FTIR Microspectroscopy
Seedlings were squashed between two BaF2 windows and rinsed thoroughly in distilled water for 2 min. The samples were then dried on the window at 37°C for 20 min. An area of 50 μm × 50 μm, halfway up the hypocotyl on the side of the central cylinder corresponding to epidermal and cortical cells, was selected for FTIR microspectroscopy. A Thermo-Nicolet Nexus spectrometer equipped with a Continuμm microscope accessory was used. Fifty interferograms were collected in transmission mode, with 8 cm−1 resolution, and co-added to improve the signal-to-noise ratio. The collected spectra were baseline corrected and normalized as described elsewhere (Robin et al., 2003).
Statistical Analysis and QTL Mapping
Standard statistical procedures, such as ANOVA, were carried out with Splus. The complete dataset was included in an ANOVA to determine the specific effects of genotype. In this analysis, we were able to quantify heritability in the broad sense of the term (from the genetic variance/total phenotypic variance ratio; variances calculated from the genotype and residual variances). All QTL analyses were performed with the Unix version of QTL Cartographer 1.14 (Basten et al., 1994, 2000). QTL were detected with the original set of 38 microsatellite markers and the genetic map obtained with MAPMAKER 3.0 in a previous study (Loudet et al., 2002; www.inra.fr/qtlat). We used standard procedures (Loudet et al., 2003) and present results for composite interval mapping (CIM). LOD significance thresholds were estimated for all traits, from permutation test analyses (1,000 permutations; overall error level: 5%), as previously described (Churchill and Doerge, 1994). The additive effects of the QTL detected were estimated from CIM results as the mean effect of replacing both Shahdara alleles at the locus studied by Bay-0 alleles. Thus, for QTL to have a positive effect, the Bay-0 allele must increase the trait value. The contribution of each identified QTL to total phenotypic variance (R2) was estimated by variance component analysis. For FTIR microspectroscopy data, each wavenumber was considered as a single trait. QTL detection was carried out for the mean values of the 10 most heritable traits.
This work was supported by GABI-GENOPLANTE (contract no. AF 2001 091).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Herman Höfte (herman.hofte@versailles.inra.fr).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.079384.
References
- Basten CJ, Weir BS, Zeng Z-B (1994) Zmap—a QTL cartographer. In C Smith, JS Gavora, B Benkel, J Chesnais, W Fairfull, JP Gibson, BW Kennedy, EB Burnside, eds, Proceedings of the 5th World Congress on Genetics Applied to Livestock Production: Computing Strategies and Software. Organizing Committee, 5th World Congress on Genetics Applied to Livestock Production, Guelph, Ontario, Canada, pp 65–66
- Basten CJ, Weir BS, Zeng Z-B (2000) QTL Cartographer, Version 1.14. North Carolina State University, Department of Statistics, Raleigh, NC
- Bonin CP, Reiter W-D (2000) A bifunctional epimerase-reductase acts downstream of the MUR1 gene product and completes the de novo synthesis of GDP-l-fucose in Arabidopsis. Plant J 21: 445–454 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Chory J (2004) Genomics tools for QTL analysis and gene discovery. Curr Opin Plant Biol 7: 132–136 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Maloof JN, Lutes J, Dabi T, Redfern JL, Trainer GT, Werner JD, Asami T, Berry CC, Weigel D, et al (2002) Quantitative trait loci controlling light and hormone response in two accessions of Arabidopsis thaliana. Genetics 160: 683–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burget EG, Reiter WD (1999) The mur4 mutant of Arabidopsis is partially defective in the de novo synthesis of uridine diphospho l-arabinose. Plant Physiol 121: 383–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burget EG, Verma R, Molhoj M, Reiter WD (2003) The biosynthesis of l-arabinose in plants: molecular cloning and characterization of a Golgi-localized UDP-d-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis. Plant Cell 15: 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush MS, Marry M, Huxham IM, Jarvis MC, McCann MC (2001) Developmental regulation of pectic epitopes during potato tuberisation. Planta 213: 869–880 [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtin CM, Delcour JA (1998) Physicochemical and bread-making properties of low molecular weight wheat-derived arabinoxylans. J Agric Food Chem 46: 4066–4073 [Google Scholar]
- De La Torre F, Sampedro J, Zarra I, Revilla G (2002) AtFXG1, an Arabidopsis gene encoding alpha-l-fucosidase active against fucosylated xyloglucan oligosaccharides. Plant Physiol 128: 247–255 [PMC free article] [PubMed] [Google Scholar]
- Estelle MA, Somerville CR (1987) Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol Gen Genet 206: 200–206 [Google Scholar]
- Fry SC, York WS, Albersheim P, Darvill A, Hayashi T, Joseleau J-P, Kato Y, Lorences EP, Maclachlan GA, McNeil M, et al (1993) An unambiguous nomenclature for xyloglucan-derived oligosaccharide. Physiol Plant 89: 1–3 [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Hofte H (1997) Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol 114: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillet-Claude C, Birolleau-Touchard C, Manicacci D, Rogowsky PM, Rigau J, Murigneux A, Martinant JP, Barriere Y (2004) Nucleotide diversity of the ZmPox3 maize peroxidase gene: relationships between a MITE insertion in exon 2 and variation in forage maize digestibility. BMC Genet 16: 5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Ulrich SE, Chirat S, Menteur S, Jestin L, Sarrafi A, Hayes PM, Jones BL, Blake TK, Wesenberg DM, et al (1995) Mapping of β-glucan content and β-glucanase activity loci in barley grain and malt. Theor Appl Genet 91: 921–927 [DOI] [PubMed] [Google Scholar]
- Hazen SP, Hawley RM, Davis GL, Henrissat B, Walton JD (2003) Quantitative trait loci and comparative genomics of cereal cell wall composition. Plant Physiol 132: 263–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- His I, Driouich A, Nicol F, Jauneau A, Hofte H (2001) Altered pectin composition in primary cell walls of korrigan, a dwarf mutant of Arabidopsis deficient in a membrane-bound endo-1,4-beta-glucanase. Planta 212: 348–358 [DOI] [PubMed] [Google Scholar]
- Jones L, Milne JL, Ashford D, McQueen-Mason SJ (2003) Cell wall arabinan is essential for guard cell function. Proc Natl Acad Sci USA 100: 11783–11788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacurakova M, Smith AC, Gidley MJ, Wilson RH (2002) Molecular interactions in bacterial cellulose composites studied by 1D FT-IR and dynamic 2D FT-IR spectroscopy. Carbohydr Res 337: 1145–1153 [DOI] [PubMed] [Google Scholar]
- Kianian SF, Phillips RL, Rines HW, Fulcher RG, Webster FH, Stuthman DD (2000) Quantitative trait loci influencing beta-glucan content in oat (Avena sativa, 2n=6x=42). Theor Appl Genet 101: 1039–1048 [Google Scholar]
- Lerouxel O, Choo TS, Seveno M, Usadel B, Faye L, Lerouge P, Pauly M (2002) Rapid structural phenotyping of plant cell wall mutants by enzymatic oligosaccharide fingerprinting. Plant Physiol 130: 1754–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cordero I, Caplan J, Molhoj M, Reiter WD (2004) Molecular analysis of 10 coding regions from Arabidopsis that are homologous to the MUR3 xyloglucan galactosyltransferase. Plant Physiol 134: 940–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F (2002) Bay-0 x Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet 104: 1173–1184 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Krapp A, Daniel-Vedele F (2003) Quantitative trait loci analysis of water and anion contents in interaction with nitrogen availability in Arabidopsis thaliana. Genetics 163: 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubberstedt T, Klein D, Dally A, Westhoff P (1998) QTL mapping in testcrosses of flint lines of maize: III. Comparison across populations for forage traits. Crop Sci 38: 1278–1289 [Google Scholar]
- Lubberstedt T, Melchinger AE, Klein D, Degenhardt H, Paul C (1997) QTL mapping in testcrosses of European flint lines of maize. 2. Comparison of different testers for forage quality traits. Crop Sci 37: 1913–1922 [Google Scholar]
- Madson M, Dunand C, Li X, Verma R, Vanzin GF, Caplan J, Shoue DA, Carpita NC, Reiter WD (2003) The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15: 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann MC, Hammouri MK, Wilson RH, Belton PS, Roberts K (1992) Fourier transform infrared microspectroscopy is a new way to look at plant cell walls. Plant Physiol 100: 1940–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechin V, Argillier O, Menanteau V, Barriere Y, Mila I, Pollet B, Lapierre C (2000) Relationship of cell wall composition to in vitro cell wall digestibility of maize inbred line stems. J Sci Food Agric 80: 574–580 [Google Scholar]
- Mouille G, Robin S, Lecomte M, Pagant S, Hofte H (2003) Classification and identification of Arabidopsis cell wall mutants using Fourier-transform infrared (FT-IR) microspectroscopy. Plant J 35: 393–404 [DOI] [PubMed] [Google Scholar]
- Pauly M, Albersheim P, Darvill A, York WS (1999) Molecular domains of the cellulose/xyloglucan network in the cell walls of higher plants. Plant J 20: 629–639 [DOI] [PubMed] [Google Scholar]
- Pauly M, Eberhard S, Albersheim P, Darvill A, York WS (2001) Effects of the mur1 mutation on xyloglucans produced by suspension-cultured Arabidopsis thaliana cells. Planta 214: 67–74 [DOI] [PubMed] [Google Scholar]
- Pena MJ, Ryden P, Madson M, Smith AC, Carpita NC (2004) The galactose residues of xyloglucan are essential to maintain mechanical strength of the primary cell walls in Arabidopsis during growth. Plant Physiol 134: 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K (1999) Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284: 1976–1979 [DOI] [PubMed] [Google Scholar]
- Perrin RM, Jia Z, Wagner TA, O'Neill MA, Sarria R, York WS, Raikhel NV, Keegstra K (2003) Analysis of xyloglucan fucosylation in Arabidopsis. Plant Physiol 132: 768–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling E, Hofte H (2003) Feedback from the wall. Curr Opin Plant Biol 6: 611–616 [DOI] [PubMed] [Google Scholar]
- Reiter WD, Chapple C, Somerville CR (1997) Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J 12: 335–345 [DOI] [PubMed] [Google Scholar]
- Robert S, Mouille G, Höfte H (2004) The mechanism and regulation of cellulose synthesis in primary walls: lessons from cellulose deficient Arabidopsis mutants. Cellulose 11: 351–364 [Google Scholar]
- Robin S, Lecomte M, Höfte H, Mouille G (2003) A procedure for the clustering of cell wall mutants in the model plant Arabidopsis based on Fourier-transform infrared (FT-IR) spectroscopy. J Appl Stat 30: 669–681 [Google Scholar]
- Scheible WR, Pauly M (2004) Glycosyltransferases and cell wall biosynthesis: novel players and insights. Curr Opin Plant Biol 7: 285–295 [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Johnson KL, Currie G, Bacic A (2000) The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell 12: 1751–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats WG, McCartney L, Knox JP (2001) In-situ analysis of pectic polysaccharides in seed mucilage and at the root surface of Arabidopsis thaliana. Planta 213: 37–44 [DOI] [PubMed] [Google Scholar]
- Wolyn DJ, Borevitz JO, Loudet O, Schwartz C, Maloof J, Ecker JR, Berry CC, Chory J (2004) Light-response quantitative trait loci identified with composite interval and eXtreme array mapping in Arabidopsis thaliana. Genetics 167: 907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York W, Darvill A, McNeil M, Albersheim P (1985) Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol 118: 3–40 [Google Scholar]
- Zablackis E, Huang J, Muller B, Darvill AG, Albersheim P (1995) Structure of plant cell walls. 39. Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol 107: 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zykwinska AW, Ralet MC, Garnier CD, Thibault JF (2005) Evidence for in vitro binding of pectin side chains to cellulose. Plant Physiol 139: 397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]