Abstract
Clinical isolate Acinetobacter baumannii CLA-1 was resistant to a series of antibiotic molecules, including carbapenems. Cloning and expression of the β-lactamase gene content of this isolate in Escherichia coli DH10B identified a chromosome-encoded oxacillinase, OXA-40, that differed by one or two amino acid changes from OXA-24, -25, and -26 and an AmpC-type cephalosporinase. The OXA-40 β-lactamase had a mainly narrow-spectrum hydrolytic profile, but it included ceftazidime and imipenem. Its activity was resistant to inhibition by clavulanic acid, tazobactam, sulbactam, and, like most of the other carbapenem-hydrolyzing oxacillinases, NaCl. OXA-40 had an FGN triad replacing a YGN motif at class D β-lactamase (DBL) positions 144 to 146. Site-directed DNA mutagenesis leading to a Phe-to-Tyr change at DBL position 144 in OXA-40 gave a mutant enzyme with increased hydrolytic activity against most β-lactams, including imipenem. Conversely, with a gene encoding the narrow-spectrum oxacillinase OXA-1 as the template, a nucleotide substitution leading to a Tyr-to-Phe change in the YGN motif of OXA-1 gave a mutant enzyme with decreased hydrolytic activity without an increase in carbapenem-hydrolyzing activity. Thus, the Phe residue in the FGN motif was not associated with carbapenem-hydrolyzing activity by itself but instead was associated with weak overall hydrolytic activity. Finally, this Phe residue in OXA-40 explained resistance to inhibition by NaCl whereas a Tyr residue in motif YGN was related to susceptibility to NaCl.
Acinetobacter baumannii is an important cause of nosocomial infections such as pneumonia, septicemia, urinary tract infections, and wound infections (6). In many cases, carbapenems have become the drugs of choice for treatment of infections due to multiple-drug-resistant A. baumannii strains (6). Nevertheless, there are growing reports of carbapenem resistance in this species. The early reports described A. baumannii isolates with β-lactamase-independent carbapenem resistance (13, 16, 30), but the most recent reports have described β-lactamase-mediated resistance (1-3, 8, 10, 15, 22, 25, 26, 28, 31). In rare cases, IMP-like and VIM-like metallo-β-lactamases (Ambler class B) have been described in Southeast Asian and Italian isolates of the species (1, 12, 25, 26, 31).
Five oxacillinases (Ambler class D) with carbapenem-hydrolyzing activity have been characterized recently in that species, several strains of which were responsible for nosocomial outbreaks (3, 8, 10, 15, 22). OXA-23 (also named ARI-1 [15, 22, 28]) and OXA-27 (3), have 99% amino acid identity, whereas they have only 60% identity with a second group of oxacillinases consisting of OXA-24, -25, and -26, which differ by a few amino acid substitutions (3, 10).
We have characterized the genetics and biochemistry of the carbapenem-hydrolyzing oxacillinase OXA-40, which belongs to the subgroup of oxacillinases containing OXA-24, -25, and -26. Like the other carbapenem-hydrolyzing oxacillinases, OXA-40 has an FGN motif in place of the classical YGN motif of oxacillinases (14, 19). To analyze the role of this Phe residue in carbapenem-hydrolyzing activity and in resistance to NaCl, we performed a series of site-directed mutagenesis experiments.
(This work was previously presented in part [L. Poirel, C. Héritier, and P. Nordmann, 12th Eur. Congr. Clin. Microbiol. Infect. Dis., abstr. O384, 2002].)
MATERIALS AND METHODS
Bacterial strains and plasmids.
A. baumannii clinical isolate CLA-1 was from a pulmonary brush of a Portuguese patient hospitalized for septic shock at the Bicêtre hospital (Le Kremlin-Bicêtre, France). He had been transferred from the Mirandelo hospital (near Porto, Portugal), where he had been hospitalized previously in an intensive care unit for a cranial injury. This strain was identified by the API 20NE system (bioMérieux, Marcy l'Etoile, France). Escherichia coli reference strain DH10B and plasmid pBK-CMV (carrying a kanamycin resistance marker) (Stratagene, Amsterdam, The Netherlands) were used for cloning and site-directed mutagenesis experiments. Salmonella enterica serotype Typhimurium strain RGN238 is a reference strain that produces the oxacillinase OXA-1 (gift from R. Labia) (21). Strains A. baumannii CIP7034T (Institut Pasteur strain collection) and E. coli K-12 (gift from P. Plesiat) were used as reference strains for endonuclease I-CeuI digestion.
Antimicrobial agents and MIC determinations.
The antimicrobial agents used and their sources have been referenced elsewhere (23). Antibiotic-containing disks were used for detection of antibiotic susceptibility with Mueller-Hinton agar plates and a disk diffusion assay (Sanofi Diagnostics Pasteur, Marnes-La-Coquette, France) (www.sfm.fr). MICs were determined by an agar dilution technique as previously reported (23), and results were interpreted in accordance with the guidelines of the NCCLS (20).
Cloning experiments.
Whole-cell DNA of A. baumannii CLA-1 was extracted as previously described (23). Since the oxacillinase genes are often located in integrons (19), primers for detection of class 1 integrons (primer INT2F, located within the integrase gene of class 1 integrons, and a primer [3′-CS] located in the 3′ conserved segment [24]) were used to generate fragments by PCR with whole-cell DNA of A. baumannii CLA-1 as the template. A cloning experiment was then performed with BamHI-digested whole-cell DNA of A. baumannii CLA-1 and BamHI-restricted plasmid pBK-CMV, followed by expression in E. coli DH10B, as described previously (24). Antibiograms obtained by disk diffusion were performed with E. coli DH10B strains harboring recombinant plasmids, and the sizes of the plasmid inserts were determined by restriction analysis (27). Recombinant plasmids pOXA-40 and pABC-1 were retained for further analysis.
Site-directed mutagenesis.
Since the identified oxacillinase contained an FGN triad in place of the more common YGN at class D β-lactamase (DBL) positions 144 to 146 (14), a site-directed mutagenesis protocol was used as described by the manufacturer (Quick Change site-directed mutagenesis kit; Stratagene) for a nucleotide that substituted a Phe residue for the Tyr residue at DBL position 144. Recombinant plasmid pOXA-40 was used as the template with primers F144Y and F144Yrev to generate recombinant plasmid pOXA-40-Tyr (Table 1).
TABLE 1.
Primers used in this study
| Primer | Sequence (5′ to 3′)a | Reference |
|---|---|---|
| F144Y | GCGGGTTAATTATGGAAATACAAATATTGGAACACAGGTCG | This study |
| F144Yrev | CGACCTGTGTTCCAATATTTGTATTTCCATAATTAACCCGC | This study |
| PreOXA1A | AGCCGTTAAAATTAAGCCC | 4 |
| PreOXA1B | CTTGATTGAAGGGTTGGGCG | 4 |
| Y144F | CTCAAGGATTTTGATTTTGGAAATCAAGACTTCTTCGG | This study |
| Y144Frev | CCAAAATCAAAATCCTTGAGATAATTCTTG | This study |
| A | AGAGTTTGATCHTGGYTYAGA | 5 |
| B | ACGGYTACCTTGTTACGACTTC | 5 |
| Oxa-imp1 | GCAAATAMAGAATATGTSCC | This study |
| Oxa-imp2 | CTCMACCCARCCRGTCAACC | This study |
H is A, T, or C; Y is C or T; S is G or C; M is A or C; R is A or G.
Conversely, the effects of a nucleotide change leading to the change from a Tyr residue to a Phe residue at DBL position 144 were investigated by using as the template the gene encoding OXA-1, a reference narrow-spectrum oxacillinase that does not possess significant carbapenem-hydrolyzing activity (19). The blaOXA-1 gene was PCR amplified with primers PreOXA1A and PreOXA1B (Table 1) from blaOXA-1-containing whole-cell DNA of S. enterica serotype Typhimurium. This 911-bp fragment, encompassing the blaOXA-1 gene, was cloned into the SmaI site of plasmid pBK-CMV, giving recombinant plasmid pOXA-1. Plasmid pOXA-1 was then used as a template with primers Y144F and Y144Frev (Table 1), giving rise to recombinant pOXA-1-Phe.
DNA sequencing and protein analysis.
The nucleotide changes induced by site-directed mutagenesis experiments were checked by sequence analysis. Both strands of the cloned DNA fragments from recombinant plasmids were sequenced with an Applied Biosystems sequencer (ABI 373). The nucleotide and deduced protein sequences were analyzed with software available over the Internet at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov).
Plasmid analysis and hybridizations.
Extraction of plasmid DNA from A. baumannii CLA-1 was attempted by two different methods (24). To search for a possible chromosomal location of the oxacillinase gene, we used the endonuclease I-CeuI (New England Biolabs, Ozyme, Saint-Quentin-en-Yvelines, France), which digests a 26-bp sequence in rrn genes for the 23S large-subunit rRNA (18). After digestion, separation of the resulting fragments by pulsed-field gel electrophoresis was performed with a CHEF-DRII apparatus (29). The sizes of endonuclease I-CeuI-generated fragments were determined by comparison with those of E. coli K-12 (18).
After transfer onto a nylon membrane (Hybond N+; Amersham Pharmacia Biotech, Orsay, France) by the Southern technique (28), endonuclease I-CeuI-generated DNA fragments of A. baumannii CLA-1 and CIP7034T were UV cross-linked (Stratalinker; Stratagene) for 2 min. They were hybridized with two different probes: a 1,504-bp PCR-generated probe for 16S and 23S rRNA genes (Roche Diagnostics, Meylan, France) (primers A and B [Table 1]) and a 495-bp PCR-generated probe (primers Oxa-imp1 and Oxa-imp2 [Table 1]) for detection of the genes encoding carbapenem-hydrolyzing oxacillinases. Labeling and revelation were performed as previously reported (24).
β-Lactamase purification.
Cultures of E. coli DH10B harboring recombinant plasmids pOXA-40 and pOXA-40-Tyr were grown overnight at 37°C in 4 liters of Trypticase soy (TS) broth containing amoxicillin (100 μg/ml) and kanamycin (30 μg/ml). Protein extracts were then obtained as previously detailed (24). These extracts were subjected to purification steps including ion-exchange chromatography with Q-Sepharose followed by S-Sepharose columns as previously described (23). Elution of the β-lactamase activities was realized with NaCl at a concentration of 50 mM. The peaks of β-lactamase activity were concentrated by using Centrisart-C30 spin columns (Sartorius, Göttingen, Germany) and dialyzed with 100 mM phosphate buffer (pH 7.0).
Kinetic parameters.
Purified β-lactamases were used for kinetic measurements performed at 30°C in 100 mM sodium phosphate (pH 7.0) (24). kcat and Km values were determined by analyzing β-lactam hydrolysis under initial-rate conditions with a UV spectrophotometer as previously described (24).
The 50% inhibitory concentrations (IC50s) of clavulanic acid, tazobactam, sulbactam, and NaCl were determined (24).Various concentrations of these inhibitors were preincubated with purified enzymes for 3 min at 30°C to determine the concentrations that decreased the rate of hydrolysis of 100 μmol of cephalothin by 50%.
Specific activities of protein extracts and of purified β-lactamases from E. coli DH10B harboring pOXA-40 and pOXA-40-Tyr were obtained as described previously (4) with 100 μM cephalothin as the substrate. The degree of protein purification was estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis (27). The protein content was measured by the Bio-Rad DC protein assay. β-Lactamase specific activities for several β-lactams were also determined with extracts of E. coli DH10B harboring plasmids pOXA-40, pOXA-40-Tyr, pOXA-1, and pOXA-1-Phe. In those cases, overnight cultures were obtained in 10 ml of TS broth containing amoxicillin (100 μg/ml) and kanamycin (30 μg/ml) and extracts were obtained as previously described (24).
IEF analysis.
Isoelectric focusing (IEF) analysis was performed with an ampholine polyacrylamide gel (pH 3.5 to 9.5) as described previously (24) with extracts of A. baumannii CLA-1 and E. coli DH10B strains harboring recombinant plasmids and with purified β-lactamases OXA-40 and OXA-40-Tyr.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been submitted to the EMBL/GenBank nucleotide sequence database and assigned accession no AF509241.
RESULTS
Antibiotic susceptibility and β-lactamases of A. baumannii CLA-1.
Isolate CLA-1 had high levels of resistance to penicillins, cephalosporins, and carbapenems (Table 2). Addition of clavulanic acid and tazobactam did not significantly decrease the MICs of ticarcillin and piperacillin (Table 2), indicating that a Bush group 2b or 2be β-lactamase was not likely to be solely involved (11). A preliminary experiment showed that the A. baumannii CLA-1 extract had significant imipenem-hydrolyzing activity (50 mU/mg of protein). Antibiotic susceptibility testing showed that A. baumannii CLA-1 was also resistant to gentamicin, tobramycin, netilmicin, fluoroquinolones, sulfonamides, and tetracycline and susceptible to amikacin, dibekacin, kanamycin, rifampin, and colistin (data not shown). An extract of A. baumannii CLA-1 subjected to IEF analysis gave two β-lactamases with pIs of 8.6 and 9.4 (data not shown).
TABLE 2.
MICs of β-lactams for A. baumannii clinical isolate CLA-1; E. coli DH10B harboring recombinant plasmids pOXA-40, pOXA-40-Tyr, pOXA-1, and pOXA-1-Phe; and E. coli reference strain DH10B
| β-Lactam(s)a | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|
| A. baumannii CLA-1 (OXA-40) | E. coli DH10B (pOXA-40) (OXA-40) | E. coli DH10B (pOXA-40-Tyr) (OXA-40-Tyr) | E. coli DH10B (pOXA-1) (OXA-1) | E. coli DH10B (pOXA-1-Phe) (OXA-1-Phe) | E. coli DH10B | |
| Amoxicillin | >512 | >512 | >512 | >512 | >512 | 4 |
| Amoxicillin + CLA | >512 | >512 | >512 | 128 | 128 | 4 |
| Ticarcillin | >512 | >512 | >512 | >512 | >512 | 4 |
| Ticarcillin + CLA | >512 | >512 | >512 | 256 | 256 | 4 |
| Piperacillin | >512 | 256 | >512 | 128 | 16 | 1 |
| Piperacillin + TZB | >512 | 128 | >512 | 128 | 16 | 1 |
| Cephalothin | >512 | 8 | 32 | 8 | 4 | 2 |
| Cefuroxime | >512 | 8 | 16 | 8 | 4 | 2 |
| Ceftazidime | >512 | 0.5 | 0.06 | 0.25 | 0.12 | 0.06 |
| Cefotaxime | >512 | 0.12 | 0.12 | 0.12 | 0.12 | 0.12 |
| Cefepime | 64 | 0.25 | 0.5 | 0.5 | 0.12 | 0.06 |
| Cefpirome | 256 | 0.25 | 0.25 | 1 | 0.25 | 0.06 |
| Moxalactam | 256 | 0.5 | 0.5 | 0.12 | 0.12 | 0.06 |
| Aztreonam | 128 | 0.12 | 0.06 | 0.12 | 0.12 | 0.12 |
| Imipenem | 256 | 2 | 4 | 0.5 | 0.25 | 0.06 |
| Meropenem | 256 | 0.5 | 0.5 | 0.12 | 0.12 | 0.06 |
CLA, clavulanic acid at a fixed concentration of 2 μg/ml; TZB, tazobactam at a fixed concentration of 4 μg/ml.
Cloning and sequencing of blaOXA-40 and surrounding sequences and β-lactamase expression in E. coli.
PCR amplification with primers annealing at the ends of class 1 integrons and whole-cell DNA of A. baumannii CLA-1 as the template failed. Recombinant plasmids were obtained after cloning of BamHI-restricted DNA of A. baumannii CLA-1 into pBK-CMV and expression in E. coli. Antibiotic susceptibility testing revealed that these E. coli clones expressed either (i) a β-lactamase whose activity included imipenem resistance and which was resistant to inhibitors or (ii) a cephalosporinase-type enzyme (data not shown). Recombinant plasmids pOXA-40 and pABC-1 were isolated from clones with each of these antibiotic resistance phenotypes that contained 2.5- and 2-kb inserts, respectively.
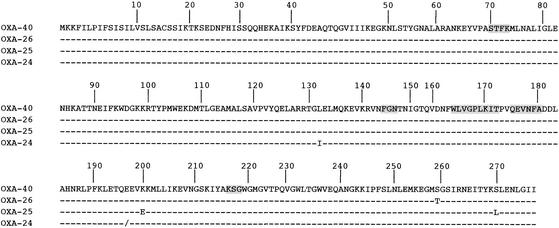
DNA sequence analysis of the 2.5-kb insert of pOXA-40 identified an 828-bp open reading frame (ORF), named blaOXA-40, encoding a 275-amino-acid protein (Fig. 1). The G+C content of this ORF was 35%, which is not within the expected range of the G+C content of A. baumannii genes (39 to 47%) (6). Within the deduced protein encoded by this ORF, a serine-threonine-phenylalanine-lysine tetrad (S-T-F-K) was found at positions 70 to 73 (DBL numbering [14]) (Fig. 1). Two out of the four structural elements characteristic of DBLs were found: W-X-X-X-X-L-X-I-X at DBL positions 164 to 172 and Q-X-X-X-X at DBL positions 176 to 180 (Fig. 1) (19). The third element, K-T-G at positions 216 to 218, was replaced by a K-S-G motif as in OXA-24, -25, and -26 but not as in OXA-23 and -27. The fourth structural element, YGN (DBL positions 144 to 146), found in most oxacillinases was replaced by an FGN motif as in all of the carbapenem-hydrolyzing oxacillinases. OXA-40 had one or two amino acid substitutions compared to OXA-24, -25, and -26 (Fig. 1).
FIG. 1.
Comparison of the amino acid sequence of OXA-40 to those of the most closely related oxacillinases, OXA-24, -25, and -26 (3, 10). Dashes indicate identical amino acids. The numbering is in accordance with the DBL numbering system (14). The motifs usually conserved among DBLs are shaded.
Analysis of the DNA sequences surrounding the blaOXA-40 gene revealed 100% identity with 20 and 40 bp of the DNA sequences upstream and downstream of blaOXA-25 and blaOXA-26, respectively, whereas the further 40-bp sequences located downstream were different. Additionally, further upstream of the blaOXA-40 gene, there was a 500-bp sequence that was identical to that encoding part of the 16S rRNA gene of A. baumannii, suggesting the chromosomal location of blaOXA-40.
Sequence analysis of the insert of pABC-1 that conferred a cephalosporinase phenotype identified an ORF that encodes a protein that has a one-amino-acid substitution compared to the sequence of the only known AmpC-type enzyme of A. baumannii AbRYC52763/97(data not shown) (9), thus identifying the second β-lactamase gene from A. baumannii CLA-1.
IEF analysis revealed that E. coli DH10B(pOXA-40) produces a β-lactamase with a pI of 8.6, like that found in extracts of A. baumannii CLA-1, a pI value higher than that of β-lactamases OXA-24, -25, and -26 (3, 10). In addition, extracts of E. coli DH10B(pABC-1) contained a β-lactamase with a pI value of 9.4 that was identical to one of those found in extracts of A. baumannii CLA-1 and to that of the AmpC enzyme of A. baumannii AbRYC52763/97 (9).
MICs of β-lactams for E. coli DH10B(pOXA-40) showed resistance to penicillins not antagonized by clavulanic acid and tazobactam addition and a slightly decreased susceptibility to carbapenems (Table 2). A similar pattern of resistance to β-lactams had been found for E. coli expressing OXA-23 and OXA-24 (7, 10). (MICs for E. coli expressing OXA-25, -26, -27 have not been published [3].)
Genetic location of blaOXA-40
Plasmid extraction of A. baumannii CLA-1 failed. However, extraction of plasmid DNA from Acinetobacter strains may be difficult. Thus, the location of blaOXA-40 was determined by using the endonuclease I-CeuI technique. We generated five fragments from restricted DNAs of A. baumannii CLA-1, six from the A. baumannii CIP7034T reference strain (2, 200, 500, 150, 60, and 40 Mb), and seven from the E. coli K-12 reference strain (data not shown). The DNA probe for rRNA genes hybridized with the endonuclease I-CeuI-generated fragments, except for the 2,200-Mb fragment. The blaOXA-40 probe hybridized only with the 500-Mb fragment of strain CLA-1, indicating the chromosomal location of this gene (data not shown).
Biochemical properties of β-lactamase OXA-40.
After purification from extracts of E. coli DH10B(pOXA-40), the specific activity of OXA-40 against cephalothin was 2 U/mg of protein and its purification factor was 37-fold. This relatively weak activity is similar to those of many purified oxacillinases (4, 19, 23, 24). Protein purity was estimated to be >90% (data not shown). OXA-40 has a narrow-spectrum hydrolytic profile including mostly penicillins (Table 3). Hydrolysis of oxacillin was detected as for OXA-25 and -26 (3), whereas it was not detected for OXA-24 (10). Hydrolysis of imipenem was low in level, whereas hydrolysis of meropenem was not detected. Hydrolysis of meropenem has been found for OXA-24, -25, and -26, although at a level lower than that of imipenem (3, 10). Additionally, OXA-40 retained some hydrolytic activity against ceftazidime (Table 3).
TABLE 3.
Kinetic parameters of purified β-lactamase OXA-40 and mutant enzyme OXA-40-Tyra
| Antibiotic | OXA-40
|
OXA-40-Tyr
|
||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (nM−1 · s−1) | kcat (s−1) | Km (μM) | kcat/Km (nM−1 · s−1) | |
| Benzylpenicillin | 5 | 23 | 220 | 30 | 9 | 3,000 |
| Ampicillin | 5 | 220 | 20 | 40 | 180 | 220 |
| Ticarcillin | 1 | 60 | 20 | 10 | 65 | 170 |
| Piperacillin | 1 | 23 | 50 | 15 | 40 | 350 |
| Cephalothin | 3 | 72 | 50 | 5 | 120 | 30 |
| Cephaloridine | 5 | 1,000 | 5 | 15 | 950 | 15 |
| Cefotaxime | NDb | —c | — | ND | — | — |
| Ceftazidime | 20 | 2,500 | 10 | ND | — | — |
| Cefepime | ND | — | — | 2 | 3,000 | 1 |
| Cefpirome | ND | — | — | 1 | 620 | 2 |
| Aztreonam | ND | — | — | ND | — | — |
| Imipenem | 0.1 | 6.5 | 15 | 0.5 | 1 | 360 |
| Meropenem | ND | — | — | — | — | — |
| Oxacillin | 2 | 876 | 3 | 6 | 155 | 40 |
| Cloxacillin | ND | — | — | 35 | 780 | 50 |
Data are the means of three independent experiments. Standard deviations were within 10% of the means.
ND, no detectable hydrolysis (<0.0.01 s−1).
—, not determinable.
Studies of activity inhibition, as measured by determination of IC50s, showed that OXA-40 was weakly inhibited by clavulanic acid (300 μM), tazobactam (180 μM), and sulbactam (190 μM), as found for most of the oxacillinases (17, 19). These IC50s were higher than those found for partially purified oxacillinases with carbapenem-hydrolyzing activity (3, 10), which were 6- to 100-fold lower, depending on the β-lactam inhibitors. OXA-40 activity was not inhibited by NaCl (IC50, 3 M) whereas NaCl is a good inhibitor of most oxacillinases (11, 17, 19).
Site-directed mutagenesis leading to a Tyr or a Phe residue at DBL position 144 and consequences for MICs and β-lactamase activity.
The role of the Phe residue at DBL position 144 of OXA-40 was first investigated by generating plasmid pOXA-40-Tyr, which contained the exact same insert as pOXA-40 except for a T-to-A change leading to the Phe-to-Tyr change at DBL position 144 of the encoded β-lactamase. Comparison of the MICs of β-lactams for E. coli DH10B(pOXA-40-Tyr) to those for E. coli DH10B(pOXA-40) showed similar resistance profiles, except for a decrease in the ceftazidime MIC for E. coli DH10B(pOXA-40-Tyr) (Table 2). The levels of imipenem resistance of the two strains were similar (Table 2).
The specific activities for several β-lactams were determined with extracts of E. coli DH10B harboring pOXA-40 and pOXA-40-Tyr. For all substrates, specific activities for extracts of E. coli DH10B(pOXA-40-Tyr) were higher than those for E. coli DH10B(pOXA-40) (Table 4).
TABLE 4.
Specific activities of oxacillinases OXA-40, OXA-40-Tyr, OXA-1, and OXA-1-Phe from extracts of E. coli DH10B harboring recombinant plasmids
| Substrate | Sp act (mU/mg of protein)a
|
|||
|---|---|---|---|---|
| OXA-40 | OXA-40-Tyr | OXA-1 | OXA-1-Phe | |
| Benzylpenicillin | 120 | 1,400 | 550 | NDb |
| Amoxicillin | 20 | 100 | 850 | ND |
| Piperacillin | 30 | 350 | 850 | ND |
| Cephalothin | 10 | 30 | 35 | 15 |
| Imipenem | 10 | 35 | 1.5 | 1.5 |
Standard deviations were within 20% of the means (means of three independent determinations are shown).
ND, not determinable (nonlinear hydrolysis curve).
To compare further the hydrolytic activity of OXA-40-Tyr to that of OXA-40, purification of OXA-40-Tyr was also performed. The specific activity of OXA-40-Tyr against cephalothin was 5.3 U/mg of protein, and its purification factor was 79-fold. Protein purity was estimated to be >90% (data not shown). The catalytic efficacy of OXA-40-Tyr was higher than that of OXA-40 for all substrates except cephalothin and ceftazidime (Table 3). The catalytic efficacy of OXA-40-Tyr for imipenem was 24-fold higher than that of OXA-40 because of both higher hydrolytic activity and greater affinity (Table 3). Kinetic parameters for cloxacillin could be determined for OXA-40-Tyr, whereas no values could be obtained for OXA-40 (Table 3). Inhibition of β-lactamase activities showed that the IC50s of tazobactam and sulbactam for OXA-40-Tyr and OXA-40 were similar whereas the IC50 of clavulanic acid for OXA-40-Tyr (1 mM) was threefold greater than that for OXA-40. The Phe-to-Tyr substitution in OXA-40-Tyr compared to OXA-40 restored its susceptibility to NaCl inhibition (IC50, 40 mM).
To evaluate further the role of the Phe residue at DBL position 144 in conferring carbapenem-hydrolyzing activity, we performed a Tyr-to-Phe residue change at DBL position 144 of the amino acid sequence of OXA-1, an oxacillinase that does not possess significant carbapenem-hydrolyzing activity (19). A site-directed mutagenesis experiment with pOXA-1, containing the blaOXA-1 gene, as the template was then performed to obtain recombinant plasmid pOXA-1-Phe, which encodes OXA-1-Phe with a Tyr-to-Phe substitution at DBL position 144. Specific activity of extracts of E. coli DH10B(pOXA-1) for the β-lactams studied was detected, although that for imipenem was feeble (Table 4). The specific activity of extracts of E. coli DH10B(pOXA-1-Phe) for cephalothin was lower than that of extracts of E. coli DH10B(pOXA-1) (Table 4). The Tyr-to-Phe substitution at DBL position 144 led to a decrease in the overall hydrolytic activity of OXA-1-Phe, mirroring the results obtained with OXA-40 and OXA-40-Tyr. Extracts of E. coli DH10B producing OXA-1 and OXA-1-Phe gave the same low-level activity against imipenem (Table 4). Inhibition of β-lactamase activities showed that the IC50s of NaCl for extracts of E. coli DH10B expressing OXA-1 and OXA-1-Phe were 50 and 2,000 mM, respectively, mirroring the IC50s of NaCl for OXA-40-Tyr and OXA-40, respectively.
DISCUSSION
This work characterizes the genetics and biochemistry of a carbapenem-hydrolyzing oxacillinase, OXA-40, produced by a multiple-drug-resistant A. baumannii clinical isolate from Portugal. OXA-40 had a few amino acid changes compared to OXA-24, -25, and -26, belonging to the same subgroup of carbapenem-hydrolyzing oxacillinases. While this work was in progress, others (F. Lopez-Otsoa, L. Gallego, K. Towner, L. Tysall, D. Livermore, and N. Woodford, 12th Eur. Congr. Clin. Microbiol. Infect. Dis., abstr. P1442, 2002) reported the same β-lactamase from A. baumannii isolates from Bilbao (northwestern Spain), and OXA-24 to -26 were from Spain (Madrid) and Belgium (Ghent) (3, 10). Thus, a subgroup of carbapenem-hydrolyzing oxacillinases may have spread in A. baumannii, at least in Europe.
The chromosomal location of the blaOXA-40 gene has been determined clearly as it has been suggested for the genes encoding OXA-24, -25, and -26 (3, 10). The DNA sequences located immediately up- and downstream of the oxacillinase genes encoding OXA-25, -26, -40 were the same, suggesting a common origin of these β-lactamase genes, whereas a clonal relationship was not identified for the OXA-24- and OXA-25-producing A. baumannii isolates (3). The mechanism of transfer of these oxacillinase genes remains unknown, since none of these genes has been found to be bracketed by class 1 integron or insertion sequences.
The two groups of carbapenem-hydrolyzing oxacillinases (OXA-23 and -27 on the one hand and OXA-24, -25, -26, and -40 on the other) have an FGN triad instead of the common YGN motif at DBL positions 144 to 146. Contrary to expected results (3, 10, 15), a Phe-to-Tyr substitution at DBL position 144 in OXA-40 gave a mutant enzyme, OXA-40-Tyr, that had stronger hydrolytic activity against most β-lactams, including imipenem. Additionally, a Tyr-to-Phe substitution at DBL position 144 in the narrow-spectrum oxacillinase OXA-1 did not provide additional carbapenem-hydrolyzing activity but instead gave a mutant enzyme with weaker overall hydrolytic activity. These experiments indicated that the Phe residue at DBL position 144 is not, by itself, responsible for carbapenem-hydrolyzing activity and that this residue, in the place of a Tyr residue, confers weak overall hydrolytic activity on carbapenem-hydrolyzing oxacillinases. The weak activity of these enzymes may explain why they have remained undetected for a long time in carbapenem-resistant A. baumannii isolates and why carbapenem resistance in A. baumannii may result from combined antibiotic resistance mechanisms (1, 8).
Resistance to inhibition by NaCl was also found in OXA-25 and -26 (3), whereas susceptibility to inhibition by NaCl was surprisingly reported for the structurally related oxacillinase OXA-24 (10). We have shown that resistance to inhibition and, conversely, susceptibility to inhibition by NaCl are related to the presence of Phe and Tyr residues at DBL position 144, respectively. Substitution of a Tyr residue for a more neutral Phe residue may prevent NaCl from binding the active site of oxacillinases, which involves at least the amino acid residues at DBL positions 144 to 146 (19).
Finally, whereas detection of carbapenem-hydrolyzing oxacillinases is difficult in multidrug-resistant A. baumannii isolates, it remains to be determined if these strains are a reservoir of carbapenem-hydrolyzing genes spreading to other gram-negative rods. In that respect, OXA-23 has been recently identified in Proteus mirabilis isolates (7).
Acknowledgments
This work was funded by a grant from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA), Université Paris XI.
We thank C. Bizet for the gift of the A. baumannii reference strain.
REFERENCES
- 1.Afzal-Shah, M., and D. M. Livermore. 1998. Worldwide emergence of carbapenem-resistant Acinetobacter spp. J. Antimicrob. Chemother. 41:576-577. [DOI] [PubMed] [Google Scholar]
- 2.Afzal-Shah, M., H. E. Villar, and D. M. Livermore. 1999. Biochemical characteristics of a carbapenemase from an Acinetobacter baumannii isolate collected in Buenos Aires, Argentina. J. Antimicrob. Chemother. 43:127-131. [DOI] [PubMed] [Google Scholar]
- 3.Afzal-Shah, M., N. Woodford, and D. M. Livermore. 2001. Characterization of OXA-25, OXA-26, and OXA-27, molecular class D β-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 45:583-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aubert, D., L. Poirel, J. Chevalier, S. Léotard, J.-M. Pagès, and P. Nordmann. 2001. Oxacillinase-mediated resistance to cefepime and susceptibility to ceftazidime in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 45:1615-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avidor, B., Y. Kletter, S. Abulafia, Y. Golan, M. Ephros, and M. Giladi. 1997. Molecular diagnosis of cat scratch disease: a two step approach. J. Clin. Microbiol. 35:1924-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergogne-Bérézin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnet, R., H. Marchandin, C. Chanal, D. Sirot, R. Labia, C. De Champs, E. Jumas-Bilak, and J. Sirot. 2002. Chromosome-encoded class D β-lactamase OXA-23 in Proteus mirabilis. Antimicrob. Agents Chemother. 46:2004-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bou, G., G. Cervero, M. A. Dominguez, C. Quereda, and J. Martinez-Beltran. 2000. Characterization of a nosocomial outbreak caused by a multiresistant Acinetobacter baumannii strain with a carbapenem-hydrolyzing enzyme: high-level carbapenem resistance in A. baumannii is not due solely to the presence of β-lactamases. J. Clin. Microbiol. 38:3299-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bou, G., and J. Martinez-Beltran. 2000. Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 44:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bou, G., A. Oliver, and J. Martinez-Beltran. 2000. OXA-24, a novel class D beta-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 44:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu, Y. W., M. Afzal-Shah, E. T. Houang, M. I. Palepou, D. J. Lyon, N. Woodford, and D. M. Livermore. 2001. IMP-4, a novel metallo-β-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 45:710-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark, R. B. 1996. Imipenem resistance among Acinetobacter baumannii: association with reduced expression of a 33-36 kDa outer membrane protein. J. Antimicrob. Chemother. 38:245-251. [DOI] [PubMed] [Google Scholar]
- 14.Couture, F., J. Lachapelle, and R. C. Lévesque. 1992. Phylogeny of LCR-1 and OXA-5 with class A and class D β-lactamases. Mol. Microbiol. 6:1693-1705. [DOI] [PubMed] [Google Scholar]
- 15.Donald, H. M., W. Scaife, S. G. Amyes, and H. K. Young. 2000. Sequence analysis of ARI-1, a novel OXA β-lactamase responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob. Agents Chemother. 44:196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gehrlein, M., H. Leying, W. Cullmann, S. Wendt, and W. Opferkuch. 1991. Imipenem resistance in Acinetobacter baumannii is due to altered penicillin binding protein. Chemotherapy 37:405-412. [DOI] [PubMed] [Google Scholar]
- 17.Ledent, P., X. Raquet, B. Joris, J. Van Beeumen, and J.-M. Frère. 1993. A comparative study of class-D β-lactamases. Biochem. J. 292:555-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, S. L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. USA 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naas, T., and P. Nordmann. 1999. OXA-type β-lactamases. Curr. Pharm. Design 5:865-879. [PubMed] [Google Scholar]
- 20.NCCLS. 2001. Performance standards for antimicrobial susceptibility testing. Eleventh informational supplement. M100-S11. NCCLS, Wayne, Pa.
- 21.Ouelette, M., L. Bissonnette, and P. H. Roy. 1987. Precise insertions of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 β-lactamase genes. Proc. Natl. Acad. Sci. USA 84:7378-7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paton, R., R. S. Miles, J. Hood, and S. G. B. Amyes. 1993. ARI-1: β-lactamase mediated imipenem resistance in Acinetobacter baumannii. Int. J. Antimicrob. Agents 2:81-88. [DOI] [PubMed] [Google Scholar]
- 23.Philippon, L. N., T. Naas, A.-T. Bouthors, V. Barakett, and P. Nordmann. 1997. OXA-18, a class D clavulanic acid-inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2188-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poirel, L., D. Girlich, T. Naas, and P. Nordmann. 2001. OXA-28, an extended-spectrum variant of OXA-10 β-lactamase from Pseudomonas aeruginosa and its plasmid- and integron-located gene. Antimicrob. Agents Chemother. 45:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poirel, L., and P. Nordmann. 2002. Acquired carbapenem-hydrolyzing β-lactamases and their genetic support. Curr. Pharm. Biotechnol. 3:117-127. [DOI] [PubMed] [Google Scholar]
- 26.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of bla(IMP) allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Scaife, W., H. K. Young, R. H. Paton, and S. G. Amyes. 1995. Transferable imipenem resistance in Acinetobacter species from a clinical source. J. Antimicrob. Chemother. 36:585-586. [DOI] [PubMed] [Google Scholar]
- 29.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urban, C., E. S. Go, K. S. Meyer, N. Mariano, and J. J. Rahal. 1995. Interactions of sulbactam, clavulanic acid and tazobactam with penicillin binding proteins of imipenem-resistant and susceptible Acinetobacter baumannii. FEMS Microbiol. Lett. 125:193-197. [Google Scholar]
- 31.Yum, J. H., K. Yi, H. Lee, D. Yong, K. Lee, J. M. Kim, G. M. Rossolini, and Y. Chong. 2002. Molecular characterization of metallo-β-lactamase-producing Acinetobacter baumannii and Acinetobacter genomospecies 3 from Korea: identification of two new integrons carrying the bla (VIM-2) gene cassettes. J. Antimicrob. Chemother. 49:837-840. [DOI] [PubMed] [Google Scholar]