Abstract
Zidovudine resistance (ZDV-R) is associated with classic genotypic changes at codons 41, 67, 70, 210, 215, and 219 of the human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) gene as well as with the multinucleoside resistance (MNR) complexes (Q151M MNR complex; 6-bp insertion/A62V complex). In addition, enhanced resistance to ZDV in the context of the classic ZDV mutations plus the M184V mutation has been associated with additional mutations at positions 208, 211, 214, and 333. In this study we investigated phenotypic ZDV-R determined by a recombinant virus assay (Antivirogram; Virco) in 223 clinical samples in relation to the above genotypic changes. 150 out of 223 clinical samples had the M184V mutation. Phenotypic ZDV-R ranged from 0.3- to 5,338-fold. Sixteen samples (15 with high ZDV-R ranging from 90- to 3,571-fold) contained MNR-associated patterns. Analysis of classic mutational patterns broadly demonstrated increasing ZDV-R with increasing number of ZDV mutations. A comparable correlation was obtained when ZDV-R was analyzed only relative to the T215Y/F mutation. Site-directed mutagenesis experiments investigating the influence of the additional mutations H208Y, R211K, and L214F on ZDV-R resulted in a 7.4- or 21-fold increase in ZDV-R when the R211K/L214F or H208Y/R211K/L214F mutations, respectively, were added to a highly ZDV-R virus. In the clinical sample data set we analyzed, the combination of R211K/L214F appeared most frequently. The H208Y change was detected only in highly ZDV-R viruses, whereas the G333E/D change was distributed equally. All changes were independent of the M184V mutation. A 2.4- or 8-fold increase in ZDV-R was observed in the clinical samples with high ZDV-R containing the R211K/L214F or H208Y/R211K/L214F mutations, respectively. We have shown that the combination of the additional mutations H208Y, R211K, and L214F in HIV-1 RT may influence ZDV-R and should be considered when assessing ZDV-R.
Zidovudine (ZDV) (3′-azido-3′-deoxythymidine) was the first nucleoside analogue used in the antiretroviral therapy of human immunodeficiency virus type 1 (HIV-1)-infected patients (5). After long-term treatment with ZDV monotherapy, virus variants with reduced sensitivity to ZDV were isolated (17). Point mutations in the reverse transcriptase (RT) gene of HIV-1 at codon 41 (11); codons 67, 70, 215, and 219 (16); and codon 210 (6, 9) were identified in clinical isolates and correlated with ZDV resistance (ZDV-R) by site-directed mutagenesis (SDM) studies. In clinical isolates from individuals treated with ZDV, these mutations appear in a distinct order and result in various levels of ZDV-R (13).
A mutation at codon 184 (1, 26) in which methionine is replaced by valine (or transiently isoleucine) is selected by lamivudine (3TC) (l-2′,3′-dideoxy-3′-thiacytidine) and leads to high-level 3TC resistance and to resensitization to ZDV in a genotypically ZDV-R background (1, 26). These are the standard mutations currently included in assessment of ZDV-R.
Combination therapy with ZDV and 3TC can lead to virus strains dually resistant to both drugs (21). Additional mutations at positions 208, 211, and 214 (S. D. Kemp and S. Bloor for the FASP Resistance Study Group, Prog. Abstr. Int. Workshop HIV Drug Resist. Treat. Strategies Erad. 1997, abstr. 11, 1997) and position 333 (12; Kemp and Bloor, Prog. Abstr. Int. Workshop HIV Drug Resist. Treat. Strategies Erad. 1997) were identified as playing a role in this dual resistance. The change at position 208 from histidine to tyrosine has also been reported to occur during treatment with foscarnet (20).
Treatment with combinations of nucleoside reverse transcriptase inhibitors (NRTIs) may result in the selection of other genotypic patterns associated with cross-resistance within this drug class. These include the Q151M multinucleoside resistance (MNR) complex with changes at codons 62, 75, 77, 116, and 151 (10, 23, 24) and a 6-bp insertion between amino acids 68 and 70 observed by a number of groups (3, 22; J. M. Cherrington, P. D. Lamy, N. A. Margot, and A. S. Mulato, abstract from the Eleventh International Conference on Antiviral Research 1998, Antivir. Res. 37:A41, 1998; L. Ross, M. Johnson, N. Graham, M. Shaefer, M. Griswold, and M. St. Clair, Prog. Abstr. 5th Conf. Retrovir. Opportun. Infect., abstr. 679, p. 207, 1998; M. Stürmer, B. Morgenstern, B. Nolte, J. Braner, A. Berger, S. Bader, H. W. Doerr, S. Staszewski, V. Miller, B. A. Larder, S. D. Kemp, and S. Bloor, Abstr. 6th Eur. Conf. Clin. Aspects Treat. HIV Infect., abstr. 429, p. 67-68, 1997) that has recently been associated with an MNR phenotype in combination with the classic ZDV mutations and also a mutation at codon 62 (A62V) (4, 18, 28).
At present, estimating the level of phenotypic ZDV-R based on genotype is complex. In this analysis we investigated the influence of the classic ZDV mutation patterns and the role of the additional mutations 208, 211, 214, and 333 on phenotypic ZDV-R.
MATERIALS AND METHODS
Sample preparation.
EDTA plasma was stored frozen at −80°C until use.
SDM experiments.
SDM was carried out as described previously (8). Briefly, mutations were generated in the RT gene of HXB2, a wild-type laboratory HIV-1 strain, with the QuikChange SDM kit (Stratagene Cloning Systems, La Jolla, Calif.).
Determination of phenotypic resistance.
Phenotypic drug resistance was tested using the Antivirogram as previously described (7). Phenotypic resistance was expressed as relative increase in the 50% inhibitory concentration (IC50) compared to a wild-type control included in each experiment.
Sequence determination.
RT genotypes were determined as previously described (14) or as follows. Viral RNA was extracted from patients' plasma with a QIAamp viral RNA kit (Qiagen GmbH, Hilden, Germany) according to the user's manual with slight modifications. The QIAamp spin columns containing 30 μl of preheated diethyl pyrocarbonate-treated water were incubated at 80°C for 5 min before centrifugation. The complete RNA from 10 μl of eluate was reverse transcribed using 50 U of murine leukemia virus reverse transcriptase (GeneAmp RNA PCR Kit; Perkin-Elmer, Norwalk, Conn.). The reaction mixture (20 μl) contained 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 5 mM MgCl2, a 1 mM concentration of each deoxynucleoside triphosphate (dNTP), 0.75 μM antisense primer RSB03 (TCTTGGGCCTTATCTATTCCAT [22]), and 20 U of RNase inhibitor and was incubated 30 min at 42°C and 5 min at 99°C. Afterwards, this was added to 80 μl of first-round PCR mix containing the following final concentrations: 50 mM OptiPerform-KOH pH 9.2 (InViTek, Berlin, Germany), 16 mM (NH4)2SO4, 0.01% Tween 20, 2.5 mM MgCl2, a 0.25 mM concentration of each dNTP, 0.2 μM concentrations of primers RSB05 (AATTGGAAGAAATCTGTTGACTCAG [22]) and RSB03, and 0.04 U of CombiPol (InViTek). The samples were incubated for 2 min at 94°C and then underwent 35 cycles with 94°C for 30 s, 59°C for 30 s, and 68°C for 3 min, with a final extension step at 68°C for 10 min. Five microliters of the first-round PCR product were added to 95 μl of second-round PCR mix. The inner primers RSB1(UP) (GTAAAACGACGGCCAGTGAAGAATTCTGTTGACTCAGATTGG [22]) and RSB1(REV) (CAGGAAACAGCTATGACTCTATGCTGCCCTATTTCTAA [22]) amplify a 671-bp fragment of RT including amino acids 1 to 192; RSB2(UP) (GTAAAACGACGGCCAGTATGACAAAAATCTTAGAGC [22]) and RSB2(REV) (CAGGAAACAGCTATGACCCCATGTTTCCTTTTGTATG [22]) amplify a 739-bp fragment including amino acids 171 to 391. Both pairs contain the M13 universal primer sequence [RSB1(UP) and RSB2(UP) (underlined)] or the M13 reverse primer sequence [RSB1(REV) and RSB2(REV) (underlined)]. The second-round PCR mix contained 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 2.5 mM MgCl2, a 0.2 mM concentration of each dNTP, 0.125 μM concentrations of primers RSB1(UP) and RSB1(REV) or 0.15 μM concentrations of primers RSB2(UP) and RSB2(REV), and 25 mU of Taq polymerase (Boehringer, Mannheim, Germany). Cycle conditions for both pairs were 95°C for 3 min and then 35 cycles with 90°C for 30 s, 56°C for 30 s, and 72°C for 1 min, with a final extension step of 72°C for 10 min. Five microliters of second-round PCR product were purified using the reagent pack system (Amersham-Pharmacia Biotech, Freiburg, Germany) as stated in the user manual. The sequencing reaction was performed using the thermosequenase fluorescence-labeled primer cycle sequencing kit (Amersham-Pharmacia Biotech) with 1.5 pmol of Cy5-labeled M13 UP or REV, respectively, and 1.7 μl of purified DNA for each nucleotide. The samples were incubated at 95°C for 3 min, followed by 25 cycles with 95°C for 30 s and 65°C for 30 s. After adding the stop solution the samples were denatured at 94°C for 2 min and analyzed using the automated sequencer ALF Express (Amersham-Pharmacia Biotech) with a ReproGel long-read gel (Amersham-Pharmacia Biotech). All cycling steps were done using a Perkin-Elmer 9600 thermocycler.
RESULTS
Sample overview.
A total of 223 samples from 172 patients attending our outpatient clinic were analyzed. These patients had received various antiretroviral treatment regimens, including non-NRTIs and protease inhibitors, often associated with a history of therapy failure. The median duration of ZDV treatment was 134 weeks (n = 202; range, 5 to 448 weeks), and the median duration of 3TC treatment was 56 weeks (n = 181; range, 2 to 147 weeks). The patient population was in a relatively advanced stage of HIV infection, with a median CD4 count of 105 cells/mm3 (n = 180; range 1 to 1,300 cells/mm3) and a median viral load of 5.21 log10 HIV-1 RNA copies/ml (n = 165; range 2.62 to 6.70 log10 HIV-1 RNA copies/ml). According to the Centers for Disease Control and Prevention classification from 1993, 76 (40.6%) patients were in stage 2B at the sample collection date and 106 (56.7%) patients were in stage 3 (n = 187).
ZDV-R ranged from 0.3- to 5,338-fold increase in IC50 compared to wild-type. Of the 223 samples, 73 were wild type at position 184 (Table 1). We investigated four different patterns based on ZDV-R-associated mutations at positions 41, 210, and 215, selected because of the association of changes at these positions with high-level ZDV-R. Samples without changes at position 41 and 215 were defined as pattern 1; samples with changes at either 41 or 215 were defined as pattern 2; samples with changes at 41 and 215 but not at 210 were defined as pattern 3; and samples with changes at 41, 210, and 215 were defined as pattern 4. Changes at positions 67, 70, and 219 were not considered in this analysis.
TABLE 1.
Summary and prevalence of genotypic patterns in sample population
| Genotypic pattern | No. of samples (%) | Phenotypic range for ZDV-R (fold) |
|---|---|---|
| M184 | 73 (32.74) | 0.3-3,571 |
| V184 | 150 (67.26) | 0.3-5,338 |
| Pattern 1 | 41 (18.39) | 0.3-2,674 |
| Pattern 2 | 35 (15.70) | 0.3-2,564 |
| Pattern 3 | 51 (22.87) | 1-5,338 |
| Pattern 4 | 96 (43.05) | 4-3,571 |
| H208Y | 28 (12.56) | 1.3-1,515 |
| R211K/L214F | 141 (63.23) | 0.3-3,571 |
| G333E/D | 41 (18.39) | 1-1,589 |
| Q151 MDR pattern | 6 (2.69) | 90-2,674 |
| 6-bp insert with A62 | 7 (3.14) | 9-3,571 |
| 6-bp insert with V62 | 3 (1.35) | 2,107-2,729 |
| Total | 223 (100.00) | 0.3-5,338 |
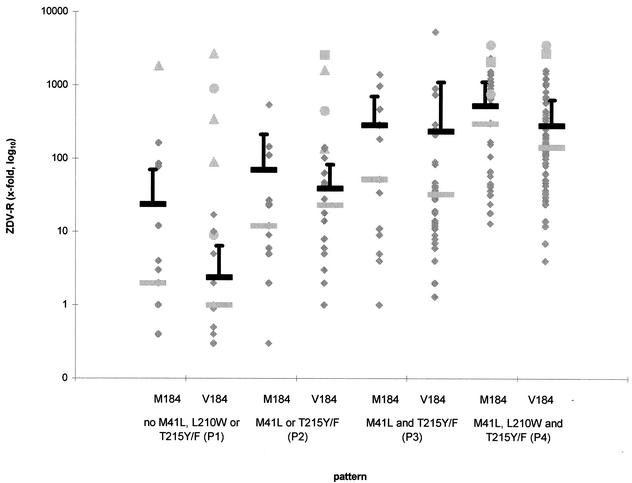
With increasing number of ZDV-associated mutations, ZDV-R increased from pattern 1 to pattern 4 (Fig. 1). The mean and median values in samples with wild type at position 184 were 23.7 and 2 (0.4- to 163-fold) for pattern 1; 70 and 12 (0.3- to 538-fold) for pattern 2; 285.5 and 52 (1- to 1,400-fold) for pattern 3; and 524.7 and 300 (13- to 2,334-fold) for pattern 4.
FIG. 1.
Phenotypic resistance to ZDV dependent on mutational patterns as comparisons of IC50 with that of a wild-type strain. Samples are shown as rhombi, triangles represent samples with the Q151M MNR complex, squares represent samples with the 6-bp insert/A62V, and circles represent samples with the 6-bp insert alone. Mean and median values are shown as dark and light bars, respectively. Error bars represent standard deviation values. Negative error bar values are not shown because they are below zero.
In samples with the M184V mutation these values were nearly the same or lower: 2.4 and 1 (0.3- to 17-fold), 38.9 and 23 (1- to 141-fold), 236.1 and 32.5 (1.3- to 5,338-fold), and 283 and 143 (4- to 1,592-fold).
In six samples the Q151M MNR complex was found with a ZDV-R range of 90- to 2,674-fold. Four of these samples had none of the classic ZDV-associated mutations, and in two samples ZDV-associated mutations were found at positions 67, 210, and 215. A 6-bp insert between position 67 and 69 was found in 10 samples with a ZDV-R range of 9- to 3,571-fold, with 9 of 10 samples being highly ZDV-R (449- to 3,571-fold). In seven samples there was only the 6-bp insert, and in three samples the 6-bp insert was found together with the A62V mutation (Table 1).
Correlation of genotype and ZDV-R.
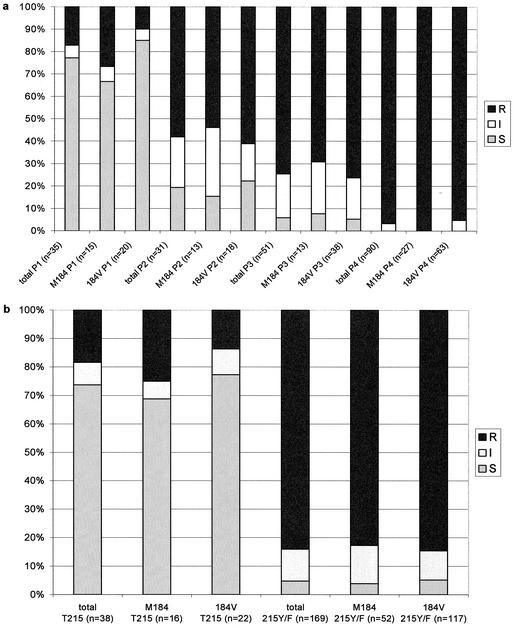
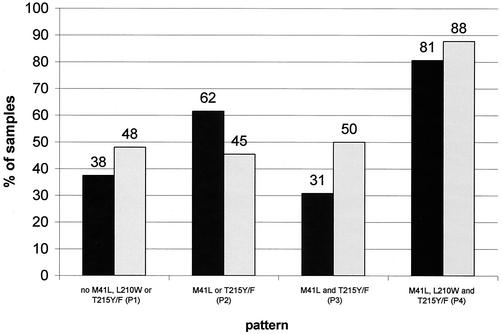
Instead of correlating the precise IC50 phenotypic change with the genotypic patterns, we classified the samples into sensitive (IC50 change, <4-fold), intermediate (IC50 change, ≥4- and <10-fold), and resistant (IC50 change, ≥10-fold) as given by the phenotypic test (Antivirogram). This analysis was done with and without the inclusion of the M184V change (Fig. 2a). The proportion of resistant phenotypes increased with the complexity of the genotypic patterns (17.1% in pattern 1, 58.1% in pattern 2, 74.5% in pattern 3, and 96.7% in pattern 4). Except for pattern 1 these data are independent of the M184V change (in pattern 1, 26.7% of resistant samples had the wild type at position 184, versus 10.0% with mutation at position 184). Because the T215Y/F change is considered a key mutation in the development of ZDV-R (2), we reanalyzed the sample population but correlated the appearance of the T215Y/F mutation instead of the genotypic patterns described above (Fig. 2b). In samples with wild type at position 215 the proportion of resistant samples was 25.0% without the M184V mutation, compared to 13.6% with the M184V change. In samples with the T215Y/F change the proportion of resistant samples was equal irrespective of position 184 (82.7% without the M184V mutation compared to 84.6% with the M184V change). Of the samples with a resistant phenotype with wild type at position 215, seven of seven had the K70R change, six of seven had the K219Q/E change, and five of seven had the D67N change (data not shown). Of the samples with a sensitive phenotype and wild type at position 215, 4 of 28 had the K70R change, 1 of 28 had the K219Q/E change, and 2 of 28 had the D67N change (data not shown).
FIG. 2.
(a) Percentage of ZDV-sensitive (S), -intermediate (I), and -R samples dependent on mutational patterns and the M184V change. (b) Percentage of ZDV-sensitive (S), -intermediate (I), and -R samples dependent on the T215Y/F and the M184V change.
SDM experiments.
Genetic mapping experiments identified the region encompassing amino acids 35 to 228 of the RT to be involved in dual resistance to ZDV and 3TC. Changes in the RT at positions 208, 211, and 214 have been identified as possible polymorphisms for dual ZDV-3TC resistance (Kemp and Bloor, Prog. Abstr. Int. Workshop HIV Drug Resist. Treat. Strategies Erad. 1997).
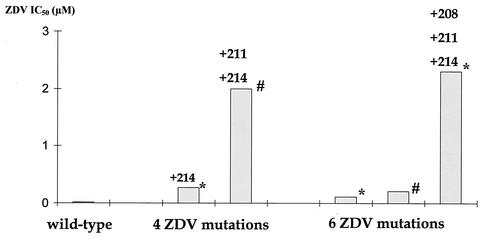
We examined the influence of H208Y, R211K, and L214F on ZDV-R in a ZDV-R virus by SDM. Adding the R211K/L214F (double mutant) combination to a virus carrying the M41L, L210W, T215Y, and K219N change (quadruple ZDV mutant) increased ZDV-R 7.5-fold (Fig. 3). The presence of the H208Y/R211K/L214F (triple mutant) combination in a virus carrying the M41L, D67N, K70R, L210W, T215Y, and K219E change (sextuple ZDV mutant) increased ZDV-R 21-fold. Compared to a wild-type virus the quadruple ZDV mutant had a 13.5-fold increase in ZDV-R and the sextuple ZDV mutant a 5.5-fold increase in ZDV-R. Adding the double mutant combination to the quadruple ZDV mutant and the triple mutant combination to the sextuple ZDV mutant increased ZDV-R 100- and 115-fold compared to wild-type virus, respectively.
FIG. 3.
SDM experiments to evaluate the effect of additional polymorphisms on ZDV-R. The virus strain with four ZDV mutations had the M41L, L210W, T215Y, K219E(*) or K219N (#), and M184V changes. In addition the virus strain with six ZDV mutations had the D67N and K70R change. Mutations added by SDM are shown on top of the bars.
Additional mutations in the clinical sample set.
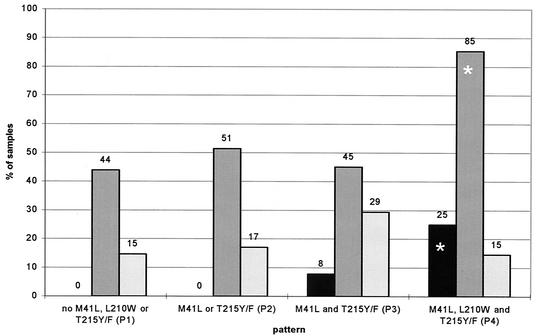
We looked for the prevalence of these mutations in our sample population. H208Y was found in 28 of 223 samples, with a ZDV-R range of 1.3- to 1,515-fold (Table 1). Presence of both 41 and 215 mutations appeared to be a prerequisite for the appearance of H208Y (Fig. 4), but it appeared independently of M184V (data not shown). The higher percentage of samples with the H208Y change in pattern 4 (25%) was statistically significant (P = 0.02; χ2-test). 19 samples had the M184V mutation, nine were wild-type at position 184.
FIG. 4.
Distribution of additional polymorphisms H208Y (black bars), R211K/L214F (dark grey bars), and G333E/D (light grey bars) in the sample population dependent on ZDV mutational patterns. Percentages are given above each bar. Asterisks highlight statistical significance (χ2-test).
The prevalence of the double mutant combination was considerably higher, detected in 141 of 223 samples, with a ZDV-R range of 0.3- to 3,571-fold (Table 1). Again this combination appeared independently of mutation at position 184 (Fig. 5); 43 of 141 samples were wild type at position 184, and 98 had the M184V mutation. Although there seemed to be a trend towards higher percentage of this double mutation in samples with the M184V change, these findings were not statistically significant (P > 0.38; χ2-test). The double-mutant combination was found with and without classic ZDV-associated mutations but appeared more frequently (82 of 96 samples) with ZDV-associated mutations at positions 41, 210, and 215 (Fig. 4). This observation reached statistical significance (P < 0.01; χ2-test).
FIG. 5.
Distribution of samples carrying the R211K/L214F double mutation dependent on ZDV mutational patterns (see above) and the M184V change. Samples wild type at position 184 are shown as black bars, and samples with the M184V mutation are shown as light grey bars. Percentages are given above each bar.
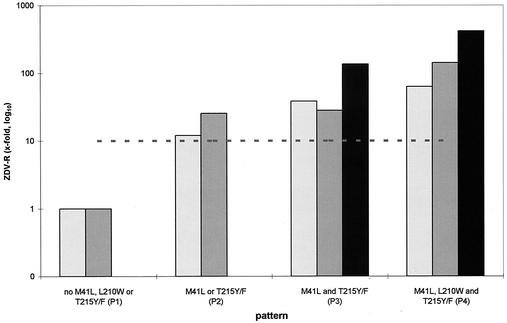
To confirm the results of the SDM, 207 clinical samples without any MNR pattern were analyzed (Fig. 6). The presence of the double-mutant combination in the background of genotypic highly ZDV-R viruses (pattern 4) increased ZDV-R 2.4-fold, and the triple-mutant combination increased ZDV-R 8-fold. These values were lower than the results obtained using SDM, but the trend of increasing ZDV-R was comparable.
FIG. 6.
Phenotypic ZDV-R for samples carrying no polymorphisms or combinations of the additional polymorphisms H208Y, R211K, and L214F dependent on ZDV mutational patterns. Black bars represent samples carrying the H208Y/R211K/L214F triple combination, dark grey bars represent samples carrying the R211K/L214F combination, and light grey bars represent samples carrying none of the above-mentioned combinations. The dotted line represents a 10-fold increase in ZDV-R.
The additional mutation G333E/D has recently been described in the context of dual ZDV-3TC resistance (12). In our sample population, the G333E/D change was found in 41 of 223 samples with a ZDV-R range of 1- to 1,589-fold (Table 1). The G333E/D change appeared independent of the mutational pattern (P > 0.05; χ2-test) (Fig. 4) and the M184V mutation (18 with wild-type, 23 with the M184V change; data not shown). The G333E/D change also appeared in conjunction with the 6-bp insert or the Q151 M MNR complex, as well as in samples without any ZDV-associated mutations.
DISCUSSION
In this report we compare phenotypic ZDV-R and genotypic changes in the HIV-1 RT in a large cross-sectional set of clinical HIV-1 isolates. In this analysis of samples from patients treated with different antiretroviral regimens, we showed that ZDV-R could not be easily deduced just from examining patterns of ZDV-associated mutations. Higher mean and median values for increasing numbers of ZDV-associated mutations were found at the level of the whole sample population, but a similar resistance range within mutation patterns 1 to 4 made it impossible to estimate ZDV-R for an individual sample. Similarly, the described reversal of ZDV-R with the appearance of the M184V mutation-associated with 3TC resistance (1, 26) was seen only at the population level. The overlapping error bars in Fig. 1 indicate the missing statistical significance. The Q151M MNR complex with at least the Q151M mutation or the 6-bp insert in combination with the A62V mutation caused high-level ZDV-R independent of ZDV mutations in our sample population. High-level ZDV-R could also be seen with the 6-bp insert alone.
The prediction of the exact degree of ZDV-R based on genotypic data in our sample population is not possible for samples with ZDV-associated mutations. Only the proportion of samples with no ZDV-associated mutations and no Q151M MNR complex or 6-bp inserts could be predicted to be ZDV sensitive in the phenotypic assay. This supports results published earlier by our group (B. Morgenstern, M. Stürmer, B. Nolte, A. Berger, S. Bader, H. W. Doerr, S. Staszewski, and V. Miller, Abstr. 6th Eur. Conf. Clin. Aspects Treat. HIV Infect., abstr. 466, p. 777, 1997). When using the classification into sensitive, intermediate, and resistant isolates, there was a good correlation with the genotypic patterns and the phenotypic data. This is in accordance with the results of other groups (2, 15; F. Schneider, J. M. Plesseria, L. Stuyver, C. Lambert, E. Fontaine, P. Kirpach, D. Ninove, V. Arendt, R. Hemmer, and J. C. Schmit, Prog. Abstr. 5th Conf. Retrovir. Opportun. Infect., abstr. 333, p. 142, 1998). Our analysis shows that correlation between genotype and ZDV-R investigating only the 215 position is sufficient for 75 to 85% of our samples, but the inclusion of all mutations responsible for ZDV-R, the M184V change, and MNR mutations will increase the predictive value of the genotypic data.
The influence of the known six ZDV-associated mutations on ZDV-R is undisputed, but the relative influence of these mutations to the phenotype is not clear. Virus strains containing mutations at positions 41 and 215 are known to be highly ZDV-R (13). The percentage of appearance of the L210W mutation and the relative increase of ZDV-R has been the subject of debate. Hooker et al. (9) found this mutation in 7 of 22 patients undergoing long-term ZDV therapy and reported an up-to-sixfold increase in ZDV-R when the L210W mutation was added to a virus strain containing at least the M41L and T215Y mutation. In contrast Harrigan et al. (6) reported the L210W mutation only in a minority of patients after prolonged ZDV therapy (21 of 105 patients with more than 2 years of ZDV therapy). The increase of ZDV-R due to adding the L210W mutation to virus strains with at least the M41L and T215Y mutation was twofold. Our data showed a relatively high proportion of samples carrying the M41L, L210W, and T215Y/F mutations (96 of 223 samples). In comparing the mean and median values of patterns containing M41L and T215Y/F with and without L210W, an increase in ZDV-R of 1.5- and 5.2-fold, respectively, was observed.
Other polymorphisms like the Q151M MNR complex (10, 23, 24) or the 6-bp insert (4, 18, 28), both responsible for resistance to all known NRTIs, were found in a small proportion of samples (9 of 223) at about the same range as reported previously (27). All samples had high ZDV-R as well as the expected cross-resistance to other NRTIs (data not shown). We also found seven samples carrying a 6-bp insert without the A62V mutation showing high ZDV-R except in one sample (ninefold). This sample is the only one showing cross-resistance, while the other six samples were sensitive or only resistant to some NRTIs (data not shown). These findings are consistent with the data from Mas et al., who reported the role of the 6-bp insert in the development of ZDV-R (19).
Other polymorphisms at positions 208, 211, 214, and 333 reported to be involved in ZDV-3TC double resistance were included in this analysis to investigate the role of these mutations in the development of ZDV-R in the clinical setting. All analyzed polymorphisms were detected in samples ranging from ZDV sensitive to highly resistant and appeared with and without the standard ZDV-associated mutations except for the H208Y change which appeared only in samples with at least the M41L and T215Y/F changes. These findings do not support a direct influence of these polymorphisms on ZDV-R. On the other hand the H208Y mutation and the double mutation R211K/L214F were found mainly associated with ZDV-associated mutations at positions 41, 210, and 215. These observations reached statistical significance and correspond with the SDM results, in which these mutations increased ZDV-R when added as double (R211K/L214F) or triple (H208Y/R211K/L214F) combinations to a highly ZDV-R background. Although the increase in ZDV-R has been observed mainly in already highly ZDV-R virus isolates and therefore would not be clinically relevant, it will be helpful to consider the double and triple mutation in the interpretation of ZDV-R.
Three mechanisms of phenotypic resistance development were described (reviewed in reference 25). Mutations at positions 65, 151, and 184 lead to the ability to discriminate between the NRTI and the corresponding dNTP by the RT. Changes at position 74 and 89 are involved in the repositioning of the template/primer complex, and the ZDV-associated mutations at positions 41, 67, 70, 215, and 219 as well as the 6-bp insert (19) are reported to increase the phosphorolytic removal of the incorporated ZDV. We would suspect a stabilization mechanism that could be explained by the structural vicinity of the mutations at positions 208, 211, and 214 to the ZDV-associated mutations L210W and T215Y/F. As described for the L210W mutation (29), H208Y, R211K, and L214F may stabilize the three-dimensional structure of the enzyme because of the interactions of the aromatic side chains. These mutations are likely involved in the compensation of the possible loss of fitness caused by the T215Y/F mutation. The exact mechanism responsible for the action of these polymorphisms needs to be further investigated.
REFERENCES
- 1.Boucher, C. A., N. Cammack, P. Schipper, R. Schuurman, P. Rouse, M. A. Wainberg, and J. M. Cameron. 1993. High-level resistance to (−) enantiomeric 2′-deoxy-3′-thiacytidine in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 37:2231-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, P., J. C. Schmit, V. Arendt, J. M. Plesseria, C. Lambert, E. Fontaine, P. Kirpach, D. Ninove, F. Schneider, and R. Hemmer. 1997. Drug resistance mutations as predictors of phenotypic zidovudine resistance in HIV-1 infection. AIDS 11:1528-1529. [PubMed] [Google Scholar]
- 3.De Antoni, A., A. Foli, J. Lisziewicz, and F. Lori. 1997. Mutations in the pol gene of human immunodeficiency virus type 1 in infected patients receiving didanosine and hydroxyurea combination therapy. J. Infect. Dis. 176:899-903. [DOI] [PubMed] [Google Scholar]
- 4.De Jong, J. J., J. Goudsmit, V. V. Lukasov, M. E. Hillebrand, E. Baan, R. Huismans, S. A. Danner, J. H. Ten Veen, F. De Wolf, and S. Jurriaans. 1999. Insertion of two amino acids combined with changes in reverse transcriptase containing tyrosine-215 of HIV-1 resistant to multiple nucleoside analogs. AIDS 13:75-80. [DOI] [PubMed] [Google Scholar]
- 5.Fischl, M. A., D. D. Richman, M. H. Grieco, M. S. Gottlieb, P. A. Volberding, O. L. Laskin, J. M. Leedom, J. E. Groopman, D. Mildvan, R. T. Schooley, G. G. Jackson, D. T. Durack, D. King, and The ZDV Collaborative Working Group. 1987. The efficacy of azidothymidine (ZDV) in the treatment of patients with AIDS and AIDS-related complex. N. Engl. J. Med. 317:185-191. [DOI] [PubMed] [Google Scholar]
- 6.Harrigan, P. R., I. Kinghorn, S. Bloor, S. D. Kemp, I. Najera, A. Kohli, and B. A. Larder. 1996. Significance of amino acid variation at human immunodeficiency virus type 1 reverse transcriptase residue 210 for zidovudine susceptibility. J. Virol. 70:593-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hertogs, K., M. P. De Béthune, V. Miller, T. Ivens, P. Schel, A. Van Cauwenberge, C. Van Den Eynde, V. Van Gerwen, H. Azijn, M. Van Houtte, F. Peeters, S. Staszewski, M. Conant, S. Bloor, S. Kemp, B. Larder, and R. Pauwels. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant HIV-1 isolates of patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hertogs, K., S. Bloor, V. De Vroey, C. Van Den Eynde, P. Dehertogh, A. Van Cauwenberge, M. Stürmer, T. Alcorn, S. Wegner, M. Van Houtte, V., Miller, and B. Larder. 2000. A novel human immunodeficiency virus type 1 reverse transcriptase mutational pattern confers phenotypic lamivudine resistance in the absence of mutation 184V. Antimicrob. Agents Chemother. 44:568-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hooker, D., G. Tachedjian, A. E. Soloman, A. D. Gurusinghe, S. Land, C. Birch, J. L. Anderson, B. M. Roy, E. Arnold, and N. J. Deacon. 1996. An in vivo mutation from leucine to tryptophan at position 210 in human immunodeficiency virus type 1 reverse transcriptase contributes to high-level resistance to 3′-azido-3′-deoxythymidine. J. Virol. 70:8010-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iversen, A. K. N., R. W. Shafer, K. Wehrly, M. A. Winters, J. I. Mullins, B. Chesebro, and T. C. Merigan. 1996. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral therapy. J. Virol. 70:1086-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kellam, P., C. A. B. Boucher, and B. A. Larder. 1992. Fifth mutation in human immunodeficiency virus type 1 reverse transcriptase contributes to the development of high-level resistance to zidovudine. Proc. Natl. Acad. Sci. USA 89:1934-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kemp, S. D., C. Shi, S. Bloor, P. R. Harrigan, J. W. Mellors, and B. A. Larder. 1998. A novel polymorphism at codon 333 of human immunodeficiency virus type 1 reverse transcriptase can facilitate dual resistance to zidovudine and l-2′,3′-dideoxy-3′-thiacytidine. J. Virol. 72:5093-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larder, B. A. 1994. Interactions between drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase. J. Gen. Virol. 75:951-957. [DOI] [PubMed] [Google Scholar]
- 14.Larder, B. A., A. Kohli, P. Kellam, S. D. Kemp, M. Kronick, and R. D. Henfrey. 1993. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature 365:671-673. [DOI] [PubMed] [Google Scholar]
- 15.Larder, B. A., A. Kohli, S. Bloor, S. D. Kemp, P. R. Harrigan, R. T. Schooley, J. M. A. Lange, K. N. Pennington, M. H. St. Clair, and the Protocol 34.225-02 Collaborative Group. 1996. Human immunodeficiency virus type 1 drug susceptibility during zidovudine (ZDV) monotherapy compared with ZDV plus 2′,3′-dideoxyinosine or ZDV plus 2′,3′-dideoxycytidine combination therapy. J. Virol. 70:5922-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larder, B. A., and S. D. Kemp. 1989. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (ZDV). Science 246:1155-1158. [DOI] [PubMed] [Google Scholar]
- 17.Larder, B. A., G. Darby, and D. D. Richman. 1989. HIV with reduced sensitivity to zidovudine (ZDV) isolated during prolonged therapy. Science 243:1731-1734. [DOI] [PubMed] [Google Scholar]
- 18.Larder, B. A., S. Bloor, S. D. Kemp, K. Hertogs, R. L. Desmet, V. Miller, M. Stürmer, S. Staszewski, J. Ren, D. K. Stammers, D. I. Stuart, and R. Pauwels. 1999. A family of insertion mutations between codons 67 and 70 of human immunodeficiency virus type 1 reverse transcriptase confer multinucleoside analog resistance. Antimicrob Agents Chemother. 43:1961-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mas a., M. Parera, C. Briones, V. Soriano, M. A. Martínez, E. Domingo, and L. Menéndez-Arias. 2000. Role of a dipeptide insertion between codons 69 and 70 of HIV-1 reverse transcriptase in the mechanism of AZT resistance. EMBO J. 21:5752-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellors, J. W., H. Z. Bazami, R. F. Schinazi, B. M. Roy, Y. Hsiou, E. Arnold, J. Weir, and D. L. Mayers. 1995. Novel mutations in reverse transcriptase of human immunodeficiency virus type 1 reduce susceptibility to foscarnet in laboratory and clinical isolates. Antimicrob Agents Chemother. 39:1087-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, V., A. Phillips, C. Rottmann, S. Staszewski, R. Pauwels, K. Hertogs, M. P. De Béthune, S. D. Kemp, S. Bloor, P. R. Harrigan, and B. A. Larder. 1998. Dual resistance to zidovudine (ZDV) and lamivudine (3TC) in patients treated with ZDV/3TC combination therapy: association with therapy failure. J. Infect. Dis. 177:1521-1532. [DOI] [PubMed] [Google Scholar]
- 22.Miller, V., M. Stürmer, S. Staszewski, B. Gröschel, K. Hertogs, M. P. De Béthune, R. Pauwels, P. R. Harrigan, S. Bloor, S. D. Kemp, and B. A. Larder. 1998. The M184V mutation in HIV-1 reverse transcriptase (RT) conferring lamivudine resistance does not result in broad cross-resistance to nucleoside analogue RT inhibitors. AIDS 12:705-712. [DOI] [PubMed] [Google Scholar]
- 23.Shafer, R. W., A. K. N. Iversen, M. A. Winters, E. Aguiniga, D. A. Katzenstein, T. C. Merigan, and the AIDS Clinical Trials Group 143 Virology Team. 1995. Drug resistance and heterogeneous long-term virologic responses of human immunodeficiency virus type 1-infected subjects to zidovudine and didanosine combination therapy. J. Infect. Dis. 172:70-78. [DOI] [PubMed] [Google Scholar]
- 24.Shirasaka, T., M. F. Kavlick, T. Ueno, W. Y. Gao, E. Kojima, M. L. Alcaide, S. Chokekijchai, B. M. Roy, E. Arnold, R. Yarchoan, and H. Mitsuya. 1995. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc. Natl. Acad. Sci. USA 92:2398-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sluis-Cremer, N., D. Arion, and M. A. Parniak. 2000. Molecular mechanisms of HIV-1 resistance to nucleoside reverse transcriptase inhibitors (NRTIs). Cell. Mol. Life Sci. 57:1408-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tisdale, M., S. D. Kemp, N. R. Parry, and B. A. Larder. 1993. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc. Natl. Acad. Sci. USA 90:5653-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Vaerenbergh, K., K. Van Laethem, J. Albert, C. A. Boucher, B. Clotet, M. Floridia, J. Gerstoft, B. Hejdeman, C. Nielsen, C. Pannecouque, L. Perrin, M. F. Pirillo, L. Ruiz, J. C. Schmit, F. Schneider, A. Schoolmeester, R. Schuurman, H. J. Stellbrink, L. Stuyver, J. Van Lunzen, B. Van Remoortel, E. Van Wijngaerden, S. Vella, M. Witvrouw, S. Yerly, E. De Clercq, J. Destmyer, and A. M. Vandamme. 2000. Prevalence and characteristics of multinucleoside-resistant human immunodeficiency virus type 1 among European patients receiving combinations of nucleoside analogues. Antimicrob Agents Chemother. 44:2109-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winters, M. A., K. L. Coolley, Y. A. Girard, D. J. Levee, H. Hamdan, R. W. Shafer, D. A. Katzenstein, and T. C. Merigan. 1998. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J. Clin. Investig. 102:1769-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yahi, N., C. Tamalet, C. Tourrès, N. Tivoli, and J. Fantini. 2000. Mutation L210W of HIV-1 reverse transcriptase in patients receiving combination therapy. J. Biomed. Sci. 7:507-513. [DOI] [PubMed] [Google Scholar]