Abstract
A base-modified nucleoside analogue, β-d-N4-hydroxycytidine (NHC), was found to have antipestivirus and antihepacivirus activities. This compound inhibited the production of cytopathic bovine viral diarrhea virus (BVDV) RNA in a dose-dependant manner with a 90% effective concentration (EC90) of 5.4 μM, an observation that was confirmed by virus yield assays (EC90 = 2 μM). When tested for hepatitis C virus (HCV) replicon RNA reduction in Huh7 cells, NHC had an EC90 of 5 μM on day 4. The HCV RNA reduction was incubation time and nucleoside concentration dependent. The in vitro antiviral effect of NHC was additive with recombinant alpha interferon-2a and could be prevented by the addition of exogenous cytidine and uridine but not of other natural ribo- or 2′-deoxynucleosides. When HCV RNA replicon cells were cultured in the presence of increasing concentrations of NHC (up to 40 μM) for up to 45 cell passages, no resistant replicon was selected. Similarly, resistant BVDV could not be selected after 20 passages. NHC was phosphorylated to the triphosphate form in Huh7 cells, but in cell-free HCV NS5B assays, synthetic NHC-triphosphate (NHC-TP) did not inhibit the polymerization reaction. Instead, NHC-TP appeared to serve as a weak alternative substrate for the viral polymerase, thereby changing the mobility of the product in polyacrylamide electrophoresis gels. We speculate that incorporated nucleoside analogues with the capacity of changing the thermodynamics of regulatory secondary structures (with or without introducing mutations) may represent an important class of new antiviral agents for the treatment of RNA virus infections, especially HCV.
Hepatitis C virus (HCV) is one of the most important Flaviviridae infections in humans and is responsible for the second most common cause of viral hepatitis. Presently, nearly 2% of the U.S. population, and an estimated 170 million people worldwide, are HCV carriers (2). The only approved therapy for chronic hepatitis C is alpha interferon (IFN-α), either alone or in combination with ribavirin. Anemia is the most common adverse effect associated with ribavirin treatment, and neuropsychiatric adverse effects of IFN-α lead to premature cessation of therapy in 10 to 20% of patients (9, 13).
As additional treatment options are urgently needed, there is an ongoing search for more potent antiviral compounds with fewer adverse effects. However, the search for improved antiviral agents is hampered by the limited and cumbersome propagation of HCV in vitro (4). Therefore, surrogate models such as the HCV RNA replicon that replicates in the human hepatoma cell line Huh7 have been developed (6, 29). Improved versions of these HCV replicons contain adaptive mutations (25), and their use has facilitated the evaluation of candidate anti-HCV drugs.
Bovine viral diarrhea virus (BVDV) is one of the best characterized members of the Flaviviridae family and has one of the largest RNA genomes (12.5 kb) in this family (8). This virus has the remarkable property of existing as noncytopathic and cytopathic (cpBVDV) biotypes, with cpBVDV strains showing insertions or viral genome rearrangements at the junction site between nonstructural protein 2 (NS2) and NS3 (32). BVDV may provide a surrogate model for HCV, both for the molecular study of viral proteins (33) and for the evaluation of antiviral compounds (3, 7, 47).
In the search for therapeutic agents, any element that is essential for viral (or replicon) RNA replication may be considered a drug discovery target. Such elements can be either viral proteins (NS2-NS3 protease, NS3-NS4A serine proteinase, NS3 RNA helicase, or RNA-dependent RNA polymerase [3, 24, 34, 36]), viral cis-acting RNA elements (internal ribosomal entry site [IRES] antisense oligonucleotides or ribozymes [1, 26]), or cellular elements (IFN-α, ribavirin, and IMP dehydrogenase [EC 1.1.1.205] inhibitors [18, 31, 41, 43]). Current knowledge of the human genome, combined with array technology and pathogen infection models, will likely lead to more defined host-pathogen-related targets for future drug design (17, 23). Today, however, the most successful classes of antiviral compounds with clinical utility in combat against other human viral pathogens (human immunodeficiency virus type 1 [HIV-1], hepatitis B virus [HBV], herpes simplex virus [HSV], and cytomegalovirus) are the protease, the nonnucleoside analogue, and the nucleoside analogue inhibitors. As the latter class of compounds is crucial in controlling herpesvirus, HIV-1, and HBV infections, it is likely and anticipated that nucleoside analogue inhibitors will also play an important role in the successful treatment of HCV.
The present report describes how a surrogate BVDV acute infection system and a persistent HCV replicon RNA system in Huh7 cells resulted in the discovery of a base- modified ribonucleoside analogue, β-d-N4-hydroxycytidine (NHC).
MATERIALS AND METHODS
Synthesis of NHC and NHC-TP.
synthesis of NHC and NHC-triphosphate (NHC-TP) will be described elsewhere (K. A. Watanabe et al., unpublished data). [3H]NHC was custom synthesized by radiolabeling at the 5 position of the base (13 Ci/mmol) (Moravek, Brea, Calif.).
Anti-BVDV quantitative real-time RT-PCR (Q-RT-PCR) assay.
Madin-Darby bovine kidney (MDBK) cells (ATCC CCL22) were grown in Dulbecco's modified Eagle's medium-F12 (Gibco/BRL, Gaithersburg, Md.) supplemented with 10% heat-inactivated horse serum (Gibco/BRL). The cells were tested for BVDV contamination by reverse transcriptase (RT) PCR as described previously (45). cpBVDV was kindly provided by Ruben Donis, University of Nebraska. cpBVDV was plaque purified three times on MDBK cell monolayers prior to large-scale virus stock preparation.
Cells were seeded in 96-well plates at 5 × 103 cells/well and incubated for 1 h (for the experiments on exponentially growing cells) or for 72 h (for experiments on confluent cells). The monolayer was infected with cpBVDV at a multiplicity of infection of 0.02 PFU/cell. After 45 min of infection, the viral inoculum was removed and the cells were washed twice with culture medium. Medium or medium containing test compound was added to these cells, followed by incubation for 24 h (exponentially growing cells) or 72 h (confluent cell monolayer). This difference in incubation time was determined after studying the dynamics of virus release in cell supernatant; there was an approximately equal amount of virus (copies/ml) produced in 24 h on exponentially growing cells as there was in 72 h on a confluent cell monolayer (data not shown). Cell culture medium was collected and clarified by centrifugation (2 min, 3,000 × g, room temperature), and viral RNA was prepared (QIAamp viral RNA mini kit; Qiagen, Valencia, Calif.). Viral RNA was detected by Q-RT-PCR. Primers and probe were designed for the BVDV NS5B region by using Primer Express software (Applied Biosystems, Foster City, Calif.), the sense primer 5′-AGCCTTCAGTTTCTTGCTGATGT-3′, probe 5′-6FAM-AAATCCTCCTAACAAGCGGGTTCCAGG-TAMRA-3′, and the antisense primer 5′-TGTTGCGAAAGGACCAACAG-3′. A standard curve with 1-log-diluted BVDV RNA from infected MDBK cells as template was generated, and it showed a linear range of at least 6 log (data not shown). Ribavirin (1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxyamide; Schering-Plough, Raritan, N.J.) was used as a positive control in these experiments.
BVDV single-cycle replication assay.
MDBK cells (3 × 105 cells per 20-mm-diameter well) were infected with cpBVDV in triplicate at a multiplicity of infection of 2.5 PFU/cell. At 1 h after infection, the virus inoculum was removed and the monolayers were washed twice with MDBK cell medium (1 ml). MDBK cell medium supplemented with 0, 1, 2, 5, 10, or 20 μM NHC was added to the cultures. At 24 h postinfection, the cultures were harvested and frozen at −70°C. The cultures were thawed at 37°C and sonicated at 0°C. Cell debris was removed by centrifugation at 1,000 × g for 10 min at 4°C, and virus yields in the supernatant fluids were measured by plaque assay.
Plaque assay for BVDV.
MDBK cell monolayers (3 × 105 cells per 20-mm-diameter well) were infected with 100 μl of 10-fold serial dilutions of cpBVDV suspended in medium. One hour after infection, the inoculum was removed, the monolayers were washed once with MDBK cell medium, and 1 ml of MDBK cell medium overlay containing 1% methylcellulose was added to the monolayers. As most plaques were counted at the 10−4 dilution of virus suspension, the concentrations of NHC that might be carried over in the virus suspension were well below that necessary to inhibit virus replication. Plaques were counted at 48 to 72 h postinfection.
BVDV resistance studies.
MDBK cells were seeded at 4 × 105 cells per 35-mm-diameter dish, infected with cpBVDV at 0.01 to 0.05 PFU/cell in 0.5 ml of medium, and incubated for 1 h at 37°C. The virus inoculum was removed, the cultures were washed twice in 1× phosphate-buffered saline, and medium containing 0, 2, 5, 10, 15, or 20 μM NHC was added. To generate resistant virus stocks, BVDV was grown in the presence of increasing concentrations of NHC. The cultures were incubated at 37°C until complete cytopathic effect (CPE) was achieved. Virus suspensions were generated by scraping the infected cells into the medium and freezing the samples at −70°C. The samples were thawed and sonicated. Cell debris was removed by centrifugation at 1,000 × g for 10 min at 4°C, and virus yields in the supernatant fluids were determined by plaque assays. Virus suspensions from samples that showed complete CPE and had titers of >103 were used to infect fresh MDBK cell monolayers.
HCV replicon assay.
HCV replicon RNA-containing Huh7 cells (clone A cells; Apath, LLC, St. Louis, Mo.) were kept at exponential growth in Dulbecco's modified Eagle's medium (high glucose, no pyruvate) containing 10% fetal bovine serum, 1× nonessential amino acids, penicillin-streptomycin-glutamine (100 U/ml, 100 μg/ml, and 0.292 mg/ml, respectively), and G418 (500 to 1,000 μg/ml). Antiviral assays were performed in the same medium without G418. Cells were seeded in a 96-well plate at 1,000 cells per well, and test compounds were added immediately after seeding. Incubation times differed according to the type of experiment. At the end of the incubation step, total cellular RNA was isolated (RNeasy 96 kit; Qiagen). Replicon RNA and an internal control (TaqMan rRNA control reagents; Applied Biosystems) were amplified in a single-step multiplex RT-PCR protocol as recommended by the manufacturer. The HCV primers and probe were designed with Primer Express software (Applied Biosystems) and covered highly conserved 5′-untranslated region (UTR) sequences (sense, 5′-AGCCATGGCGTTAGTA(T/A)GAGTGT-3′, and antisense, 5′-TTCCGCAGACCACTATGG-3′; probe, 5′-FAM-CCTCCAGGACCCCCCCTCCC-TAMRA-3′).
To express the antiviral effectiveness of a compound, the threshold RT-PCR cycle of the test compound was subtracted from the average threshold RT-PCR cycle of the no-drug control (ΔCtHCV). A ΔCt of 3.3 equals a 1-log reduction (equal to the 90% effective concentration [EC90]) in replicon RNA levels. The cytotoxicity of the test compound could also be expressed by calculating the ΔCtrRNA values. The ΔΔCt specificity parameter could then be introduced (ΔCtHCV − ΔCtrRNA), in which the levels of HCV RNA are normalized for the rRNA levels and calibrated against the no-drug control. Recombinant alpha interferon-2a (IFN-α-2a) (Roferon-A; Hoffmann-La Roche, Inc., Nutley, N.J.) served as a positive control.
Northern and Western blots.
HCV replicon cells were grown in the presence of 65 μM NHC for 4 days. For Northern blot analysis, total RNA was extracted and manipulated according to the instruction manual in the NorthernMax-Gly system for Northern blotting (Ambion, Austin, Tex.). Minus-strand RNA probes were prepared by using a T7-RNA polymerase with the incorporation of [α-32P]UTP (Maxiscript in vitro transcription kit; Ambion).
For Western blotting, cells were washed twice, trypsinized, collected by centrifugation (14,000 × g, 1 min), and resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (40) to a concentration of 104 cells/μl. Cellular proteins were separated by 8% polyacrylamide gel electrophoresis and electroblotted onto a BA85 nitrocellulose membrane (Schleicher and Schuell, Keene, N.H.). The membrane was blocked overnight in 1% blocker casein (Pierce, Rockford, Ill.)-0.1% Tween 20. The monoclonal antibody 5B-3B1 (kindly provided by Darius Moradpour, Freiburg, Germany), targeting an epitope in the NS5B protein, was diluted 1:10,000 in blocking solution and incubated with the membrane for 2 h with agitation. The membrane was washed four times with TNET (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.1 mM EDTA, 0.1% Tween 20) and incubated with 1:1,000× diluted secondary antibody (goat anti-mouse immunoglobulin G-alkaline phosphatase; Pierce) in TNET for 2 h. The blot was then washed four times with TNET and developed with One Step Nitro Blue Tetrazolium-BCIP (5-bromo-4-chloro-3-indolylphosphate) (Pierce). The reaction was stopped by washing with 10 mM Tris-HCl (pH 7.5)-100 mM EDTA.
NS5B enzyme assay.
The 21-amino-acid carboxy-terminal truncated NS5B protein was cloned from the HCV replicon cells, modified with a six-His-terminal tail, expressed in a prokaryotic expression vector (pQE60; Qiagen), and subsequently purified over a Talon cobalt affinity resin column (Clontech, Palo Alto, Calif.). Purification was monitored by SDS-PAGE and Western blotting. The resulting purified protein was dialyzed overnight against 50 mM sodium phosphate (pH 8.0)-300 mM sodium chloride-0.5% Triton X-100-50% glycerol-2 mM dithiothreitol. The dialysate maintained consistent activity for more than 6 months when stored at −20°C. Protein was quantified with the Coomassie Plus protein assay reagent (Pierce) by using a bovine serum albumin standard from the same supplier. This purified protein was then used in the RNA polymerase activity assays with a purified T7-derived synthetic RNA molecule as template, as described previously (39). NS5B assays used 200 ng of negative-strand IRES RNA as template, 200 ng of purified NS5B protein, 20 U of Anti-RNase (Ambion), 500 μM GTP, 5 μM ATP and CTP, 2 μM UTP, 10 μCi [α-32P]UTP (800 Ci/mmol and 20 mCi/ml; all nucleotides were obtained from Amersham Biosciences, Piscataway, N.J.), 50 mM HEPES-NaOH (pH 7.5), 1 mM MgCl2, 0.75 mM MnCl2, and 2 mM dithiothreitol in a final volume of 20 μl. Reactions were performed at 27°C for 2 h; these conditions were essentially as described previously and resulted in an endpoint detection assay (28, 30, 39). The reaction was terminated by the addition of 80 μl of 5 mM EDTA-0.1% SDS. The resulting solution was extracted with 300 μl of Trizol-LS (Invitrogen Life Technologies, Carlsbad, Calif.) and 80 μl of chloroform. RNA was precipitated from the aqueous phase by the addition of 5 μg of glycogen (Ambion), 1/10 volume of 5 M ammonium acetate, and 1 volume of 2-propanol. This solution was stored at −20°C for 1 h, and the RNA was collected by centrifugation at 13,100 × g. The RNA pellet was washed with 70% EtOH and briefly air dried. RNA pellets were resuspended in 5 μl of Gel Loading Buffer II (95% formamide, 0.025% xylene cyanol, 0.025% bromophenol blue, 18 mM EDTA, 0.025% SDS; Ambion) and heated to 100°C for 5 min. Samples were directly placed on ice for 2 min and loaded on a 5% denaturing PAGE sequencing gel. Electrophoresis proceeded at 60 W constant power until the bromophenol blue was eluted. The gel was then exposed for 1 h, and data was collected from a Molecular Dynamics model 625E phosphorimager (Amersham Biosciences).
Uptake of NHC in HCV replicon cells.
Confluent replicon cells were incubated with 10 μM [3H]NHC (500 dpm/pmol) for specific time periods, after which the cell layers were washed with cold phosphate-buffered saline. Nucleotides were extracted by overnight incubation at −20°C in 60% MeOH (1 ml), followed by 1-h incubation (200 μl, 60% methanol) on ice. Extracts were combined, dried, resuspended in water, and analyzed by high-performance liquid chromatography on a Phenomenex 5-μm-diameter C18 (250 by 4.6 mm) column. The mobile phase consisted of buffer A (25 mM ammonium acetate with 5 mM tetrabutyl ammonium phosphate (TBAP), pH 7.0) and buffer B (methanol). Detection was done at 238 nm. Elution was performed using a multistage linear gradient of buffer B from 0 to 40% for 60 min at a flow rate of 1 ml/min. Radioactivity was analyzed with the 500TR Radiometric Flo-One radiochromatography analyzer (Packard Instrument Company, Inc., Meriden, Conn.). NHC-TP concentrations were calculated by reference to the standard curves obtained by analyzing chemically synthesized NHC-TP under similar conditions.
Cytotoxicity assays.
MDBK, Huh7 cells, HCV replicon cells, HepG2 cells, or human peripheral blood monocyte (PBM) cells (5 × 104 per well) were seeded in 96-well plates in the presence of increasing concentrations of the test compound and incubated in a 5% CO2 incubator at 37°C. After a 3-day incubation for HepG2 cells or a 5-day incubation for PBM cells, cell viability was measured in a colorimetric assay using the CellTiter 96 AQueous One Solution cell proliferation assay (Promega, Madison, Wis.). Toxicity levels were expressed as the concentration that inhibited cell growth by 50% (IC50).
Mitochondrial toxicity testing.
Low-passage-number HepG2 cells were seeded at 5,000 cells/well in collagen-coated 96-well plates. Test compounds were added to the medium to obtain a final concentration of 10 μM. On culture day 7, cellular nucleic acids were extracted (RNeasy 96 kit; Qiagen). The mitochondrial cytochrome C oxidase subunit II (COXII) gene and the β-actin gene were amplified from 5 μl of sample with a multiplex quantitative PCR protocol as described previously (44). Lactic acid concentrations in the culture medium were determined using the d-lactic acid-l-lactic acid UV method (R-Biopharm GmbH, D-64293 Darmstadt, Germany).
Evaluation of toxicity in mice.
Six-week-old female Swiss mice (SWR/J; Charles River Laboratory, Wilmington, Mass.) were injected intraperitoneally (i.p.) with 300 μl at doses of 0, 3, 10, 33, or 100 mg/kg of body weight/day for 6 days. Compound was dissolved in pyrogen-free, sterile 0.85% NaCl solution. Animals were monitored every day for weight (loss or gain), ruffled fur, and mortality up to 24 days after the end of treatment. The statistical significance of changes in animal weight was evaluated by one-way analysis of variance using Sigma Stat version 2.0 (SPSS Sciences, Chicago, Ill.). A P of <0.05 was deemed statistically significant. All experiments were conducted under the approval of the Institutional Animal Care and Use Committee (IACUC) of the Department of Veteran Affairs, Atlanta, Ga.
RESULTS
Anti-BVDV activity.
During the course of evaluating our nucleoside analogue library for the inhibition of cpBVDV replication, we discovered that NHC (Fig. 1) exhibits potent inhibition of viral RNA production in MDBK cell supernatant. NHC showed a dose-dependent inhibition of cpBVDV RNA production, with an EC90 of 5.4 ± 0.9 μM (Fig. 2A). In these experiments, d-ribavirin, included as a positive control, had an EC90 of 3.5 ± 1.5 μM. Both NHC and ribavirin showed significant toxicity in MDBK cells in exponential growth phase (IC50 = 7 and 5 μM, respectively) but were essentially not toxic when confluent cells were used (IC50 > 75 μM). On such a monolayer of MDBK cells, NHC had an EC90 of 3.4 ± 0.08 μM while ribavirin showed an EC90 of 6.8 ± 0.08 μM against cpBVDV.
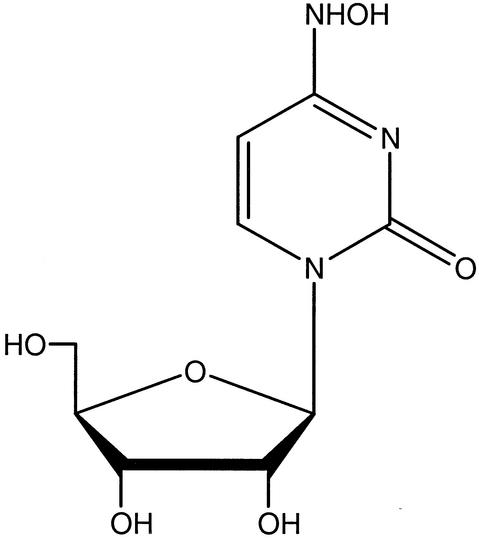
FIG. 1.
Chemical structure of NHC.
FIG. 2.
Antiviral effect of NHC on cpBVDV. EC90s mentioned in the text are deduced from the graphs. Error bars for all data points are included but are very often less than the size of the symbol. (A) Dose-dependent viral RNA yield reduction in MDBK cell supernatant. Cell supernatant fluids were harvested 24 h (on exponentially growing cells) or 72 h (on confluent monolayer cells) postinfection, and viral RNA levels were quantified by Q-RT-PCR. The zero value corresponds to no drug added to the medium. ▿, ribavirin (confluent cells); •, ribavirin (exponentially growing cells); ▪, NHC (confluent cells); ○, NHC (exponentially growing cells); ▾, control compound showing no activity. VL, viral load; S.D., standard deviation. (B) Dose-dependent reduction of viral RNA in a yield reduction assay. cpBVDV was recovered from a single-cycle infection assay, and the virus yield was titrated by means of a plaque assay. •, NHC; ▪, NN-DNJ (47).
The dose-dependent reduction of viral RNA after treatment with NHC and N-nonyl-deoxynojirimycin (NN-DNJ) (47) was also confirmed in a yield reduction assay (Fig. 2B). cpBVDV was recovered from a single-cycle infection assay, and the virus yield was titrated by plaque assay. NHC had an EC90 of 2 μM, while the EC90 for NN-DNJ was approximately 67 μM. Note that NN-DNJ (and other glucosidase inhibitors) inhibits viral replication by reducing the infectivity of newly synthesized virus particles with only a slight effect on viral RNA synthesis in single-cycle virus yield assays (22). The results of the yield assay were in agreement with those of the Q-RT-PCR assay, which measured BVDV RNA levels.
Dynamics of the replicon RNA levels.
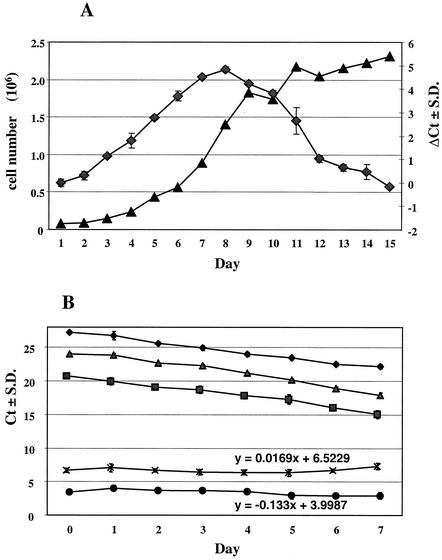
As considerable incubation time- dependent fluctuations in the amounts of HCV RNA in replicon cells were observed (37), we designed a time course experiment in which the cellular nucleic acids and the HCV RNA dynamics in the replicon cells were studied in detail. The dynamics of the HCV RNA levels increased in parallel with the cell count (Fig. 3A). At day 7, a plateau level was reached and then there was a steady decrease of HCV RNA as the cells became confluent. Furthermore, exponentially growing cells were seeded in 96-well plates and kept in culture until a confluent monolayer of cells was visually observed. Cells were harvested daily for 7 days, and the levels of nucleic acids were determined (Fig. 3B). Based on the amplification results of the rRNA gene, it was calculated that HCV replicon cells produced five cell generations (equivalent to five PCR cycles) in 7 days, which corresponds to a doubling time of approximately 28 h. Both the rRNA and HCV RNA levels followed slopes similar to those for the rRNA gene, as further demonstrated by the slope of the ΔCt curves. In these experiments, the selected seeding and assay conditions (see Materials and Methods) stabilized the HCV RNA levels over a period of at least 7 days, a prerequisite for reliable compound evaluation in this system.
FIG. 3.
Dynamics of the cellular and HCV RNA levels. Ct, Q-RT-PCR threshold cycle; ΔCt: subtraction of the one PCR threshold cycle from another; S.D., standard deviation. (A) HCV replicon cells were seeded at 104 cells per well in a 24-well plate. Cells were harvested daily and counted, and total RNA was extracted. ♦, ΔCtHCV(day x−day 0); ▴, number of viable cells per well. (B) Monitoring of the cellular DNA and RNA levels over a 7-day period. Exponentially growing HCV replicon cells were seeded at 103 cells/well in a 96-well plate and harvested daily, and total RNA was extracted and quantified. ♦, rRNA gene (DNA); ▪, rRNA; ▴, HCV; ×, ΔCt(rRNA gene−rRNA); •, ΔCt(HCV−rRNA).
Anti-HCV RNA effect of NHC on HCV replicon cells.
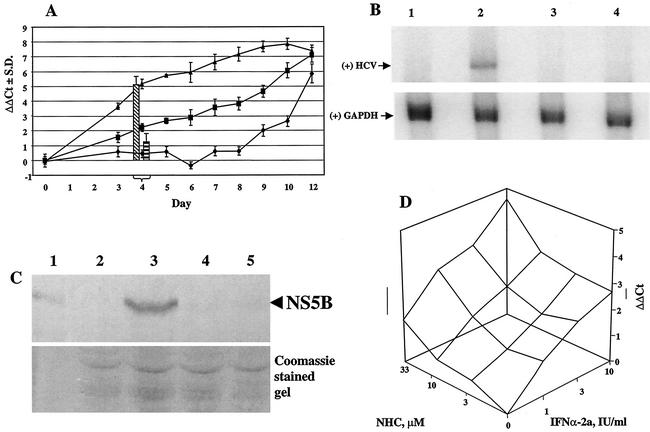
HCV RNA levels were evaluated in the presence of NHC over a period of 12 days (Fig. 4A). The ΔΔCt values for HCV in the absence of any drug (ΔΔCt(HCV Ct−rRNA Ct−no-drug Ct)) were close to zero for up to 8 days, indicating that the HCV RNA level was constant. However, from day 9 onwards, when the cells reached confluency, a decrease in intracellular HCV RNA was observed, as indicated by an increase in the ΔΔCt value. In parallel, cells were treated with 10 or 100 μM NHC, which resulted in an ongoing reduction in HCV RNA levels (Fig. 4A). In the 10 μM experiment, the slope of the curve did not change for up to 8 days and hence a new steady-state level of the intracellular RNA ratio (HCV/rRNA) was not reached. Clearly, treatment with NHC resulted in a ΔΔCt value of ≥3.3 at day 8 (or an EC90 of 10 μM at day 8). Based on similar experiments (data not shown), including dose response curves for NHC and with harvesting at day 4, we concluded that this nucleoside analogue showed the following antiviral and selectivity index characteristics: (i) an EC50 of 0.7 μM and EC90 of 5 μM, calculated from ΔCt(HCV Ct−no-drug Ct), and (ii) an EC50 of 1.6 μM and EC90 of 65 μM, calculated from ΔΔCt(HCV Ct−rRNA Ct−no-drug Ct).
FIG. 4.
Antiviral effect of NHC on HCV RNA levels. (A) Dynamics of the antiviral effect of NHC over a 12-day treatment period are shown. ΔΔCt(HCV Ct−rRNA Ct−no-drug Ct); ♦, no treatment; ▪, NHC at 10 μM; ▴, NHC at 100 μM. The antiviral activities at day 4 of IFN-α-2a (100 IU/ml [left bar]) and ribavirin (100 μM [right bar]) are given as references. S.D., standard deviation. (B) Cells were treated for 4 days, followed by RNA extraction and Northern blot analysis. The upper gel shows the positive-strand (+) HCV RNA with a hybridization signal at approximately 8 kb. The lower gel shows the control hybridization of GAPDH mRNA at approximately 1.4 kb. Lane 1, parental Huh7 cell line; lane 2, HCV replicon cells, untreated; lane 3, 100 IU of IFN-α-2a/ml; lane 4, 65 μM NHC. (C) Cells were treated for 4 days, followed by protein extraction and Western blot analysis. The upper gel shows the α-NS5B reactivity of proteins transferred to the membrane; the lower gel is a Coomassie-stained gel illustrating equal amounts of protein in each lane (total cellular protein equivalent of 5 × 104 cells per lane was loaded). Lane 1, recombinant HCV NS5B protein; lane 2, parental Huh7 cell line; lane 3, HCV replicon cells, untreated; lane 4, 100 IU of IFN-α-2a/ml; lane 5, 65 μM NHC. (D) HCV replicon cells were incubated with NHC and IFN-α-2a, either alone or in different combinations. After 94 h of incubation, total RNA was extracted and amplified in multiplex conditions. Analysis was performed by using Combostat software (Jasper, Ga.) (5).
As a comparison for the day 4 antiviral effect of NHC, IFN-α-2a (100 IU/ml) and ribavirin (100 μM) were tested four times and average ΔΔCt values of 5.07 ± 0.67 and 1.22 ± 0.67, respectively, were obtained (Fig. 4A). Based on the ΔΔCt values of the dose response curves obtained at day 4, IFN-α-2a showed an EC90 of 4.5 IU/ml while ribavirin resulted in an EC90 of greater than 100 μM (data not shown).
Two other lines of evidence illustrate the antiviral effect of NHC. In Northern blot analysis, a 4-day treatment of HCV replicon cells at 65 μM resulted in reduction below the limit of detection of positive-strand HCV, a condition that was similar to the treatment of these cells with 100 IU of IFN-α-2a/ml (Fig. 4B). By Western blot analysis, no NS5B protein could be detected under similar conditions (Fig. 4C). In addition, NHC was tested in combination with IFN-α-2a and after 96-h incubation, HCV RNA and rRNA were extracted and amplified (Fig. 4D). The results indicate that the antiviral activity of NHC is at least additive with that of IFN-α-2a (5).
The antiviral effect of NHC could be prevented by the simultaneous addition of cytidine or uridine to the cell culture medium. Other natural nucleosides (guanosine, adenosine) or 2′-deoxynucleosides (2′-deoxycytidine, 2′-deoxyuridine, 2′-deoxyadenosine, 2′-deoxyguanosine, thymidine) had no effect on the antiviral activity of NHC (Fig. 5A). Similarly, the reduction in rRNA levels observed with NHC at 50 μM was also prevented by the addition of cytidine and uridine. The prevention of antiviral activity by cytidine or uridine was concentration dependent, with cytidine being more effective than uridine (Fig. 5B).
FIG. 5.
Prevention and rebound of the anti-HCV RNA effect of NHC. HCV replicon cells were treated for 4 days with or without simultaneous incubation of natural nucleosides or deoxynucleosides at 50 μM. (A) Plot showing the differences in Ct outcomes in the different settings with 50 μM NHC. ▴, ΔCt(rRNA Ct−no-drug Ct); •, ΔCt(HCV Ct−no-drug Ct). No, incubation with NHC but no competitive nucleosides added; C, cytidine; G, guanosine; A, adenosine; U, uridine; C,G,A,U, mix of the four nucleosides each at 25 μM; dU, 2′-deoxyuridine; dC, 2′-deoxycytidine; dA, 2′-deoxyadenosine; dG, 2′-deoxyguanosine, T, thymidine; dC,dG,dA,T, mix of the four deoxynucleosides each at 25 μM. (B) Dose-dependent antiviral prevention by adding natural nucleosides to the culture medium already containing 100 μM NHC. ▪, uridine; ♦, cytidine. (C) Rebound effect of HCV replicon after removal of NHC from culture supernatant. ♦, no-drug control; ▪, 33 μM NHC; •, 100 μM NHC; ▴, 100 IU of IFN-α-2a/ml. S.D., standard deviation.
As stabilized conditions were established up to 8 days for HCV RNA replicon dynamics (Fig. 3B and 4A), we designed an experiment in which the rebound of the replicon after cessation of treatment could be studied (Fig. 5C). HCV replicon cells were treated for 4 days with NHC or IFN-α-2a in G418-free medium, after which the medium was replaced with compound-free fresh assay medium. Initially, a 1-day extension of the antiviral effect was observed (increase of ΔΔCt value up to day 5), indicating that the intracellular active metabolite of NHC had a sufficiently long half-life to support the antiviral action. However, under all the conditions tested, the HCV RNA replicon rebounded from day 6 onwards. Interestingly, by the last day of possible testing (day 8), the replicon RNA levels had not returned to pretreatment baseline levels. This suggests that treatment with NHC and IFN-α-2a might have a durable effect on the replicon copy numbers after cessation of therapy. These experiments were repeated under G418 conditions, and similar results were obtained (data not shown).
NHC was essentially inactive against HIV-1 in human PBM cells (EC90 = 82.4 μM) and HBV in HepG2 cells (EC90 > 100 μM) as determined by using assays previously described (42, 44). NHC had modest activities against HSV type 1 (F strain) and HSV type 2 (G strain) in a plaque assay using Vero cells, with EC50s of 26 and 22 μM, respectively (data not shown).
Intracellular metabolism.
HCV replicon cells were treated with 10 μM 3H-labeled NHC, and intracellular nucleotide levels were determined after 1-, 2-, and 8-h incubations. In triplicate experiments, NHC was rapidly converted into the mono-, di-, and triphosphate, reaching up to 71.12 ± 22.66 pmol of NHC-TP/106 cells after 8-h incubation (details on cellular pharmacology will be published elsewhere [B. I. Hernandez-Santiago et al., unpublished data]). As high levels of NHC-TP in HCV replicon cells were detected, it is anticipated that NHC-TP is the molecular form responsible for the antiviral activity in the replicon system.
Effect of NHC-TP activity on the HCV NS5B enzyme.
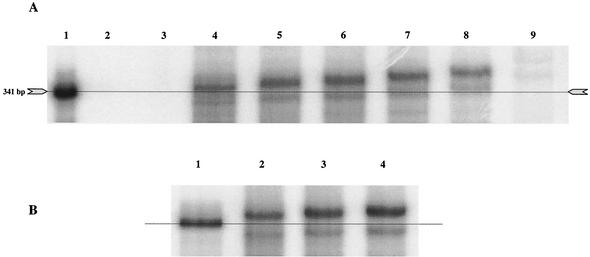
Chemically synthesized NHC-TP was used to study the potential inhibition of the in vitro polymerization reaction of the NS5B protein (Fig. 6). A T7-derived RNA template consisting of the negative-strand 5′ UTR of HCV was used in assay conditions essentially as previously described (39). NHC did not appear to inhibit the polymerization reaction when added to the standard buffer conditions, even at concentrations of 40 μM (EC50s > 40 μM) and with a 2-h incubation time. Since this NS5B assay is considered an endpoint assay, the results in Fig. 6A do not allow us to draw conclusions about the kinetics and the possibility of NHC being a competitive inhibitor. There was, however, a concentration-dependent increase in the apparent molecular weight of the product compared to that of the template when analyzed under standard PAGE conditions (7 M urea-5% acrylamide) (Fig. 6A). Under reaction conditions without CTP (Fig. 6A, lane 9), a very faint but distinct polymerization product was observed, suggesting that NHC-TP could act as a weak alternative substrate. Incorporation of NHC-TP instead of CTP should increase the molecular weight of the polymerization product by 16 (one extra oxygen) for each event, but this could not explain the observed electrophoretic shift. If all C residues were replaced by NHC, the molecular weight should have changed by 1,584 (16 × 99) at the maximum. Therefore, it was postulated that the PAGE conditions did not completely denature the 5′ UTR stem-loop structures and that the polymerization product might be migrating as a result of secondary structure and not molecular size. PAGE analysis of the products under more stringent denaturation conditions (7 M urea, 45% [vol/vol] formamide, 5% acrylamide) showed that there were almost no differences in migration between reactions with and without NHC (Fig. 6B). Taken together, these results suggest that, when NHC-TP is incorporated, the secondary and/or tertiary stability of the IRES is changed, which can be visualized under certain PAGE conditions.
FIG. 6.
Cell-free HCV NS5B polymerization reactions in the presence of NHC-TP. Synthetic 5′-UTR IRES template was incubated under conditions supporting NS5B enzyme polymerization in the presence of increasing concentrations of NHC-TP. (A) Gel electrophoresis conditions: 7 M urea-5% acrylamide. [α-32P]UMP-labeled positive-strand RNA template (artificial RNA derived from an SP6 template) used as size marker (lane 1), polymerization reactions without negative-strand RNA template (lane 2), and polymerization reactions including negative-strand RNA template but with a deficient enzyme (site-directed mutagenesis of the active center from G317D318D319 to G317A318A319) (lane 3) are shown. Full-length polymerization products in the absence of NHC-TP (lane 4), in 5 μM NHC-TP (lane 5), in 10 μM NHC-TP (lane 6), in 20 μM NHC-TP (lane 7), in 40 μM NHC-TP (lane 8), and in 40 μM NHC-TP and the absence of CTP (lane 9) are shown. (B) Gel electrophoresis conditions: 7 M urea-45% (vol/vol) formamide-5% acrylamide. [α-32P]UMP-labeled positive-strand RNA template (artificial RNA derived from an SP6 template) used as size marker is shown (lane 1). Full-length polymerization products in the absence of NHC-TP (lane 2), in 10 μM NHC-TP (lane 3), and in 20 μM NHC-TP (lane 4) are shown.
Attempts to select NHC-resistant HCV replicon and BVDV.
To examine whether NHC treatment permitted the selection of resistant replicon mutants, replicon-containing cells were maintained in exponential-growth conditions in the presence of a constant concentration of G418 (500 μg/ml) and increasing concentrations of NHC (Fig. 7A). This treatment schedule resulted in significant cell death as the concentration of compound was increased. The maximum tolerated concentration of NHC under these conditions was 40 μM. Huh7 replicon cells from several passages were tested for sensitivity to the antiviral activity of NHC. No reduction in sensitivity was observed for any passage tested (Fig. 7B). For example, cells collected at passage 45 were seeded side by side with untreated cells and tested for sensitivity in the presence of 100 μM NHC. These results showed that (i) the HCV replicon in the selected cells is still sensitive to the compound (no change in EC90) and (ii) the initial reductions in rRNA levels appeared to be less prominent in the passage-45-selected cells. In addition, the entire NS5B gene was sequenced from the replicon at passage 45 and there was only 1 nucleotide change detected compared to that for HCV RNA obtained from untreated cells (H375CAC → CAT). These results suggest that, instead of selecting for a resistant HCV replicon, cells with increased resistance to NHC were selected.
FIG. 7.
Long-term treatment of HCV replicon cells with NHC. (A) HCV replicon cells were incubated for the indicated passage number in the presence of G418 (500 μg/ml). The cells were passaged at a ratio of 1:3 during the whole incubation period. (B) Results of an antiviral sensitivity assay are shown. Untreated and NHC-pretreated HCV replicon cells were seeded in a 96-well plate at 103 cells per well in presence or absence of 100 μM NHC and incubated for 96 h. The ratio HCV RNA to rRNA levels were determined by Q-RT-PCR. ▪, ΔCt(rRNA Ct−rRNA-untreated Ct); ♦, ΔCt(HCV Ct−rRNA-untreated Ct). S.D., standard deviation. (C) cpBVDV was grown in the presence of increasing concentrations of compound for 20 passages. (D) cpBVDV, harvested at passage 20, was tested in a 24-h virus yield assay. ▪, passage 20 virus; ♦, control virus.
Similarly, cpBVDV was also passaged in the presence of increasing concentrations of NHC in MDBK cells (Fig. 6C and D). At concentrations of NHC above 5 μM, the virus-associated CPE was delayed by 24 to 48 h and often did not reach completion. At each passage, the concentration of NHC was increased to the next highest concentration unless the virus yields dropped below 103 PFU/ml. When this occurred, virus suspensions from samples grown at the next lowest concentration of compound were used to infect MDBK cells for the next passage (Fig. 7C). The virus preparation at passage 20 was tested for reduced sensitivity in a 24-h virus yield assay in comparison with that for untreated cpBVDV. Both viral pools exhibited a similar sensitivity profile when tested against increasing concentrations of NHC (Fig. 7D). Thus, attempts to select NHC-resistant cpBVDV were unsuccessful.
Toxicity of NHC.
In standard 3- or 4-day toxicity assays in parental Huh7 cells, HCV replicon cells, HepG2 cells, or human PBM cells, NHC did not show any remarkable toxicity (IC50s > 100 μM). However, in several of the experiments performed with Q-RT-PCR, a modest reduction in rRNA levels could be detected at concentrations greater than 50 μM (for examples, see Fig. 5A and 7B), indicating some toxicity at the molecular level.
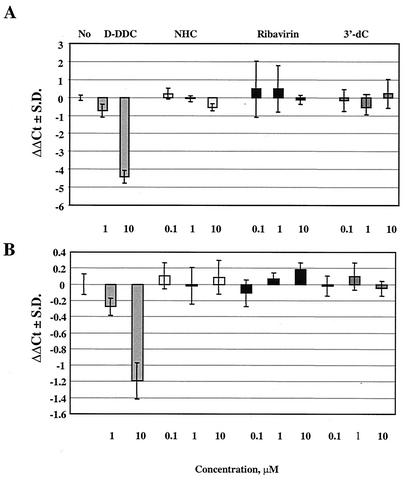
As the intracellular metabolism studies showed that the compound was phosphorylated to the NHC-TP form, we questioned whether this metabolite could potentially interfere with the synthesis of mitochondrial nucleic acids (mitDNA and mitRNA). Therefore, HepG2 cells were treated with 0.1, 1, or 10 μM of NHC for 7 days and mitochondrial COXII RNA and DNA levels were determined relative to the concentrations of β-actin DNA (Fig. 8) (44). The results demonstrated that, whereas β-d-2′,3′-dideoxycytidine (D-DDC) results in a significant reduction in mitDNA levels (and as a consequence of this, also in mitRNA levels), NHC did not change the mitDNA or mitRNA levels at the highest concentration tested. In addition, the 7-day cell supernatant for the 10 μM experiment was tested for the presence of total d- and l-lactic acid and the following results were obtained: 0.61, 1.74, 3.54, and 1.72 μg/μl for control medium (no cells), the no-drug control, D-DDC, and NHC, respectively. Thus, no increase in lactic acid production was observed after incubation with NHC.
FIG. 8.
Relative mitDNA and mitRNA levels. HepG2 cells were exposed for 7 days to 0.1, 1, or 10 μM concentrations of compound, and the ratio of mitochondrial to nuclear nucleic acids was determined. D-DDC, ribavirin, and 3′-deoxycytidine (3′-dC) were included as controls. (A) Ratio of COXII DNA to β-actin DNA levels; ΔΔCt(COXII Ct−β-actin Ct−no-drug control Ct). (B) Ratio of COXII to β-actin RNA levels (RT-PCR).
To determine the potential toxicity of NHC in small animals, five groups of six 6-week-old female Swiss mice (SWR/J) were treated i.p. at doses of 0, 3, 10, 33, or 100 mg/kg/day for 6 days (Fig. 9). All the animals survived the 6-day treatment and the 24-day monitoring period. A statistically significant decrease in weight gain was observed in the 100 mg/kg/day group during the treatment period. However, all animals recovered when treatment was stopped, and at the end of the second week (day 14), their weight was similar to that of mice in the no-drug control treatment group. The no-observed-effect dose level for NHC in this experiment was 33 mg/kg/day.
FIG. 9.
NHC i.p. treatment in mice. Mice were injected i.p. with NHC on day 0 to day 5 (ip treatment), and changes in weight were monitored on the indicated days. ♦, 0 mg/kg/day; ▴, 3 mg/kg/day; •, 10 mg/kg/day; ▪, 33 mg/kg/day; ×, 100 mg/kg/day; *, significantly different from control (P < 0.05). For clarity, standard deviations (S.D.) are only shown for the 0-mg/kg/day and 100-mg/kg/day treatment groups.
DISCUSSION
We describe for the first time a base-modified nucleoside analogue with the capacity to reduce HCV replicon RNA levels in Huh7 cells. The data on NHC can be summarized as follows. (i) In an acute BVDV infection system, EC90s of 5.4 ± 0.9 and 3.4 ± 0.08 μM on exponential-phase and confluent cells, respectively, were obtained. (ii) In a persistent HCV replication system in Huh7 cells, an EC90 of 5 μM (based on ΔCtHCV) was obtained after 96-h incubation, the results further confirmed by Northern and Western blotting. (iii) The compound interacted at least additively with IFN-α-2a. (iv) The antiviral effect could be prevented only by the addition of uridine or cytidine to culture medium. (v) High levels of NHC-TP were formed in HCV replicon cells. (vi) NHC-TP was unable to inhibit the polymerization reaction in in vitro NS5B assays. (vii) NHC-TP served with low efficiency as an alternative substrate to CTP. (viii) Attempts to raise resistant HCV replicon RNA or BVDV RNA were unsuccessful. (ix) Although modest reductions in rRNA levels in HCV replicon cells were observed, there were no obvious toxicities in cell proliferation assays or in mitochondrial functions (no changes in DNA, RNA, or lactic acid levels at 10 μM for 7 days). (x) Finally, the no-effect dose level in mice after 6 days i.p. was 33 mg/kg/day.
The compound appears to be RNA virus specific (hepacivirus and pestivirus) and was essentially inactive against HIV-1, HBV, and HSV. The mechanism by which the compound inhibits virus replication is unclear. NHC clearly interacts with cytoplasmic RNA metabolism (e.g., the HCV RNA replicon), although no direct inhibition of the NS5B polymerase was demonstrated in the endpoint detection assay. There are several other possibilities of action, either through direct interaction with viral proteins (e.g., NS3-encoded helicase protein), through interaction with host cell proteins (replicon RNA binding, translation, or nucleotide metabolism), or indirectly through the incorporation and disruption of important secondary or tertiary structures of HCV RNA.
Our experiments showed that NHC-TP is not an inhibitor of the NS5B polymerase but that it did act as an alternative substrate with very low efficiency. As NHC-TP is formed intracellularly at levels in the range of 71 ± 22.7 pmol/106 cells after 8 h of incubation, they could compete with the intracellular CTP levels (range, 77 ± 6.98 pmol/106 cells in PBM cells [14]) and occasionally get incorporated into HCV replicon RNA. Such base-modified HCV RNA templates could subsequently suffer from the changed stability of IRES stem-loop structures, leading to the reduction of ribosome entry, differences in translation initiation and elongation, changes in the recognition of the amino acid-tRNA molecules, and changes in the secondary structure of the 3′ UTR (16), thereby affecting the initiation of replication. Furthermore, the processivity and reliability of copying the viral template can be changed, resulting in an increased mutagenicity rate (because NHC behaves as either cytosine or uracil [27]) leading to a C → U transition and forcing the replicon into “error catastrophe,” essentially as has been described previously for ribavirin (10, 11). Previously, NHC has been found to be a powerful mutagen of the base-analog type in Escherichia coli K12, Salmonella enterica serovar Typhimurium, and Neurospora crassa and results mainly in AT-to-GC transitional alterations (12, 19, 20, 21, 38). The β-d-N4-aminocytidine, a nucleoside analogue with an exceptionally high mutagenic activity at least 2 orders of magnitude greater than that of NHC (35) has been found inactive in the HCV replicon system (data not shown), suggesting that compounds with a mutagenic character in bacteria cannot directly be seen as viral mutagens in eukaryotic cells.
Another possible mechanism of action of NHC can be as an antimetabolite. A typical example for this group of compounds is ribavirin. Ribavirin (43) is a broad-spectrum agent against a variety of RNA viruses (HCV, respiratory syncytial virus, lassa fever virus) through several mechanisms, including the inhibition of viral RNA transcription, elongation, cap formation, and mutagenicity. The major event occurring in cells exposed to this compound is the intracellular activation by the adenosine kinase to ribavirin-monophosphate (46) and inhibition of the IMP dehydrogenase enzyme in a substrate-type-dependent manner, resulting in depletion of the GTP and dGTP pools (31). However, in our assays, we showed an antiviral effect of ribavirin in the BVDV system (EC90 = 3.5 ± 1.5 μM) (Fig. 2A), but there was no specific antiviral effect in the HCV replicon RNA assay, as measured by the ΔΔCt method (EC90 > 100 μM) (Fig. 4A). NHC could act in a way similar to ribavirin but with a more profound effect on the HCV replicon RNA levels. In this hypothesis, NHC metabolites act with a cellular enzyme involved in the de novo synthesis of purine (most likely not GTP, as the activity was not reversed by purines) or pyrimidine triphosphates, resulting in the specific depletion of such natural NTPs. For comparison, a molecule closely related to NHC, the 2′-deoxy analogue of NHC in its 5′-monophosphate form (dNHC-TP), was reported to be a strong inhibitor of thymidylate synthase (TS) (EC 2.1.1.45) (15). When this compound was tested in the replicon system together with the TS inhibitors β-d-2′-deoxy-5-fluorouridine and β-d-5-fluorouridine, all three compounds were found inactive (ΔΔCt at day 4; EC90 > 100 μM) (data not shown), suggesting that TS inhibition does not influence HCV replicon levels. This is not surprising, since the thymidine-related pathway typically does not lead to the precursors needed in RNA synthesis.
In summary, we describe a base-modified nucleoside analogue NHC with anti-BVDV and anti-HCV activities. The detailed mechanism of action remains to be determined, but our results suggest that NHC acts as a weak alternative substrate for the viral polymerase.
Acknowledgments
We thank Darius Moradpour for providing us with the anti-NS5B antibody and Paul Black and Jason Grier for assistance in mice experiments.
R.F.S. is supported in part by the Department of Veterans Affairs. He is a founder of and consultant to Pharmasset Ltd., and his particulars have been reviewed by Emory University's Conflict of Interest Committee. His group received no funding from Pharmasset to perform this work.
REFERENCES
- 1.Alt, M., S. Eisenhardt, M. Serwe, R. Renz, J. W. Engels, and W. H. Caselmann. 1999. Comparative inhibitory potential of differently modified antisense oligodeoxynucleotides on hepatitis C virus translation. Eur. J. Clin. Investig. 29:868-876. [DOI] [PubMed] [Google Scholar]
- 2.Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341:556-562. [DOI] [PubMed] [Google Scholar]
- 3.Baginski, S. G., D. C. Pevear, M. Seipel, S. C. Sun, C. A. Benetatos, S. K. Chunduru, C. M. Rice, and M. S. Collett. 2000. Mechanism of action of a pestivirus antiviral compound. Proc. Natl. Acad. Sci. USA 97:7981-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 5.Belen'kii, M. S., and R. F. Schinazi. 1994. Multiple drug effect analysis with confidence interval. Antivir. Res. 25:1-11. [DOI] [PubMed] [Google Scholar]
- 6.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 7.Branza-Nichita, N., D. Durantel, S. Carrouée-Durantel, R. A. Dwek, and N. Zitzmann. 2001. Antiviral effect of N-butyldeoxynojirimycin against bovine viral diarrhea virus correlates with misfolding of E2 envelope proteins and impairment of their association into E1-E2 heterodimers. J. Virol. 75:3527-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collett, M. S., D. K. Anderson, and E. Retzel. 1988. Comparisons of the pestivirus bovine viral diarrhea virus with members of the flaviviridae. J. Gen. Virol. 69:2637-2643. [DOI] [PubMed] [Google Scholar]
- 9.Collier, J., and R. Chapman. 2001. Combination therapy with interferon-alpha and ribavirin for hepatitis C: practical treatment issues. BioDrugs 15:225-238. [DOI] [PubMed] [Google Scholar]
- 10.Crotty, S., C. E. Cameron, and R. Andino. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. USA 98:6895-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crotty, S., D. Maag, J. J. Arnold, W. Zhong, J. Y. Lau, Z. Hong, R. Andino, and C. E. Cameron. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6:1375-1379. [DOI] [PubMed] [Google Scholar]
- 12.de Serres, F. J., and H. E. Brockman. 1993. Comparison of the spectra of genetic damage in N4-hydroxycytidine-induced ad-3 mutations between nucleotide excision repair-proficient and -deficient heterokaryons of Neurospora crassa. Mutat. Res. 285:145-163. [DOI] [PubMed] [Google Scholar]
- 13.Di Bisceglie, A. M., J. McHutchison, and C. M. Rice. 2002. New therapeutic strategies for hepatitis C. Hepatology 35:224-231. [DOI] [PubMed] [Google Scholar]
- 14.Di Pierro, D., B. Tavazzi, C. F. Perno, M. Bartolini, E. Balestra, R. Calio, B. Giardina, and G. Lazzarino. 1995. An ion-pairing high-performance liquid chromatographic method for the direct simultaneous determination of nucleotides, deoxynucleotides, nicotinic coenzymes, oxypurines, nucleosides, and bases in perchloric acid cell extracts. Anal. Biochem. 231:407-412. [DOI] [PubMed] [Google Scholar]
- 15.Felczak, K., A. Miazga, J. Poznanski, M. Bretner, T. Kulikowski, J. M. Dzik, B. Golos, Z. Zielinski, J. Ciesla, and W. Rode. 2000. 5-Substituted N(4)-hydroxy-2′-deoxycytidines and their 5′-monophosphates: synthesis, conformation, interaction with tumor thymidylate synthase, and in vitro antitumor activity. J. Med. Chem. 43:4647-4656. [DOI] [PubMed] [Google Scholar]
- 16.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geiss, G. K., M. C. An, R. E. Bumgarner, E. Hammersmark, D. Cunningham, and M. G. Katze. 2001. Global impact of influenza virus on cellular pathways is mediated by both replication-dependent and -independent events J. Virol. 75:4321-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janion, C. 1978. The efficiency and extent of mutagenic activity of some new mutagens of base-analogue type. Mutat. Res. 56:225-234. [DOI] [PubMed] [Google Scholar]
- 20.Janion, C. 1984. Some problems of mutagenesis induced by base analogues. Acta Biochim. Pol. 31:183-192. [PubMed] [Google Scholar]
- 21.Janion, C., and B. W. Glickman. 1980. N4-hydroxycytidine: a mutagen specific for AT to GC transitions. Mutat. Res. 72:43-47. [DOI] [PubMed] [Google Scholar]
- 22.Jordan, R., O. V. Nikolaeva, L. Wang, B. Conyers, A. Mehta, R. A. Dwek, and T. M. Block. 2002. Inhibition of host ER glucosidase activity prevents golgi processing of virion-associated bovine viral diarrhea virus E2 glycoproteins and reduces infectivity of secreted virions. Virology 295:10-19. [DOI] [PubMed] [Google Scholar]
- 23.Kato-Maeda, M., Q. Gao, and P. M. Small. 2001. Microarray analysis of pathogens and their interaction with hosts. Cell. Microbiol. 3:713-719. [DOI] [PubMed] [Google Scholar]
- 24.Kim, J. L., K. A. Morgenstern, J. P. Griffith, M. D. Dwyer, J. A. Thomson, M. A. Murcko, C. Lin, and P. R. Caron. 1998. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6:89-100. [DOI] [PubMed] [Google Scholar]
- 25.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, P. A., L. M. Blatt, K. S. Blanchard, K. S. Bouhana, P. A. Pavco, L. Bellon, and J. A. Sandberg. 2000. Pharmacokinetics and tissue distribution of a ribozyme directed against hepatitis C virus RNA following subcutaneous or intravenous administration in mice. Hepatology 32:640-646. [DOI] [PubMed] [Google Scholar]
- 27.Les, A., L. Adamowicz, and W. Rode. 1993. Structure and conformation of N4-hydroxycytosine and N4-hydroxy-5-fluorocytosine. A theoretical ab initio study. Biochim. Biophys. Acta 1173:39-48. [DOI] [PubMed] [Google Scholar]
- 28.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 30.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markland, W., T. J. McQuaid, J. Jain, and A. D. Kwong. 2000. Broad-spectrum antiviral activity of the IMP dehydrogenase inhibitor VX- 497: a comparison with ribavirin and demonstration of antiviral additivity with alpha interferon. Antimicrob. Agents Chemother. 44:859-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendez, E., N. Ruggli, M. S. Collett, and C. M. Rice. 1998. Infectious bovine viral diarrhea virus (strain NADL) RNA from stable cDNA clones: a cellular insert determines NS3 production and viral cytopathogenicity. J. Virol. 72:4737-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nam, J. H., J. Bukh, R. H. Purcell, and S. U. Emerson. 2001. High-level expression of hepatitis C virus (HCV) structural proteins by a chimeric HCV/BVDV genome propagated as a BVDV pseudotype. J. Virol. Methods 97:113-123. [DOI] [PubMed] [Google Scholar]
- 34.Narjes, F., M. Brunetti, S. Colarusso, B. Gerlach, U. Koch, G. Biasiol, D. Fattori, R. De Francesco, V. G. Matassa, and C. Steinkuhler. 2000. Alpha-ketoacids are potent slow binding inhibitors of the hepatitis C virus NS3 protease. Biochemistry 39:1849-1861. [DOI] [PubMed] [Google Scholar]
- 35.Negishi, K., C. Harada, Y. Ohara, K. Oohara, N. Nitta, and H. Hayatsu. 1983. N4-aminocytidine, a nucleoside analog that has an exceptionally high mutagenic activity. Nucleic Acids Res. 11:5223-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pallaoro, M., A. Lahm, G. Biasiol, M. Brunetti, C. Nardella, L. Orsatti, F. Bonelli, S. Orru, F. Narjes, and C. Steinkuhler. 2001. Characterization of the hepatitis C virus NS2/3 processing reaction by using a purified precursor protein. J. Virol. 75:9939-9946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Popowska, E., and C. Janion. 1975. The metabolism of N4-hydroxycytidine—a mutagen for Salmonella typhimurium. Nucleic Acids Res. 2:1143-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reigadas, S., M. Ventura, L. Sarih-Cottin, M. Castroviejo, S. Litvak, and T. Astier-Gin. 2001. HCV RNA-dependent RNA polymerase replicates in vitro the 3′ terminal region of the minus-strand viral RNA more efficiently than the 3′ terminal region of the plus RNA. Eur. J. Biochem. 268:5857-5867. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schinazi, R. F., R. M. J. Lloyd, M. H. Nguyen, D. L. Cannon, A. McMillan, N. Ilksoy, C. K. Chu, D. C. Liotta, H. Z. Bazmi, and J. W. Mellors. 1993. Characterization of human immunodeficiency viruses resistant to oxathiolane-cytosine nucleosides. Antimicrob. Agents Chemother. 37:875-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sidwell, R. W., J. H. Huffman, G. P. Khare, L. B. Allen, J. T. Witkowski, and R. K. Robins. 1972. Broad-spectrum antiviral activity of Virazole: 1-beta-d-ribofuranosyl- 1,2,4-triazole-3-carboxamide. Science 177:705-706. [DOI] [PubMed] [Google Scholar]
- 44.Stuyver, L. J., S. Lostia, M. Adams, J. S. Mathew, B. S. Pai, J. P. Grier, P. M. Tharnish, Y. Choi, Y. Chong, H. Cho, C. K. Chu, M. J. Otto, and R. F. Schinazi. 2002. Antiviral activities and cellular toxicities of 2′,3′-dideoxy-2′,3′-didehydrocytidine analogues. Antimicrob. Agents Chemother. 46:3854-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sullivan, D. G., and R. K. Akkina. 1995. A nested polymerase chain reaction assay to differentiate pestiviruses. Virus Res. 38:231-239. [DOI] [PubMed] [Google Scholar]
- 46.Willis, R. C., D. A. Carson, and J. E. Seegmiller. 1978. Adenosine kinase initiates the major route of ribavirin activation in a cultured human cell line. Proc. Natl. Acad. Sci. USA 75:3042-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zitzmann, N., A. S. Mehta, S. Carrouee, T. D. Butters, F. M. Platt, J. McCauley, B. S. Blumberg, R. A. Dwek, and T. M. Block. 1999. Imino sugars inhibit the formation and secretion of bovine viral diarrhea virus, a pestivirus model of hepatitis C virus: implications for the development of broad spectrum anti-hepatitis virus agents. Proc. Natl. Acad. Sci. USA 96:11878-11882. [DOI] [PMC free article] [PubMed] [Google Scholar]