INTRODUCTION
Classical genetic approaches to gene identification rely on disruption of a gene leading to a recognizable phenotype. This approach continues to be an extremely successful one, yielding mutations that result in overt phenotypes reflecting the function of the corresponding gene. Not all genes, however, can be uncovered by mutagenesis for two main reasons. First, many genes are functionally redundant, sharing overlapping functions with other genes that may or may not be related at the sequence level. Mutation of a functionally redundant gene is not likely to lead to an easily recognizable phenotype, because one or more other family members can provide the same function. Analysis of systematic gene knockouts has revealed that a significant percentage of yeast genes have no obvious phenotype when disrupted, despite testing under a wide range of growth conditions (Ross-MacDonald et al., 1999). Therefore, it is likely that disruption of many plant genes will not result in an easily identifiable phenotype. Second, many genes function at multiple stages of development. Mutations in these genes may lead to early lethality or may be highly pleiotropic, which can mask the role of a gene in a specific pathway.
A second common method of gene identification is based on expression patterns. In this case, genes that are expressed in spatially, temporally, or conditionally restricted patterns are isolated, traditionally via some type of differential screening approach. Recently, the differential mRNA display technique (Reuber and Ausubel, 1995) has increased the sensitivity of this approach. Additionally, recent developments with microarray and gene chip technologies allow the expression profiles of many genes to be analyzed (Richmond and Somerville, 2000). Nevertheless, these approaches are ultimately limited by the source of tissue used to isolate the RNA probe. Genes that are expressed transiently, at low levels, or in a small number of cells are unlikely to be identified.
In recent years, techniques to identify genes have been developed that utilize random integration of reporter gene constructs. This approach has been called enhancer detection in Drosophila (reviewed in Bellen, 1999) and has also proven to be an extremely powerful tool in mouse developmental biology. In this review, I describe how enhancer detection systems have been adapted to plant biology so as to add to the arsenal of gene identification techniques available to the plant biology community.
Several different types of “trapping” systems have been developed. The major difference among such systems lies in the reporter gene construct that is used. All these systems have been designed for gene identification, and the generic term “gene trap” is therefore used to refer to them collectively, regardless of the specific reporter gene construct.
BACKGROUND
A system that allows gene activity to be monitored by creating gene fusions with a reporter gene was first used in bacterial genetics >20 years ago (Casadaban and Cohen, 1979). Random insertions of a lacZ reporter gene into the Escherichia coli chromosome were generated. These created gene fusions that could be used to monitor the expression of individual genes. In this way, genes could be identified based on their pattern of expression over time or under a variety of conditions in the absence of a mutant phenotype or any sequence information. Because the sequence of the inserted DNA was known, it provided a “tag” for easy isolation of the chromosomal gene. This approach has been modified for use in a number of species and has been extensively exploited in Drosophila and mouse genetics (reviewed in Bellen, 1999).
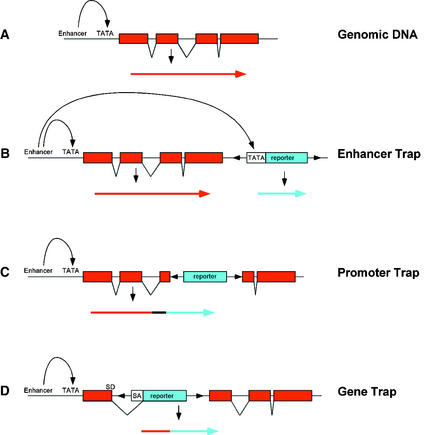
Reporter genes can be used to construct three basic types of gene trap: enhancer trap, promoter trap, and gene trap (Figure 1). Each type is able to respond to cis-acting regulatory sequences at the site of insertion. In an enhancer trap (Figure 1B), the reporter gene is fused to a minimal promoter, typically containing a TATA box and transcription start site, that is unable to drive reporter gene expression alone but can be activated by neighboring enhancer elements. Promoter traps and gene traps contain a promoterless reporter gene so that expression can occur only when the insertion is within a transcriptional unit and in the correct orientation (Figures 1C and 1D). Expression of a promoter trap reporter gene requires that it be inserted into an exon, leading to a transcriptional fusion (Figure 1C). In contrast, gene trap constructs contain one or more splice acceptor sequences preceding the reporter gene (Figure 1D), which allow expression if insertion occurs in an intron. Splicing from the splice donor sites in the chromosomal gene to the splice acceptor sites in the reporter gene results in fusion of upstream exon sequences to the reporter gene. In addition to transcriptional fusions, promoter trap and gene trap insertions can also create translational fusions, which may provide information about protein localization.
Figure 1.
Structure of Enhancer, Gene, and Promoter Trap Vectors.
(A) A generic chromosomal gene with exons (boxes) and introns (lines).
(B) Enhancer trap construct. The minimal promoter of the reporter gene (TATA) is activated by a chromosomal enhancer element, resulting in expression of the reporter gene.
(C) Promoter trap construct. The promoterless reporter gene can be expressed when insertion occurs in an exon so as to result in a transcriptional fusion.
(D) Gene trap construct. The promoterless reporter gene contains splice acceptor (SA) sequences. Expression of the reporter gene occurs upon its insertion into an intron. Splicing from the chromosomal splice donor (SD) site to the SA sequence results in creation of a transcriptional fusion.
Arrows in each panel represent the transcripts that are produced as a consequence of insertion.
Each type of reporter gene construct has its own advantages. Because enhancer traps do not have the same constraints on expression as promoter and gene traps, which must insert within a gene and in the correct orientation, enhancer trap insertions lead to a higher frequency of reporter gene expression. However, expressed promoter or gene traps are more likely to cause gene disruption than are expressed enhancer traps. Also, because enhancers can activate gene expression at considerable distances, the genes controlling reporter gene expression may be more easily identified in promoter or gene trap insertions than in enhancer trap insertions.
Why Use Gene Traps?
Gene traps provide a powerful tool for gene identification. Genes are identified based on reporter gene expression; therefore, a mutant phenotype is not required. This advantage allows identification of two classes of genes that are not easily amenable to classic genetic analysis: functionally redundant genes and genes that have functions at multiple developmental stages. The simultaneous identification of mutations in two redundant genes has in fact only rarely been attained (e.g., Wilhelmi and Preuss, 1996; Aida et al., 1997), and most mutations in redundant genes have thus gone undetected by traditional means. In addition, disruption of a gene might result in a subtle phenotype that could go undetected. However, if a gene trap insertion disrupts a gene, then the expression pattern might suggest the type of phenotype to look for and may lead to observation of phenotypes that could otherwise be overlooked.
On the other hand, a mutation in a gene that is required at multiple stages of development is likely to result in lethality, whereas pleiotropic genes may result in complex phenotypes that are difficult to approach experimentally. Gene traps allow genes to be identified based solely on expression pattern, so that loss-of-function mutations are not mandatory. Although a gene trap insertion may disrupt gene function, the disruption per se is not necessary for gene identification. Therefore, functionally redundant genes whose expression patterns meet screening criteria can be identified in the absence of an easily recognizable phenotype. Moreover, essential genes can be identified based on reporter gene expression, moreover, in viable heterozygotes, even if disruption causes lethality. Likewise, genes that have roles at multiple developmental stages can be identified based on expression, even if mutations occur within pleiotropic genes.
To use gene traps, a collection of individuals is generated that contains a reporter gene integrated randomly into the genome. Various strategies for obtaining efficient reporter gene expression have been utilized (see below). Each insertion is maintained as a separate line, which can subsequently be screened for reporter gene expression. Screens can be designed to identify genes that are expressed in specific cells or tissues, at specific developmental stages, or in response to an environmental stimulus.
Gene Traps in Flies and Mice
An enhancer trap system was first described in Drosophila 13 years ago (O'Kane and Gehring, 1987). The authors used P-element–mediated transformation to introduce the reporter lacZ gene into the genome. The lacZ gene was driven by the weak, constitutive P-element promoter, so that its expression could not be detected by staining for β-galactosidase activity. Insertions of this construct into the genome conferred many different cell- and tissue-specific lacZ expression patterns, demonstrating that cis-acting regulatory elements could be detected using this approach. Second-generation systems used transposition rather than transformation to recover fly lines containing P-element insertions and a plasmid rescue cassette for rapid isolation of flanking genomic DNA (Bellen et al., 1989; Bier et al., 1989; Grossniklaus et al., 1989; Wilson et al., 1989).
At about the same time, traps were described in mice (Allen et al., 1988; Kothary et al., 1988). Initially, both enhancer and promoter traps were tested (Gossler et al., 1989). Because enhancers can act over large distances, it has been argued that the genes associated with an enhancer might be more difficult to identify with enhancer traps than promoter or gene traps (Skarnes, 1990). This argument might especially hold for organisms with large genomes and low gene density. For this reason, work in mouse has tended to use promoter or gene trap vectors. Gene trap constructs are typically introduced into totipotent embryonic stem cells by using a retroviral vector. Embryonic stem cells that subsequently test positive for retroviral integration are subsequently transferred to the mouse germline. The gene trap vector commonly used in mouse contains a lacZ reporter gene with one or more splice acceptor (SA) sequences (reviewed in Rossant and Hopkins, 1992; Brennan and Skarnes, 1999). If insertion occurs in an intron, then splicing will create a transcriptional fusion between the adjacent exon sequence and lacZ. In practice, use of SA sequences increases the frequency of insertions that result in reporter gene expression by 10- to 100-fold (Gossler et al., 1989; Skarnes, 1993).
GENE TRAPS IN PLANTS
Summary of Plant Systems
The first-generation gene trap systems in plants were designed to determine how frequently T-DNA insertions integrated into genes. Early experiments were performed by transforming tobacco protoplasts with a T-DNA containing a promoterless antibiotic resistance gene adjacent to one border (André et al., 1986; Teeri et al., 1986). Recovery of a transformed plant relied on the generation of a gene fusion that led to expression of the antibiotic resistance gene. Therefore, this approach was limited to detection of gene fusions that were expressed in regenerating tissues. Further modifications of the experimental approach incorporated a second selectable marker in the T-DNA, so that transformed plants could be regenerated and subsequently screened for expression of the promoterless antibiotic resistance gene (Koncz et al., 1989; Herman et al., 1990). The next step was inclusion of the β-glucuronidase (gusA or uidA) reporter gene that could be easily visualized by histochemical staining (Fobert et al., 1991; Kertbundit et al., 1991; Topping et al., 1991). This advance allowed spatial and temporal expression patterns to be visualized. Constructs that contained either a promoterless gusA gene or a gusA gene driven by a minimal promoter were used. More recently, transposable elements have been used to deliver enhancer or gene traps into plant genomes (Fedoroff and Smith, 1993; Klimyuk et al., 1995; Sundaresan et al., 1995).
Choice of Insertion Vehicle
The two main alternatives for delivery of enhancer and gene trap reporters into plant genomes are T-DNAs and transposable elements. There are advantages and disadvantages to each, which are summarized below.
T-DNA
T-DNA–mediated transformation is a common way to generate transgenic plants. In plants in which transformation efficiency is reasonably high, the use of T-DNAs allows large collections of independent insertions to be quickly generated. Because T-DNAs are not known to insert with site specificity, it should be possible to saturate the genome with T-DNA insertions (Azpiroz-Leehan and Feldmann, 1997). In fact, large T-DNA collections have been generated in Arabidopsis (Feldmann and Marks, 1987; Bouchez et al., 1993; Campisi et al., 1999; Krysan et al., 1999; Weigel et al., 2000). Systematic efforts are now underway to use these collections for “reverse genetic” screens to identify insertions in any cloned gene (McKinney et al., 1995; Winkler et al., 1998; Krysan et al., 1999). The use of T-DNAs for insertional mutagenesis is limited, however, to those plant species that can be easily transformed by T-DNA.
Significantly, multiple T-DNA insertions often occur in a single plant, both in multiple copies per locus and in multiple loci (Bechtold et al., 1993; Lindsey et al., 1993). This multiplicity is a potential problem when delivering enhancer or gene trap reporter genes, because multiple insertions may complicate interpretation of expression patterns. Additionally, T-DNA insertion events can often be complex, generating T-DNA repeats in direct or inverted orientations, with occasional rearrangements of adjacent chromosomal DNA (Ohba et al., 1995; Nacry et al., 1998; Laufs et al., 1999). These complexities may result in reporter gene expression from promoters introduced along with the T-DNA rather than from chromosomal promoters. Complex insertions may also complicate subsequent molecular analyses, making it difficult to isolate the chromosomal genes driving reporter gene expression. Finally, genomic T-DNA insertions are generally stable, so that remobilization is not readily possible, as it is for transposable elements (see below).
T-DNA vectors have been used by a number of different groups to deliver gene trap constructs into plants. Enhancer, promoter, and gene trap reporter genes have been used, and expression of reporter genes has been efficient with the reporter gene positioned at either the left (Lindsey et al., 1993) or right (Campisi et al., 1999) border of the T-DNA.
Transposable Elements
Transposable elements are routinely used to perform insertional mutagenesis. In species that do not have active, well-characterized transposable element systems, heterologous elements can be utilized (reviewed in Osborne et al., 1991). In this case, the elements are introduced into the plant genome by T-DNA–mediated transformation and subsequently mobilized. Insertional mutagenesis with transposable elements offers several advantages over T-DNAs. Although integrated transposable elements in the absence of transposase (see below), like integrated T-DNAs, are stable, transposable element insertions can be selectively destabilized upon expression of transposase. In this way, remobilization can generate germinal revertants so as to verify that a phenotype is indeed caused by insertion of the transposon. Somatic revertant sectors can also be generated for clonal analysis, which is useful for studying the cell-autonomous nature of the given gene product. Finally, because some eukaryotic elements preferentially transpose to closely linked locations (Greenblatt, 1984), derivative alleles can be generated by remobilizing an element (Das and Martienssen, 1995).
The maize Ac/Ds and En/Spm transposable elements have been developed for use in heterologous species. The behavior of these elements has been extensively studied, and they have been modified for efficient transposition in plants such as tobacco, tomato, and Arabidopsis (reviewed in Osborne and Baker, 1995). To date, only the Ac/Ds system has been used for enhancer or gene trap systems, although it is possible that other elements will also prove useful for this purpose. An important consideration when choosing a transposable element system is copy number. The Ac/Ds system has been chosen due to its low copy number (Bancroft et al., 1992). Elements that have a tendency to amplify, such as En/Spm (Aarts et al., 1995), potentially complicate interpretation of reporter gene expression patterns.
The Ac/Ds transposable element system is a two-component system with autonomous (Ac) and nonautonomous (Ds) components. The Ac element encodes a transposase that binds to the terminal inverted repeat ends of both Ac and Ds elements, catalyzing their transposition to new locations in the genome. Ds elements are most often derivatives of Ac that have lost the ability to produce a transposase but retain the terminal inverted repeats. The Ac transposase, when produced in trans, is able to recognize the ends of Ds elements and catalyze their movement to new chromosomal locations. The use of a two-component system allows for stable insertions to be generated, because the autonomous element can be segregated away from the insertion. Such insertions, moreover, can subsequently be remobilized by introducing the transposase via genetic crosses. Remobilization can allow somatic and germinal revertants and derivative alleles to be isolated.
As in maize, Ac/Ds elements demonstrate a marked preference for transposition to genetically linked locations in Arabidopsis and other plants (Greenblatt, 1984; Dooner and Belachew, 1989; Jones et al., 1990; Dooner et al., 1991; Osborne et al., 1991; Bancroft and Dean, 1993; Carroll et al., 1995; Machida et al., 1997). Although this feature can be advantageous at times, it may be problematic when the goal is to generate random insertions throughout the genome. Schemes have been designed to enrich for transposition events that are not linked to the donor locus (Sundaresan et al., 1995).
Choice of Reporter Gene
β-Glucuronidase
The bacterial gusA (uidA) gene is a commonly used reporter gene in plants. The GUS protein is quite stable, and it retains activity when it is fused to other proteins (Jefferson et al., 1987). GUS activity can be detected by histochemical staining using a variety of substrates that are commercially available. Unfortunately, most of the substrates are expensive, and the histochemical stains are destructive, so that GUS assays cannot be performed on live tissue. Detection of GUS activity is very sensitive (Jefferson et al., 1987; Lindsey et al., 1993), however, and activity can even be detected in single cells. The cost of GUS substrate prohibits its use for large plants or for large-scale screens on many plants. Because Arabidopsis plants are quite small, however, individuals can be stained in small volumes of substrate solution.
Green Fluorescent Protein
The jellyfish green fluorescent protein (GFP) has recently been used as a reporter gene in plants, although modifications were required to obtain GFP proteins that fluoresce efficiently in Arabidopsis (Haseloff and Amos, 1995; Siemering et al., 1996; Haseloff et al., 1997). Because GFP is fluorescent, it can be directly detected by illumination; no substrate is required, and detection is therefore relatively inexpensive, provided the appropriate light source is available. Detection of GFP activity is also nondestructive, can be performed in live cells, and can be monitored over time. Recent modifications have improved the sensitivity of GFP (Haseloff et al., 1997), making it a useful reporter for gene trap systems.
Other Options
Other reporter genes might also be useful. Lc is a member of the maize R gene family of Myc-like transcription factors, regulates anthocyanin biosynthesis, and is particularly promising as a reporter gene. Expression of Lc in heterologous plants leads to anthocyanin accumulation (Lloyd et al., 1992; Goldsbrough et al., 1996). This system is thus advantageous because detection does not require expensive substrate and is nondestructive. This type of visual reporter is especially attractive for screens of large, field-grown plants, for which illumination for GFP detection might be impractical. However, expression of high levels of the Lc-encoded transcription factor may have phenotypic effects. For example, high levels of expression of Lc in Arabidopsis lead to an increase in the number of trichomes formed.
Transposable Elements: Tagging Strategies
When transposable elements are used for insertional mutagenesis, careful consideration should be given to the strategy used to recover new insertions. The two most common types of systems, relying either on selection for excision or on selection for transposition, are described below, using the Ac/Ds system as an example. In both approaches, Ac and Ds elements are introduced into the genome via T-DNA–mediated transformation, and single-locus T-DNA insertions are identified. Typically, the Ac and Ds elements are introduced on separate T-DNA constructs, but they can also be introduced on the same construct if a counter-selectable marker (see below) is used. A single T-DNA approach might be advantageous in plants for which crossing is laborious. Only a few transgenic plants need to be generated, because homozygous transformants provide the starting parental material. Most often, the Ac element is modified by removal of one of the terminal inverted repeat ends so that it is unable to transpose and simply provides a source of transposase. Mutagenesis is initiated by intercrossing homozygous Ac and Ds parents. During development of an F1 plant that is heterozygous for both elements, the Ds element will transpose. F1 plants are selfed to recover F2 plants in which the Ds element has transposed and segregated away from Ac. Recovery of plants that no longer have an autonomous element (Ac) allows the new Ds insertions to be stabilized. The Ac element can be either selected against or screened out (see below).
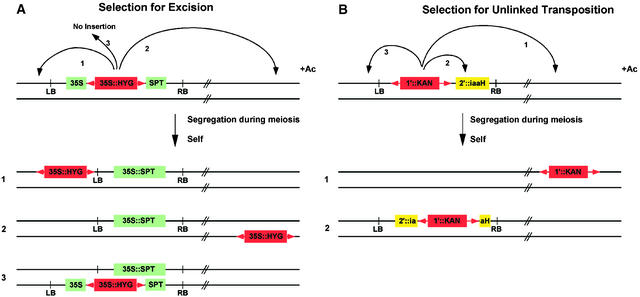
Selection for Excision
Transposon mutagenesis is initiated from a donor Ds element previously introduced into the genome by transformation. In a system that selects for excision, as shown in Figure 2A, the donor Ds element, carrying a ubiquitously expressed antibiotic resistance gene, arrives within a T-DNA construct to the plant genome by Agrobacterium-mediated transformation. Within the T-DNA, the Ds element resides in the untranslated leader of a second ubiquitously expressed antibiotic resistance gene. Transformed plants are therefore sensitive to the second antibiotic because the presence of the Ds element prevents expression of the resistance gene. Upon mobilization, the Ds element excises, resulting in expression of the second antibiotic resistance gene (Jones et al., 1989). Seedlings that are resistant to both antibiotics therefore reflect a transposition event wherein the Ds element has excised from its T-DNA location and reinserted in another chromosomal location (Figure 2A). Because excision of a Ds element is not always followed by reinsertion (Bancroft and Dean, 1993; Long et al., 1993), it is important to select for the presence of the Ds element. A host of different antibiotic resistance genes are available for use (reviewed in Osborne and Baker, 1995). Resistance genes against herbicides that can be applied to soil-grown seedlings have also been used (DeBlock et al., 1987; Tissier et al., 1999), thereby reducing the labor involved in the screening process.
Figure 2.
Transposable Element Tagging Strategies.
(A) Selection for excision. The donor Ds element, shown in red, carries an antibiotic resistance gene, 35S::HYG, and is inserted in the untranslated leader of a second antibiotic resistance gene, 35S::SPT, which resides on a T-DNA. In the presence of Ac, Ds excises, restoring the 35S::SPT gene and resulting in resistance to streptomycin. F2 plants that are resistant to streptomycin and hygromycin contain a Ds element that has excised from its original location in the T-DNA. Transposition to linked locations (class 1) or unlinked locations (class 2) is recovered. Class 3 plants, which do not contain a transposed element, are recovered when the Ds element has excised in the egg but not sperm (or vice versa).
(B) Selection for transposition. The donor Ds element, shown in red, carries an antibiotic resistance gene, 1′::KAN, and is adjacent to a counterselectable marker gene, 2′::iaaH, within a T-DNA. In the presence of Ac, the Ds element excises. F2 plants that are resistant to both kanamycin and naphthalene acetamide (NAM) carry a Ds element but not an iaaH gene. This selection enriches for recovery of unlinked transpositions. Linked transpositions (class 3) are not recovered due to counterselection against the iaaH gene. Linked transpositions that inactivate the iaaH gene (class 2) are recovered because plants are NAM resistant.
LB, left border of T-DNA; RB, right border of T-DNA.
In general, selection for excision is very effective, although not always ideal. One problem is that excision events occurring late in the development of the F1 plants (after the male and female lineages have diverged) are present in the male but not the female gametes, or vice versa. When this situation occurs, an F2 plant that is resistant to both antibiotics does not necessarily contain a transposed element (Figure 2A, class 3; Long et al., 1993). In addition, because >50% of transpositions occur to linked locations (Greenblatt, 1984; Dooner and Belachew, 1989; Jones et al., 1990; Dooner et al., 1991; Osborne et al., 1991; Bancroft and Dean, 1993; Carroll et al., 1995; Machida et al., 1997), many of the transposed elements recovered using this scheme are linked to the donor locus (Figure 2A, class 1). To obtain broad coverage throughout the genome, many transposition events must therefore be isolated.
When the goal is to target a gene that is linked to the donor locus, this system works extremely well. A number of genes have been cloned in Arabidopsis and other plants based on this “directed tagging” approach (e.g., Jones et al., 1994), and efforts are under way to systematically map donor T-DNA loci for use in directed tagging of linked genes (Smith et al., 1996; Long et al., 1997; http://www.jic.bbsrc.ac.uk/staff/caroline-dean/dslaunch.htm).
Selection for Transposition
To select for unlinked transposition events (Figure 2B), the donor Ds element, carrying a ubiquitously expressed antibiotic resistance gene, is introduced into plants via T-DNA transformation. A ubiquitously expressed counterselectable marker gene, whose presence can be selected against, is also present on the T-DNA. F2 seedlings that are resistant to both positive and negative selective agents will therefore contain a Ds element that has transposed to a new location and segregated away from the T-DNA. Using this system, linked transposition events are selected against, thereby enriching for insertions that are not linked to the donor. This scheme allows for efficient recovery of insertions throughout the genome (Sundaresan et al., 1995; Parinov et al., 1999). Counterselectable marker genes are typically conditional, acting by conversion of an innocuous compound into one that is toxic to plant cells. Several different counterselectable marker genes have been used in plants, including indoleacetamide hydrolase (iaaH; Klee et al., 1987), cytosine deaminase (codA; Stougaard, 1993), and cytochrome P450SU1 (SU1; O'Keefe et al., 1994). iaaH confers sensitivity to naphthalene acetamide (NAM); codA confers sensitivity to 5-fluorocytosine; and SU1 confers sensitivity to the sulfonylurea proherbicide R7402.
One problem with this system is that linked transpositions within the donor T-DNA may disrupt the counterselectable marker. This type of insertion leads to the recovery of double-resistant seedlings that do not have a useful transposed Ds element (Figure 2B, class 2). In practice, this occurs in 5 to 15% of double-resistant seedlings (Sundaresan et al., 1995; Parinov et al., 1999). This problem might be avoided by including two copies of a counterselectable marker within the T-DNA; however, this might also lead to cosuppression, which could inactivate the counterselectable marker.
Although Ds elements exhibit no known insertion site specificity, transposition of unlinked Ds elements does not result in an entirely random distribution. Parinov et al. (1999) have determined the map location of 356 Ds insertions. They amplified sequences flanking the insertions and determined map positions using the characterized Arabidopsis genomic sequence. Ds elements were inserted on all five Arabidopsis chromosomes, with a higher frequency of insertions found on chromosomes 2 and 4 (likely reflecting the greater amount of sequence information available for those two chromosomes at the time of the study; Lin et al., 1999; Mayer et al., 1999). However, apparent transposition “hot spots” were observed. A major bias for insertion near the nucleolus organizer regions on chromosomes 2 and 4 was seen, and several other regions also appeared to contain more insertions than would be expected for random distribution (Parinov et al., 1999). Additionally, preferential insertion near the 5′ ends of genes was observed (Parinov et al., 1999). Preferential insertion into the 5′ ends of genes has also been observed for other transposable elements, including Drosophila P elements, and may reflect the ability of elements to insert into “open” chromatin (Spradling et al., 1995).
Cold Spring Harbor Gene Trap System
A gene trap system that uses selection for transposition was developed at Cold Spring Harbor Laboratory (Sundaresan et al., 1995). This system uses the Ac/Ds transposable elements and a positive/negative selection for transposition. Both enhancer trap and gene trap reporter gene constructs have been developed. The transposase in the immobilized Ac element is driven by the cauliflower mosaic virus (CaMV) 35S promoter. The Ac is introduced into plants by T-DNA–mediated transformation, and an iaaH gene, driven by the ubiquitously expressed 2′ promoter is also present on the Ac T-DNA. The Ds elements were constructed by replacing sequences of the maize Ac element with an NPTII gene driven by the ubiquitously expressed 1′ promoter and a GUS reporter gene. The GUS gene lies at the 3′ end of the element, relative to the Ac sequence (GenBank accession number for the Ac sequence is X05424). The GUS gene in the enhancer trap element (DsE) is fused to a minimal promoter from the CaMV 35S gene. This region of the promoter has no detectable activity unless chromosomal enhancer elements are nearby (Benfey et al., 1989). The GUS gene in the gene trap element (DsG) is promoterless and contains three SA sites in each of three reading frames, fused upstream of the initial ATG codon. This construct allows GUS expression via transcriptional and translational fusion if the DsG element inserts in an intron. Additionally, naturally occurring splice donor sites in the 3′ end of the Ds element (Wessler et al., 1987; Nussaume et al., 1995) allow splicing and expression if insertion occurs in an exon. In reality, only two of the three SA sites appear to be used (Nussaume et al., 1995), but reporter gene expression in transposants occurs with reasonably high efficiency (Sundaresan et al., 1995). Each Ds element was subcloned into a binary T-DNA vector for introduction into Arabidopsis by Agrobacterium-mediated transformation. The counterselectable iaaH gene, driven by the ubiquitously expressed 2′ promoter, was included on each T-DNA, thereby allowing both the Ac and the donor Ds loci to be selected against after mobilization.
Transposition is initiated by crossing parental plants homozygous for Ac with plants homozygous for one of the Ds elements. F1 plants are allowed to self, and F2 seeds are collected. F2 seeds are germinated on media containing NAM and kanamycin. Plants that are resistant to both NAM and kanamycin contain a Ds element somewhere in the genome, but that has segregated away from the Ac element and the donor Ds T-DNA. This selection enriches for transposition events that are not linked to the original Ds location. The selection is quite robust, although the need for selection to be done in media rather than soil makes the process labor intensive. Double-resistant F2 seedlings are transferred to soil and allowed to self-pollinate, and F3 seeds are collected. Screens for GUS expression are performed on F3 plants.
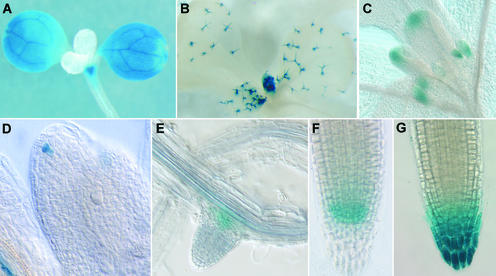
In a screen of 2000 transposants, 32% of DsG insertions and 45% of DsE insertions were found to exhibit GUS activity in seedlings (Martienssen, 1998). A variety of different expression patterns was identified, including those showing organ, tissue, or cell type specificity. A few representative expression patterns are shown in Figure 3. Surprisingly few transposants showed ubiquitous expression in all cell types (P.S. Springer, Q. Gu, D. Bush, C. Yordan, and R. Martienssen, unpublished results). This observation may indicate that even so-called housekeeping functions required in all plant cells are likely supplied by gene family members with distinct domains of expression.
Figure 3.
GUS Reporter Gene Expression Patterns in Representative Enhancer and Gene Trap Lines.
Expression is evident, as follows:
(A) In cotyledons and shoot apex but not leaves.
(B) In trichomes.
(C) In stipules and leaf tips.
(D) In a single cell at the tip of leaf primordium.
(E) In a lateral root primoridium.
(F) In a root tip.
(G) In a root cap.
Gene Traps Provide Tools for Plant Development
Gene Identification
Perhaps the most exciting use of gene traps is in the identification of genes with specific patterns of expression that are differentially regulated. Reporter gene expression that is cell or tissue specific, or temporally or conditionally regulated, may be identified. As described above, gene traps allow the identification of genes that are not amenable to classical genetic analysis. Therefore, novel genes are likely to be found in any gene trap screen. Screens have in fact been successful in identifying genes specifically expressed in lateral roots (Malamy and Benfey, 1997), developing embryos (Topping et al., 1994; Topping and Lindsey, 1997), and shoot apices (Springer et al., 1995; P.S. Springer and R. Martienssen, unpublished results). Conditional screens have also been performed to identify genes regulated by nematode infections (Barthels et al., 1997; Favery et al., 1998) and in response to abiotic stress (M. Rojas-Pierce, E. Bray, and P.S. Springer, unpublished results). To date, only a few plant genes have been cloned and characterized using gene traps. However, these cases provide examples of identification of both essential and redundant genes. Two specific examples are described below.
The PROLIFERA (PRL) gene was identified as a gene trap DsG insertion that showed GUS activity in dividing cells. PRL encodes a protein that is related to MCM7 (Springer et al., 1995), a member of the MCM gene family found in all eukaryotes and required for the initiation of DNA replication (reviewed in Kearsey and Labib, 1998). Expression in dividing cells is consistent with this predicted role in cell division. Disruption of PRL by the DsG element led to megagametophyte and embryo lethality. Arrest of both megagametophytes and embryos occurred at variable stages of development (Springer et al., 1995, 2000). There are many embryo-lethal mutations in Arabidopsis that have variable phenotypes, which makes it difficult to determine the cause of lethality (Meinke, 1991). However, the GUS expression pattern in dividing cells suggested a role in cell division before the gene was cloned.
An enhancer trap DsE insertion was identified in the AGL8 gene (Gu et al., 1998), which had previously been described as a member of a large family of MADS box genes related to AGAMOUS (Mandel and Yanofsky, 1995). The DsE insertion in the 5′ untranslated leader of AGL8 caused a loss-of-function mutation that resulted in a failure of the silique to elongate after fertilization. This failure caused the developing silique to be shortened and crowded with seeds. AGL8 was therefore renamed FRUITFULL (FUL) to reflect this phenotype (Gu et al., 1998). The defect in silique elongation in ful mutants was consistent with its expression pattern in the carpel valves. However, FUL was also expressed in the shoot apical meristem and upregulated in meristems undergoing a transition to flowering, suggesting a possible role in the reproductive transition. Indeed, ful mutants showed a slight delay in flowering time (Ferrándiz et al., 2000). However, when combined with mutations in related MADS box genes APETALA1 (AP1) and CAULIFLOWER (CAL), the ful mutation causes a complete failure to flower (Ferrándiz et al., 2000). Thus, FUL appears to act redundantly with AP1 and CAL to promote the transition to flowering (Ferrándiz et al., 2000). This function was suggested by the expression pattern and was only uncovered by analysis of the triple mutant.
Identification of Markers
An equally important use of gene traps is to identify tissue- or cell-specific expression patterns. Such expression patterns can then be used as markers to identify particular cells or tissues. Markers are useful tools for developmental analysis, although relatively few have been described in plants. Markers that are expressed in distinct patterns during development can be used to examine normal patterns of development and to characterize mutant phenotypes. Specifically, markers are useful for determining when changes in cell fate occur during morphogenesis and for following cell lineages during development.
Molecular markers are particularly effective tools for analysis of mutant phenotypes. Because developmental mutations often disrupt cellular identity, it can be difficult to determine which specific defects are caused by a gene disruption. The availability of a collection of different cell type–specific markers is very useful for determining cellular identity in mutant tissues. Markers are also useful for identifying very early alterations in developmental pattern. It is sometimes possible to trace a mutant phenotype to a defect that occurs at an early stage in development, perhaps before deviation from normal morphology or anatomy can be detected. Gene traps in plants have been used quite extensively in this capacity. Gene trap patterns have been used to study epidermal patterning (Berger et al., 1998a, 1998b), lateral root initiation (Smith and Fedoroff, 1995; Malamy and Benfey, 1997), leaf development (Tsukaya and Uchimiya, 1997), flower development (Roe et al., 1997; Liljegren et al., 2000), and embryogenesis (Topping et al., 1994; Topping and Lindsey, 1997; Willemsen et al., 1998). Therefore, even in the absence of prior molecular information, gene trap expression patterns can be enormously useful.
Promoter Identification and Ectopically Expressed Genes
In addition to gene identification, gene traps allow for the identification of promoters that drive specific expression. Once identified, specific promoters can be used to drive the ectopic expression of experimental genes to examine patterns of development. An example of this was demonstrated recently by Tsugeki and Fedoroff (1999). The authors isolated a root cap–specific promoter in an enhancer trap line. This promoter was then used to drive expression of a diphtheria toxin gene (DT-A) to examine the developmental consequences of ablating root cap cells.
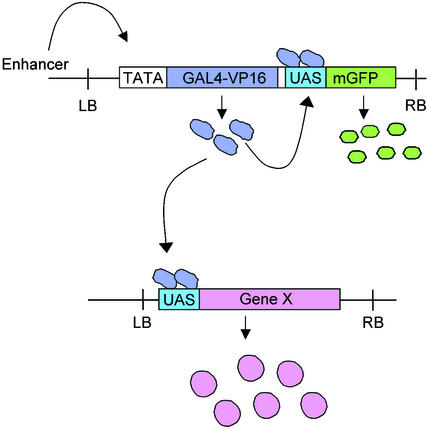
An elegant modification of this approach makes use of the yeast transcriptional activator GAL4 as a reporter gene. The GAL4 system was first used in Drosophila (Brand and Perrimon, 1993) and has been adapted by Jim Haseloff for use in Arabidopsis (http://www.plantsci.cam.ac.uk/Haseloff/IndexGAL4.html). In this system, a T-DNA containing a modified yeast GAL4 gene (GAL4-VP16) fused to the minimal CaMV 35S promoter and a modified GFP (mGFP) gene driven by the GAL4 upstream activating sequence (UAS) are transformed into Arabidopsis (Figure 4). When GAL4-VP16 is positioned under control of a chromosomal enhancer, expression can be visualized by GFP fluorescence, because GAL4 activates GFP expression from the UAS. A battery of enhancer trap lines expressing this GAL4-VP16 fusion in different patterns has been generated. This system can then be used to express a gene of interest ectopically in many different patterns upon fusion to UAS elements. Genetic crosses are used to bring the ectopic gene under control of the GAL4-VP16 activator, expressed in a particular pattern that is visualized by GFP. This system can also be used to genetically ablate specific cells or morphological regions by expressing a toxin gene in the same way.
Figure 4.
Gene Misexpression Using the GAL4 System.
A modified GAL4 gene (GAL4-VP16) fused to a minimal promoter that responds to chromosomal enhancer elements is introduced into transgenic plants on a T-DNA vector. The T-DNA also contains an mGFP gene fused to UAS sequences. GFP reports GAL4-VP16 expression, because GAL4 controls transcription of GFP through the UAS element. Misexpression of Gene X is achieved by crossing plants containing a UAS::Gene X fusion to a plant with an individual GAL4 enhancer trap insertion. GAL4 activates Gene X, causing it to be expressed in the same pattern as the GFP reporter. LB, left border of T-DNA; RB, right border of T-DNA.
Identification of Targets of Regulatory Genes
A potentially powerful use of gene traps is for the identification of downstream genes in a regulatory cascade. This approach has been used very effectively in Drosophila to identify target genes that are regulated by the Antennapedia gene (Wagner-Bernholz et al., 1991). The rationale is to identify candidate gene trap lines in which the reporter gene is expressed in an appropriate pattern that overlaps the expression domains of the regulatory gene of interest. Gene trap lines can be crossed to mutants that have lost regulatory gene function or to transgenic plants that ectopically express the regulatory gene. Expression patterns of the reporter that are altered in mutant and transgenic backgrounds can indicate genes that are potentially controlled by the regulatory gene. We have used this approach to identify potential downstream targets of the KNAT1 homeobox gene (P.S. Springer, B. Shuai, G. Chuck, S. Hake, and R. Martienssen, unpublished data).
Genomics Resources
A variety of gene trap lines exist that contain single Ds element insertions in the genome (Sundaresan et al., 1995; Martienssen, 1998; Parinov et al., 1999). To identify the gene that controls reporter gene expression, one can use various polymerase chain reaction (PCR) techniques to isolate genomic DNA flanking the insertions (Martienssen, 1998). Thermal asymmetric interlaced PCR is particularly effective for amplifying flanking DNA sequences (Liu et al., 1995; Tsugeki et al., 1996). Once a small region of flanking sequence is known, then database searches can be performed to identify corresponding genomic DNA sequences and candidate genes. Because the Arabidopsis genome will soon be completely sequenced (Meinke et al., 1998), there is a high probability that even a small region of flanking sequence is sufficient to identify a corresponding genomic sequence. Such identification immediately yields the map position of the corresponding Ds insertion. Furthermore, because the genome sequence is being systematically annotated, predicted genes within the region of the insertion can also be identified, which allows for the identification of candidate genes driving reporter gene expression. In the case of gene trap insertions (which result in transcriptional fusions), 5′ RACE (for random amplification of cDNA ends)–PCR can also be used to amplify exon sequence upstream of the insertion (Skarnes et al., 1992; Springer et al., 1995). This approach works well for genes that are relatively abundantly expressed. Because RACE-PCR yields exon sequences, candidate genes are directly identified, and cDNA libraries can be screened directly with PCR product probes.
A number of groups are systematically amplifying and sequencing genomic DNA flanking random gene trap insertions in Arabidopsis. Databases containing this sequence information have been generated (Martienssen, 1998; Parinov et al., 1999). The eventual inclusion of expression data and phenotypic information will be an important component of these databases. This sequence information can be used for reverse genetic screens to identify insertions in previously cloned genes and members of gene families. Together, the sequence, expression, and phenotypic data will contribute to current efforts to determine the function of all genes in the Arabidopsis genome.
Do Reporter Gene Patterns Accurately Reflect Chromosomal Gene Expression?
An important consideration is whether reporter gene expression patterns accurately mimic expression of endogenous genes. This question is often asked about gene traps. In Drosophila and mouse, a vast majority of enhancer and gene trap patterns accurately reflect endogenous gene expression (reviewed in Bellen, 1999). There is no reason a priori to believe that the situation is different in plants. However, whereas gene traps have been extensively used as markers for developmental analysis in plants, there are relatively few published descriptions of isolation and characterization of genes controlling reporter gene expression. In most of the cases that have been reported, reporter gene expression does indeed accurately reflect the overall pattern of plant gene expression, although not necessarily the transcript abundance (Smith and Fedoroff, 1995; Di Laurenzio et al., 1996; Gu et al., 1998; Springer et al., 2000; Vielle-Calzada et al., 2000). In fact, gene traps are much more likely to accurately reflect expression than promoter–reporter gene fusions that are extensively used for expression analysis, because all regulatory elements should be in place in a gene trap insertion (with the exception of those that are disrupted by the insertion). In contrast, promoter–reporter gene fusions may not include regulatory elements such as those that are in introns or 3′ to the transcribed region. In addition, position effects that can cause transgenes to be incorrectly expressed are not a problem with gene traps, because the controlling elements remain in their natural context in the chromosome. The ability of promoter–reporter gene fusions to accurately report gene expression has recently been questioned, and the use of promoter–reporter gene fusions as the sole determinant of gene expression has been deemed insufficient (Taylor, 1997). In contrast, gene traps are quite likely to accurately reflect endogenous gene expression.
Nonetheless, examples in which expression of a reporter gene and the chromosomal gene do not correlate have been reported. In one example, an enhancer trap insertion that was expressed in a root cap–specific pattern was identified (Tsugeki and Fedoroff, 1999). When the flanking gene was cloned, however, it showed a different expression pattern, leading the authors to conclude that the insertion had disrupted a promoter element needed for proper expression of the GUS reporter. In another example, a T-DNA insertion into the tobacco genome resulted in expression of the GUS reporter in a seed coat–specific pattern (Fobert et al., 1994). No transcribed gene could be found in the region of the insertion, and the region was not conserved in related species, leading the authors to conclude that the insertion had activated a cryptic promoter. However, it is possible that this particular promoter drives expression of a noncoding RNA or a small peptide that might not have been detected by the methods used. We have observed a similar situation with a DsG insertion in one of our transposant lines (P.S. Springer and R. Martienssen, unpublished results).
CONCLUDING REMARKS
Gene traps have proven to be highly useful tools in plant developmental biology. Their largest contribution has been in the generation of tissue- and cell type–specific markers. Gene identification has been slower in coming, and in some cases, the genes regulating reporter gene expression have been surprisingly difficult to identify. Nonetheless, as the sequence of the Arabidopsis genome is completed, gene traps are likely to play an increasingly important role in the next phase of Arabidopsis genomics—determining gene function.
Acknowledgments
I thank R. Martienssen for many useful discussions; J. Haseloff, E. Bray, and B. Aronson for helpful comments on the manuscript; D. Holding and M. Geisler for contributions to Figure 3; and J. Haseloff for contributing Figure 4. Research on gene traps in the author's laboratory is supported by National Science Foundation Grant Nos. IBN-9712927 and IBN-9875371 and a grant from the Southwest Consortium on Plant Genetics and Water Resources.
References
- Aarts, M.G.M., Corzaan, P., Stiekema, W.J., and Pereira, A. (1995). A two-element Enhancer-Inhibitor transposon system in Arabidopsis thaliana. Mol. Gen. Genet. 247, 555–564. [DOI] [PubMed] [Google Scholar]
- Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, N.D., Cran, D.G., Barton, S.C., Hettle, S., Reik, W., and Surani, M.A. (1988). Transgenes as probes for active chromosomal domains in mouse development. Nature 333, 852–855. [DOI] [PubMed] [Google Scholar]
- André, D., Colau, D., Schell, J., Van Montagu, M., and Hernalsteens, J.-P. (1986). Gene tagging in plants by a T-DNA insertion mutagen that generates APH(3′)II-plant gene fusions. Mol. Gen. Genet. 204, 512–518. [Google Scholar]
- Azpiroz-Leehan, R., and Feldmann, K.A. (1997). T-DNA insertion mutagenesis in Arabidopsis: Going back and forth. Trends Genet. 13, 152–156. [DOI] [PubMed] [Google Scholar]
- Bancroft, I., and Dean, C. (1993). Transposition pattern of the maize element Ds in Arabidopsis thaliana. Genetics 134, 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft, I., Bhatt, A.M., Sjodin, C., Scofield, S., Jones, J.D.G., and Dean, C. (1992). Development of an efficient two-element transposon tagging system in Arabidopsis thaliana. Mol. Gen. Genet. 233, 449–461. [DOI] [PubMed] [Google Scholar]
- Barthels, N., et al. (1997). Regulatory sequences of Arabidopsis drive reporter gene expression in nematode feeding structures. Plant Cell 9, 2119–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Ser. III Sci. Vie 316, 1194–1199. [Google Scholar]
- Bellen, H.J. (1999). Ten years of enhancer detection: Lessons from the fly. Plant Cell 11, 2271–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen, H.J., O'Kane, C.J., Wilson, C., Grossniklaus, U., Pearson, R.K., and Gehring, W.J. (1989). P-element–mediated enhancer detection: A versatile method to study development in Drosophila. Genes Dev. 3, 1288–1300. [DOI] [PubMed] [Google Scholar]
- Benfey, P.N., Ren, L., and Chua, N.H. (1989). The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue-specific expression patterns. EMBO J. 8, 2195–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, F., Haseloff, J., Schiefelbein, J., and Dolan, L. (1998. a). Positional information in root epidermis is defined during embryogenesis and acts in domains with strict boundaries. Curr. Biol. 8, 421–430. [DOI] [PubMed] [Google Scholar]
- Berger, F., Linstead, P., Dolan, L., and Haseloff, J. (1998. b). Stomata patterning on the hypocotyl of Arabidopsis thaliana is controlled by genes involved in the control of root epidermis patterning. Dev. Biol. 194, 226–234. [DOI] [PubMed] [Google Scholar]
- Bier, E., Vaessin, H., Shepherd, S., Lee, K., McCall, K., Barbel, S., Ackerman, L., Carretto, R., Uemura, T., Grell, E., Jan, L.Y., and Jan, Y.N. (1989). Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 3, 1273–1287. [DOI] [PubMed] [Google Scholar]
- Bouchez, D., Camilleri, C., and Caboche, M. (1993). A binary vector based on Basta resistance for in planta transformation of Arabidopsis thaliana. C. R. Acad. Sci. Ser. III Sci. Vie 316, 1188–1193. [Google Scholar]
- Brand, A.H., and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Brennan, J., and Skarnes, W.C. (1999). Gene trapping in mouse embryonic stem cells. Methods Mol. Biol. 97, 123–138. [DOI] [PubMed] [Google Scholar]
- Campisi, L., Yang, Y., Yi, Y., Heilig, E., Herman, B., Cassista, A.J., Allen, D.W., Xiang, H., and Jack, T. (1999). Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 17, 699–707. [DOI] [PubMed] [Google Scholar]
- Carroll, B.J., Klimyuk, V.I., Thomas, C.M., Bishop, G.J., Harrison, K., Scofield, S.R., and Jones, J.D.G. (1995). Germinal transpositions of the maize element Dissociation from T-DNA loci in tomato. Genetics 139, 407–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban, M.J., and Cohen, S.N. (1979). Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: In vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. USA 76, 4530–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, L., and Martienssen, R.A. (1995). Site-selected transposon mutagenesis at the hcf106 locus in maize. Plant Cell 7, 287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBlock, M., Botterman, J., Vandewiele, M., Dockx, J., Thoen, C., Gossele, V., RaoMovva, N., Thompson, C., Van Montagu, M., and Leemans, J. (1987). Engineering herbicide resistance in plants by expression of a detoxifying enzyme. EMBO J. 6, 2513–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio, L., Wysocka-Diller, J., Malamy, J.E., Pysh, L., Helariutta, Y., Freshour, G., Hahn, M.G., Feldmann, K.A., and Benfey, P.N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86, 423–433. [DOI] [PubMed] [Google Scholar]
- Dooner, H.K., and Belachew, A. (1989). Transposition of the maize element Ac from the bz-m2 (Ac) allele. Genetics 122, 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H.K., Keller, J., Harper, E., and Ralston, E. (1991). Variable patterns of transposition of the maize element Activator in tobacco. Plant Cell 3, 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery, B., Lecomte, P., Gil, N., Bechtold, N., Bouchez, D., Dalmasso, A., and Abad, P. (1998). RPE, a plant gene involved in early developmental steps of nematode feeding cells. EMBO J. 17, 6799–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff, N.V., and Smith, D.L. (1993). A versatile system for detecting transposition in Arabidopsis. Plant J. 3, 273–289. [DOI] [PubMed] [Google Scholar]
- Feldmann, K.A., and Marks, M.D. (1987). Agrobacterium-mediated transformation of germinating seeds of Arabidopsis thaliana: A non-tissue culture approach. Mol. Gen. Genet. 208, 1–9. [Google Scholar]
- Ferrándiz, C., Gu, Q., Martienssen, R., and Yanofsky, M.F. (2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127, 725–734. [DOI] [PubMed] [Google Scholar]
- Fobert, P.R., Miki, B.L., and Iyer, V.N. (1991). Detection of gene regulatory signals in plants revealed by T-DNA–mediated fusions. Plant Mol. Biol. 17, 837–851. [DOI] [PubMed] [Google Scholar]
- Fobert, P.R., Labbé, H., Cosmopoulos, J., Gottlob-McHugh, S., Ouellet, T., Hattori, J., Sunohara, G., Iyer, V.N., and Miki, B.L. (1994). T-DNA tagging of a seed coat–specific cryptic promoter in tobacco. Plant J. 6, 567–577. [DOI] [PubMed] [Google Scholar]
- Goldsbrough, A.P., Tong, Y., and Yoder, J.I. (1996). Lc as a non-destructive visual reporter and transposition excision marker gene for tomato. Plant J. 9, 927–933. [Google Scholar]
- Gossler, A., Joyner, A.L., Rossant, J., and Skarnes, W.C. (1989). Mouse embryonic stem cells and reporter constructs to detect developmentally regulated genes. Science 244, 463–465. [DOI] [PubMed] [Google Scholar]
- Greenblatt, I.M. (1984). A chromosome replication pattern deduced from pericarp phenotypes resulting from movements of the transposable element Modulator in maize. Genetics 108, 471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., Bellen, H.J., Wilson, C., and Gehring, W.J. (1989). P-element–mediated enhancer detection applied to the study of oogenesis in Drosophila. Development 107, 189–200. [DOI] [PubMed] [Google Scholar]
- Gu, Q., Ferrándiz, C., Yanofsky, M.F., and Martienssen, R. (1998). The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125, 1509–1517. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., and Amos, B. (1995). GFP in plants. Trends Genet. 11, 328–329. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, L., Jacobs, A., Van Montagu, M., and Depicker, A. (1990). Plant chromosome/marker gene fusion assay for study of normal and truncated T-DNA integration events. Mol. Gen. Genet. 224, 248–256. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.A., Thomas, C.M., Hammond-Kosack, K.E., Balint-Kurti, P.J., and Jones, J.D.G. (1994). Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266, 789–793. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G., Carland, F.M., Maliga, P., and Dooner, H.K. (1989). Visual detection of transposition of the maize element Activator (Ac) in tobacco seedlings. Science 244, 204–207. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G., Carland, F., Lim, E., Ralston, E., and Dooner, H.K. (1990). Preferential transposition of the maize element Activator to linked chromosomal locations in tobacco. Plant Cell 2, 701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey, S.E., and Labib, K. (1998). MCM proteins: Evolution, properties, and role in DNA replication. Biochim. Biophys. Acta 1398, 113–136. [DOI] [PubMed] [Google Scholar]
- Kertbundit, S., De Greve, H., Deboeck, F., Van Montagu, M., and Hernalsteens, J.-P. (1991). In-vivo random β-glucuronidase gene fusions in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 88, 5212–5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee, H.J., Horsch, R.B., Hinchee, M.A., Hein, M.B., and Hoffmann, N.L. (1987). The effects of overproduction of two Agrobacterium tumefaciens T-DNA auxin biosynthetic gene products in transgenic petunia plants. Genes Dev. 1, 86–96. [Google Scholar]
- Klimyuk, V.I., Nussaume, L., Harrison, K., and Jones, J.D.G. (1995). Novel GUS expression patterns following transposition of an enhancer trap Ds element in Arabidopsis. Mol. Gen. Genet. 249, 357–365. [DOI] [PubMed] [Google Scholar]
- Koncz, C., Martini, N., Mayerhofer, R., Koncz-Kalman, Z., Körber, H., Redei, G.P., and Schell, J. (1989). High-frequency T-DNA–mediated gene tagging in plants. Proc. Natl. Acad. Sci. USA 86, 8467–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothary, R., Clapoff, S., Brown, A., Campbell, R., Peterson, A., and Rossant, J. (1988). A transgene containing lacZ inserted into the dystonia locus is expressed in neural tube. Nature 335, 435–437. [DOI] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs, P., Autran, D., and Traas, J. (1999). A chromosomal paracentric inversion associated with T-DNA integration in Arabidopsis. Plant J. 18, 131–139. [DOI] [PubMed] [Google Scholar]
- Liljegren, S.J., Ditta, G.S., Eshed, Y., Savidge, B., Bowman, J.L., and Yanofsky, M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. [DOI] [PubMed] [Google Scholar]
- Lin, X., et al. (1999). Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 402, 761–768. [DOI] [PubMed] [Google Scholar]
- Lindsey, K., Wei, W., Clarke, M.C., McArdle, H.F., Rooke, L.M., and Topping, J.F. (1993). Tagging genomic sequences that direct transgene expression by activation of a promoter trap in plants. Transgenic Res. 2, 33–47. [DOI] [PubMed] [Google Scholar]
- Liu, Y.-G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Lloyd, A.M., Walbot, V., and Davis, R.W. (1992). Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258, 1773–1775. [DOI] [PubMed] [Google Scholar]
- Long, D., Swinburne, J., Martin, M., Wilson, K., Sundberg, E., Lee, K., and Coupland, G. (1993). Analysis of the frequency of inheritance of transposed Ds elements in Arabidopsis after activation by a CaMV 35S promoter fusion to the Ac transposase gene. Mol. Gen. Genet. 241, 627–636. [DOI] [PubMed] [Google Scholar]
- Long, D., Goodrich, J., Wilson, K., Sundberg, E., Martin, M., Puangsomlee, P., and Coupland, G. (1997). Ds elements on all five Arabidopsis chromosomes and assessment of their utility for transposon tagging. Plant J. 11, 145–148. [DOI] [PubMed] [Google Scholar]
- Machida, C., Onouchi, H., Koizumi, J., Hamada, S., Semiarti, E., Torikai, S., and Machida, Y. (1997). Characterization of the transposition pattern of the Ac element in Arabidopsis thaliana using endonuclease I-SceI. Proc. Natl. Acad. Sci. USA 94, 8675–8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., and Yanofsky, M.F. (1995). The Arabidopsis AGL8 MADS box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1. Plant Cell 7, 1763–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen, R.A. (1998). Functional genomics: Probing plant gene function and expression with transposons. Proc. Natl. Acad. Sci. USA 95, 2021–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, K., et al. (1999). Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature 402, 769–777. [DOI] [PubMed] [Google Scholar]
- McKinney, E.C., Ali, N., Traut, A., Feldmann, K.A., Belostotsky, D.A., McDowell, J.M., and Meagher, R.B. (1995). Sequence-based identification of T-DNA insertion mutations in Arabidopsis: Actin mutants act2-1 and act4-1. Plant J. 8, 613–622. [DOI] [PubMed] [Google Scholar]
- Meinke, D.W. (1991). Embryonic mutants in Arabidopsis thaliana. Dev. Genet. 12, 382–392. [Google Scholar]
- Meinke, D.W., Cherry, J.M., Dean, C., Rounsley, S.D., and Koornneef, M. (1998). Arabidopsis thaliana: A model plant for genome analysis. Science 282, 662–682. [DOI] [PubMed] [Google Scholar]
- Nacry, P., Camilleri, C., Courtial, B., Caboche, M., and Bouchez, D. (1998). Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis. Genetics 149, 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussaume, L., Harrison, K., Klimyuk, V., Martienssen, R., Sundaresan, V., and Jones, J.D. (1995). Analysis of splice donor and acceptor site function in a transposable gene trap derived from the maize element Activator. Mol. Gen. Genet. 249, 91–101. [DOI] [PubMed] [Google Scholar]
- Ohba, T., Yoshioka, Y., Machida, C., and Machida, Y. (1995). DNA rearrangement associated with the integration of T-DNA in tobacco: An example for multiple duplications of DNA around the integration target. Plant J. 7, 157–164. [DOI] [PubMed] [Google Scholar]
- O'Kane, C.J., and Gehring, W.J. (1987). Detection in situ of genomic regulatory elements in Drosophila. Proc. Natl. Acad. Sci. USA 84, 9123–9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe, D.P., Tepperman, J.M., Dean, C., Leto, K.J., Erbes, D.L., and Odell, J.T. (1994). Plant expression of a bacterial cytochrome P450 that catalyzes activation of a sulfonylurea pro-herbicide. Plant Physiol. 105, 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne, B.I., and Baker, B. (1995). Movers and shakers: Maize transposons as tools for analyzing other plant genomes. Curr. Opin. Cell Biol. 7, 406–413. [DOI] [PubMed] [Google Scholar]
- Osborne, B.I., Corr, C.A., Prince, J.P., Hehl, R., Tanksley, S.D., McCormick, S., and Baker, B. (1991). Ac transposition from a T-DNA can generate linked and unlinked clusters of insertions in the tomato genome. Genetics 129, 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parinov, S., Sevugan, M., Ye, D., Yang, W.-C., Kumaran, M., and Sundaresan, V. (1999). Analysis of flanking sequences from Dissociation insertion lines: A database for reverse genetics in Arabidopsis. Plant Cell 11, 2263–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber, T.L., and Ausubel, F.M. (1995). Differential mRNA display. Methods Cell Biol. 49, 431–440. [PubMed] [Google Scholar]
- Richmond, T., and Somerville, S. (2000). Chasing the dream: Plant EST microarrays. Curr. Opin. Plant Biol. 3, 108–116. [DOI] [PubMed] [Google Scholar]
- Roe, J.L., Nemhauser, J.L., and Zambryski, P.C. (1997). TOUSLED participates in apical tissue formation during gynoecium development in Arabidopsis. Plant Cell 9, 335–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant, J., and Hopkins, N. (1992). Of fin and fur—Mutational analysis of vertebrate embryonic development. Genes Dev. 6, 1–13. [DOI] [PubMed] [Google Scholar]
- Ross-MacDonald, P., et al. (1999). Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402, 413–418. [DOI] [PubMed] [Google Scholar]
- Siemering, K.R., Golbik, R., Sever, R., and Haseloff, J. (1996). Mutations that suppress the thermosensitivity of green fluorescent protein. Curr. Biol. 6, 1653–1663. [DOI] [PubMed] [Google Scholar]
- Skarnes, W.C. (1990). Entrapment vectors: A new tool for mammalian genetics. Bio/Technology 8, 827–831. [DOI] [PubMed] [Google Scholar]
- Skarnes, W.C. (1993). The identification of new genes: Gene trapping in transgenic mice. Curr. Opin. Biotechnol. 4, 684–689. [DOI] [PubMed] [Google Scholar]
- Skarnes, W.C., Auerbach, B.A., and Joyner, A.L. (1992). A gene trap approach in mouse embryonic stem cells: The lacZ reporter is activated by splicing, reflects endogenous gene expression, and is mutagenic in mice. Genes Dev. 6, 903–918. [DOI] [PubMed] [Google Scholar]
- Smith, D.L., and Fedoroff, N.V. (1995). LRP1, a gene expressed in lateral and adventitious root primordia of Arabidopsis. Plant Cell 7, 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D.L., Yanai, Y., Liu, Y.-G., Ishiguro, S., Okada, K., Shibata, D., Whittier, R.F., and Fedoroff, N.V. (1996). Characterization and mapping of Ds-GUS–T-DNA lines for targeted insertional mutagenesis. Plant J. 10, 721–732. [DOI] [PubMed] [Google Scholar]
- Spradling, A.C., Stern, D.M., Kiss, I., Roote, J., Laverty, T., and Rubin, G.M. (1995). Gene disruptions using P transposable elements: An integral component of the Drosophila genome project. Proc. Natl. Acad. Sci. USA 92, 10824–10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, P.S., McCombie, W.R., Sundaresan, V., and Martienssen, R.A. (1995). Gene trap tagging of PROLIFERA, an essential MCM2-3-5–like gene in Arabidopsis. Science 268, 877–880. [DOI] [PubMed] [Google Scholar]
- Springer, P.S., Holding, D.R., Groover, A., Yordan, C., and Martienssen, R.A. (2000). The essential Mcm7 protein PROLIFERA is localized to the nucleus of dividing cells during the G1 phase and is required maternally for early Arabidopsis development. Development 127, 1815–1822. [DOI] [PubMed] [Google Scholar]
- Stougaard, J. (1993). Substrate-dependent negative selection in plants using a bacterial cytosine deaminase gene. Plant J. 3, 755–761. [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D.G., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9, 1797–1810. [DOI] [PubMed] [Google Scholar]
- Taylor, C.B. (1997). Promoter fusion analysis: An insufficient measure of gene expression. Plant Cell 9, 273–275. [Google Scholar]
- Teeri, T.H., Herrera-Estrella, L., Depicker, A., Van Montagu, M., and Palva, E.T. (1986). Identification of plant promoters in situ by T-DNA–mediated transcriptional fusions to the npt-II gene. EMBO J. 5, 1755–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier, A.F., Marillonnet, S., Klimyuk, V., Patel, K., Torres, M.A., Murphy, G., and Jones, J.D. (1999). Multiple independent defective Suppressor-mutator transposon insertions in Arabidopsis: A tool for functional genomics. Plant Cell 11, 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topping, J.F., and Lindsey, K. (1997). Promoter trap markers differentiate structural and positional components of polar development in Arabidopsis. Plant Cell 9, 1713–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topping, J.F., Wei, W., and Lindsey, K. (1991). Functional tagging of regulatory elements in the plant genome. Development 112, 1009–1019. [DOI] [PubMed] [Google Scholar]
- Topping, J.F., Agyeman, F., Henricot, B., and Lindsey, K. (1994). Identification of molecular markers of embryogenesis in Arabidopsis thaliana by promoter trapping. Plant J. 5, 895–903. [DOI] [PubMed] [Google Scholar]
- Tsugeki, R., and Fedoroff, N.V. (1999). Genetic ablation of root cap cells in Arabidopsis. Proc. Natl. Acad. Sci. USA 96, 12941–12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugeki, R., Kochieva, E.Z., and Fedoroff, N.V. (1996). A transposon insertion in the Arabidopsis SSR16 gene causes an embryo-defective lethal mutation. Plant J. 10, 479–489. [DOI] [PubMed] [Google Scholar]
- Tsukaya, H., and Uchimiya, H. (1997). Genetic analyses of the formation of the serrated margin of leaf blades in Arabidopsis: Combination of a mutational analysis of leaf morphogenesis with the characterization of a specific marker gene expressed in hydathodes and stipules. Mol. Gen. Genet. 256, 231–238. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada, J.-P., Baskar, R., and Grossniklaus, U. (2000). Delayed activation of the paternal genome during seed development. Nature 404, 91–94. [DOI] [PubMed] [Google Scholar]
- Wagner-Bernholz, J.T., Wilson, C., Gibson, G., Schuh, R., and Gehring, W.J. (1991). Identification of target genes of the homeotic gene Antennapedia by enhancer detection. Genes Dev. 5, 2467–2480. [DOI] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler, S.R., Baran, G., and Varagona, M. (1987). The maize transposable element Ds is spliced from RNA. Science 237, 916–918. [DOI] [PubMed] [Google Scholar]
- Wilhelmi, L.K., and Preuss, D. (1996). Self-sterility in Arabidopsis due to defective pollen tube guidance. Science 274, 1535–1537. [DOI] [PubMed] [Google Scholar]
- Willemsen, V., Wolkenfelt, H., de Vrieze, G., Weisbeek, P., and Scheres, B. (1998). The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development 125, 521–531. [DOI] [PubMed] [Google Scholar]
- Wilson, C., Pearson, R.K., Bellen, H.J., O'Kane, C.J., Grossniklaus, U., and Gehring, W.J. (1989). P-element–mediated enhancer detection: An efficient method for isolating and characterizing developmentally regulated genes in Drosophila. Genes Dev. 3, 1301–1313. [DOI] [PubMed] [Google Scholar]
- Winkler, R.G., Frank, M.R., Galbraith, D.W., Feyereisen, R., and Feldmann, K.A. (1998). Systematic reverse genetics of transfer-DNA–tagged lines of Arabidopsis: Isolation of mutations in the cytochrome P450 gene superfamily. Plant Physiol. 118, 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]