Abstract
Rat cDNA encoding a 372-amino-acid peroxin was isolated, primarily by functional complementation screening, using a peroxisome-deficient Chinese hamster ovary cell mutant, ZPG208, of complementation group 17. The deduced primary sequence showed ∼25% amino acid identity with the yeast Pex3p, thereby we termed this cDNA rat PEX3 (RnPEX3). Human and Chinese hamster Pex3p showed 96 and 94% identity to rat Pex3p and had 373 amino acids. Pex3p was characterized as an integral membrane protein of peroxisomes, exposing its N- and C-terminal parts to the cytosol. A homozygous, inactivating missense mutation, G to A at position413, in a codon (GGA) for Gly138 and resulting in a codon (GAA) for Glu was the genetic cause of peroxisome deficiency of complementation group 17 ZPG208. The peroxisome-restoring activity apparently required the full length of Pex3p, whereas its N-terminal part from residues 1 to 40 was sufficient to target a fusion protein to peroxisomes. We also demonstrated that Pex3p binds the farnesylated peroxisomal membrane protein Pex19p. Moreover, upon expression of PEX3 in ZPG208, peroxisomal membrane vesicles were assembled before the import of soluble proteins such as PTS2-tagged green fluorescent protein. Thus, Pex3p assembles membrane vesicles before the matrix proteins are translocated.
INTRODUCTION
Membrane biogenesis and its regulation are one of the major foci in modern molecular cell biology (Schatz and Dobberstein, 1996). The peroxisome has been widely used as a model intracellular organelle suitable for studies using mammals and yeast (Erdmann et al., 1997; Fujiki, 1997; Subramani, 1997; Waterham and Cregg, 1997). Some human fatal genetic disorders such as Zellweger syndrome are linked to peroxisomal malfunction and failure of peroxisome biogenesis (Lazarow and Moser, 1995; Fujiki, 1997). Genetic heterogeneity has been found in subjects with these peroxisome biogenesis disorders (PBDs), comprising 13 different complementation groups (CGs) (Shimozawa et al., 1992; Moser et al., 1995; Poulos et al., 1995; Kinoshita et al., 1998; Shimozawa et al., 1998a,b). Understanding of peroxisome biogenesis has significantly progressed, mainly based on findings of topogenic signals and peroxins required for peroxisomal protein import (Erdmann et al., 1997; Fujiki, 1997; Subramani, 1997; Waterham and Cregg, 1997). Generally accepted models include peroxisomal soluble as well as membrane proteins being encoded by nuclear genes, translated on free polyribosomes in the cytosol, most of which, if not all, are posttranslationally translocated to preexisting peroxisomes (Lazarow and Fujiki, 1985). Recent evidence suggests the involvement of endoplasmic reticulum (ER) in peroxisomal membrane biogenesis in yeast (Titorenko and Rachubinski, 1998).
We identified 16 CGs in mammals by CG analysis between Chinese hamster ovary (CHO) cell mutants and fibroblasts from PBD patients (Fujiki, 1997; Kinoshita et al., 1998; Ghaedi et al., 1999a). Therefore, mammalian peroxisome biogenesis probably requires at least 16 genes or their products. We isolated a novel CG of CHO cell mutants, ZPG208 and ZPG209 (Ghaedi et al., 1999b). These mutants are apparently defective in peroxisome membrane assembly, as are the ZP119 cells (Kinoshita et al., 1998) and the cells of patients (Shimozawa et al., 1998a) of CG-G, CG-J, and CG-D (Honsho et al., 1998). More recently, we isolated peroxin cDNA, PEX19, by functional phenotype complementation assay, using ZP119 (Matsuzono et al., 1999). Thus, peroxisome assembly-defective CHO cell mutants are useful to investigate molecular and cellular mechanisms involved in peroxisome biogenesis and for elucidation of primary defects of PBD (Fujiki, 1997; Okumoto and Fujiki, 1997; Okumoto et al., 1998b; Otera et al., 1998; Tamura et al., 1998). Using another approach, i.e., expressed sequence tagging search on a human DNA database using yeast peroxin genes, we identified the human orthologue of Yarrowia lipolytica PEX16 (Honsho et al., 1998). PEX16 (Honsho et al., 1998; South and Gould, 1999) and PEX19 (Matsuzono et al., 1999) are responsible for PBDs of CG-D and CG-J, respectively. While the present work was in progress, human PEX3 was cloned using a homology search and yeast PEX3 (Kammerer et al., 1998; Soukupova et al., 1999). It interacts with Pex19p (Soukupova et al., 1999). A potential region responsible for targeting of human (Kammerer et al., 1998; Soukupova et al., 1999) and yeast (Baerends et al., 1996; Wiemer et al., 1996) Pex3p has also been reported. However, a PEX3-defective phenotype has not been described for mammalian cells. Moreover, contrary to extensive investigation of the import of soluble proteins, molecular mechanisms involved in assembly of peroxisomal membrane vesicles are not well understood.
We isolated PEX3 cDNAs from rat, human, and Chinese hamster, primarily by genetic phenotype complementation screening using ZPG208. We also identified an inactivating, missense transition mutation in the PEX3 gene of ZPG208 cells. We found that Pex3p is involved at the initial stage in peroxisome membrane assembly, before the import of matrix protein. Topogenic and functional analyses of Pex3p are also discussed.
MATERIALS AND METHODS
Rat Liver cDNA Library and Search for Complementing cDNA
The rat (Rn) liver cDNA library, containing unidirectionally inserted cDNA under the cytomegalovirus (CMV) promoter in a ZAP Express predigested vector (Stratagene, La Jolla, CA), has been described previously (Okumoto and Fujiki, 1997; Okumoto et al., 1998b). We screened the cDNA library by functional complementation assay, using a CHO cell mutant, ZPG208, as described (Okumoto et al., 1998b; Tamura et al., 1998). Among the cDNA pools examined, a positive one (C8), containing 6000 clones that restored peroxisomes in ZPG208, was further divided into subpools and screened. While complementing cDNA cloning was in progress, using C8 subpools consisting of 300 clones per pool, we identified a human expressed sequence tag clone, AA305508, that showed good homology (47% identity) to PEX3 of Saccharomyces cerevisiae (Hoehfeld et al., 1991). We then asked whether the cDNA pool C8 might contain a plasmid corresponding to human PEX3. We used PCR primers, sense (5′-AAGATGCTGAGGTCTGTATG-3′; potential initiation codon, underlined) and antisense (5′-GGCTCTCGGAATTCAGTTGC-3′), containing nucleotide residues at positions −3 to 17 and 246–265, respectively, of the expressed sequence tag clone AA305508. A PCR product of the expected size was obtained. Thereby, full-length RnPEX3 cDNA cloning was facilitated by colony hybridization of the C8, using as a probe the 268-bp human (Hs) PEX3 cDNA PCR product. Three clones, pBK-CMV-1–3, were isolated and separately transfected into ZPG208 cells. Numerous green fluorescent protein (GFP)-positive punctates, presumably peroxisomes, were observed in respective transfectants (our unpublished results), thus indicating that PEX3 is a potential complementing gene. An EcoRΙ–XhoΙ cDNA fragment of each of the three cDNA clones was directly sequenced. The nucleotide sequence of both strands was determined by the dideoxy-chain termination method, using various oligo-DNA primers, RnPEX3 Internal F and RnPEX3 Full R1 (Table 1), and a Dye-terminator DNA sequence kit (Applied Biosystems, Foster City, CA). Alignment was done using a GENETYX-Mac program (Software Development, Tokyo, Japan). pBK-CMV-1–3 contained the same open reading frame (ORF); pBK-CMV-2 was named pBK-CMV-RnPEX3 (see RESULTS).
Table 1.
Synthetic oligonucleotide primers used
| Codea | Sequence (5′ to 3′) | Underlined |
|---|---|---|
| ClPEX3 Full F | CAGAGTCTGAAGATGCTGAGATCAATG | |
| ClPEX3 Internal F1 | TTACCTGGATAATGCAACAGTTGGAA | |
| ClPEX3 Full R | TCTTCTTGAAGGAAGAAAGTCATTTCTCCAGTTGTTG | |
| ClPEX3 Internal FR1 | AGTTAGATACACCTGATGAAGTAGACTGCTGATGAA | |
| RnPEX3 Internal F | TCAACTCGGAGAGTCTGACAGCT | |
| RnPEX3 F1 | TGGGATCCTGCTGAGATCAATGTGGAATTTTCTGAA | BamHI site |
| RnPEX3 F1/Sal1-Hyb | GGTGGTCGACGCTGAGATCAATGTGGAATT | SalI site |
| RnPEX3 F1.1 | TTGGGATCCGCATCTTCCTGGGCACAGTCC | BamHI site |
| RnPEX3 F2 | GCTTGGGATCCAATATGGACAGAAAAAACTTAGAG | BamHI site |
| RnPEX3 F3/Sal1-Hyb | GTGGGTCGACTTTCACAAGAAGTATTGTA | SalI site |
| RnPEX3 Full R1 | TAAGGTGCTAAATGAATCTA | |
| RnPEX3 Internal R2 | TACAATACTTCTTGTGAAACTTATTATC | |
| RnPEX3 R1 | CTCTGGGCCCCAATCATTTCTCCAGTTGTTGG | ApaI site |
| RnPEX3 R1/NcoI | CATGCCATGGGGCATTTCTCCAGTTGTTG | NcoI site |
| RnPEX3 R1/NheI | GGTAGCTAGCTTTCTCCAGTTGTTGG | NheI site |
| RnPEX3 R1/NotI-Hyb | CCCAGCGGCCGCTCATTTCTCCAGTTGTTG | NotI site |
| RnPEX3 R2 | CTCTGGGCCCCAATCACTGCTGAGTGGGTCGGAAGAACT | ApaI site |
| RnPEX3 R2/NcoI | CATGCCATGGGCTGCTCAGTGGGTCGGA | NcoI site |
| RnPEX3 R2/NotI-Hyb | CCCAGCGGCCGCTCACTGCTCAGTGGGTCG | NotI site |
| RnPEX3 R40/NcoI | CAGTCCATGGGTATTTCTCTAAGTTTTTTCTGTCCA | NcoI site |
| RnPEX3 R40/NotI-Hyb | CCACGCGGCCGCCTATATTTCTCTAAGTTT | NotI site |
| HsPEX19 F/SalI | ACGCGTCGACGGCCGCCGCTGAGGAAGG | SalI site |
| HsPEX19 R/NotI | TCATTAGCGGCCGCCTCGAGTCATCACATGATCAGACACTG | NotI site |
F and R, forward and reverse primers, respectively.
Screening of Human and Chinese Hamster cDNA Libraries
Full-length HsPEX3 cDNA was isolated by colony hybridization on a human liver cDNA library (Tamura et al., 1998) in pCMVSPORT (Life Technologies, Rockville, MD) with the 268-bp HsPEX3 cDNA (see above) as probe. One positive clone was isolated from a subpool, F7-16, and its nucleotide sequence, named pCMVSPORT·HsPEX3, had 1428 bp and contained a 1119-bp ORF encoding a 373-amino-acid polypeptide. Approximately 3.3 × 105 independent colonies of a cDNA library from wild-type CHO-K1 cells in pSPORT I (Otera et al., 1998) were screened using the 32P-labeled RnPEX3 (0.35-kb PCR product and a pair of primers, RnPEX3 F1 and RnPEX3 Internal R2), by hybridization and washing at 37 and 55°C, respectively. Three positive clones were isolated; a shorter one was subcloned into pBluescript II SK(−) (Stratagene) at the SalI–NotI site and sequenced using oligonucleotide primers ClPEX3 Internal F1 and ClPEX3 Internal FR1. The SalΙ–NotΙ fragment of Chinese hamster (Cl) PEX3 was subcloned into the pCMVSPORT Ι vector.
Transfection of PEX3
The BamHΙ–XhoΙ fragment of RnPEX3 in pBK-CMV vector (Okumoto et al., 1998b) was ligated into the BamHΙ–XhoΙ sites of an expression vector, pcDNA3.1/Zeo(+), containing the Zeocin gene (Invitrogen, Carlsbad, CA). ZPG208 cells were transfected with pcDNA3.1/Zeo·RnPEX3 by lipofection (Tamura et al., 1998; Shimizu et al., 1999). Stable transformants were selected in the presence of 250 μg/ml Zeocin (Invitrogen) and were examined for peroxisomes using import of peroxisome targeting signal type 2-tagged GFP (PTS2-GFP) (Ghaedi et al., 1999b). One of the transformants highly expressing GFP in peroxisomes, cloned by the limiting dilution method, was termed 208P3. Eleven other groups of CHO cell mutants as well as fibroblasts derived from peroxisome-deficient patients were similarly transfected with pcDNA3.1/Zeo·RnPEX3. Transfection HsPEX3 and ClPEX3 was likewise done.
Morphological Analysis
PTS2-GFP in TKaG2-derived cells such as ZPG208 that had been grown on a cover glass was observed, without cell fixation (Ghaedi et al., 1999b), under a Carl Zeiss (Thornwood, NY) Axioskop FL microscope using a number 17 filter. Peroxisomes in CHO cells and human fibroblasts were assessed by indirect immunofluorescence light microscopy. Antibodies used were rabbit antibodies to rat liver catalase (Tsukamoto et al., 1990), human catalase (Shimozawa et al., 1992), PTS1 peptide (Otera et al., 1998), 70-kDa peroxisomal integral membrane protein (PMP70) (Tsukamoto et al., 1990), and Pex14p (Shimizu et al., 1999), as well as goat anti-rat catalase antibody (Okumoto et al., 1998b). Anti-Pex3p antibody was raised in rabbits by immunizing with a synthetic peptide comprising the C terminus, an 18-amino-acid sequence of human Pex3p (see Figure 2, dashed underline) supplemented with Gly-Cys at the N terminus that had been linked to keyhole limpet hemocyanin (Tsukamoto et al., 1991). Antigen–antibody complex was detected using Texas Red-labeled sheep anti-rabbit immunoglobulin G (IgG) antibody (Cappel, Durham, NC) or donkey anti-goat IgG antibody conjugated to rhodamine (Chemicon, Pittsburgh, PA).
Figure 2.
Amino acid sequence alignment of PEX3 protein from three mammalian species. Deduced amino acid sequence of rat (Rn) Pex3p was compared with those of Pex3p from Chinese hamster (Cl) and human (Hs). The dash indicates a space. Identical amino acids between species are shaded. The primary sequences with higher hydrophobicity are overlined. The sequence used for chemical synthesis of Pex3p peptide is shown by the dashed underline. The solid arrowhead indicates the position of mutation in pex3 mutants ZPG208 and ZPG209 (see Figure 4).
Mutation Analysis
Total RNA was obtained from ZPG208 and ZPG209 cells, using an RNeasy kit (Qiagen, Hilden, Germany). Reverse transcription (RT)-PCR was performed using 5 μg of total RNA, Superscript reverse transcriptase (Life Technologies), and a pair of ClPEX3-specific PCR primers: ClPEX3 Full F and Full R (Table 1). The RT-PCR product was cloned into the pGEM-T Easy vector (Promega, Madison, WI) and sequenced. ZPG208-derived PEX3 cDNA was inserted into pCMVSPORT and transfected to CHO cells by lipofection.
Expression of Epitope-tagged Pex3p
Tagging of epitopes, flag, and tandem hemagglutinin (HA–HA, influenza virus hemagglutinin) to the N and C terminus, respectively, of RnPex3p was done as follows. The full length of RnPEX3 was amplified using a pair of primers: RnPEX3 F1 and RnPEX3 R1/NheΙ. The PCR product was digested with BamHΙ and NheΙ and then ligated into the BamHΙ–NheΙ sites, upstream of a double-HA tag sequence, of pBluescript II SK(−)·HsPEX16-HA (Honsho et al., 1998). BamHΙ–ApaΙ fragment of the pBluescript II SK(−)·RnPEX3-HA was inserted into the pUcD2SRαHyg·flag-RnPEX12 (Okumoto et al., 1998b), in place of the RnPEX12 cDNA. All plasmid constructs were assessed by sequence analysis. Pex3p-HA and flag-Pex3p were detected using rabbit anti-HA antibody and mouse anti-flag antibody (M2; Scientific Imaging Systems, New Haven, CT), in cells that had been fixed with 4% paraformaldehyde and then permeabilized with either 25 μg/ml digitonin or 0.1% Triton X-100 (Motley et al., 1994; Okumoto and Fujiki, 1997; Okumoto et al., 1998b). Antigen–antibody complex was detected using fluorescein isothiocyanate-labeled sheep anti-rabbit IgG antibody (Cappel) or sheep anti-mouse IgG antibody (Amersham Pharmacia Biotech, Tokyo, Japan) and Texas Red-labeled goat anti-rabbit IgG antibody (Leinco Technologies, Ballwin, MO).
Protease Protection Assay
The postnuclear supernatant (PNS) fraction of CHO-K1 cells transfected with flag-RnPEX3-HA was treated with several different concentrations of proteinase K, in the presence and absence of 1% Triton X-100 for 30 min on ice. The reaction was terminated with 1 mM phenylmethylsulfonyl fluoride and then by precipitation using trichloroacetic acid (Shimizu et al., 1999). The resulting whole-cell proteins were analyzed by SDS-PAGE. Pex3p, Pex14p, and acyl-coenzyme A (CoA) oxidase (AOx), a matrix protein, were assessed by immunoblot with antibodies to flag, HA, Pex14p (Shimizu et al., 1999), and AOx, respectively.
Subcellular Fractionation
Subcellular fractionation of rat liver and CHO cells was done as described (Tsukamoto et al., 1991; Miura et al., 1992; Kinoshita et al., 1998; Shimizu et al., 1999). Each fraction was separated by SDS-PAGE and electrophoretically transferred onto a polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA). Pex3p and peroxisomal marker proteins, including 3-ketoacyl-CoA thiolase (Tsukamoto et al., 1990) and Pex13p (Toyama et al., 1999), were probed with the respective antibodies and then visualized using the ECL Western blotting detection reagent (Amersham Pharmacia Biotech). For determination of intraperoxisomal localization, the peroxisomal fraction was diluted with 20 mM HEPES-KOH, pH 7.6. Membrane and soluble fractions were separated by centrifugation for 30 min at 100,000 × g. Sodium carbonate treatment (Fujiki et al., 1982a) and Triton X-114 extraction (Bodier, 1981) were done as described.
Construction of Pex3p Fusion with enhanced GFP (EGFP)
cDNAs encoding fusion proteins of the wild-type Pex3p and various truncated mutants with EGFP were constructed as follows: the ORF coding for the full-length Pex3p and its truncated mutants comprising amino acid residues 16–372, 1–312, and 1–40 were generated by PCR strategy, with pPBK-CMV·RnPEX3 as the template and primer-pairs designed for creating BamHI and NcoI at 5′ and 3′ tails, RnPEX3 F1 and RnPEX3 R1/NcoI, RnPEX3 F1.1 and RnPEX3 R1/NcoI, RnPEX3 F1 and RnPEX3 R2/NcoI, and RnPEX3 F1 and RnPEX3 R40/NcoI, respectively. Each of BamHI and NcoI fragments of the resulting PCR products was inserted into the BamHI and NcoI sites of pEGFP vector upstream of an EGFP gene (Clontech, Palo Alto, CA). The BamHI–ApaI fragments containing the ORF encoding Pex3p-EGFP fusion proteins were inserted into the pUcD2Hyg·flag-RnPEX12 (Okumoto et al., 1998b) in place of the RnPEX12 cDNA.
Yeast Two-Hybrid Assay
Maintenance and transformation of yeast cells, using the Proquest two-hybrid system (Life Technologies), were done according to the manufacturer's protocol. The ORFs for the full-length rat Pex3p and its truncated mutants were amplified by PCR with pBK-CMV·RnPEX3 as the template and primers (see below) introducing the SalΙ and NotΙ sites at 5′ and 3′ sites, respectively. Primers used were sense primer, RnPEX3 F1/SalI-hyb, and antisense, RnPEX3 R1/NotI-hyb, for the full length, amino acid residues 1–372 (Table 1). PCR for PEX3 variants encoding amino acid residues 1–312 and 1–40 were done with RnPEX3 F1/SalI-hyb as a forward primer and RnPEX3 R2/NotI-hyb and RnPEX3 R40/NotI-hyb as reverse primers, respectively. A PEX3 mutant for residues 110–372 was amplified with a forward primer, RnPEX3 F3/SalI-hyb, and a reverse primer, RnPEX3 R1/NotI-hyb. The ORF for Pex3p-G138E was amplified using primers RnPEX3 F1/SalI-hyb and RnPEX3 R1/NotI-hyb and ZPG208-derived PEX3 as the template. The resulting PCR products were excised with SalΙ and NotΙ. The fragments were separately inserted into the SalΙ–NotΙ sites downstream of the GAL4 DNA-binding domain in the pDBLeu plasmid or the GAL4-activating domain in the pPC86 plasmid. For the constructs encoding fusions with human Pex19p, PCR amplification was done with primers HsPEX19 F/SalI and HsPEX19 R/NotI using pUcD2Hyg·HsPEX19 (Matsuzono et al., 1999) as the template. The resulting product was inserted into pDBLeu and pPC86 plasmids, as described for PEX3.
Cotransformation of two hybrid vectors into S. cerevisiae MaV203 (Matα, leu2-3, 112, trp1-901, his3Δ200, ade2-101, gal4Δ, gal80Δ, SPAL10::URA3, GAL1::lacZ, HIS3UAS GAL1::HIS3@LYS2, can1R, cyh2R) was done according to instructions of the manufacturer. Individual transformants were screened for their potential to grow on synthetic complete medium lacking tryptophan, leucine, and histidine by expression of their chromosomal HIS3 gene. The transformants were also assayed for β-galactosidase activity of the lacZ marker gene.
Coimmunoprecipitation Assay
To verify the findings in vivo, we did a coimmunoprecipitation assay. Human Pex3p and Pex19p were separately synthesized in a rabbit reticulocyte cell-free translation system (Miyazawa et al., 1989) using in vitro transcripts of HsPEX3 and HsPEX19 (Matsuzono et al., 1999), in the presence and absence of 1.2 mCi/ml [35S]methionine and [35S]cysteine (Amersham Pharmacia Biotech), respectively. [35S]Pex3p and Pex19p were incubated overnight at 4°C and subjected to immunoprecipitation with anti-Pex19p antibody (Matsuzono et al., 1999), as described (Miyazawa et al., 1989).
Other Methods
In vitro transcription and translation (Miyazawa et al., 1989) were done as described. Immunoprecipitation of [35S]Pex3p and catalase latency assay with digitonin were done as described (Tsukamoto et al., 1990). Protein assay was done using a Bio-Rad protein assay kit. Cell resistance to the 12-(1′-pyrene)dodecanoic acid/long-wavelength UV light (P12/UV) and 9-(1′-pyrene)nonanol/UV (P9OH/UV) treatments was determined under conditions of 2 μM, 1.5 min and 6 μM, 2 min (Shimozawa et al., 1992), respectively.
RESULTS
Cloning of a Rat PEX3 cDNA
We used a transient expression assay as a cDNA cloning strategy (Tsukamoto et al., 1995; Okumoto and Fujiki, 1997; Okumoto et al., 1998b; Tamura et al., 1998) to search for a complementing cDNA of a CG17 CHO cell mutant, ZPG208, defective in peroxisome assembly (Ghaedi et al., 1999b) (Figure 1a). A rat liver cDNA library divided into small pools was transfected to ZPG208. Peroxisome-restoring positive cDNA clones were isolated by searching for a punctate fluorescent pattern of PTS2-GFP in cells, presumably restoring peroxisomal import. One combined pool (C8) yielded several peroxisome-restored cells of ZPG208 in a single dish (Figure 1b). After a third round of screening, i.e. at the step of 300 clones per pool screening, final cDNA cloning was done by a colony hybridization method, using a 268-bp HsPEX3 cDNA (see MATERIALS AND METHODS) as probe. One positive clone, named pBK-CMV·PEX3, was isolated, which restored peroxisomal import of PTS2-GFP in ZPG208 (our unpublished data). The cDNA portion of pBK-CMV·PEX3, sequenced on both strands, indicated that the cDNA was 1952 bp in length with an ORF encoding a protein consisting of 372 amino acids (Figure 2). The calculated molecular mass of its deduced amino acid sequence was 42,209 Da. The amino acid sequence showed 25, 32, and 33% identity with those of Pex3p from S. cerevisiae (Hoehfeld et al., 1991), Hansenula polymorpha (Baerends et al., 1996), and Pichia pastoris (Wiemer et al., 1996), respectively. Thus we termed this cDNA rat PEX3, RnPEX3. (The GenBank database accession number for rat PEX3 is AB035306.) RnPEX3 complemented peroxisomal import of PTS2-GFP in ZPG208 (Figure 1c). Human (Hs) and Chinese hamster (Cl) PEX3 cDNA were cloned by colony hybridization from human and Chinese hamster cDNA libraries. Both HsPEX3 and ClPEX3 encoded 373-amino-acid Pex3p, with 94 and 97% identity to rat Pex3p at a deduced amino acid sequence level, whereas rat Pex3p was shorter by one amino acid, at alignment position 222 (Figure 2). (The GenBank database accession numbers for human and Chinese hamster PEX3 are AB035307 and AB035308, respectively.) Pex3p apparently contained at least two hydrophobic segments, thereby suggesting that Pex3p is a membrane protein (Figure 2, overlines).
Figure 1.
Restoration of peroxisomes in CHO mutant cells. (a and f) Peroxisome-deficient ZPG208 cells; (b), peroxisome-restored ZPG208 after lipofection with a combined pool (C8) of rat cDNA library; (c) ZPG208 transfected with pBKCMV·RnPEX3 plasmid; d, e, and g) 208P3 cells, stable RnPEX3-transformants of ZPG208 cells; (h) ZPG209 was transfected with pCMVSPORT·HsPEX3. (a–c) Fluorescent micrograph of GFP-expressing ZPG208. Cells were stained with antisera to catalase (d and h), PTS1 (e), and PMP70 (f and g), respectively. Magnification, 630×; bar, 20 μm.
PEX3 Restored Peroxisome Biogenesis in ZPG208
Several phenotypic abnormalities attributable to peroxisome deficiency, such as impaired import of both matrix and membrane proteins, were found in ZPG208 (Ghaedi et al., 1999b). To determine whether RnPEX3 could correct these mutant phenotypes, a stable RnPEX3-transformant of ZPG208, named 208P3, was isolated. PTS1 proteins were noted in numerous vesicular structures, presumably peroxisomes, when stained with antibodies to catalase (Figure 1d), PTS1 (Figure 1e), and 3-ketoacyl-CoA thiolase, a PTS2 protein (our unpublished data). Numerous PMP70-positive particles were detected in 208P3 cells (Figure 1g), whereas peroxisomal membrane remnants were not discernible in ZPG208 (Ghaedi et al., 1999b) (Figure 1f). These results strongly suggested that 208P3 cells had morphologically normal peroxisomes, as seen in the wild-type CHO-K1 cells. When another CHO cell mutant, ZPG209 of the same CG as ZPG208, was transfected with human PEX3 (HsPEX3), catalase was likewise localized in peroxisomes (Figure 1h), thus demonstrating that HsPEX3 is functional in CHO cells.
In peroxisome-deficient cells, peroxisomal proteins mislocalized to the cytosol are rapidly degraded or are not converted to mature forms, despite normal synthesis (Tsukamoto et al., 1990; Shimozawa et al., 1992; Okumoto et al., 1997). In the digitonin titration assay, nearly 60% of the catalase activity was latent at the digitonin concentration of 100 μg/ml in the wild-type cells (Figure 3A). In ZPG208 cells, nearly full activity of catalase was detected at 100 μg/ml digitonin, with the same latency profile as lactate dehydrogenase, a cytosolic enzyme (Okumoto et al., 1998b; Tamura et al., 1998) (our unpublished results); hence catalase was present in the cytosol. This is consistent with our earlier observations using several CGs of CHO mutants (Tsukamoto et al., 1990; Shimozawa et al., 1992; Okumoto et al., 1998b; Tamura et al., 1998). In 208P3 cells, catalase showed almost the same latency as in wild-type CHO-K1 cells, thereby demonstrating restoration of peroxisome biogenesis. A moderately higher level of total catalase activity in ZPG208 and 208P3 cells, compared with that of CHO-K1, apparently reflects cell size (Table 2).
Figure 3.
Complementation of biogenesis of peroxisomal proteins. (A) Latency of catalase activity in CHO-K1, ZPG208, and 208P3 cells. Circles, CHO-K1; triangles, ZPG208; squares, 208P3. Relative free catalase activity is expressed as a percentage of the total activity measured in the presence of 1% Triton X-100 (Tsukamoto et al., 1990). The results represent a mean of duplicate assays. (B) Biogenesis of peroxisomal proteins. Subcellular fractions (20-μg aliquot each) from CHO-K1, ZPG208, and 208P3 were subjected to SDS-PAGE and transferred to a polyvinylidene difluoride membrane. Immunoblot analysis was done using rabbit antibodies to peroxisomal AOx (upper panel), thiolase (middle panel), and PMP70(lower panel). Cell types are indicated at the top. Lanes 1, 4, and 7, postnuclear supernatant (PNS) fraction; lanes 2, 5, and 8, organellar fraction (Pell) derived from PNS; lanes 3, 6, and 9, cytosolic fraction (Sup) from PNS. Arrows show AOx components A–C; P and M designate a larger precursor and mature protein of thiolase, respectively. Open arrowheads indicate the thiolase precursor; the arrowhead shows PMP70. Dots in the top panel indicate nonspecific bands (Tamura et al., 1998).
Table 2.
Properties of wild-type CHO-K1, ZPG208, and RnPEX3-transformed ZPG208 (208P3) cells
| Cell | Peroxisome | Total catalase activity (mU/106 cells) | Catalase latency (%) | P9OH/UV (%) | P12/UV (%) |
|---|---|---|---|---|---|
| CHO-K1 | + | 5.8 | 61 | 0 | 76 |
| ZPG208 | − | 8.0 | 5 | 81 | 0 |
| 208P3 | + | 8.2 | 55 | 0 | 72 |
Catalase latency represents peroxisomal catalase, calculated from Figure 3A, as described (Tsukamoto et al., 1990). For determination of P12/UV or P9OH/UV resistance, 200 or 1 × 105 cells were inoculated into 60-mm dishes and selected (Shimozawa et al., 1992). The numbers of colonies were counted in triplicate experiments and expressed as percentages of that of unselected control.
AOx, the first enzyme of the peroxisomal fatty acid β-oxidation system, is synthesized as a 75-kDa polypeptide (A component) and is proteolytically converted into 53- and 22-kDa polypeptides (B and C components, respectively) in peroxisomes (Miyazawa et al., 1987; Miyazawa et al., 1989; Tsukamoto et al., 1990). All three polypeptide components were evident in CHO-K1 cells, exclusively in the organelle fraction, presumably in peroxisomes, as assessed by immunoblotting (Figure 3B, top panel, lanes 1–3), whereas AOx protein was under the detectable level in ZPG208, probably because of rapid degradation (Tsukamoto et al., 1990; Ghaedi et al., 1999b) (Figure 3B, top panel, lanes 4–6). The three components of AOx were found in particulate fractions in 208P3, as in CHO-K1 cells (Figure 3B, top panel, lanes 7–9), indicative of proper import and proteolytic conversion of AOx. Peroxisomal 3-ketoacyl-CoA thiolase, the third enzyme of the peroxisomal β-oxidation system, is synthesized as a larger precursor with an amino-terminal presequence, which contains PTS2 (Osumi et al., 1991; Swinkels et al., 1991) and is converted to a mature form in peroxisomes (Hijikata et al., 1987; Tsukamoto et al., 1990; Miura et al., 1994; Tsukamoto et al., 1994a). In wild-type CHO-K1 cells, only the matured thiolase was detected and mostly in the PNS as well as organelle fractions, presumably in peroxisomes, with a little in the cytosol (Figure 3B, middle panel, lanes 1–3), thereby reflecting rapid processing of the precursor form. In ZPG208 cells, only the larger precursor was found in particulate and soluble fractions (Figure 3B, middle panel, lanes 4–6), implying the absence of processing activity. The thiolase precursor in the membrane pellet may be due to nonspecific binding to some organelles. Physiological implications remain to be clarified. 208P3 cells showed only the mature form of thiolase, as in CHO-K1, demonstrating the complementation of PTS2 protein import and processing (Figure 3B, middle panel, lanes 7–9). PMP70 was absent in ZPG208 (Ghaedi et al., 1999b) (Figure 3B, bottom panel, lanes 4–6), presumably by a rapid degradation as in the pex19 CHO mutant ZP119 (Kinoshita et al., 1998). Another peroxisomal membrane protein such as Pex13p was also under the detectable level in ZPG208 (see Figure 5A). Appearance of PMP70 in the particulate fraction of 208P3, at a similar level as in CHO-K1, also suggested the restored biogenesis of peroxisomal membrane proteins (Figure 3B, bottom panel, lanes 1–3 and 7–9).
Figure 5.
Subcellular localization of Pex3p. (A) Upper panel, size comparison of in vitro transcription–translation products of human, Chinese hamster, and rat PEX3 cDNAs and subcellular localization of Pex3p in CHO cells. The [35S]Methionine- and [35S]cysteine-labeled in vitro transcription–translation product of each PEX3 and subcellular fractions of CHO cells were subjected to SDS-PAGE and transferred to a polyvinylidene difluoride membrane. Autoradiography for detection of 35S-labeled protein was exposed for 5 d; immunodetection was done with rabbit anti-Pex3p peptide antibody. Lanes 1–3, in vitro transcription–translation product (1 μl) of human, Chinese hamster, and rat PEX3, respectively; lanes 4–6, 7–9, 10–12, and 13–15, PNS, organellar, and cytosolic fractions (20 μg each) of CHO-K1 and 208P3, a stable RnPEX3 transfectant of ZPG208, ZPG208, and a pex2 mutant, Z65, a typical peroxisome membrane remnant-positive CHO mutant (Tsukamoto et al., 1991; Shimozawa et al., 1992; Tsukamoto et al., 1994b), respectively. A composite of two separate experiments is represented. The arrow indicates the migration of human and Chinese hamster Pex3p; the arrowhead shows rat Pex3p. Lower panel, expression level of Pex13p and Pex14p in pex3 mutant ZPG208. Subcellular fractions (30 μg each) of CHO-K1 and pex3 mutant ZPG208 and 208P3 were analyzed by immunoblot using antibodies to Pex13p and Pex14p. Exposure for detection of Pex13p was several times longer than that for Pex14p. (B) Isopycnic subcellular fractionation. A light mitochondrial fraction of rat liver was fractionated by isopycnic sucrose density gradient ultracentrifugation. The gradient was collected into 36 tubes. After marker enzyme and protein assays, an equal volume (15 μl, 2 μl for detection of catalase) of odd-numbered fractions plus fraction 24 was analyzed by immunoblot. Upper panel, distribution of marker enzymes: catalase for peroxisomes; glutamate dehydrogenase, mitochondria; esterase, microsomes; and N-acetyl-β-glucosaminidase, lysosomes. Lower panel, immunoblots with antisera against Pex3p, Pex14p, PMP70, catalase, and AOx. Arrows, AOx components A–C. Results are presented in the direction of lower to higher density of sucrose, from left to right.
P9OH, incorporated into plasmalogens at an early stage of synthesis, produces active oxygen by UV irradiation (Morand et al., 1990). Cell culture in the presence of P9OH, followed by short exposure to UV, kills wild-type CHO cells but not peroxisome-defective mutants. Conversely, P12/UV treatment specifically kills peroxisome-defective cells grown in P12-added medium upon UV irradiation, because of lack of synthesis of plasmalogen, an oxygen radical scavenger (Zoeller et al., 1988). 208P3 cells were resistant to P12/UV treatment and sensitive to P9OH/UV, similar to CHO-K1 cells (Table 2). In contrast, mutant ZPG208 cells were resistant to P9OH/UV treatment and sensitive to P12/UV (Ghaedi et al., 1999b) (Table 2). Taken together, these results demonstrated that RnPEX3 restored peroxisome biogenesis in ZPG208.
At 3 d after RnPEX3 transfection, CG17 CHO mutants, ZPG208 and ZPG209, were mostly complemented for peroxisome assembly, whereas none of the other CGs of peroxisome-deficient CHO cell mutants showed peroxisomes (Table 3). Furthermore, RnPEX3 was introduced into fibroblasts from patients with PBD, such as Zellweger syndrome, of CGs, D and G of Gifu University (Gifu, Japan) (Shimozawa et al., 1992; Poulos et al., 1995) and CG-VI of the Kennedy–Krieger Institute (Baltimore, MD) (Shimozawa et al., 1992) that were distinct from CHO mutants. As expected, none of the PBD fibroblasts was morphologically restored for peroxisome assembly (Table 3). Collectively, these results demonstrate that Pex3p is the peroxisome biogenesis factor only for CG17.
Table 3.
Complementation of CHO cell mutants and PBD patient fibroblasts by RnPEX3
| CHO mutant | Peroxisomepositive | Patient fibroblasts | Peroxisomepositive | Gene | |
|---|---|---|---|---|---|
| ZPG208 | + | PEX3 | |||
| ZPG209 | + | ||||
| ZP124 | − | (A | VIII) | ||
| B | (VII) | − | PEX10 | ||
| ZP92 | − | (C | IV) | PEX6 | |
| D | (IX) | − | PEX16 | ||
| Z24/ZP107 | − | (E | I) | PEX1 | |
| Z65 | − | (F | X) | PEX2 | |
| ZP139 | − | (II) | PEX5 | ||
| ZP109 | − | (III) | PEX12 | ||
| VI | − | ||||
| G | − | ||||
| ZP128 | − | (H) | PEX13 | ||
| ZP119 | − | J | − | PEX19 | |
| ZPG207 | − | R | XI | − | PEX7 |
| ZP110 | − | PEX14 | |||
| ZP114 | − | ||||
| ZP126 | − | ||||
Peroxisome-deficient CHO mutants (Fujiki, 1997; Tateishi et al., 1997; Kinoshita et al., 1998), including ZPG208 and ZPG209 (Ghaedi et al., 1999b), CG-A ZP124 (Ghaedi et al., 1999a), PEX6-defective ZP92 (Shimozawa et al., 1992; Tsukamoto et al., 1995), PEX1-defective ZP107 (Okumoto et al., 1997; Tamura et al., 1998), PEX2-impaired Z65 (Tsukamoto et al., 1991, 1994b), PEX5-impaired ZP139 (Otera et al., 1998), PEX12-defective ZP109 (Okumoto et al., 1997, 1998b; Okumoto and Fujiki, 1997), PEX13-impaired ZP128 (Toyama et al., 1999), PEX19-impaired ZP119 (Kinoshita et al., 1998; Matsuzono et al., 1999), PEX14-deficient ZP110 (Tateishi et al., 1997; Shimizu et al., 1999), ZP114 (Tateishi et al., 1997), and ZP126 (Ghaedi et al., 1999a), as well as patient fibroblasts of several CGs shown not to be represented by CHO mutants (Ghaedi et al., 1999b), i.e., CGs B (patient PBDB-01) (Okumoto et al., 1998a), D (patient PBDD-01) (Honsho et al., 1998), and G (patient 2) (Poulos et al., 1995; Ghaedi et al., 1999b) of Gifu University (Gifu, Japan) and CG-VI (Shimozawa et al., 1992) of Kennedy-Krieger Institute (Baltimore, MD), were transfected with pcDNA3.1/Zeo·RnPEX3 and examined for peroxisome assembly by immunostaining with antisera to rat and human catalase, respectively, at 4 d after transfection. PEX7-defective ZPG207 (Ghaedi et al., 1999b) and fibroblasts from a patient with rhizomelic chondrodysplasia punctata (Ghaedi et al., 1999b) were likewise transfected and stained with anti-rat thiolase antibody. Cells of CG in parenthesis were not used in this experiment. +, peroxisomes were complemented; −, not complemented.
Dysfunction of Pex3p in CHO Mutants
PEX3 in Mutants.
To investigate dysfunction of Pex3p in ZPG208, we determined the nucleotide sequence of Pex3p cDNA isolated from ZPG208 by RT-PCR. In the six independent cDNA clones isolated, all showed a point mutation at position 413 of a codon for Gly138 (GGA) to a codon for Glu138 (GAA), termed ClPEX3G138E (Figure 4A), strongly suggesting a homozygous mutation. Therefore, dysfunction of the Pex3p caused by a missense mutation is most likely to be the primary defect in the mutant ZPG208. It is noteworthy that the mutation site, G138E, was located in the interior of the hydrophobic segment (see Figure 2). The same homozygous missense mutation was found in ZPG209 (our unpublished results).
Figure 4.
Mutation of PEX3 in ZPG208 cells. (A) Mutation analysis of PEX3 in ZPG208. Partial sequence and deduced amino acid sequence of PEX3 cDNA isolated from CHO-K1 (left) and a mutant ZPG208 (right) are shown. A point mutation at nucleotide residue 413 changes a codon (GGA) for Gly138 (Figure 2, solid arrowhead) to a codon (GAA) for Glu138. (B) Transfection of mutated Chinese hamster PEX3. ZPG208 cells were transfected with Chinese hamster PEX3, ClPEX3 (a), or ZPG208-derived PEX3, ClPEX3G138E (b). A fluorescence micrograph of intracellular localization of PTS2-GFP is shown. Magnification, 630×; bar 20 μm.
Complementation of Protein Transport by ClPEX3.
When mutant ZPG208 cells were transfected with wild-type ClPEX3 cDNA, PTS2-GFP was found in numerous particles, thereby indicating complementation of peroxisomal protein import (Figure 4B, a), as was the case with RnPEX3 (see Figure 1). To assess the impaired function of Pex3p in ZPG208, ZPG208-derived PEX3 cDNA, ClPEX3G138E, was transfected back to the mutant cells. PTS2-GFP was present in the cytosol, in a diffuse manner, in ClPEX3G138E-transfected ZPG208 (Figure 4B, b), hence demonstrating dysfunction of the mutated form of Pex3p. Moreover, ZPG209, the same CG mutant as ZPG208, showed cytosolic PTS2-GFP in transfectants of ClPEX3G138E (our unpublished results), confirming the impaired function of ClPEX3G138E.
Taken together, we conclude that dysfunction of Pex3p caused by a missense mutation is the primary defect in impaired peroxisome biogenesis in CG17 CHO mutants ZPG208 and ZPG209.
Intracellular Localization of Pex3p
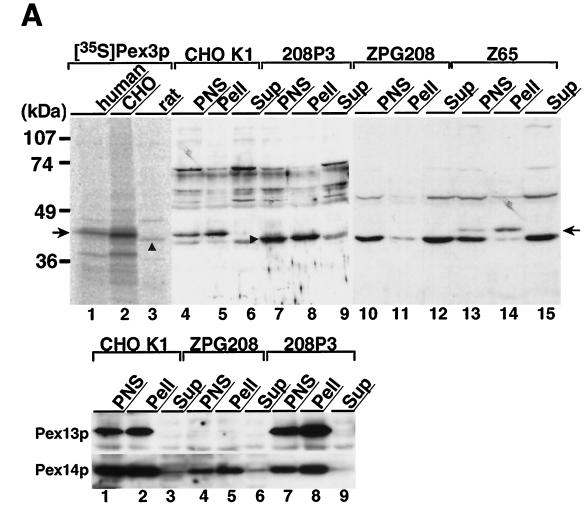
The C-terminal peptide of human Pex3p (residues 355–372) was used to raise rabbit antibody. This antibody reacted only with a single protein with an apparent molecular mass of ∼42 kDa, nearly the same size as the calculated one, in immunoblot of rat liver homogenates (our unpublished results), indicating that the antibody is specific. The mobility in SDS-PAGE of Pex3p synthesized in vitro by coupled transcription–translation of ClPEX3 was indistinguishable from that of Pex3p detected by immunoblot of subcellular fractions of CHO-K1, i.e. PNS and organellar fractions (Figure 5A, upper panel, compare lane 2 and lanes 4 and 5, arrow), thereby indicating that a cloned ClPEX3 encodes bona fide Pex3p associated with organelles. This result implies the synthesis of Pex3p at its final size, consistent with a general rule for peroxisomal proteins (Lazarow and Fujiki, 1985). Human Pex3p synthesized in vitro also showed a similar mobility in SDS-PAGE (Figure 5A, lane 1), whereas rat Pex3p showed a slightly higher mobility (lane 3, arrowhead).
Intracellular localization of Pex3p was also investigated by subcellular fractionation of 208P3 cells stably expressing rat Pex3p. Pex3p was detected in the PNS fraction and then exclusively recovered in the organellar fraction, not in the cytosol (Figure 5A, upper panel, lanes 7–9, arrowhead), as was the case for endogenous Pex3p in CHO-K1 (lanes 4–6). Peroxisomal membrane remnants, called peroxisomal ghosts, are found in most CHO cell mutants impaired in peroxisome biogenesis (Shimozawa et al., 1992) such as a pex2 mutant, Z65 (Tsukamoto et al., 1991), as well as in PBD patient fibroblasts (Santos et al., 1988). Pex3p was detected in Z65 cells and fractionated in the membrane fraction (Figure 5A, lanes 13–15, arrow), whereas it was not discernible in a pex3, ZPG208 (lanes 10–12), possibly because of a rapid degradation. This implies that Pex3p is localized in endomembranes in Pex2 cells, presumably in peroxisomal remnants where PMP70 is targeted (see below). Biogenesis of other peroxisomal membrane proteins such as Pex13p (Toyama et al., 1999) and Pex14p was investigated. Pex14p was detectable in the membrane fraction of ZPG208 (Figure 5A, lower panel, lanes 4–6) but was less in amount compared with the level in CHO-K1 and 208P3 (lanes 1–3 and 7–9). In contrast, Pex13p was not detectable in ZPG208 (lanes 4–6). Pex12p, another peroxin integrated to peroxisomal membranes (Okumoto and Fujiki, 1997), was likewise below the detection level (our unpublished results). In CHO-K1 as well as 208P3 cells, Pex13p and Pex14p were detected in organelle fractions (lanes 1–3 and 7–9). It may be that Pex14p locates in peroxisome-related membrane vesicles not morphologically detectable or in some endomembranes by mistargeting. In contrast, Pex13p appears to be degraded. The results imply that the stability of peroxisomal membrane proteins in the pex3 mutant may vary from one protein to another.
Upon further fractionation of the light mitochondrial fraction from rat liver by sucrose density gradient centrifugation, Pex3p was detected as a single band and cosedimented with peroxisomal marker enzymes catalase and AOx as well as peroxisomal integral membrane proteins Pex14p (Shimizu et al., 1999) and PMP70, thereby indicating that Pex3p is a peroxisomal protein (Figure 5B). This is consistent with morphological observations (see below). In addition, the distribution of Pex3p on the gradient was different from that of marker enzymes, glutamate dehydrogenase for mitochondria, esterase for microsomes, and N-acetyl-β-glucosaminidase for lysosomes, thus confirming the peroxisomal location of Pex3p.
The subcellular localization of Pex3p was also determined by immunofluorescent microscopy with Pex3p tagged at its N terminus with an epitope flag. In wild-type CHO-K1 cells transfected with flag-HsPEX3, Pex3p was detected in a punctate staining pattern with use of an anti-flag antibody (Figure 6A, a). The pattern was superimposable on that obtained using an anti-catalase antibody (Figure 6A, b), thereby demonstrating that flag-Pex3p was targeted to peroxisomes. Similar results were obtained when flag-RnPEX3 was expressed in CHO-K1 cells and stained with an anti-flag antibody (our unpublished results). The flag-tagged Pex3p fully restored peroxisome assembly in ZPG208, as efficiently as did Pex3p, and was colocalized with catalase, indicating that the N-terminal tagging did not interfere with function of Pex3p (our unpublished results). These results were interpreted to mean that flag-Pex3p was translocated to peroxisomes. Endogenous Pex3p was barely detectable using an anti-Pex3p antibody in CHO-K1 (our unpublished results). PEX2-defective Z65, (Tsukamoto et al., 1991) representing a typical pex phenotype with normal import of membrane proteins (Shimozawa et al., 1992), was transfected with flag-RnPEX3-HA. Flag-Pex3p-HA was colocalized with PMP70 in peroxisomal ghosts (Figure 6A, c and d). Thus, translocation of Pex3p does not appear to be impaired in these mutant cells, consistent with the notion that import of peroxisomal membrane proteins is normal in mutants with peroxisomal remnants. Collectively, the data demonstrate peroxisomal localization of Pex3p.
Figure 6.
Intracellular localization of Pex3p. (A) Morphological analysis. Flag-HsPEX3 was expressed in CHO-K1 cells. (a and b) CHO-K1 cells stained with antibodies to flag and catalase, respectively; (c and d) PEX2-impaired, peroxisome remnant-positive Z65 was transfected with flag-RnPEX3-HA and stained with antibodies to HA and PMP70, respectively. Magnification, 630×; bar, 20 μm. (B) Intraperoxisomal localization of Pex3p. Immunoblotting was done with antibodies specific for Pex3p, Pex12p, PMP22, and catalase. Lane 1, rat liver peroxisomes (10 μg); lanes 2 and 3, soluble and membrane fractions after the sodium carbonate treatment (Fujiki et al., 1982b) of peroxisomes (25 μg); lanes 4 and 5, soluble and membrane fractions after the Triton X-114 treatment (Bodier, 1981) of peroxisomes (25 μg).
Hydropathy analysis of Pex3p suggested that Pex3p apparently contains at least two hydrophobic segments (see Figure 2, overlines). Pex3p was not extractable with 50 mM HEPES-KOH, pH 7.6, from freshly isolated rat liver peroxisomes (our unpublished results). The integrity of Pex3p was verified by extraction with 0.1 M sodium carbonate, pH 11.5 (Fujiki et al., 1982a), and treatment with 1% Triton X-114 (Bodier, 1981) (Figure 6B). Pex3p was not extractable with sodium carbonate, as was the case for peroxisomal integral membrane proteins PMP22 (Fujiki et al., 1984) and Pex12p (Okumoto and Fujiki, 1997; Okumoto et al., 1998b) (Figure 6B, lane 3), in contrast to a matrix enzyme, catalase (lane 2), thereby strongly suggesting that Pex3p is an integral membrane protein. Upon treatment with Triton X-114, Pex3p as well as PMP22 and Pex12p were recovered in a detergent phase, and catalase was recovered in an aqueous phase (lanes 4 and 5), thereby indicating that Pex3p is an integral membrane protein.
Membrane Topology of Pex3p
Topology of Pex3p in peroxisomal membranes was investigated using a differential cell permeabilization procedure, in which detection of Pex3p was done using antibodies to epitope tags. When ZPG208 cells transfected with flag-RnPEX3-HA encoding both N- and C-terminally epitope-tagged Pex3p were treated with Triton X-100, which solubilizes all cellular membranes, both flag-Pex3p-HA and catalase were detected in particulates, in a superimposable manner, thus indicating localization of Pex3p in peroxisomes (Figure 7, A and B, a and b) and consistent with findings described above. Hence, flag and HA tagging does not affect localization and function of Pex3p. When ZPG208 cells expressing flag-RnPex3p-HA were permeabilized with 25 μg/ml digitonin, under conditions in which plasma membranes are selectively permeabilized and intraperoxisomal proteins are inaccessible to exogenous antibodies (Okumoto and Fujiki, 1997; Okumoto et al., 1998b). Flag-Pex3p-HA was observed in a punctate staining pattern with use of an anti-flag antibody, whereas almost no staining of cells was noted using an anti-catalase antibody (Figure 7A, c and d). Similar punctate staining was discernible using an anti-HA antibody (Figure 7B, c and d). The data strongly suggest that both N- and C-terminal parts of Pex3p are exposed to the cytosol. The same results regarding Pex3p topology were obtained by expression of flag-Pex3p-HA in CHO-K1 cells (our unpublished results).
Figure 7.
Topology of Pex3p. (A and B) Morphological analysis. Transmembrane topology of Pex3p was determined by the differential cell permeabilization method. ZPG208 cells transfected with flag-RnPEX3-HA were fixed and then treated with 0.1% Triton X-100 (A and B, a and b) or 25 μg/ml digitonin (A and B, c and d) under conditions in which plasma membranes are specifically permeabilized (Okumoto and Fujiki, 1997; Okumoto et al., 1998b). Cells were stained with antibodies to flag (A, a and c), catalase (A and B, b and d), and HA (B, a and b). Note that Pex3p was detected after both types of treatments. Magnification, 630×; bar, 20 μm. (C) Protease treatment. The PNS fraction of CHO-K1 cells (1 × 106 each) transfected with flag-RnPEX3-HA was treated with proteinase K in the absence (lanes 1–4) or presence (lane 5) of Triton X-100. The amount of proteinase K used is indicated at the top. After terminating the reaction with phenylmethylsulfonyl fluoride, cell proteins were recovered by precipitation with trichloroacetic acid and analyzed by SDS-PAGE followed by immunoblot using antibodies specific for flag, HA, Pex14p, and AOx. Dots indicate nonspecific bands (Tamura et al., 1998), as in Figure 3B. The upward open arrowhead shows a cleavage product of AOx, migrating just below the AOx-B (Miura et al., 1992). Note that both N- and C-terminal parts of Pex3p were cleaved off by protease treatment, under which a matrix enzyme, AOx, was protected from the digestion.
Transmembrane topology of Pex3p was also determined by means of protease treatment. When the PNS fraction of CHO-K1 cells expressing flag-RnPex3p-HA was treated with proteinase K, Pex3p was detected in immunoblots using antibodies to flag and HA, before treatment with proteinase K (Figure 7C, lane 1). With 25 μg/ml the protease Pex3p was hardly detectable using the flag or HA antibody (lane 2). After digestion of the PNS fraction with 50 and 100 μg/ml proteinase K, Pex3p was no longer visible (lanes 3 and 4). Similarly, Pex3p was completely digested when adding 1% Triton X-100 (lane 5). Pex14p, a peroxin integrated to peroxisomal membranes and exposing its N- and C-terminal regions to the cytosol (Shimizu et al., 1999), was likewise digested by proteinase K, as assessed with antibody to Pex14p C-terminal peptide (Shimizu et al., 1999). Under such conditions, AOx, a matrix enzyme, was fully protected from digestion (lanes 2–4), whereas pretreatment with Triton X-100 abolished the protease protection (lane 5). Similar results were obtained using flag-RnPex3p-HA–expressing, peroxisome-restored ZPG208 cells (our unpublished results). Collectively, we conclude that both N- and C-terminal parts of Pex3p are exposed to the cytosol.
Kinetics of Peroxisome Biogenesis
We investigated kinetics of peroxisome assembly with respect to membrane vesicle formation as well as soluble protein import. ZPG208 originally expressing PTS2-GFP was transfected with flag-RnPEX3-HA and monitored under a fluorescent microscope. RnPex3p was detectable by immunoblot with anti-Pex3p antibody at 12 h after the transfection and reached a steady level at ∼24 h after transfection (Figure 8B). RnPex3p was also morphologically visible by staining with an anti-HA antibody in several punctate structures in cells at 12 h, whereas PTS2-GFP was diffused in the cytoplasm and apparently in the nucleus as well (Figure 8A, a with arrow and e), as seen in the untransfected cells (Figure 1a). At 18 h, Pex3p became more clearly visible in punctate structures in part of the transfected cells, possibly representing assembled peroxisomal membranes, where PTS2-GFP was not discernible in a punctate manner (Figure 8A, b with arrow and f). Several PMP70-positive particles in a cell were discernible at 18 h (Figure 8A, j, arrowheads). At 24 h, Pex3p-positive vesicles that had increased in number were colocalized with PTS2-GFP in a superimposable manner (Figure 8A, c and g). This was interpreted to mean that part of the assembled peroxisomal membrane vesicles imported PTS2-GFP. At 24 h, PMP70-positive particles, similar in number to Pex3p-carrying ones, were visible, thereby demonstrating reestablishment of membrane assembly (Figure 8A, k). In contrast, catalase was imported into peroxisomes (Figure 8A, m–p), superimposable with PTS2-GFP-positive vesicles (our unpublished results), only at 36 h after RnPEX3 transfection, thereby indicating that catalase is imported at a slower rate, compared with the case of PTS2. Collectively, peroxisomal membrane vesicles containing Pex3p as well as PMP70 are likely to form before the import of matrix proteins. The import kinetics of matrix proteins appears to be variable.
Figure 8.
Kinetics of peroxisome biogenesis. (A) ZPG208 cells expressing PTS2-GFP were transfected with pUcD2Hyg·flag-RnPEX3-HA and then monitored by fluorescent microscope. (a–h) Pex3p and PTS2-GFP were detected in the same set of cells at indicated time; (i–p) PMP70 and catalase were from the other sets, each with PTS2-GFP (our unpublished data). (a–d) Pex3p was visualized using rabbit anti-HA antibody and Texas Red-labeled goat anti-rabbit IgG antibody; (e–h) PTS2-GFP; (i–l and m–p) PMP70 and catalase were detected with rabbit antibodies to rat PMP70 and catalase, respectively, and a second antibody, Texas Red-labeled goat anti-rabbit IgG antibody. (a, e, i, and m) Twelve hours after the transfection; (b, f, j, and n) 18 h; (c, g, k, and o) 24 h; (d, h, l, and p) 36 h. Arrows in a and b indicate a cell showing several Pex3p-positive particles; arrowheads in j designate PMP70-containing particles. Note that Pex3p, but not PTS2-GFP and catalase, is already in vesicular structures at 12 and 18 h (see RESULTS). Magnification, 630×; bar, 20 μm. (B) Expression of rat Pex3p in ZPG208. Pex3p (arrowhead) was detected by immunoblotting of flag-RnPEX3-HA-transfected ZPG208 lysates (2 × 105 cells each) with anti-Pex3p antibody, at the indicated times, where cell-doubling time was 18 h.
Functional and Topogenic Regions of Pex3p
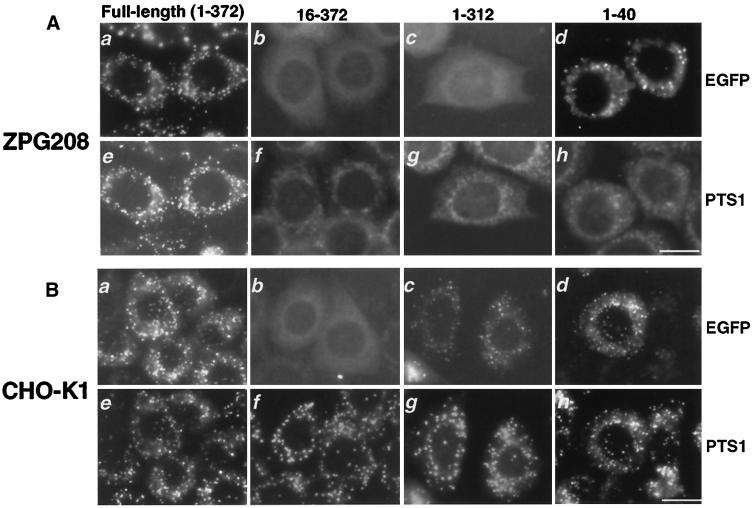
To elucidate structural and functional aspects of Pex3p, we constructed various truncated Pex3p variants by C-terminally fusing to EGFP and expressed them in ZPG208 and CHO-K1 cells. Full-length Pex3p was functional as a peroxin in restoring peroxisome biogenesis in ZPG208 and targeted to peroxisomes (Figure 9, A and B, a and e). ZPG208-derived, full-length but functionally inactive Pex3p was translocated to peroxisomes, as assessed by colocalization with PTS1 proteins, when expressed in CHO-K1 (our unpublished results). However, the endogenous Pex3p mutant was not found in ZPG208 on immunoblots, presumably because of a degradation. Pex3p variants truncated at the N-terminal portion, such as those of residues 1–15 (Figure 9, A and B, b and f) as well as 1–30, 1–109, and 1–150 (Table 4), were all biologically inactive and were apparently localized in the cytoplasm. In contrast, Pex3p with residues 1–312, i.e., with deletion of the C-terminal 60 amino acids, was localized in peroxisomes, as assessed with PTS1, although the biological activity was eliminated (Figure 9, A and B, c and g). Another mutant deleted from residue 204 to the C terminus was also inactive but was still targeted to peroxisomes (Table 4). All of the truncation mutants used for fusion with EGFP were N-terminally tagged with flag and likewise expressed in ZPG208. These Pex3p mutants did not restore the impaired assembly of peroxisomes (our unpublished data), consistent with results described above. A Pex3p variant with only the N-terminal sequence 1–40 directed EGFP to peroxisomes when expressed in CHO-K1 (Figure 9B, d and h). This 40-amino-acid Pex3p protein was targeted to many vesicular structures in ZPG208, where no complementation was evident (Figure 9A, d and h). Interestingly, not only full-length Pex3p-EGFP, similar to flag-Pex3p (see Figure 6A), but also that of residues 1–40 were targeted to peroxisomal remnants in pex2 Z65 (our unpublished results). Taken together, it is apparent that nearly full-length Pex3p is required for biological activity, whereas peroxisome-targeting information resides in N-terminal residues 1–40, consistent with the findings in humans (Kammerer et al., 1998; Soukupova et al., 1999) and yeast (Wiemer et al., 1996) Pex3p.
Figure 9.
Functional and topogenic regions of Pex3p. N-terminally flag-tagged and C-terminally EGFP-fused rat Pex3p and its variants were verified for peroxisome-restoring activity in ZPG208 and intracellular localization in wild-type CHO-K1. (A) ZPG208 was transfected with cDNAs encoding fused proteins encompassing Pex3p with amino acid residues of the full-length, 1–372 (a and e), 16–372 (b and f), 1–312 (c and g), and 1–40 (d and h). Peroxisome-forming activity was verified by import of Pex3p-EGFP, in which peroxisomes were assessed by immunostaining of PTS1. Bar, 20 μm. (B) The same set of the full-length and truncated variants of Pex3p-EGFP fusion protein were also expressed in CHO-K1; peroxisomal localization of Pex3p-EGFP assessed by GFP fluorescence was confirmed by immunostaining of PTS1. Bar, 20 μm.
Table 4.
Peroxin activity and intracellular localization of Pex3p variants
| Flag-Pex3p-EGFP residues | Peroxisome-restoring activity | Localization |
|---|---|---|
| Full-length, 1–372 | + | Peroxisome |
| 16–372 | − | Cytosol |
| 31–372 | − | Cytosol |
| 110–372 | − | Cytosol |
| 151–372 | − | Cytosol |
| 1–312 | − | Peroxisome |
| 1–203 | − | Peroxisome |
| 1–40 | − | Peroxisome |
N-terminally flag tagged and C-terminally EGFP-fused full-length Pex3p and truncated variants were verified for peroxisome-restoring activity in ZPG208 and intracellular localization in wild-type CHO-K1 cells, as in Figure 9. +, complemented; −, not complemented.
Identification of Pex19p as a Binding Partner of Pex3p
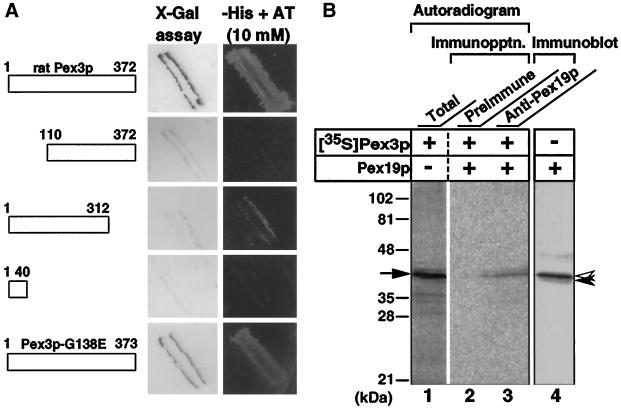
To determine whether Pex3p interacts with mammalian peroxins, we did the yeast two-hybrid assay. Pex19p, a farnesylated protein required for peroxisome assembly in CG-J (Matsuzono et al., 1999), gave positive β-galactosidase activity and yeast growth in His−/3-aminotriazole (AT)+ medium (Figure 10A). In contrast, the other peroxins, including Pex16p (Honsho et al., 1998), Pex14p (Shimizu et al., 1999), Pex13p (Toyama et al., 1999), Pex11pα (Abe et al., 1998), and Pex11pβ (Abe and Fujiki, 1998), as well as the RING peroxins Pex2p (Tsukamoto et al., 1991), Pex10p (Okumoto et al., 1998a), and Pex12p (Okumoto et al., 1998b) resulted in negative findings (our unpublished results). To search for a region responsible for the interaction with Pex19p, a Pex3p variant with amino acid residues 1–312 gave a weak signal (Figure 10A), whereas all of the other mutants, including those with residues 110–372 (Figure 10A), 151–372 and 1–203 (our unpublished results), and 1–40 (Figure 10A) did not interact with Pex19p. Interestingly, ZPG208-derived Pex3p-G138E was positive, both in β-galactosidase activity and yeast growth in His−/AT+ medium. Therefore, the interaction apparently requires nearly full-length Pex3p.
Figure 10.
Physical interaction of Pex3p with Pex19p. (A) Yeast two-hybrid assay. Host strain MaV203 was transformed with two plasmids, one encoding the DNA binding domain (BD) of Gal4p fused to either the full-length or truncated forms comprising residues 110–372, 1–312, and 1–40 of rat Pex3p or ZPG208-derived full-length Pex3p, and the other encoding the Gal4p transactivation domain (TA) fused to the full-length Pex19p. Transformants were tested for β-galactosidase expression. Color intensities of these strains after the β-galactosidase filter assay are shown. The BD fused to the full-length Pex3p showed no self-activation. Transformants were also verified for growth on synthetic complete medium without histidine (−His) in the presence of 10 mM AT (+AT). (B) Coimmunoprecipitation of Pex3p with Pex19p. Pex3p and Pex19p, indistinguishable in size in SDS-PAGE, were separately synthesized in a cell-free translation system in the presence and absence of [35S]methionine and [35S]cysteine, respectively. After incubation of [35S]Pex3p with Pex19p, immunoprecipitation (Immunopptn.) was done using an anti-Pex19p antibody. [35S]Pex3p was detected using a Fujix BAS1500 Bio-Imaging Analyzer (Fuji Photo Film, Tokyo, Japan) at an exposure for 16 h; Pex19p was visualized by immunoblot. Lane 1, [35S]Pex3p (1 μl of translation product); lanes 2 and 3, immunoprecipitates from 10 μl each of translation products [35S]Pex3p and Pex19p, with preimmune serum and anti-Pex19p antibody, respectively; lane 4, Pex19p (10 μl) detected by immunoblot with anti-Pex19p antibody. The arrow indicates Pex3p; solid and open arrowheads designate Pex19p with and without modification by farnesylation, respectively (Matsuzono et al., 1999).
To confirm the findings in the two-hybrid assay, cell-free synthesized rat 35S-Pex3p was incubated with the in vitro transcription–translation product of human PEX19. Immunoprecipitation of Pex19p gave rise to concomitant recovery of 35S-Pex3p, whereas that with the preimmune serum showed no protein band (Figure 10B, lanes 1–3). It is noteworthy that cell-free synthesized Pex19p was detected as two bands on immunoblots: one representing the farnesylated Pex19p (solid arrowhead) and the other (open arrowhead) for the nonmodified one (lane 4), as described (Matsuzono et al., 1999). Conversely, 35S-Pex19p coimmunoprecipitated with Pex3p when using anti-Pex3p antibody (our unpublished results). Collectively, the results demonstrate that Pex3p specifically binds to Pex19p.
DISCUSSION
The CG17 CHO cell mutants ZPG208 and ZPG209 are defective in import of both matrix and membrane proteins, similar to the phenotype of a pex19 mutant, ZP119 (Kinoshita et al., 1998). Peroxisomal remnants were seen in 10 other CGs of CHO cell mutants (Zoeller et al., 1989; Shimozawa et al., 1992; Okumoto et al., 1997; Tateishi et al., 1997; Otera et al., 1998; Ghaedi et al., 1999a; Toyama et al., 1999) and 9 CGs of fibroblasts from PBD patients (Santos et al., 1992; Wendland and Subramani, 1993; Shimozawa et al., 1998a), excluding CG-G (Shimozawa et al., 1998a), PEX16-deficient CG-D (Honsho et al., 1998), and PEX19-defective CG-J (Kinoshita et al., 1998; Shimozawa et al., 1998a; Matsuzono et al., 1999). In the present work, we isolated a rat Pex3p cDNA by functional complementation of ZPG208. Expression of the full-length RnPEX3 fully restored the impaired peroxisome biogenesis, including membrane vesicle assembly, in ZPG208 and ZPG209. We delineated the homozygotic mutant PEX3 allele from ZPG208 and ZPG209: a one-base transition, G413 to A in a codon for Gly138, resulted in Glu138. Pex3p with G138E was not functionally active in complementing impaired peroxisome biogenesis in ZPG208. Accordingly, PEX3 is responsible for the peroxisome biogenesis of CG17 and is the 12th gene to be identified to date in mammals (Table 3). None of the fibroblasts from patients with PBD of 13 CGs was complemented, indicating that the PEX3 gene is not the causal gene of human peroxisome-defective disorders of the CGs so far classified. ZPG208 and ZPG209 are thus the first pex3 mutants to be identified in mammals. It is noteworthy that yeast Pex3p expression complemented peroxisome biogenesis in respective pex3 mutants of S. cerevisiae (Hoehfeld et al., 1991), H. polymorpha (Baerends et al., 1996), and P. pastoris (Wiemer et al., 1996), where pex3 cells of H. polymorpha and P. pastoris were apparently absent from peroxisomal structures (Baerends et al., 1996; Wiemer et al., 1996).
Upon transfection of RnPEX3 into ZPG208 devoid of peroxisomal remnants, most striking was the formation of morphologically recognizable peroxisomal membrane vesicles, apparently preceding the import of matrix proteins such as PTS1 and PTS2 proteins and catalase. Dysfunction of Pex3p caused impaired membrane assembly, resulting in the mutant phenotype defect of matrix protein import and used for mutant screening. Pex3p can be classified as a peroxin essential for the assembly of peroxisome membranes. Very recently, Pex16p and Pex19p were also shown to function in assembly of peroxisome vesicles in mammals (Honsho et al., 1998; Matsuzono et al., 1999; South and Gould, 1999), as was the case for Pex19p in yeast (Snyder et al., 1999). Mutation of human Pex16p (Honsho et al., 1998; South and Gould, 1999) and Pex19p (Matsuzono et al., 1999) severely affected peroxisome assembly in CG-D (CG-IX) and CG-J patients with Zellweger syndrome. Accordingly, Pex16p and Pex19p can also be categorized into this group of peroxins. We demonstrated in the present study that the membrane assembly process(es) involving integration of Pex3p is temporally differentiated from the import of soluble proteins during peroxisome biogenesis. Moreover, import of PTS2 and catalase at a different rate in PEX3-transfected ZPG208 implies temporally differential translocation of matrix proteins into peroxisomal membrane vesicles. Similar types of protein import, distinct between membrane polypeptides and soluble proteins, have been observed in pex16 and pex19 mutant cells, upon expression of complementing cDNAs PEX16 and PEX19, respectively (Honsho et al., 1998; Matsuzono et al., 1999; South and Gould, 1999). Therefore, it is most likely that Pex3p functions as an essential factor required for the translocation process of membrane protein and/or membrane vesicle assembly, possibly in a concerted manner with other peroxins such as Pex16p and Pex19p. Taken together, our results provide evidence that peroxisomes may form de novo and do not have to arise from preexisting, morphologically recognizable peroxisomes. At such an early stage of peroxisome assembly, ER may be involved, as was suggested for Pex2p and Pex16p, both initially residing in ER, in Y. lipolytica (Titorenko and Rachubinski, 1998). However, no direct evidence for the involvement of ER in peroxisome assembly has been noted in mammalian cells. Accordingly, several issues, including those regarding roles of the peroxins Pex3p, Pex16p, and Pex19p in assembly of membrane vesicles as well as translocons for membrane polypeptides and soluble matrix proteins, remain to be addressed.
We found that Pex3p interacts with Pex19p both in vivo and in vitro. However, it is unclear whether the interaction is direct or is mediated by a factor(s), if any, present in the assay used. Such Pex3p–Pex19p binding was recently found in S. cerevisiae (Goette et al., 1998) and P. pastoris (Snyder et al., 1999) as well as in human cells (Soukupova et al., 1999). It is interesting to note that Pex3p with G138E also interacts with Pex19p in vivo, in yeast two-hybrid assays. Although Pex3p-G138E was under the detectable level in the mutant ZPG208, this mutant Pex3p appears to be stable in S. cerevisiae used for the binding assay, as in CHO-K1 cells. We interpreted this observation to mean that the undetectable level of Pex3p, presumably because of a rapid turnover, is the cause of the phenotype of pex3 ZPG208. Pex3p may function in peroxisomal membrane assembly at an early stage of peroxisome assembly by interacting with Pex19p but not with Pex16p. Pex19p is a farnesylated peroxin, partly if not all residing on the peroxisome membrane, exposing its N-terminal portion to the cytosol (Matsuzono et al., 1999). However, it is not clear whether Pex3p is a prerequisite peroxin for Pex19p to be localized to and/or anchored on peroxisomal membranes. Because several truncated Pex3p mutants, including that consisting of only the N-terminal 40 amino acid residues, were properly translocated to peroxisomal membranes despite the lack of binding to Pex19p, as verified in yeast two-hybrid assays, Pex3p does not appear to require Pex19p for targeting. It is possible that interaction of Pex3p and Pex19p leads to assembly of potential membrane vesicles, which then function as protein import-competent machinery, comprising at least Pex14p (Fransen et al., 1998; Shimizu et al., 1999; Will et al., 1999) and Pex13p (Liu et al., 1999; Shimozawa et al., 1999; Toyama et al., 1999) and possibly including the RING family Pex2p (Tsukamoto et al., 1991), Pex10p (Okumoto et al., 1998a; Warren et al., 1998), and Pex12p (Chang et al., 1997; Okumoto and Fujiki, 1997; Okumoto et al., 1998b). It is also noteworthy that Pex3p may function upstream of Pex19p in P. pastoris (Snyder et al., 1999). All these findings suggest that assembly processes of peroxisome membrane vesicles are mediated by Pex3p, Pex16p, and Pex19p in mammals. The assembled membrane vesicles import other membrane components, including those for a potential matrix protein import machinery, to form “premature peroxisomes,” which are capable of importing matrix proteins. The matured peroxisomes then divide so that progeny peroxisomes emerge. Pex11p is more likely involved in proliferation and division of peroxisomes (Abe and Fujiki, 1998; Abe et al., 1998; Schrader et al., 1998) (I. Abe and Y. Fujiki, unpublished observation). Further investigation, using ZPG208 and ZPG209 together with PEX3, should shed light on molecular mechanisms involved in peroxisome biogenesis, especially with respect to membrane vesicle assembly at the initial stage of peroxisome biogenesis.
Pex14p has been characterized as a convergent component of potential peroxisomal import machinery of soluble proteins such as PTS1 and PTS2 (Albertini et al., 1997; Fransen et al., 1998; Shimizu et al., 1999; Will et al., 1999). Peroxisomal membrane ghosts are discernible in CHO pex14 mutants, where the matrix protein import is severely impaired (Shimizu et al., 1999). The presence of Pex14p in the pex3 mutant is consistent with that of Pex14p found in a pex16 Zellweger patient fibroblasts (South and Gould, 1999) (M. Honsho and Y. Fujiki, unpublished observation) and a pex19 mutant (Matsuzono et al., 1999) (Y. Matsuzono and Y. Fujiki, unpublished observation). This may mean that Pex14p in the sedimentable membrane fraction functions, in addition to a role as a convergent component of the translocon for soluble proteins, as a factor, including a scaffold for the interaction and assembly of several peroxins such as Pex3p, Pex16p, and Pex19p. These are all required for peroxisome membrane biogenesis. Pex14p may thus regulate an early stage of peroxisome biogenesis. However, it is also equally possible that contrary to Pex12p, Pex13p, and PMP70, Pex14p is simply one of the membrane proteins that is stable in cell mutants lacking morphologically recognizable peroxisomal membrane vesicles. Physiological consequences of Pex14p-positive membranes found in pex3, pex16, and pex19 mutants remain to be determined.
With respect to the membrane topology of Pex3p, we concluded from several lines of morphological and biochemical evidence that both N- and C-terminal parts are oriented to the cytosol. Contrary to this observation, Soukupova et al. (1999) suggested that myc-tagged human Pex3p faces its C-terminal region to the cytoplasm and the N-terminal part to the matrix side of peroxisomes, when expressed in cultured human skin fibroblasts, as is the case for S. cerevisiae Pex3p (Hoehfeld et al., 1991). It is less likely that Pex3p has a different topology depending on cell types, even in mammalian cells, although different epitope tags may affect the Pex3p topology. We showed that topogenic information of Pex3p resides at the N-terminal region and comprises residues at positions 1–40, in good agreement with the observation by other investigators on human Pex3p (Kammerer et al., 1998; Soukupova et al., 1999) and P. pastoris Pex3p (Wiemer et al., 1996). Baerends et al. (1996) noted that the N-terminal sequence, 1–16, of H. polymorpha Pex3p targeted catalase lacking functional PTS1 to ER and nuclear membranes, thus inferring ER-mediated peroxisome assembly. The highly conserved residues at 9–15, LKRHKKK (proposed consensus residues in boldface letters) of human Pex3p are likely to be responsible for such targeting activity. Pex3p (1–40)-GFP was targeted not only to peroxisomes in the wild-type CHO-K1 cells but also to peroxisomal remnants in CHO mutants defective in PTS1 and PTS2 import, which implies that the N-terminal 40-amino-acid sequence is sufficient for localization of Pex3p to peroxisomal membranes. A potential receptor recognizing such membrane topogenic signals may be investigated if one makes use of Pex3p and its variants together with the pex3 mutant.
ACKNOWLEDGMENTS
We thank M. Honsho for helpful comments and discussion and other members of our laboratory for stimulating discussion. M. Ohara provided language assistance. This work was supported in part by a CREST grant (to Y.F.) from the Japan Science and Technology Corporation and a grant-in-aid for scientific research (09044094 to Y.F.) from The Ministry of Education, Science, Sports, and Culture of Japan.
Abbreviations used:
- AOx
acyl-coenzyme A oxidase
- AT
3-aminotriazole
- CG
complementation group
- CHO
Chinese hamster ovary
- CoA
coenzyme A
- EGFP
enhanced green fluorescent protein
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- HA
hemagglutinin
- IgG
immunoglobulin G
- ORF
open reading frame
- P9OH
9-(1′-pyrene) nonanol
- P12
12-(1′-pyrene) dodecanoic acid
- PBD
peroxisome biogenesis disorder
- PMP70
70-kDa peroxisomal integral membrane protein
- PNS
postnuclear supernatant
- PTS
peroxisomal targeting signal
- RT
reverse transcription
REFERENCES
- Abe I, Fujiki Y. cDNA cloning and characterization of a constitutively expressed isoform of the human peroxin Pex11p. Biochem Biophys Res Commun. 1998;252:529–533. doi: 10.1006/bbrc.1998.9684. [DOI] [PubMed] [Google Scholar]
- Abe I, Okumoto K, Tamura S, Fujiki Y. Clofibrate-inducible, 28-kDa peroxisomal integral membrane protein is encoded by PEX11. FEBS Lett. 1998;431:468–472. doi: 10.1016/s0014-5793(98)00815-1. [DOI] [PubMed] [Google Scholar]
- Albertini M, Rehling P, Erdmann R, Girzalsky W, Kiel JAKW, Veenhuis M, Kunau W-H. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell. 1997;89:83–92. doi: 10.1016/s0092-8674(00)80185-3. [DOI] [PubMed] [Google Scholar]
- Baerends RJS, Rasmussen SW, Hilbrands RE, van der Heide M, Faber KN, Reuvekamp PTW, Klei JAKW, Cregg JM, van der Klei IJ, Veenhuis M. The Hansenula polymorpha PER9 gene encodes a peroxisomal membrane protein essential for peroxisome assembly and integrity. J Biol Chem. 1996;271:8887–8894. doi: 10.1074/jbc.271.15.8887. [DOI] [PubMed] [Google Scholar]
- Bodier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- Chang C-C, Lee W-H, Moser H, Valle D, Gould SJ. Isolation of the human PEX12 gene, mutated in group 3 of the peroxisome biogenesis disorders. Nat Genet. 1997;15:385–388. doi: 10.1038/ng0497-385. [DOI] [PubMed] [Google Scholar]
- Erdmann R, Veenhuis M, Kunau W-H. Peroxisomes: organelles at the crossroads. Trends Cell Biol. 1997;7:400–407. doi: 10.1016/S0962-8924(97)01126-4. [DOI] [PubMed] [Google Scholar]
- Fransen M, Terlecky SR, Subramani S. Identification of a human PTS1 receptor docking protein directly required for peroxisomal protein import. Proc Natl Acad Sci USA. 1998;95:8087–8092. doi: 10.1073/pnas.95.14.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y. Molecular defects in genetic diseases of peroxisomes. Biochim Biophys Acta. 1997;1361:235–250. doi: 10.1016/s0925-4439(97)00051-3. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Fowler S, Shio H, Hubbard AL, Lazarow PB. Polypeptide and phospholipid composition of the membrane of rat liver peroxisomes: comparison with endoplasmic reticulum and mitochondrial membranes. J Cell Biol. 1982a;93:103–110. doi: 10.1083/jcb.93.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982b;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Rachubinski RA, Lazarow PB. Synthesis of a major integral membrane polypeptide of rat liver peroxisomes on free polysomes. Proc Natl Acad Sci USA. 1984;81:7127–7131. doi: 10.1073/pnas.81.22.7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaedi K, Itagaki A, Toyama R, Tamura S, Matsumura T, Kawai A, Shimozawa N, Suzuki Y, Kondo N, Fujiki Y. Newly identified Chinese hamster ovary cell mutants defective in peroxisome assembly represent complementation group A of human peroxisome biogenesis disorders and one novel group in mammals. Exp Cell Res. 1999a;248:482–488. doi: 10.1006/excr.1999.4412. [DOI] [PubMed] [Google Scholar]
- Ghaedi K, Kawai A, Okumoto K, Tamura S, Shimozawa N, Suzuki Y, Kondo N, Fujiki Y. Isolation and characterization of novel peroxisome biogenesis-defective Chinese hamster ovary cell mutants using green fluorescent protein. Exp Cell Res. 1999b;248:489–497. doi: 10.1006/excr.1999.4413. [DOI] [PubMed] [Google Scholar]
- Goette K, Girzalsky W, Linkert M, Baumgart E, Kammerer S, Kunau W-H, Erdmann R. Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol Cell Biol. 1998;18:616–628. doi: 10.1128/mcb.18.1.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M, Ishii N, Kagamiyama H, Osumi T, Hashimoto T. Structural analysis of cDNA for rat peroxisomal 3-ketoacyl-CoA thiolase. J Biol Chem. 1987;262:8151–8158. [PubMed] [Google Scholar]
- Hoehfeld J, Veenhuis M, Kunau WH. PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J Cell Biol. 1991;114:1167–1178. doi: 10.1083/jcb.114.6.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honsho M, Tamura S, Shimozawa N, Suzuki Y, Kondo N, Fujiki Y. Mutation in PEX16 is causal in the peroxisome-deficient Zellweger syndrome of complementation group D. Am J Hum Genet. 1998;63:1622–1630. doi: 10.1086/302161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer S, Holzinger A, Welsch U, Roscher AA. Cloning and characterization of the gene encoding the human peroxisomal assembly protein Pex3p. FEBS Lett. 1998;429:53–60. doi: 10.1016/s0014-5793(98)00557-2. [DOI] [PubMed] [Google Scholar]
- Kinoshita N, Ghaedi K, Shimozawa N, Wanders RJA, Matsuzono Y, Imanaka T, Okumoto K, Suzuki Y, Kondo N, Fujiki Y. Newly identified Chinese hamster ovary cell mutants are defective in biogenesis of peroxisomal membrane vesicles (peroxisomal ghosts), representing a novel complementation group in mammals. J Biol Chem. 1998;273:24122–24130. doi: 10.1074/jbc.273.37.24122. [DOI] [PubMed] [Google Scholar]
- Lazarow PB, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- Lazarow PB, Moser HW. Disorders of peroxisome biogenesis. In: Scriver CR, Beaudet AI, Sly WS, Valle D, editors. The Metabolic Basis of Inherited Disease. New York: McGraw-Hill; 1995. pp. 2287–2324. [Google Scholar]
- Liu Y, Bjoerkman J, Urquhart A, Wanders RJA, Crane DI, Gould SJ. PEX13 is mutated in complementation group 13 of the peroxisome-biogenesis disorders. Am J Hum Genet. 1999;65:621–634. doi: 10.1086/302534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzono Y, Kinoshita N, Tamura T, Shimozawa N, Hamasaki M, Ghaedi K, Wanders RJA, Suzuki Y, Kondo K, Fujiki Y. Human PEX19: cDNA cloning by functional complementation, mutation analysis in a patient with Zellweger syndrome and potential role in peroxisomal membrane assembly. Proc Natl Acad Sci USA. 1999;96:2116–2121. doi: 10.1073/pnas.96.5.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura S, Kasuya-Arai I, Mori H, Miyazawa S, Osumi T, Hashimoto T, Fujiki Y. Carboxyl-terminal consensus Ser-Lys-Leu-related tripeptide of peroxisomal proteins functions in vitro as a minimal peroxisome-targeting signal. J Biol Chem. 1992;267:14405–14411. [PubMed] [Google Scholar]
- Miura S, Miyazawa S, Osumi T, Hashimoto T, Fujiki Y. Post-translational import of 3-ketoacyl-CoA thiolase into rat liver peroxisomes in vitro. J Biochem. 1994;115:1064–1068. doi: 10.1093/oxfordjournals.jbchem.a124458. [DOI] [PubMed] [Google Scholar]
- Miyazawa S, Hayashi H, Hijikata M, Ishii N, Furuta S, Kagamiyama H, Osumi T, Hashimoto T. Complete nucleotide sequence of cDNA and predicted amino acid sequence of rat acyl-CoA oxidase. J Biol Chem. 1987;262:8131–8137. [PubMed] [Google Scholar]
- Miyazawa S, Osumi T, Hashimoto T, Ohno K, Miura S, Fujiki Y. Peroxisome targeting signal of rat liver acyl-coenzyme A oxidase resides at the carboxy terminus. Mol Cell Biol. 1989;9:83–91. doi: 10.1128/mcb.9.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morand OH, Allen L-AH, Zoeller RA, Raetz CRH. A rapid selection for animal cell mutants with defective peroxisomes. Biochim Biophys Acta. 1990;1034:132–141. doi: 10.1016/0304-4165(90)90066-6. [DOI] [PubMed] [Google Scholar]
- Moser AB, et al. Phenotype of patients with peroxisomal disorders subdivided into sixteen complementation groups. J Pediatr. 1995;127:13–22. doi: 10.1016/s0022-3476(95)70250-4. [DOI] [PubMed] [Google Scholar]
- Motley A, Hettema E, Distel B, Tabak H. Differential protein import deficiencies in human peroxisome assembly disorders. J Cell Biol. 1994;125:755–767. doi: 10.1083/jcb.125.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto K, Bogaki A, Tateishi K, Tsukamoto T, Osumi T, Shimozawa N, Suzuki Y, Orii T, Fujiki Y. Isolation and characterization of peroxisome-deficient Chinese hamster ovary cell mutants representing human complementation group III. Exp Cell Res. 1997;233:11–20. doi: 10.1006/excr.1997.3552. [DOI] [PubMed] [Google Scholar]
- Okumoto K, Fujiki Y. PEX12 encodes an integral membrane protein of peroxisomes. Nat Genet. 1997;17:265–266. doi: 10.1038/ng1197-265. [DOI] [PubMed] [Google Scholar]
- Okumoto K, Itoh R, Shimozawa N, Suzuki Y, Tamura S, Kondo N, Fujiki Y. Mutation in PEX10 is the cause of Zellweger peroxisome deficiency syndrome of complementation group B. Hum Mol Genet. 1998a;7:1399–1405. doi: 10.1093/hmg/7.9.1399. [DOI] [PubMed] [Google Scholar]
- Okumoto K, et al. PEX12, the pathogenic gene of group III Zellweger syndrome: cDNA cloning by functional complementation on a CHO cell mutant, patient analysis, and characterization of Pex12p. Mol Cell Biol. 1998b;18:4324–4336. doi: 10.1128/mcb.18.7.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi T, Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T. Amino-terminal presequence of the precursor of peroxisomal 3-ketoacyl-CoA thiolase is a cleavable signal peptide for peroxisomal targeting. Biochem Biophys Res Commun. 1991;181:947–954. doi: 10.1016/0006-291x(91)92028-i. [DOI] [PubMed] [Google Scholar]
- Otera H, et al. Peroxisome targeting signal type 1 (PTS1) receptor is involved in import of both PTS1 and PTS2: studies with PEX5-defective CHO cell mutants. Mol Cell Biol. 1998;18:388–399. doi: 10.1128/mcb.18.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos A, Christodoulou J, Chow CW, Goldblatt J, Paton BC, Orii T, Suzuki Y, Shimozawa N. Peroxisomal assembly defects: clinical, pathologic, and biochemical findings in two patients in a newly identified complementation group. J Pediatr. 1995;127:596–599. doi: 10.1016/s0022-3476(95)70121-4. [DOI] [PubMed] [Google Scholar]
- Santos MJ, Hoefler S, Moser AB, Moser HW, Lazarow PB. Peroxisome assembly mutations in humans: structural heterogeneity in Zellweger syndrome. J Cell Physiol. 1992;151:103–112. doi: 10.1002/jcp.1041510115. [DOI] [PubMed] [Google Scholar]
- Santos MJ, Imanaka T, Shio H, Small GM, Lazarow PB. Peroxisomal membrane ghosts in Zellweger syndrome—aberrant organelle assembly. Science. 1988;239:1536–1538. doi: 10.1126/science.3281254. [DOI] [PubMed] [Google Scholar]
- Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- Schrader M, Reuber BE, Morrell JC, Jimenez-Sanchez G, Obie C, Stroh TA, Valle D, Schroer TA, Gould SJ. Expression of PEX11β mediates peroxisome proliferation in the absence of extracellular stimuli. J Biol Chem. 1998;273:29607–29614. doi: 10.1074/jbc.273.45.29607. [DOI] [PubMed] [Google Scholar]
- Shimizu N, et al. The peroxin Pex14p: cDNA cloning by functional complementation on a Chinese hamster ovary cell mutant, characterization, and functional analysis. J Biol Chem. 1999;274:12593–12604. doi: 10.1074/jbc.274.18.12593. [DOI] [PubMed] [Google Scholar]
- Shimozawa N, et al. Genetic basis of peroxisome-assembly mutants of humans, Chinese hamster ovary cells and yeast: identification of a new complementation group of peroxisome-biogenesis disorders apparently lacking peroxisomal-membrane ghosts. Am J Hum Genet. 1998a;63:1898–1903. doi: 10.1086/302142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimozawa N, et al. Peroxisome biogenesis disorders: identification of a new complementation group distinct from peroxisome-deficient CHO mutants and not complemented by human. PEX13. Biochem Biophys Res Commun. 1998b;243:368–371. doi: 10.1006/bbrc.1997.8067. [DOI] [PubMed] [Google Scholar]
- Shimozawa N, et al. Nonsense and temperature-sensitive mutations in PEX13 are the cause of complementation group H of peroxisome biogenesis disorders. Hum Mol Genet. 1999;8:1077–1083. doi: 10.1093/hmg/8.6.1077. [DOI] [PubMed] [Google Scholar]
- Shimozawa N, Tsukamoto T, Suzuki Y, Orii T, Fujiki Y. Animal cell mutants represent two complementation groups of peroxisome-defective Zellweger syndrome. J Clin Invest. 1992;90:1864–1870. doi: 10.1172/JCI116063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder WB, Faber KN, Wenzel TJ, Koller A, Leuers GH, Rangell L, Keller GA, Subramani S. Pex19p interacts with Pex3p and Pex10p and is essential for peroxisome biogenesis in Pichia pastoris. Mol Biol Cell. 1999;10:1745–1761. doi: 10.1091/mbc.10.6.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukupova M, Sprenger C, Gorgas K, Kunau W-H, Dodt G. Identification and characterization of the human peroxin PEX3. Eur J Cell Biol. 1999;78:357–374. doi: 10.1016/S0171-9335(99)80078-8. [DOI] [PubMed] [Google Scholar]
- South ST, Gould SJ. Peroxisome synthesis in the absence of preexisting peroxisomes. J Cell Biol. 1999;144:255–266. doi: 10.1083/jcb.144.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S. PEX genes on the rise. Nat Genet. 1997;15:331–333. doi: 10.1038/ng0497-331. [DOI] [PubMed] [Google Scholar]
- Swinkels BW, Gould SJ, Bodnar AG, Rachubinski RA, Subramani S. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 1991;10:3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura S, Okumoto K, Toyama R, Shimozawa N, Tsukamoto T, Suzuki Y, Osumi T, Kondo N, Fujiki Y. Human PEX1 cloned by functional complementation on a CHO cell mutant is responsible for peroxisome-deficient Zellweger syndrome of complementation group I. Proc Natl Acad Sci USA. 1998;95:4350–4355. doi: 10.1073/pnas.95.8.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi K, Okumoto K, Shimozawa N, Tsukamoto T, Osumi T, Suzuki Y, Kondo N, Okano I, Fujiki Y. Newly identified Chinese hamster ovary cell mutants defective in peroxisome biogenesis represent two novel complementation groups in mammals. Eur J Cell Biol. 1997;73:352–359. [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA. The endoplasmic reticulum plays an essential role in peroxisome biogenesis. Trends Biochem Sci. 1998;23:231–233. doi: 10.1016/s0968-0004(98)01226-2. [DOI] [PubMed] [Google Scholar]
- Toyama R, Mukai S, Itagaki A, Tamura S, Shimozawa N, Suzuki Y, Kondo N, Wanders RJA, Fujiki Y. Isolation, characterization, and mutation analysis of PEX13-defective Chinese hamster ovary cell mutants. Hum Mol Genet. 1999;8:1673–1681. doi: 10.1093/hmg/8.9.1673. [DOI] [PubMed] [Google Scholar]
- Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T, Osumi T. Characterization of the signal peptide at the amino terminus of the rat peroxisomal 3-ketoacyl-CoA thiolase precursor. J Biol Chem. 1994a;269:6001–6010. [PubMed] [Google Scholar]
- Tsukamoto T, Miura S, Fujiki Y. Restoration by a 35K membrane protein of peroxisome assembly in a peroxisome-deficient mammalian cell mutant. Nature. 1991;350:77–81. doi: 10.1038/350077a0. [DOI] [PubMed] [Google Scholar]
- Tsukamoto T, et al. Peroxisome assembly factor-2, a putative ATPase cloned by functional complementation on a peroxisome-deficient mammalian cell mutant. Nat Genet. 1995;11:395–401. doi: 10.1038/ng1295-395. [DOI] [PubMed] [Google Scholar]
- Tsukamoto T, Shimozawa N, Fujiki Y. Peroxisome assembly factor 1: nonsense mutation in a peroxisome-deficient Chinese hamster ovary cell mutant and deletion analysis. Mol Cell Biol. 1994b;14:5458–5465. doi: 10.1128/mcb.14.8.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Yokota S, Fujiki Y. Isolation and characterization of Chinese hamster ovary cell mutants defective in assembly of peroxisomes. J Cell Biol. 1990;110:651–660. doi: 10.1083/jcb.110.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren DS, Morrell JC, Moser HW, Valle D, Gould SJ. Identification of PEX10, the gene defective in complementation group 7 of the peroxisome-biogenesis disorders. Am J Hum Genet. 1998;63:347–359. doi: 10.1086/301963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Cregg J. Peroxisome biogenesis. Bioessays. 1997;19:57–66. doi: 10.1002/bies.950190110. [DOI] [PubMed] [Google Scholar]
- Wendland M, Subramani S. Presence of cytoplasmic factors functional in peroxisomal protein import implicates organelle-associated defects in several human peroxisomal disorders. J Clin Invest. 1993;92:2462–2468. doi: 10.1172/JCI116854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemer EAC, Luers GH, Faber KN, Wenzel T, Veenhuis M, Subramani S. Isolation and characterization of Pas2p, a peroxisomal membrane protein essential for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. J Biol Chem. 1996;271:18973–18980. doi: 10.1074/jbc.271.31.18973. [DOI] [PubMed] [Google Scholar]
- Will GK, Soukupova M, Hong X, Erdmann KS, Kiel JAKW, Dodt G, Kunau W-H, Erdmann R. Identification and characterization of the human orthologue of yeast Pex14p. Mol Cell Biol. 1999;19:2265–2277. doi: 10.1128/mcb.19.3.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoeller RA, Allen L-AH, Santos MJ, Lazarow PB, Hashimoto T, Tartakoff AM, Raetz CRH. Chinese hamster ovary cell mutants defective in peroxisome biogenesis. Comparison to Zellweger syndrome. J Biol Chem. 1989;264:21872–21878. [PubMed] [Google Scholar]
- Zoeller RA, Morand OH, Raetz CRH. A possible role for plasmalogens in protecting animal cells against photosensitized killing. J Biol Chem. 1988;263:11590–11596. [PubMed] [Google Scholar]