Abstract
Aberrant retinoid signaling in human cancers is extending from the nucleus to the cytoplasm. Recently, we have demonstrated frequent epigenetic inactivation of a retinoic acid receptor (RAR), RARβ2, in nasopharyngeal carcinoma (NPC). To further explore targets contributing to aberrant retinoid signaling in NPC, the expression of cellular retinol-binding proteins (CRBPs), cellular retinoic acid-binding proteins (CRABPs), RARs, and retinoid X receptors (RXRs) was examined. Apart from RARβ2, transcriptional silencing of two CRBPs, CRBPI and CRBPIV, was observed in NPC cell lines and xenografts. Hypermethylation of CRBPI and CRBPIV CpG islands was found to be closely correlated with the loss of expression. Treatment with the DNA methyltransferase inhibitor, 5-aza-2′-deoxycytidine, resulted in reexpression of CRBPI and CRBPIV gene expression in NPC cell lines. Both CRBPI and CRBPIV hypermethylations were also observed in 43/48 (87.8%) and 26/48 (54.2%) primary NPC tumors, respectively. Here, we reported for the first time that CRBPIV was transcriptionally inactivated by promoter hypermethylation in human cancer. Simultaneous methylation of CRBPI, CRBPIV, and RARβ2 was commonly found in NPC primary tumors. Our findings implied that epigenetic disruption of the CRBPs, CRBPI and CRBPIV, is important in NPC tumorigenesis and may contribute to the loss of retinoic acid responsiveness in cancer.
Keywords: CRBPIV, CRBPI, retinoid, hypermethylation, nasopharyngeal carcinoma
Introduction
Retinol, or vitamin A, is indispensable for embryonic development, growth, vision, and survival of vertebrates [1]. The vitamin A metabolite, retinoic acid, regulates multiple biologic processes, including cell proliferation and differentiation, by modulating the rate of transcription of numerous target genes. The effects of retinoic acid are mediated by at least two types of proteins: cellular retinoid-binding proteins and nuclear retinoid receptors. The cellular retinoid-binding proteins, including CRBPs and cellular retinoic acid-binding proteins (CRABPs), belong to a family of cytosolic proteins binding small hydrophobic ligands [2]. CRBPs are proposed to facilitate the formation of retinol esters for storage or conversation of retinol to retinoic acid through a retinal intermediate. These proteins may also participate in the entry of retinol into the cell [3]. The function of CRABPs is to protect retinoids in vivo from other cellular proteins, transform bound retinoid into specific biologic compounds, and modulate the concentration of free retinoic acid available to nuclear retinoid receptors [4]. Retinoid receptors function as ligand-inducible transcription factors. Two subtypes of retinoid receptor exist, namely, retinoic acid receptors (RARs) and retinoid X receptors (RXRs). RARs are activated by all-trans-retinoic acid, and 9-cis-retinoic acid activates both RARs and RXRs. RARs form heterodimers with RXRs, which regulate transcription by binding at the retinoic acid response elements (RAREs) on the promoter regions of various target genes [5].
Aberrant retinoid signaling in cancer has been firstly highlighted by the leukemogenic role of the promyelocytic leukemia-RARα fusion protein [6]. Frequent reduced expression and aberrant promoter methylation of RARβ2 in solid cancers have also been reported [7–10]. Aside from retinoid receptors, transcriptional silencing of a cellular retinoid-binding protein, CRBPI, by promoter hypermethylation is common in human cancers [11]. In NPC, only mild growth inhibition was observed when treating with 13-cis retinoic acid, although retinoid was used in chemotherapy and chemoprevention of head and neck cancer [12]. These data implied that defects in retinoid signaling may occur in NPC. Our previous study has demonstrated that transcription silencing of RARβ2 by promoter hypermethylation is common in both NPC cell lines and primary tumors [13]. To explore targets contributing to aberrant retinoid signaling in NPC, we investigated the expression of retinoid signaling molecules (CRBPs, CRABPs, RARs, and RXRs) comprehensively. Besides RARβ2, we have firstly discovered the silencing of two cellular retinol-binding proteins (CRBPs), CRBPI and CRBPIV, by promoter hypermethylation in NPC cells. Our findings demonstrated that hypermethylation of CRBPs is common in NPC and may contribute to the disruption of retinoid signaling in this cancer.
Materials and Methods
Cell Lines, Xenografts, and Primary Tumors
Five NPC cell lines (C666-1, CNE1, CNE2, HK1, and HONE1), three xenografts (X2117, X666, and XNPC8), an immortalized nasopharyngeal epithelial cell line (NP69), and two samples of normal nasopharyngeal epithelial outgrowth cultures (NP1 and NP2) were included in our study [14,15]. The NPC cell lines were maintained in RPMI 1640 medium supplemented with 10% FBS, whereas NP69 was grown in keratinocyte serum-free medium (Invitrogen, Carlsbad, CA). Forty-nine cases of archived paraffin-embedded primary NPC tumors were retrieved from the pathology bank of the Department of Anatomical and Cellular Pathology at The Chinese University of Hong Kong (Hong Kong, SAR, China). The neoplastic cells were isolated from each tumor sample by microdissection and subjected to DNA extraction as described previously [16]. The histologic diagnoses of the specimens were confirmed by a pathologist. All of these tumors were classified as undifferentiated carcinomas. The male-to-female ratio of patients was 4.8:1. The age range was 36 to 68 years (mean 52 years). The clinical disease stage of patients is from stages II to IV (UICC staging classification).
Reverse Transcription Polymerase Chain Reaction (RT-PCR)
The mRNA expression of CRBP (CRBPI, CRBPII, CRBPIII, and CRBPIV), CRABP (CRABPI and CRABPII), RAR (RARα, RARβ2, and RARγ), and RXR (RXRα, RXRβ, and RXRγ) genes in NPC cell lines, xenografts, and pool samples from normal nasopharyngeal epithelia (normal NP) was examined by RT-PCR analysis. Total RNA of nasopharyngeal samples was isolated using TRIZOL reagent (Invitrogen) according to the manufacturer's protocol. Two micrograms of total RNA from each sample was subjected to cDNA synthesis using Superscript II reverse transcriptase (Invitrogen). The primers and conditions for RT-PCR analysis were listed in Table 1. The transcript of GAPDH was amplified as control.
Table 1.
PCR Primer Sequences and Conditions for RT-PCR and Real-Time Quantitative RT-PCR.
| Genes | Sequences | Product Size (bp) | Annealing (°C) | Cycles |
| RT-PCR | ||||
| CRBPI | Forward: 5′-AAACTGGCTCCAGTCACTCC-3′ | 610 | 65 | 35 |
| Reverse: 5′-CAGGTCACTTTATTGGCATGG-3′ | ||||
| CRBPII | Forward: 5′-CAGAATGGAACCTGGGAGAT-3′ | 352 | 60 | 35 |
| Reverse: 5′-AGGTCAGCTCCAGGTACAGC-3′ | ||||
| CRBPIII | Forward: 5′-CGGAGAGAAGCCAAGATCCC-3′ | 239 | 67 | 35 |
| Reverse: 5′-TGGGACGCCAGACCTTGACC-3′ | ||||
| CRBPIV | Forward: 5′-TCCACATCCAGCAGCAGAGCC-3′ | 182 | 67 | 35 |
| Reverse: 5′-GGACAGGTTTATTGAAGCTGAGC-3′ | ||||
| CRABPI | Forward: 5′-CCTTGCGAGCTCAGAGTGT-3′ | 721 | 62 | 35 |
| Reverse: 5′-TTTGAGACACGTCTAACCAGTTT-3′ | ||||
| CRABPII | Forward: 5′-CCAACTTCTCTGGCAACTGG-3′ | 400 | 62 | 35 |
| Reverse: 5′-TAGACCCTGGTGCACACAAC-3′ | ||||
| RARα | Forward: 5′-GTCTTTGCCTTCGCCAACCAG-3′ | 333 | 60 | 35 |
| Reverse: 5′-GCCCTCTGAGTTCTCCAACA-3′ | ||||
| RARβ2 | Forward: 5′-TGCAAGGGAGATCATGTTTG-3′ | 1360 | 60 | 35 |
| Reverse: 5′-TTATTGCACGAGTGGTGACTG-3′ | ||||
| RARγ | Forward: 5′-TTCGAGATGCTGAGCCCTAGCTTCC-3′ | 351 | 62 | 35 |
| Reverse: 5′-CATGCCCACTTCAAAGCACTTCTGC-3′ | ||||
| RXRα | Forward: 5′-TTCTCCACCCAGGTGAACTC-3′ | 805 | 62 | 35 |
| Reverse: 5′-GGGTGAAAAGCTGTTTGTCG-3′ | ||||
| RXRβ | Forward: 5′-CTTTCTCTCAGGGGCTTCCT-3′ | 521 | 62 | 35 |
| Reverse: 5′-ACCCCATGGAAGAACTGATG-3′ | ||||
| RXRγ | Forward: 5′-CAGGAAAGCACTACGGGGTA-3′ | 798 | 62 | 35 |
| Reverse: 5′-GCTGTTCCGGATACTTCTGC-3′ | ||||
| GAPDH | Forward: 5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′ | 983 | 65 | 35 |
| Reverse: 5′-CATGTGGGCCATGAGGTCCACCAC-3′ | ||||
| Real-time QRT-PCR | ||||
| CRBPI | Forward: 5′-GGGAAGGAGTTTGAGGAGGA-3′ | 105 | 60 | |
| Reverse: 5′-TTCTCACCCTTCTGCACACA-3′ | ||||
| CRBPIV | Forward: 5′-TCCACATCCAGCAGCAGAGCC-3′ | 182 | 65 | |
| Reverse: 5′-GGACAGGTTTATTGAAGCTGAGC-3′ | ||||
| CRABPI | Forward: 5′-AGGTCGGAGAAGGCTTTGAG-3′ | 104 | 60 | |
| Reverse: 5′-AGAAGAGTTTGCGTGCAGTG-3′ | ||||
| RARβ2 | Forward: 5′-CAGGAGAAAGCTCTCAAAGCA-3′ | 117 | 60 | |
| Reverse: 5′-CTTGGGACGAGTTCCTCAGA-3′ | ||||
| β-Actin | Forward: 5′-CGCGAGAAGATGACCCAGAT-3′ | 98 | 60 | |
| Reverse: 5′-GTACGGCCAGAGGCGTACAG-3′ | ||||
Real-Time RT-PCR
The expression of CRBPI, CRBPIV, and RARβ2 was quantitatively examined in nasopharyngeal epithelial cells by real-time RT-PCR analysis. By using the SYBR Green RT-PCR Kit (Applied Biosystems, Foster City, CA), PCR products were stained with SYBR Green and analyzed using an I-cycler (Bio-Rad, Hercules, CA). The PCR primers and conditions were listed in Table 1. All reactions were performed in triplicate. Relative mRNA of the target gene of each NPC line was normalized with β-actin and calculated using the 2[-ΔΔC(T)] method, which compares the mRNA amount of each sample to that of normal NP.
Bisulfite Sequencing
To investigate the de novo methylation pattern of the 5′ CpG island of CRBPI and CRBPIV in NPC, five NPC cell lines, three xenografts, and two normal nasopharyngeal epithelial cell growths (NP1 and NP2) were subjected to bisulfite sequencing. Genomic DNA were modified by bisulfite treatment using the CpGenome DNA Modification Kit (Intergen, New York, NY). For the CRBPI and CRBPIV genes, the DNA sequence of the putative promoter and exon 1 were obtained from the UCSC Genome Browser, and the CpG islands of these genes were then identified. The criteria of a 5′ CpG island are: GC content > 60%, ratio of CpG to GpC > 0.6, and minimum length of 200 bp. The PCR primers are listed in Figure 2. For bisulfite sequencing, PCR amplification was performed on 100 ng of bisulfite-modified genomic DNA. The amplified fragments were cloned, and 8 to 10 clones of each sample were sequenced with a Big Dye Terminator Reaction Mix and analyzed by a ABI 3100 Genetic Analyzer (Applied Biosystems).
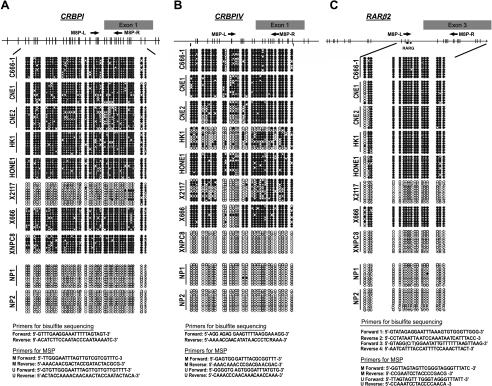
Figure 2.
Bisulfite sequencing of the CRBPI, CRBPIV, and RARβ2 5′ CpG islands in NPC cell lines, xenografts, and normal nasopharyngeal epithelial outgrowths (NP1 and NP2). Each row represents an individual subclone. Open circles, unmethylated CpG sites; closed circles, methylated CpG sites. The nucleotide position of CpG sites (grey bar) is indicated at the top. The primer sequences of bisulfite sequencing and MSP analysis are listed at the bottom. RARE, retinoic acid responsive element.
Methylation-Specific PCR (MSP)
The promoter methylation status of CRBPI, CRBPIV, and RARβ2 in the primary tumors was investigated by MSP as described previously [17]. Genomic DNA of the microdissected specimens were modified by bisulfite treatment. Primer sequences for both methylated and unmethylated alleles of the genes were shown in Figure 2. One microliter of bisulfite-modified DNA from the samples was subjected to PCR amplification. In vitro methylated DNA (IVD) (Intergen) served as a control for methylated sequences, whereas DNA from peripheral blood lymphocytes (PBLs) served as a control for unmethylated sequences. All MSP reactions were duplicated. Twenty-five microliters of PCR products was loaded onto a 10% nondenaturing polyacrylamide gel, stained with ethidium bromide, and visualized under UV illumination.
5-Aza-2′-deoxycytidine Treatment
To examine the correlation of promoter hypermethylation and expression of RARβ2, CRBPI, and CRBPIV genes, NPC cell lines were subject to 5-aza-2′-deoxycytidine treatment. Cells were plated and incubated for 4 days with 0, 1, 3, and 5µM 5-aza-2′-deoxycytidine (Sigma Chemical Co., St. Louis, MO). The medium and the drug were replaced every 24 hours, and cells were harvested for total RNA and DNA extraction after 4 days.
Results
Aberrant Expression of CRBPI, CRBPIV, CRABPI, and RARβ2 in NPC
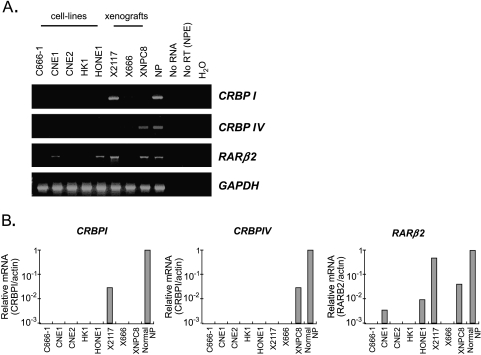
In this study, we have comprehensively examined the transcription of the CRBP (CRBPI, CRBPII, CRBPIII, and CRBPIV), CRABP (CRABPI and CRABPII), RAR (RARα, RARβ2, and RARγ) and RXR (RXRα, RXRβ, and RXRγ) genes in NPC cell lines, xenografts, and pool samples from normal nasopharyngeal epithelia (normal NP) by RT-PCR analysis. Among the 12 genes examined, CRBPII and CRABPI showed no expression in normal nasopharyngeal epithelial cells. The finding suggested that these two genes are not involved in the retinoid signaling in this cell type. We have also found the expression of seven genes (CRBPIII, CRABPII, RARα, RARγ, RXRα, RXRβ, and RXRγ) in all NPC samples as well as normal NP. As shown in Figure 1A, only the expression of three retinoid signaling molecules, including RARβ2, CRBPI, and CRBPIV, was lost in most of the NPC cell lines and xenografts. Consistent with our previous report, expression of RARβ2 was lost in 60% of NPC cell lines (3/5; C666-1, CNE2, and HK1) and an NPC xenograft (X666). The complete loss of RARβ2 expression in these NPC samples has also been confirmed by real-time RT-PCR analysis. The quantitative analysis has further demonstrated a dramatic downregulation of RARβ2 expression in the other two NPC cell lines, CNE1 and HONE1 (Figure 1B).
Figure 1.
Detection of CRBPI, CRBPIV, and RARβ2 expression in NPC cell lines, xenografts, and normal nasopharyngeal epithelia (NP) by (A) RT-PCR analysis and (B) real-time RT-PCR analysis.
The CRBPI transcripts were found to be lost in 5/5 (100%) NPC cell lines (C666-1, CNE1, CNE2, HK1, and HONE1) and 2/3 xenografts (2/3; X666 and XNPC8) by both conventional and quantitative RT-PCR analysis. Downregulation of CRBPI transcript (35-fold) was also detected in the xenograft X2117 (Figure 1B). For the CRBPIV gene, no expression has also been detected in all NPC cell lines and 2/3 xenografts (X2117 and X666), whereas downregulation of the gene was observed in the xenograft XNPC8 (Figure 1B). Our results demonstrated that loss of CRBPI and CRBPIV expression was frequently detected in NPC cell lines and xenografts.
Methylation Status of CRBPI and CRBPIV Promoters in NPC Cell Lines and Xenografts
Aberrant promoter hypermethylation is closely related to the loss of gene expression of cancer-related genes in human cancer. To assess whether the loss of CRBPI and CRBPIV expression in NPC was correlated with promoter hypermethylation, the methylation status of the 5′ CpG island of the genes was determined by bisulfite genomic sequencing.
For CRBPI gene, a 414-bp fragment from the 5′ CpG island was amplified and a total of 38 CpG dinucleotides was investigated (Figure 2A). Dense methylation was observed in seven NPC samples (C666-1, CNE1, CNE2, HK1, HONE1, X666, and XNPC8) without CRBPI expression. Partial methylation was found in a xenograft (X2117) that showed downregulation of CRBPI expression. No aberrant methylation was found in the two normal nasopharyngeal epithelial samples (NP1 and NP2). For CRBPIV, 29 CpG dinucleotides within its 5′-promoter region were investigated (Figure 2B). The 5′ CpG island of CRBPIV, especially the CpG sites within exon 1, exclusively methylated in all seven NPC lines (C666-1, CNE1, CNE2, HK1, HONE1, X2117, and X666) incapable of expressing CRBPIV. No methylation was observed in CRBPIV-expressing NPC xenograft (XNPC8) and normal nasopharyngeal epithelial cells (NP1 and NP2). The results suggest a strong correlation of transcriptional silencing of CRBPI and CRBPIV with dense methylation of their promoters.
In addition to CRBPI and CRBPIV, we also examined the methylation pattern of RARβ2 promoter as described previously [9,10,13]. Dense methylation of RARβ2 promoter was found in five NPC cell lines (C666-1, CNE1, CNE2, HK1, and HONE1) and an NPC xenograft (X666), which showed either absent or dramatic reduction in RARβ2 expression. However, no methylation was found in XNPC8, although it showed a reduced expression of RARβ2. Methylated sequences were rarely found in RARβ2-expressing samples, including an NPC xenograft (X2117) and normal nasopharyngeal epithelial outgrowths (NP1 and NP2). The findings confirmed that promoter hypermethylation is the major mechanism for RARβ2 inactivation in NPC [13].
Reactivating CRBPI and CRBPIV Expression after 5-Aza-2′-deoxycytidine Treatment
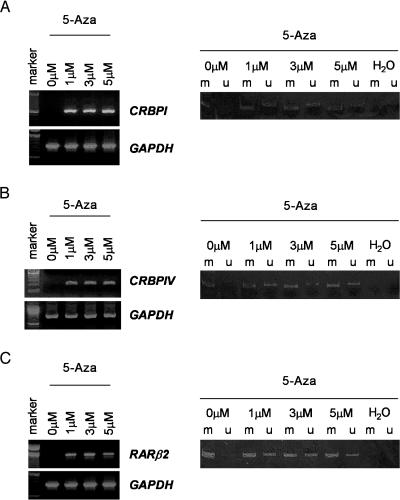
Because promoter methylation status is intimately associated with CRBPI and CRBPIV, we sought to ask whether demethylation of the CRBPI and CRBPIV promoters is required for restoration of their expression. For this purpose, we treated the NPC cell line HK1, which showed transcriptional silencing and promoter hypermethylation of CRBPI and RARβ2 with the demethylation agent, 5-aza-2′-deoxycytidine. After treatment, unmethylated alleles of CRBPI and RARβ2 were detected by MSP analysis, whereas reexpression of these genes was observed. (Figure 3, A and C). Reexpression and demethylation of the CRBPIV gene were also detected in the 5-aza-2′-deoxycytidine-treated NPC cell line C666-1 in which dense methylation and transcriptional inactivation of CRBPIV were found (Figure 3B).
Figure 3.
(A) Reactivation of CRBPI expression in NPC cell line (HK1) treated with 5-aza-2′-deoxycytidine. Left panel: The reexpression of CRBPI in HK1 after 5-aza-2′-deoxycytidine treatment. Right panel: The methylation status of CRBPI promoter of HK1 cells after demethylation. MSP analysis demonstrated that the unmethylated sequence was detected in samples treated with 5-aza-2′-deoxycytidine. (B) Reexpression of CRBPIV and detection of unmethylated sequences in NPC cell line C666-1 treated with 5-aza-2′-deoxycytidine. (C) After 5-aza-2′-deoxycytidine treatment, RARβ2 transcripts and unmethylated allele was detected in the NPC cell line HK1.
Promoter Hypermethylation of CRBPI and CRBPIV in Primary NPC Tumors
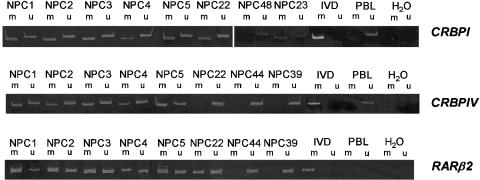
To assess the prevalence of promoter hypermethylation of CRBPI and CRBPIV in primary NPC tumors, we examined 49 primary NPC samples by MSP. The results of MSP analysis showed remarkable correlation with bisulfite genomic sequencing in cell lines and xenografts. Collectively, aberrant promoter hypermethylation of CRBPI and CRBPIV was found in 43/49 (87.8%) and 26/48 (54.2%) of primary NPCs, respectively. We have correlated the methylated status of each of the genes with the stage of the NPC patient. No significant correlation between methylation of these genes and stage was found (data not shown). Forty-six samples (93.9%) showed aberrant methylation of either CRBPI or CRBPIV. The representative examples of MSP analysis of CRBPI and CRBPIV of NPC primary tumors are shown in Figure 4. Among 49 primary tumors examined, 23 (46.9%) cases showed promoter hypermethylation of both CRBPI and CRBPIV genes (Table 2). Taken together, our results demonstrated high frequencies of CRBP promoter hypermethylation in NPC cell lines, xenografts, and primary tumors.
Figure 4.
Representative examples of MSP analysis of CRBPI, CRBPIV, and RARβ2 in primary NPC tumors. The PCR products in lane m indicate the presence of methylated templates of the genes; the PCR products in lane u show the presence of unmethylated alleles. In vitro methylated DNA (IVD) and peripheral blood lymphocytes (PBLs) served as methylated and unmethylated controls, respectively. H2O, negative control.
Table 2.
Methylation of RARβ, CRBPI, and CRBPIV in Primary Tumors of NPC.
| Promoter Methylation | Methylated Cases/Total Number | Percentage |
| CRBP1 | 43/49 | 87.8 |
| CRBPIV | 26/48 | 54.2 |
| RARβ | 37/47 | 78.7 |
| CRBPI and CRBPIV | 23/49 | 46.9 |
| RARβ and CRBPI | 34/49 | 69.4 |
| RARβ and CRBPIV | 23/49 | 46.9 |
| RARβ, CRBPI, and CRBPIV | 21/49 | 42.9 |
| CRBPI or CRBPIV | 46/49 | 93.9 |
| RARβ or CRBPI | 46/49 | 93.9 |
| RARβ or CRBPIV | 40/49 | 81.9 |
| RARβ or CRBPI or CRBPIV | 47/49 | 95.9 |
To elucidate the relationship of promoter hypermethylation of CRBPI, CRBPIV, and RARβ2, the methylation status of RARβ2 was also determined in the 49 primary tumors by MSP. Hypermethylation of RARβ2 was detected in 37/47 (78.7%) of primary tumors. No significant correlation of RARβ2 methylation and stage of patients was found. Similar findings have also reported in our previous study [13]. Forty-seven primary tumors (95.9%) showed methylation of at least one of the CRBPI, CRBPIV, and RARβ2 genes (Table 2). Promoter methylation of all three genes was found in 1/3 xenografts, 5/5 cell lines, and 21/49 (42.9%) primary tumors. The findings showed that the concurrent simultaneous inactivation of the CRBPI, CRBPIV, and RARβ2 genes by hypermethylation is relatively common in NPC.
Discussion
In the present study, we have screened for aberrant expression of the components of the retinoic acid pathway (cellular retinoid-binding proteins and nuclear retinoid receptors) in NPC. In addition to the well-studied RARβ2 gene, two retinoid signaling components, CRBPI and CRBPIV, were found to be common targets for epigenetic inactivation. A high frequency of CRBPI and CRBPIV promoter hypermethylation (88% and 54%, respectively) was detected in primary NPC tumors. The findings suggest that loss of CRBPI and CRBPIV expression is important in NPC development. Inactivation of these CRBP genes may result in the loss of cellular retinoic acid homeostasis, inability to uptake natural retinol, and synthesis of retinoic acid.
CRBPI is the most well-known retinoid-binding protein, and it is postulated to regulate the uptake and intracellular fate of retinol. This protein draws retinol from the bloodstream into cells, solubilizes retinol and retinal, and protects cells from membranolytic retinoid action [18]. Thus, loss of CRBPI would be expected to compromise retinol uptake, providing a growth advantage to cancer cells. Additionally, the absence of CRBPI may also compromise retinoic acid synthesis through the conversion of retinol to retinyl ester or retinoic acid [18,19]. This conversion includes several enzymatic reactions that probably function through recognition of retinoid-binding proteins [20]. In breast cancer, loss of CRBPI expression was as frequent in ductal carcinoma in situ as in invasive lesion, suggesting that it is a relatively early event in carcinogenesis [21]. A functional study has shown that downregulation of CRBP blocks differentiation and promotes the growth of SV40-transformed breast epithelial cells [22]. Recently, Esteller et al. [11] have also demonstrated that CRBPI was commonly silenced by promoter hypermethylation in various human malignancies, including lymphoma (60%), colon cancer (57%), gastric cancer (42%), liver cancer (30%), and breast cancer (19%). The extremely high incidence of CRBPI methylation (87.8%) suggested that the gene may play a role in NPC tumorigenesis though the disruption of the retinoid signaling pathway.
CRBPIV is newly identified and belongs to a clearly distinct CRBP superfamily with a relatively different mode of retinol-binding activity [23]. In this study, we have provided the first evidence that transcription silencing of CRBPIV by aberrant methylation is involved in the tumorigenesis of human cancers. The CRBPIV gene was not only frequently inactivated in NPC, our preliminary study also detected CRBPIV promoter hypermethylation in multiple human cancer cell lines, including colon cancer (2/5), prostatic cancer (2/5), and ovarian cancer (4/13) (personal observation). The findings implied that CRBPIV is a common target for the disruption of retinoid signaling pathway in human cancers. However, the exact function of this retinol-binding protein is still not known yet. To elucidate the function of CRBPIV in NPC tumorigenesis, depletion of CRBPIV expression in an immortalized nasopharyngeal cell line NP69 by siRNA is ongoing.
In this study, we have further confirmed a high frequency of RARβ2 methylation in primary NPC tumors (78.7%). Our data also demonstrated the relatively common simultaneous inactivation of CRBPI, CRBPIV, and RARβ2 in NPC cell lines, xenografts, and primary tumors. Although the underlying biologic significance of this observation is largely unknown, the findings may indicate that inactivation of all these genes—CRBPI, CRBPIV, and RARβ2—is necessary to completely abolish the retinoid signaling pathway in NPC. Further functional studies to investigate their roles in retinoid signaling in nasopharyngeal epithelial cells are needed. A recent study has also reported the relatively common simultaneous epigenetic inactivation of CRBPI and RARβ2 in human cancers [11]. The authors proposed that CRBPI may have a function independent of its retinol-binding ability. Nevertheless, these findings indicated that inactivation of the CRBPs, CRBPI and CRBPIV, as well as loss of RARβ2, are important in the development of human cancers.
Taken together, in this study, CRBPI, CRBPIV, and RARβ2 gene silencing by promoter hypermethylation was frequently observed in NPC. The disruption of these genes in the majority of NPC samples may explain the resistance of retinoic acid treatment in this cancer type. Besides, RARβ2, CRBPI, and CRBPIV are novel candidate targets involving in aberrant retinoid signaling in human cancers. Our findings first demonstrated that epigenetic inactivation of multiple CRBPs is involved in the tumorigenesis of NPC.
Acknowledgements
This work was carried out within the Hong Kong Cancer Genetics Research Group and was supported by the Kadoorie Charitable Foundations and the Hong Kong Research Grant Council (CUHK 4067/02M, 4071/02M, 410/03M, and HKUST 2/03C).
Abbreviations
- CRBP
cellular retinol-binding protein
- CRABP
cellular retinoic acid-binding protein
- RAR
retinoic acid receptor
- RXR
retinoid X receptor
References
- 1.Blomhoff R, Green MH, Green JB, Berg T, Norum KR. Vitamin A metabolism: new perspectives on absorption, transport, and storage. Physiol Rev. 1991;71:951–990. doi: 10.1152/physrev.1991.71.4.951. [DOI] [PubMed] [Google Scholar]
- 2.Newcomer ME. Retinoid binding proteins: structural determinants important for function. FASEB J. 1995;9:229–239. doi: 10.1096/fasebj.9.2.7781925. [DOI] [PubMed] [Google Scholar]
- 3.Napoli JL. Retinoic acid biosynthesis and metabolism. FASEB J. 1996;10:993–1001. doi: 10.1096/fasebj.10.9.8801182. [DOI] [PubMed] [Google Scholar]
- 4.Fiorella PD, Giguère V, Napoli JL. Expression of cellular retinoic acid-binding protein (type II) in Escherichia coli: characterization and comparison to cellular retinoic acid-binding protein (type I) J Biol Chem. 1993;268:21545–21552. [PubMed] [Google Scholar]
- 5.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 6.Zelent A. Translocation of the RAR alpha locus to the PML or PLZF gene in acute promyelocytic leukaemia. Br J Haematol. 1994;86:451–460. doi: 10.1111/j.1365-2141.1994.tb04773.x. [DOI] [PubMed] [Google Scholar]
- 7.Jing Y, Bleiweiss IJ, Waxman S, Zelent A, Mira-y-Lopez R. Defective expression of cellular retinol-binding protein type I and retinoic acid receptors α2, β2, and γ2 in human breast cancer cells. FASEB J. 1996;10:1064–1070. doi: 10.1096/fasebj.10.9.8801168. [DOI] [PubMed] [Google Scholar]
- 8.Xu XC, Sozzi G, Lee JS, Lee JJ, Pastorino U, Pilotti S, Kurie JM, Hong WK, Lotan R. Suppression of retinoic acid receptor β in non-small cell lung cancer in vivo: implication for lung cancer development. J Natl Cancer Inst. 1997;89:624–629. doi: 10.1093/jnci/89.9.624. [DOI] [PubMed] [Google Scholar]
- 9.Widschwendter M, Berger J, Hermann M, Müller HM, Amberger A, Zeschnigk M, Widschwendter A, Abendstein B, Zeimet AG, Daxenbichler G, Marth C. Methylation and silencing of the retinoic acid receptor-β2 gene in breast cancer. J Natl Cancer Inst. 2000;92:826–832. doi: 10.1093/jnci/92.10.826. [DOI] [PubMed] [Google Scholar]
- 10.Sirchia SM, Ferguson AT, Sironi E, Subramanyan S, Orlandi R, Sukumar S, Sacchi N. Evidence of epigenetic changes affecting the chromatin state of the retinoic acid receptor β2 promoter in breast cancer cells. Oncogene. 2000;19:1556–1563. doi: 10.1038/sj.onc.1203456. [DOI] [PubMed] [Google Scholar]
- 11.Esteller M, Guo M, Moreno V, Peinado MA, Capella G, Galm O, Baylin SB, Herman JG. Hypermethylation-associated inactivation of the cellular retinol-binding protein 1 gene in human cancer. Cancer Res. 2002;62:5902–5905. [PubMed] [Google Scholar]
- 12.Lam PK, To EWH, Chan ESY, Liew CT, Lung IWH, King WK. In vitro inhibition of head and neck cancer-cell growth by human recombinant interferon-α and 13-cis retinoic acid. Br J Biomed Sci. 2001;58:226–229. [PubMed] [Google Scholar]
- 13.Kwong J, Lo KW, To KF, Teo PM, Johnson PJ, Huang DP. Promoter hypermethylation of multiple genes in nasopharyngeal carcinoma. Clin Cancer Res. 2002;8:131–137. [PubMed] [Google Scholar]
- 14.Lo KW, Kwong J, Hui AB, Chan SY, To KF, Chan AS, Chow LS, Teo PM, Johnson PJ, Huang DP. High frequency of promoter hypermethylation of RASSF1A in nasopharyngeal carcinoma. Cancer Res. 2001;61:3877–3881. [PubMed] [Google Scholar]
- 15.Tsao SW, Wang X, Liu Y, Cheung YC, Feng H, Zheng Z, Wong N, Yuen PW, Lo AK, Wong YC, Huang DP. Establishment of two immortalized nasopharyngeal epithelial cell lines using SV40 large T and HPV16E6/E7 viral oncogenes. Biochim Biophys Acta. 2002;1590:150–158. doi: 10.1016/s0167-4889(02)00208-2. [DOI] [PubMed] [Google Scholar]
- 16.Lo KW, Teo PM, Hui AB, To KF, Tsang YS, Chan SY, Mak KF, Lee JC, Huang DP. High resolution allelotype of microdissected primary nasopharyngeal carcinoma. Cancer Res. 2000;60:3348–3353. [PubMed] [Google Scholar]
- 17.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR. A novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noy N. Retinoid-binding proteins: mediators of retinoid action. Biochem J. 2000;348:481–495. [PMC free article] [PubMed] [Google Scholar]
- 19.Ross AC. Overview of retinoid metabolism. J Nutr. 1993;123:346–350. doi: 10.1093/jn/123.suppl_2.346. [DOI] [PubMed] [Google Scholar]
- 20.Napoli JL. Biosynthesis and metabolism of retinoic acid: roles of CRBP and CRABP in retinoic acid: roles of CRBP and CRABP in retinoic acid homeostasis. J Nutr. 1993;123:362–366. doi: 10.1093/jn/123.suppl_2.362. [DOI] [PubMed] [Google Scholar]
- 21.Kuppumbatti YS, Bleiweiss IJ, Mandeli JP, Waxman S, Mira-y-Lopez R. Cellular retinol-binding protein expression and breast cancer. J Natl Cancer Inst. 2000;92:475–480. doi: 10.1093/jnci/92.6.475. [DOI] [PubMed] [Google Scholar]
- 22.Kuppumbatti YS, Rexer B, Nakajo S, Nakaya K, Mira-y-Lopez R. CRBP suppresses breast cancer cell survival and anchorage-independent growth. Oncogene. 2001;20:7413–7419. doi: 10.1038/sj.onc.1204749. [DOI] [PubMed] [Google Scholar]
- 23.Fotti C, Calderone V, Ramazzina I, Zanotti G, Berni R. Ligand binding and structural analysis of a human putative cellular retinol-binding protein. J Biol Chem. 2002;277:41970–41977. doi: 10.1074/jbc.M207124200. [DOI] [PubMed] [Google Scholar]