Abstract
A complex class 1 integron, In34, found in a conjugative plasmid from a multidrug-resistant Klebsiella pneumoniae strain isolated in 1997 at a hospital in Sydney, Australia, was shown to have a backbone related to that of In2, which belongs to the In5 family. In In34, the aadB gene cassette replaces the aadA1a cassette in In2, and two additional resistance genes, dfrA10 and aphA1, that are not part of a gene cassette are present. The aphA1 gene is in a Tn4352-like transposon that is located in the tniA gene. The dfrA10 gene lies adjacent to a 2,154-bp DNA segment, known as the common region, that contains an open reading frame predicting a product of 513 amino acids (Orf513). Orf513 is 66 and 55% identical to the products of two further open reading frames that, like the common region, are found adjacent to antibiotic resistance genes. A 27-bp conserved sequence was found at one end of each type of common region. The loss of dfrA10 due to homologous recombination between flanking direct repeats and incorporation of the excised circle by homologous recombination were demonstrated. Part of In34 is identical to the sequenced portion of In7, which is from a multidrug-resistant Escherichia coli strain that had been isolated 19 years earlier in the same hospital. In34 and In7 are in plasmids that contain the same six resistance genes conferring resistance to ampicillin, chloramphenicol, gentamicin, kanamycin, neomycin, tobramycin, trimethoprim, and sulfonamides, but the plasmid backbones appear to be unrelated, suggesting that translocation of a multiple-drug-resistance-determining region as well as horizontal transfer may have occurred.
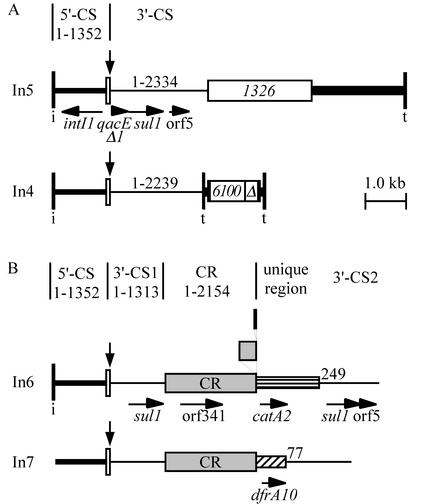
The roles of gene cassettes and class 1 integrons in the acquisition and spread of antibiotic resistance genes are well established (20, 35, 36), and over 75 gene cassettes that include antibiotic resistance genes have been reported (19, 27, 35; S. R. Partridge, unpublished data). In addition to their ability to acquire new resistance genes, class 1 integrons are either transposons (34) or defective transposon derivatives that have a variety of backbone structures that fall into two main groups, the In5 and In4 families (7, 18, 32, 33). Typically they are bounded by 25-bp imperfect inverted repeats designated IRi and IRt (Fig. 1A), and the defective transposons can be viewed as derived from the backbone structure of transposon Tn402 (In16) by acquisition of the 3′ conserved (3′-CS) segment and of insertion sequence elements, followed by deletion and rearrangement, leading to the loss of genes that determine transposition functions (7, 33). These defective transposons can move if transposition enzymes are supplied in trans (31, 38).
FIG. 1.
Structures of class 1 integrons. (A) In4 and In5, representing the main backbone types; (B) class 1 integrons containing the common region. IRi and IRt are shown as vertical bars labeled i and t, respectively, and the attI1 site is shown as a narrow open box. The CR is represented as a shaded box, and insertion sequences are represented as numbered open boxes, with Δ indicating a partial copy of IS6100. The extents of the 5′-CS (medium-thickness lines), the 3′-CS (thin lines), and the tni module (thick line) present in published sequences are indicated, with the numbers referring to positions in standard versions of these sequences. Unique regions are shown as hatched boxes, and genes are indicated by horizontal arrows. The shaded boxes above the end of the complete CR in In6 represent duplications of the final 393 and 28 bp of the CR. The cassettes present at the positions marked by vertical arrows are as follows: In5, aacA(IIa); In4, aacC1-orfE-aadA2-cmlA1; In6, aacA4-aadA2; In7, aadB. The sequences used are from GenBank accession nos. M29695 and U38230 and standard sequences for In5; U12338 for In4; sequences compiled from U38230L06822, U04277, U04278, and X68227, together with standard and unpublished sequences, for In6; and L06418 for In7.
A further unusual class 1 integron backbone type was originally identified in two integrons, In6 and In7, in the early 1990s (19, 21, 30, 39). These integrons (Fig. 1B) contain the 5′-CS and part of the 3′-CS (bp 1 to 1313; here designated 3′-CS1) flanking one or more gene cassettes. Following 3′-CS1 is a region of 2,154 bp that is common to In6 and In7 and is designated the common region (CR) to distinguish it from the CSs. Adjacent to the CR is a unique region that does not resemble a gene cassette but that includes an antibiotic resistance gene, catA2 (chloramphenicol resistance, formerly catII) in In6 (39) and dfrA10 (trimethoprim resistance, formerly dhfrX) in In7 (30). Beyond the unique region is a second partial copy of the 3′-CS, here designated 3′-CS2, which starts at bp 249 of the standard 3′-CS in In6 (21) and at bp 77 in In7 (30), suggesting that deletion accompanied the acquisition of the DNA segments containing the resistance genes. Next to the right-hand end of the complete CR in In6 are two adjacent partial copies of the CR, corresponding to the last 393 bp and 28 bp, respectively (Fig. 1B), and comparison of the four boundaries in In6 and In7 allowed this end of the CR to be defined unambiguously (Fig. 2A).
FIG. 2.
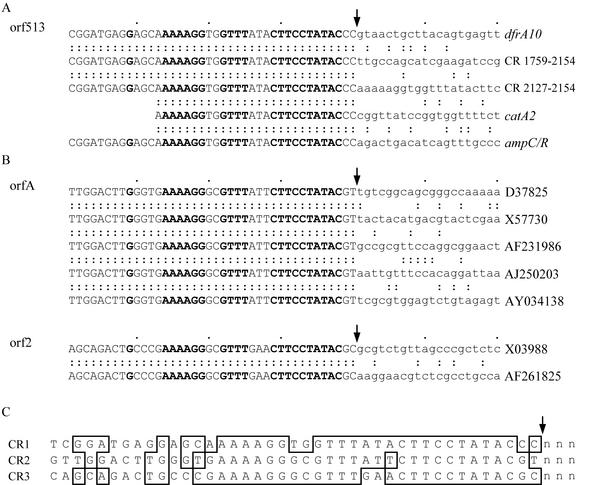
Right-hand boundary of the CR. (A) Sequences downstream of orf513 in In6, In7, and pSAL-1 are aligned to show the boundary between CR and unique sequences or CR duplications. (B) Alignment of sequences downstream of orfA and orf2 showing the equivalent boundaries. Uppercase letters represent CR sequences (CR1, orf513; CR2, orfA; CR3, orf2), and lowercase letters represent unique sequences, with vertical arrows indicating the positions of the boundaries. Colons indicate identical bases, and bases in boldface type are conserved in all three CRs. The sequences used in panel A are listed in Fig. 1 plus pSAL-1 integron (GenBank accession no. AJ237702), and the sequences in panel B were obtained from the GenBank accession numbers indicated on the right-hand side. (C) Comparison of the boundaries of the three CRs. The sequences at the right-hand boundaries of CR1, CR2, and CR3 are aligned, with identical nucleotides boxed.
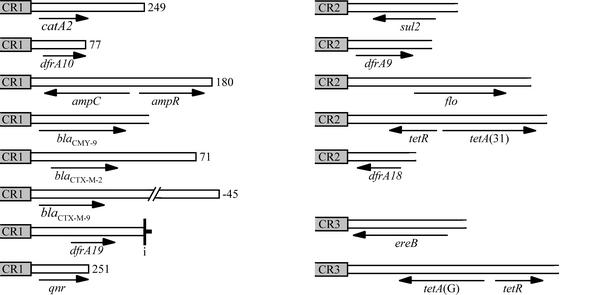
A number of other class 1 integrons containing the CR adjacent to various antibiotic resistance genes have since been reported or deposited in GenBank (Fig. 1). These genes include blaDHA-1 in the integron in pSAL-1 from Salmonella enterica serovar Enteritidis (GenBank accession no. AJ237702 [42]), blaCTX-M-9 in In60 of pMSP071 from Escherichia coli (AF174129 [37]), dfrA19 (formerly dfrA18) in pIncF1/97 from S. enterica serovar Typhimurium (AJ310778 [43]), blaCMY-9 in pCMXR1 from E. coli (AB061794 [16]), blaCTX-M-2 in InS21 in pS21 from S. enterica serovar Infantis (AJ311891.2 [15]) and in In35 in pMAR-12 from Proteus mirabilis (AY079169 [2]), and qnr in pMG252 from Klebsiella pneumoniae (AY070235 [41]). In some of these sequences, a second copy of the 3′-CS (3′-CS2) with a deletion at the beginning is also present, but in In60 part of a 59-base element (59-be) precedes bp 1 of the 3′-CS. In two cases the origin of the unique fragment that contains the antibiotic resistance gene has been identified as a portion of a bacterial chromosome. The region containing the ampC and ampR (blaDHA-1) genes in the pSAL-1 integron appears to originate from the region containing the ampC and ampR genes in the chromosome of Morganella morganii (42). Similarly, the 2,185-bp unique region of InS21 that includes the blaCTX-M-2 gene (15) is 99% identical to a region from Kluyvera ascorbata that includes the kluA-1 β-lactamase (GenBank accession no. AJ272538 [24]), as noted by Oliver et al. (28). This suggests that continuous regions of chromosomal DNA have been incorporated adjacent to the CR.
Generally, only the central portion of class 1 integrons containing the CR has been sequenced, and the structure of the complete integron is not known. The extent of 3′-CS2 beyond the sul1 gene has been determined only for In6 (18), which lacks the IRt end, and whether 3′-CS2 is normally associated with the types of structures found in In4- and In5-type class 1 integrons has not been established. In the study described here we have determined the complete sequence of a class 1 integron containing the CR from a conjugative plasmid found in a multidrug-resistant K. pneumoniae strain isolated at Royal North Shore Hospital in Sydney, Australia, in 1997. Errors in our previous reports of the sequence of the CR (39) were identified and corrected. Movement of the dfrA10 gene by homologous recombination was also examined.
MATERIALS AND METHODS
Bacterial strains and plasmids.
DH5α (supE44 ΔlacU169 [φ80 lacZ ΔM15] hsdR17 recA1 endA1 gyrA96 thi-1 relA1) was used to propagate plasmids. UB1637Rf is a spontaneous rifampin-resistant mutant of UB1637 (F− his lys trp recA56 rpsL) (13) generated for use as a recipient in conjugation experiments. The Rec+ strain AB1157 (F− proA2 his-4 thr-1 leuB6 thi-1 argE3 lacY1 galK2 ara-14 xyl-5 mtl-1 tsx-33 rpsL31 sup-37 λ−) (14) was used for homologous recombination experiments. Strain 24120497A is a K. pneumoniae clinical strain isolated at Royal North Shore Hospital, Sydney, Australia, in 1997. The plasmids used in the study are listed in Table 1. pRMH760 was transferred from 24120497A to UB1637Rf and then into DH5α by conjugation. Fragments of pRMH760 were recovered by ligation of restriction digests of pRMH760 to pUC19 digested with the same enzyme. Transformants were either selected on ampicillin plates and then patched onto plates containing other antibiotics to which pRMH760 confers resistance or were selected directly on such plates. Subclones of pRMH760 fragments were constructed in the same way. Bacteria were routinely cultured at 37°C in Luria-Bertani (LB) medium or on LB agar. Mueller-Hinton agar was used to select for resistance to trimethoprim and sulfamethoxazole. The following antibiotics (Sigma) were added at the indicated concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 25 μg ml−1; gentamicin, 4 μg ml−1; kanamycin, 50 μg ml−1; neomycin, 50 μg ml−1; rifampin, 100 μg ml−1; streptomycin, 25 μg ml−1; spectinomycin, 25 μg ml−1; sulfamethoxazole, 100 μg ml−1; trimethoprim, 25 μg ml−1; tetracycline, 10 μg ml−1; and tobramycin, 12.5 μg ml−1.
TABLE 1.
Plasmids
| Plasmid | Antibiotic resistance phenotypea | Description | Reference |
|---|---|---|---|
| pDG0101 | Apr Gmr Kmr Sur Tbr Tpr | 7.6-kb BamHI fragment of pDGO100 in pBR322 | 17 |
| pDGO109 | Apr Gmr Kmr Tbr | 2.1- and 1.3-kb BamHI-HindIII fragments of pDGO101 in pBR322 | 17 |
| pRMH760 | Apr Cmr Gmr Kmr Nmr Sur Tbr Tpr | Large conjugative plasmid from K. pneumoniae 24120497A | This work |
| pRMH761 | Apr Kmr Nmr | 8.8-kb BamHI fragment of pRMH760 in pUC19 | This work |
| pRMH763 | Apr | 5.8-kb BamHI-HindIII fragment of pRMH761 in pUC19 | This work |
| pRMH766 | Apr Tpr Sur | 8.1-kb SphI fragment of pRMH760 in pUC19 | This work |
| pRMH769 | Apr Kmr Nmr | 1.8-kb PstI fragment of pRMH760 in pUC19 | This work |
| pRMH770 | Apr Tpr | 1.9-kb PstI fragment of pRMH760 in pUC19 | This work |
| pRMH774 | Apr | 7.3-kb PstI fragment of pRMH760 in pUC19 | This work |
| pRMH776 | Apr | 2.1-kb PstI fragment of pRMH760 in pUC19 | This work |
| pRMH777 | Apr Gmr Kmr Tbr | Large PstI fragment of pRMH760 in pUC19 | This work |
| pRMH780 | Apr Gmr Kmr Tbr | 2.7-kb BamHI-PstI fragment of pRMH777 in pUC19 | This work |
| pRMH784 | Apr Gmr Kmr Tbr | 6.7-kb PstI-SalI fragment of pRMH777 in pUC19 | This work |
| pRMH793 | Apr | 2.3-kb HindIII fragment of pRMH766 in pUC19 | This work |
| pRMH795 | Apr Sur | 5.0-kb EcoRI fragment of pRMH766 in pUC19 | This work |
| pRMH830 | Apr Cmr Gmr Smr Spr Sur | ∼7.5-kb SphI fragment of pSa in pUC19 | This work |
| pRMH858 | Apr Cmr Smr Spr Sur | 4.8-kb BamHI fragment of R388::Tn1405 in pACYC184 | 31 |
| pRMH912 | Apr Cmr Smr Spr Sur Tpr | Insertion of 4.0 kb from pRMH766 into pRMH858 | This work |
Abbreviations: Ap, ampicillin; Cm, chloramphenicol; Gm, gentamicin; Km, kanamycin; Nm, neomycin; Sm, streptomycin; Sp, spectinomycin; Su, sulfamethoxazole; Tb, tobramycin; Tp, trimethoprim.
DNA isolation and restriction mapping.
Plasmid DNA for restriction analysis and cloning was isolated by an alkaline lysis method (3). Restriction enzymes were used in accordance with the instructions of the manufacturers. Fragments were separated by electrophoresis on 1% (wt/vol) agarose gels and visualized by staining with ethidium bromide. DNA fragments were purified from agarose gels by using an UltraClean DNA purification kit (Mo Bio Laboratories Inc.). EcoRI-digested bacteriophage SPP1 DNA (Geneworks) was used as a size marker. Plasmid DNA for sequencing was purified by using a Wizard maxiprep kit (Promega).
DNA sequencing and analysis.
Automated sequencing was performed by the sequencing facility at the Department of Biological Sciences, Macquarie University Sydney, on an ABI PRISM 377 sequencer by using the Big Dye system. DNA sequences were assembled by using MacVector (version 6.5) software and AssemblyLIGN software (Oxford Molecular). GenBank searches were performed by using the BLASTN and FastA programs available through WebANGIS (Australian National Genomic Information Service). Programs in the Genetics Computer Group (GCG) Wisconsin Package (version 8.1.0) were used via WAG (WebANGIS GCG) to align and analyze the DNA sequences. The DNA sequence of at least one strand of the cloned fragments was determined. Both strands were sequenced where ambiguities arose or where the sequence differed from published sequences. Three differences between the IS1353 sequences from In34 and In2 were traced to errors in the In2 sequence (7). The two open reading frames (ORFs) of IS1353, orfA and orfB, proposed previously now form a single ORF, orfAB. The presence of 4 extra bases in the In34 CR sequence was found to be due to errors in the In6 and In7 sequences (39). The relevant database entries (In2, GenBank accession nos. U40482, U42226, and AF071413; In6, GenBank accession no. L06822.3; In7, GenBank accession no. L06418.2) have been corrected.
Acquisition and loss of resistance genes.
NcoI-digested pRMH766 was run on an agarose gel; and the 4.0-kb fragment was cut out, purified, and religated to create a circular molecule. The ligation mix was transformed into AB1157(pRMH858) by electroporation. Plasmid DNA from colonies selected with trimethoprim was mapped by digestion with PstI and NcoI, and one plasmid with the circle incorporated was designated pRMH912. To facilitate the detection of rare deletants, plasmid DNA prepared from AB1157(pRMH912) grown without trimethoprim was digested with XhoI to cut the parental plasmid and transformed into DH5α. Transformants were selected with chloramphenicol and screened for trimethoprim resistance. Plasmid DNA from trimethoprim-sensitive colonies was mapped with PstI and NcoI.
Nucleotide sequence accession numbers.
The sequence of In34 has been submitted to GenBank under accession no. AY123253. An additional In7 (pDGO100) sequence has been added to GenBank accession no. L06418.
RESULTS
pRMH760 and In34.
A large plasmid, pRMH760, from a multiply drug resistant K. pneumoniae strain isolated in 1997 at Royal North Shore Hospital, Sydney, Australia, was transferable to E. coli by conjugation and conferred resistance to ampicillin, chloramphenicol, gentamicin, kanamycin, neomycin, sulfamethoxazole, tobramycin, and trimethoprim. The resistances to streptomycin, spectinomycin, and tetracycline also present in the original clinical strain were not transferred. Fragments of pRMH760 containing the antibiotic resistance determinants were cloned, mapped, and partially sequenced to identify the genes. Six resistance genes, aadB, sul1, dfrA10, aphA1, catA1, and blaTEM, were clustered together in a region of about 45 kb. All but the catA1 and blaTEM genes lay within the boundaries of a class 1 integron, designated In34, which was completely sequenced (Fig. 3A).
FIG. 3.
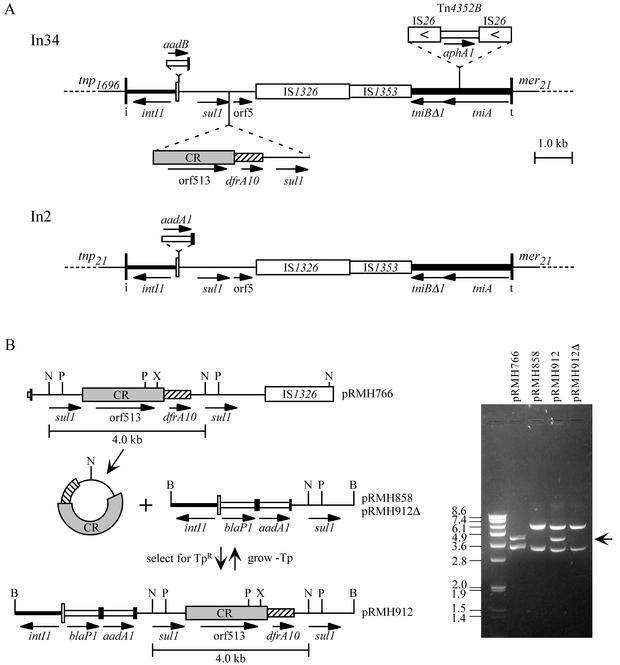
Structure of In34 and relationship to In2. (A) Structures of In34 and In2. The features are as described in the legend to Fig. 1, with gene cassettes represented as open boxes and an adjacent filled box (the 59-be). The aphA1 region is shown as a narrow open box. tnp21, tnp1696, and mer21 represent the tnp regions of Tn21 and Tn1696 and the mer region of Tn21, respectively. (B) Movement of CR1-dfrA10 by homologous recombination. The line diagrams show the inserts in the named plasmids, and the gel shows NcoI digests of these plasmids. The 4.0-kb NcoI fragment that forms the circular molecule and that is acquired and lost by homologous recombination is indicated in the line diagram (bar) and on the gel (horizontal arrow). The sizes of markers are given in kilobases, and restriction enzyme site abbreviations are as follows: B, BamHI; N, NcoI; P, PstI; and X, XhoI.
In34 is bounded by IRi and IRt and includes a complete copy of the 5′-CS with the strong version of the Pc promoter (11) and a single gene cassette, aadB, conferring resistance to gentamicin, kanamycin, and tobramycin (9). However, In34 contains two partial copies of the 3′-CS separated by the 2,154-bp CR and a region containing the dfrA10 trimethoprim resistance determinant. The sequence of In34 from the BamHI site in the 5′-CS to the PstI site in 3′-CS2 was identical to the corrected sequence of In7 from pDGO100 (GenBank accession no. L06418.2). 3′-CS2 of In34 extends to bp 2025 of the standard 3′-CS and is followed by the insertion sequence IS1326 with IS1353 within it. This configuration is also found in In5-like class 1 integron In2 (GenBank accession no. AF071413 [7]), which constitutes the central region of Tn21 (Fig. 3A).
The similarity between In34 and In2 continues through the truncated tni region to IRt, except that a composite transposon flanked by a duplication of 8 bp of the target sequence is inserted in the tniA gene of In34, 1,393 bp from the IRt end. The transposon consists of directly repeated copies of the insertion sequence IS26 surrounding a central region containing the kanamycin and neomycin resistance determinant aphA1 [aph(3′)-Ia]. It is almost identical to Tn4352 from plasmid NTP16 (GenBank accession no. M20306 [44]). In In34 two additional C residues are present in the central region adjacent to the first copy of IS26, and this transposon variant has been designated Tn4352B.
The sequences immediately adjacent to In34 were also determined. At the IRi end the sequence matched the region immediately adjacent to the IRi end of In4 in the mercury resistance transposon Tn1696 (GenBank accession no. U12338 [32]). The sequence adjacent to the IRt end of In34 was identical to that found adjacent to the IRt end of In2 in Tn21 (GenBank accession no. AF071413 [26]). Thus, In34 is flanked by the tnp region of Tn1696 and the mer region of Tn21 (Fig. 3B). These modules have not previously been found in the same transposon.
Relationship of In34 to In7.
In7 is part of multiresistance plasmid pDGO100 from an E. coli strain isolated in 1978 at the same Sydney hospital as pRMH760 (17). pDGO100 is also transferable and confers resistance to the same antibiotics as pRMH760. Minor differences between In34 and the available sequence of In7 all lay in the CR and were traced to errors in the In7 sequence which were corrected (GenBank accession no. L06418.2). These differences change the reading frame near the end of the previously reported ORF, orf341 (39), extending the predicted protein to 513 amino acids. The ORF was renamed orf513. Other recent database entries that include the CR (GenBank accession nos. AB061794, AY049746, AJ310778, AF174129, AJ311891.2, and AY079169) contain the extended orf513 reading frame.
The catA1, blaTEM, and aphA1 resistance genes were also found in pDGO100, indicating that pDGO100 and pRMH760 contain six resistance genes in common. However, restriction digests indicated that the plasmid backbones of pRMH760 and pDGO100 are different (data not shown), and previous studies have shown that pDGO100 contains two class 1 integrons, with the dfrA7 cassette in the second one (8). Although the part of the tni module found in In34 is present in pDGO100, it did not include Tn4352, indicating that this transposon was acquired more recently by In34.
Homologues of the CR.
Database searches revealed that Orf513, the predicted product of orf513, is 66% identical (80% similar) to the predicted product of an ORF designated orfA from a plasmid found in an E. coli strain from cattle (10) and in the multidrug resistance region of an SXT-type conjugative transposon from Vibrio cholerae (23). Orf513 is also 55% identical to the predicted product of orf2 found in the multidrug resistance region in the SGI1 genomic island associated with the clone of S. enterica serovar Typhimurium DT104 commonly designated ACSSuT, which is resistant to a core group of antibiotics including ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and trimethoprim (5, 6). An alignment is reported by Boyd et al. (4).
To examine whether orfA and orf2 are part of regions equivalent to the CR and if they are found adjacent to antibiotic resistance genes, we searched for copies of the DNA sequence downstream of the ORFs. A boundary equivalent to the right-hand end of the CR was identified in each case (Fig. 2B), and the adjacent region included an antibiotic resistance gene (Fig. 4). Thus, these elements, now designated CR1 (orf513), CR2 (orfA), and CR3 (orf2), appear to have been important in mobilizing several of the resistance genes found in gram-negative organisms. A comparison of the sequences near the right-hand boundaries of the three CRs revealed a conserved region close to the boundary (Fig. 2B) that is likely to be recognized by Orf513 and its relatives.
FIG. 4.
Antibiotic resistance genes adjacent to CRs. The ends of the CRs associated with orf513 (CR1), orfA (CR2), and orf2 (CR3) are shaded boxes; and the unique regions are open boxes, with antibiotic resistance genes indicated by horizontal arrows. For unique regions adjacent to CR1, a closed end indicates the end of the unique region; and the adjacent number is the position of the first base of 3′-CS2 in the standard 3′-CS, or the −45 indicates 45 bp of a 59-be preceding bp 1 of the standard 3′-CS. In one case IRi marks this boundary. The sequences used here are from the GenBank accession numbers listed in Fig. 1 plus the following: ampC/R, GenBank accession no. AJ237702; blaCMY-9, AB061794; blaCTX-M-2, AJ311891; blaCTX-M-9, AF174129; dfrA19, AJ310778; qnr, AY070235; sul2, D37825; dfrA9, X57730; flo, AF231986; tetR/A (31), AJ250203; dfrA18, AY034138; ereB, X03988; tetR/A(G), AF261825.
Role of homologous recombination.
Homologous recombination between directly repeated sequences can lead to the loss of one copy of the repeated sequence together with the intervening region. The excised fragment would be circular and could subsequently be reinserted into any copy of the repeat sequence to re-create the original configuration. As wild-type bacterial strains are generally proficient in homologous recombination, this suggests a potential route for the movement of the segments containing CR1 and adjacent antibiotic resistance genes, which are flanked by two copies of part of the 3′-CS. The excised circular molecule could recombine with the 3′-CS in any integron and give rise to new integron configurations. To examine this possibility a 4.0-kb NcoI fragment from pRMH766 containing CR1, dfrA10, and 3′-CS sequences (Fig. 3B) was circularized and introduced into Rec+ strain AB1157 containing pRMH858, a plasmid with a cloned integron fragment that includes the relevant part of the 3′-CS (31). Rare trimethoprim-resistant transformants were recovered, and plasmid DNAs from them gave the predicted digestion patterns for pRMH858 that had acquired the CR1-dfrA10-sul1 circle (pRMH912; Fig. 3B). Deletion of the same fragment and the loss of trimethoprim resistance were also detected at very low frequencies after the growth of AB1157(pRMH912) in the absence of trimethoprim (Fig. 3B).
DISCUSSION
The plasmid pRMH760 studied here confers resistance to the same set of antibiotics as pDGO100, which is the source of In7 (17), and the overlapping portions of the sequenced regions of In34 and In7 are identical. The identification of two closely related integrons on different plasmids from E. coli and K. pneumoniae strains, isolated almost 20 years apart in 1978 and 1997 at the same hospital, respectively, illustrates how complex structures containing antibiotic resistance genes can persist in a local environment. The fact that the dfrA10 gene has not been seen in surveys of dfr genes conducted in the United Kingdom (40) and South Africa (1) indicates that it is not a widely distributed gene, adding weight to the argument for persistence. It has, however, recently been found in isolates of S. enterica serovar Agona from poultry in Belgium (GenBank accession no. AY049746 [4]), indicating that it may have been prevalent but undetected in other countries or has begun to spread globally.
In34 appears to be part of a transposon made up of the tnp module of Tn1696 and the mer module of Tn21, and it is possible that movement of the integron to a new plasmid location occurred as part of this larger transposon structure. The tnp1696-In34-mer21 transposon could have been created by recombination within the 5′-CS or 3′-CS of two transposons, one related to Tn1696 and the other related to Tn21. Such an event may be possible in plasmids such as pDGO100 if one of the two integrons that they contain is situated in a Tn1696-like transposon and the other is situated in a Tn21-like transposon. Alternatively, plasmids with two integrons could arise by an equivalent recombination event between integrons in two separate plasmids. Further examination of the structures of both In7 and pDGO100 may shed light on this question.
If In34 and possibly In7 are originally derived from In2 (Fig. 3), this is likely to have involved several steps, each of which may have occurred by homologous recombination. For example, recombination in the 5′-CS and 3′-CS flanking the gene cassettes could have replaced the gene cassette. The central region of In7 and In34 containing the dfrA10 gene, CR1, and a partial 3′-CS may also have been acquired by homologous recombination, as demonstrated in this study. It can also be lost as demonstrated here and in the recent study of Boyd et al. (4), and the loss of chloramphenicol resistance from pSa (22) has been shown to be due to the loss of CR1, the catA2 gene, and one copy of the sul1 gene from In6 (25). In In34 the CR1-associated structure is found within an integron of the In5 family; but movement of the CR1-containing central region by homologous recombination, as demonstrated here, should permit the resistance gene associated with CR1 to move into other types of class 1 integrons that contain the 3′-CS. Indeed, the same central region that includes the dfrA10 gene was recently found to be associated with an In4-like integron (Fig. 1A) in an S. enterica serovar Agona strain (4); and In6, which includes CR1 with catA2, is part of a third type of integron in which 3′-CS2 ends at bp 2119, with unidentified sequence beyond it (18).
Overall, homologous recombination is likely to contribute extensively to the movement of antibiotic resistance genes between plasmids. Even Tn4352 was not necessarily acquired by In34 by transposition, although it is flanked by an 8-bp duplication, consistent with its having originally taken up this position by transposition. Restriction maps in several publications suggest that a Tn4352-like transposon is associated with a number of Tn21-like transposons such as Tn1935, and the map of Tn1935 suggests that the Tn4352-like transposon is in a similar position in the tni region of the integron (12). In Tn21-derived transposon TnSF1 (GenBank accession no. AF188331) a version of Tn4352 is found in the same orientation as it is in In34 and it has the same right-hand boundary with tniA. An IS26 with the same right-hand boundary is also found in multidrug resistance plasmid pHCM1 from S. enterica serovar Typhi (29). Hence, the original event leading to the introduction of Tn4352 at this position, acquisition either of IS26 or of the transposon, may have occurred only once, followed by the global distribution of integrons containing it. It could have subsequently spread to further class 1 integrons such as In34 via homologous recombination in the flanking tni regions. pRMH760 can also potentially lose and regain the aphA1 gene by homologous recombination between the flanking insertion sequence.
CR1 has now been found in association with several different antibiotic resistance genes (Fig. 4), but it is always in a class 1 integron, as shown in Fig. 1B. In all cases the boundary between 3′-CS1 and CR1 is identical, indicating that CR1 may have entered the 3′-CS only once. Only the right-hand end appears to be active and able to incorporate new sequences adjacent to it. This step is likely to be catalyzed by Orf513. Resolution of the complex structure that this event would create appears to involve a less specific event that in most cases has occurred within the beginning of the 3′-CS and that has led to the loss of part of the 3′-CS. The similarity of Orf513 to OrfA and Orf2 led us to look for other common features and to examine the proximity of orfA and orf2 to antibiotic resistance genes. In both cases a discrete end equivalent to the right-hand end of CR1 was identified, and various resistance genes were found adjacent to this end (Fig. 2 and 4). The short conserved region found near this end (Fig. 2B) is likely to be recognized by Orf513 and its relatives. Although the mechanism by which such a one-ended system picks up new genes remains to be established experimentally, the weak similarity of Orf513, OrfA, and Orf2 proteins to the transposase of IS801 reported previously (10) may shed some light on this process. This deserves further investigation.
Acknowledgments
We thank Kylie Parisis for isolation of pRMH760 and pRMH761 and Clarence Fernandez for providing 24120497A and strains containing pDGO100.
S.R.P. was supported by grant 192108 from the Australian National Health and Medical Research Council.
REFERENCES
- 1.Adrian, P. V., K. P. Klugman, and S. G. B. Aymes. 1995. Prevalence of trimethoprim resistant dihydrofolate reductase genes identified with oligonucleotide probes in plasmids from isolates of commensal faecal flora. J. Antimicrob. Chemother. 35:497-508. [DOI] [PubMed] [Google Scholar]
- 2.Arduino, S. M., P. H. Roy, G. A. Jacoby, B. E. Orman, S. A. Pineiro, and D. Centron. 2002. blaCTX-M-2 is located in an unusual class 1 integron (In35) which includes Orf513. Antimicrob. Agents Chemother. 46:2303-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, D., A. Cloeckaert, E. Chaslus-Dancla, and M. R. Mulvey. 2002. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob. Agents Chemother. 46:1714-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briggs, C. E., and P. M. Fratamico. 1999. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob. Agents Chemother. 43:846-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, H. J., H. W. Stokes, and R. M. Hall. 1996. The integrons In0, In2, and In5 are defective transposon derivatives. J. Bacteriol. 178:4429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burnside, J. M., and D. J. Groot Obbink. 1996. Plasmid pDGO100 contains a second integron with the trimethoprim resistance gene dfrA7 as the inserted cassette. Plasmid 35:67-70. [DOI] [PubMed] [Google Scholar]
- 9.Cameron, F. H., D. J. Groot Obbink, V. A. Ackerman, and R. M. Hall. 1986. Nucleotide sequence of the AAD(2") aminoglycoside adenyltransferase determinant aadB. Evolutionary relationship of this region with those surrounding aadA in R538-1 and dhrfII in R388. Nucleic Acids Res. 14:8625-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cloeckaert, A., S. Baucheron, G. Flaujac, S. Schwarz, C. Kehrenberg, J.-L. Martel, and E. Chaslus-Dancla. 2000. Plasmid-mediated florphenicol resistance encoded by the floR gene in Escherichia coli isolated from cattle. Antimicrob. Agents Chemother. 44:2858-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colonna, B., M. Bernardini, G. Micheli, F. Maimone, M. Nicoletti, and M. Caslino. 1988. The Salmonella wien virulence plasmid pZM3 carries Tn1935, a multiresistance transposon containing a composite IS1936-kanamycin resistance element. Plasmid 20:221-231. [DOI] [PubMed] [Google Scholar]
- 13.de la Cruz, F., and J. Grinsted. 1982. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J. Bacteriol. 151:222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Witt, S. K., and E. A. Adelberg. 1962. The occurrence of a genetic transposition in a strain of Escherichia coli. Genetics 47:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Conza, J., J. A. Ayala, P. Power, M. Mollerach, and G. Gutkind. 2002. Novel class 1 integron (InS21) carrying blaCTX-M-2 in Salmonella enterica serovar Infantis. Antimicrob. Agents Chemother. 46:2257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doi, Y., N. Shibata, K. Shibayama, K. Kamachi, H. Kurokawa, K. Yokoyama, T. Yagi, and Y. Arakawa. 2002. Characterization of a novel plasmid-mediated cephalosporinase (CMY-9) and its genetic environment in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 46:2427-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groot Obbink, D. J., L. J. Ritchie, F. H. Cameron, J. S. Mattick, and V. P. Ackerman. 1985. Construction of a gentamycin resistance gene probe for epidemiological studies. Antimicrob. Agents Chemother. 28:96-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall, R. M., H. J. Brown, D. E. Brookes, and H. W. Stokes. 1994. Integrons found in different locations have identical 5′ ends but variable 3′ ends. J. Bacteriol. 176:6286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall, R. M., and C. M. Collis. 1998. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updates 1:109-119. [DOI] [PubMed] [Google Scholar]
- 20.Hall, R. M., and C. M. Collis. 1995. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15:593-600. [DOI] [PubMed] [Google Scholar]
- 21.Hall, R. M., and H. W. Stokes. 1990. The structure of a partial duplication in the integron of plasmid pDGO100. Plasmid 23:76-79. [DOI] [PubMed] [Google Scholar]
- 22.Hedges, R. W., and N. Datta. 1971. fi− R factors giving choramphenicol resistance. Nature 234:220-221. [Google Scholar]
- 23.Hochhut, B., Y. Lotfi, D. Mazel, S. M. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humeniuk, C., G. Arlet, V. Gautier, P. Grimont, R. Labia, and A. Philippon. 2002. β-Lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46:3045-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ireland, C. R. 1983. Detailed restriction enzyme map of crown gall-suppressive IncW plasmid pSa, showing ends of deletion causing chloramphenicol sensitivity. J. Bacteriol. 155:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazel, D., and J. Davies. 1999. Antibiotic resistance in microbes. Cell Mol. Life Sci. 56:742-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliver, A., J. C. Pérez-Díaz, T. M. Coque, F. Baquero, and R. Cantón. 2001. Nucleotide sequence and characterization of a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-10) isolated in Spain. Antimicrob. Agents Chemother. 45:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 30.Parsons, Y., R. M. Hall, and H. W. Stokes. 1991. A new trimethoprim resistance gene, dhfrX, in the In7 integron of plasmid pDG0100. Antimicrob. Agents Chemother. 35:2436-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Partridge, S. R., H. J. Brown, and R. M. Hall. 2002. Characterization and movement of the class 1 integron known as Tn2521 and Tn1405. Antimicrob. Agents Chemother. 46:1288-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rådström, P., O. Sköld, G. Swedberg, J. Flensburg, P. H. Roy, and L. Sundström. 1994. Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu, and the retroelements. J. Bacteriol. 176:3257-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 36.Recchia, G. D., and R. M. Hall. 1997. Origins of the mobile gene cassettes found in integrons. Trends Microbiol. 5:389-394. [DOI] [PubMed] [Google Scholar]
- 37.Sabate, M., F. Navarro, E. Miro, S. Campoy, B. Mirelis, J. Barbe, and G. Prats. 2002. Novel complex sul1-type integron in Escherichia coli carrying blaCTX-M-9. Antimicrob. Agents Chemother. 46:2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sinclair, M. I., and B. W. Holloway. 1982. A chromosomally located transposon in Pseudomonas aeruginosa. J. Bacteriol. 151:569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stokes, H. W., C. Tomaras, Y. Parsons, and R. M. Hall. 1993. The partial 3′-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid 30:39-50. [DOI] [PubMed] [Google Scholar]
- 40.Towner, K. J., G. I. Carter, H.-K. Young, and S. G. B. Amyes. 1991. Detection of novel trimethoprim resistance determinants in the United Kingdom using biotin-labelled DNA probes. Epidemiol. Infect. 106:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tran, J. H., and G. A. Jacoby. 2002. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. USA 99:5638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verdet, C., G. Arlet, G. Barnaud, P. H. Lagrange, and A. Philippon. 2000. A novel integron in Salmonella enterica serovar enteritidis, carrying the blaDHA-1 gene and its regulator gene ampR, originated from Morganella morganii. Antimicrob. Agents Chemother. 44:222-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villa, L., P. Visca, F. Tosini, C. Pezzella, and A. Carattoli. 2002. Composite integron array generated by insertion of an ORF341-type integron within a Tn21-like element. Microb. Drug Resist. 8:1-8. [DOI] [PubMed] [Google Scholar]
- 44.Wrighton, C. J., and P. Strike. 1987. A pathway for the evolution of the plasmid NTP16 involving the novel kanamycin resistance transposon Tn4352. Plasmid 17:37-45. [DOI] [PubMed] [Google Scholar]