Abstract
Background
The USA300 strain of community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) can cause severe infection and is increasingly recognized as a cause of community outbreaks. In 2004, an outbreak was identified in the Calgary Health Region (CHR).
Methods
MRSA isolates were identified with standard methods at a central regional laboratory and typed via pulsed-field gel electrophoresis (PFGE). Isolates were tested by PCR for mecA, Panton–Valentine leukocidin (PVL), SCCmec, and spa genes. Cases were defined as such if a clinical isolate of the USA300 strain was noted between January 1 and September 30, 2004, and the patient had lived or traveled in CHR within 2 years before symptom onset. Demographic, clinical and risk data on all such cases were collected from several sources for statistical analysis. A case was defined as high-risk if the patient had a history of drug use, homelessness or incarceration.
Results
Of 40 isolates with the USA300 PFGE pattern, all tested positive for PVL, SCCmec type IVa and spa type 008. Almost all infections (39/40, 98%) involved skin and soft tissues, except for 1 death from necrotizing hemorrhagic pneumonia; a notable proportion (38%) required hospital admission or intravenous antimicrobial therapy. The outbreak centred on the high-risk population in CHR (70%; risk ratio 169.4, 95% confidence interval 86.1–333.0).
Interpretation
People with histories of illicit drug use, homelessness or recent incarceration were at highest risk for infection with CA-MRSA. The emergence and spread of this virulent strain has important implications for treatment and public health in Canada.
In 2004, an investigation of an outbreak of community-acquired (CA) methicillin-resistant Staphylococcus aureus (MRSA) infections caused by the USA300 strain in the Calgary Health Region identified that those at highest risk had histories of illicit drug use, homelessness or recent incarceration.
The USA300 strain of methicillin-resistant Staphylococcus aureus (MRSA) has been increasingly associated with multiple outbreaks in the United States involving prison inmates, men who have sex with men, military recruits and, most recently, professional athletes.1–4 This strain possesses the small mobile staphylococcal cassette chromosome (SCC) mec type-IV genetic element, which contains the mecA resistance gene, with or without other antibiotic resistance determinants.5 This element is more easily transferred to other strains of S. aureus than the larger SCCmec elements (types I–III) prevalent in health care–acquired MRSA strains.5 USA300 strains may also possess the Panton–Valentine leukocidin determinant (PVL), a virulence factor associated with primary skin infections and pneumonia.6 The emergence of these CA-MRSA strains and their outbreak potential is thought to pose a threat to public health.1,3,4
In May through July 2004, a physician working at Corrections Facility A, a local corrections facility in Calgary, Alberta, noted an increased number of MRSA-positive skin infections among recently admitted inmates. A similar observation was made by physicians working in microbiology laboratories and hospitals in the Calgary Health Region (CHR). The cause of the infections was determined to be the USA300 strain of CA-MRSA (labelled CMRSA10 under Canadian nomenclature). We describe the results of our outbreak investigation demonstrating the introduction and dissemination of the USA300 strain into a marginalized population in the CHR, and describe hypotheses for its transmission within this population.
Methods
In the laboratory, isolates of MRSA were identified on mannitol salt agar screen plates supplemented with oxacillin (6 mg/L), confirmed by detection of the mecA and nuc genes via a polymerase chain reaction (PCR) assay.7 Susceptibilty testing was done with VITEK (bioMerieux, Inc. Haselwood, Mo.) or Kirby Bauer methods, plus a D-test for clindamycin susceptibility.8 Typing was done with pulsed-field gel electrophoresis (PFGE) after DNA extraction and digestion with Sma1 in a standard protocol.9 All isolates were further tested to confirm methicillin resistance and for the presence of PVL with a triplex PCR assay, which simultaneously detects lukS-PV, lukF-PV and mecA genes.10 The MRSA isolates were typed for SCCmec with a multiplex PCR SCCmec typing assay into subtypes I, II, III, IVa, IVb, IVc, IVd and V.11 Isolates were typed for spa (a polymorphism of the S. aureus protein-A gene),12,13 and a representative selection of isolates was further tested with multilocus sequence typing (MLST).14 The identification of MRSA isolates matching the USA300 strain was based on the similarity of PFGE patterns to control strains provided by the National Microbiology Laboratory of the Public Health Agency of Canada.
For case findings and definitions, we used laboratory reports provided by the regional laboratory of USA300 MRSA isolates to identify potential cases; in those with more than one positive isolate, the first report was used. This laboratory processes all specimens submitted from community physicians, clinics, hospitals and long-term care facilities in the CHR. A study case was defined as a patient with a positive isolate for the USA300 strain of MRSA between Jan. 1 and Sept. 30, 2004; a clinical infection; and a history of residence in or travel to the CHR during the 2 years before symptom onset. We used a history of one of the following during the 2 years before symptom onset to assign patients a high-risk status: illicit drug use (i.e., heroin, cocaine, crack cocaine, or abuse of morphine or other prescription opiates); incarceration in Alberta anytime since Jan. 1, 2002; or homelessness (having no fixed address, or staying in homeless shelters). Patients without any such history were identified as low-risk.
For demographic, clinical and risk information, we used chart reviews of medical records at hospitals, primary care clinics and a local corrections facility; an electronic search of the provincial Corrections Services admission database, HIV and hepatitis C registries, and hospital admission databases; and interviews with patients and their primary care providers. The information collected on potential risk factors included the presence of comorbid disease and histories of incarceration, use of group-living community facilities (detoxification centres or homeless shelters), travel, drug-use behaviours and types of drugs used, wound-care behaviours and previous hospital admissions.
We used EpiData version 3.0 (The EpiData Association, Denmark) to collect data, which was then analyzed with EpiINFO6 version 6.04 (US Centers for Disease Control and Prevention) and SPSS version 12.0.1. To compare medians, we used the Wilcoxon rank–sum test; to compare proportions, the χ2 test or Fisher's exact test, as appropriate.
We calculated the cumulative incidence of USA300 MRSA infection using CHR population estimates from Alberta Registry population data (from the Calgary Health Region internal Web site; accessed 2005 Aug. 10): 15 540 for the high-risk population (6000 homeless persons, 3540 injection drug users and 6000 individuals admitted to the local corrections facility at least once per year) and 1 127 825 for the low-risk population.15 Relative risks of infection were calculated with 95% confidence intervals (CIs).
To identify people with USA300 infections that were potentially health care–acquired, we used criteria from the Canadian Nosocomial Infections Surveillance Program (i.e., patients with infections that developed ≥ 3 days after admission to hospital, or who stayed in a hospital or resided in a long-term care facility at any time during the 12 months before symptom onset).16
Results
Laboratory results
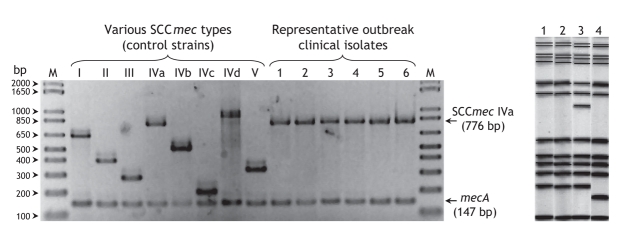
Between Jan. 1 and Sept. 30, 2004, we identified 42 patients with laboratory isolates of the USA300 strain of MRSA. (In comparison, between Jan. 1, 2002 and Dec. 31, 2003, only 7 individuals with this strain were identified.) Antimicrobial susceptibility profiles revealed the strains were uniformly resistant to oxacillin, erythromycin and ciprofloxacin, but susceptible to gentamicin, clindamycin, trimethoprim–sulfamethoxazole, tetracycline and vancomycin. None of the isolates exhibited inducible clindamycin resistance on the D-test. All 42 isolates tested positive for the PVL determinant, carried the SCCmec type-IVa genetic element (Fig. 1, left panel) and were of the same spa type. Selected isolates were also determined to have identical MLST types (sequence type 8). PFGE analysis (Fig. 1, right panel) divided isolates into 3 patterns, all belonging to a single clonal group identical to USA300.17
Fig. 1: Typing results for outbreak-related clinical isolates of methicillin-resistant Staphylococcus aureus (MRSA) in the Calgary Health Region (CHR). Left panel: Staphylococcal cassette chromosome mec (SCCmec) typing results. Lanes 1–6, representative outbreak-associated clinical isolates; lane M, molecular size markers, 1 Kb Plus DNA Ladder (Invitrogen). Control strains used: types I (NCTC10442), II (N315), III (85/ 2082), IVa (CA05), IVb (8/6–3P), IVc (MR108), IVd (JCSC 4469) and V (WIS [WBG8318]-JCSC3624), corresponding to distinct amplicon sizes of 613, 398, 280, 776, 493, 200, 881 and 325 bp, respectively. In addition, a 147-bp mecA gene (methicillin-resistant determinant) amplicon was present in all strains. Note: bp = base pairs. Right panel: Pulsed-field gel electrophoresis patterns of USA 300 strains of MRSA circulating in the CHR between Jan. 1 and Sept. 30, 2004. Lane 1, pattern A (31 of 40 cases, 78%); lane 2, USA300 reference strain pattern; lane 3, pattern B (7 cases, 18%); lane 4, pattern C (2 cases, 5%).
Epidemiologic results
Of these 42 isolates, 40 met the case definition (95%; Fig. 2. Excluded isolates were from screening swabs or CHR nonresidents). Twenty-eight of these 40 cases (70%) had a history of illicit drug use, homelessness and/or incarceration (i.e., were at high risk). Interviews were less likely to occur with people at high (6/28, 21%) than at low risk (11/12, 92%, p < 0.001). The cumulative incidence of USA300 infection among high-risk cases (240.2 per 100 000 population) was found to be higher than that of low-risk cases (1.4 per 100 000). The relative risk of infection in high-risk compared with low-risk cases was 169.4 (95% CI 86.1–333.0).
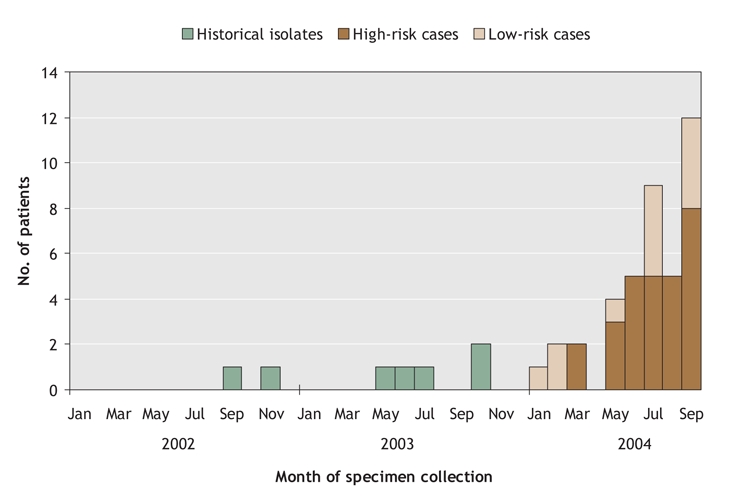
Fig. 2: Number of cases that furnished the USA300 strain of methicillin-resistant Staphylococcus aureus (MRSA) in the Calgary Health Region by month of specimen collection, from Jan. 1, 2004 through Sept. 30, 2004 (n = 40; 7 historical isolates were included for comparison). A further 310 infections were identified through ongoing surveillance up to Dec. 31, 2005 (8–34 cases per month).
Characteristics of these 40 cases are presented in Table 1. Almost all (39/40, 98%) were treated for skin and soft-tissue infections (SSTI) of varying severity. One person (3%) presented with rapidly progressive hemorrhagic necrotizing pneumonia, and died from this infection. Patients classified as being at high risk were more likely to have hepatitis C infection (14/28 [50%] v. 0/12, p < 0.05).
Table 1
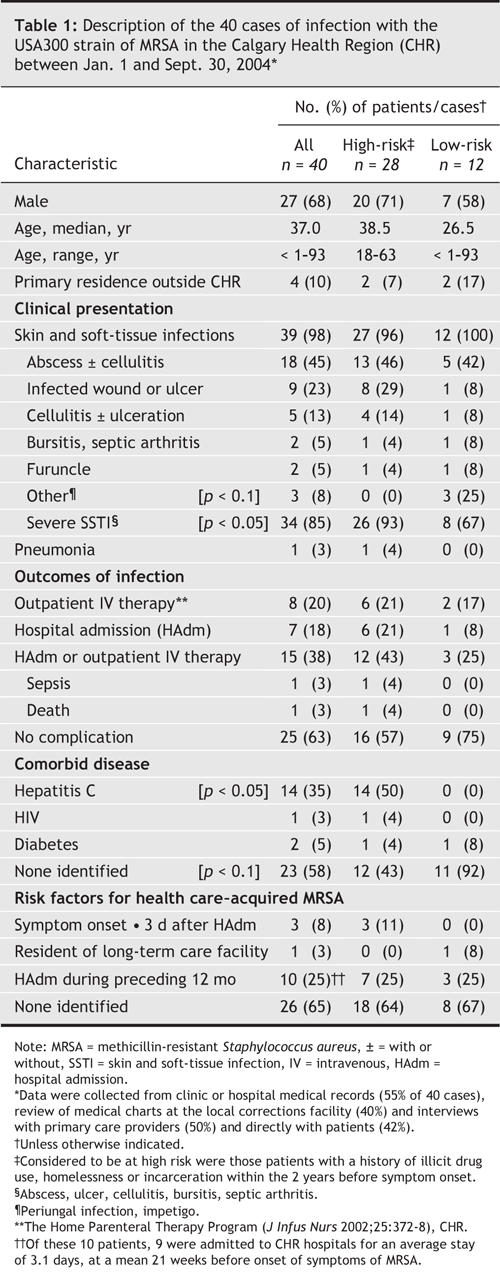
Fourteen (35%) of the 40 patients with USA300 MRSA infections met criteria for their case be considered a potential health care–acquired infection. Three of these (7.5%) had onset of infection ≥ 3 days after admission to hospital (range 6–44 d). Although 10 (25%) had been admitted to a hospital during the 12 months before the onset of infection, most of these patients had short hospital stays or were in units at low risk of MRSA infection; of these, one had an infection appear 10 days after discharge from a surgical ward adjacent to a ward where another patient was being treated for an infection with USA300.
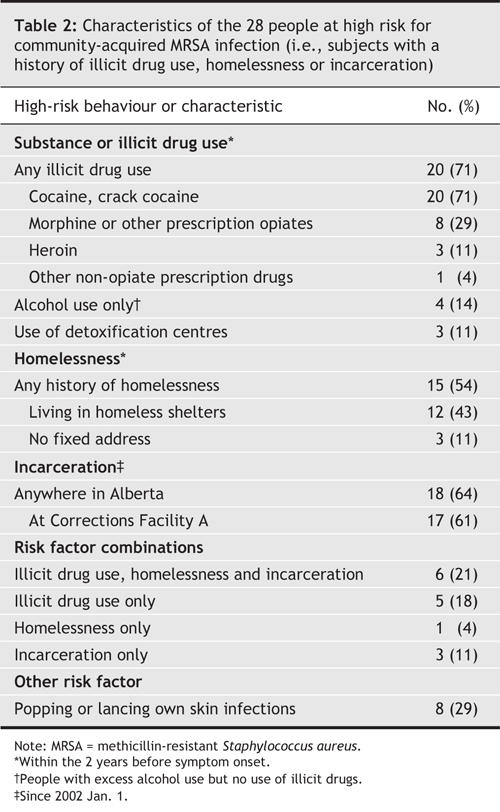
Additional characteristics of the 28 high-risk cases are presented in Table 2. Eighteen (64%) of these patients had a history of recent incarceration in the Alberta corrections system, the majority having passed through local Correction Facility A (range, 1–20 incarcerations/person). Furthermore, of the 18 patients who had been incarcerated (for a range of 5–11 weeks), 3 had onset of their infection during their incarceration.
Table 2
Interpretation
CA-MRSA infections are being reported with increasing frequency, particularly those with the USA300 strain.1,18,19 We report an outbreak of infections by the USA300 strain of MRSA, centred on an urban CHR population with histories of illicit drug use, homelessness or recent incarceration. We used a strain-based case definition and described the clinical features and hypotheses for transmission of a single virulent strain of CA-MRSA. A single central laboratory performed all microbiologic testing, which allowed the identification of all patients who were infected with the USA300 strain and a direct estimation of population incidence.
Because people in drug-using or homeless populations may tend to seek medical care at a later stage of infection,20 the CHR's burden of illness from this strain of MRSA may be even greater than we have documented. Our ability to interview high-risk patients was limited, which may also have led to underreportage of the characteristics of this group (e.g., the percentage who reported drug use).
The overall population prevalence of MRSA colonization in the CHR is probably low, in keeping with that recently reported in the United States.21 Outbreaks or a high prevalence of CA-MRSA infections have, however, been previously found in marginalized populations (prison inmates,4 drug-using and homeless people,22,23 and low-income communities24), where these strains may become endemic and difficult to control.25 Ongoing surveillance of CA-MRSA (and USA300) in the CHR has identified a further 310 infections up to the end of 2005 (8–34 cases/mo), with at least 73% occurring in the high-risk or transient population, which suggests that the strain has become endemic in this setting. Transmission within these populations has been attributed to factors that are environmental (e.g., crowded living conditions, such as in homeless shelters or jails), social (e.g., belonging to drug-using networks) and behavioural (e.g., poor personal hygiene).23,26–29 Lancing one's own skin infections, with the accompanying possibility of environmental contamination and transmission to close contacts, may also be a route of spread in the CHR high-risk population. Given that 71% of the patients whose cases met our inclusion criteria reported use of cocaine or crack cocaine, transmission may also be increased by the more frequent use of these drugs, drug use in settings such as crack houses, or sharing contaminated drugs or drug paraphernalia (e.g., needles, crack pipes).23,27,28 Although none of 7 cultures that we did on 2 samples of crack cocaine seized by police in Calgary grew CA-MRSA, 4 of 12 crack pipes that we tested had results positive for methicillin-sensitive S. aureus, which suggests that pipes may be a potential vector.
The majority of infections were community-acquired. Possible nosocomial acquisition may be explained by mitigating factors in each case, or by confounding from high rates of hospitalization.20,30 The infections identified in this outbreak were severe, with a high proportion (38%) of cases requiring intravenous antimicrobial therapy or hospital admission, and one death from a rapidly progressing necrotizing pneumonia.6,31 The severity of the infections and the potential consequences of delays in the initiation of appropriate treatment led to recommendations for empiric antibiotic treatment of skin and soft-tissue infections in the high-risk population in the CHR.
Of concern was the identification of 12 cases outside of the high-risk population. This suggests that the USA300 strain may be present in the general population, as has been recently documented.19
Control measures have included education of health care providers and the high-risk population, and recommendations for improved infection control in homeless shelters, group living settings and the local corrections facility. Despite these measures, this strain has now become endemic in this marginalized population in the CHR. We feel it is critical for Canadian physicians to be aware of our experience in the CHR with this strain of CA-MRSA which, with its considerable morbidity and mortality, quickly became endemic in a marginalized population and has the potential for inappropriate initial antibiotic selection, which may adversely affect outcomes.
@ See related article page 161
Acknowledgments
The authors would like to acknowledge the contribution of the following individuals: L. Ward, Infection Prevention and Control, Calgary Health Region; J. McClure, Calgary Laboratory Services; E. Smith and I. Grigoriu, Alberta Corrections; D. Gravel, Nosocomial and Occupational Infections, Public Health Agency of Canada; L. Panaro, Canadian Field Epidemiology Program; and K. Hiramatsu and T. Ito, Jutendo University, Tokyo, for provision of control strains for typing.
Footnotes
Published at www.cmaj.ca on June 27, 2006.
This article has been peer reviewed.
Contributors: Mark Gilbert was responsible for the design of the data collection tool and interviews, and data collation, epidemiologic analysis and preliminary interpretation for the outbreak investigation. Judy MacDonald and John Conly initiated, organized and coordinated the investigation, contributed to the study design and data analysis, and were responsible for the final interpretation of all aspects of the investigation. Jennifer Siushansian and Kevin Laupland contributed to the study design and epidemiologic analysis. Kunyan Zhang, Sameer Elsayed, Dan Gregson, Tom Louie, Marie Louie and Michael Mulvey contributed microbiologic or molecular typing data, molecular characterization of the strains and the interpretation of these data. John Gillespie, Virginia Wheeler, Diane Nielsen and Karen Hope were responsible for the identification of cases, acquisition of data and implementation of control measures. Agnes Honish and Gloria Keays contributed to the study design and epidemiologic analysis, and helped coordinate the investigation at the provincial level. Mark Gilbert, Judy MacDonald and John Conly drafted the initial manuscript. All of the authors revised the manuscript for critical content and approved the final version to be published.
Dr. Mark Gilbert is a Field Epidemiologist with the Canadian Field Epidemiology Program at the Public Health Agency of Canada. His interests include research in communicable disease epidemiology, and in health issues related to marginalized populations, particularly sexual minority and substance-using populations.
Competing interests: None declared.
Correspondence to: Dr. John Conly, Department of Medicine, Centre for Antimicrobial Resistance, University of Calgary, Calgary Health Region, and Calgary Laboratory Services, Calgary AB T2N 2T9; fax 403 944-1095; jconly@ucalgary.ca
REFERENCES
- 1.Chambers HF. Community-associated MRSA–resistance and virulence converge. N Engl J Med 2005;352:1485-7. [DOI] [PubMed]
- 2.Ellis MW, Hospenthal DR, Dooley DP, et al. Natural history of community-acquired methicillin-resistant Staphylococcus aureus colonization and infection in soldiers. Clin Infect Dis 2004;39:971-9. [DOI] [PubMed]
- 3.Kazakova SV, Hageman JC, Matava M, et al. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N Engl J Med 2005;352:468-75. [DOI] [PubMed]
- 4.Centers for Disease Control and Prevention (CDC). Methicillin-resistant Staphylococcus aureus infections in correctional facilities — Georgia, California, and Texas, 2001–2003. MMWR Morb Mortal Wkly Rep 2003;52:992-6. [PubMed]
- 5.Hiramatsu K, Cui L, Kuroda M, et al. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol 2001;9:486-93. [DOI] [PubMed]
- 6.Gillet Y, Issartel B, Vanhems P, et al. Association between Staphylococcus aureus strains carrying gene for Panton–Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 2002;359:753-9. [DOI] [PubMed]
- 7.Hussain Z, Stoakes L, Massey V, et al. Correlation of oxacillin MIC with mecA gene carriage in coagulase-negative staphylococci. J Clin Microbiol 2000;38:752-4. [DOI] [PMC free article] [PubMed]
- 8.Jorgensen JH, Crawford SA, McElmeel ML, et al. Detection of inducible clindamycin resistance of staphylococci in conjunction with performance of automated broth susceptibility testing. J Clin Microbiol 2004;42:1800-2. [DOI] [PMC free article] [PubMed]
- 9.Mulvey MR, Chui L, Ismail J, et al. Development of a Canadian standardized protocol for subtyping methicillin-resistant Staphylococcus aureus using pulsed-field gel electrophoresis. J Clin Microbiol 2001;39:3481-5. [DOI] [PMC free article] [PubMed]
- 10.Zhang K, McClure J-A, Lau V, et al. A new triplex PCR assay for detection of the staphylococcal virulence gene Panton–Valentine leukocidin (PVL) and simultaneous discrimination of MRSA (methicillin-resistant Staphylococcus aureus) From MSSA (methicillin-sensitive Staphylococcus aureus) [abstract C-148]. Abstracts of the 105th General Meeting of American Society for Microbiology; 2005 June 5–9, Atlanta. Washington: American Society for Microbiology; 2005.
- 11.Zhang K, McClure JA, Elsayed S, et al. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicilllin-resistant Staphylococcus aureus. J Clin Microbiol 2005;43:5026-33. [DOI] [PMC free article] [PubMed]
- 12.Harmsen D, Claus H, Witte W, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol 2003;41:5442-8. [DOI] [PMC free article] [PubMed]
- 13.Shopsin B, Gomez M, Montgomery SO, et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol 1999;37:3556-63. [DOI] [PMC free article] [PubMed]
- 14.Enright MC, Day NP, Davies CE, et al. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 2000;38:1008-15. [DOI] [PMC free article] [PubMed]
- 15.Guenter CD, Fonseca K, Nielsen DM, et al. HIV prevalence remains low among Calgary's needle exchange program participants. Can J Public Health 2000;91:129-32. [DOI] [PMC free article] [PubMed]
- 16.Simor AE, Ofner-Agostini M, Bryce E, et al. The evolution of methicillin-resistant Staphylococcus aureus in Canadian hospitals: 5 years of national surveillance. CMAJ 2001;165(1):21-6. [PMC free article] [PubMed]
- 17.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 1995;33:2233-9. [DOI] [PMC free article] [PubMed]
- 18.Mulvey M, MacDougall L, Cholin B, et al. Community-associated methicillin-resistant Staphylococcus aureus, Canada. Emerg Infect Dis 2005;11:844-50. [DOI] [PMC free article] [PubMed]
- 19.King MD, Humphrey BJ, Wang YF, et al. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann Intern Med 2006;144:309-17. [DOI] [PubMed]
- 20.Palepu A, Tyndall MW, Leon H, et al. Hospital utilization and costs in a cohort of injection drug users. CMAJ 2001;165:415-20. [PMC free article] [PubMed]
- 21.Graham PL III, Lin SX, Larson EL. A US population-based survey of Staphylococcus aureus colonization. Ann Intern Med 2006;144:318-25. [DOI] [PubMed]
- 22.Colombo C, Zink R, Ruef C. Persistence of a methicillin-resistant Staphylococcus aureus clone in a drug-use network. Clin Infect Dis 2003;37:990-1. [DOI] [PubMed]
- 23.Daly P, Bryce EA, Buxton J. Reply to Dr. Charlebois et al. Clin Infect Dis 2002;34:425-33). Clin Infect Dis 2002;35:1135. [DOI] [PubMed]
- 24.Fleisch F, Zbinden R, Vanoli C, et al. Epidemic spread of a single clone of methicillin-resistant Staphylococcus aureus among injection drug users in Zurich, Switzerland. Clin Infect Dis 2001;32:581-6. [DOI] [PubMed]
- 25.Charlebois ED, Bangsberg DR, Moss NJ, et al. Population-based community prevalence of methicillin-resistant Staphylococcus aureus in the urban poor of San Francisco. Clin Infect Dis 2002;34:425-33. [DOI] [PubMed]
- 26.Young DM, Harris HW, Charlebois ED, et al. An epidemic of methicillin-resistant Staphylococcus aureus soft tissue infections among medically underserved patients. Arch Surg 2004;139:947-51. [DOI] [PubMed]
- 27.Bassetti S, Battegay M. Staphylococcus aureus infections in injection drug users: risk factors and prevention strategies. Infection 2004;32:163-9. [DOI] [PubMed]
- 28.Quagliarello B, Cespedes C, Miller M, et al. Strains of Staphylococcus aureus obtained from drug-use networks are closely linked. Clin Infect Dis 2002;35:671-7. [DOI] [PubMed]
- 29.Turabelidze G, Lin M, Wolkoff B, et al. Personal hygiene and methicillin-resistant Staphylococcus aureus Infection. Emerg Infect Dis 2006;12:423-7. [DOI] [PMC free article] [PubMed]
- 30.Kushel MB, Vittinghoff E, Haas JS. Factors associated with the health care utilization of homeless persons. JAMA 2001;285:200-6. [DOI] [PubMed]
- 31.Francis JS, Doherty MC, Lopatin U, et al. Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton–Valentine leukocidin genes. Clin Infect Dis 2005;40:100-7. [DOI] [PubMed]