Abstract
The polar growth of plant cells depends on the secretion of a large amount of membrane and cell wall materials at the growing tip to sustain rapid growth. Small GTP-binding proteins, such as Rho-related GTPases from plants and ADP-ribosylation factors (ARFs), have been shown to play important roles in polar growth via regulating intracellular membrane trafficking. To investigate the role of membrane trafficking in plant development, a Dissociation insertion line that disrupted a putative ARF GTPase-activating protein (ARFGAP) gene, AT2G35210, was identified in Arabidopsis (Arabidopsis thaliana). Phenotypic analysis showed that the mutant seedlings developed isotropically expanded, short, and branched root hairs. Pollen germination in vitro indicated that the pollen tube growth rate was slightly affected in the mutant. AT2G35210 is specifically expressed in roots, pollen grains, and pollen tubes; therefore, it is designated as ROOT AND POLLEN ARFGAP (RPA). RPA encodes a protein with an N-terminal ARFGAP domain. Subcellular localization experiments showed that RPA is localized at the Golgi complexes via its 79 C-terminal amino acids. We further showed that RPA possesses ARF GTPase-activating activity and specifically activates Arabidopsis ARF1 and ARF1-like protein U5 in vitro. Furthermore, RPA complemented Saccharomyces cerevisiae glo3Δ gcs1Δ double mutant, which suggested that RPA functions as an ARFGAP during vesicle transport between the Golgi and the endoplasmic reticulum. Together, we demonstrated that RPA plays a role in root hair and pollen tube growth, most likely through the regulation of Arabidopsis ARF1 and ARF1-like protein U5 activity.
The polar growth of cells as exemplified in root hairs and pollen tubes is a common phenomenon in plants. The tip growth of root hairs and pollen tubes is due to the deposition of cell membranes and wall materials at a restricted tip area of the plasma membrane (Schnepf, 1986). Recent genetic studies in Arabidopsis (Arabidopsis thaliana) have shown that small GTPases, such as ADP-ribosylation factors (ARFs), Rho-related GTPases from plants (ROPs), and Ras-related in brain, play important roles during the polar growth of root hairs and pollen tubes (Zheng and Yang, 2000; Cheung et al., 2002; Gu et al., 2004; Preuss et al., 2004). For example, ROP2 GTPase is a positive regulator of both root hair initiation and tip growth. Dominant-negative GDP-bound ROP2 reduces the number of hair-forming sites and leads to shorter, wavy hairs, whereas the expression of constitutively active GTP-bound ROP2 inhibits and depolarizes root hair tip growth. ROP2 overexpression results in longer root hair with multiple tips (Jones et al., 2002). Similarly, ARF1, a Golgi-localized protein, is required for root hair tip growth by modulating ROP2 function in Arabidopsis (Xu and Scheres, 2005). In addition, ARF1 also affects the localization of the auxin efflux carrier PINFORMED2 (PIN2) with slow kinetics, resulting in changes in auxin polar transport (Xu and Scheres, 2005).
Vesicular transport in mammalian cells and yeast (Saccharomyces cerevisiae) is regulated by a large family of RAS-related GTPases (Novick and Zerial, 1997; Schimmoller et al., 1998; Chavrier and Goud, 1999; Der and Balch, 2000; Zerial and McBride, 2001). Genetic and biochemical analyses have shown that these small GTPases are essential for delivering transport vesicles to specific endomembrane compartments or to the plasma membrane. Small GTPases, such as ARFs, play roles in the protein traffic between the Golgi apparatus and the endoplasmic reticulum (ER) in tobacco (Nicotiana tabacum) and Arabidopsis cultured cells (Takeuchi et al., 2002). They are active in their GTP-bound form generated from the inactive GDP-bound form catalyzed by guanine-nucleotide exchange factors for ARF GTPases, such as GNOM ARF GTPase in Arabidopsis (Steinmann et al., 1999). Inactivation of ARFs depends on the hydrolysis of GTP to GDP mediated by ARF GTPase-activating proteins (ARFGAPs). Dominant inhibitory mutants of ARF1 block ER-to-Golgi transport and trigger disassembly of the Golgi apparatus (Dascher and Balch, 1994). VASCULAR NETWORK DEFECTIVE3 (VAN3), a class I ARFGAP, is located in a subpopulation of the trans-Golgi network (TGN) and is involved in the formation of a leaf vascular network by regulating auxin signaling via a TGN-mediated vesicle transport system (Koizumi et al., 2005).
ARFGAPs, characterized by the presence of a zinc-finger motif and a conserved Arg residue within the ARFGAP domain, consist of a large gene family with 15 members in Arabidopsis (Vernoud et al., 2003). Compared to ARFs, little is known about the function of ARFGAPs in plants, except for VAN3 (Koizumi et al., 2005). Here, using biochemical and forward genetics approaches, we show that ROOT AND POLLEN ARFGAP (RPA), a class II ARFGAP, activates the GTPase activity of Arabidopsis ARF1 and ARF1-like protein U5 in vitro. Loss of RPA function results in the formation of aberrant root hairs and slow growth of pollen tubes. These results suggest that RPA plays an essential role in root hair development and pollen tube elongation in Arabidopsis.
RESULTS
Identification of Mutants in Putative ARFGAP Genes
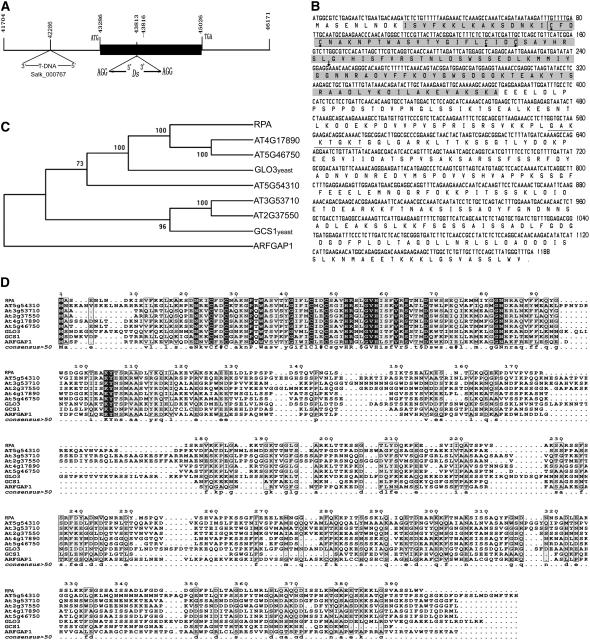
To study the role of ARFGAP in plant development, we identified a mutant sgt3118 in which the Dissociation (Ds) element was inserted into a putative ARFGAP gene, At2g35210 (also named AtAGD10; Vernoud et al., 2003) in our gene-enhancer trap collections (Parinov et al., 1999). Analysis of the Ds flanking sequences indicated that the Ds element was inserted into the third exon of At2g35210 and caused 3-bp duplication at the insertion site (Fig. 1A). Southern-blot analysis with the 750-bp fragment of the 5′ end of the Ds as a probe showed that there is only one Ds copy in sgt3118 (Supplemental Fig. 1). Consistently, the kanamycin resistance conferred by the NPTII gene within the Ds showed a typical 3:1 (kanr:kans = 558:185) segregation in the selfed progeny, indicating that there is only a single Ds insertion into the At2G35210 gene in the mutant. BLAST analyses further identify a T-DNA insertion line, SALK_000767, in which the T-DNA was inserted into the promoter 1,000 bp upstream of the ATG codon of AT2G35210, as confirmed by sequencing of the flanking sequences (Fig. 1A).
Figure 1.
Molecular characterization of the RPA (AT2G35210) gene. A, Scheme depicts the Ds insertion and T-DNA insertion sites. The black box indicates the predicted ORF of the RPA gene. The Ds insertion caused 3-bp duplication (AGG) in the sgt3118 mutant. B, ORF sequence of RPA and its predicted peptide sequence. The Ds insertion site is indicated with an arrow and the ARFGAP domain is shown in the shaded box; the boxed amino acids within the shaded boxes show the zinc-finger motif and the four Cys residues are marked with a black dot below them. The underline shows the putative ATP/GTP-binding motif. C, Phylogenetic tree of RPA protein with its homologs from Arabidopsis, yeast, and animal. D, Alignment of RPA protein with its homologs from Arabidopsis and other organisms. Identical amino acids are shown with white letters on black boxes and similar ones are boxed. RPA, Arabidopsis RPA protein; AT4G17890, AT5G46750, AT5G54310, AT3G53710, AT2G37550, Arabidopsis homologs of RPA; GLO3 and GCS1, yeast homologs; ARFGAP1, rat homolog.
The full-length cDNA of At2G35210 is 1,475 bp in length with an open reading frame (ORF) of 1,188 bp (Fig. 1B); it encodes a peptide of 395 amino acids (Fig. 1B) with a molecular mass of 43.08 kD and a pI of 9.57. Sequence comparison of the cDNA and genomic fragment revealed that there are seven exons and six introns in the AT2G35210 gene. In AT2G35210, an N-terminal CXXCX16CXXC motif in the ARFGAP domain represents a typical zinc-finger structure (Fig. 1B), which is also present in the PF01412 family of plants (http://www.sanger.ac.uk/cgi-bin/Pfam/getacc?PF01412), ARFGAP1 in rats, and GROWTH COLD SENSITIVE1 (GCS1) and GCS1P-LIKE ORF3 (GLO3) in yeast (Cukierman et al., 1995). In the ARFGAP region, there are three REV interacting domains (residues 22–41, 41–58, and 62–83). In addition, there is one domain, GAKKTGKT, that matches the consensus [A/G]-X4-G-K-[S/T] of the potential ATP-/GTP-binding motif (the P-loop; PS00017; Fig. 1B).
Sequence analysis revealed that AT2G35210 belongs to the class II ARFGAP family with six members (AT5G54310, AT3G53710, AT2G37550, AT4G17890, AT5G46750, and AT2G35210; also named AtAGD5–10) in Arabidopsis (Vernoud et al., 2003). Class II ARFGAPs are characterized by the presence of a single ARFGAP domain (residues 10–126) at their N terminus and no other discernible motifs. Phylogenetic analysis showed that AT2G35210, AT4G17890, and AT5G46750 were in the same clade (Fig. 1C), indicating that AT2G35210 is closer to AT4G17890 (AtAGD8) and AT5G46750 (AtAGD9) than to the other members of this family. In addition, AT2G35210 is closer to the yeast homologs GLO3 and GCS1 than to the animal homolog ARFGAP1 (Fig. 1, C and D). These analyses suggest that AT2G35210 could be a putative ARFGAP in Arabidopsis.
AT2G35210 Is Only Expressed in Roots and Pollen
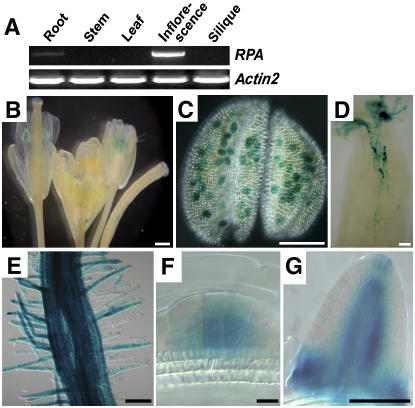
To determine the expression pattern of AT2G35210 in Arabidopsis, reverse transcription (RT)-PCR analysis was performed. AT2G35210 expression was detected strongly in inflorescences and at a lower level in roots (Fig. 2A). This result indicated that AT2G35210 is specifically expressed in flowers and roots, which suggests that AT2G35210 might play a role in flower and root development.
Figure 2.
Expression pattern of the RPA gene. RNA was extracted from tissues as indicated and subjected to RT-PCR analysis (A) and the RPA expression pattern was revealed by the GUS reporter in Pro:RPA-GUS:Ter transgenic plants (B–G). A, RT-PCR result showing RPA expression in inflorescence and root, not in other tissues as indicated. Actin2 is used as an internal control. B, Inflorescence shows GUS staining in the anther. C, Micrograph of an anther showing GUS staining in pollen grains. D, Micrograph of a pollinated pistil showing GUS staining in pollen tubes. E, Micrograph showing GUS activity in root and root hair. The intensity of the staining does not reflect the expression level. F, Micrograph showing GUS staining in lateral root primordial cells. G, Micrograph showing GUS staining in a lateral root. Scale bars, 500 μm (B–E); 10 μm (F); and 100 μm (G).
To determine the cellular expression pattern of AT2G35210, Pro:AT2G35210-β-glucuronidase (GUS):Ter was constructed and transformed to Arabidopsis. Seedlings, roots, leaves, and inflorescences of independent transgenic lines were checked for GUS activity. As a result, GUS activity was observed specifically in mature pollen grains (Fig. 2, B and C) and pollen tubes within the style (Fig. 2D). In addition, GUS activity was also detected in roots, especially in root hairs (Fig. 2, E and F), lateral root primordia (Fig. 2F), and lateral roots (Fig. 2G). No GUS activity could be detected in other tissues (data not shown). The GUS pattern is consistent with RT-PCR analysis. These data indicated that AT2G35210 is expressed in pollen grains, pollen tubes, and roots, including root tips and root hairs. Therefore, we designated AT2G35210 as RPA.
RPA Is Localized in Golgi Complexes via Its 79 C-Terminal Amino Acids
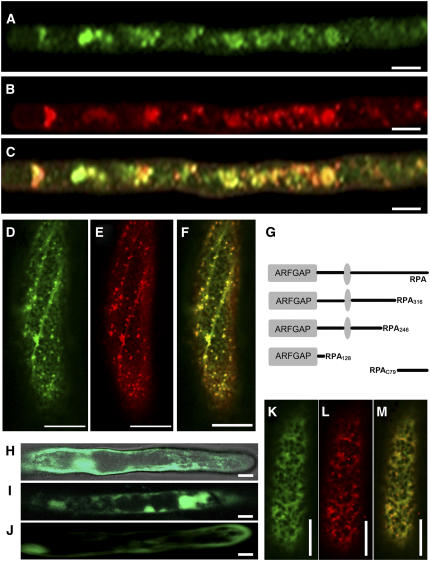
Because ARFGAP is a key regulatory component in vesicle formation for membrane transport, we investigated whether RPA protein is involved in the secretory system. The P35S:RPA-enhanced green fluorescent protein (EGFP) construct and the P35S:EGFP control vector were introduced into Arabidopsis via Agrobacterium-mediated transformation. Confocal laser-scanning microscopy on transgenic root hairs showed that RPA-EGFP was detected as punctuate structures scattered within the cytoplasm (Fig. 3A), whereas the EGFP control showed homogeneous localization. To determine the precise subcellular localization of RPA-EGFP, transgenic roots were stained with the Golgi marker BODIPY FL C5 ceramide (B–22650; Molecular Probes; Fig. 3B). Confocal laser-scanning microscopy showed that most of the RPA-EGFP fluorescence colocalized with the Golgi marker (Fig. 3C). These results suggested that RPA mainly resides in or associates with the Golgi apparatus. To further confirm its Golgi localization, the P35S:RPA-EGFP construct was cobombarded to onion (Allium cepa) epidermal cells together with the P35S:N-sialyltransferase (N-ST)-monomeric red fluorescent protein (mRFP) construct coding for the Golgi-localized N-ST fused to mRFP (Lee et al., 2002). Twenty hours later, the RPA-EGFP fluorescence signal appeared as punctuate structures (Fig. 3D), as observed in transgenic root hairs. N-ST-mRFP also displayed a similar spotty pattern (Fig. 3E). The RPA-EGFP and N-ST-mRFP signals colocalized to the same spots when the two images were merged (Fig. 3F). These results indicated that the RPA protein was localized to the Golgi complexes.
Figure 3.
Subcellular localization of RPA-EGFP protein. A, Root hair of Arabidopsis transformed with P35S:RPA-EGFP showing spotty distribution of RPA-EGFP within the cytoplasm. B, The same root hair shown in A stained with Golgi marker BODIPY FL C5 ceramide (B-22650; Molecular Probes), showing the distribution of the Golgi apparatus. C, Merged image of A and B showing the colocalization of RPA-EGFP and the BODIPY FL C5 ceramide signal. D, Onion epidermal cell cobombarded with P35S:RPA-EGFP and P35S:N-ST-mRFP showing punctuated distribution of RPA-EGFP (green). E, The same cell as in D showing the distribution the Golgi marker protein N-ST (red). F, Merged image of D and E showing the colocalization of RPA-EGFP and N-ST-mRFP, indicating that RPA is localized at the Golgi complexes. G, Drawing showing the construction of truncated RPA-EGFP fusions: RPA128 (1–128 amino acids), RPA246 (1–246 amino acids), and RPA316 (1–316 amino acids). The gray oval indicates putative ATP/GTP-binding motif. H, Root hair transformed with the P35:RPA128-EGFP construct, showing RPA128-EGFP localization. I, Root hair transformed with the P35:RPA246-EGFP construct, showing RPA246-EGFP localization. J, Root hair transformed with the P35:RPA316-EGFP construct, showing RPA316-EGFP localization distributed throughout the cell. K, Onion epidermal cell cobombarded with P35S:RPAC79-EGFP and P35S:N-ST-mRFP constructs, showing RPAC79-EGFP localization (green). L, The same cell as in K, showing localization of Golgi marker protein N-ST-mRFP (red). M, Merged image of K and L, showing colocalization of RPAC79-EGFP and N-ST-mRFP. Scale bars, 10 μm.
To further study which part of the RPA protein determines its subcellular localization in the cell, we fused three truncated RPA peptides: RPA128 (residues 1–128), RPA246 (1–246), and RPA316 (1–316; Fig. 3G) to the N terminus of EGFP, respectively, and introduced them into Arabidopsis. The fluorescence of the three truncated RPA proteins was distributed throughout the whole cell, including the nucleus (Fig. 3, H–J). These results indicated that all three truncated RPA proteins lacking the 79 C-terminal amino acids could not be targeted to Golgi complexes correctly. This suggests that the 79 C-terminal residues of the RPA are responsible for its Golgi localization in the cell. To confirm this possibility, the 79 C-terminal amino acids of RPA were fused to EGFP to yield the P35S:RPAC79-EGFP construct, which was then cobombarded with the P35S:N-ST-mRFP construct to onion epidermal cells. As we expected, RPAC79-EGFP colocalized with N-ST-mRFP (Fig. 3, K–M), demonstrating that the Golgi localization of RPA protein was indeed determined by its 79 C-terminal amino acids.
RPA Possesses ARF GTPase-Activating Activity and Specifically Activates ARF1 and U5 in Vitro
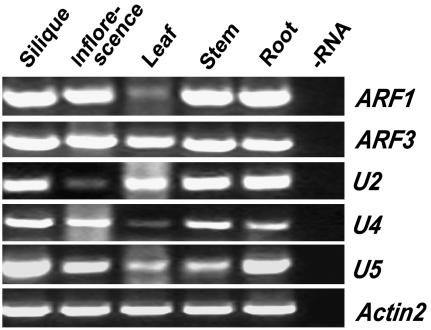
To investigate whether RPA has ARF GTPase-activating activity as suggested by its homology to mammalian ARFGAP1, an in vitro ARF GTPase-activating activity assay was carried out with its potential substrates. The selection of putative ARF substrates was made based on their subcellular localization, expression pattern, and sequence homology. Among the ARF proteins, ARF1, ARF2, and ARF3 are associated with the Golgi complexes. ARF1 and ARF3 represent different phylogenetic branches of the ARF family and have been used previously for ARFGAP assays. In addition, several ARF-like proteins have been predicted to play a role in vesicle trafficking. Therefore, we chose ARF1, ARF3, and ARF-Likes (ARLs) as potential substrates for RPA. We first analyzed their expression pattern with RT-PCR. All of them showed a broad expression pattern throughout the plant, with higher expression levels in roots and inflorescences, except U2 (Fig. 4). At least their expression pattern overlaps with RPA in inflorescences and roots.
Figure 4.
RT-PCR analysis of members of the ARF gene family. The top five images show the expression of five members of the ARF gene family in organs as indicated. Lane −RNA, RT-PCR amplification without addition of RNA as a control for DNA contamination. The bottom image shows ACTIN2 expression as an internal PCR amplification and template control.
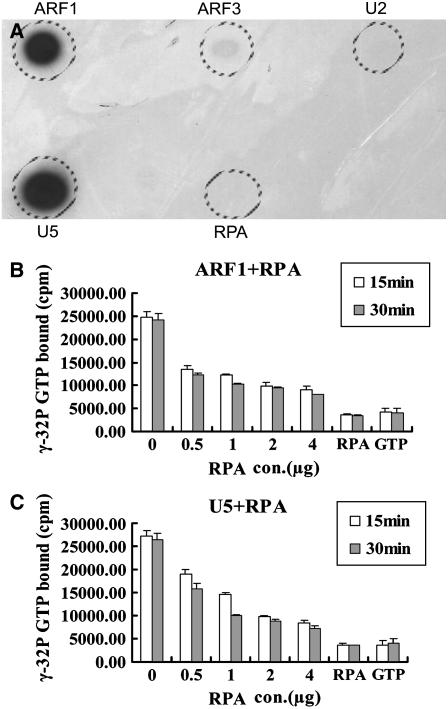
To further narrow down the number of candidate substrates, we used the stable GTP-binding property of ARFs (Ding et al., 1996) to facilitate our substrate selection. We fused the N terminus of ARF1, ARF3, U2, and U5 proteins to a poly-His tag and produced in Escherichia coli because it was difficult to obtain soluble, full-length recombinant proteins. Such truncated forms of ARFs have previously been used successfully in GAP activity assays in humans (Paris et al., 1997) and Arabidopsis (Rikke et al., 2000). In addition, His-6-RPA was also produced in E. coli for the assay. The GTP-binding assay of the in vitro-expressed His-6-ARF1Δ17, His-6-ARF3Δ17, His-6-U5Δ17, and His-6-U2Δ16 fusion proteins was performed. The result showed that ARF1 and U5 have strong GTP-binding capability (Fig. 5A), ARF3 has very weak GTP-binding activity (Fig. 5A), and U2 and RPA have no detectable GTP-binding capability (Fig. 5A). Finally, ARF1, ARF3, and U5 were used for the ARFGAP assay.
Figure 5.
ARF GTPase-activating activity assay of RPA protein. A, Autoradiograph showing GTP binding to proteins as indicated. Note strong GTP-binding activity of ARF1 and U5; very weak binding for ARF3. B, GTPase-activating activity of RPA on ARF1 GTPase assessed by hydrolysis of α-32P-GTP-ARF1. RPA, No ARF1 protein added; GTP, no protein added. C, GTPase-activating activity of RPA on U5 protein assessed by hydrolysis of GTP bound to U5. RPA, No U5 protein added; GTP, no protein added.
ARF GTPases do not exhibit intrinsic GTPase activity (Kahn and Gilman, 1986) and, therefore, rely on the GTPase-activating activity of ARFGAPs to switch from the active GTP-bound form to the inactive GDP-bound form. To investigate whether RPA possesses GTPase-activating activity that mediates effective ARF1-GTP, ARF3-GTP, and U5-GTP hydrolysis, His-6-ARF1Δ17, His-6-ARF3Δ17, and His-6-U5Δ17 were first incubated with [γ-32P]GTP, respectively, for 30 min, then His-6-RPA protein was added and incubated for another 15 or 30 min. As the incubation time and amount of His-6-RPA protein increased, the total [γ-32P] bound to putative substrate His-6-ARF1Δ17 declined (Fig. 5B); this result suggested that the GTP bound to the substrate was hydrolyzed by His-6-RPA protein and the hydrolysis is dependent on the presence of the protein in a dosage- and time-dependent manner. Similarly, His-6-RPA protein could also activate the GTPase activity of the His-6-U5Δ17 (Fig. 5C) with similar efficiency as that to His-6-ARF1Δ17. However, His-6-RPA protein could not activate His-6-ARF3Δ17 (data not shown). In a control experiment with either RPA alone or GTP as a substrate, only a residual amount of radioactivity was retained on the membrane (Fig. 5, B and C). These data demonstrated that RPA is indeed able to activate ARF1 and the ARF1-like protein U5 GTPase activity in vitro.
Complementation of the Yeast glo3Δ gcs1Δ Mutant with RPA
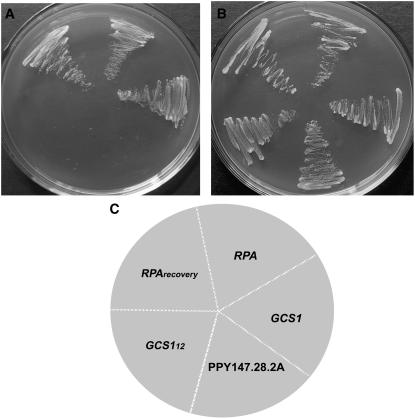
Previous studies showed that GLO3 and GCS1 are two ARFGAPs of yeast. In vitro assay indicated that efficient retrieval transport from the Golgi to the ER requires these two proteins (Poon et al., 1996, 1999). To obtain more insight into RPA function and to further verify that RPA is an ARFGAP involved in the vesicle transport between Golgi and ER, we carried out a yeast mutant complementation test with RPA. The glo3Δ gcs1Δ double mutant, which contains a temperature-sensitive gcs1 allele (gcs1-28), could only grow at 22°C to 25°C, but not at 37°C (Poon et al., 1996). glo3Δ gcs1Δ cells transformed with RPA driven by the GCS1 promoter grew well at 37°C, whereas glo3Δ gcs1Δ cells transformed with the control vector did not grow at 37°C (Fig. 6A), and they could all grow at 23°C (Fig. 6B). Furthermore, plasmid recovery experiments showed that there is no loss of constructs during the complementation (Fig. 6). As a positive control, glo3Δ gcs1Δ cells transformed with GCS1 grew well at 37°C (Fig. 6A). These data showed that RPA exhibited sufficient ARFGAP activity and rescued the temperature-sensitive phenotype of the yeast glo3Δ gcs1Δ mutant, suggesting that RPA could serve as an ARFGAP for yeast vesicle transport between the Golgi and ER.
Figure 6.
Functional complementation of glo3Δ gcs1Δ mutant in yeast by RPA. A, glo3Δ gcs1Δ mutant cell transformed with RPA and the GCS1 positive control could grow at 37°C, whereas the cells transformed with GCS112 (12 amino acids) could not grow. The PPY147.28.2A cells containing a temperature-sensitive GCS1 allele also could not grow at 37°C. B, All yeast cells could grow at the permissive temperature 23°C. C, Schematic drawing depicts the position of yeast cells transformed with genes as indicated. RPA, RPA gene; RPArecovery, RPA plasmid was reisolated from single yeast colony and retransformed into the mutant; GCS1, GCS1 gene; GCS112, GCS1 fragment containing 12 amino acids from the N terminus; PPY147.28.2A, control cells not transformed.
Root Hair Development Was Altered in the rpa Mutant
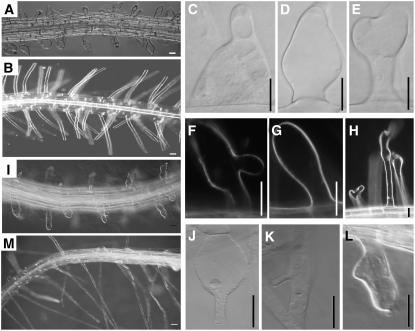
To investigate the biological function of the putative ARFGAP gene, a phenotypic analysis was carried out on F2 individuals homozygous for the Ds insertion. The sgt3118 mutant seedlings, named rpa-1, displayed several aberrant root hair phenotypes, including bulged (Fig. 7, A, C, D, and E), branched (Fig. 7, E, F, and H), and shorter root hair (Fig. 7A) compared with that of the Landsberg erecta (Ler) wild type (Fig. 7B). With PCR analysis, we identified the homozygous SALK_000767 line, named rpa-2, and the root hairs of rpa-2 also showed bulged (Fig. 7, I and J), branched (Fig. 7, K and L), and shorter root hair (Fig. 7I). Furthermore, the root hair phenotype of rpa-1 could be rescued by the RPA gene (Fig. 7M). This evidence indicated that the root hair phenotype is indeed caused by the loss of function of the RPA gene.
Figure 7.
The root hair phenotype of rpa-1 and rpa-2. The rpa mutant root showing deformed root hairs in rpa-1 (A, C–H) and rpa-2 (I–L), compared to that of Ler wild type (B). The aberrant root hair phenotype in rpa-1 was complemented by the P35S:RPA gene (M). Scale bars, 20 μm.
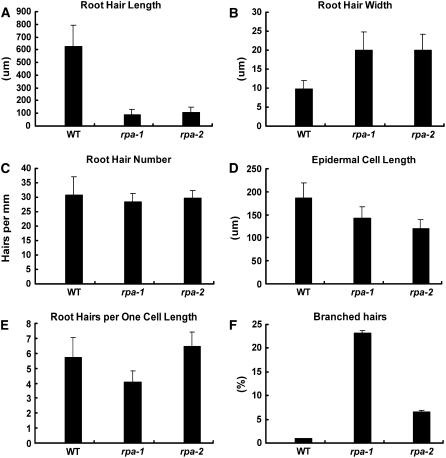
To quantify these differences, we measured root hair length and width, root hair number per millimeter of root length, epidermal cell length, and branched root hair percent in rpa-1 and rpa-2 lines and their wild-type counterparts (Fig. 8). Microscopic observation showed that rpa-1 and rpa-2 have shorter root hairs than the wild type, with only one-seventh the length of wild-type root hairs (approximately 628 μm; Fig. 8A), whereas the width of both rpa-1 and rpa-2 are 2 times that of wild-type hairs (9.75 μm; Fig. 8B). When the number of root hairs per millimeter were measured, rpa-1 and rpa-2 have less hairs compared to that of the Ler wild type (approximately 31; Fig. 8C). The length of the mutant epidermal cells is about three-fourths of wild type for rpa-1 (Fig. 8D) and two-thirds for rpa-2 (approximately 186.7 μm; Fig. 8D); when the root hair density was corrected for epidermal cell length, rpa-1 still has about three-fourths of the root hairs of wild type (Fig. 8E), whereas rpa-2 has slightly more root hairs per epidermal cell length compared to that of Ler wild type. In addition, rpa-1 has about 23% branched hairs and rpa-2 has 6.47% branched hairs, whereas Ler wild type only has <1% branched hairs (Fig. 8F). These data indicate that rpa-1 has a slightly more severe phenotype than rpa-2 because rpa-2 is a weak allele where trace amounts of RPA transcripts were detectable, but not in rpa-1 (Supplemental Fig. 2). Together, the mutant phenotype of rpa-1 and rpa-2 suggests that RPA plays a role in root hair polar growth.
Figure 8.
Quantitative analysis of root hair phenotypes. A, Mean length of 50 root hairs from 10 siblings for each genotype. B, Mean width of 50 root hairs from 10 siblings for each genotype. C, Mean number of root hairs/millimeter of root. Visible hairs were counted in a 1-mm section of mature root and only those observed from above were counted. D, Mean epidermal cell length (mm) obtained from 50 cells (both atrichoblasts and trichoblasts) from 10 siblings for each genotype. E, Number of root hairs per one cell length. The number of hairs/millimeter root length was multiplied by the corresponding epidermal cell length to give the mean number of hairs per unit cell length. F, Percentage of branched hairs obtained from 10 siblings for each genotype. WT, Arabidopsis ecotype Ler; rpa-1, sgt3118 mutant in Ler background; rpa-2, SALK_000767 mutant in Col background.
Pollen Tube Growth Is Slightly Affected in the rpa-1 Mutant
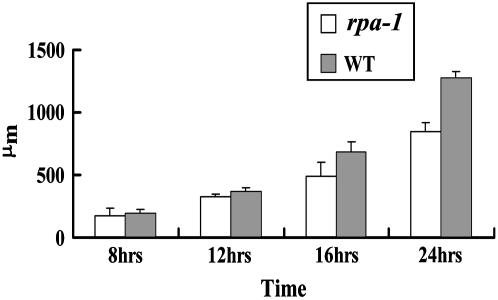
Because the loss of RPA function affected the polar growth of root hairs, we investigated whether it also affected the polar growth of pollen tubes in rpa-1. After 8-h germination, about 25.0% for rpa-1 mutant pollen and 26.8% for Ler wild-type pollen had germinated. As the incubation time reached 16 h, the pollen germination rate reached 72.2% for rpa-1 pollen grains and 76.5% for Ler wild-type pollen grains. These results suggested that the loss of RPA function had no effect on pollen germination. To further investigate whether pollen tube polar growth was affected, the length of germinated pollen tubes was measured microscopically. There was little difference between Ler wild type and rpa-1 in pollen tube length at 8 h after germination and both were about 200 μm in length (Fig. 9). As the incubation time increased, the difference in pollen tube length between Ler wild type and rpa-1 became obvious. After 16-h incubation, the pollen tubes of Ler wild type were more than 700 μm in length and that of rpa-1 was only about 500 μm, 200 μm shorter than that of wild-type pollen tubes (Fig. 9). When the incubation time was extended to 24 h, the rpa-1 pollen tubes were 500 μm shorter than that of Ler wild type. This suggested that the loss of RPA function indeed had an effect on pollen tube elongation.
Figure 9.
In vitro pollen germination assay. The pollen tube lengths of rpa-1 and Ler wild type were measured at different time points as indicated. Note the difference in pollen tube length becomes obvious after 12 h of growth.
Taken together, we concluded that the root hair and pollen tube phenotypes were caused by loss of function of the RPA gene, suggesting that RPA plays a role in root hair development and pollen tube elongation.
DISCUSSION
RPA, a Functional ARFGAP of ARF1 and U5, Plays a Role in Root Hair Development in Arabidopsis
In this article, we described the phenotypic and molecular characterization of a novel ARFGAP in Arabidopsis. RPA belongs to the class II ARFGAP family (Vernoud et al., 2003) and is homologous to mammalian ARFGAP1 and the yeast GLO3 and GCS1. Previous studies have shown that the mammalian ARFGAP1 is localized at the Golgi complexes and possesses GTPase-activating activity (Cukierman et al., 1995); GLO3 and GCS1 are two yeast ARFGAPs, which are also localized at Golgi complexes and mediate the retrograde vesicular transport from the Golgi network to the ER (Poon et al., 1996, 1999). In yeast, either the glo3Δ or the gcs1Δ single-mutant cell is viable, but the glo3Δ gcs1Δ double mutant is lethal; therefore, the GLO3 and GCS1 ARFGAPs provide an overlapping essential function for the vesicular transport between the Golgi and the ER (Poon et al., 1999). Because RPA complemented the temperature-sensitive gcs1 allele (gcs1–28) in the glo3Δ gcs1Δ double-mutant background, we conclude that RPA acts as a functional ARFGAP in yeast to allow the conditional glo3Δ gcs1Δ double mutant to grow at 37°C. Similarly, the mammalian ARFGAP1 has also been shown to provide sufficient ARFGAP activity by complementing the glo3Δ gcs1Δ temperature-sensitive mutation (Cukierman et al., 1995; Poon et al., 1999). RPA-EGFP colocalizes with the Golgi marker protein N-ST-mRFP, indicating that RPA is mainly localized at the Golgi apparatus. All of these data suggest that RPA is a functional ARFGAP that is involved in vesicle transport between the Golgi and ER complexes in Arabidopsis.
In Arabidopsis, 21 putative ARF and ARL GTPases and 15 ARFGAPs have been identified based on sequence homology (Vernoud et al., 2003). Little is known about the interactions between the ARF and ARFGAP protein in plants. Here we provided biochemical evidence that RPA activates ARF1 and U5 GTPase activity in vitro, not ARF3 and U2, although we could not exclude that RPA also activates other ARFs or ARLs because we did not check all 21 potential substrates. It is known that ARF1 in Arabidopsis is involved in the retrograde vesicular transport from Golgi to ER and plays a role in root hair development via the regulation of ROP2 subcellular localization and action (Pimpl et al., 2000; Lee et al., 2002; Xu and Scheres, 2005), whereas the function of U5 remains unknown. Taken together, we concluded that RPA plays a role in the vesicular transport between the Golgi and the ER through the activation of ARF1 GTPase activity that is essential for polar tip growth of root hairs.
Recently, it was shown that ARF1 plays a role in the subcellular localization of the auxin efflux carrier PIN2 in root cells (Xu and Scheres, 2005). Previous studies showed that another ARFGAP in Arabidopsis, VAN3, localizes in a subpopulation of the TGN and functions in a vein pattern formation by regulating auxin signaling via a TGN-mediated vesicle transport system (Koizumi et al., 2005). OsAGAP, an ARFGAP in rice (Oryza sativa), may be involved in the mediation of plant root development by regulating auxin level (Zhuang et al., 2005). Therefore, it is also possible that RPA functions in controlling polar growth of root hairs and roots via its regulation on ARF1 GTPase activity.
RPA May Have a Role in Male Gametophyte Competitiveness
Our data showed that the RPA gene is highly expressed in flowers. GUS reporter analysis indicated that RPA is expressed in pollen grains and pollen tubes. Nevertheless, the homozygous rpa mutant is completely fertile and its pollen grains have the same competitiveness as wild type. A simple explanation would be genetic redundancy due to the presence of a large family of functionally overlapping genes. In vitro pollen germination assays showed that the rpa pollen tubes grow slower than wild type. Because the difference in growth becomes significant only after 12 h of germination, it is longer than the time (about 5–7 h) required for pollen tubes to reach ovules in Arabidopsis pistils. It is therefore reasonable to explain the fertile phenotype. Such defects in pollen tube growth may not be detrimental to plants having short styles, such as Arabidopsis, but may be detrimental to plants having long styles or long distances between ovules and the site of pollen germination, such as maize (Zea mays). Indeed, mutation in ROP2 does affect male competitiveness in maize, where pollen tubes have to travel a long distance to reach the ovules (Arthur et al., 2003). Therefore, RPA might provide a competitive advantage for male gametophytes in plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotypes Ler and Columbia (Col) were grown on soil at 22°C ± 2°C in a greenhouse with a 16-h light/8-h dark cycle. Seeds were surface sterilized with 20% commercial bleach for 5 min, 70% ethanol for another 5 min, then rinsed four times with sterile water and germinated onto a Murashige and Skoog plate (Murashige and Skoog, 1962) with or without antibiotics; 50 mg L−1 kanamycin (Sigma-Aldrich) or 20 mg L−1 hygromycin (Roche) were supplemented for kanamycin or hygromycin selection, respectively. Transformation was done via Agrobacterium tumefaciens-mediated vacuum infiltration (Bechtold and Pelletier, 1998). Selection of transgenic plants and GUS staining were performed as described previously (Sundaresan et al., 1995). The SALK T-DNA insertion lines in Arabidopsis ecotype Col background were obtained from the Arabidopsis Biological Resource Center (Ohio State University).
In Vitro Pollen Germination Assay
Pollen grains were collected from freshly dehiscent anthers of Ler wild type and sgt3118 at the end of stage 12 of flower development (Smyth et al., 1990) and dabbed on slides that were covered with solid pollen germination medium containing 1 mm CaCl2, 1 mm H3BO4, 1 mm MgSO4·7H2O, 1 mm KNO3, 0.25 mm Suc, and 1% agarose (Fan et al., 2001). Slides were incubated at 25°C to 28°C with 100% moisture. Pollen germination ratio and pollen tube length were determined using a Zeiss skop II microscope.
RT-PCR Analysis
Total RNA was extracted from roots, stems, leaves, inflorescences, siliques, and 7-d-old seedlings using TRIzol reagent (Invitrogen). One microgram of total RNA was used as a template and reverse transcribed with alfalfa mosaic virus reverse transcriptase (TaKaRa) in the presence of oligo(dT) primers. One microliter of the RT product was then used as a template in subsequent PCR reactions with 95°C for 1 min, 30 cycles of 94°C for 30 s, annealing for 30 s according to specified temperature, 72°C for 90 s, and, finally, 72°C for 10 min. PCR products were analyzed with 1% agarose gel.
Molecular Cloning
For the construction of the Pro:RPA-GUS:Ter reporter gene, the 4.8-kb genomic fragment of the RPA was separated into three parts, amplified separately with primer pairs 3118P-5′/3118P-3′, 3118G-5′/3118G-3′, and 3118T-5′/3118T-3′ (Table I), then cloned into the binary vector pBI101 (CLONTECH) to give rise to the Pro:RPA-GUS:Ter fusion gene.
Table I.
List of primers
| Construct | Primer | Sequence |
|---|---|---|
| Pro:RPA-GUS:Ter construct | 3118P-5′ | 5′-GTAAGCTTCTTTTAGTTTTATTCTAGTCG-3′ |
| 3118P-3′ | 5′-CTGGATCCTGTAGGGATGTGAAACAATAT-3′ | |
| 3118G-5′ | 5′-GTCGGATCCATGGCGTCTGAGAATCTGAATG-3′ | |
| 3118G-3′ | 5′-GCAGGATCCCAAACCCATAAGCTGGAAG-3′ | |
| 3118T-5′ | 5′-GTCGAGCTCTCTGTTGCTTCCAGCTTATG-3′ | |
| 3118T-3′ | 5′-AGAGAGCTCAAATCTCAAATGACAACC-3′ | |
| RPA-EGFP construct | RPA-BamHI-5′ | 5′-GTCGGATCCATGGCGTCTGAGAATCTGAATG-3′ |
| RPA-KpnI-3′ | 5′-GCAGGTACCCAAACCCATAAGCTGGAAG-3′ | |
| RPA-128AA-KpnI-3′ | 5′-CTTGGTACCTCCTCAGCCTTGCTTTTTGC-3′ | |
| RPA-246AA-KpnI-3′ | 5′-CTTGGTACCCTGTTTTGAACATTGTCCGC-3′ | |
| RPA-316AA-KpnI-3′ | 5′-GTTGGTACCTTTCCAAAGTACTGAGCAGAGG-3′ | |
| RPA-79AA-NcoI-5′ | 5′-CAACCATGGACAACAACTCTGCTGATC-3′ | |
| RPA-79AA-NcoI-3′ | 5′-CAACCATGGAACCCATAAGCTGGAAGC-3′ | |
| His-RPA construct | RPA-EcoRI-5′ | 5′-GTGAATTCCATGGCGTCTGAGAATCTGAATG-3′ |
| RPA-XhoI-3′ | 5′-GTACTCGAGCCAAACCCATAAGCTGGAAGCA-3′ | |
| His-ARF construct | ARF1-BamHI-5′ | 5′-GAGGATCCATGCGTATTCTGATGGTTGGT-3′ |
| ARF1-XhoI-3′ | 5′-GATCTCGAGCTAAGCCTTGTTTGCGATGT-3′ | |
| U5-BamHI-5′ | 5′-GAGGACCATGAGAATTCTGATGGTTGGT-3′ | |
| U5-XhoI-3′ | 5′-GATCTCGAGCAGCAAATCAACTCCACCACG-3′ | |
| ARF3-BamHI-5′ | 5′-GAGGATCCGCTCGAATCCTCGTCCTCGGT-3′ | |
| ARF3-NotI-3′ | 5′-GAGCGGCCGCTTAGCCACTTCCCGACTTCA-3′ | |
| U2-BamHI-5′ | 5′-GAGGATCCATGCGTATCCTCATGGTGGGT-3′ | |
| U2-XhoI-3′ | 5′-GATCTCGAG CTAAGCCTTGTTTGCGATGT-3′ | |
| PGCS1-RPA construct | EcoRI-Gcs1-5′ | 5′-CTCGAATTCAGAAACATAAGATGGAATCC-3′ |
| Gcs1 Lig RPA | 5′-ACCCGCAGGCGTAAACTCAAAG-3′ | |
| RPA-BspE1-3′ | 5′-ATCTCCGGA TCAAACCCATAAGCTGGAAG-3′ |
For the complementation test of the rpa-1 mutant, the RPA ORF was amplified by PCR with gene-specific primers and cloned downstream of the 35S promoter in the binary vector pGD1301 to yield the pGD1301-RPACDS construct. At the same time, the 4.8-kb genomic sequence of the RPA gene was also cloned into pGD1301, giving rise to the pGD1301-RPA construct.
To produce proteins for biochemical assay, the RPA ORF was amplified by PCR with gene-specific primers RPA-EcoRI-5′/RPA-XhoI-3′ (Table I) and then subcloned into the pET28b (Novagen) vector between EcoRI and XhoI to generate His-tagged RPA. Similarly, several potential substrates of RPA, such as ARF1, ARF3, U5, and U2, were selected and His-tagged proteins were produced in Escherichia coli. Briefly, the truncated version of ARF1 (18–181 amino acids), ARF3 (18–182 amino acids), U5 (18–165 amino acids), and U2 (17–185 amino acids) were amplified by PCR with primer pairs: ARF1-BamHI-5′/ARF1-XhoI-3′, U5-BamHI-5′/U5-XhoI-3′, U2-BamHI-5′/U2-XhoI-3′ (Table I), respectively. The PCR products were subcloned into the pET28a expression vector between BamHI and XhoI; ARF3 (18–182 amino acids) was amplified with primer pair ARF3-BamHI-5′/ARF3-NotI-3′ (Table I) and cloned into the pET28a vector between BamHI and NotI. The His-RPA protein was purified by a nickel column under denaturing conditions and renatured by dialysis. The His-tagged ARF1, ARF3, U2, and U5 proteins were purified by nickel-nitrolotriacetic acid spin columns under native conditions according to manufacturer's recommendation (Qiagen).
Subcellular Localization of RPA
The ORF of RPA was obtained through RT-PCR with the gene-specific primer pair RPA-BamHI-5′/RPA-KpnI-3′ (Table I) and cloned into the pEGFP vector (CLONTECH) between the BamHI and KpnI sites, then the RPA-EGFP fragment was subcloned downstream of the 35S constitutive promoter into pCAMBIA1301 between BamHI and XbaI to give rise to P35S:RPA-EGFP. Similarly, RPA128-EGFP, RPA246-EGFP, RPA316-EGFP, and RPAC79-EGFP were constructed with gene-specific primers (Table I).
The plasmids P35S:RPA-EGFP and P35S:N-ST:mRFP were cobombarded into onion (Allium cepa) cells using the PDS-100/He Biolistik particle delivery system (Bio-Rad). The bombarded samples were incubated at 22°C for 24 h on Murashige and Skoog plates. Fluorescent signals were observed using the Zeiss LSM510 META laser-scanning microscope at the Center for Developmental Biology.
GTP-Binding Assay of ARFs
One microgram of gel-purified ARFs were spotted onto nitrocellulose membrane (Pall Gelman Science) and treated with hybridization solution (50 mm Tris-HCl, 5 mm MgCl2, 0.3% Tween 20, 0.5 mm EDTA, 5 mm dithiothreitol [DTT], and 0.3% bovine serum albumin, pH 7.5) for 30 min at 30°C and then transferred to new hybridization solution containing [α-32P]GTP (10−9 m) for another 15 min at 30°C. The membrane was washed with hybridization solution without bovine serum albumin three times and finally autoradiography was performed.
ARF GTPase-Activating Activity Assay
ARF GTPase-activating activity was detected according to Goldberg (1999) with slight modification. ARF1 and U5 (4 μg) were first loaded with [γ-32P]GTP (10−9 m) in HEPES buffer (20 mm, pH 7.5, 50 mm MgCl2, and 1 mm DTT), incubated for 30 min at 30°C, and RPA was then added to a final volume of 60 μL; the incubation time was extended for another 30 min at 30°C. Twenty microliters of the mixture were transferred onto nitrocellulose membrane and washed three times with washing buffer (20 mm HEPES, pH 7.5, 5 mm MgCl2, 1 mm DTT, 0.5 mm EDTA, and 0.3% Tween 20), for 5 min each. Finally, γ-32P retained on the membrane was counted by liquid scintillation.
Complementation of glo3Δ gcs1Δ Mutant with RPA
The temperature-sensitive glo3Δ gcs1Δ (Saccharomyces cerevisiae) mutant strain PPY147.28.2a and the plasmid derived from YEP352 used for the transformation were kindly provided by Professor Gerald C. Johnston (Dalhousie University). The GCS1 gene was replaced by RPA (residues 13–395) according to Poon et al. (1999) using the primer combination EcoRI-Gcs1-5′/Gcs1 Lig RPA/ RPA-BspE1-3′. The resulting gene product consists of the first 12 amino acids of GCS1 protein and amino acids 13 to 395 of RPA protein. For the recovery experiment, plasmid DNA was isolated from a single yeast (Saccharomyces cerevisiae) colony. Yeast transformation was performed according to the protocol of CLONTECH.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AT2G35210.
Supplementary Material
Acknowledgments
We thank Professor G.C. Johnston (Dalhousie University, Halifax, Nova Scotia, Canada) for kindly providing the glo3Δgcs1Δ strain and the YEP352 vector, and Professor Kang Chong (Institute of Botany, Chinese Academy of Sciences, China) for support with experimental materials; Professor Inhwan Hwang (Pohang University of Science and Technology, Korea) for providing us with P35S:N-ST-mRFP; and Dr. Dongqiao Shi (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing) for technical assistance in confocal microscopy. We also thank the Arabidopsis Biological Resource Center, Ohio State University, for providing the T-DNA insertion lines.
This work was supported by the BAI REN JI HUA program, Chinese Academy of Sciences, and the National Science Foundation of China (grant no. 30425030).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Wei-Cai Yang (wcyang@genetics.ac.cn).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.077818.
References
- Arthur KM, Vejlupkova Z, Meeley RB, Fowler JE (2003) Maize ROP2 GTPase provides a competitive advantage to the male gametophyte. Genetics 165: 2137–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Chavrier P, Goud B (1999) The role of ARF and Rab GTPases in membrane transport. Curr Opin Cell Biol 11: 466–475 [DOI] [PubMed] [Google Scholar]
- Cheung AY, Chen CYH, Glaven RH, Graaf BHJ, Vidali L, Hepler PK, Wu HM (2002) Rab2 GTPase regulates vesicle trafficking between the endoplasmic reticulum and the Golgi bodies and is important to pollen tube growth. Plant Cell 14: 945–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukierman E, Huber I, Rotman M, Cassel D (1995) The ARF1 GTPase activating protein: zinc finger motif and Golgi complex localization. Science 270: 1999–2002 [DOI] [PubMed] [Google Scholar]
- Dascher C, Balch WE (1994) Dominant inhibitory mutants of ARF1 block endoplasmic reticulum to Golgi transport and trigger disassembly of the Golgi apparatus. J Biol Chem 269: 1437–1448 [PubMed] [Google Scholar]
- Der CJ, Balch WE (2000) GTPase traffic control. Nature 405: 800–804 [DOI] [PubMed] [Google Scholar]
- Ding M, Vitale N, Tsai SC, Adamik R, Moss J, Vaughan M (1996) Characterization of a GTPase-activating protein that stimulates GTP hydrolysis by both ADP-ribosylation factor (ARF) and ARF-like proteins. J Biol Chem 271: 24005–24009 [DOI] [PubMed] [Google Scholar]
- Fan LM, Wang YF, Wang H, Wu WH (2001) In vitro Arabidopsis pollen germination and characterization of the inward potassium currents in Arabidopsis pollen grain protoplasts. J Exp Bot 52: 1603–1614 [PubMed] [Google Scholar]
- Goldberg J (1999) Structural and functional analysis of the ARF1-ARF1GAP complex reveals a role for coatomer in GTP hydrolysis. Cell 96: 893–902 [DOI] [PubMed] [Google Scholar]
- Gu Y, Wang Z, Yang Z (2004) ROP/RAC GTPase: an old new master regulator for plant signaling. Curr Opin Plant Biol 7: 527–536 [DOI] [PubMed] [Google Scholar]
- Jones MA, Shen JJ, Fu Y, Li H, Yang Z, Grierson CS (2002) The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14: 763–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Gilman AG (1986) The protein cofactor necessary for ADP ribosylation of Gs by cholera toxin is itself a GTP binding protein. J Biol Chem 261: 7906–7911 [PubMed] [Google Scholar]
- Koizumi K, Naramoto S, Sawa S, Yahara N, Ueda T, Nakano A, Sugiyama M, Fukuda H (2005) VAN3 ARF-GAP-mediated vesicle transport is involved in leaf vascular network formation. Development 132: 1699–1711 [DOI] [PubMed] [Google Scholar]
- Lee MH, Min MK, Lee YJ, Jin JB, Shin DH, Kim DH, Lee KH, Hwang I (2002) ADP-ribosylation factor 1 of Arabidopsis plays a critical role in intracellular trafficking and maintenance of endoplasmic reticulum morphology in Arabidopsis. Plant Physiol 129: 1507–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Novick P, Zerial M (1997) The diversity of Rab proteins in vesicle transport. Curr Opin Cell Biol 9: 496–504 [DOI] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, Ye D, Yang W-C, Kumaran M, Sundaresan V (1999) Analysis of flanking sequences from Dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 11: 2263–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris S, Beraud-Dufour S, Robineau S, Bigay J, Antonny B, Chabre M, Chardin P (1997) Role of protein-phospholipid interaction in the activation of ARF1 by the guanine nucleotide exchange factor. J Biol Chem 272: 22221–22226 [DOI] [PubMed] [Google Scholar]
- Pimpl P, Movafeghi A, Coughlan S, Denecke J, Jillmer S, Robinson DG (2000) In situ localization and in vitro induction of plant COPI-coated vesicles. Plant Cell 12: 2219–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon PP, Cassel D, Spang A, Rotman M, Pick E, Singer RA, Johnston GC (1999) Retrograde transport from the yeast Golgi is mediated by two ARF1GAP proteins with overlapping function. EMBO J 18: 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon PP, Wang X, Rotman M, Huber I, Cukierman E, Cassel D, Singert RA, Johnston GC (1996) Saccharomyces cerevisiae Gcs1 is an ADP-ribosylation factor GTPase-activating protein. Proc Natl Acad Sci USA 93: 10074–10077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss ML, Serna J, Falbel TG, Bednarek SY, Nielsen E (2004) The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. Plant Cell 16: 1589–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikke BJ, Karin LA, Gitte IF, Henrik BN, Jim H, Hans MJ, John M, Karen S (2000) Promiscuous and specific phospholipids binding by domains in ZAC, a membrane-associated Arabidopsis protein with an ARF1 GAP zinc finger and a C2 domain. Plant Mol Biol 44: 799–814 [DOI] [PubMed] [Google Scholar]
- Schimmoller F, Simon I, Pfeffer SR (1998) Rab GTPases, directors of vesicle docking. J Biol Chem 273: 22161–22164 [DOI] [PubMed] [Google Scholar]
- Schnepf E (1986) Cellular polarity. Annu Rev Plant Physiol 37: 23–47 [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, Gälweiler L, Palme K, Jürgens G (1999) Coordinated polar localization of auxin efflux carrier PINI by GNOM ARF GEF. Science 286: 316–318 [DOI] [PubMed] [Google Scholar]
- Sundaresan V, Springer PS, Volpe T, Haward S, Jones JDG, Dean C, Ma H, Martienssen RA (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Ueda T, Yahara N, Nakano A (2002) ARF1 GTPase plays roles in the protein traffic between the endoplasmic reticulum and the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J 31: 499–515 [DOI] [PubMed] [Google Scholar]
- Vernoud V, Horton AC, Yang Z, Nielsen E (2003) Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol 131: 1191–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Scheres B (2005) Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR1 function in epidermal cell polarity. Plant Cell 17: 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M, McBride H (2001) Rab proteins as membrane organizers. Nat Cell Biol 2: 107–117 [DOI] [PubMed] [Google Scholar]
- Zheng Z-L, Yang Z (2000) The Rop GTPase: an emerging signaling switch in plants. Plant Mol Biol 44: 1–9 [DOI] [PubMed] [Google Scholar]
- Zhuang X, Xu Y, Chong K, Lan L, Xue Y, Xu Z (2005) OsAGAP, an ArfGAP from rice, regulates root development mediated by auxin in Arabidopsis. Plant Cell Environ 28: 147–156 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.