Abstract
Avoparcin was used as a feed additive in New Zealand broiler production from 1977 until June 2000. We report here on the effects of the usage and discontinuation of avoparcin on the prevalence of vancomycin-resistant enterococci (VRE) in broilers. Eighty-two VRE isolates were recovered from poultry fecal samples between 2000 and mid-2001. VRE isolates were only obtained from broiler farms that were using, or had previously used, avoparcin as a dietary supplement. Of these VRE isolates, 73 (89%) were VanA-type Enterococcus faecalis and nine (11%) were VanA-type Enterococcus faecium. All E. faecalis isolates were found to have an identical or closely related pulsed-field gel electrophoresis (PFGE) pattern of SmaI-digested DNA and were susceptible to both ampicillin and gentamicin. The PFGE patterns of the nine E. faecium isolates were heterogeneous. All VRE contained both the vanA and ermB genes, which, regardless of species or PFGE pattern, resided on the same plasmid. Eighty-seven percent of the VRE isolates also harbored the tet(M) gene, while for 63 and 100%, respectively, of these isolates, the avilamycin and bacitracin MICs were high (≥256 μg/ml). Five of eight vancomycin-resistant E. faecalis isolates recovered from humans in New Zealand revealed a PFGE pattern identical or closely related to that of the E. faecalis poultry VRE isolates. Molecular characterization of Tn1546-like elements from the VRE showed that identical transposons were present in isolates from poultry and humans. Based on the findings presented here, a clonal lineage of VanA-type E. faecalis dominates in VRE isolated from poultry and humans in New Zealand.
Since their discovery in 1986 (34, 43), vancomycin-resistant enterococci (VRE) have emerged as a major cause of nosocomial infection. In the United States, the first VRE isolate was found in 1987 (37), and by 1997, more than 15% of nosocomial enterococcal infections in United States hospitals were due to VRE (14). Surveillance of VRE in the United States has shown that Enterococcus faecium is the dominant species identified (24). This is also the situation in Europe, where VanA-type E. faecium is the predominant phenotype isolated from animal, human, or environmental sources (3, 7, 11, 12, 18, 21, 22, 32, 38, 46). It is unclear why this particular species makes up the majority of VRE isolated.
A possible explanation for the emergence and spread of VRE in Europe has been the use of the growth promoter avoparcin in animal husbandry. Avoparcin is a glycopeptide produced by Streptomyces candidus and is closely related to vancomycin. The discovery in 1993, of VanA-type VRE in food animals in England (7), led to the postulation that food animals might be a potential reservoir for resistance genes. While the extent to which animal sources play a contribution to human infections remains a matter of debate, the use of avoparcin has been associated with an increase in the occurrence of VRE in broilers and pigs (6). Due to the potential for spread of resistance through the food chain, a European Union-wide ban was imposed in 1997 on avoparcin use in animal husbandry. Since this discontinuation, a decrease in the prevalence of VRE in Danish poultry has been observed (4); however, this trend has not been seen in Norway (10).
In New Zealand, VRE are uncommonly recovered from human sources, with only about 16 isolates being recovered prior to 2002 (unpublished data). In this country avoparcin has been used in poultry as a prophylactic for necrotic enteritis caused by Clostridium perfringens since 1977. Between 1999 and 2000, 2,195 kg of avoparcin was used. Because of the withdrawal of avoparcin from the market by the manufacturer in 1999, the use of avoparcin was discontinued in June 2000. We report here on the effect of usage and discontinuation of avoparcin on the prevalence of VRE in poultry animals and report the association of a distinct clone of vancomycin-resistant E. faecalis with both poultry and humans in New Zealand.
MATERIALS AND METHODS
Bacterial isolates.
Poultry VRE were isolated from broilers from four poultry suppliers as described below. Human strains used in this study were isolates from the culture collections of the Institute of Environmental Science and Research: Health, Communicable Disease Centre, Porirua, New Zealand; Auckland District Health Board; and the Canterbury District Health Board. E. faecium BM4147, which contains Tn1546 on plasmid pIP816, was used in this study as a positive control in the long-template PCR amplification and digestion of Tn1546. E. faecalis ATCC 29212 and E. faecium ATCC 19434 were used to generate intragenic probes for species identification. E. faecalis JH2-2 (25) and E. faecium GE-1 (20) were recipient strains used in conjugation experiments.
Sampling and isolation of VRE from poultry.
In April 2000, fecal samples were obtained from 66 individual broilers from four suppliers, A, B, C, and D in New Zealand. One year later (2001), 10 additional samples from each of four farms, A1, A2, A3, and A4, managed by original supplier A were received (40 samples in total). For each fecal sample, a pea-sized scoop was emulsified in 10-ml Streptococcus faecalis medium (Bacto SF medium; Difco Laboratories, Detroit, Mich.). After incubation for 48 h at 35°C, dilutions were made to 10−3 and 100 μl was spread onto bile esculin azide (BEA) agar plates (Becton Dickinson and Co., Sparks, Md.) to obtain single colonies. All BEA agar plates were incubated for 48 h at 35°C. The natural ability of enterococci to grow in the presence of bile and to hydrolyze esculin to form black colonies was used for their selection.
Antimicrobial susceptibility testing.
Any black esculin-positive colonies growing on BEA agar plates were assumed to be enterococci. One hundred enterococcus-like colonies were chosen randomly from each sample and inoculated onto brain heart infusion (BHI) agar plates (Becton Dickinson and Co.) containing 32 μg of vancomycin/ml. All plate contents were incubated at 37°C and examined for growth after 24 h. Any positive growth was assumed to represent VRE. Vancomycin, teicoplanin, gentamicin, and ampicillin MICs for these isolates were obtained using Mueller-Hinton agar (Difco) plates and E-test strips (AB BIODISK, Solna, Sweden) according to the manufacturer's instructions. All VRE were also examined for their sensitivity to erythromycin (Sigma-Aldrich Chemicals, St. Louis, Mo.), tetracycline (Sigma), avilamycin (Eli Lilly & Co., Indianapolis, Ind.), and zinc bacitracin (50,000 IU/g; Aldrich Chemical Co., Milwaukee, Wis.). The MICs of these antibiotics were determined by following the National Committee for Clinical Laboratory Standards (NCCLS) guidelines (36).
PCR amplification.
All primer sequences and specificities used in this study are listed in Table 1. For species identification, intragenic probes for efaA and aac(6′)-Ii were generated by PCR from E. faecalis ATCC 29212 and E. faecium ATCC 19434, respectively. An intragenic probe for vanA was generated from E. faecium BM4147, while probes for ermB and tet(M) were generated from an erythromycin- and tetracycline-resistant clinical VRE isolate. All PCR products were sequenced to ensure homology with the published sequence for these genes. Amplification of vanX was carried out using the primers VanX1 and VanX2. PCRs were performed in 100-μl volumes with 1 U of Taq DNA polymerase (Roche Molecular Biochemicals, Mannheim, Germany) in accordance with the manufacturer's instructions and the PCR program described previously (30). DNA template was prepared by dissolving a bacterial colony in 50 μl of water and freeze-thawing at −20°C. PCR products were purified with a PCR purification kit (Roche). Tn1546-like elements were amplified using an Expand Long Template PCR system (Roche) and the conditions recommended by the manufacturer. The primers used were ORF1-F1 and VanY-R1 (Table 1). Amplification consisted of one cycle at 94°C for 2 min and 10 cycles at 94°C for 10 s, 65°C for 30 s, and 68°C for 8 min. This was followed by 20 cycles at 94°C for 10 s, 65°C for 30 s, and 68°C for 8 min (with the elongation time increased by 10 s per cycle) and a final cycle at 68°C for 7 min.
TABLE 1.
PCR primers used in this study
| Primer | Sequence (5′ → 3′) | Specificity | Reference |
|---|---|---|---|
| efaAF | CGTTAGCTGCTTGCGGGAATC | efaA | 40 |
| efaAR | CCATACTACGTTTATCGACAC | ||
| aac(6′)-IiF | GCGGTAGCAGCGGTAGACCAAG | aac(6′)-Ii | 19 |
| aac(6′)-IiR | GCATTTGGTAAGACACCTACG | ||
| VanABF | GTAGGCTGCGATATTCAAAGC | vanA | 8 |
| VanAR | CGATTCAATTGCGTAGTCCAA | ||
| ermB-1 | CATTTAACGACGAAACTGGC | ermB | 28 |
| ermB-2 | GGAACATCTGTGGTATGGCG | ||
| TetM-FW | ACAGAAAGCTTATTATATAAC | tet(M) | 5 |
| TetM-RV | TGGCGTGTCTATGATGTTCAC | ||
| VanX1 | ACTTGGGATAATTTCACCGG | vanX | 27 |
| VanX2 | TGCGATTTTGCGCTTCATTG | ||
| ORF1-F1 | AATCTTCATTAAAGCTACCTGTCCG | Tn1546 | 17 |
| VanY-R1 | TATCTCATAACGAAGATTAGTCGGC |
Genomic DNA preparation and restriction enzyme digestion.
Genomic DNA embedded in agarose was prepared essentially as described by Keis et al. (30) for the preparation of clostridial genomic DNA, except that all steps were carried out aerobically and that bacteria were grown to an optical density at 650 nm of 0.6 in 10 ml of BHI broth. Genomic DNA was extracted from enterococci using the following method: bacterial cultures were grown overnight in 20 ml of BHI broth. Cells were pelleted and washed in 10 ml of Tris-EDTA (TE) buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) and were resuspended in 200 μl of TE-glucose buffer (50 mM glucose, 25 mM Tris-HCl [pH 8.0], and 10 mM EDTA) containing 12.5 mg of lysozyme/ml and 125 U of mutanolysin/ml. The suspension was incubated for 2 h at 37°C before addition of N-lauroyl sarcosine to a final concentration of 8% and 4 μg of RNase A/ml and incubation for a further 15 min at 37°C. To this mixture, 4 μg of pronase/ml was added and the solution was incubated for 30 min at 37°C. TE buffer was used to bring the volume up to 600 μl. The DNA was extracted twice with buffer-saturated phenol, three times with phenol-chloroform-isoamyl alcohol (25:24:1), and once with chloroform-isoamyl alcohol (24:1). DNA was precipitated with 1 volume of isopropanol and 1/10 volume of 3 M sodium acetate, pH 5.2, washed twice with 70% ethanol, air dried, and resuspended in 100 μl of TE buffer.
Genomic DNA embedded in agarose was digested with the restriction endonucleases SmaI (Roche) and I-CeuI (New England Biolabs, Inc., Beverly, Mass.) by equilibrating slices of DNA plugs (10 by 2 mm) three times for 15 min in 100 μl of the restriction enzyme buffer recommended by the manufacturer before adding 10 U of the enzyme. DNA plugs digested with SmaI were preincubated overnight at 4°C prior to digestion at 25°C for 4 h. I-CeuI digests were incubated at 37°C for 4 h. For reproducibility, the I-CeuI-digested DNA plugs were treated after digestion with 0.5 mg of proteinase K/ml in EDTA-N-lauroyl sarcosine buffer (0.5 M EDTA [pH 9.0], 1% N-lauroyl sarcosine) as described previously (31). Long-template PCR products were digested with 10 U of ClaI (Roche), in the buffer recommended by the manufacturer, for 3 h at 37°C.
PFGE and agarose gel electrophoresis.
Pulsed-field gel electrophoresis (PFGE) was performed by contour-clamped homogeneous electric field electrophoresis using the CHEF-DRIII system (Bio-Rad Laboratories, Richmond, Calif.). Agarose plugs were equilibrated three times in 0.5× Tris-borate-EDTA buffer (0.45 mM Tris-borate, 1 mM EDTA) for 15 min prior to electrophoresis. Gels were routinely run at 6 V/cm, 14°C, at an included angle of 120°, on a 1.2% agarose gel (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, England) with pulse times of 5 to 25 s for 22 h. The Low Range PFG Marker (New England Biolabs, Inc.) containing lambda concatemers and lambda-digested HindIII fragments was used as a size standard. Long-template PCR products digested with ClaI were electrophoresed for 2 h at 70 V in 0.8% LE agarose (Roche).
Southern hybridization.
The DNA from enterococcal colonies was bound to nitrocellulose filters as follows: overnight cultures, grown in BHI broth were stamped onto Hybond-N+ nylon membranes (Amersham) overlaid on BHI agar and were incubated for 24 h at 37°C. Colonies were fixed and lysed by soaking the membranes in denaturation solution (0.5 M NaOH, 1.5 M NaCl) for 40 min and by boiling twice in 0.5% sodium dodecyl sulfate for 20 min. Membranes were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and blotted dry. DNA fragments separated by PFGE were transferred onto Hybond-N+ nylon membrane using a VacuGene (Pharmacia-LKB) vacuum blotting system, as described previously (31).
Radiolabeled PCR products were prepared by incorporation of [α-32P]dCTP-labeled deoxyribonucleotides (Amersham) using the RTS Radprime DNA labeling kit (Gibco BRL Life Technologies, Gaithersburg, Md.). Prehybridization and hybridization were carried out in the same hybridization buffer (16) at 65°C for 18 to 24 h. After hybridization, membranes were washed stringently as described by Keis et al. (31).
DNA sequencing and analysis.
PCR products were sequenced directly. Sequencing reactions were carried out by using a PRISM ready reaction DyeDeoxy terminator cycle sequencing kit (Applied Biosystems Inc., Warrington, United Kingdom) and a model ABI377 automated DNA sequencer (Applied Biosystems). DNA sequences were aligned using the LaserGene application software Megalign (DNASTAR, Inc.) for Apple Macintosh computers, which employed the algorithm CLUSTALV.
Conjugation and plasmid-curing experiments.
Potential donor strains were mated with various recipient strains to determine whether antimicrobial resistance could be transferred. The recipient strains were E. faecalis JH2-2 (25) and E. faecium GE-1 (20), which are resistant to fusidic acid and rifampin. Transfer in broth and transfer on plates were carried out as described earlier by Christie et al. (15) and Carias et al. (13), respectively. Transconjugants were selected on BHI agar plates containing vancomycin (32 μg/ml), rifampin (50 μg/ml), and fusidic acid (25 μg/ml).
Plasmid curing was carried out by subculturing an overnight culture grown in BHI broth at 37°C for 28 days.
RESULTS
Isolation and characterization of VRE isolates from poultry.
In 2000, a total of 500 enterococci were isolated from the feces of 66 individual broilers from four poultry suppliers in New Zealand. One of these suppliers (supplier A) was confirmed to have used avoparcin for an extended period of time, whereas the remaining three suppliers (B, C, and D) had never used avoparcin as a feed additive. No VRE were isolated from suppliers B, C, and D; however, 18 VRE out of a total of 200 enterococcus isolates tested were cultured from supplier A broiler fecal samples. All 18 VRE isolates had high-level resistance to vancomycin and teicoplanin (MICs ≥ 256 μg/ml) but were susceptible to ampicillin and gentamicin. With species-specific intragenic probes, two of the VRE isolates were identified as E. faecium and the remaining 16 as E. faecalis. Colony hybridization with a PCR product specific for the vanA gene demonstrated that all VRE contained this gene. Furthermore, all VRE were PCR positive for vanX and exhibited the guanine base pair variation in the vanX gene at position 8234 (data not shown) as previously reported for poultry isolates (26).
Forty additional samples were received in 2001 from four farms, A1, A2, A3, and A4, belonging to supplier A. Four hundred enterococci were tested in 2001. In farm A1, no resistant enterococci were detected. In farm A2, 14 of 100 enterococci tested were found to be vancomycin resistant. DNA hybridization, using species-specific probes, identified two VRE isolates as E. faecium and the remaining 12 as E. faecalis. In farm A3, 10 of 100 enterococci were found to be VRE, with five vancomycin-resistant E. faecalis and five vancomycin-resistant E. faecium isolates cultured. Forty VRE isolates were cultured from farm A4. All VRE from this farm were E. faecalis. In total, 7 E. faecium and 57 E. faecalis vancomycin-resistant isolates were cultured from the four farms. All 64 VRE isolates revealed high-level resistance to vancomycin and teicoplanin (MICs ≥ 256 μg/ml) but were susceptible to ampicillin and gentamicin. All contained the vanA gene.
All 82 VRE from both 2000 and 2001 were found to be resistant to erythromycin; MICs ranged from 50 to ≥256 μg/ml. All contained the ermB gene. Seventy-one of the VRE isolates were found to be resistant to tetracyline (MICs ≥ 20 μg/ml), and colony hybridization showed that all of these contained the tetracycline resistance gene tet(M). For 52 of the VRE isolates, avilamycin MICs were ≥ 256 μg/ml, and for all 82 VRE isolates, bacitracin MICs were ≥ 256 μg/ml (data not shown).
DNA fingerprinting of VRE from poultry.
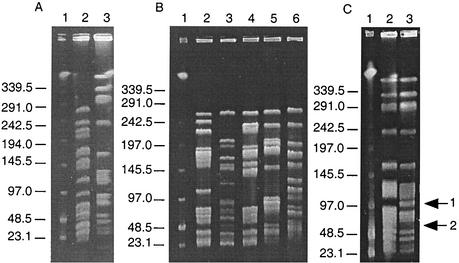
DNA fingerprinting of the poultry VRE was carried out by comparing the SmaI digestion patterns obtained after PFGE, using the criteria described by Tenover et al. (42), for interpreting chromosomal DNA restriction patterns produced by PFGE. In total, seven PFGE patterns were found among the 82 VRE isolated in 2000 and 2001 from supplier A. VRE isolated in 2000 exhibited one of two PFGE patterns, depending on the species to which the isolates belonged. All 16 E. faecalis isolates had an identical PFGE pattern (designated 1a), whereas both E. faecium isolates had the same PFGE pattern (designated 2) (Fig. 1A). A further five PFGE patterns (designated 3 to 7) were identified for E. faecium isolates recovered in 2001 (Fig. 1B) and appeared quite unrelated, with differences of more than seven bands. Both E. faecium isolates from farm A2 belonged to PFGE pattern 3, while at farm A3 a total of four PFGE patterns were identified, namely, 4, 5, 6, and 7 for the five E. faecium strains (Fig. 1B). In contrast to the vancomycin-resistant E. faecium isolates, for which no identical PFGE patterns were found from different farms, all vancomycin-resistant E. faecalis strains from farms A2, A3, and A4 had the same DNA fingerprint (Fig. 1C). Of 12 randomly chosen vancomycin-resistant E. faecalis isolates from farm A4, one belonged to PFGE pattern 1a, while the remaining 11 had a closely related PFGE pattern (designated 1b). This contained one additional band, and two fragments were absent when compared to pattern 1a (Fig. 1C).
FIG. 1.
Representative SmaI PFGE patterns among VRE isolated from broilers. (A) VRE isolated in 2000. Lane 1, lambda DNA ladder standard; lane 2, PFGE pattern 2; lane 3, PFGE pattern 1a. (B) Vancomycin-resistant E. faecium isolated in 2001. Lane 1, lambda DNA ladder standard; lane 2, PFGE pattern 7; lane 3, PFGE pattern 6; lane 4, PFGE pattern 5; lane 5, PFGE pattern 4, lane 6, PFGE pattern 3. (C) Vancomycin-resistant E. faecalis isolated in 2001. Lane 1, lambda DNA ladder standard; lane 2, PFGE pattern 1a; lane 3, PFGE pattern 1b. Arrow 1 indicates the presence of an additional band, while arrow 2 denotes the absence of two bands when compared with PFGE pattern 1a. Sizes indicated are in kilobases.
Location of the resistance genes.
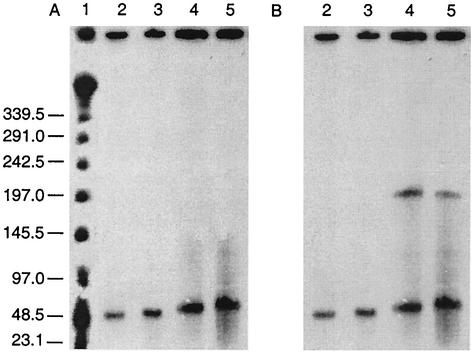
I-CeuI is a restriction enzyme that recognizes a specific site in the 23S rRNA operon of bacterial genomes and cleaves only chromosomal DNA. Hybridization of I-CeuI-digested DNA and nondigested DNA can therefore determine whether a gene is present on the chromosome or is plasmid borne. I-CeuI digestion of genomic DNA was completed on each of the seven poultry VRE PFGE patterns. In all isolates, the same band in both digested and uncut lanes hybridized to both the vanA and ermB gene probes (Fig. 2 shows a representative pattern for two VRE isolates). In all PFGE patterns, the vanA and ermB genes appeared to be plasmid borne, hybridizing to bands varying in size from 44.5 to 83.7 kb (Table 2). In PFGE pattern 2 a second plasmid band (approximately 204.7 kb) hybridized to the ermB probe (Fig. 2B).
FIG. 2.
Hybridization with vanA (A) and ermB (B) probes to I-CeuI-digested genomic DNA of two VRE isolates. Two lanes of genomic DNA from each isolate are shown. The first in each pair was not digested with I-CeuI, while the second was incubated with the enzyme. Lane 1, lambda DNA ladder standard; lanes 2 and 3, poultry PFGE pattern 1a; lanes 4 and 5, poultry PFGE pattern 2. Sizes indicated are in kilobases.
TABLE 2.
Locations and sizes of hybridized I-CeuI fragments probed with vanA, ermB, and tet(M) gene products
| Strain | PFGE pattern | I-CeuI fragment (length in kb) hybridized with antibiotic resistance genes
|
||
|---|---|---|---|---|
| vanA | ermB | tet(M) | ||
| 5A-13 | 1a | Pa (61.0) | P (61.0) | P (55.5) |
| 1B-10-96 | 1b | P (66.3) | P (66.3) | P (79.3) |
| 7-11-7 | 2 | P (62.8) | P (62.8, 204.7) | P (204.7) |
| 10-9A | 3 | P (44.5) | P (44.5) | Cb |
| 11-3A | 4 | P (52.8) | P (52.8) | P (82.8) |
| 12-9VP | 5 | P (68.7) | P (68.7) | C |
| 12-8VP | 6 | P (75.6) | P (75.6) | NDc |
| 12-7VP | 7 | P (83.7) | P (83.7) | C |
P, plasmid.
C, chromosome.
ND, not detected.
The location of the tet(M) gene was also investigated by using I-CeuI digestion (data not shown). In PFGE patterns 1a, 1b, 2, and 4, the resistance genes appeared to be located on plasmids ranging in size from 51.9 to 204.7 kb (Table 2). In PFGE pattern 2, the tet(M) probe hybridized to the same large 204.7-kb fragment that had hybridized to the ermB probe. The tet(M) genes appeared to be chromosomally located in PFGE patterns 3, 5, and 7, with hybridization occurring to I-CeuI-digested bands greater than 339.5 kb. No hybridization was seen with PFGE pattern 6.
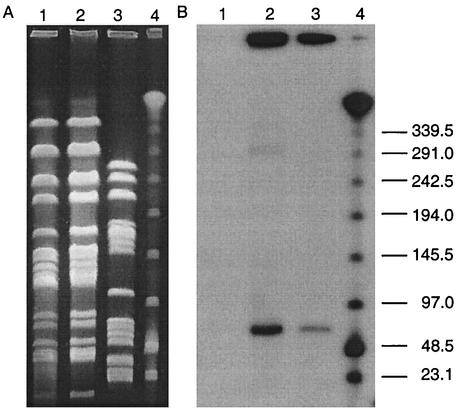
The ability of VRE isolates, representative of the different PFGE patterns, to transfer resistance by conjugation was assessed by using both broth and plate mating. Transfer of erythromycin and vancomycin resistance was demonstrated for an E. faecium isolate (12-9VP) belonging to PFGE pattern 5. The resulting transconjugant was designated JH2-129. Transfer occurred in broth mating with a frequency of conjugation of 9.0 × 10−10 per recipient to E. faecalis JH2-2 but not to E. faecium GE-1 (<10−10 per recipient). No transfer was observed using plate mating for any of the VRE isolates. Transfer of the plasmid from donor to transconjugant was confirmed by PFGE and hybridization. The ermB and vanA probes hybridized to a SmaI fragment of approximately 68.7 kb in both the donor and transconjugant, illustrating the presence of a transferable element (Fig. 3). It is also worth noting that four vancomycin-resistant E. faecium isolates, PFGE patterns 3, 4, 6, and 7, became vancomycin susceptible after subculturing twice on BHI agar plates without vancomycin. When further examined, these isolates no longer contained either the vanA or ermB gene (data not shown), suggesting the loss of the resistance plasmid. The stability of resistance in the vancomycin-resistant E. faecalis PFGE pattern 1a was tested by subculturing a representative strain through a series of fresh BHI broth daily for 28 days. No loss of resistance was seen after this time, indicating stability of the plasmid.
FIG. 3.
Conjugative transfer of vancomycin resistance. (A) SmaI macrorestriction patterns of strains involved in conjugative transfer of vancomycin resistance; (B) corresponding Southern blot hybridized with a vanA gene probe. Lane 1, recipient strain JH2-2; lane 2, transconjugant strain JH2-129; lane 3, donor strain 12-9VP; lane 4, lambda DNA ladder standard. Sizes are indicated in kilobases.
Comparison of poultry VRE with clinical VRE in New Zealand.
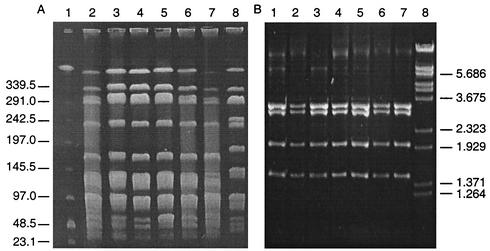
We chose 12 recent human clinical VRE isolates for a comparative analysis with our poultry VRE. Of these 12 isolates, eight were VanA-type E. faecalis and four were VanA-type E. faecium. All clinical isolates were resistant to gentamicin; MICs of ≥256 μg/ml were recorded, while only the four E. faecium isolates were resistant to ampicillin (MICs ≥ 256 μg/ml). For six of the clinical VRE isolates, bacitracin MICs were ≥ 256 μg/ml. The 12 clinical isolates were a heterogeneous group with seven distinct PFGE patterns. However, 5 of the 12 isolates, all VanA-type E. faecalis, belonged to a single PFGE pattern, indistinguishable from or closely related to the dominant VanA-type E. faecalis pattern found in poultry (Fig. 4A, lanes 4 to 8). PCR amplification and DNA sequencing of the vanX gene in the 12 human VRE isolates revealed that the base pair variation at position 8234 was guanine in all eight E. faecalis isolates. Thymine was found in position 8234 in the four E. faecium isolates. Colony hybridization revealed that all clinical isolates also contained the ermB gene conveying erythromycin resistance. The location of the resistance genes was examined in one PFGE pattern 1 clinical isolate, and the ermB and vanA genes were found to hybridize to a plasmid band of approximately 61 kb (data not shown).
FIG. 4.
Comparison of poultry and human VRE in New Zealand. (A) PFGE of SmaI macrorestriction patterns in vancomycin-resistant E. faecalis. Lane 1, lambda DNA ladder standard; lanes 2 and 3, poultry isolates; lanes 4 to 8, human clinical isolates. (B) Long-template PCR ClaI RFLP patterns of Tn1546 elements. Lane 1, E. faecium BM4147; lanes 2 and 3, poultry VRE isolates; lanes 4 to 7, human clinical isolates; lane 8, lambda DNA cut with BstEII. ClaI fragments are 3,228, 2,968, 2,009, and 1,471 bp. Marker sizes are indicated in kilobases.
ClaI RFLP analysis of Tn1546 elements.
To further examine the relationship between our poultry and human VRE isolates, ClaI restriction fragment length polymorphism (RFLP) analysis of Tn1546 elements was carried out. Identical Tn1546 types among human and animal isolates infer horizontal transfer from a common resistance gene pool. Tn1546 was amplified from E. faecium BM4147 and was digested with ClaI. Four restriction fragments were obtained, and these were consistent with the predicted sizes of 3,228, 2,968, 2,009, and 1,471 bp from the DNA sequence. This method was applied to all 12 human isolates plus one poultry isolate from each of the seven PFGE patterns. For two E. faecium isolates, one poultry and one human, no long-template PCR product was obtained after repeated attempts. Digestion of the remaining 17 positive long-template PCR products gave identical results to BM4147 (Fig. 4B). This observation suggests a degree of conservation between the Tn1546 elements in New Zealand VRE isolates of animal and human origin.
DISCUSSION
This is the first published report of VRE in food animals (in this case broilers) in New Zealand. Resistance to vancomycin in the 82 VRE strains isolated (73 E. faecalis and 9 E. faecium strains) was always due to the presence of the vanA gene and occurred only in enterococci recovered from the poultry supplier that had previously used avoparcin. The dominance of VanA-type E. faecalis in our isolates is unusual, as in Europe VanA-type E. faecium appears to prevail among animals, including poultry, that are fed avoparcin (2, 33). In fact, European studies have found that, in VRE isolated from human, animal or environmental sources, the vanA genotype and species E. faecium predominate (3, 7, 12, 18, 21, 22, 32, 38, 46). For example, of 90 VRE strains isolated from poultry carcasses in Norway, none were E. faecalis (11). Likewise, a study on vancomycin susceptibility in Denmark among E. faecium and E. faecalis isolates found 221 vancomycin-resistant E. faecium and only 2 vancomycin-resistant E. faecalis strains (3). It should be noted that speciation was carried out on our isolates using species-specific probes, rather than by biochemical methods, which can sometimes give misleading results (41). All 73 vancomycin-resistant E. faecalis strains included in the present study revealed PFGE patterns ranging from identical to closely related, indicating some degree of clonality among the study isolates of this species. Conversely, the vancomycin-resistant E. faecium isolates revealed a range of distinct PFGE patterns that appeared to dominate at particular farms. This is similar to the findings of Aarestrup et al. (1), who also found heterogeneous E. faecium PFGE patterns among broilers.
In our studies, the continued occurrence of VRE in poultry from supplier A 1 year after avoparcin was eliminated as a food additive is not a new finding. As previously documented in pigs in Denmark (1), this may happen if the genes responsible for resistance occur on DNA elements containing genes encoding resistance to other antimicrobials that continue to be included in the animal's diet. All VRE isolates from New Zealand poultry contained both vanA and ermB genes, which, regardless of species or PFGE pattern, were found residing together on the same plasmid. This genetic linkage of erythromycin and vancomycin resistance has been seen previously in both animal (1, 9) and human (35, 44) isolates. The macrolide tylan is still used in New Zealand poultry animals, and its use could theoretically coselect for vancomycin-resistant bacteria. However, it seems that the farms from which the 2001 samples were taken had not used tylan in the previous 6 months but had used zinc bacitracin. Could it be that the use of zinc bacitracin in the diet is the coselecting force or that VRE clones are somehow better adapted for environmental survival (23) and/or subsequent recolonization of new animals? In this context, vancomycin resistance was lost upon subculturing without selection pressure in four E. faecium isolates but was retained in all vancomycin-resistant E. faecalis isolates. After subculturing of one E. faecalis strain continuously for 28 days, no loss of plasmid was seen. It is possible that the dominance of this vancomycin-resistant E. faecalis clone is due to its ability to retain the resistance plasmid in the absence of glycopeptide (i.e., avoparcin) selection pressure. It is also worth noting that the low frequency of transfer of vancomycin resistance seen in this study is likely to add to the dominance of this clonal lineage of vancomycin-resistant E. faecalis. Future studies are aimed at establishing the selection factors involved in plasmid maintenance in a glycopeptide-free environment. These data may provide answers as to why this vancomycin-resistant E. faecalis clone dominates in VRE isolated in New Zealand.
Although VRE presently appear uncommon in hospitalized patients in New Zealand, a striking factor of recorded cases has been the dominance of VanA-type E. faecalis among the isolates. Of the eight vancomycin-resistant E. faecalis strains of human origin that we have studied, five appear to be indistinguishable from or genetically related to the dominant poultry vancomycin-resistant E. faecalis clone. Indistinguishable PFGE patterns have previously been seen in vancomycin-resistant E. faecium from a Dutch farmer and one of his turkeys (45) and in a Danish woman who harbored an isolate very similar to the VRE clone that is predominantly found in Danish pigs (29). In the present study, both human and animal isolates that were typeable through RFLP of the Tn1546 element had an identical arrangement of genes, suggesting a common resistance gene pool and perhaps indicating horizontal transfer among different hosts. In a study by Schouten et al. (39), geographical association of different transposon types was suggested to be a result of differences in meat consumption between European countries. Because New Zealand is geographically isolated, it is not surprising that the resistance genes appear to have a common origin. Identical Tn1546 types have previously been found in humans and animals (17, 47-49). Because an identical VRE clone is found in poultry and retail chicken portions (unpublished data), current studies are aimed at investigating the prevalence of fecal VRE colonization in healthy humans in New Zealand.
Acknowledgments
This work was funded by an Otago Medical Research Foundation Grant and by an Otago Research Grant awarded to G.M.C. and J.M.B.S.
We thank the poultry industry of New Zealand for their cooperation in this study.
REFERENCES
- 1.Aarestrup, F. M. 2000. Characterization of glycopeptide-resistant Enterococcus faecium (GRE) from broilers and pigs in Denmark: genetic evidence that persistence of GRE in pig herds is associated with coselection by resistance to macrolides. J. Clin. Microbiol. 38:2774-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aarestrup, F. M. 1995. Occurrence of glycopeptide resistance among Enterococcus faecium isolates from conventional and ecological poultry farms. Microb. Drug Resist. 1:255-257. [DOI] [PubMed] [Google Scholar]
- 3.Aarestrup, F. M., P. Ahrens, M. Madsen, L. V. Pallesen, R. L. Poulsen, and H. Westh. 1996. Glycopeptide susceptibility among Danish Enterococcus faecium and Enterococcus faecalis isolates of animal and human origin and PCR identification of genes within the VanA cluster. Antimicrob. Agents Chemother. 40:1938-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aarestrup, F. M., A. M. Seyfarth, H. D. Emborg, K. Pedersen, R. S. Hendriksen, and F. Bager. 2001. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 45:2054-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aminov, R. I., N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bager, F., M. Madsen, J. Christensen, and F. M. Aarestrup. 1997. Avoparcin used as a growth promoter is associated with the occurrence of vancomycin-resistant Enterococcus faecium on Danish poultry and pig farms. Prev. Vet. Med. 31:95-112. [DOI] [PubMed] [Google Scholar]
- 7.Bates, J., Z. Jordens, and J. B. Selkon. 1993. Evidence for an animal origin of vancomycin-resistant enterococci. Lancet 342:490-491. [DOI] [PubMed] [Google Scholar]
- 8.Bell, J. M., J. C. Paton, and J. Turnidge. 1998. Emergence of vancomycin-resistant enterococci in Australia: phenotypic and genotypic characteristics of isolates. J. Clin. Microbiol. 36:2187-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boerlin, P., A. Wissing, F. M. Aarestrup, J. Frey, and J. Nicolet. 2001. Antimicrobial growth promoter ban and resistance to macrolides and vancomycin in enterococci from pigs. J. Clin. Microbiol. 39:4193-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borgen, K., G. S. Simonsen, A. Sundsfjord, Y. Wasteson, Ø. Olsvik, and H. Kruse. 2000. Continuing high prevalence of VanA-type vancomycin-resistant enterococci on Norwegian poultry farms three years after avoparcin was banned. J. Appl. Microbiol. 89:478-485. [DOI] [PubMed] [Google Scholar]
- 11.Borgen, K., M. Søorum, Y. Wasteson, and H. Kruse. 2001. VanA-type vancomycin-resistant enterococci (VRE) remain prevalent in poultry carcasses 3 years after avoparcin was banned. Int. J. Food Microbiol. 64:89-94. [DOI] [PubMed] [Google Scholar]
- 12.Butaye, P., L. A. Devriese, and F. Haesebrouck. 2001. Differences in antibiotic resistance patterns of Enterococcus faecalis and Enterococcus faecium strains isolated from farm and pet animals. Antimicrob. Agents Chemother. 45:1374-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carias, L. L., S. D. Rudin, C. J. Donskey, and L. B. Rice. 1998. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J. Bacteriol. 180:4426-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention. 1998. Summary of notifiable diseases—United States, 1997. Morb. Mortal. Wkly. Rep. 46:1-87. [PubMed] [Google Scholar]
- 15.Christie, P. J., R. Z. Korman, S. A. Zahler, J. C. Adsit, and G. M. Dunny. 1987. Two conjugation systems associated with Streptococcus faecalis plasmid pCF10: identification of a conjugative transposon that transfers between S. faecalis and Bacillus subtilis. J. Bacteriol. 169:2529-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Descheemaeker, P. R., S. Chapelle, L. A. Devriese, P. Butaye, P. Vandamme, and H. Goossens. 1999. Comparison of glycopeptide-resistant Enterococcus faecium isolates and glycopeptide resistance genes of human and animal origins. Antimicrob. Agents Chemother. 43:2032-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devriese, L. A., M. Ieven, H. Goossens, P. Vandamme, B. Pot, J. Hommez, and F. Haesebrouck. 1996. Presence of vancomycin-resistant enterococci in farm and pet animals. Antimicrob. Agents Chemother. 40:2285-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donabedian, S., J. W. Chow, D. M. Shlaes, M. Green, and M. J. Zervos. 1995. DNA hybridization and contour-clamped homogeneous electric field electrophoresis for identification of enterococci to the species level. J. Clin. Microbiol. 33:141-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eliopoulos, G. M., C. Wennersten, S. Zighelboim-Daum, E. Reiszner, D. Goldmann, and R. C. Moellering, Jr. 1988. High-level resistance to gentamicin in clinical isolates of Streptococcus (Enterococcus) faecium. Antimicrob. Agents Chemother. 32:1528-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gambarotto, K., M. C. Ploy, F. Dupron, M. Giangiobbe, and F. Denis. 2001. Occurrence of vancomycin-resistant enterococci in pork and poultry products from a cattle-rearing area of France. J. Clin. Microbiol. 39:2354-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gambarotto, K., M. C. Ploy, P. Turlure, C. Grélaud, C. Martin, D. Bordessoule, and F. Denis. 2000. Prevalence of vancomycin-resistant enterococci in fecal samples from hospitalized patients and nonhospitalized controls in a cattle-rearing area of France. J. Clin. Microbiol. 38:620-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasman, H., and F. M. Aarestrup. 2002. tcrB, a gene conferring transferable copper resistance in Enterococcus faecium: occurrence, transferability, and linkage to macrolide and glycopeptide resistance. Antimicrob. Agents Chemother. 46:1410-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hospital Infections Program. 1999. National Nosocomial Infections Surveillance (NNIS) System report, data summary from January 1990-May 1999, issued June 1999. Am. J. Infect. Control 27:520-532. [DOI] [PubMed] [Google Scholar]
- 25.Jacob, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen, L. B. 1998. Differences in the occurrence of two base pair variants of Tn1546 from vancomycin-resistant enterococci from humans, pigs, and poultry. Antimicrob. Agents Chemother. 42:2463-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen, L. B., P. Ahrens, L. Dons, R. N. Jones, A. M. Hammerum, and F. M. Aarestrup. 1998. Molecular analysis of Tn1546 in Enterococcus faecium isolated from animals and humans. J. Clin. Microbiol. 36:437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen, L. B., N. Frimodt-Møller, and F. M. Aarestrup. 1999. Presence of erm gene classes in gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol. Lett. 170:151-158. [DOI] [PubMed] [Google Scholar]
- 29.Jensen, L. B., A. M. Hammerum, R. L. Poulsen, and H. Westh. 1999. Vancomycin-resistant Enterococcus faecium strains with highly similar pulsed-field gel electrophoresis patterns containing similar Tn1546-like elements isolated from a hospitalized patient and pigs in Denmark. Antimicrob. Agents Chemother. 43:724-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keis, S., C. F. Bennett, V. K. Ward, and D. T. Jones. 1995. Taxonomy and phylogeny of industrial solvent-producing clostridia. Int. J. Syst. Bacteriol. 45:693-705. [DOI] [PubMed] [Google Scholar]
- 31.Keis, S., J. T. Sullivan, and D. T. Jones. 2001. Physical and genetic map of the Clostridium saccharobutylicum (formerly Clostridium acetobutylicum) NCP 262 chromosome. Microbiology 147:1909-1922. [DOI] [PubMed] [Google Scholar]
- 32.Klare, I., H. Heier, H. Claus, G. Böhme, S. Marin, G. Seltmann, R. Hakenbeck, V. Antanassova, and W. Witte. 1995. Enterococcus faecium strains with vanA-mediated high-level glycopeptide resistance isolated from animal foodstuffs and fecal samples of humans in the community. Microb. Drug Resist. 1:265-272. [DOI] [PubMed] [Google Scholar]
- 33.Klare, I., H. Heier, H. Claus, R. Reissbrodt, and W. Witte. 1995. vanA-mediated high-level glycopeptide resistance in Enterococcus faecium from animal husbandry. FEMS Microbiol. Lett. 125:165-171. [DOI] [PubMed] [Google Scholar]
- 34.Leclercq, R., E. Derlot, J. Duval, and P. Courvalin. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319:157-161. [DOI] [PubMed] [Google Scholar]
- 35.Leclercq, R., E. Derlot, M. Weber, J. Duval, and P. Courvalin. 1989. Transferable vancomycin and teicoplanin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 33:10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 37.Sahm, D. F., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schouten, M. A., J. A. Hoogkamp-Korstanje, J. F. Meis, and A. Voss. 2000. Prevalence of vancomycin-resistant enterococci in Europe. Eur. J. Clin. Microbiol. Infect. Dis. 19:816-822. [DOI] [PubMed] [Google Scholar]
- 39.Schouten, M. A., R. J. Willems, W. A. Kraak, J. Top, J. A. Hoogkamp-Korstanje, and A. Voss. 2001. Molecular analysis of Tn1546-like elements in vancomycin-resistant enterococci isolated from patients in Europe shows geographic transposon type clustering. Antimicrob. Agents Chemother. 45:986-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh, K. V., T. M. Coque, G. M. Weinstock, and B. E. Murray. 1998. In vivo testing of an Enterococcus faecalis efaA mutant and use of efaA homologs for species identification. FEMS Immunol. Med. Microbiol. 21:323-331. [DOI] [PubMed] [Google Scholar]
- 41.Teixeira, L. M., R. R. Facklam, A. G. Steigerwalt, N. E. Pigott, V. L. Merquior, and D. J. Brenner. 1995. Correlation between phenotypic characteristics and DNA relatedness within Enterococcus faecium strains. J. Clin. Microbiol. 33:1520-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uttley, A. H., C. H. Collins, J. Naidoo, and R. C. George. 1988. Vancomycin-resistant enterococci. Lancet i:57-58. [DOI] [PubMed]
- 44.Uttley, A. H., R. C. George, J. Naidoo, N. Woodford, A. P. Johnson, C. H. Collins, D. Morrison, A. J. Gilfillan, L. E. Fitch, and J. Heptonstall. 1989. High-level vancomycin-resistant enterococci causing hospital infections. Epidemiol. Infect. 103:173-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van den Bogaard, A. E., L. B. Jensen, and E. E. Stobberingh. 1997. Vancomycin-resistant enterococci in turkeys and farmers. N. Engl. J. Med. 337:1558-1559. [DOI] [PubMed] [Google Scholar]
- 46.van den Braak, N., A. van Belkum, M. van Keulen, J. Vliegenthart, H. A. Verbrugh, and H. P. Endtz. 1998. Molecular characterization of vancomycin-resistant enterococci from hospitalized patients and poultry products in The Netherlands. J. Clin. Microbiol. 36:1927-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Werner, G., I. Klare, and W. Witte. 1997. Arrangement of the vanA gene cluster in enterococci of different ecological origin. FEMS Microbiol. Lett. 155:55-61. [DOI] [PubMed] [Google Scholar]
- 48.Willems, R. J., J. Top, N. van den Braak, A. van Belkum, D. J. Mevius, G. Hendriks, M. van Santen-Verheuvel, and J. D. van Embden. 1999. Molecular diversity and evolutionary relationships of Tn1546-like elements in enterococci from humans and animals. Antimicrob. Agents Chemother. 43:483-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodford, N., A. M. Adebiyi, M. F. Palepou, and B. D. Cookson. 1998. Diversity of VanA glycopeptide resistance elements in enterococci from humans and nonhuman sources. Antimicrob. Agents Chemother. 42:502-508. [DOI] [PMC free article] [PubMed] [Google Scholar]