Abstract
Although the physiological functions of phytochrome A (PhyA) are now known, the distribution of endogenous PhyA has not been examined. We have visualized endogenous PhyA apoprotein (PHYA) by immunolabeling cryosections of pea tissue, using PHYA-deficient mutants as negative controls. By this method, we examined the distribution of PHYA in different tissues and changes in its intracellular distribution in response to light. In apical hook cells of etiolated seedlings, PHYA immunolabeling was distributed diffusely in the cytosol. Exposure to continuous far-red (cFR) light caused a redistribution of the immunolabeling to the nucleus, first detectable after 1.5 hr and greatest at 4.5 hr. During this time, the amounts of spectrally active phytochrome and PHYA did not decline substantially. Exposure to continuous red (cR) light or to a brief pulse of red light also resulted in redistribution of immunolabeling to the nucleus, but this occurred much more rapidly and with a different pattern of intranuclear distribution than it did in response to cFR light. Exposures to cR light resulted in loss of immunolabeling, which was associated with PHYA degradation. These results indicate that the light-induced intracellular location of PHYA is wavelength dependent and imply that this is important for PhyA activity.
INTRODUCTION
The phytochrome family of plant photoreceptors regulates various molecular and cellular processes of plant development in response to the light environment (Quail et al., 1995). Phytochromes are soluble chromoproteins that convert photoreversibly between two spectrally distinct forms when sequentially absorbing red (R) and far-red (FR) light, and this interconversion occurs immediately both in vivo and in vitro (Butler et al., 1959). Phytochromes are encoded by a small gene family (phytochrome genes PHYA to PHYE in Arabidopsis; Sharrock and Quail, 1989; Clack et al., 1994). Studies with mutants deficient in specific phytochromes have shown that phytochrome A (PhyA) and phytochrome B (PhyB) have distinct action spectra for the photoinduction of seed germination (Shinomura et al., 1996) and distinct fluence and wavelength requirements for expression of the chlorophyll a/b binding protein gene (CAB) (Hamazato et al., 1997). The fundamental molecular basis for these differences is of great interest but has not been elucidated. Recent genetic and molecular analyses have defined differences in PhyA and PhyB activities with respect to interacting factors and signaling intermediates. Those studies suggest that PhyA and PhyB signals are transduced by overlapping signal transduction pathways (Deng and Quail, 1999).
To complement such approaches, one must also consider the ways in which the concentration, photochemical activity, and localization of each phytochrome are regulated in tissues and cells (Pratt, 1994). In tissues, this regulation may affect the transmission of the light signal from the site of photoperception to the responsive organ. An analysis of the way in which photoreceptors are redistributed within the cell in response to light treatment may provide information about the early steps in signal transduction (Sakamoto and Nagatani, 1996; Yamaguchi et al., 1999). The distribution of phytochrome in plants has been analyzed previously by various methods, including spectrophotometry (Furuya and Hillman, 1964), microbeam irradiation (Haupt, 1970), immunochemistry (Pratt, 1994), and techniques using reporter genes (Komeda et al., 1991; Adam et al., 1994, 1996; Somers and Quail, 1995). Each of these techniques has certain advantages and limitations (see Pratt, 1994; Nagatani, 1997).
Several early immunochemical studies used anti-phytochrome antibodies for immunohistochemical visualization of the phytochrome apoprotein in situ. In etiolated monocot seedlings, phytochrome was found to be most abundant in the apex and was also detected in the apical cells of roots (Pratt and Coleman, 1971, 1974). In etiolated seedlings of dicot species, phytochrome was most abundant in the subepidermal cortical cells of the hook region in etiolated seedlings of pea and soybean (Saunders et al., 1983; Cope and Pratt, 1992). Phytochrome was also detectable in the ovular tissue in the developing embryo of peanut (Thompson et al., 1992). The subcellular localization of phytochrome was also examined in several of these studies. In completely etiolated seedlings, phytochrome appeared to be diffusely distributed throughout the cytosol (Pratt and Coleman, 1974). When etiolated seedlings were briefly exposed to R light, the phytochrome became localized to small areas within the cytosol (Mackenzie et al., 1975). However, these findings must be reexamined with methods that detect phytochrome unambiguously and discriminate among the different phytochrome species.
Reporter protein techniques provide one convenient approach for investigating the distribution of proteins in vivo. For example, translational fusions with β-glucuronidase (GUS) or the green fluorescent protein (GFP) have been widely used to examine the subcellular localization of various proteins (Restrepo et al., 1990; Chalfie et al., 1994). This technique has been applied in the study of phytochrome distribution by Sakamoto and Nagatani (1996), who fused fragments of Arabidopsis PhyB apoprotein (PHYB) to a GUS reporter protein in transgenic Arabidopsis plants. Histochemical staining of GUS activity indicated nuclear localization of the fusion protein. More recently, a phenotypic analysis of transgenic Arabidopsis plants expressing a fusion of full-length Arabidopsis PHYB and GFP showed that the fusion protein was both spectrally and biologically functional in vivo and accumulated in the nucleus in a light-dependent manner (Yamaguchi et al., 1999). A parsley PhyA apoprotein (PHYA)–GFP fusion transiently expressed in parsley protoplasts was constitutively cytosolic (Kircher et al., 1999a), whereas both biologically active rice PhyA–GFP and tobacco PhyB–GFP expressed in transgenic tobacco plants showed nuclear import dependent on the quality of the light (Kircher et al., 1999b).
Because GFP fluorescence does not require any cofactor or substrate, it can be used to observe the subcellular localization of fusion proteins in vivo. However, during such fusion protein studies, one often has to use a strong, nonnative promoter, such as cauliflower mosaic virus 35S, to detect clear labels from the reporter. This introduces the possibility that the distribution of the reporter labeling may differ from that of the authentic protein (Fukuda et al., 1997; Kircher et al., 1999a). Moreover, the stability of the reporter protein may differ from that of the target protein, so that the effect of rapid turnover of the target protein may not be detectable (Li et al., 1998). In contrast, immunohistochemical and immunocytochemical techniques allow direct examination of the native distribution of endogenous proteins. To date, however, these techniques have not been used for specific phytochromes, largely because of the requirement for both phytochrome type–specific monoclonal antibodies and the corresponding phytochrome apoprotein–deficient mutants to serve as negative controls.
In this study, we have analyzed the tissue-specific distribution and subcellular localization of PHYA in etiolated wild-type pea seedlings by using immunohistochemical analysis and monoclonal anti-phytochrome antibodies (Nagatani et al., 1984, 1987; Abe et al., 1985; Lumsden et al., 1985; Shinomura et al., 1996) and the PHYA-deficient fun1-1 (FR unresponsive) mutant (Weller et al., 1997) as a negative control. Our results definitively demonstrate that native PHYA is redistributed to the nucleus in response to irradiation with continuous R or FR light. We also show that the kinetics of PHYA import and the pattern of distribution within the nucleus differ under R and FR light.
RESULTS
Specific Detection of PHYA by Immunohistochemical Techniques
From our library of anti-phytochrome antibodies, we tested 25 monoclonal anti-PHYA antibodies for immunogold labeling of cryosections from wild-type pea and the PHYA-deficient fun1-1 mutant. This test distinguished three groups of monoclonal antibodies (Table 1). Nine monoclonal antibodies showed PHYA-specific binding in the wild type but not in the PHYA-deficient mutant (fun1-1) on cryosections from the hook region of 5-day-old etiolated seedlings; moreover, all of these gave a similar staining pattern. The specificity of selected antibodies and reproducibility of the labeling were confirmed with a second mutant (fun1-2; data not shown). Three monoclonal antibodies produced immunogold labeling in both the wild type and fun1-1, although the intensity of the label of the wild type was stronger than that of fun1-1 in all cases. The remaining 13 monoclonal antibodies did not show any label bound to the sample sections. On the basis of these results, we selected the anti-PHYA antibody mAA01, raised against Arabidopsis PHYA, for use in all subsequent experiments because it showed the greatest affinity for PHYA in the cryosections.
Table 1.
Determining the Specificity of Various Monoclonal Anti-PHYA Antibodies by Immunohistochemistry on Cryosections of Etiolated Pea Seedlings
| Immunogold Labelinga
|
||
|---|---|---|
| Wild Type | PHYA-Deficient Mutant fun1-1 | Monoclonal Anti-PHYA Antibodiesb |
| + | − | mAA01, mAP09, mAP10, mAP18, mAP20, mAP21, mAP31, mAP35, mAR08, |
| + | + | mAA02, mAP05, mAP23 |
| − | + | Nil |
| − | − | mAP13, mAP14, mAP16, mAP19, mAP22, mAP25, mAP28, mAP29, mAP30, mAP32, mAP33, mAP34, mAR07 |
Immunogold labeling was observed using bright-field microscopy. Cryosections of the hook region of 5-day-old wild-type and mutant pea seedlings were treated with each of the monoclonal antibodies and gold-conjugated anti–mouse IgG. The immunogold labeling was enhanced by silver staining. (+), label was detectable; (−), label was undetectable.
mAA, anti–Arabidopsis PHYA monoclonal antibody; mAP, anti–pea PHYA monoclonal antibody; mAR, anti–rye PHYA monoclonal antibody.
Detection of PHYA in Developing Seedlings Grown in Darkness
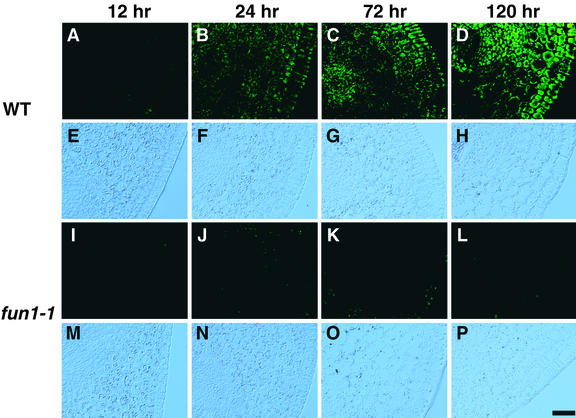
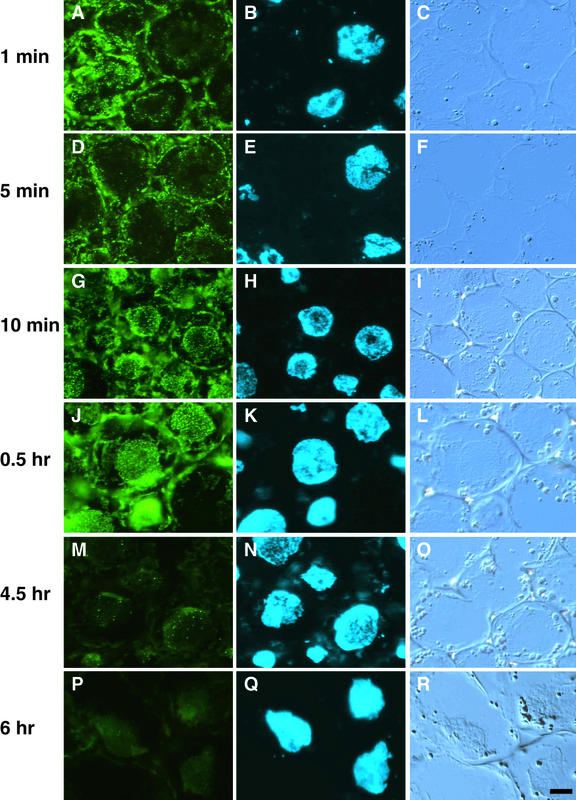
We first monitored the increase in immunochemically detectable PHYA during germination of pea seeds in darkness (Figure 1). PHYA-associated fluorescein isothiocyanate (FITC) labeling was not detectable in embryos for the first 12 hr after the onset of imbibition (Figure 1A). During this time, no morphological or histological signs of germination were apparent. By 24 hr, a faint FITC labeling was observable in the hook region of the germinating embryo (Figure 1B). The intensity of FITC labeling was stronger but still relatively weak after 48 hr of imbibition, and some variation in intensity of the labeling was seen among sections from different plants (data not shown). At this time, the radicle was <10 mm long and the plumule had not yet emerged from the seed coat. At 72 hr, the intensity of FITC labeling in the hook region had increased markedly (Figure 1C). In these seedlings, epicotyls were ∼8 mm long, and roots extended between 20 and 45 mm. Uneven distribution of PHYA among different tissues within the epicotyl was clearly observed during the period from 72 to 120 hr (Figures 1C and 1D). Using an identical procedure for immunostaining and the same period of exposure with the charge-coupled device camera, we detected no nonspecific fluorescence in the fun1-1 mutant (Figures 1I to 1L).
Figure 1.
Detection of PHYA during Early Development of Pea Seedlings in Darkness.
Distribution of PHYA in the hook region of embryos and seedlings was visualized by immunofluorescence of FITC using monoclonal antibody mAA01. The content of immunochemically detectable PHYA increases from the onset of imbibition, and (A) to (D) and (I) to (L) show epifluorescence microscopy of FITC. The positions of the tissue sections in (A) to (D) and (I) to (L) are evident from Nomarski differential interference contrast images in (E) to (H) and (M) to (P).
(A) and (E) Wild type after imbibition for 12 hr.
(B) and (F) Wild type after imbibition for 24 hr.
(C) and (G) Wild type after imbibition for 72 hr.
(D) and (H) Wild type after imbibition for 120 hr.
(I) and (M) fun1-1 after imbibition for 12 hr.
(J) and (N) fun1-1 after imbibition for 24 hr.
(K) and (O) fun1-1 after imbibition for 72 hr.
(L) and (P) fun1-1 after imbibition for 120 hr.
WT, wild type. 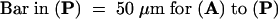 .
.
Tissue-Specific Distribution of PHYA in Etiolated Seedlings
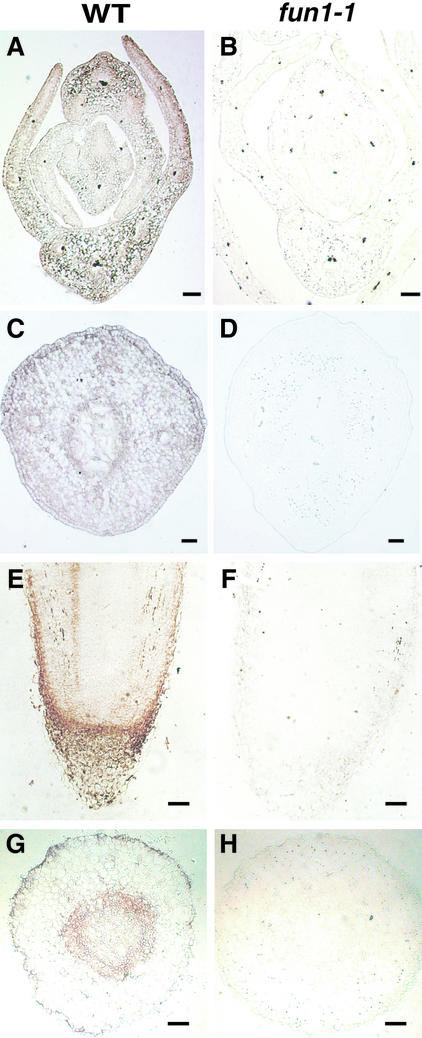
To examine the distribution of PHYA in different organs and tissues of 5-day-old etiolated seedlings, we used immunogold labeling (Figure 2). Immunogold labeling (which was enhanced with silver) was abundant in primary leaves and decreased toward the apical meristem (Figure 2A). The intensity of immunogold labeling was stronger in the epidermis than in the mesophyll and was weaker in vascular tissue in leaves (Figure 2A). In the epicotyl hook, the epidermis and inner abutting cells were stained more deeply than other regions (Figure 2C). The immunogold labeling decreased in intensity toward the inner side of the cortex. Although xylem was not stained, a cell layer a few cells wide surrounding the vascular tissue was deeply stained (Figure 2C). This included the innermost layer of the cortex, the endodermis, the pericycle, and phloem elements.
Figure 2.
Distribution of PHYA in Etiolated Pea Seedlings.
Tissue-specific distribution of PHYA in 5-day-old seedlings was visualized by silver-enhanced immunogold labeling with monoclonal antibody mAA01. The label appears as a brownish color. (A), (C), (E), and (G) show the wild type (WT); (B), (D), (F), and (H) show the PHYA-deficient mutant (fun1-1).
(A) and (B) are cross-sections of the shoot apex.
(C) and (D) are cross-sections of the hook.
(E) and (F) are longitudinal sections of the root tip.
(G) and (H) are cross-sections of root tip.
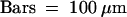 .
.
In roots, the greatest intensity of immunogold labeling was observed in the root cap and the epidermis of the root tip (Figure 2E). Younger root cap cells were deeply stained, but newly developed cortex cells were only weakly stained, even though both cells are adjacent to the root apical meristem. In the cortex of the root, the region close to the meristem showed relatively strong staining, but the underlying region of cells undergoing vacuolization was distinguished by much weaker labeling. As occurred in the hook region of the epicotyl, an inner cell layer a few cells wide surrounding the vascular tissue was also deeply stained (Figure 2G). The vascular tissue itself was weakly stained with label. In all cases, no nonspecific immunogold labeling was detected in the fun1-1 mutant with the use of identical procedures for staining and observation (Figures 2B, 2D, 2F, and 2H).
Light-Induced Changes in Subcellular Localization of PHYA
The immunofluorescence detection method also enabled us to detect PHYA within single cells by microscopy. This allowed us to investigate the effect of different irradiation conditions on the subcellular localization of PHYA. The distribution of FITC-labeled PHYA was examined by using fluorescent microscopy and Nomarski differential interference contrast images (Figures 3 to 5). The position of the nuclei in the images was determined by using Hoechst33258 label, which fluoresces at a wavelength distinct from FITC (Figures 3 to 5).
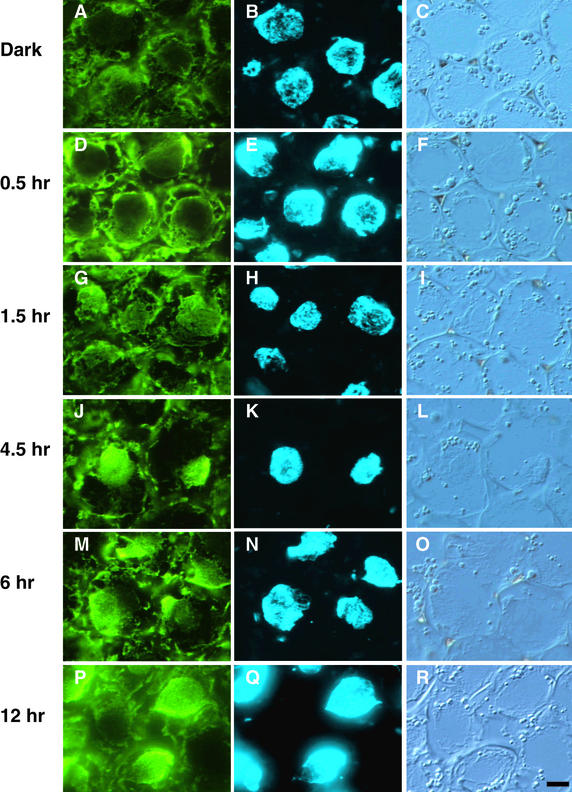
Figure 3.
cFR Light–Induced Nuclear Localization of PHYA.
The subcellular localization of PHYA in wild-type pea hook cells exposed to cFR light was visualized by immunofluorescence of FITC using monoclonal antibody mAA01. (A), (D), (G), (J), (M), and (P) show epifluorescence images of FITC; the nuclei in (A), (D), (G), (J), (M), and (P) were visualized with epifluorescence images of H33258 in (B), (E), (H), (K), (N), and (Q), respectively. The positions of these respective cells are evident from Nomarski differential interference contrast images (C), (F), (I), (L), (O), and (R).
(A) to (C) Dark control.
(D) to (F) After 0.5 hr under cFR illumination.
(G) to (I) After 1.5 hr under cFR illumination.
(J) to (L) After 4.5 hr under cFR illumination.
(M) to (O) After 6 hr under cFR illumination.
(P) to (R) After 12 hr under cFR illumination.
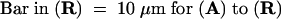 .
.
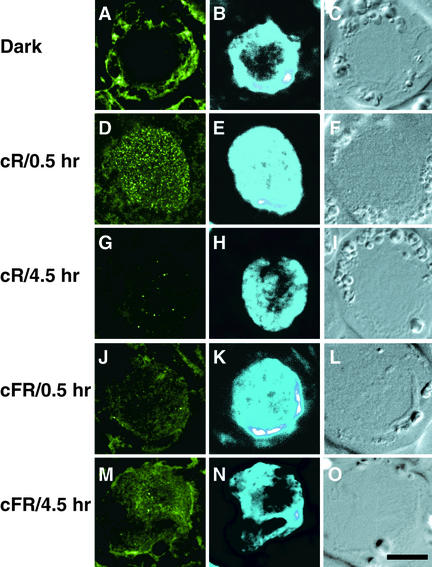
Figure 5.
Intranuclear Localization of PHYA.
PHYA-associated FITC labeling in optical sections taken 1.0 μm from the nuclei was visualized with confocal laser scanning microscopy in (A), (D), (G), (J), and (M). The nuclei of these cells are evident from epifluorescence microscopy of H33258 images in (B), (E), (H), (K), and (N), respectively. The positions of these respective cells are evident from Nomarski differential interference contrast images (C), (F), (I), (L), and (O).
(A) to (C) Dark control.
(D) to (F) After 0.5 hr under cR illumination.
(G) to (I) After 4.5 hr under cR illumination.
(J) to (L) After 0.5 hr under cFR illumination.
(M) to (O) After 4.5 hr under cFR illumination.
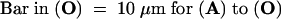 .
.
In apical hook cells of 5-day-old etiolated seedlings, PHYA was diffusely distributed in the cytosol (Figure 3A). However, when etiolated seedlings of the same age were exposed to continuous far-red (cFR) or continuous red (cR) light, a strong PHYA-associated FITC labeling appeared in the nucleus (Figures 3J, 3K, 4G, and 4H). At that time, FITC labeling in the cytosol was still detectable. However, the kinetics of this redistribution differed under cFR (Figure 3) and cR (Figure 4) light. Under cFR irradiation, the FITC labeling only became distinct in the nucleus after 1.5 hr (Figures 3G and 3H) and gradually accumulated in the nucleus over the next 3 hr (Figures 3J and 3K), the nuclear accumulation of PHYA being greatest after 4.5 hr (Figure 3J) and remaining at that level after 6 (Figure 3M), 12 (Figure 3P), and 24 hr (data not shown) of exposure to cFR light. In contrast, under cR irradiation, FITC labeling formed many speckles throughout the cytosol after 1 min (Figure 4A) and continued through 5 min (Figure 4D), at which time some FITC labeling appeared around the nuclei (Figures 4A, 4B, 4D, and 4E). Accumulation of FITC labeling within the nucleus was first obvious as many small speckles after 10 min in cR light (Figures 4G and 4H), and a similar distribution was observable at 20 min (data not shown), 0.5 hr (Figures 4J and 4K), and 1.5 hr (data not shown). At this time, FITC labeling showed relatively uniform distribution in the cytosol (Figures 4G and 4J). The FITC labeling in both the nucleus and the cytosol of etiolated seedlings decreased after the first 1.5 hr of cR irradiation. Weak labeling was still visible as a few speckled forms after 4.5 hr of cR light (Figure 4M) but was undetectable after cR light exposures of 6 (Figure 4P) or 12 hr (data not shown).
Figure 4.
cR Light–Induced Nuclear Localization and Degradation of PHYA.
The subcellular localization of PHYA in wild-type pea hook cells exposed to cR light was visualized by immunofluorescence of FITC using monoclonal antibody mAA01. (A), (D), (G), (J), (M), and (P) show epifluorescence images of FITC; the nuclei in (A), (D), (G), (J), (M), and (P) were visualized with epifluorescence images of H33258 in (B), (E), (H), (K), (N), and (Q), respectively. The positions of these respective cells are evident from Nomarski differential interference contrast images (C), (F), (I), (L), (O), and (R).
(A) to (C) After 1 min under cR illumination.
(D) to (F) After 5 min under cR illumination.
(G) to (I) After 10 min under cR illumination.
(J) to (L) After 0.5 hr under cR illumination.
(M) to (O) After 4.5 hr under cR illumination.
(P) to (R) After 6 hr under cR illumination.
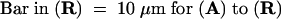 .
.
We also examined the effects of shorter irradiations with R or FR light on the subcellular change of PHYA distribution. In 5-day-old etiolated seedlings returned to darkness after irradiation with a 5-sec pulse of R light, the nuclear localization of FITC labeling showed kinetics similar to those seen in seedlings maintained under cR (Figure 4). However, no marked change of subcellular localization of FITC labeling was detected at 1.5 and 6 hr after 1 or 10 min of FR irradiation. Once again, no nonspecific FITC labeling fluorescence was detected in the fun1-1 mutant when identical procedures for staining and observation were used (data not shown).
We also examined the intranuclear distribution of PHYA, using confocal laser scanning microscopy (Figure 5). Different distribution patterns of PHYA were observed in nuclei under cR or cFR light. FITC labeling was present in the cytosol of cells from dark-grown plants (Figure 5A). After 0.5 hr of cR irradiation, the FITC labeling appeared within the nucleus as many small speckles (Figures 5D and 5E). At that time, a few speckles of the FITC labeling were also apparent in the cytosol (Figure 5D). In contrast, after 0.5 hr of cFR irradiation, weak FITC labeling and only a few speckles were observed in nuclei (Figures 5J and 5K). After 4.5 hr under cR light, only a few speckles remained in both the nuclei and the cytosol (Figure 5G). However, FITC labeling could be detected in nuclei after 4.5 hr under cFR light (Figures 5M and 5N). We observed a relatively uniform pattern of FITC labeling (Figure 5M), unlike the conspicuous speckling seen under cR light (Figure 5D). At the times when the nuclear accumulation was significant, FITC labeling in the cytosol was also detectable (Figure 5M).
Protein Gel Blot Analysis and Spectrophotometric Measurement in Vivo
To assess the effect of these light treatments on the relative amount of PHYA, immunoblotting and spectrophotometric measurements were taken. In the apical 2 to 3 cm of the epicotyl, the amount of PHYA detectable by immunoblotting did not change substantially during 12 hr of cFR irradiation (Figure 6A). However, the quantity of PHYA in the same area of the epicotyl gradually decreased during 0 to 4.5 hr of cR irradiation; after 6 hr, the label was undetectable (Figure 6A). On the basis of serial dilution of extracts, we estimated that after 4.5 hr of cR irradiation, seedlings contained <5% of the amount of PHYA seen in dark-grown seedlings. In contrast, the amount of PHYA did not change substantially during 4.5 hr of cFR irradiation (Figure 6B). Consistent with these observations, we also found that there was no significant loss of photoreversible phytochrome in comparable apical segments taken from seedlings exposed to cFR for 4 hr, whereas a marked decrease in the amount of photoreversible phytochrome was detected after 4 hr of cR irradiation (Table 2).
Figure 6.
Immunoblot Detection of PHYA.
(A) PHYA was detected by immunoblotting in wild-type seedlings after light treatment. Five-day-old etiolated seedlings were exposed to cR light (top) and cFR light (bottom) for 0.5, 1.5, 4.5, 6, or 12 hr. The shoot apex region was harvested for protein extraction. Lanes were loaded with 10 μg of protein.
(B) Extracts from seedlings irradiated for 0 hr (top), 4.5 hr in cR light (center), and 4.5 hr in cFR light (bottom) containing 10 μg of protein were serially diluted with 4 volumes of extraction buffer (final concentration per extract 2 μg of protein). All experiments were conducted with mAA01.
Table 2.
In Vivo Spectrophotometric Measurement of Phytochrome in Pea Seedlings
| Continuous Light Treatment | Duration (hr) | ΔΔ A (730–800 nm)a |
|---|---|---|
| Dark | — | 12.7 ± 0.3 |
| Far-red | 4 | 12.0 ± 0.4 |
| Red | 4 | 1.6 ± 0.2 |
n = 5.
DISCUSSION
Immunochemically Specific Detection of PHYA
Many previous studies have reported tissue or subcellular localization of phytochrome by immunostaining. In most of these studies, however, the particular molecular species of phytochrome that were stained could not be clearly defined (Pratt, 1994). The one exception to date is the report of the nuclear localization of PHYB (Sakamoto and Nagatani, 1996) in which a PHYB-deficient mutant was used as a negative control. In the earlier studies, the antibodies most likely reacted mainly to PHYA (Nagatani, 1997), because this is the predominant species of phytochrome in dark-grown plants (Somers et al., 1991; Nagatani et al., 1993; Weller et al., 1997). However, artifactual staining can often occur during the immunostaining process, and study of a mutant specifically deficient in the antigen in question is crucial to evaluate the specificity of the antibodies.
We have used a PHYA-deficient pea mutant (fun1-1) as a negative control to distinguish PHYA-specific immunolabeling from nonspecific binding of antibody and to thereby select PHYA-specific monoclonal antibodies (Table 1). We confirmed the reproducibility of the labeling with two different staining procedures, immunogold and immunofluorescence (Figures 1 and 2), and applied this technique to investigate the localization of PHYA both at the tissue level and within the cell.
Tissue-Specific Distribution of PHYA in Etiolated Seedlings
Because phytochrome is a soluble protein (Butler et al., 1959), methods of sample preparation for immunohistochemical staining should minimize the potential for movement of the phytochrome during the assay (Pratt and Coleman, 1974). Therefore, we adopted a cryosectioning method in fresh tissue. Rapid freezing of the specimen after sampling ensured that the antigen was immediately stabilized in ice. An additional advantage of this method is that sections are cut prior to fixation with aldehyde, which allows the fixation to be completed much more quickly than when intact tissue is used. A third advantage of the cryosectioning method is that it eliminates several steps needed for paraffin or plastic sectioning (dehydration with organic reagent, infiltration into plastic embedding material, and heat treatment for polymerization of the embedding material), which could potentially interfere with antigenicity (Boonstra et al., 1987). We have, therefore, assumed that this method allows detection of PHYA in its original location and without the loss of antigenicity.
The content of PHYA in cells of the epicotyl hook reached an immunohistochemically detectable level 24 hr after the onset of imbibition (Figure 1). This is consistent with previous findings that the content of PHYA in crude extract from pea embryonic axes increases during imbibition in darkness (Konomi et al., 1987). Those authors reported a 30-fold increase over the first 12 hr of imbibition and, on the basis of ELISA results, estimated the quantity at 12 hr to be ∼0.2 mg per axis of PHYA. In our study, we first detected FITC labeling associated with PHYA in the hook region of the germinating embryo after 24-hr imbibition (Figure 1B), with an increased label detected after 72 hr (Figure 1C). This indicates that the content of PHYA continued to increase between 12 and 72 hr. Synthesis of photoreversible phytochrome has been observed in germinating pea axis (McArthur and Briggs, 1970). Therefore, the increase in PHYA content revealed by immunostaining is very likely to truly reflect the synthesis of spectrally active PhyA.
In 5-day-old etiolated seedlings, PHYA was present in young leaves (Figure 2A), in the hook region of the epicotyl (Figure 2C), and in the root tip (Figures 2E and 2G). Almost all cells that we observed were stained to some extent, except for xylem elements, although the intensity of staining varied considerably. This distribution corresponds to that reported for spectrophotometrically detectable phytochrome in 3-day-old etiolated pea seedlings (Furuya and Hillman, 1964). Given a lack of evidence that any substantial quantity of nonchromophoric phytochrome is present, the pattern of PHYA distribution revealed by immunostaining very likely truly reflects the distribution of spectrally active PhyA in those organs.
The preferential distribution of PHYA in the epidermis of young leaves, the epicotyl hook region, and root tips (Figure 2) is also partially consistent with other previous reports, but some differences are apparent. For example, in tobacco, phytochrome was observed in the stem epidermis when tissue prints were immunostained (Jordan et al., 1995), whereas in dark-grown pea and soybean, phytochrome was not detected in the epidermis of the apical hook, except in the guard cells (Saunders et al., 1983; Cope and Pratt, 1992). In dark-grown seedlings of monocots, phytochrome was detected in epidermal cells of the lower region of the coleoptile and primary leaves in some tribes (Pratt and Coleman, 1971, 1974). The reason for these differences is not known, although radically different patterns of phytochrome distribution in the shoots of different grass tribes had been previously reported (Pratt and Coleman, 1974). However, other studies suggest that light perception in the epidermis may play an important role in modulating elongation growth. For example, in pea, phytochrome regulation of stem elongation may occur partly through modulation of the amount of indole-3-acetic acid in the epidermis (Behringer and Davies, 1992). Also, the elongation response of maize epidermal cells to cFR irradiation is different in the coleoptile and mesocotyl, and this difference is correlated with differences in the light-regulated reorganization of actin microfilaments (Waller and Nick, 1997).
A layer surrounding the vascular tissue—comprising the innermost layer of the cortex, the endodermis, the pericycle, and phloem elements—was deeply stained in the hook region of the epicotyl and the root tip (Figures 2C and 2G). This observation also has some precedent. Endogenous phytochrome was detected in vascular tissue of tobacco stems by immunodetection on tissue prints (Jordan et al., 1995). When PhyA was overexpressed in the vascular ring, which corresponds to phloem and the surrounding companion cells, a dwarf phenotype was elicited that was related to a reduction in gibberellin concentrations (Jordan et al., 1995). This finding was interpreted to suggest that PhyA acting in the vascular tissue might regulate stem elongation by way of affecting gibberellin metabolism or transport (Jordan et al., 1995). Our observations are consistent with the possibility that photoregulation of stem elongation may occur in the epidermis and the vascular tissue. Because PHYA is also clearly present in the epidermis and the vascular tissue of the root tip, perhaps it has a similar role in controlling root growth.
Among all tissues examined, immunogold labeling was greatest in the root cap (Figure 2E). In etiolated grass seedlings, a high concentration of phytochrome was also observed in the root cap (Pratt and Coleman, 1971, 1974). In pea, photoactive phytochrome in the root apex was detected by spectrophotometry (Briggs and Siegelman, 1965), and in dark-grown seedlings of tobacco and Arabidopsis, a PHYA promoter fused to the GUS gene was strongly active in the root tip (Adam et al., 1994; Somers and Quail, 1995). In maize, PHYA1 mRNA was abundant in the root cap, and its expression was downregulated by very-low-fluence R light (Johnson et al., 1991). These data all show that PHYA is synthesized and accumulates in the root tip, including in the cap. Several other observations suggest that the immunochemically detectable PHYA in the root tip is likely to be physiologically active. In maize, light-induced gravitropic curving in roots is a very-low-fluence response, occurring in response to R, FR, or blue light (Feldman and Briggs, 1987). This response, which is very similar to other very-low-fluence responses that are known to be mediated by PhyA (Shinomura et al., 1996), strongly implies that PhyA has a role in roots. Many aspects of root development are regulated by light, including root extension, geosensitivity, and lateral root production (Furuya and Torrey, 1964); moreover, in species that show light sensitivity in the root, the region of light perception lies within the root cap (Feldman, 1984). Light-regulated changes in mRNA expression and in the activities of proteins within the root cap have also been reported (Feldman et al., 1988). Therefore, we speculate that the root cap might be one of the sites within the root that is sensitive for PhyA activity.
Finally, we observed that in general, PHYA was more abundant in tissues at an early stage of differentiation than in the meristematic region. For example, primary leaves gave a relatively weak immunogold labeling, which became even weaker toward the meristematic region (Figure 2A). In the root, the apical meristem was also more weakly stained than was the root cap and epidermis in the root tip (Figure 2E). In previous reports, phytochrome has been shown to be most abundant in relatively young, rapidly expanding cells recently derived from meristems, rather than in meristems themselves (Pratt, 1994). Meanwhile, in the area adjacent to the meristematic region in the root, the younger root cap cells were deeply stained, but the newly developed cortex cells were weakly stained (Figure 2E). These observations, together with the heterogeneous distribution of PHYA in various tissues, suggest the possibility that the different phytochrome content in different tissues and cell types is established at an early stage of differentiation.
Light-Regulated Nuclear Import of Phytochromes
FITC labeling associated with PHYA is uniformly distributed throughout the cytoplasm in hook cells of 5-day-old pea seedlings kept in complete darkness (Figure 3A). A uniform cytoplasmic distribution of immunochemically detectable phytochrome has been reported previously for etiolated seedlings of several species (Pratt, 1994). When 5-day-old etiolated pea seedlings are exposed to cFR or cR irradiation, however, the FITC labeling in the hook cells appears in the nucleus, and the kinetics of this appearance differ according to the light treatment (Figures 3 and 4). This striking change in localization is not associated with a marked change in the amount of detectable PHYA on immunoblots (Figure 6). Thus, the decrease of PHYA in the cytoplasm after irradiation does not reflect enhanced degradation of the protein, and the appearance of PHYA in the nucleus is not the result of enhanced synthesis. Therefore, we suggest that detectable PHYA in the nucleus must reflect import to the nucleus in response to exposure to cFR or cR irradiation. In the period during which nuclear accumulation of PHYA was visible, FITC labeling was still detectable in the cytoplasm (Figures 3J, 3M, 3P, 4G, and 4J). Therefore, apparently a subpopulation of PHYA is not translocated to the nucleus, or some of the PHYA imported to the nucleus is subsequently exported—or both.
Of the three major differences between the effects of cFR and cR irradiation on the subcellular localization of PHYA, the first concerns the time at which nuclear accumulation is maximal. Time-course experiments show that nuclear localization under cR illumination is maximal after 10 to 30 min of exposure to cR light (Figure 4G) and is still visible after 1.5 hr (data not shown). In contrast, under cFR light, the nuclear localization of PHYA becomes evident only after 1.5 hr and reaches a maximum after 4.5 hr (Figures 3G and 3J).
The second difference concerns the distribution pattern of the PHYA in the nucleus at the time of maximum labeling. After 10 to 30 min of exposure to cR irradiation, FITC labeling was visualized as very dense speckling in the nucleus and somewhat less dense staining in the cytoplasm (Figures 4G, 4J, and 5D). At that time, the FITC labeling under cFR light showed cytoplasmic distribution, and only a few speckles were detected in the nucleus (Figures 3D and 5J). In contrast, after 4.5 hr of cFR exposure, FITC labeling showed a relatively uniform pattern of distribution with few speckles in the nucleus (Figures 3J and 5M). By that time, the FITC labeling under cR light was much reduced, although a few speckles were still visible (Figures 4M and 5G); they were undetectable after 6 hr under cR (Figure 4P). We did not characterize the speckling in more detail. However, its appearance was clearly dependent on light and, therefore, may represent an important event in the fate of PHYA after phototransformation.
The third difference between the effects of cFR and cR light exposure was the stability of PHYA. Nuclear accumulation of PHYA was clearly visible for at least 12 hr under cFR light (Figure 3P) without substantial changes in the quantity of detectable PHYA visible on the immunoblot (Figure 6) or of photoreversible phytochrome (Table 2). However, PHYA was visible in neither the nucleus nor the cytoplasm after exposure to 6 hr of cR light (Figure 4). The amounts of detectable PHYA on immunoblots (Figure 6) and of spectrally photoreversible phytochrome (Table 2) had decreased by that time. Therefore, we conclude that the loss of PHYA-associated FITC labeling under cR light resulted from enhanced degradation of the apoprotein. These observations are consistent with studies on oat, in which phytochrome also became gradually undetectable by immunostaining during cR irradiation (Mackenzie et al., 1978), and with many previous reports of FR light–absorbing phytochrome (Pfr)–specific degradation of PhyA (Clough et al., 1999). However, the site at which PHYA is degraded is still not determined. From our results, we speculate that degradation occurs either in both the nucleus and the cytosol or only in the cytosol, in which case nuclear PHYA would be exported before degradation.
Furthermore, the phenomenon of sequestering, in which phytochrome rapidly associates with amorphous cytoplasmic structures after a brief treatment with R light (Mackenzie et al., 1975), has been speculated to be an early step in degradation of the photoreceptor (Pratt, 1994). In our study, FITC labeling was visualized as very dense speckling and subsequently decayed in both the nuclei and the cytoplasm under cR light. Further examination is necessary to determine whether the speckling has the same structural basis in both the nucleus and the cytosol, whether it corresponds to the previously reported phenomenon of sequestering, and whether it is involved in PHYA degradation.
The kinetics of the nuclear redistribution of the FITC label in response to a brief R light pulse were very similar to those seen in response to cR. In contrast, brief FR light treatments given to dark-grown seedlings caused no substantial change in the localization of FITC labeling. These observations are consistent with the suggestion that the change in subcellular localization of phyA may be dependent on its conversion to Pfr. Both cR light and pulses of R light are equally effective for photoconversion, whereas a pulse of FR light is predicted to produce only a very low proportion of Pfr. However, detailed analyses of fluence and fluence-rate dependence will be necessary to determine whether differences in effectiveness for photoconversion of PhyA in R and FR light can account for differences in its subcellular redistribution.
Kircher et al. (1999b) recently reported that a single 5-min pulse of R or FR light followed by 15 min of darkness was sufficient to induce detectable nuclear accumulation of rice PhyA–GFP in transgenic tobacco. The authors observed speckling of the GFP signal in the cytoplasm within 2 min of pulse irradiation. The GFP signal was subsequently seen as dense speckling in the nucleus under both cR and cFR light and was visible in the cytosol under cR but not cFR light (Kircher et al., 1999b). The rice PhyA–GFP fusion protein also showed a strong speckled distribution in nuclei of light-grown transgenic tobacco (Kircher et al., 1999b). These results differ from ours in the kinetics, distribution pattern, and protein stability under cR and cFR irradiation. Some of these differences may reflect different behaviors of monocot and dicot PhyA. They may also, in part, relate to differences in the protein detected (native PHYA versus overexpressed GFP fusion) and in the detection method (immunostaining with FITC label in sections versus reporter protein of GFP in living cells).
There is also strong evidence that PhyB undergoes light-induced nuclear import. A biologically active Arabidopsis PhyB–GFP fusion protein localized to the nucleus in transgenic Arabidopsis under cR light (Yamaguchi et al., 1999). R light pulses induced nuclear localization of biologically active tobacco PhyB–GFP in transgenic tobacco, but this was reversible by exposure to FR light (Kircher et al., 1999b). In addition, important amounts of endogenous PhyB are detected in nuclei isolated from light-grown Arabidopsis leaves (Sakamoto and Nagatani, 1996) as well as from light-grown pea seedlings (A. Nagatani, unpublished result). Fusion proteins between spectrally inactive C-terminal fragments of PHYB and GUS localize to the nucleus, suggesting the existence of a functional nuclear localization signal in the PHYB sequence (Sakamoto and Nagatani, 1996). Although such a signal has not been conclusively identified, some putative signals are present. These same putative signals are also present in the PHYA sequence (Nagatani, 1997).
The light stimuli that elicit nuclear localization of PhyA (cFR) or PhyB (cR) also result in partial deetiolation of etiolated seedlings through the high-irradiance response mode (Hartmann, 1966; Mancinelli, 1994). Individual responses include inhibition of stem elongation, expansion of cotyledons and true leaves, and induction of some photosynthetic genes. Analysis of phytochrome mutants has shown that the cFR light–induced responses are mediated almost entirely through PhyA, whereas the responses to cR light are mainly mediated through PhyB (Quail et al., 1995). Hence, perhaps both PhyA and PhyB enter the nucleus to induce these responses. In accordance with this, structural/functional analyses of mutated PhyA and PhyB suggest common signal transduction mechanisms for those two major phytochrome species (Quail et al., 1995; Ni et al., 1998). It is intriguing that the kinetics of the light-induced nuclear import of PHYA (Figure 4) and those of PhyB (Yamaguchi et al., 1999) are very similar, although the former are elicited with FR light and the latter with R light. This prompts speculation as to whether another common mechanism regulates the nucleo/cytoplasmic partitioning of these phytochromes.
On the other hand, R light induces nuclear translocation of both PHYA and PhyB–GFP, but nuclear accumulation of PHYA (Figure 4) reaches a maximum earlier than does that of PHYB (Kircher et al., 1999b; Yamaguchi et al., 1999). During the time in which PhyB–GFP accumulates in the nuclei in cR light, PHYA becomes undetectable (Figure 4) as a result of degradation (Figure 6 and Table 2). Recently, Parks and Spalding (1999) reported the sequential and coordinated action of PhyA and PhyB during stem elongation of Arabidopsis under R light. We speculated that the sequential physiological response might be related to the different time kinetics of nucleo/cytoplasmic partitioning of these phytochromes. Additionally, nuclear localization of PhyA–GFP was induced by both R and FR light, in contrast with the R light–induced nuclear localization of PhyB–GFP, which could be reversed by subsequent FR irradiation (Kircher et al., 1999b). We showed here that nuclear localization of PHYA is detectable by shorter irradiations with cR (Figure 4) than with cFR (Figure 3) light. Furthermore, we observed different distribution patterns and stability of PHYA under cR and cFR light (Figures 3 to 5). These observations indicate the possibility that phytochrome species–specific and light stimuli–specific regulating mechanisms may exist. Regulated nuclear translocation of signaling proteins with the appropriate stimulation has been observed in eukaryotes, including plants, and mechanisms have been identified that control this activity (Nagatani, 1998; Smith and Raikhel, 1999).
Recently, phytochrome-interacting molecules have been isolated by using the yeast two-hybrid assay, and their subcellular localization has been investigated. One such protein, PKS1 (for phytochrome kinase substrate 1), which binds to both PhyA and PhyB, is a kinase substrate phosphorylated in a phytochrome-dependent manner, which is supposed to be a negative regulator of PhyB signaling. PKS1–GFP fusion proteins are cytoplasmic (Fankhauser et al., 1999). Another molecule, PIF3 (for phytochrome-interacting factor 3), which also binds to both PhyA and PhyB, is a basic helix-loop-helix protein and potential transcriptional regulator; it is indispensable for normal signal transduction of phytochrome. PIF3 is suggested to have nuclear localization activity (Ni et al., 1998; Halliday et al., 1999). More recently, a phytochrome-interacting molecule, NDPK2 (for nucleoside diphosphate kinase 2), has been reported to have a kinase activity and is involved in the response of both PhyA and PhyB. NDPK2–GFP fusion proteins are localized in both the nuclei and the cytoplasm (Choi et al., 1999). Considering the light-dependent subcellular partitioning of phytochrome and its possible interaction with those molecules in the cytoplasm or the nucleoplasm, phytochrome may have multiple site-specific signaling mechanisms within the cell.
METHODS
Plant Material and Growth Conditions
Seeds of wild-type pea (Pisum sativum cv Torsdag) and the isogenic phyA-deficient mutants fun1-1 and fun1-2 (for far-red unresponsive; Weller et al., 1997) were imbibed in water for 6 hr in darkness and grown on vermiculite saturated with water at 23°C in darkness.
Light Treatments
Five-day-old etiolated seedlings were exposed to continuous far-red light (cFR) or continuous red (cR) light for various lengths of time. After light treatment, plants were immediately cryoembedded for immunostaining or frozen in liquid nitrogen for immunoblotting. R light was obtained from fluorescent tubes (FL-20S Re-66; Toshiba, Tokyo, Japan) filtered through 3-mm-thick red acrylic (Acrylight K5-102; Mitsubishi Rayon, Tokyo, Japan), 3-mm-thick scattering filter (Acrylight K5-001E; Mitsubishi Rayon), and 3-mm-thick white glass. The light intensity measured at 660 nm was 55 μmol m−2 sec−1. FR light was obtained from fluorescent tubes (FL-20S FR-74; Toshiba) filtered through 3-mm-thick FR acrylic (Deraglass A-900; Asahi Kasei, Osaka, Japan) and 3-mm-thick white glass. The light intensity measured at 750 nm was 42 μmol m−2 sec−1.
Preparation of Antibodies
We used 21 monoclonal antibodies raised against pea PhyA apoprotein (PHYA)—mAP05 (Nagatani et al., 1984); mAP09 (Lumsden et al., 1985); mAP10 (Abe et al., 1985); mAP13, mAP14, mAP16, mAP18, mAP19, mAP20, mAP21, mAP22, mAP23, and mAP25 (Nagatani et al., 1987); mAP28, mAP29, mAP30, mAP31, mAP32, mAP33, mAP34, and mAP35 (Nagatani et al., 1984)—two monoclonal antibodies raised against rye PHYA—mAR07 and mAR08 (Nagatani et al., 1987)—and two monoclonal antibodies raised against Arabidopsis PHYA—mAA01 and mAA02 (Shinomura et al., 1996). For the initial screening, culture supernatants of the hybridomas were used. For subsequent experiments, the Arabidopsis anti-PHYA IgG mAA01 was prepared from ascites fluid.
Cryosectioning and Immunostaining
Tissues excised from pea seedlings were submerged in cryoembedding material (Tissu Mount; Shiraimatsu Kikai, Osaka, Japan) in a small cup. The specimen in the cup was then rapidly frozen by placing on an aluminum block (San mag, Tokyo, Japan) cooled with liquid nitrogen. Until cryoembedding was complete, the etiolated tissues were handled only under green safelight. Cryosections 8- to 10-μm-thick were made with a cryomicrotome (CM 1900; Leica Instruments, Nussloch, Germany) adjusted to −20°C and mounted on the silane-coated slide. The section on the slide was immediately air dried, fixed with 4% formaldehyde freshly prepared from paraformaldehyde in 0.1 M phosphate buffer, pH 7.0, and rinsed with 0.1 M phosphate buffer.
Immunostaining was performed with monoclonal anti-phytochrome antibodies and gold-conjugated (particle size 10 nm) anti–mouse IgG antibody (AuroProbe EM; Amersham Pharmacia Biotech., Uppsala, Sweden) combined with a silver enhancement kit (IntenSE M; Amersham Pharmacia Biotech.) or fluorescein isothiocyanate (FITC)–conjugated anti–mouse IgG (Amersham Pharmacia Biotech.). Before incubation with antibodies, the sections on the slides were treated with Block Ace (Dainippon Pharmaceutical, Osaka, Japan) to prevent nonspecific binding. Culture supernatants of hybridomas were used without dilution. IgG (33.1 mg/mL) was diluted 1:1250, and anti–mouse IgG antibody was diluted 1:20 in PBS containing 0.1% Block Ace. PBS containing 0.05% Tween 20 (Bio-Rad) was used for rinsing. The specimens on the slides were treated as follows: (1) 1 hr in Block Ace, (2) incubation overnight at 4°C in anti-PHYA antibody, (3) three rinses, (4) incubation for 2 hr in anti–mouse IgG antibody, and (5) three more rinses. Sections labeled with gold-conjugated antibody were subsequently treated with silver enhancement solution for 10 min. Sections that were labeled with FITC-conjugated antibody were treated with Bisbenzimide Hoechst 33258 (Wako Pure Chemical Industries, Osaka, Japan) to visualize the nucleus.
After immunostaining, sections were embedded in Perma Fluor (Immunon; Shandon/Lipshaw, Pittsburgh, PA) under a cover slip and viewed by optical microscopy (model AX70; Olympus Optical, Tokyo, Japan). Cubes of dichroic mirror and filter combinations were used for fluorescence observation (U-MNIB for detection of FITC and U-MNUA for detection of Hoechst33258; Olympus Optical). Images were obtained with a cooled color charge-coupled device (CCD) camera (model C5801-01; Hamamatsu Photonics, Shizuoka, Japan). Confocal laser scanning microscopy combined a confocal unit in which an argon laser at 488-nm excitation and multiDM filter (model CSU10; Yokogawa Electric Corporation, Tokyo, Japan) were used with optical microscopy (model BX60; Olympus Optical). Confocal images were obtained with a CCD camera (model C4742-95; Hamamatsu Photonics).
Immunoblotting
Shoot apical segments ∼3 cm long were harvested and homogenized to powder in liquid nitrogen by using a mortar. Phytochrome extraction buffer (Nagatani et al., 1993) was added to the powder (2 mL per gram of tissue) and allowed to sit for 15 min at room temperature. After centrifugation (27.6g for 20 min at 4°C), the supernatant was retained, and saturated ammonium sulfate solution was added (2:3 v/v) before 30 min of incubation on ice. The precipitated material was collected by centrifugation (27.6g for 20 min at 4°C) and resuspended in phytochrome extraction buffer. Protein concentrations were determined by using Protein Assay Kits (Bio-Rad). All procedures described above were performed under dim green safe light (Nagatani et al., 1989). After size fractionation by SDS-PAGE in 8.0% ProSieve GTG gel (Pierce, Rockford, IL), the proteins were blotted onto nitrocellulose membrane (Bio-Rad). For immunoblot analysis, blots were incubated with the monoclonal antibody mAA01 as a primary antibody, followed by horseradish peroxidase–conjugated anti–mouse IgG (Organon Teknika Corp., West Chester, PA) as a secondary antibody. PHYA was visualized by using the Chemluminescence Plus Protein Gel-Blotting Detection System (Amersham Pharmacia Biotech) according to the manufacturer's instructions.
Spectrophotometry
Spectrophotometric measurements of phytochrome were performed as previously described (Weller et al., 1995), except that the apical segments were ∼2.5 cm long.
Acknowledgments
We thank Drs. Akihiko Nakano and Chieko Saito for support with confocal laser scanning microscopy and Drs. Yasuji Fukuda, Peter H. Quail, Anne-Marie Lambert, and Anne-Catherine Schmit for helpful advice. This work was supported in part by a grant from the Program for Promotion of Basic Research Activity for Innovative Biosciences to M.F. The experiments were performed under Hitachi Advanced Research Laboratory Project No. B2023.
References
- Abe, H., Yamamoto, K.T., Nagatani, A., and Furuya, M. (1985). Characterization of green tissue–specific phytochrome isolated immunochemically from pea seedlings. Plant Cell Physiol. 26, 1387–1399. [Google Scholar]
- Adam, E., Szell, M., Szekeres, M., Schäfer, E., and Nagy, F. (1994). The developmental and tissue-specific expression of tobacco phytochrome-A genes. Plant J. 6, 283–293. [Google Scholar]
- Adam, E., Kozma-Bognar, L., Kolar, C., Schäfer, E., and Nagy, F. (1996). The tissue-specific expression of a tobacco phytochrome B gene. Plant Physiol. 110, 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer, F.J., and Davies, P.J. (1992). Indole-3-acetic acid levels after phytochrome-mediated changes in the stem elongation rate of dark- and light-grown Pisum seedlings. Planta 188, 85–92. [DOI] [PubMed] [Google Scholar]
- Boonstra, J., Maurik, P.V., and Verkleij, A.J. (1987). Immunogold labelling of cryosections and cryofractures. In Cryotechniques in Biological Electron Microscopy, R.A. Steinbrecht and K. Zierold, eds (Heidelberg, Germany: Springer-Verlag), pp. 216–230.
- Briggs, W.R., and Siegelman, H.W. (1965). Distribution of phytochrome in etiolated seedlings. Plant Physiol. 40, 934–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, W.L., Norris, K.H., Siegelman, H.W., and Hendricks, S.B. (1959). Detection, assay, and preliminary purification of the pigment controlling photoresponsive development of plants. Proc. Natl. Acad. Sci. USA 45, 1703–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie, M., Euskirchen, G., Ward, W.W., and Prasher, D.C. (1994). Green fluorescent protein as a marker for gene expression. Science 263, 802–805. [DOI] [PubMed] [Google Scholar]
- Choi, G., Yi, H., Lee, J., Kwon, Y.-K., Soh, M.S., Shin, B., Luka, Z., Hahn, T.-R., and Song, P.-S. (1999). Phytochrome signaling is mediated through nucleoside diphosphate kinase 2. Nature 401, 610–613. [DOI] [PubMed] [Google Scholar]
- Clack, T., Mathews, S., and Sharrock, R.A. (1994). The phytochrome apoprotein family in Arabidopsis is encoded by five genes: The sequences and expression of PHYD and PHYE. Plant Mol. Biol. 25, 413–427. [DOI] [PubMed] [Google Scholar]
- Clough, R.C., Jordan-Beebe, E.T., Lohman, K.N., Marita, J.M., Walker, J.M., Gatz, C., and Vierstra, R.D. (1999). Sequences within both the N- and C-terminal domains of phytochrome A are required for PFR ubiquitination and degradation. Plant J. 17, 155–167. [DOI] [PubMed] [Google Scholar]
- Cope, M., and Pratt, L.H. (1992). Intracellular redistribution of phytochrome in etiolated soybean (Glycine max L.) seedlings. Planta 188, 115–122. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., and Quail, P.H. (1999). Signalling in light-controlled development. Semin. Cell Dev. Biol. 10, 121–129. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., Yeh, K.C., Lagarias, J.C., Zhang, H., Elich, T.D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284, 1539–1541. [DOI] [PubMed] [Google Scholar]
- Feldman, L.J. (1984). Regulation of root development. Annu. Rev. Plant Physiol. 35, 223–242. [DOI] [PubMed] [Google Scholar]
- Feldman, L.J., and Briggs, W.R. (1987). Light-regulated gravitropism in seedling roots of maize. Plant Physiol. 83, 241–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, L.J., Piechulla, B., and Sun, P.S. (1988). Light-regulated protein and mRNA synthesis in root caps of maize. Plant Mol. Biol. 11, 27–34. [PubMed] [Google Scholar]
- Fukuda, M., Gotoh, Y., and Nishida, E. (1997). Interaction of MAP kinase with MAP kinase kinase: Its possible role in the control of nucleocytoplasmic transport of MAP kinase. EMBO J. 16, 1901–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya, M., and Hillman, W.S. (1964). Observations on spectrophotometrically assayable phytochrome in vivo in etiolated Pisum seedlings. Planta 63, 31–42. [Google Scholar]
- Furuya, M., and Torrey, J.G. (1964). The reversible inhibition by red and far-red light of auxin-induced lateral root initiation in isolated pea roots. Plant Physiol. 39, 987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday, K.J., Hudson, M., Ni, M., Quin, M., and Quail, P.H. (1999). poc1: An Arabidopsis mutant perturbed in phytochrome signaling because of a T-DNA insertion in the promoter of PIF3, a gene encoding a phytochrome-interacting bHLH protein. Proc. Natl. Acad. Sci. USA 96, 5832–5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazato, F., Shinomura, T., Hanzawa, H., Chory, J., and Furuya, M. (1997). Fluence and wavelength requirements for Arabidopsis CAB gene induction by different phytochromes. Plant Physiol. 115, 1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, K.M. (1966). A general hypothesis to interpret ‘high energy phenomena’ of photomorphogenesis on the basis of phytochrome. Photochem. Photobiol. 5, 349–366. [Google Scholar]
- Haupt, W. (1970). Localization of phytochrome in the cell. Physiol. Vég. 8, 551–563. [Google Scholar]
- Johnson, E.M., Pao, L.I., and Feldman, L.J. (1991). Regulation of phytochrome message abundance in root caps of maize. Plant Physiol. 95, 544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, E.T., Hatfield, P.M., Hondred, D., Talon, M., Zeevaart, J.A.D., and Vierstra, R.D. (1995). Phytochrome A overexpression in transgenic tobacco. Plant Physiol. 107, 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, S., Wellmer, F., Nick, P., Rügner, A., Schäfer, E., and Harter, K. (1999. a). Nuclear import of the parsley bZIP transcription factor CPRF2 is regulated by phytochrome photoreceptors. J. Cell Biol. 144, 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schäfer, E., and Nagy, F. (1999. b). Light quality–dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11, 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda, Y., Yamashita, H., Sato, N., Tsukaya, H., and Naito, S. (1991). Regulated expression of a gene-fusion product derived from the gene for phytochrome I from Pisum sativum and uidA gene from E. coli in transgenic Petunia hybrida. Plant Cell Physiol. 32, 737–743. [Google Scholar]
- Konomi, K., Abe, H., and Furuya, M. (1987). Changes in the content of phytochrome I and II apoproteins in embryonic axes of pea seeds during imbibition. Plant Cell Physiol. 28, 1443–1451. [Google Scholar]
- Li, X., Zhao, X., Fang, Y., Jiang, X., Duong, T., Fan, C., Huang, C.-C., and Kain, S.R. (1998). Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 273, 34970–34975. [DOI] [PubMed] [Google Scholar]
- Lumsden, P.J., Yamamoto, T.Y., Nagatani, A., and Furuya, M. (1985). Effect of monoclonal antibodies on the in vitro PFR dark reversion of pea phytochrome. Plant Cell Physiol. 26, 1313–1322. [Google Scholar]
- Mackenzie, J.M., Jr., Coleman, R.A., Briggs, W.R., and Pratt, L.H. (1975). Reversible redistribution of phytochrome within the cell upon conversion to its physiologically active form. Proc. Natl. Acad. Sci. USA 72, 799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie, J.M., Jr., Briggs, W.R., and Pratt, L.H. (1978). Intracellular phytochrome distribution as a function of its molecular form and of its destruction. Am. J. Bot. 65, 671–676. [Google Scholar]
- Mancinelli, A.L. (1994). The physiology of phytochrome action. In Photomorphogenesis in Plants, 2nd ed, R.E. Kendrick and G.H.M. Kronenberg, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 105–140.
- McArthur, J.A., and Briggs, W.R. (1970). Phytochrome appearance and distribution in embryonic axis and seedling of Alaska peas. Planta 91, 146–154. [DOI] [PubMed] [Google Scholar]
- Nagatani, A. (1997). Spatial distribution of phytochromes. J. Plant Res. 110, 123–130. [DOI] [PubMed] [Google Scholar]
- Nagatani, A. (1998). Regulated nuclear targeting. Curr. Opin. Plant Biol. 1, 470–474. [DOI] [PubMed] [Google Scholar]
- Nagatani, A., Yamamoto, K.T., Furuya, M., Fukumoto, T., and Yamashita, A. (1984). Production and characterization of monoclonal antibodies which distinguish different surface structures of pea (Pisim sativum cv. Alaska) phytochrome. Plant Cell Physiol. 25, 1059–1068. [Google Scholar]
- Nagatani, A., Lumsden, P.J., Konomi, K., and Abe, H. (1987). Application of monoclonal antibodies to phytochrome studies. In Phytochrome and Photoregulation in Plants, M. Furuya, ed (Tokyo: Academic Press), pp. 95–114.
- Nagatani, A., Kendrick, R.E., Koorneef, M., and Furuya, M. (1989). Partial characterization of phytochrome I and II in etiolated and de-etiolated tissues of a photomorphogenetic mutant (lh) of cucumber (Cucumis sativus L.) and its isogenic wild type. Plant Cell Physiol. 30, 667–674. [Google Scholar]
- Nagatani, A., Reed, J.W., and Chory, J. (1993). Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 102, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667. [DOI] [PubMed] [Google Scholar]
- Parks, B.M., and Spalding, E.P. (1999). Sequential and coordinated action of phytochrome A and B during Arabidopsis stem growth revealed by kinetic analysis. Proc. Natl. Acad. Sci. USA 96, 14142–14146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt, L.H. (1994). Distribution and localization of phytochrome within the plant. In Photomorphogenesis in Plants, 2nd ed, R.E. Kendrick and G.H.M. Kronenberg, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 163–185.
- Pratt, L.H., and Coleman, R.A. (1971). Immunochemical localization of phytochrome. Proc. Natl. Acad. Sci. USA 10, 2431–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt, L.H., and Coleman, R.A. (1974). Phytochrome distribution in etiolated grass seedlings as assayed by an indirect antibody-labelling method. Am. J. Bot. 61, 195–202. [Google Scholar]
- Quail, P.H., Boylan, M.T., Parks, B.M., Short, T.W., Xu, Y., and Wagner, D. (1995). Phytochromes: Photosensory perception and signal transduction. Science 268, 675–680. [DOI] [PubMed] [Google Scholar]
- Restrepo, M.A., Freed, D.D., and Carrington, J.C. (1990). Nuclear transport of plant potyviral proteins. Plant Cell 2, 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, K., and Nagatani, A. (1996). Nuclear localization activity of phytochrome B. Plant J. 10, 859–868. [DOI] [PubMed] [Google Scholar]
- Saunders, M.J., Cordonnier, M.-M., Palevitz, B.A., and Pratt, L.H. (1983). Immunofluorescence visualization of phytochrome in Pisum sativum L. epicotyls using monoclonal antibodies. Planta 159, 545–553. [DOI] [PubMed] [Google Scholar]
- Sharrock, R.A., and Quail, P.H. (1989). Novel phytochrome sequences in Arabidopsis thaliana: Structure, evolution, and differential expression of plant regulatory photoreceptor family. Genes Dev. 3, 1745–1757. [DOI] [PubMed] [Google Scholar]
- Shinomura, T., Nagatani, A., Hanzawa, H., Kubota, M., Watanabe, M., and Furuya, M. (1996). Action spectra for phytochrome A– and B–specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93, 8129–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H.M.S., and Raikhel, N.V. (1999). Protein targeting to the nuclear pore. What can we learn from plants? Plant Physiol. 119, 1157–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers, D.E., and Quail, P.H. (1995). Temporal and spatial expression patterns of PHYA and PHYB genes in Arabidopsis. Plant J. 7, 413–427. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Sharrock R.A., Tepperman, J.M., and Quail, P.H. (1991). The hy3 long hypocotyl mutant of Arabidopsis is deficient in phytochrome B. Plant Cell 3, 1263–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, L.K., Burgess, C.L., and Skinner, E.N. (1992). Localization of phytochrome during peanut (Arachis hypogaea) gynophore and ovule development. Am. J. Bot. 79, 828–832. [Google Scholar]
- Waller, F., and Nick, P. (1997). Response of actin microfilaments during phytochrome-controlled growth of maize seedlings. Protoplasma 200, 154–162. [Google Scholar]
- Weller, J.L., Nagatani, A., Kendrick, R.E., Murfet, I.C., and Reid, J.B. (1995). New lv mutants of pea are deficient in phytochrome B. Plant Physiol. 108, 525–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller, J.L., Murfet, I.C., and Reid, J.B. (1997). Pea mutants with reduced sensitivity to far-red light define an important role for phytochrome A in day-length detection. Plant Physiol. 114, 1225–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, R., Nakamura, M., Mochizuki, N., Kay, S.A., and Nagatani, A. (1999). Light-dependent translocation of a phytochrome B–GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]