Abstract
delayed dehiscence1 is an Arabidopsis T-DNA mutant in which anthers release pollen grains too late for pollination to occur. The delayed dehiscence1 defect is caused by a delay in the stomium degeneration program. The gene disrupted in delayed dehiscence1 encodes 12-oxophytodienoate reductase, an enzyme in the jasmonic acid biosynthesis pathway. We rescued the mutant phenotype by exogenous application of jasmonic acid and obtained seed set from previously male-sterile plants. In situ hybridization studies showed that during the early stages of floral development, DELAYED DEHISCENCE1 mRNA accumulated within all floral organs. Later, DELAYED DEHISCENCE1 mRNA accumulated specifically within the pistil, petals, and stamen filaments. DELAYED DEHISCENCE1 mRNA was not detected in the stomium and septum cells of the anther that are involved in pollen release. The T-DNA insertion in delayed dehiscence1 eliminated both DELAYED DEHISCENCE1 mRNA accumulation and 12-oxophytodienoate reductase activity. These experiments suggest that jasmonic acid signaling plays a role in controlling the time of anther dehiscence within the flower.
INTRODUCTION
Dehiscence is the terminal function of the anther and results in the release of pollen grains to enable pollination, fertilization, and seed set to occur (Goldberg et al., 1993, 1995). Dehiscence involves the breakage of the anther wall at a specific site that runs the length of the anther (Keijzer, 1987; Bonner and Dickinson, 1989; Goldberg et al., 1993, 1995; Beals and Goldberg, 1997). This site forms an indentation, or “notch,” between the locules of each theca. The stomium, which is derived from the outer L1 layer of the stamen primordium, and the septum, which is derived from the primordium L2 layer, are two specialized cell types present within the notch region (Goldberg et al., 1993). In solanaceous plants, the septum is designated as either the circular cell cluster (e.g., tobacco; Koltunow et al., 1990), the intersporangial septum (e.g., tomato; Bonner and Dickinson, 1989), the hypodermal stomium (e.g., sweet pepper; Horner and Wagner, 1992), or the oxalate package (D'Arcy et al., 1996). The mechanisms responsible for the differentiation of the stomium and septum from L1 and L2 cells positioned between each pair of locules are not known.
The program of events that leads to the breakage of the stomium and pollen release begins early in anther development and involves a number of processes. First, septum and stomium cells must differentiate from precursor cells between the locules of the anther. Second, septum and stomium cells must undergo a developmentally timed cell-degeneration program (tobacco [Koltunow et al., 1990; Beals and Goldberg, 1997] and Arabidopsis Arabidopsis [Sanders et al., 1999]). The septum degenerates before the stomium, thereby creating a bilocular anther that establishes the stomium as the future site of anther wall breakage and pollen release. Targeted ablation of the stomium results in anthers that fail to dehisce, indicating that active processes must occur within these cells to enable pollen release (Beals and Goldberg, 1997). Third, expansion of the endothecial layer and deposition of fibrous lignified bands in the walls of the endothecial cells are required for dehiscence (Sanders et al., 1999). Finally, events leading to anther dehiscence must be coordinated precisely with pollen differentiation, floral development, and flower opening. The mechanisms that control and coordinate all of these developmental activities within the flower are not known.
We performed genetic screens to identify Arabidopsis male-fertility mutants, using T-DNA and ethyl methanesulfonate mutagenesis (Sanders et al., 1999). One of our goals was to identify mutants that have defects in the anther dehiscence program. In our screens, and those of others, Arabidopsis male-fertility mutants were isolated that are either delayed in the timing of anther dehiscence or in which dehiscence fails to occur (Dawson et al., 1993; Goldberg et al., 1993; Park et al., 1996; Sanders et al., 1999). Here, we report the isolation and characterization of a gene disrupted in the dehiscence mutant, delayed dehiscence1. delayed dehiscence1 was identified in a T-DNA mutagenesis screen, and the mutant phenotype was found to cosegregate with a complex T-DNA insertion (Sanders et al., 1999). The DELAYED DEHISCENCE1 gene encodes 12-oxophytodienoate reductase, an enzyme in the jasmonic acid biosynthesis pathway. 12-Oxophytodienoate reductase enzymatic activity is absent in delayed dehiscence1 plants. In wild-type plants, DELAYED DEHISCENCE1 mRNA accumulates specifically in the pistil, petals, and stamen filaments of the flower before the initiation of the anther dehiscence program. Mutant flowers do not contain DELAYED DEHIS-CENCE1 mRNA. The delayed dehiscence1 phenotype was rescued by exogenous application of jasmonic acid, suggesting that jasmonic acid plays a role in controlling the timing of anther dehiscence.
RESULTS
delayed dehiscence1 Is Defective in the Timing of Anther Dehiscence
We previously identified an anther dehiscence mutant, delayed dehiscence1, from a T-DNA mutagenesis screen for Arabidopsis male-fertility mutants (Sanders et al., 1999). Table 1 lists the general characteristics of the delayed dehiscence1 segregating line and mutant phenotype. The defect in delayed dehiscence1 plants is specific for anther dehiscence, because pollen grains within the mutant anthers are capable of fertilization (Sanders et al., 1999).
Table 1.
Phenotypic Characteristics of the delayed dehiscence1 Mutant
| Characterization | Description |
|---|---|
| Phenotypea | Male-sterile mutants; anthers do not dehisce at flower opening but dehisce in senescing flowers |
| Pollena | Capable of pollination and seed set; seeds within rare siliques generate plants with 100% mutant phenotype; DAPI staining of nuclei in pollen showed normal development |
| Segregationa,b | Recessive mutation |
| Kanamycin selectionc,d | 3:1 KanR:KanS segregation ratio indicating a single T-DNA insertion with a functional neomycin- phosphotransferase gene |
Sanders et al. (1999). Seed sown from three rare siliques produced 16, 17, and 19 plants, all of which were male sterile.
A 3:1 ratio of wild-type to male-sterile mutants. One sample phenotypic segregation was 114 wild-type : 35 mutant (χ2 analysis, P = between 0.9 and 0.5).
T-DNA construct used for mutagenesis contains the neomycin-phosphotransferase gene, which confers kanamycin resistance (Feldmann and Marks, 1987).
One sample segregation was 339 KanR: 113 KanS (χ2 analysis, P = between 1.0 and 0.975).
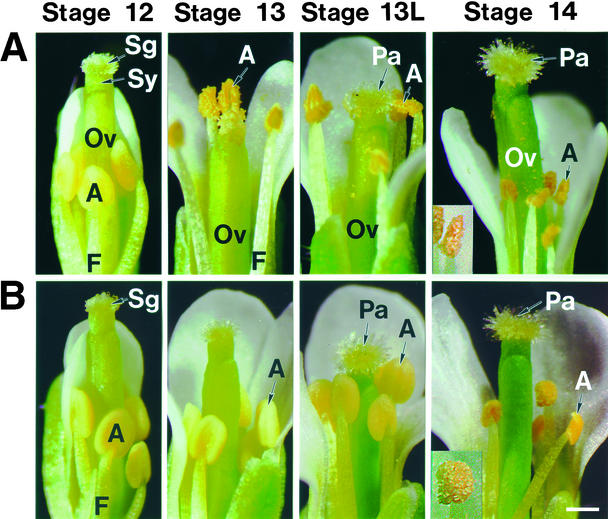
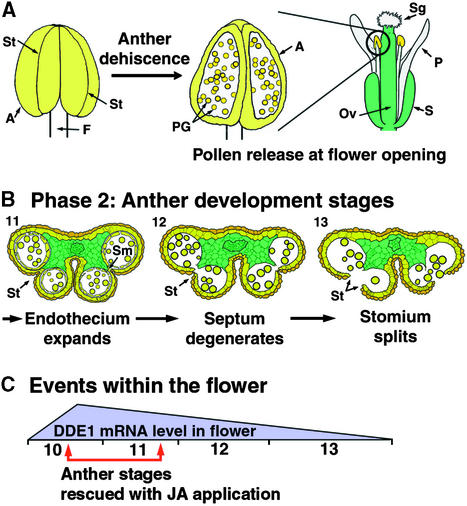
Figure 1 shows a series of flowers from a single inflorescence for wild-type and delayed dehiscence1 plants (Figures 1A and 1B, respectively); these stages of anther development have been described previously (Sanders et al., 1999). In wild-type plants, anthers dehisced at the time of flower opening, after the filaments had elongated to position the anthers in line with the stigmatic papilla (Figure 1A, stages 12 and 13). Pollen release occurred before the full expansion of the stigmatic papilla (Figure 1A, late stage 13). Following successful pollination, the pistil elongated to generate a silique, which contained the developing seeds. The initial stages of silique expansion and floral senescence are shown in Figure 1A, stage 14. The other floral organs—sepals, petals, and stamens—senesced and fell away from the expanding silique (data not shown).
Figure 1.
Dehiscence and Senescence of Wild-Type and delayed dehiscence1 Anthers.
A developmental series of flowers from young to old within a single inflorescence were photographed using a dissecting microscope. Stages 12, 13, late-13 (13L), and 14 of anther development are shown. These stages were defined in Sanders et al. (1999). Inserts of stage 14 anthers were magnified ×2.25 and enhanced digitally. The photographs represent a series of six or seven flowers from wild-type or delayed dehiscence1 inflorescences, respectively.
(A) Wild-type flowers. Floral bud (anther stage 12). Flower at anthesis (anther stage 13). Older open flower (late anther stage 13L). Initial stages of senescence in floral structures after seed set (anther stage 14).
(B) delayed dehiscence1 flowers, representing the same stages shown for the wild type in (A).
A, anther; F, filament; Ov, ovary; Pa, stigmatic papilla; Sg, stigma; Sy, style.  .
.
In contrast to those of wild-type plants, delayed dehiscence1 anthers did not dehisce at the time of flower opening (Figure 1B, stage 13). Mutant anthers did not dehisce and release pollen grains until the initiation of floral senescence (Figure 1B, stage 14). All other events, including floral development, floral organ senescence, and vegetative development, were similar to events in wild-type plants (Figures 1A and 1B; data not shown). These data show that anther dehiscence in delayed dehiscence1 occurred too late for successful pollination and resulted in a male-sterile phenotype.
Stomium and Septum Differentiation Is Similar in delayed dehiscence1 and Wild-Type Anthers
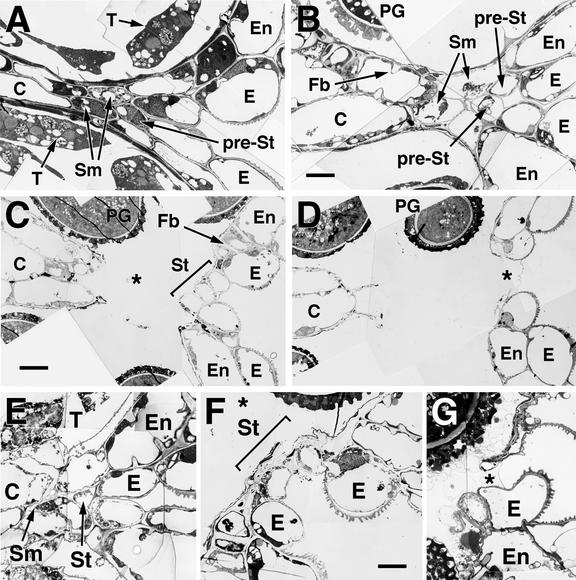
We used the transmission electron microscope (TEM) to characterize Arabidopsis anther dehiscence at the level of cellular events occurring within the anther notch. The resolution of the TEM enabled us to identify the Arabidopsis stomium and septum cell types. Figure 2 shows TEM micrographs of anther dehiscence in wild-type and delayed dehiscence1 anthers.
Figure 2.
Development and Degeneration of the Stomium Region in Wild-Type and delayed dehiscence1 Anthers.
Flowers were fixed and embedded in Spurr's epoxy resin, from which ultrathin sections were prepared and stained for TEM as described in Methods. Anther stages are described in Sanders et al. (1999).
(A) to (D) Stomium region development and dehiscence in wild-type anthers.
(A) Stage 10 anther.
(B) Early stage 12 anther.
(C) Stage 12 bilocular anther. The asterisk indicates the space generated by the degeneration of the septum cells. Stomium cells have begun to degenerate.
(D) Early stage 13 anther. The asterisk indicates the space created by the degeneration of the stomium cells. The only connection between the anther walls at this stage is the cuticle left behind by the degenerated stomium cells.
(E) to (G) Stomium region development and dehiscence in delayed dehiscence1 anthers.
(E) delayed dehiscence1 stomium region from a stage 11 anther. The tapetum is further advanced in its degeneration than that shown in (A) for wild-type anthers.
(F) delayed dehiscence1 stomium region from a stage 13 anther. Bilocular anther created by degeneration of the septum cells. The asterisk indicates the space generated by the degeneration of the septum cells. Stomium cells have begun to degenerate.
(G) delayed dehiscence1 stomium region from a stage 14 anther. The asterisk indicates the space created by the degeneration of the stomium cells. The only connection between the anther walls at this stage is the cuticle.
C, connective; E, epidermis; En, endothecium; Fb, fibrous bands; PG, pollen grain; pre-St, pre-stomium cell; Sm, septum; St, stomium; T, tapetum.  ; b
; b ;
; 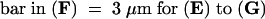 .
.
At stage 10 of development in wild-type anthers (Sanders et al., 1999), all cell types destined to become the stomium and septum were identified within the notch region (Figure 2A). At this stage, the anther has a four-locule structure and the locules contain developing microspores or male gametophytes (Sanders et al., 1999). The prestomium cell was smaller and lacked the large vacuole present in the contiguous epidermal cells (Figure 2A). The septum cells, adjacent and subepidermal to the stomium, were also small and cytoplasmically dense, in contrast to the contiguous endothecium and the connective cells that contained large vacuoles. Tapetal cells, although degenerating, were still present within the anther locules (Figure 2A).
From stages 10 to 12 in wild-type anthers, the dehiscence program occurred and resulted in pollen release (Figure 1). The tapetum degenerated and, by stage 12, was no longer present within the locules (Figure 2B). The cells of the endothecium and connective became enlarged, and lignified fibrous bands were deposited within the walls of these cells (Figure 2B). In addition, the septum cells degenerated, resulting in a bilocular anther (Figures 2B and 2C). At stage 12, the mature stomium was observed as three cells that were noticeably smaller than neighboring epidermal cells (Figure 2C, stage 12). Degeneration of the stomium cells was similar to that observed for septum cells, and by stage 13, only a cuticle remnant was left bridging the site of anther wall breakage and pollen release (Figures 2C and 2D).
The TEM analysis of the delayed dehiscence1 notch region, shown in Figures 2E to 2G, indicated that at the cellular level, stomium and septum cell differentiation and degeneration events were not detectably different from those of wild-type anthers (Figures 2C and 2F). However, delayed dehiscence1 stomium cells degenerated at a later time in development than stomium cells in wild-type anthers (cf. Figures 2C and 2D to Figures 2F and 2G, respectively). Together, these data show that dehiscence involves the differentiation and degeneration of septum and stomium cells within the anther notch and that stomium degeneration in delayed dehiscence1 is delayed relative to that of the wild type.
A Complex T-DNA Insertion Cosegregates with the delayed dehiscence1 Mutant Phenotype
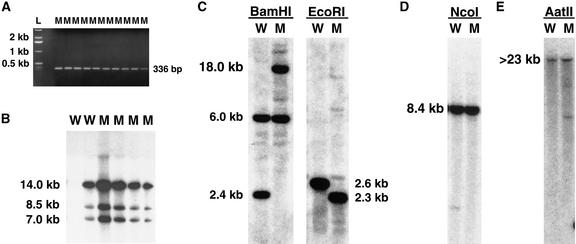
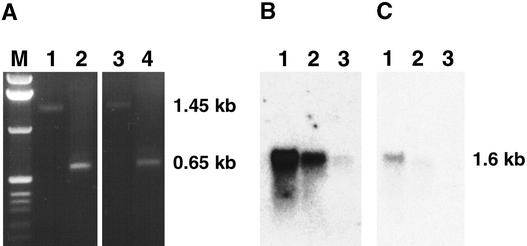
We performed polymerase chain reaction (PCR) amplification with T-DNA–specific primers and analyzed genomic blots with T-DNA probes to determine whether a T-DNA insert was linked with the delayed dehiscence1 phenotype (Figure 3). Table 1 shows that the delayed dehiscence1 phenotype segregated in a 3:1 ratio of wild-type to mutant plants, indicating that it was a recessive mutation. In addition, the delayed dehiscence1 line segregated in a 3:1 KanR:KanS ratio, which suggested a single T-DNA insertion with an active neomycin-phosphotransferase gene (Table 1). T-DNA–specific PCR primers were used to show that the T-DNA cosegregated with the mutant phenotype. Figure 3A shows the expected 336-bp product in 10 delayed dehiscence1 mutants. The 336-bp product was not detected in nontransgenic wild-type DNA but was observed in some of the plants with a wild-type phenotype in the delayed dehiscence1 segregating population. These plants were hemizygous for the T-DNA insertion (data not shown). A T-DNA insert was detected in all 106 delayed dehiscence1 mutants tested using primers that generated either the 336-bp fragment (Figure 3A) or a 823-bp product (data not shown; see Methods), indicating close linkage between the T-DNA insertion and the mutant phenotype.
Figure 3.
Detection of T-DNA and Plant Flanking Sequence in delayed dehiscence1 and Wild-Type Plants.
(A) PCR amplification of T-DNA–specific products in delayed dehiscence1 mutants. Genomic DNA was isolated from individual delayed dehiscence1 plants and used as a template to determine the presence of a T-DNA insertion. PCR amplification with primers specific to the T-DNA generated a 336-bp product in all 106 mutant plants studied (see Methods). The results from 10 individual delayed dehiscence1 mutants are shown here.
(B) Detection of T-DNA insert in plants that segregate for the delayed dehiscence1 phenotype. Genomic DNA (1 μg) was isolated from individual wild-type and mutant plants, digested with EcoRI, fractionated on a 0.7% agarose gel, and blotted to a Nytran filter. The filter was hybridized with a right border T-DNA probe (see Methods). Exposure time of autoradiogram was 14 hr. Bands at 7, 8.5, and 14 kb were observed in all mutant plants and in hemizygous wild-type plants but not in homozygous wild-type plants. A single copy T-DNA would be expected to produce one hybridizing fragment in this DNA gel blot (Behringer and Medford, 1992).
(C) Identification of an RFLP between wild-type and delayed dehiscence1 plants. Genomic DNA (2 μg) was isolated from wild-type and delayed dehiscence1 plants by using a CsCl purification procedure (see Methods), digested with BamHI or EcoRI, fractionated on a 0.6% agarose gel, and blotted to a Nytran filter. The filters were hybridized with the 974-bp plant sequence that flanked the T-DNA left border (see Methods). Positions of the BamHI and EcoRI sites and of the 974-bp fragment in the DELAYED DEHISCENCE1 region are shown in Figure 4. Exposure time was 12 hr.
(D) and (E) Comparison of plant genomic DNA flanking the T-DNA insertion site in delayed dehiscence1 and wild-type plants.
(D) Genomic DNA (2 μg) was isolated from wild-type and delayed dehiscence1 plants (see Methods), digested with NcoI, fractionated on a 0.6% agarose gel, and blotted to a Nytran filter. The filter was hybridized with a genomic DNA sequence probe that contained exons 1 and 2 of delayed dehiscence1 (see Methods and Figure 4). Exposure time of autoradiogram was 21 hr.
(E) Genomic DNA (2 μg) was isolated from wild-type and delayed dehiscence1 plants (see Methods), digested with AatII, fractionated on a 0.6% agarose gel, and blotted to a Nytran filter. The filter was hybridized with a cDNA sequence probe that contained exons 3 and 4 of delayed dehiscence1 (see Methods and Figure 4). Exposure time of autoradiogram was 21 hr.
L, marker lane; M, delayed dehiscence1 mutant; W, wild-type plant.
DNA gel blots were hybridized with T-DNA–specific probes to determine the structure of the T-DNA insert in the delayed dehiscence1 segregating population. Figure 3B shows the results obtained by hybridizing the T-DNA right border probe to mutant and wild-type plant DNAs from a delayed dehiscence1 segregating population. delayed dehis-cence1 mutants contained three EcoRI T-DNA fragments that were 7, 8.5, and 14 kb in length (Figure 3B). Hybridization of mutant DNA with the T-DNA left border showed seven SalI T-DNA fragments that were 15, 13, 10, 9, 7, 5.5, and 1.25 kb in length (data not shown). A single T-DNA insert would be expected to produce one EcoRI band and one SalI band when hybridized with right border and left border T-DNA probes, respectively (Behringer and Medford, 1992). Wild-type plants showed either the same complex EcoRI or SalI fragments as delayed dehiscence1 mutants or no hybridization, indicating that some wild-type plants were hemizygous for the T-DNA insertion. Twenty-five delayed dehiscence1 mutant plants showed the same EcoRI and SaLI T-DNA fragment patterns (data not shown). Taken together, these results indicated the presence of multiple T-DNAs in the delayed dehiscence1 mutant within a single locus.
Isolation of Mutant and Wild-Type Genomic Clones Containing the T-DNA Insertion Site
To characterize the gene disrupted by T-DNA, a genomic library was constructed using delayed dehiscence1 DNA and screened for clones that contained the T-DNA left and right border termini (see Methods). Eighty-one phage clones were identified and purified through three successive rounds of hybridization. Individual clones were screened by hybridization to wild-type Arabidopsis genomic DNA gel blots to determine whether they contained plant DNA sequences (data not shown). One lambda clone isolated from the delayed dehiscence1 genomic library contained a 974-bp plant DNA sequence that flanked a T-DNA left border. This region was cloned, labeled, and hybridized to a genomic blot that contained both wild-type and delayed dehiscence1 genomic DNAs. Figure 3C shows that a restriction fragment length polymorphism (RFLP) between wild-type DNA and the delayed dehiscence1 mutant was identified by using two different restriction enzymes. For example, a 2.4-kb BamHI fragment in wild-type DNA was increased to 18 kb after insertion of the T-DNA in the delayed dehiscence1 locus. The RFLP in the EcoRI DNA gel blot (Figure 3C) caused the wild-type DNA EcoRI fragment size to decrease from 2.6 to 2.3 kb because of an EcoRI site present within the T-DNA left border (Behringer and Medford, 1992). Taken together, these results indicate that we isolated a plant DNA sequence flanking the T-DNA insertion in delayed dehiscence1 plants.
The 974-bp delayed dehiscence1 plant sequence flanking the T-DNA was used to screen a wild-type Arabidopsis genomic library to isolate the corresponding wild-type DNA region and identify the gene disrupted by the T-DNA insertion. Twenty-four clones were identified and purified to single isolates (data not shown). One 18-kb clone, designated F7, contained the 974-bp plant DNA sequence and the complete DELAYED DEHISCENCE1 gene (data not shown).
A Single Gene Is Disrupted by the T-DNA Insertion in delayed dehiscence1
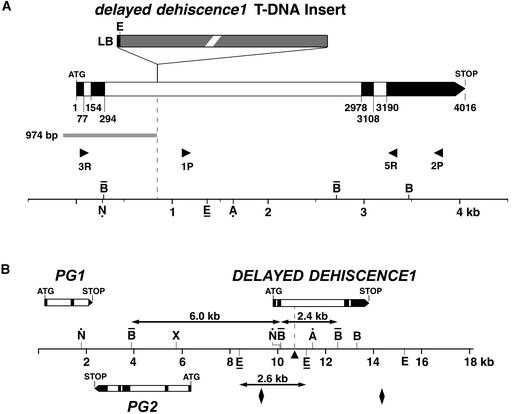
We sequenced both the 974-bp DNA fragment from the delayed dehiscence1 genomic library and the entire 18-kb wild-type genomic DNA insert to identify genes in the region of the T-DNA insertion (GenBank accession number AF218257). Sequence information from the 974-bp fragment identified the insertion site of the T-DNA left border within the delayed dehiscence1 mutant allele and positioned the T-DNA insertion within the 18-kb sequence, as illustrated in Figures 4A and 4B. DNA sequence analysis also suggested several possible exons at the region of the T-DNA insertion site and a putative structure for the gene disrupted by the T-DNA. We designated the putative wild-type gene disrupted by the T-DNA as DELAYED DEHISCENCE1. PCR primers were designed using sequence information from the exons surrounding the T-DNA insertion site to amplify a partial cDNA (Figure 4A, primer 3R; see Methods). cDNAs were amplified from inflorescence mRNA, cloned, and sequenced (see Methods). Figures 4A and 4B show the DELAYED DEHISCENCE1 gene structure determined by comparison of the genomic and cDNA sequences. The DELAYED DEHIS-CENCE1 gene consists of four exons with a coding sequence (ATG→STOP) of 1176 bp that specifies a 391–amino acid protein sequence (shown in Figure 5). The 974-bp flanking sequence contained exons 1 and 2, and sequence analysis showed that the delayed dehiscence1 T-DNA insertion was in the second intron (Figure 4A). Two other putative genes, PG1 and PG2, were identified within the 18-kb wild-type DNA sequence (GenScan; see Methods) and are illustrated in Figure 4B. PG1 and PG2 are upstream of the T-DNA insertion site in the delayed dehiscence1 mutant.
Figure 4.
Schematic Representation of the DELAYED DEHISCENCE1 Gene Region in the Arabidopsis Genome.
The black blocks represent exons, including the ATG and STOP codons. The white blocks represent introns. The BamHI and EcoRI restriction enzyme sites that generated the bands detected in Figure 3C are highlighted (BamHI, overhead bar: 2.4- and 6.0-kb bands; EcoRI, underline: 2.6-kb band). The restriction enzyme sites that generated the bands detected in Figures 3D and 3E are highlighted (NcoI and AatII, black dot). The 18-kb genomic DNA sequence containing the DELAYED DEHISCENCE1 gene is GenBank accession number AF218257.
(A) The DELAYED DEHISCENCE1 gene structure and T-DNA insertion site in delayed dehiscence1. The nucleotide positions for the exons in DELAYED DEHISCENCE1 are designated from the ATG. The T-DNA left border insertion site is 848 bp downstream of the ATG (where A is 1). The T-DNA insertion (gray block with black border, not drawn to scale) is a complex concatamer. The 974 bp of plant DNA upstream of the T-DNA left border was isolated from a delayed dehiscence1 genomic clone and is shown as a gray block. This sequence represents nucleotides 9683 to 10,656 in the 18-kb DELAYED DEHISCENCE1 DNA sequence (GenBank accession number AF218257). The primers used for the amplification of cDNA products (3R, 3′ RACE (rapid amplification of cDNA ends) primer, 33 bp downstream of the ATG within exon 1; 5R, 5′ RACE primer, 3347 bp downstream of the ATG within exon 4; see Methods) and genomic DNA PCR (1P and 2P, 1100 and 3821 bp downstream of the ATG, respectively; see Methods) are indicated as arrowheads (not to scale). The base of the arrowhead (first primer nucleotide) denotes the position of the primer within the 18-kb DNA sequence. Only the EcoRI restriction site at the left-border (LB) terminus is shown in the T-DNA insertion.
(B) The 18-kb genomic region containing the delayed dehiscence1 gene. The GenScan-MIT program predicted two putative genes in this region (PG1 and PG2; see Methods). These sequences did not identify known genes or expressed sequence tags in GenBank searches. The diamonds delineate the region shown in (A), and the triangle indicates the position of the T-DNA insertion. Only the three EcoRI sites that span the DELAYED DEHISCENCE1 gene are shown. The region of the Arabidopsis genome (ecotype Columbia) that contains the DELAYED DEHISCENCE1 gene has been released to the GenBank database (chromosome 2, BAC F5K7, accession number AC006413, 106,716 bp). There is a 1.2-kb insertion in the second intron in our Wassilewskija (Ws) ecotype DELAYED DEHISCENCE1 sequence that is not present in the Columbia ecotype sequence for this region. In the Columbia ecotype, this 1.2-kb sequence appears to be present within chromosome 1 (BAC F10B6, GenBank accession number AC006917). A cDNA clone has recently been isolated from Arabidopsis that represents the DELAYED DEHISCENCE1 transcript (GenBank accession number AF132212). Two other closely related 12-oxophytodienoate reductase-like genes have also been identified through the Arabidopsis Genome Sequencing Project that may encode related enzymes (GenBank accession numbers 3482915 and AB010695).
A, AatII; B, BamHI; E, EcoRI; N, NcoI; X, XhoI.
Figure 5.
Amino Acid Sequence Alignment of DELAYED DEHISCENCE1 and OPR Proteins.
The amino acid sequence alignment was generated using ClustalW and MacBoxShade (see Methods). The DELAYED DEHISCENCE1 (DDE1) amino acid sequence was translated from the DELAYED DEHISCENCE1 coding sequence (GenBank accession number AF218257). The OPR1 and OPR2 amino acid sequences were obtained from GenBank accession number ATU92460 (Biesgen and Weiler, 1999). The amino acids in DELAYED DEHISCENCE1 that are identical to amino acids in both OPR1 and OPR2 proteins are shaded. Gaps generated by the alignment are indicated as dots.
Analysis of the delayed dehiscence1 974-bp plant flanking sequence showed that the DNA sequence immediately 5′ to the T-DNA insertion was the same as in wild-type DNA (data not shown). To determine whether the T-DNA insertion had disrupted plant sequences more distally 5′ to the insertion site, nontransgenic wild-type and mutant DNAs were digested with NcoI, gel blotted, and hybridized with a 5′ DELAYED DEHISCENCE1 probe containing exons 1 and 2 (Figures 3D and 4). As Figure 3D shows, the wild-type 8.4-kb NcoI DNA fragment upstream of the T-DNA insertion site in delayed dehiscence1 had not been altered.
To address the possibility of a sequence alteration 3′ to the T-DNA insertion site, PCR primers were designed specifically for the DELAYED DEHISCENCE1 genomic sequence downstream of the T-DNA left border. A PCR primer pair, 1P and 2P, in which primer 1P was located 243 bp downstream of the left border insertion (illustrated in Figure 4B), was successful in generating the corresponding wild-type 2.7-kb fragment from the delayed dehiscence1 mutant DNA (data not shown). This result demonstrated that the T-DNA insertion did not generate a 3′ deletion in the delayed dehiscence1 gene. We also confirmed that genomic DNA further 3′ to the mutated delayed dehiscence1 gene was not disrupted. DNA gel blots containing nontransgenic wild-type and mutant DNA digested with AatII were hybridized with a 3′ DELAYED DEHISCENCE1 probe containing exons 3 and 4 (Figures 3E and 4). The same >23-kb AatII fragment was detected in both delayed dehiscence1 mutant and nontransgenic wild-type plants (Figure 3E).
Taken together, these results indicate that no rearrangements or deletions were detected upstream or downstream of the T-DNA insertion in delayed dehiscence1 mutants and suggest that the T-DNA in delayed dehiscence1 affects only the DELAYED DEHISCENCE1 gene at its site of insertion.
The Gene Disrupted in delayed dehiscence1 Encodes an Enzyme in the Jasmonic Acid Biosynthesis Pathway
Homology searches with DELAYED DEHISCENCE1 nucleotide and translated amino acid sequences identified the Arabidopsis 12-OXOPHYTODIENOATE REDUCTASE genes, OPR1 and OPR2, as a strong match (Biesgen and Weiler, 1999). The amino acid alignment of the three OPR family members is shown in Figure 5. The structures and the homology relationships of the OPR1, OPR2, and DELAYED DEHISCENCE1 genes are shown in Table 2. 12-Oxophytodienoate reductase (OPR) is an enzyme in the jasmonic acid biosynthesis pathway, as shown in Figure 6A. The OPR enzymatic step converts 12-oxophytodienoic acid (OPDA) into 3-oxo-2-(2′-Z-pentenyl)-cyclopentane-1-octanoic acid (OPC-8:0) (Figure 6A; Schaller and Weiler, 1997a). The conserved structure of DELAYED DEHISCENCE1 and the OPR genes indicated a gene-family relationship in which DELAYED DEHISCENCE1 was the more divergent member (Figure 5 and Table 2). The DELAYED DEHISCENCE1 protein has ∼50% identity (67% similarity) with the OPR1 and OPR2 peptides. The region of conservation was spread throughout the protein (Figure 5). The DELAYED DEHISCENCE1 gene was also identified as being related to OPR1 and OPR2 in an Arabidopsis Genome Project DNA sequence release (chromosome 2, 106,716 bp, BAC F5K7, accession number AC006413). These results suggest that the DELAYED DEHISCENCE1 gene is a divergent member of the Arabidopsis 12-OXOPHYTODIENOATE REDUCTASE gene family and encodes an enzyme in the jasmonic acid biosynthesis pathway.
Table 2.
Comparison of Arabidopsis DELAYED DEHISCENCE1 and 12-OXOPHYTODIENOATE REDUCTASE Gene Family Membersa
| Coding Sequence (ATG to STOP)b
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Number of Exons | Number of Amino Acids | Exon | Intron | Exon | Intron | Exon | Intron | Exon | % Identity to OPR1c |
| OPR1 | 3 | 372 | — | — | 218 | 96 | 308 | 75 | 593 | 100 |
| OPR2 | 4 | 374 | 83 | 79 | 141 | 93 | 308 | 83 | 593 | 89 |
| DDE1 | 4 | 391 | 77 | 76 | 141 | 2683 | 131 | 81 | 827 | 50 |
GenBank accession number AF218257 for the DELAYED DEHISCENCE1 18-kb gene region. GenBank accession no. ATU92460 for the 12-OXOPHYTODIENOATE REDUCTASE genes, OPR1 and OPR2 (Biesgen and Weiler, 1999). The DELAYED DEHISCENCE1 gene was recently identified in the Arabidopsis sequencing project. It is present on the chromosome 2 BAC F5K7 (GenBank accession no. AC006413) and was annotated in a conceptual translation as a putative OPR enzyme (GenBank accession no. AAD19764).
As defined in this table, the first exon begins at the ATG codon and the last exon ends at the STOP codon. Nucleotide lengths of introns are in italics.
Sequence comparison by GenBank BlastP (Blast 2 Sequences at NCBI; http://www.ncbi.nlm.nih.gov). Percent identity to OPR1 at amino acid level.
Figure 6.
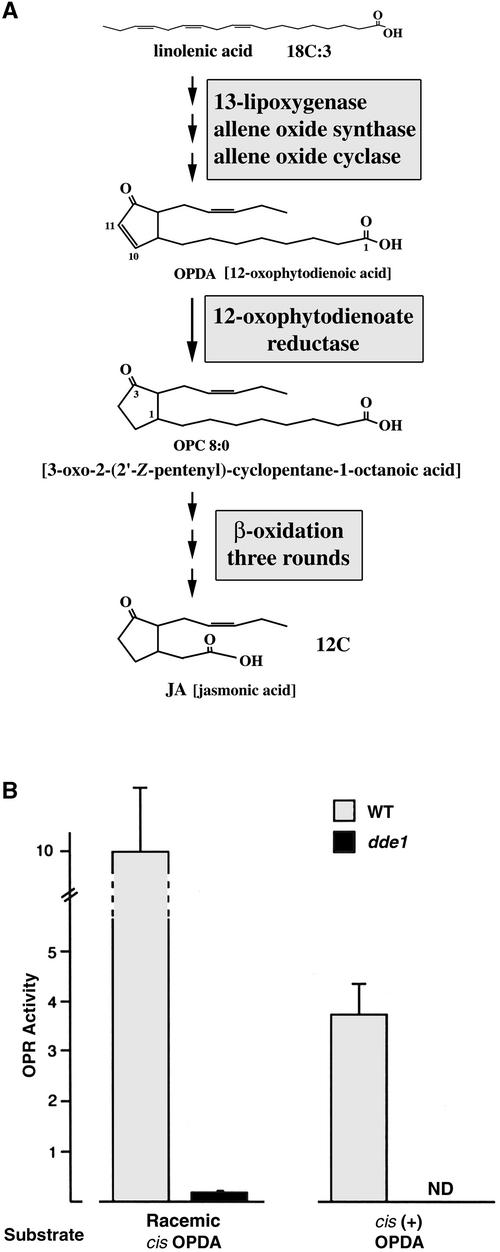
Biosynthesis of Jasmonic Acid and 12-Oxophytodienoate Reductase Enzymatic Activity in Wild-Type and delayed dehiscence1 Plants.
(A) Jasmonic acid biosynthesis pathway (modified from Schaller and Weiler, 1997a; Weiler, 1997). Linolenic acid (18:3) is converted to the cyclic molecule 12-oxophytodienoic acid (OPDA) after three enzymatic steps that occur in the chloroplast. In the third step, allene oxide cyclase produces a specific stereochemical form of OPDA (Schaller and Weiler, 1997b). The cytoplasmic enzyme OPR converts OPDA into 3-oxo-2-(2′-Z-pentenyl)-cyclopentane-1-octanoic acid (OPC-8:0). The generation of jasmonic acid from OPC-8:0 occurs in the peroxisomes after three rounds of β-oxidation.
(B) Absence of OPR enzyme activity in vegetative tissues of delayed dehiscence1. delayed dehiscence1 (dde1) and wild-type (WT) plants at the rosette stage were assayed for 12-oxophytodienoate reductase (OPR) activity. The conversion of OPDA to OPC-8:0 (OPR activity) was assayed by the addition of racemic cis-OPDA or cis-(+)OPDA to crude protein extracts (see Methods). The results are an average of five independent experiments. Error bars represent standard deviation of the mean of all five experiments. ND, not detected.
Exogenous Jasmonic Acid Restores Fertility to delayed dehiscence1 Plants
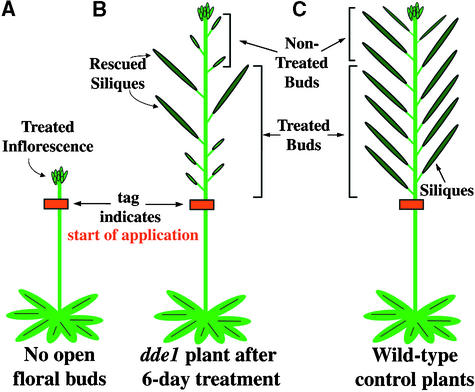
We attempted to rescue the delayed dehiscence1 male-sterile phenotype by exogenous application of jasmonic acid to determine whether a defect in jasmonic acid synthesis was responsible for the delay in anther dehiscence. Figures 7A and 7B show in a schematic diagram how the exogenous jasmonic acid (2 μmol/mL) was applied to developing inflorescences of delayed dehiscence1 plants (see Methods). Control treatments that involved no application, or water-only applications, were performed to observe background levels of delayed dehiscence1 seed set and the effect that “brushing” inflorescences might have on the pollination process.
Figure 7.
Schematic Representation of the Jasmonic Acid Rescue Experiment.
Inflorescences of delayed dehiscence1 (dde1) and wild-type plants were tagged after removal of all open flowers. Exogenous jasmonic acid was applied to the floral buds of the tagged inflorescences over a 6-day period (see Methods). After 6 to 8 days, the inflorescences were examined for siliques.
(A) delayed dehiscence1 plant tagged at start of experiment.
(B) delayed dehiscence1 plant at end of experiment. After jasmonic acid treatment, seed set was observed, as demonstrated by the presence of siliques. Siliques were found at specific positions within the developing inflorescence.
(C) Wild-type control plants at end of the experiment. Wild-type plants set seed under all experimental conditions (see Methods). Floral buds on wild-type inflorescences continued to set seed after completion of the jasmonic acid treatment.
Table 3 shows the results of two jasmonic acid application experiments (Figure 7). The application of jasmonic acid was continued for a 6-day period because we did not know (1) whether the mutant buds would respond to jasmonic acid, (2) at what developmental stage buds would be responsive to jasmonic acid, or (3) when the jasmonic acid would no longer have an effect. In experiment 1, an average of 2.3 siliques was found per jasmonic acid–treated inflorescence on delayed dehiscence1 mutants (Table 3). No siliques were observed in the nontreated or water-treated controls (Table 3). In wild-type inflorescences, the number of siliques counted in all three treatments ranged from 3.4 to 6.6. In experiment 2, an average of 2.7 siliques was found per jasmonic acid–treated inflorescence on delayed dehiscence1 mutants (Table 3). The range of siliques counted on wild-type inflorescences for all three treatments in this experiment was between 9.1 and 10.5 (Table 3). In experiment 2, as in experiment 1, no siliques were observed in the nontreated or water-treated delayed dehiscence1 controls (Table 3).
Table 3.
Rescue of delayed dehiscence1 Phenotype after Exogenous Application of Jasmonic Acida
| Experimental Treatment | No. of Inflorescences | No. of Siliquesb | No. of Unexpanded Pistils | Average No. of Buds per Inflorescence |
Average No. of Siliquesbper Inflorescence |
|---|---|---|---|---|---|
| Experiment 1c | |||||
| delayed dehiscence1 | |||||
| No treatment | 13 | 0 | 94 | 7.2 | 0 |
| Water | 15 | 0 | 105 | 7.0 | 0 |
| Jasmonic acid | 15 | 35 | 110 | 9.7 | 2.3d |
| Wild type | |||||
| No treatment | 15 | 99 | 0 | 6.6 | 6.6 |
| Water | 14 | 48 | 0 | 3.4 | 3.4 |
| Jasmonic acid | 15 | 68 | 0 | 4.5 | 4.5 |
| Experiment 2c | |||||
| delayed dehiscence1 | |||||
| No treatment | 14 | 0 | 292 | 20.8 | 0 |
| Water | 14 | 0 | 256 | 18.3 | 0 |
| Jasmonic acid | 15 | 41 | 273 | 20.9 | 2.7d |
| Wild type | |||||
| No treatment | 15 | 136 | 15 | 10.6 | 9.1 |
| Water | 15 | 201 | 28 | 15.2 | 13.4 |
| Jasmonic acid | 15 | 157 | 98 | 17.0 | 10.5 |
Application of 2 μmol/mL jasmonic acid or water to inflorescences of delayed dehiscence1 and wild-type plants as described in Methods. The procedure of McConn and Browse (1996) was followed.
The application of jasmonic acid was limited to a 6-day period. In our experiments, we did not know whether the mutant buds would respond to jasmonic acid or at what developmental stage buds would be responsive to jasmonic acid. Rescue of mutant buds could be limited to the period of jasmonic acid application or restricted to a responsive stage of development. The delayed dehiscence1 mutants responded to jasmonic acid treatment and set seed, as observed by the presence of siliques. Siliques on delayed dehiscence1 mutants were produced within a specific region of the inflorescence (see Figure 7B). By contrast, most buds set seed to generate siliques on wild-type inflorescences. Wild-type plants set seed throughout the period of experimental treatment and continued to do so after the 6-day treatment was discontinued. Therefore, in these experiments the number of siliques counted on wild-type inflorescences will overestimate the potential siliques that can be rescued on a mutant inflorescence.
Two jasmonic acid rescue experiments were performed under different conditions with plants grown in our greenhouse. Experiment 1 (August 1998) was a 6-day treatment, 10 applications, with siliques tallied after 6 days. Experiment 2 (November 1998) was a 6-day treatment, 11 applications, with siliques tallied after 8 days.
In a trial sowing, we obtained 151 progeny from jasmonic acid–rescued siliques on mutant plants. All except one showed the delayed dehiscence1 phenotype.
In both experiments, siliques induced on delayed dehiscence1 mutants were limited to a specific region of the inflorescence (see Figure 7B). There was a “lag” in the production of siliques from delayed dehiscence1–treated buds. The oldest buds did not produce siliques and were not rescued by jasmonic acid. In contrast, most buds in wild-type inflorescences set seed to generate siliques regardless of age or region (Figure 7C). Wild-type plants set seed throughout the 6-day period of experimental treatment and continued to do so after treatment was discontinued. In these experiments, therefore, the number of siliques counted on wild-type inflorescences overestimated the potential siliques that could be rescued on a mutant inflorescence.
After jasmonic acid application, seed was collected from siliques of delayed dehiscence1 mutants and sown. Progeny plants exhibited the delayed dehiscence1 sterile phenotype, confirming that they were self-progeny of the mutant (Table 3). Together, these results show that jasmonic acid can be used to rescue the delayed dehiscence1 mutant phenotype and that floral buds need to be at a specific developmental stage to be rescued. The chemical complementation of the mutant phenotype with jasmonic acid demonstrates that the T-DNA disruption of the DELAYED DEHISCENCE1 gene (12-OXOPHYTODIENOATE REDUCTASE) was responsible for the delayed dehiscence1 phenotype.
DELAYED DEHISCENCE1 mRNA Is Present in Floral and Vegetative Organs
We used PCR to determine the presence of DELAYED DEHISCENCE1 mRNA in wild-type floral and vegetative organs. Figure 8A shows that reverse transcription–PCR products were obtained for DELAYED DEHISCENCE1 in both inflorescence and seedling mRNAs. We hybridized a DELAYED DEHISCENCE1 cDNA clone and the 18-kb DELAYED DEHISCENCE1 genomic DNA clone with polysomal poly(A) mRNA gel blots to determine (1) the level of DELAYED DEHISCENCE1 mRNA in inflorescence tissue and (2) whether transcripts of the putative genes PG1 and PG2 (Figure 4B) were present in inflorescence RNA. Figure 8B shows the presence of the 1.6-kb DELAYED DEHISCENCE1 mRNA in inflorescence polysomal poly(A) mRNA. DELAYED DEHISCENCE1 transcripts were also detected at a reduced level in silique polysomal poly(A) mRNA (Figure 8B). Under our RNA gel blot conditions, the DELAYED DEHISCENCE1 cDNA would not have hybridized with related OPR1 and OPR2 mRNAs (Table 2). As shown in Figure 8C, only the DELAYED DEHISCENCE1 mRNA was detected on RNA gel blots hybridized with the 18-kb DELAYED DEHISCENCE1 genomic region. At a longer autoradiogram exposure (6 days), the hybridization signal for the 18-kb genomic region probe was equivalent to that obtained with the cDNA probe (data not shown), and no PG1 and PG2 transcripts were detected (data not shown). Taken together, these results indicate that DELAYED DEHISCENCE1 mRNA is present on polysomes in developing inflorescences.
Figure 8.
Representation of DELAYED DEHISCENCE1 mRNAs within Arabidopsis Floral and Vegetative Organs.
(A) Amplification of DELAYED DEHISCENCE1 cDNA products from inflorescence and seedling poly(A) mRNA. cDNAs generated from inflorescence polysomal poly(A) mRNA (lanes 1 and 2) and from seedling polysomal poly(A) mRNA (lanes 3 and 4) were used as templates for the generation of 5′ and 3′ RACE products (see Methods). Lanes 1 and 3 show the 3′ RACE product amplified with a gene-specific primer (3R, Figure 4A; see Methods), and lanes 2 and 4 show the 5′ RACE product amplified with a gene-specific primer (5R, Figure 4A; see Methods). Lane M shows the marker DNA fragments.
(B) and (C) Polysomal poly(A) mRNA was isolated from wild-type plants, size-fractionated by electrophoresis in formaldehyde gels, blotted to nylon filters, and hybridized with 32P-labeled probes (see Methods). Lanes 1, 2, and 3 contain 9 μg of inflorescence, 3 μg of inflorescence, and 2.25 μg of silique RNAs, respectively. (B) Hybridization with DELAYED DEHISCENCE1 cDNA. (C) Hybridization with 18-kb DELAYED DEHISCENCE1 genomic region.
DELAYED DEHISCENCE1 mRNA Is Regulated Spatially and Temporally during Floral Development
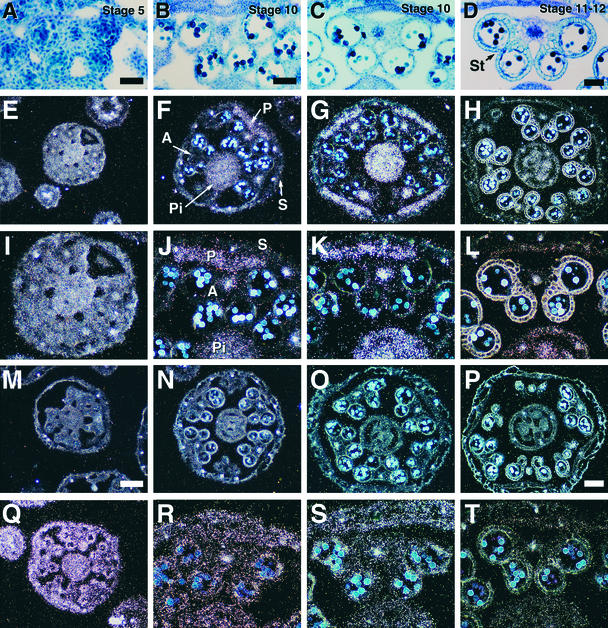
We hybridized a DELAYED DEHISCENCE1 anti-mRNA probe with transverse wild-type inflorescence sections to localize DELAYED DEHISCENCE1 mRNA within developing floral buds (see Methods). Under our in situ hybridization conditions, the DELAYED DEHISCENCE1 anti-mRNA probe does not cross-hybridize with OPR1 and OPR2 mRNAs (Table 2; data not shown).
Figure 9 shows the results of the in situ hybridization experiment. A ribosomal gene was used as a positive control for this experiment. We focused the in situ hybridization studies on stages of floral development that contained anthers undergoing dehiscence program events. Figures 9A to 9D show bright-field photographs of stages 5, 10 (early and late), and 12 of wild-type anther development (Sanders et al., 1999). Stage 5 of anther development precedes meiosis (Figure 9A), stage 10 occurs before the initiation of the anther dehiscence program (Figures 9B and 9C), and at stage 12 (Figure 9D) the anther becomes bilocular after the degeneration of the septum. After stage 12, the stomium degenerates to allow pollen release (stage 13; Figures 1A and 2D).
Figure 9.
Localization of DELAYED DEHISCENCE1 mRNA within Floral Tissues of Arabidopsis Wild-Type and delayed dehiscence1 Plants.
Inflorescences were fixed, embedded in paraffin, sliced into 10-μm-thick sections, and hybridized with DELAYED DEHISCENCE1 anti-mRNA and ribosomal anti-RNA probes as outlined in Methods. Photographs were taken by using bright-field and dark-field microscopy.
(A) to (D) Bright-field photographs of wild-type anther development stages. (A) Stage 5. Premeiotic microspore mother cells. (B) Early stage 10. (C) Late stage 10, before endothecium expansion. (D) Early stage 12. The tapetum has almost degenerated and the endothecium has expanded, but the anther still has four locules and an intact septum. These bright-field anther photographs are close-ups of anthers shown in (E) to (H).
(E) to (L) Hybridization of DELAYED DEHISCENCE1 anti-mRNA probes to sections of wild-type floral buds corresponding to anther stages shown in (A) to (D). (I) to (L) are close-ups of anthers shown in (E) to (H). Slide emulsions were exposed for 8 days, and the photographs were taken by using dark-field photography.
(M) to (P) Hybridization of DELAYED DEHISCENCE1 anti-mRNA probes to sections of delayed dehiscence1 floral buds corresponding to anther stages shown in (A) to (D). Slide emulsions were exposed for 8 days, except (N), which was exposed for 18 days.
(Q) to (T) Hybridization of ribosomal anti-RNA probes to sections of wild-type and delayed dehiscence1 floral buds corresponding to anther stages shown in (A) to (D). (Q) Wild type; (R) to (T) delayed dehiscence1. Slide emulsions were exposed for 1 hr.
A, anther; P, petal; Pi, pistil; S, sepal; St, stomium. 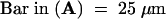 ;
; 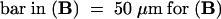 , (C), (I) to (K), (R), and (S);
, (C), (I) to (K), (R), and (S); 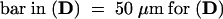 , (L), and (T);
, (L), and (T); 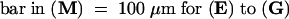 , (M) to (O), and (Q);
, (M) to (O), and (Q); 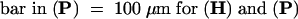 .
.
In wild-type floral sections at stage 5, the DELAYED DEHISCENCE1 mRNA was localized within all floral organs (Figures 9E and 9I). Later in anther development, DELAYED DEHISCENCE1 mRNA was localized specifically within the pistil, petal, and stamen filament (Figures 9F to 9H and 9J to 9L). Serial anther sections showed that DELAYED DEHISCENCE1 mRNA was present within the vascular region closest to the stamen filament but decreased in sections farther from the base of the anther (data not shown). At stage 10, the level of DELAYED DEHISCENCE1 mRNA increased within the pistil, petal, and stamen filament (cf. Figures 9F and 9G to Figures 9J and 9K, respectively). However, after stage 10, the DELAYED DEHISCENCE1 mRNA at these sites decreased to a low level (anther development stage 12; Figures 9H and 9L). The highest level of DELAYED DEHISCENCE1 mRNA was observed to correspond to the anther stage preceding the expansion of the endothecium and the appearance of fibrous bands (Figure 9; Sanders et al., 1999). During late anther development and after the initiation of dehiscence program events, DELAYED DEHISCENCE1 mRNA was not detected above background levels in cells of the anther, except within the vascular region contiguous to the stamen filament (Figures 9F to 9H and 9J to 9L).
DELAYED DEHISCENCE1 anti-mRNA probes were hybridized with delayed dehiscence1 floral sections to determine the level of DELAYED DEHISCENCE1 mRNA within the mutant. DELAYED DEHISCENCE1 mRNA was not detected within the cross-sections of delayed dehiscence1 floral buds, as shown in Figures 9M to 9P. As a positive control we hybridized anti-rRNA to the anther developmental stages 5, 10, and 12 (see Methods), the results of which are shown in Figures 9Q to 9T. rRNA was localized to all cells and organs of the flower and was clearly present within the cells of the anthers in our floral sections. These results show that anther sections at all developmental stages contained cells in which a positive in situ hybridization could be obtained.
Taken together, our results indicate that no DELAYED DEHISCENCE1 mRNA is present in delayed dehiscence1 mutant plants. In wild-type plants, the DELAYED DEHISCENCE1 mRNA accumulation is regulated in a spatial and temporal pattern during floral development. During the anther dehiscence program, DELAYED DEHISCENCE1 mRNA is detectable only within the vascular region of the anther.
OPR Enzymatic Activity Is Abolished in delayed dehiscence1
To determine the level of OPR enzymatic activity in the delayed dehiscence1 mutants, we assayed for the conversion of OPDA into OPC-8:0, using crude protein extracts from vegetative tissue (Figure 6A; Schaller and Weiler, 1997a, 1997b). Racemic cis-12-oxophytodienoic acid and cis(+)-12-oxophytodienoic acid, substrates for OPR, were added to extracts to measure OPR activity in vitro.
Figure 6B shows the results from the addition of alternative OPDA substrate sources to the crude protein extracts from wild-type and delayed dehiscence1 plants. The level of substrate conversion was compared between wild-type and mutant plants. When a racemic mixture of cis-OPDA was used, the level of OPR enzyme activity in delayed dehiscence1 mutants was <5% of the value obtained for the wild type (Figure 6B). When cis(+)-OPDA was the sole substrate source, no OPR enzyme activity was detected by conversion of the substrate in delayed dehiscence1 mutants (Figure 6B).
Even using gas chromatography–mass spectrometry, we found no detectable conversion of OPDA into OPC-8:0 in delayed dehiscence1 plants, indicating the absence of OPR enzyme activity. In addition, when the level of jasmonic acid was measured in delayed dehiscence1 mutants and compared with that of the wild type, the level of jasmonic acid in the delayed dehiscence1 mutants was not detectable (data not shown). Taken together, these results indicate that the delayed dehiscence1 mutants have no detectable 12-oxophytodienoate reductase activity and no detectable level of jasmonic acid. In addition, these results show that the T-DNA insertion eliminated 12-oxophytodienoate reductase enzyme activity in delayed dehiscence1 mutants and demonstrate that in the delayed dehiscence1 mutant, the T-DNA insertion disrupted a gene encoding 12-oxophytodienoate reductase, an enzyme in the octadecanoid pathway (Figure 6A).
DISCUSSION
We isolated an Arabidopsis gene, DELAYED DEHISCENCE1, that encodes 12-oxophytodienoate reductase, an enzyme in the jasmonic acid biosynthesis pathway (Figure 6A). This gene is disrupted by a T-DNA insertion in delayed dehiscence1 plants, which causes a delay in anther dehiscence and male sterility. In mutant plants, DELAYED DEHISCENCE1 mRNA is not detected, and no 12-oxophytodienoate reductase enzyme activity is observed. Application of exogenous jasmonic acid to floral buds of delayed dehiscence1 plants rescues the male sterility defect, an indication that jasmonic acid signaling, either directly or indirectly, plays a role in the anther dehiscence process.
Anther Dehiscence Requires a Coordinated Program of Events
Anther dehiscence leads to breakage of the anther wall and the release of pollen grains from the locules at flower opening (Figures 1 and 2). Figure 10 summarizes the events in the dehiscence program of the Arabidopsis anther. Two specialized cell types—the septum and the stomium—undergo sequential degeneration events that lead to development of a bilocular anther (Figures 2A to 2C and 10) and breakage of the anther wall (Figures 2C, 2D, and 10). Previously, we ablated the stomium of tobacco anthers, which generated male-sterile plants (Beals and Goldberg, 1997). These findings, together with the TEM studies in Figure 2, suggest that cell degeneration of the stomium and breakage of the anther wall are genetically programmed events that are required for dehiscence to occur. The molecular processes that control the cell degeneration events during anther dehiscence are probably initiated before this degeneration can be visualized in the septum and stomium cells.
Figure 10.
Schematic Representation of the Anther Dehiscence Program in Arabidopsis.
(A) Anther dehiscence at floral opening. Dehiscence is the terminal step in anther development that releases mature pollen grains.
(B) Transverse sections of late anther development stages that represent key steps in the anther dehiscence program. Endothecium expansion signifies the initial changes that occur in the cells of the anther wall and the start of the anther dehiscence program. Septum degeneration creates a bilocular anther. Stomium breakage opens a site in the anther wall to release the pollen grains.
(C) Events within the flower. The highest level of DELAYED DEHISCENCE1 mRNA within floral sections was determined to be at stage 10 of anther development (Figure 9). This level decreases in later floral stages (Figure 9). Preliminary experiments indicate that the period of anther development that can be rescued by jasmonic acid application is at approximately stage 10 to 11 (P.Y. Lee and P.M. Sanders, unpublished results). Both the stage at which the most DELAYED DEHISCENCE1 mRNA is found in the flower and the stage at which jasmonic acid is the most effective for delayed dehiscence1 rescue correspond to the initial stages of the dehiscence program in the anther.
The events that determine the timing of anther dehiscence must be coordinated with other developmental processes that occur within both the anther (e.g., pollen development) and the flower (e.g., flower opening, pistil receptivity, and ovary development) to ensure that successful fertilization and seed set take place (Figures 10A and 10B). The flowers of delayed dehiscence1 mutants open, are receptive to pollination, and senesce just as those on wild-type plants do (Figure 1). In delayed dehiscence1, anther dehiscence events appear similar to those in the wild type; however, a change in the timing of stomium cell degeneration causes the mutant defect to occur (Figure 2 and Sanders et al., 1999). These results show that degeneration of the stomium in delayed dehiscence1 mutant anthers is uncoupled from other dehiscence program events (e.g., septum degeneration).
The complexity of the anther dehiscence program suggests that many genes are required for pollen release. This prediction has been confirmed in our recent Arabidopsis ethyl methanesulfonate screen for sterility mutants by the finding that a large number of mutants affect the dehiscence process (Sanders et al., 1999). Many of these mutants are delayed in dehiscence with phenotypes similar to delayed dehiscence1. These data indicate that many genes participate in the dehiscence process, several of which may encode enzymes in biosynthesis (e.g., jasmonic acid) and cell degeneration pathways.
DELAYED DEHISCENCE1 Encodes an Enzyme in the Jasmonic Acid Biosynthesis Pathway
The demonstration that DELAYED DEHISCENCE1 encodes an enzyme in the octadecanoid pathway suggests a role for jasmonic acid in anther dehiscence. DELAYED DEHISCENCE1 encodes 12-oxophytodienoate reductase, a cytoplasmic enzyme that catalyzes the removal of a double bond in the first cyclic intermediate, 12-oxophytodienoic acid, in the octadecanoic pathway (Figure 6A; Vick and Zimmerman, 1984; Schaller and Weiler, 1997a). The octadecanoid pathway results in the synthesis of jasmonic acid (Figure 6A; Vick and Zimmerman, 1984; Weiler, 1997). The DELAYED DEHISCENCE1 gene is the third gene identified to encode an Arabidopsis 12-oxophytodienoate reductase–like enzyme. The related OPR1 and OPR2 genes were previously identified by Schaller et al. (1998) and Biesgen and Weiler (1999). In delayed dehiscence1 plants, we did not detect mRNA encoded by the DELAYED DEHISCENCE1 gene (Figure 9), nor did we detect 12-oxophytodienoate reductase activity (Figure 6B). Together, these results indicate that the DELAYED DEHISCENCE1–encoded enzyme is the predominant enzymatic activity responsible for conversion of OPDA in vivo (Figure 6) and, therefore, that DELAYED DEHISCENCE1 encodes the major 12-oxophytodienoate reductase enzyme in Arabidopsis.
The role that the other 12-oxophytodienoate reductase enzymes have in Arabidopsis is not known. The OPR1 and OPR2 enzymes have been reported to exhibit different substrate specificities, and Schaller et al. (1998) have suggested that OPR1 is not an enzyme in the octadecanoid pathway but may serve some unknown enzymatic function in vivo. OPR1 and OPR2 (Schaller et al., 1998; Biesgen and Weiler, 1999) and two other candidate genes recently identified in the Arabidopsis Genome Sequencing Project (GenBank accession numbers 3,482,915 and AB010695) therefore, may represent OPR genes expressed in specific regions of the plant or at specific times in development in comparison with DELAYED DEHISCENCE1. Alternatively, these genes may reflect the highly conserved nature of the 12-oxophtyodienoate reductase protein structure and encode enzymes with different functions in vivo (Schaller and Weiler, 1997b; Schaller et al., 1998).
Dehiscence Mutants Can Be Used to Dissect the Jasmonic Acid Biosynthesis and Signaling Pathways
In Arabidopsis fertility mutant screens, our laboratory and those of others have identified a large number of dehiscence-related mutants (Dawson et al., 1993; Park et al., 1996; Sanders et al., 1999). These mutants have been characterized as either delayed dehiscence mutants or as nondehiscence mutants in which anthers do not dehisce. Like delayed dehiscence1, many of these mutants are defective in the timing of dehiscence and release pollen too late for successful pollination. Many of the delayed dehiscence mutants may represent defects in jasmonic acid synthesis or perception. In contrast, non-dehiscence1 is a member of the second class of dehiscence mutants in which anthers fail to dehisce (Sanders et al., 1999). In non-dehiscence1, the anther wall layer and the connective cells degenerate and the stomium fails to break (Sanders et al., 1999). This suggests that the absence of the connective cells and the anther wall leads to a defect in the mechanical “springing” required for wall opening, in the signaling within the anther, or both (Sanders et al., 1999).
Arabidopsis anther dehiscence mutants have also been identified in screens designed to detect unrelated phenotypes. The coi1 mutant, identified by root-growth insensitivity to jasmonic acid, is defective in both pollen development and anther dehiscence (Feys et al., 1994; Dao-Xin et al., 1998). The triple fad mutant (fad3-2, fad7-2, fad8) created to be deficient in the production of trienoic acids is also defective in pollen development and anther dehiscence (McConn and Browse, 1996). Triple fad, but not coi1, can be rescued by exogenous jasmonic acid. Triple fad leads to a defect in the precursors required for jasmonic acid biosynthesis (Figure 6A); coi1, in contrast, may cause a defect in jasmonic acid perception (Dao-Xin et al., 1998). Other Arabidopsis jasmonic acid response mutants, such as jin (Berger et al., 1996) and jar (Staswick et al., 1992), and a tomato mutant, def1, which is blocked in the octadecanoid pathway (Howe et al., 1996), are fertile.
These genetic studies, along with the isolation of the DELAYED DEHISCENCE1 gene, indicate that it should be possible to identify mutants in all steps of the jasmonic acid pathway according to dehiscence-related mutant phenotypes. Similar studies have identified genes encoding brassinosteroid biosynthesis enzymes and their related mutant phenotypes (Altmann, 1998; Azpiroz et al., 1998; Choe et al., 1998, 1999a, 1999b). Mutants defective in dehiscence that cannot be rescued by jasmonic acid might identify genes downstream of the jasmonic acid “signal” (e.g., receptor genes) or genes that function independently of the jasmonic acid pathway.
Jasmonic Acid Biosynthesis Can Occur by Other Pathways
A predominant role of jasmonic acid is in the activation of signal transduction pathways in response to insect predators and pathogen attack (Wasternack and Parthier, 1997; Staswick et al., 1998; Vijayan et al., 1998). We observed a sharp increase in jasmonic acid when we wounded the leaves of wild-type plants (data not shown). Surprisingly, we also observed a slight increase in jasmonic acid after wounding in delayed dehiscence1 plants, although the level in wounded leaf tissue was at the borderline of experimental detection (data not shown).
What pathway is responsible for jasmonic acid production in wounded delayed dehiscence1 plants? Although OPR1 and OPR2 transcripts are found predominantly in the Arabidopsis root, lower levels of both transcripts are present elsewhere, including in flowers (Biesgen and Weiler, 1999). In addition, an alternative pathway has been proposed recently that may generate jasmonic acid using 16:3 trienoic acids as substrate (Weber et al., 1997; Farmer et al., 1998). These data suggest that there is an alternative source of jasmonic acid synthesis in Arabidopsis that does not rely on the 12-oxophytodienoate reductase activity of the DELAYED DEHISCENCE1–encoded protein.
Jasmonic Acid May Control the Timing of Anther Dehiscence
The chemical complementation of the delayed dehiscence1 male-sterility phenotype indicates that jasmonic acid (or a derivative), directly or indirectly, plays a role in the timing of anther dehiscence (Table 3 and Figure 7). The defect in delayed dehiscence1 anthers delays stomium degeneration, the terminal step in the anther dehiscence program. Surprisingly, late-stage anthers were not rescued by exogenous jasmonic acid application in our experiments (Figure 7B). The lag in rescue observed in our experiments suggests that exogenous jasmonic acid exerts its effect before the delayed dehiscence1 defect is apparent and that a jasmonic acid “signal” is required at a specific developmental period in the anther before anther dehiscence. Preliminary experiments treating individual, developmentally staged delayed dehiscence1 floral buds with jasmonic acid suggest that only buds containing anthers at approximately stages 10 and 11 (stage 10 is shown in Figures 9B and 9C) can be rescued and produce expanded siliques (P.Y. Lee, P.M. Sanders, and R.B.Goldberg, unpublished results). In contrast, buds with anthers at either later or earlier stages fail to respond to jasmonic acid (P.Y. Lee, P.M. Sanders, and R.B. Goldberg, unpublished results). Assuming that jasmonic acid is equally permeable at all developmental stages, these preliminary results suggest the existence of a developmental “window” in which jasmonic acid is perceived by the anther, such that target cells for the jasmonic acid signal are competent to recognize the “signal” only during this time period. This hypothesis might explain why <100% of the jasmonic acid–treated delayed dehiscence1 bud clusters (Figure 7B) were rescued in the experiments presented in Table 3. That is, only a subset of the treated buds contained anthers at stages competent to be rescued. The question remains as to how the jasmonic acid signal, when perceived at one stage in anther development, is transferred to the terminal step in anther development and results in stomium degeneration (Figures 10B and 10C).
What Is the Source of Jasmonic Acid within the Arabidopsis Flower?
If jasmonic acid is a “signal” that initiates or coordinates events during the anther dehiscence program, what is the endogenous source of jasmonic acid within the flower? One clue to answering this question can be obtained from our in situ hybridization experiments (Figure 9). In early anther development, before the differentiation of the stomium and septum and preceding the expansion of the endothecium (Sanders et al., 1999), we detected DELAYED DEHISCENCE1 mRNA throughout premeiotic anthers and in other organs of the developing floral buds (Figures 9E and 9I). By contrast, during late stages of anther development, when the dehiscence program occurs, we did not detect DELAYED DEHISCENCE1 mRNA in the stomium, septum, connective, wall layers, or epidermis of the anther (Figures 9F to 9H and 9J to 9L). Assuming that DELAYED DEHISCENCE1 mRNA is not present below the detection limit of our in situ hybridization experiments, these results suggest that the DELAYED DEHISCENCE1 gene is not expressed in the cells of the anther that play a role in dehiscence (Figures 9J to 9L). If the jasmonic acid “signal” is produced within the anther, then it must originate from jasmonic acid synthesized during the early stages of anther development.
We also observed that during the later stages of Arabidopsis flower development, DELAYED DEHISCENCE1 mRNA was present in substantial levels in the pistils, petals, and stamen filaments and that DELAYED DEHISCENCE1 mRNA levels were highest in stage 10 anthers (Figures 9F to 9H and 10C), that is, before the initiation of dehiscence program events (Figure 2). The level of DELAYED DEHISCENCE1 mRNA decreased after stage 10 and by stage 12 was also decreased within the pistil, petals, and stamen filament (Figures 9, 10B, and 10C). The accumulation profile of DELAYED DEHISCENCE1 mRNA during late floral development corresponds to the stage of anther development that can be rescued by jasmonic acid (Figure 7; data not shown). Methyl-jasmonic acid is known to exert its effect as a volatile signal in plant defense responses (Farmer and Ryan, 1990). If the DELAYED DEHISCENCE1 mRNA localization pattern predicts the expression pattern for the DELAYED DEHISCENCE1 gene (i.e., the DELAYED DEHISCENCE1–encoded 12-oxophytodienoate reductase enzyme activity), then perhaps jasmonic acid is synthesized primarily within the pistil and petals. If so, then these organs might provide the major source of “signal” for the anther within the enclosed floral bud. Alternatively, because DELAYED DEHISCENCE1 mRNA is present in stamen filaments, the filaments could also be a site of jasmonic acid synthesis and a source of the “signal” within the anther.
Exogenous jasmonic acid application is capable of restoring fertility in the delayed dehiscence1 mutant flowers during a specific period of anther development (Figure 7B). Late-stage delayed dehiscence1 floral buds were not rescued. Together, the stage of anther development responsive to the exogenous jasmonic acid and the mRNA localization pattern suggest that the jasmonic acid “signal” is required before the initiation of the anther dehiscence program (Figures 10B and 10C). Are the stomium cells the target for jasmonic acid, or is stomium cell degeneration the last step in a cascade of events initiated earlier? Our results indicate that jasmonic acid acts to time the degeneration of the stomium, but it is not known what other steps or molecules might play a part in the series of events leading to stomium degeneration.
Why Is There a Delay in Dehiscence in delayed dehiscence1 Mutants?
What causes dehiscence to be “delayed” in delayed dehiscence1 anthers? Because the T-DNA insertion knocks out the predominant 12-oxophytodienoate reductase enzyme, the level of jasmonic acid may be insufficient to “signal” the start of stomium cell degeneration at the time of wild-type anther dehiscence. In delayed dehiscence1 flowers, perhaps the threshold level of jasmonic acid is reached later in floral development. Alternative 12-oxophytodienoate reductase enzymes encoded by the other members of the OPR gene family (e.g., OPR1 and OPR2) might produce sufficient levels of jasmonic acid to allow this buildup to occur. In addition, different pathways for generating jasmonic acid in delayed dehiscence1 mutants (e.g., the hexadecanoid pathway) could also account for the “delay” observed in anther dehiscence.
The isolation of the DELAYED DEHISCENCE1 gene shows that jasmonic acid acts as a signal to control the timing of dehiscence. Our conclusion is based on the following findings: (1) a single T-DNA insertion generating a male-sterile phenotype, (2) the absence of DELAYED DEHISCENCE1 mRNA transcripts in delayed dehiscence1 inflorescence tissue, (3) the absence of DELAYED DEHISCENCE1 12-oxophytodienoate reductase enzymatic activity, and (4) the direct complementation of the delayed dehiscence1 phenotype with jasmonic acid. The anther perceives the jasmonic acid signal before the initiation of the major dehiscence program events. The precise mechanism by which jasmonic acid signaling controls the timing of anther wall breakage at the stomium remains to be determined.
METHODS
Mutant Isolation and Genetic Analysis
The male-sterile delayed dehiscence1 mutant was identified in a screen of T-DNA mutagenized Arabidopsis lines (ecotype Wassilewskija [Ws]) generated by Dr. Ken Feldmann (University of Arizona) (Feldmann and Marks, 1987; Feldmann, 1991; Forsthoefel et al., 1992). The male-sterility screen and our initial characterization of the delayed dehiscence1 phenotype were described in Sanders et al. (1999). The Feldmann line 1926 segregates for the delayed dehiscence1 phenotype and is available through the Arabidopsis Biological Resource Center (ABRC) seed catalog (http://aims.cps.msu.edu/aims).
Light Microscopy
Bright-field photographs of individual flowers were taken using a dissecting microscope (Olympus model SZH; Olympus, Lake Success, NY). Mutant and wild-type flowers were fixed overnight in FAA (3.7% formaldehyde, 50% ethanol, and 5.0% glacial acetic acid), dehydrated in a graded ethanol series (twice at 50%, 60%, 70%, 85%, and 95%; three times at 100%), embedded in Spurr's epoxy resin (Spurr, 1969; TedPella, Inc., Redding, CA), and sectioned into 1-μm-thick sections with an ultramicrotome (Sorvall model MT-600; DuPont, Wilmington, DE). Anther transverse sections were stained in 1% toluidine blue at 42°C for 1 to 2 hr. Bright-field photographs of the anther cross-sections were taken using a compound microscope (Olympus model BH2). All photographs were taken with Kodak Gold 100 film (ISO 100/21).
Transmission Electron Microscopy
Inflorescences from wild-type and delayed dehiscence1 plants were fixed, processed, and embedded in Spurr's epoxy resin as described by Owen and Makaroff (1995). Flowers were placed in individual molds before the resin blocks were cured. The floral buds were sectioned (1-μm-thick sections) with an ultramicrotome (model MT-600; Sorval) and stained in 1% toluidine blue as described above, and the region of the stomium within the sections was visualized in the light microscope. Ultrathin anther sections were then generated and placed on formavar-coated copper grids, where they were stained with lead citrate and uranyl acetate. The anther sections were observed in a JEOL electron microscope (model 100CX II; JEOL USA, Inc., Peabody, MA) at 80 kV, and pictures of the anther notch regions were taken using Kodak Electron Microscope film No. 4489.
Genomic DNA Isolation and T-DNA Insert Analysis
Leaves from individual Arabidopsis (Ws) plants were ground in liquid nitrogen, and the genomic DNA was isolated for amplification by polymerase chain reaction (PCR) (Edwards et al., 1991). The genomic DNA was amplified by PCR with T-DNA–specific primers and Taq DNA polymerase (Life Technology Gibco-BRL, Gaithersburg, MD). Two PCR primer pairs for the T-DNA construct (Feldmann and Marks, 1987) were used. Primer pair 1 amplified a 336-bp region spanning the pBR322 plasmid and right-border regions of the T-DNA (forward primer, 5′-agtgactggcgatgctgtc-3′; reverse primer, 5′-ggcggctgatacaccatc-3′). Primer pair 2 amplified an 823-bp region within the pBR322 plasmid region of the T-DNA (forward primer, 5′-tatccatcgcgtccgccatctcca-3′; reverse primer, 5′-cgccccgacacccgccaacac-3′). The annealing temperatures for the primer pairs were 60 and 56°C, respectively. The PCR profile was 25 cycles, each consisting of 94°C for 2 min, annealing temperature for 1 min, and 72°C for 1 min; for the final step, the temperature was held at 72°C for 5 min. All PCR reactions were performed in a DNA Thermal Cycler 480 (Perkin-Elmer, Branchburg, NJ).
High molecular weight genomic DNA from delayed dehiscence1 and wild-type Arabidopsis (Ws) plants was isolated by the method of Murray and Thompson (1980) for DNA gel blots and CsCl banded high molecular weight genomic DNA or that of Jofuku and Goldberg (1988) for DNA gel blots and genomic library construction. DNA (1 to 2 μg) was digested with restriction endonucleases (Life Technologies Gibco-BRL and New England Biolabs, Inc., Beverly, MA), separated by size in 0.6 or 0.8% agarose gels in 1 × TAE buffer (0.04 M Tris-acetate and 0.001 M EDTA), and transferred to Nytran membranes (Schleicher and Schuell, Keene, NH). The DNA gel blots were hybridized with T-DNA right and left border sequences (Zambryski et al., 1980) that had been labeled with 32P-dCTP by random primer synthesis (Feinberg and Vogelstein, 1983).
Genomic Library Construction and Screening
delayed dehiscence1 genomic DNA was partially digested with MboI, ligated into λGEM12 vector arms (Promega, Madison, WI), and packaged into phage (Gigapack III Gold Packaging Extract; Stratagene, La Jolla, CA) to generate a delayed dehiscence1 genomic DNA library. This library was screened with T-DNA left- and right-border probes to identify clones containing T-DNA termini. Successive rounds of screening were performed to isolate individual lambda clones. Library screening and lambda DNA isolation were performed as described by Jofuku and Goldberg (1988). The T-DNA termini clones were labeled with 32P-dCTP by random primer synthesis (Feinberg and Vogelstein, 1983) and individually hybridized to Arabidopsis wild-type genomic DNA gel blots to determine which clones contained flanking plant sequences. One clone, phage 46, contained a 974-bp plant sequence flanking the T-DNA left border. The DNA fragment containing this sequence was recloned into the plasmid pBCKS(+) (Stratagene). The 974-bp plant sequence was used to screen for the corresponding sequence in an Arabidopsis (Ws) wild-type genomic library (in the phage vector λGEM11 [Promega]; a gift from Dr. Ken Feldmann, University of Arizona). An 18-kb genomic DNA insert from an isolated wild-type phage was subcloned into a modified version of pBluescript KS+. The Stratagene pBluescript KS+ was modified by replacing the SmaI-PstI restriction enzyme sites within the multiple cloning region with a linker containing SfiI-NdeI-SfiI-AscI restriction enzyme sites.
Comparison of Mutant and Wild-Type Genomic DNA Surrounding the T-DNA Insertion Site
PCR primers designed from the 3′ end of the DELAYED DEHISCENCE1 gene were used to amplify a 2.7-kb genomic DNA region from wild type and delayed dehiscence1. The two primers were 1P (5′-gtcggtttgggtttacaaaagag-3′) and 2P (5′-tagttccttattgaatcctccactg-3′); their positions are shown in Figure 4A. The PCR profile used was 25 cycles of 94°C for 2 min, 60°C for 1 min, and 72°C for 1 min, followed by a final 72°C for 5 min.
Wild-type and delayed dehiscence1 genomic (2 μg) DNAs were digested with NcoI and AatII restriction enzymes, separated by size on a 0.6% agarose gel in 1 × TAE buffer, and transferred to Nytran membranes. The NcoI digestion was designed to separate the plant sequence flanking the T-DNA left-border terminus from the inserted T-DNA (Figure 4A). AatII was chosen to separate the plant DNA flanking the T-DNA right-border terminus from the inserted T-DNA (Figure 4A). The NcoI DNA gel blot was hybridized with DELAYED DEHISCENCE1 exons 1 and 2, the AatII DNA gel blot with exons 3 and 4 (Figure 4A). Both probes were labeled with 32P-dCTP by random primer synthesis (Feinberg and Vogelstein, 1983).
Poly(A) mRNA Isolation, RNA Gel Blots, and cDNA Synthesis
Polysomal RNA was isolated from a developmental pool of unopened floral buds and young seedlings from wild-type Arabidopsis (Ws) plants (Cox and Goldberg, 1988). Poly(A) mRNA was isolated by using PolyA Tract magnetic beads (Promega). Floral bud and young seedling poly(A) mRNAs were used to generate double-stranded cDNAs (Marathon cDNA Kit; Clontech, Palo Alto, CA).
Poly(A) mRNA was size fractionated by electrophoresis in formaldehyde gels and blotted to Nytran membranes as outlined in Koltunow et al. (1990). The RNA gel blots were hybridized with either labeled DELAYED DEHISCENCE1 cDNA or the 18-kb genomic sequence containing the DELAYED DEHISCENCE1 gene (Cox and Goldberg, 1988). Each probe was labeled with 32P-dCTP by random primer synthesis (Feinberg and Vogelstein, 1983).
Amplification of 5′ and 3′ Products of Rapid Amplification of cDNA Ends
Gene-specific primers were designed to identify 5′ and 3′ rapid amplification of cDNA ends (RACE) products. cDNA products were generated from wild-type Arabidopsis (Ws) inflorescence and young seedling double-stranded cDNAs. Plant sequences flanking the T-DNA generated the 3′ cDNA (3′RACE) primer, 5′-tctgttttcttcttacaagatgggaagattcg-3′. Plant sequences in DELAYED DEHISCENCE1 exon 4 generated the 5′ cDNA (5′RACE) primer, 5′-gca-ataatcctccaccacccgaggtatc-3′. The RACE primers were designed to match the Marathon (Clontech) cDNA amplification protocol specifications. The 3′ cDNAs were reamplified, gel-purified (GeneClean; Bio101, Vista, CA), and cloned into the pCR2.1 vector (TA Cloning; Invitrogen, Carlsbad, CA).
In Situ Hybridization Studies
Inflorescences from wild-type Arabidopsis (Ws) and delayed dehiscence1 plants were fixed, processed, and sectioned as described previously (Cox and Goldberg, 1988; Yadegari et al., 1994; Beals and Goldberg, 1997). The synthesis of single-stranded labeled RNA probes, in situ hybridization, slide washing, and exposure of the Kodak NTB-2 emulsion were performed as described by Yadegari et al. (1994) and Beals and Goldberg (1997). RNAs were labeled with 33P-dUTP (Beals and Goldberg, 1997). In situ hybridization was performed with an anti-sense probe generated from the DELAYED DEHISCENCE1 3′ cDNA clone. Sense and anti-sense 33P-rRNA probes were used as controls (Delsney et al., 1983). Bright-field and dark-field photographs were taken using an Olympus compound microscope (model BH2) with Kodak Gold 100 ASA film (ISO 100/21). The photographs were digitized by using the Adobe Photoshop (Adobe Systems Inc., San Jose, CA). The KPT-Equilizer plug-in (Metacreations Corp., Carpinteria, CA) was used to adjust the digital image to obtain the best resolution of silver grains.
Assay of 12-Oxophytodienoate Reductase Activity
DELAYED DEHISCENCE1 and wild-type plants at the rosette stage were assayed for 12-oxophytodienoate reductase activity. The DELAYED DEHISCENCE1 plant population was obtained from the sowing of self seed collected on mutant plants after jasmonic acid application and rescue. Crude protein extracts (250 μg) (extracted with 50 mM potassium phosphate buffer, pH 7.5; 100,000g supernatant) were incubated for 35 min with 5 μg of racemic cis-12-oxophytodienoic acid (OPDA) (34 μM) or 5 μg of cis (+)-OPDA and 1 mM NADPH in 0.5 mL of 50 mM potassium phosphate buffer, pH 7.5, at 30°C. The reaction was stopped by adjusting the solution to pH <3.0 with 1 M HCl and extracted with 2 volumes of ethyl acetate. Extracted OPDA and 3-oxo-2-(2′-Z-pentenyl)-cyclopentane-1-octanoic acid (OPC-8:0) were quantified as described by Schaller and Weiler (1997b). The experiment was repeated five times; the data reported are an average of the generated results. Racemic cis-OPDA was synthesized as described by Laudert et al. (1997). The (+)-enantiomer of cis-OPDA, 9S,13S-OPDA, was prepared as described by Schaller et al. (1998). Enantiomeric excess (of the 9S,13S-enantiomer over the 13R,13R-enantiomer) was 99%.
DNA Sequence Analysis
DNA was sequenced either at the UCLA Life Sciences Sequencing Facility or at Ceres, Inc. (Malibu, CA). Sequencing was initiated by commercial or gene-specific primers using ABI PCR protocols for the sequencing of plasmid templates (Perkin-Elmer/Roche Molecular, Branchburg, NJ). For the wild-type DELAYED DEHISCENCE1 18-kb genomic clone, templates were prepared using the Genome Priming System (New England BioLabs Inc., Beverly, MA). Compilation and analysis of DNA sequence data were performed with the Genetics Computer Group Wisconsin programs. Open reading frames and exon–intron junctions were identified by using NetPlantGene (Hebsgaard et al., 1996; http://genome.cbs.dtu.dk/services/NetPGene) and GenScan-MIT (http://ccr-081.mit.edu/GENSCANW.html) and were confirmed by comparing gene and cDNA sequences. DNA sequence comparisons were performed using the NCBI GenBank BLAST programs (Altschul et al., 1990, 1997; http://www.ncbi.nlm.nih.gov/). DNA and protein alignments were performed using the ClustalW program (http://bic.nus.edu.sg:8888/clustalw/clustalw.html). MacBoxShade was used to shade identical residues in the alignments (Figure 5; http://helix.nih.gov/science/boxshade.html). Primers were designed by use of Primer3 (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3.cgi).
Whole Inflorescence Jasmonic Acid Rescue Experiments
Inflorescences of recently flowering delayed dehiscence1 and wild-type plants were tagged, and all flowers with emerged petals were removed. Inflorescences were given one of three treatments—no treatment, water, or 2.0 μmol/mL jasmonic acid (J-2500; Sigma, St. Louis, MO)—twice a day (morning and night) by applying water or jasmonic acid solution to floral buds from 15 inflorescences with a small paint brush (capacity ∼800 μL). The plants were grown in a growth cabinet (16-hr-light/8-hr-dark at 24°C) for 6 days, after which treatment was discontinued. This procedure was modified from that described by McConn and Browse (1996). The inflorescences that were tagged but not treated acted as a control for the experiment. After 6 days (experiment 1) or 8 days (experiment 2), the number of expanded siliques was counted for each tagged inflorescence. Seeds from the siliques identified on delayed dehiscence1 and wild-type plants were collected and sown for analysis of fertility phenotype.
Acknowledgments
We thank Dr. Ken Feldmann (Ceres, Inc.) for allowing us to screen his Arabidopsis T-DNA lines. We thank Dr. Koen Weterings (UCLA) for his help and interaction on this project. We greatly appreciate the help of Drs. Yu Ping Lu and Ya Ping Li (Ceres, Inc.) in sequencing the 18-kb genomic clone containing the DELAYED DEHISCENCE1 gene, and we are grateful to Dr. Jon Donson (Ceres, Inc.) for a set of T-DNA primers. We also thank Margaret Kowalczyk and Brandon Le (UCLA) for help in preparing figures for this paper. P.Y.L. was supported by a Howard Hughes Medical Institute grant through the UCLA Undergraduate Biological Sciences Education Program. This research was funded by a National Science Foundation grant to R.B.G.
References
- Altmann, T. (1998). Recent advances in brassinosteroid molecular genetics. Curr. Opin. Plant Biol. 1, 378–383. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiroz, R., Wu, Y., LoCascio, J.C., and Feldmann, K.A. (1998). An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell 10, 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beals, T.P., and Goldberg, R.B. (1997). A novel cell ablation strategy blocks tobacco anther dehiscence. Plant Cell 9, 1527–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer, F.J., and Medford, J.I. (1992). A plasmid rescue technique for the recovery of plant DNA disrupted by T-DNA insertion. Plant Mol. Biol. Rep. 10, 190–194. [Google Scholar]
- Berger, S., Bell, E., and Mullet, J.E. (1996). Two methyl jasmonate–insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol. 111, 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesgen, C., and Weiler, E.W. (1999). Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10,11-reductases. Planta 208, 155–165. [DOI] [PubMed] [Google Scholar]
- Bonner, L.J., and Dickinson, H.G. (1989). Anther dehiscence in Lycopersicon esculentum Mill. I. Structural aspects. New Phytol. 113, 97–115. [DOI] [PubMed] [Google Scholar]
- Choe, S., Dilkes, B.P., Fujioka, S., Takatsuto, S., Sakurai, A., and Feldmann, K.A. (1998). The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10, 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, S., Noguchi, T., Fujioka, S., Takatsuto, S., Tissier, C.P., Gregory, B.D., Ross, A.S., Tanaka, A., Yoshida, S., Tax, F.E., and Feldmann, K.A. (1999. a). The Arabidopsis dwf7/ste1 mutant is defective in the delta7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell 11, 207–221. [PMC free article] [PubMed] [Google Scholar]
- Choe, S., Dilkes, B.P., Gregory, B.D., Ross, A.S., Yuan, H., Noguchi, T., Fujioka, S., Takatsuto, S., Tanaka, A., Yoshida, S., Tax, F.E., and Feldmann, K.A. (1999. b). The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 119, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, K.H., and Goldberg, R.B. (1988). Analysis of plant gene expression. In Plant Molecular Biology: A Practical Approach, C.H. Shaw, ed (Oxford, UK: IRL Press), pp. 1–34.
- Dao-Xin, X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- D'Arcy, W.G., Keating, R.C., and Buchmann, S.L. (1996). The calcium oxalate package or so-called resorption tissue in some angiosperm anthers. In The Anther: Form, Function and Phylogeny, W.G. D'Arcy and R.C. Keating, eds (Cambridge, UK: Cambridge University Press), pp. 159–191.
- Dawson, J., Wilson, Z.A., Aarts, M.G.M., Braithwaite, A.F., Briarty, L.G., and Mulligan, B.J. (1993). Microspore and pollen development in six male-sterile mutants of Arabidopsis thaliana. Can. J. Bot. 71, 629–638. [Google Scholar]
- Delsney, M., Cooke, R., and Penon, P. (1983). Sequence heterogeneity in radish nuclear ribosomal RNA genes. Plant Sci. Lett. 30, 107–119. [Google Scholar]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer, E.E., and Ryan, C.A. (1990). Interplant communication: Airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc. Natl. Acad. Sci. USA 87, 7713–7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer, E.E., Weber, H., and Vollenweider, S. (1998). Fatty acid signaling in Arabidopsis. Planta 206, 167–174. [DOI] [PubMed] [Google Scholar]
- Feinberg, A.P., and Vogelstein, B. (1983). A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132, 6–13. [DOI] [PubMed] [Google Scholar]
- Feldmann, K.A. (1991). T-DNA insertion mutagenesis in Arabidopsis: Mutational spectrum. Plant J. 1, 71–82. [Google Scholar]
- Feldmann, K.A., and Marks, M.D. (1987). Agrobacterium-mediated transformation of germinating seeds of Arabidopsis thaliana: A non-tissue culture approach. Mol. Gen. Genet. 208, 1–9. [Google Scholar]
- Feys, B.J., Benedetti, C.E., Penfold, C.N., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel, N.R., Wu, Y., Schulz, B., Bennett, M.J., and Feldmann, K.A. (1992). T-DNA insertion mutagenesis in Arabidopsis: Prospects and perspectives. Aust. J. Plant Physiol. 19, 353–366. [Google Scholar]
- Goldberg, R.B., Beals, T.P., and Sanders, P.M. (1993). Anther development: Basic principles and practical applications. Plant Cell 5, 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, R.B., Sanders, P.M., and Beals, T.P. (1995). A novel cell-ablation strategy for studying plant development. Philos. Trans. R. Soc. London B 350, 5–17. [DOI] [PubMed] [Google Scholar]
- Hebsgaard, S.M., Korning, P.G., Tostrup, N., Engelbrecht, J., Rouze, P., and Brunak, S. (1996). Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 24, 3439–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner, H.T., and Wagner, B.L. (1992). Association of four different calcium crystals in the anther connective tissue and hypodermal stomium of Capsicum annuum (Solanaceae) during microsporogenesis. Am. J. Bot. 79, 531–541. [Google Scholar]
- Howe, G.A., Lightner, J., Browse, J., and Ryan, C.A. (1996). An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8, 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku, K.D., and Goldberg, R.B. (1988). Analysis of plant gene structure. In Plant Molecular Biology: A Practical Approach, C.H. Shaw, ed (Oxford, UK: IRL Press), pp. 37–66.
- Keijzer, C.J. (1987). The process of anther dehiscence and pollen dispersal: 1. The opening mechanism of longitudinally dehiscing anthers. New Phytol. 105, 487–498. [DOI] [PubMed] [Google Scholar]
- Koltunow, A.M., Truettner, J., Cox, K.H., Wallroth, M., and Goldberg, R.B. (1990). Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2, 1201–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudert, D., Hennig, P., Stelmach, B.A., Müller, A., Andert, L., and Weiler, E.W. (1997). Analysis of 12-oxophytodienoic acid enantiomers in biological samples by capillary chromatography–mass spectrometry using cyclodextrin stationary phases. Anal. Biochem. 246, 211–217. [DOI] [PubMed] [Google Scholar]
- McConn, M., and Browse, J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, M.G., and Thompson, W.F. (1980). Rapid isolation of high-molecular-weight plant DNA. Nucleic Acids Res. 8, 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen, H.A, and Makaroff, C.A. (1995). Ultrastructure of microsporogenesis and microgametogenesis in Arabidopsis thaliana (L.) Heynh. ecotype Wassilewskija (Brassicaceae). Protoplasma 185, 7–21. [Google Scholar]
- Park, S.K., Yoon, Y.H., Kim, B.C., Hwang, Y.H., Chung, I.K., Nam, H.G., and Kim, D.U. (1996). Pollen of a male-sterile mutant of Arabidopsis thaliana isolated from a T-DNA insertion pool is not effectively released from the anther locule. Plant Cell Physiol. 37, 580–585. [DOI] [PubMed] [Google Scholar]
- Sanders, P.M., Bui, A.Q., Weterings, K., McIntire, K.N., Hsu, Y.C., Lee, P.Y., Truong, M.T., Beals, T.P., and Goldberg, R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant. Reprod. 11, 297–322. [Google Scholar]
- Schaller, F., and Weiler, E.W. (1997. a). Enzymes of octadecanoid biosythesis in plants: 12-Oxo-phytodienoate 10,11-reductase. Eur. J. Biochem. 245, 294–299. [DOI] [PubMed] [Google Scholar]
- Schaller, F., and Weiler, E.W. (1997. b). Molecular cloning and characterization of 12-oxophytodienoate reductase, an enzyme of the octadecanoid signaling pathway from Arabidopsis thaliana: Structural and functional relationship to yeast old yellow enzyme. J. Biol. Chem. 272, 28066–28072. [DOI] [PubMed] [Google Scholar]
- Schaller, F., Hennig, P., and Weiler, E.W. (1998). 12-Oxophytodienoate-10,11 reductase: Occurrence of two isoenzymes of different specificity against stereoisomers of 12-oxophytodienoic acid. Plant Physiol. 118, 1345–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurr, A.H. (1969). A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26, 31–43. [DOI] [PubMed] [Google Scholar]
- Staswick, P.E., Su, W., and Howell, S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89, 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Yuen, G.Y., and Lehman, C.C. (1998). Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 15, 747–754. [DOI] [PubMed] [Google Scholar]
- Vick, B.A., and Zimmerman, D.C. (1984). Biosynthesis of jasmonic acid by several plant species. Plant Physiol. 75, 458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan, P., Shockey, J., Levesque, C.A., Cook, R.J., and Browse, J. (1998). A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 7209–7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C., and Parthier, B. (1997). Jasmonate-signalled plant gene expression. Trends Plant Sci. 2, 302–307. [Google Scholar]
- Weber, H., Vick, B.A., and Farmer, E.E. (1997). Dinor-oxo-phytodienoic acid: A new hexadecanoid signal in the jasmonate family. Proc. Natl. Acad. Sci. USA 94, 10473–10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler, E.W. (1997). Octadecanoid-mediated signal transduction in higher plants. Naturwissenschaften 84, 340–349. [Google Scholar]
- Yadegari, R., de Paiva, G.R., Laux, T., Koltunow, A.M., Apuya, N., Zimmerman, J.L., Fischer, R.L., Harada, J.J., and Goldberg, R.B. (1994). Cell differentiation and morphogenesis are uncoupled in Arabidopsis raspberry embryos. Plant Cell 6, 1713–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambryski, P., Holsters, M., Kruger, K., Depicker, A., Schell, J., Montagu, M.V., and Goodman, H.M. (1980). Tumor DNA structure in plant cells transformed by A. tumefaciens. Science 209, 1385–1391. [DOI] [PubMed] [Google Scholar]