Abstract
The formation of the cell plate, a unique structure in dividing plant cells, is pivotal for cytokinesis. A mutation in the Arabidopsis KORRIGAN (KOR) gene causes the formation of aberrant cell plates, incomplete cell walls, and multinucleated cells, leading to severely abnormal seedling morphology. The mutant, designed kor1-2, was identified as a stronger allele than the previously identified kor1-1, which appears to be defective only in cell elongation. KOR1 encodes an endo-1,4-β-d-glucanase with a transmembrane domain and two putative polarized targeting signals in the cytosolic tail. When expressed in tobacco BY2 cells, a KOR1-GFP (green fluorescence protein) fusion protein was localized to growing cell plates. Substitution mutations in the polarized targeting motifs of KOR1 caused the fusion proteins to localize to the plasma membrane as well. Expression of these mutant genes in kor1-2 plants complemented only the cell elongation defect but not the cytokinesis defect, indicating that polarized targeting of KOR1 to forming cell plates is essential for cytokinesis. Our results suggest that KOR1 plays a critical role during cytokinesis.
INTRODUCTION
Plasma membranes of plant cells are encompassed by cell walls. In one aspect, the wall must be sufficiently rigid to maintain the basic structure of a cell and to provide mechanical support for the whole plant. The wall, on the other hand, must also be structurally flexible enough to allow cell growth. These seemingly contradictory requirements of rigidity and flexibility must be finely balanced for the cell wall to execute its crucial roles in structural maintenance and protection as well as regulation of plant growth and development. Whereas speculations on the biosynthesis and modification of cell walls have remained controversial for decades, recent advances in genetic analysis have provided important clues on these fundamental questions. The Arabidopsis RSW1 gene, for example, encodes the catalytic subunit of a cellulose synthase, and a conditional mutation in this locus caused a reduction of cellulose in cell walls and swollen roots at the restrictive temperature (Arioli et al., 1998). Reduced cellulose content has also been observed in several other Arabidopsis mutants, including tbr (Potikha and Delmer, 1995) and irx (Turner and Somerville, 1997). The IRX3 gene also encodes a cellulose synthase that belongs to the same family as RSW1 (Taylor et al., 1999). The characterization of the Arabidopsis mutant korrigan (kor; Nicol et al., 1998) revealed another facet of the complexity of cell walls. KOR appears to be a member of the endo-1,4-β-d-glucanase family, a class of proteins that have been suspected to be involved in modifying plant cell walls. An insertional mutation in the KOR promoter causes reduced expression of the gene and impaired expansive growth, suggesting that KOR is involved in cell elongation.
At the cellular level, although the mechanism of cell divisions is fairly conserved throughout evolution, plant cells have adopted a distinctive way to undergo cytokinesis, a process that physically separates the cytoplasm of daughter cells after nuclear division. In animal cells, cytokinesis involves the formation of a contractile acto-myosin-based ring, which pulls the parental plasma membrane inward to the center of the division plane and eventually forms a new membrane between daughter cells (reviewed in Glotzer, 1997; Field et al., 1999). In plant cells, however, this process appears to be reversed: the separation starts from the center of the division plane by forming a centrifugally growing cell plate and finishes by fusing the nascent cell plate with the parental plasma membrane at a predetermined site (Staehelin and Hepler, 1996). Cell plate formation, which has been well documented by electron microscopy, is suggested to consist of five stages. The process is mainly characterized by transport and fusion of Golgi-derived vesicles at the equatorial region to form a continuous tubulo-vesicular network (TVN), consolidation of the TVN into a fenestrated plate-like structure rich in callose, and finally an accumulation of cellulose at maturing cell plates (Samuels et al., 1995). Using biochemical approaches, investigators have characterized a number of proteins associated with or localized to the cell plate. Phragmoplastin, a dynamin-like GTPase that localizes to growing cell plates, was proposed to be involved in vesicular fusion during cytokinesis (Gu and Verma, 1996, 1997). Asada et al. (1997) characterized a tobacco kinesin-like protein that is associated with phragmoplasts and may serve as a motor molecule for microtubule translocation. Similar kinesin-related proteins have also been identified in Arabidopsis (Liu et al., 1996; Bowser and Reddy, 1997). Two mitogen-activated protein kinases identified from tobacco (Calderini et al., 1998) and alfalfa (Bogre et al., 1999) localize to phragmoplasts, and their kinase activities seem to be mitosis specific, suggestive of a regulatory role in cytokinesis. Genetic analysis has identified several cytokinesis-defective mutants (reviewed in Heese et al., 1998; see also Nickle and Meinke, 1998; Mayer et al., 1999). Thus far, the knolle mutation is the only one that has been characterized at the molecular level, and its wild-type allele encodes a target-soluble N-ethylmaleimide-sensitive factor attachment protein receptor (t-SNARE), which localizes to forming cell plate and mediates vesicle fusion during cytokinesis (Lukowitz et al., 1996; Lauber et al., 1997).
Targeting of transmembrane proteins depends largely on sorting signals residing within the cytosolic tails of the proteins, which are recognized by specific receptors. In mammalian cells, di-leucine (LL; where the leucines can also be replaced by isoleucines) and YXXΦ (where Y refers to tyrosine, X refers to any amino acid residues, and Φ refers to hydrophobic residues with a bulky side chain) motifs are among the best-understood examples. Both signals are involved in endocytosis as well as protein sorting in polarized and nonpolarized cells (Mellman et al., 1993; Mellman, 1996; Marks et al., 1997). The tyrosine-5 in the Asialogycoprotein (ASGP) receptor, for example, is critical for basolateral localization of the protein in polarized epithelial cells (Geffen et al., 1993). Similarly, the LL motif is essential for polarized targeting of low-density lipoprotein and Fc receptors in Mardis-Darby canine kidney (MDCK) cells (Matter et al., 1992, 1994; Hunziker and Fumey, 1994). Polarized targeting has recently been shown for two Arabidopsis proteins, AtPIN1 (Galweiler et al., 1998) and AtPIN2 (Muller et al., 1998), that are presumably carriers of polar auxin transport. Although both proteins contain multiple YXXΦ and LL signals, it is not known whether these motifs are functionally important for the targeting of these molecules.
Here, we report the characterization of an Arabidopsis cytokinesis-defective mutant kor1-2, which is allelic to the previously identified kor1-1 (Nicol et al., 1998). Compared with kor1-1, which appears to affect only cell elongation, kor1-2 was shown to be a stronger allele and to cause typical cytokinesis defects. This phenotypic variation is presumably due to differences in the residual expression level of KOR1 between these two mutant alleles. We also present evidence that KOR1 localizes to the forming cell plates by polarized targeting, which is essential for cytokinesis but not for cell elongation.
RESULTS
The kor1-2 Mutation Causes Defective Cytokinesis
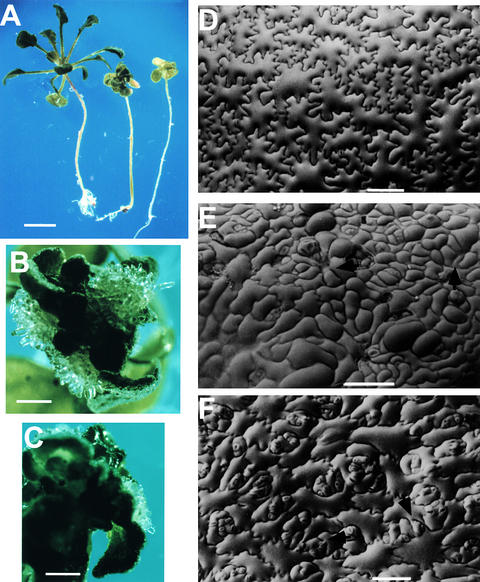
In a screen of mutants with defects in growth and development, we isolated an Arabidopsis mutant that was severely defective in shoot organogenesis. The mutant was subsequently found to be allelic to the previously identified mutant kor (Nicol et al., 1998). We named this mutant kor1-2 and henceforth refer to the previously identified allele as kor1-1. Young kor1-2 mutant seedlings were markedly smaller than wild type, but all of the kor1-2 embryonic organs, including roots, hypocotyls, and cotyledons, were well defined (Figure 1A). However, before the first pair of true leaves was fully expanded, white-colored calli were initiated from the shoot apical meristem (SAM) and petioles. Two to three weeks after germination, all kor1-2 adult organs had become transformed into yellowish green callus (Figure 1B), indicating that morphogenesis was disrupted by the mutation. Scanning electron microscopy showed that whereas wild-type hypocotyl epidermal cells were elongated and formed an organized pattern (Figure 1C), mutant cells were misshapen and irregularly arranged, often containing cell wall stubs (Figure 1D), suggesting that both cell division and cell elongation were affected by the mutation. In wild-type plants, the first pair of true leaves is initiated between the two cotyledons (Figure 1E). In contrast, only leaf-like projections were randomly produced from the SAM of kor1-2 (Figure 1F), and these were eventually transformed into calli.
Figure 1.
The Phenotype of the kor1-2 Mutant.
(A) Seven-day-old wild-type (right) and kor1-2 (left) seedlings.
(B) A five-week-old kor1-2 seedling.
(C) to (F) Scanning electron microscopy of wild-type ([C] and [E]) and kor1-2 ([D] and [F]) seedlings. (C) and (D) show hypocotyls. (E) and (F) show top views of the seedlings.
Arrows indicate incomplete cell walls. 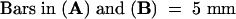 ;
; 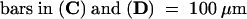 ;
; 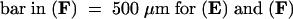 .
.
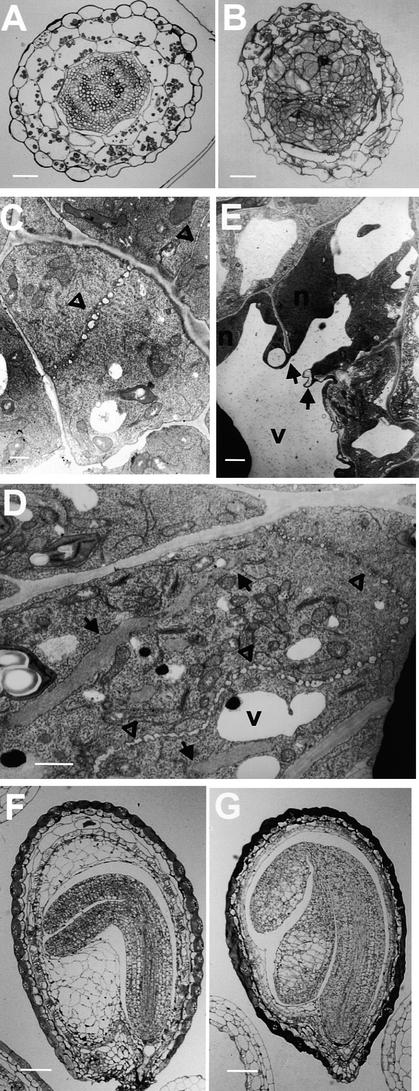
Comparison of transverse sections of wild-type and kor1-2 roots, cotyledons (data not shown), or hypocotyls (Figures 2A and 2B) revealed that unlike the wild type, the mutant did not have distinctively defined cell files. The mutant cells divided randomly and often contained incomplete cell walls. Transmission electron microscopy indicated that whereas wild-type cells formed a straight cell plate at an equatorial plane (Figure 2C), highly curved and randomly positioned cell plates were observed in kor1-2 cells (Figure 2D). Progression of the mutant cell plate was often halted in front of nuclei and vacuoles and usually terminated before reaching the parental plasma membrane. In kor1-2 cells, incomplete and fragmented walls were frequently observed, leading to multinucleated cells (Figures 2D and 2E). Although the cell division pattern was highly irregular, the preprophase band, a typical marker of the division plane, appeared to be unaffected by the mutation (data not shown).
Figure 2.
Cellular Phenotype of kor1-2.
(A) and (B) Light microscopy of transverse sections of wild-type (A) and kor1-2 (B) hypocotyls.
(C) to (E) Transmission electron microscopy of ultrathin sections from wild-type (C) and kor1-2 ([D] and [E]) roots.
(F) and (G) Light microscopy of wild-type (F) and kor1-2 (G) developing embryos.
Arrows indicate incomplete cell walls, and open triangles denote cell plates. N, nucleus; V, vacuole. 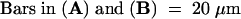 ;
; 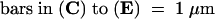 ;
; 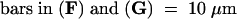 .
.
To track the origin of the mutation during embryogenesis, we collected embryos at various developmental stages from kor1-2/+ plants and examined them using light microscopy. Unexpectedly, no apparent abnormalities, including disorganized cell files, enlarged cells, and cell wall stubs, were observed until the developing embryos progressed into the cotyledon stage (cf. Figures 2F and 2G).
Genetic Analysis of the kor1-2 Mutation and Molecular Cloning of the KOR1 Gene
The kor1-2 mutant was isolated from a pool of T-DNA–transformed lines in the C24 background. Segregation analysis demonstrated that kor1-2 was a recessive mutation in a single Mendelian locus. Using cleaved amplified polymorphic sequences (CAPS) (Konieczny and Ausubel, 1993) and simple sequence length polymorphism (SSLP) markers (Bell and Ecker, 1994), we mapped the mutation to chromosome 5 between DFR (16.7 centimorgans [cM]; 94 chromosomes tested) and LFY3 (17.9 cM; 96 chromosomes tested). Recombination was not detected between the mutation and the nga129 marker (232 chromosomes tested), suggesting that the mutation is within 1 cM of nga129. Consistent with our genetic mapping data, DNA sequencing showed that nga129 was ∼60 kb north of the KOR1 gene (http://www.kazusa.or.jp/arabi). Genetic analysis also suggested that the mutation was probably tagged by the T-DNA insertion and that the mutant genome contained a single T-DNA insertion.
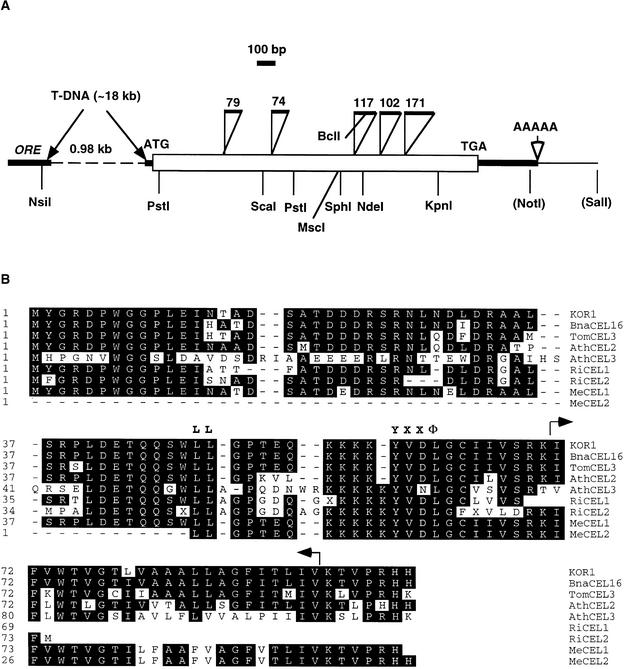
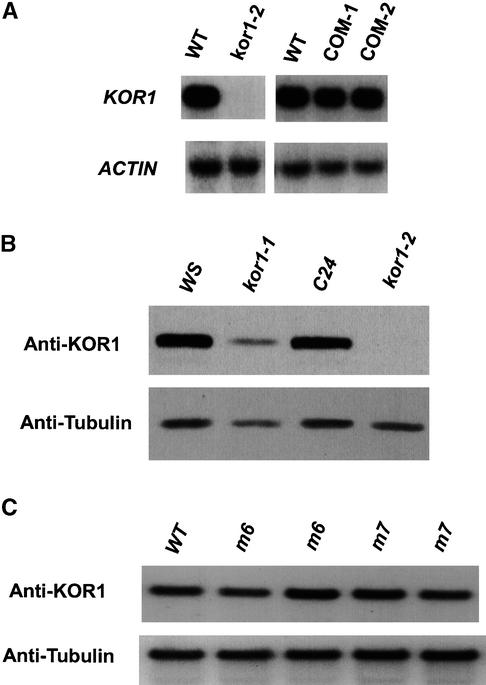
The T-DNA tag and flanking sequences were cloned and compared with wild-type genomic DNA sequences. The T-DNA insertion deleted a 1-kb DNA fragment between two adjacent transcription units, designated ORE (for oxidoreductase) and CEL (for cellulase), respectively (Figure 3A). The deletion encompassed part of the ORE 3′-untranslated region (UTR), the entire CEL promoter, and the 5′-UTR 22 bp upstream from the putative start codon. RNA gel blot analysis showed that ORE mRNA levels were unchanged in the mutant (data not shown). By contrast, neither the CEL transcript (Figure 4A) nor the encoded protein (Figure 4B) was detectable in kor1-2, indicating that the expression of CEL was disrupted by the T-DNA.
Figure 3.
Molecular Characterization of the KOR1 Gene.
(A) Schematic map of the genomic structure of the KOR1 gene. The open square indicates the coding sequence, and open triangles denote introns (numbers above refer to the sizes of introns); thick lines indicate untranslated regions, and thin lines denote untranscribed regions. The dashed line indicates the deleted sequence in the kor1-2 genome. Restriction sites (noted in parentheses) were from cloning vectors. The sequences reported here have been deposited in the GenBank database under the accession numbers AF074092 and AF074375.
(B) An alignment of the N-terminal regions of subfamily III endo-1,4-β-d-glucanases. Arrows indicate the transmembrane domain. The conserved polarized targeting signals are highlighted (LL and YXXΦ). BnaCEL16, Brassica napus CEL16; TomCEL3, tomato CEL3; AthCEL2 and 3, Arabidopsis thaliana CEL2 (T22A6.90) and CEL3 (F5I14.14), respectively; RiCEL1 and 2, rice CEL1 (C28145) and CEL2 (D46633), respectively; MeCEL1 and 2, Medicago CEL1 (AA661030) and CEL2 (AA660466), respectively. See text for the accession numbers of other sequences.
Figure 4.
Expression Pattern of KOR1 in Wild-Type and kor1 Plants.
(A) RNA gel blot analysis of the KOR1 transcript. Ten micrograms of total RNA, prepared from 7-day-old seedlings, was loaded in each lane. All blots were rehybridized with an actin probe. During the first 3 weeks after germination, KOR1 expression was not detectable by RNA gel blot analysis, RT-PCR, or protein gel blot analysis (see [B]) in kor1-2 seedlings. Residual expression, however, could be detected in 4- to 5-week-old kor1-2 seedlings (data not shown), presumably the result of a cryptic promoter in the T-DNA insertion. The residual expression in kor1-2 corresponded ∼3.0 to 3.5% of that in wild-type plants at the same growth stage (assayed by quantitative RT-PCR; see Methods). COM-1 and COM-2, kor1-2 plants carrying the ORE-KOR1 or KOR1 transgenes, respectively; WT, wild type.
(B) Analysis of KOR1 proteins by protein gel blot analysis. Whole-cell extracts were prepared from 7-day-old seedlings. Approximately 20 μg of total proteins was loaded in each lane. The blot was probed with the anti-KOR1 antibody, followed by reprobing with an anti-tubulin antibody (Amersham). WS and C24, wild-type plants of the Wassilewskija and C24 ecotypes, respectively (kor1-1 and 1-2 were in WS and C24 background, respectively).
(C) Accumulation of mutant proteins in m6 and m7 seedlings. Whole-cell extracts were prepared from 3-week-old seedlings. See (B) for other technical details. WT, wild type.
Genetic complementation demonstrated that wild-type genomic DNA fragments containing either the CEL transcription unit alone or the ORE-CEL transcription units, but not that of ORE alone, were able to completely rescue the mutant phenotype. In addition, a CEL cDNA fragment under the control of the CEL promoter also fully rescued the kor1-2 phenotype. These results indicate that the CEL transcription unit represents the KOR1 gene and that the phenotype was caused entirely by a deletion mutation in KOR1.
While this work was in progress, Nicol et al. (1998) published their studies on kor1-1. Based on the sequences (with some discrepancies; see Methods) and map positions reported, these two mutations appeared to be allelic. Genetic complementation demonstrated that they were indeed alleles. The kor1-1 mutation, also tagged by a T-DNA, was shown to cause defects in cell elongation: kor1-1 seedlings had shorter hypocotyls when germinated in the dark. Compared with kor1-2, kor1-1 appears to be a weak allele. This is consistent with molecular analysis of the two mutants: whereas the entire promoter was deleted in kor1-2, in kor1-1 ∼200 bp of the sequence upstream from the putative start codon and the rest of the transcription unit remained intact. Consequently, the KOR1 transcript level in kor1-1 was >30-fold that in kor1-2 (in 7-day-old seedlings, assayed by quantitative reverse transcription–polymerase chain reaction [RT-PCR]; see Methods). Similarly, whereas a substantial amount of the protein accumulated in kor1-1, this protein was not detected in kor1-2 under the same assay conditions (see Figure 4B). The phenotypic differences of these two alleles likely result from different expression levels of KOR1 (see Discussion).
KOR1 Encodes a Putative Membrane-Anchored Endo-1,4-β-d-Glucanase
The KOR1 cDNA sequence (OR16pep; GenBank accession numbers U37702 and S71215) and several clones of expressed sequence tags (ESTs) had been previously deposited in the databases. The KOR1 open reading frame, interrupted by five short introns (Figure 3A), encodes a putative polypeptide of 621 amino acids with a predicted molecular mass of 69.2 kD and a pI of 9.07. Sequence comparison suggested that KOR1 is a member of the E-type endo-1,4-β-d-glucanases (EGases; EC 3.2.1.4), also known as cellulases. EGases have been found in bacteria, fungi, plants, and several other lower organisms; they are generally believed to hydrolyze polysaccharides containing a 1,4-β-d-glucan backbone (Henrissat et al., 1989; Beguin, 1990). Plant EGases are presumed to modify cell wall structures during organ abscission, fruit ripening, and expansive growth (Brummell et al., 1994; Rose and Bennett, 1999). KOR1 is most similar to Brassica napus CEL16 (94% identity; accession number CAB51903), tomato CEL3 (81% identity; Brummell et al., 1997; accession number AAC49704), and two putative Arabidopsis proteins, T22A6.90 (77% identity; accession number AL078637.1) and F5I14.14 (56% identity; accession number AC001229). On the basis of phylogenetic analysis, plant EGases were classified into three groups (Brummell et al., 1997). Tomato CEL3, KOR1, Brassica CEL16, T22A6.90, and F5I14.14 belong to subfamily III. Other known members in this subfamily include the proteins encoded by several EST clones from rice, Medicago, and soybean. In contrast to those of subfamilies I and II, subfamily III EGases do not have a characteristic endoplasmic reticulum import signal sequence but instead contain a transmembrane domain (TMD) with a short N-terminal tail facing the cytoplasm (70 amino acid residues; see Figure 5). The TMD of KOR1 was identified as a highly hydrophobic region of 23 residues flanked by two and one positively charged residues at the N and C termini (Figure 3B), respectively—a typical structural feature of the type II transmembrane proteins localized to the plasma membrane (von Heijine, 1992; Munro, 1998). In its cytosolic tail, KOR1 contains two putative polarized targeting motifs, LL and YXXΦ. Both motifs are located at similar positions in all subfamily III EGases from a variety of species (Figure 3B), implying that they have functioned similarly throughout evolution. Near the C terminus, KOR1 contains two cellulase signature motifs (CSMs; see Figure 5) that in bacteria and fungi have been shown to be important for catalytic activity (Beguin, 1990; Henrissat, 1991).
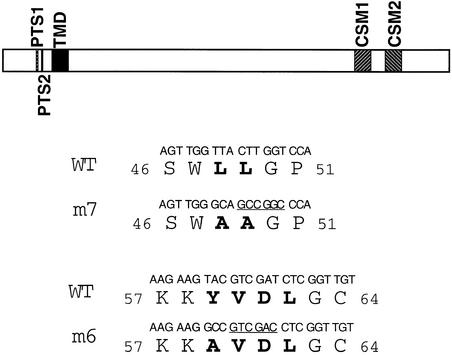
Figure 5.
Structures of Wild-Type and Mutant KOR1 Proteins.
A schematic diagram of the structural features of KOR1 (top) and DNA and amino acid sequences of wild-type and mutant KOR1 (bottom). Mutated residues are shown in boldface. Numbers refer to the positions of the amino acid residues in the protein. Underlined sequences indicate restriction enzyme sites introduced when the mutants were generated. GCCGGC, NaeI; GTCGAC, SalI; CSM, cellulase signature motif; PTS, polarized targeting signal; TMD, transmembrane domain; WT, wild type.
KOR1 mRNA is present in all examined organs/tissues, with lower levels in flowers and siliques (data not shown). In contrast to subfamily I and II EGase genes, the transcription of KOR1 and F5I14.14 (data not shown; see also Nicol et al., 1998) as well as tomato CEL3 (Brummell et al., 1997) is not upregulated by ethylene or auxin. These findings suggest that subfamily III EGases are not only structurally but also functionally distinct from other types of EGases.
Cell Cycle–Dependent Localization of KOR1-GFP Fusion Proteins to Forming Cell Plates by Polarized Targeting
To gain additional insight into KOR1 functions, we attempted to determine its subcellular localization. Antibodies against the N-terminal cytosolic tail were generated and affinity purified. In protein gel blot analysis, the antibody detected a major band with an apparent molecular mass of 72 kD in extracts prepared from wild-type but not from kor1-2 seedlings (Figure 4B). However, we were unable to reproducibly detect positive signals by immunocytology with this antibody. We also expressed in kor1-2 plants a KOR1 promoter-controlled fusion gene with the hemagglutinin gene (HA), KOR1-HA, which was able to completely rescue the mutant phenotype. Although the fusion protein could be readily detected by anti-HA antibodies in protein gel blot analyses, we encountered the same technical difficulty as for anti-KOR1 antibodies in immunocytology when we used anti-HA antibodies from various commercial sources (see Methods for more details).
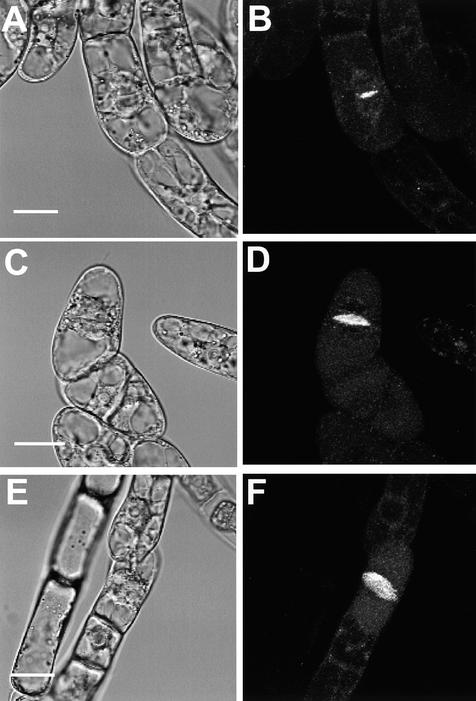
As an alternative approach, a fusion gene of KOR1 with the green fluorescent protein (GFP) gene, KOR1-GFP, was placed under the control of the XVE-inducible expression system (J. Zuo, Q.-W. Niu, and N.-H. Chua, unpublished data) and stably transformed into tobacco BY2 cells. The XVE inducible system, in principle, is similar to that of the previously published GVG system (Aoyama and Chua, 1997) but uses the regulatory domain of the human estrogen receptor and the DNA binding domain of the LexA repressor. The target promoter consists of multiple copies of the LexA operator fused to a –46 35S minimal promoter. Transcription of the target gene in the XVE system can be tightly controlled by estradiol (J. Zuo, Q.-W. Niu, and N.-H. Chua, unpublished data). In a control experiment, GFP was uniformly distributed in cytoplasm and accumulated slightly in the nucleus throughout the cell cycle. In contrast, KOR1-GFP fluorescence assumed a punctate pattern in interphase cells (Figures 6, 7A, and 7B, and data not shown), presumably indicating localization of the fusion protein to Golgi stacks or vesicles. This notion was supported by an early observation that the major fraction of tomato CEL3 comigrated with Golgi and plasma membrane markers (Brummell et al., 1997). At the onset of cytokinesis, the fusion protein was dramatically relocalized to the center of the division plane, where the cell plate was initiated (Figures 6A and 6B). As cytokinesis progressed, the fluorescence exactly followed the growth of the cell plate exactly and was eventually visible as an expanding disc or ring at later stages (Figures 6A to 6F; see also Figures 7A and 7B).
Figure 6.
Subcellular Localization of the KOR1-GFP Fusion Protein in Tobacco BY2 Cells.
(A), (C), and (E) Bright-field images of BY2 cells expressing the KOR1-GFP fusion proteins.
(B), (D), and (F) Confocal images of the same cells.
(A) and (B) A cell in the early stage of cytokinesis.
(C) to (F) Cells in late stages of cytokinesis. Note the disk-like structure in (F).
All images were taken from the transgenic cells cultured for 16 to 18 hr in the presence of 2 μM estradiol. 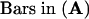 ,
, 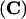 ,
, 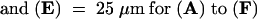 .
.
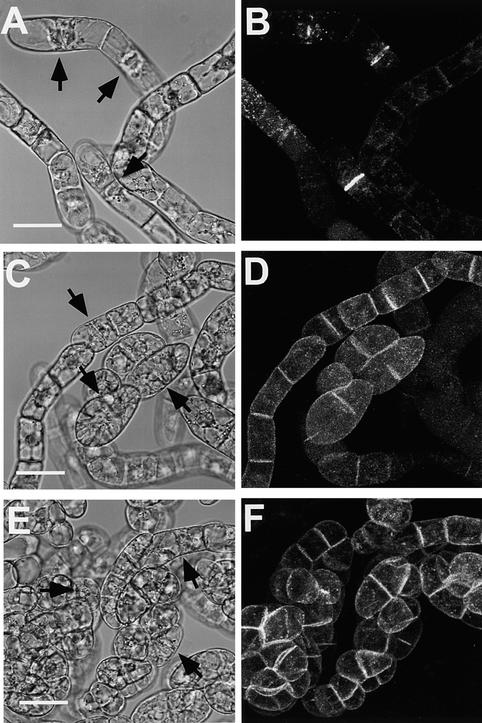
Figure 7.
Localization of m6- and m7-GFP in Tobacco BY2 Cells.
(A), (C), and (E) Bright-field images of BY2 cells expressing wild-type or mutant KOR1-GFP fusion proteins.
(B), (D), and (F) Confocal images of the same cells. Other technical details as in Figure 6.
(A) and (B) KOR1-GFP transgenic cells.
(C) and (D) m7-GFP transgenic cells.
(E) and (F) m6-GFP transgenic cells.
The cell plates are indicated by arrows. Bars in (A), (C), and (E) = 50 μm for (A) to (F).
The above observations suggested that KOR1-GFP localized to forming cell plates during cytokinesis. A critical question remaining to be addressed, however, was whether KOR1-GFP was mistargeted in BY2 cells. Like all subfamily III EGases, KOR1 contains two putative polarized targeting motifs within its cytosolic tail. If localization of KOR1-GFP to the cell plate is regulated by a polarized targeting mechanism rather than mistargeting, then mutations in these motifs would be expected to cause a depolarized targeting; otherwise, localization of the fusion proteins would not be affected by these mutations. To test this idea, we introduced substitutions into these two motifs (Figure 5), and localization of the resulting mutants tagged by GFP was monitored as before. Substitution for either tyrosine-59 by an alanine (m6-GFP) or for the LL (residues 48 and 49) by two alanines (m7-GFP) resulted in the fusion proteins being uniformly distributed to the plasma membranes with a slightly stronger GFP signal at the cell plate (cf. Figures 7A to 7B, and 7C to 7F). Two other mutants, however, in which the putative active histidine-513 of CSM1 (m1-GFP; the lumen portion) or tyrosine-2 (m13-GFP; the cytoplasmic tail) was replaced with an alanine, localized to the cell plate the same as the wild-type KOR1-GFP (data not shown). Localization of the m1- and m13-GFP fusion proteins suggested that the altered localization of m6- and m7-GFP was unlikely to be the consequence of a generally structural effect. Similar observations were made using the 35S promoter to control the transgenes (not tested for m13-GFP; data not shown). Taken together, the above results indicated that KOR1-GFP localizes to forming cell plates by a polarized targeting mechanism that involves both the LL and YXXΦ motifs.
Polarized Targeting of KOR1 Is Essential for Cytokinesis but Not for Cell Elongation
To test the functional significance of the YXXΦ and LL motifs, mutant cDNA encoding the two substitutions (m6 and m7) as well as the wild-type cDNA were placed under the control of the KOR1 promoter and transformed into kor1- 2/+ plants. As mentioned before, KOR1::KOR1 cDNA fully complemented the mutant phenotype. Transgenic plants homozygous for kor1-2 and carrying KOR1::m6 or m7 transgenes (henceforth referred to as m6 or m7 plants/seedlings) were initially screened by genetic and phenotypic analyses. As a restriction enzyme site was introduced into the mutated sites for both mutant cDNAs (see Figure 5), identity of m6 and m7 plants was further confirmed by RT-PCR, followed by the appropriate restriction digests. To eliminate the effects of over- or underexpression of the transgenes, transgenic lines that expressed m6 or m7 in quantities comparable with that of wild type were identified by protein gel blot analysis (Figure 4C) and then used for subsequent experiments.
Compared with the small-sized kor1-2, the m6 and m7 seedlings were similar in size to the wild type shortly after germination. In addition, the m6 and m7 mutants had fully elongated hypocotyls, like the wild type, when germinated in the dark (Figure 8A). These data suggested that mutations in either motif did not have detectable effects on expansive growth. However, true leaves were highly curled to form a ball-like structure, and white-colored callus appeared at or around the SAM when true leaves were initiated (Figures 8A to 8C). Growth of calli usually varied from localized colonies at or around the SAM to complete transformation of all adult organs into callus, similar to the behavior in kor1-2 seedlings. All m6 and m7 plants rarely flowered and were sterile. Scanning electron microscopy of m6 and m7 seedlings revealed an irregular cell division pattern and cell wall stubs similar to those observed in kor1-2 (cf. Figures 8D and 8F). Whereas cell expansion in kor1-2 cotyledons was substantially affected, the cell sizes of m6 and m7 plants were relatively similar to that of wild-type cells. Interestingly, all mutant cotyledons (m6, m7, and kor1-2) had more guard cells than did wild-type cotyledons, presumably because of altered cell specifications affected by the mutations. From these data, we concluded that mutations in the polarized targeting motifs affected cytokinesis but not cell elongation.
Figure 8.
Phenotypes of m6 and m7 Mutants.
(A) Four-week-old wild-type (left), m7 (middle), and m6 (right) seedlings. Seeds were germinated and cultured in the dark for 5 days and then cultured under continuous white light for an additional 3 weeks before being photographed.
(B) and (C) Callus formation in m6 (B) and m7 (C) seedlings. Images were taken from 30-day-old seedlings cultured in 16-hr-light/8-hr-dark cycles. The same observations were also made from seedlings cultured in all tested conditions, including those described in (A).
(D) to (F) Scanning electron microscopy of wild-type (D), kor1-2 (E), and m6 (F) cotyledons from 5-day-old seedlings.
Arrows indicate cell wall stubs. 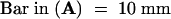 ;
; 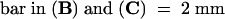 ;
; 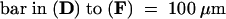 .
.
DISCUSSION
The KOR1 Activity Is Essential for Cytokinesis in Vegetative Development
In this report, we have presented several lines of evidence showing that KOR1 plays a critical role in cytokinesis in addition to cell elongation (Nicol et al., 1998). First, deletion of the entire KOR1 promoter causes a nearly null mutation of the gene, and the phenotype of the resulting mutant should thus reflect the function of its wild-type allele. At the cellular level, the mutation leads to the formation of deformed cell plates, incomplete cell walls, and multinucleated cells, a characteristic in all other known defective cytokinesis mutants (Heese et al., 1998). In an independent screen, V. Sundaresan and co-workers (V. Sundaresan, personal communication) identified a similar mutant, sgt2447. Genetic complementation demonstrated that sgt2447 (in Landsberg background) was allelic to kor1-2. Because these two alleles showed a very similar phenotype at the seedling stage as well as at the cellular level, the kor1-2 phenotype should not be allele- or ecotype-specific. Second, the kor1-2 mutation transforms all adult organs into calli, and this phenotype cannot be rescued by treatment with any known hormones (J. Zuo and N.-H. Chua, unpublished data). Thus, the formation of callus is unlikely to be due to an imbalance of endogenous hormones. Lukowitz et al. (1996) proposed that incomplete separation of dividing cells in knolle mutant cells caused miscommunication among neighboring cells, which consequently could disrupt cell specifications in the affected tissues. Likewise, the transformation of adult organs into dedifferentiated calli in kor1-2 may also be a consequence of the disruption of cell specifications. Third, based on the localization patterns of the KOR1-, m6-, and m7-GFP fusion proteins, KOR1 most probably localizes to forming cell plates during cytokinesis (see below). Finally, mutations in the putative polarized targeting motifs not only abolished the targeting specificity but also functionally impaired cytokinesis, whereas cell expansion remained normal. These observations indicate that defective cytokinesis in the original kor1-2 mutant was not a secondary consequence of deficient cell elongation but rather a primary effect of the mutation.
In knolle mutant cells, Golgi-derived vesicles are properly transported to and aligned along the division plane but fail to fuse (Lauber et al., 1997). Unlike knolle but similar to the pea cyd mutant (Liu et al., 1995), transport and fusion of vesicles are unaffected in kor1-2 cells, whereas the cell plate is randomly positioned and often terminated before reaching the parental plasma membrane, indicating that KOR1 is involved in cell plate maturation. Expression of cytokinesis mutations shows a variety of developmental and tissue specificities. In knolle (Lukowitz et al., 1996) and keule (Assaad et al., 1996), cytokinesis defects originate from the first zygotic division, whereas the cyt mutation appears to express from the heart stage (Nickle and Meinke, 1998). Compared with these pro-embryo- and embryo-defective mutants, the onset of the cyd phenotype is as late as the cotyledon stage (Liu et al., 1995). Interestingly, mutations in several other loci seem to restrict defective cytokinesis in certain tissues or cell types, including floral organs (Liu et al., 1997) and male gametophytes (Chen and McCormick, 1996; Spielman et al., 1997; Hulskamp et al., 1997). This diversity probably reflects the developmental specificity of a particular gene or mechanisms operating in different cell types. The latter rationale can be best illustrated by the fact that the mechanism used by meiotic and gametophytic cells in cytokinesis differs from that used by somatic cells (Heese et al., 1998). The kor1-2 mutation, similar to that of cyd, begins to exert apparent effects when developing embryos enter the cotyledon stage and subsequently impairs cell divisions in all adult organs. Therefore, KOR1 appears to act mainly, if not specifically, on vegetative development.
KOR1 Is Involved in Cell Plate Maturation
The primary cell wall has been described as a network of cellulose microfibrils that are cross-linked by hemicellulose, mainly xyloglucans, and reinforced by embedded pectins (Carpita et al., 1996; Pennell, 1998; Reiter, 1998). In bacteria and fungi, EGases have been demonstrated to hydrolyze 1,4-β–linked glucans, mainly cellulose (Beguin, 1990; Henrissat, 1991). Largely because of sequence homology, plant EGases were proposed to have a similar activity. An important structural difference, however, is that plant EGases do not have a cellulose binding domain (although a recently identified strawberry faEG3 contains a putative cellulose binding domain; see Trainotti et al., 1999) and thus are incapable of hydrolyzing crystalline cellulose by in vitro assays.
We propose that KOR1 functions in the assembly of new cell wall during cell plate maturation. KOR1 may be involved in relieving the strains of cellulose-xyloglucan microfibrils to coordinate the assembly of new cell walls. Alternatively, the enzyme may modify newly deposited glucans to enable their integration into new cell walls. During cell wall loosening, degradation of matrix glucans appears to be necessary for disassembly of the wall and subsequent reassembly of new walls (Cosgrove, 1997, 1999; Rose and Bennett, 1999). In response to auxin as well as to several other cues, xyloglucan, for example, undergoes substantial degradation (Fry, 1989; Hoson, 1993). Moreover, size distributions of xyloglucan appear to change rapidly and reversibly, and these events likely involve changes in the EGase activity (Hoson, 1993; Cosgrove, 1997, 1999). The EGase activity, in turn, is stimulated by oligosaccharides derived from xyloglucans (Farkas and Maclachlan, 1988). These observations suggest that the formation of new cell walls and the EGase-mediated xyloglucan turnover are tightly coupled processes. Although the EGase action may have a different physiological significance, the fact that the enzymatic cores are highly conserved from bacteria to higher plants suggests that the enzymatic mechanism of membrane-anchored EGases is probably similar to that used by members of subfamilies I and II or by lower organisms. Indeed, replacing the active residues in the CSMs with alanines affected cytokinesis and/or cell elongation (J. Zuo and N.-H. Chua, unpublished data), indicating that cleavage of 1,4-β–linked glucans is an intrinsic activity of KOR1 and is essential for its function.
A second possibility, as proposed for tomato CEL3 (Brummell et al., 1997), is that KOR1 is involved in cellulose biosynthesis. This view was inferred from the observation that a membrane-anchored EGase identified in Agrobacterium tumefaciens was shown to be essential for cellulose synthesis (Matthysse et al., 1995). The importance of cellulose biosynthesis in cytokinetic plant cells has recently been shown by a study of the cytokinesis-defective mutant cyt (Nickle and Meinke, 1998). The cyt mutant cells accumulate excessive callose, a 1,3-β–linked glucan that transiently accumulates at the cell plate of wild-type cells (Samuels et al., 1995). Because accumulation of excess callose was thought to be an indication of impaired cellulose biosynthesis, the CYT gene was suggested to play a role in cell wall biosynthesis. Moreover, inhibition of cellulose biosynthesis by the herbicide dichlobenil partially phenocopies the cellular phenotype of cyt (Nickle and Meinke, 1998) and results in abnormal cell plates remarkably similar to those observed in kor1-2 cells (Mineyuki and Gunning, 1990; Vaughn et al., 1996). In general, an inability to form a new cell wall, whether because of impaired cellulose synthesis or defective assembly of the wall, probably reduces the mechanical strength, which consequently leads to the formation of distorted cell plates in kor1-2 cells.
Polarized Targeting of KOR1 and Its Final Destination
During cytokinesis, a transient polarity may be established between the parental plasma membrane and the cell plate; hence, localization of transmembrane proteins to the cell plate may be directed by a polarized targeting mechanism. Both the YXXΦ and LL motifs have been found in the cytosolic tails of many plant transmembrane proteins, including AtPIN1, AtPIN2, and KNOLLE. AtPIN1 (Galweiler et al., 1998) and AtPIN2 (Muller et al., 1998) belong to a large family of carrier proteins and are involved in polar auxin transport. Both proteins localize to the basal end of auxin transport–competent cells, although it remains unclear whether the YXXΦ and LL motifs are indeed involved in the polarized targeting of these proteins. Interestingly, the LL motif appears to be involved in the targeting of syntaxins (reviewed in Tang and Hong, 1999), a class of proteins that includes KNOLLE. Although it is unknown whether the LL or the YXXΦ motifs (or both) are involved in the targeting of KNOLLE, localization of the protein to the cell plates is probably mediated by a polarized targeting mechanism.
By using a GFP tag, we were able to show that KOR1 localizes to forming cell plates in tobacco BY2 cells. Although these observations could be further confirmed by immunocytology, the fact that mutations in the polarized targeting motifs abolished the specificity of targeting strongly suggests that the wild-type KOR1 protein most likely localizes to the cell plate during cytokinesis. Moreover, because the subfamily III EGases are highly conserved, particularly in the N-terminal region, where the targeting signals are located (92 or 95% identity between KOR1 and tomato CEL3 or Brassica CEL16, respectively), it is unlikely that the protein localizes differently in different types of cells or tissues. More importantly, mutations that abolished polarized targeting of the GFP fusion proteins in BY2 cells indeed affected cytokinesis rather than cell elongation, providing strong support for the notion that KOR1 localizes to forming cell plates in vivo by polarized targeting.
KOR1 contains two typical polarized targeting signals, a feature shared by many polarized targeted proteins. These two types of signals function cooperatively in most cases (Mellman, 1996; Kirchhausen et al., 1997) and can function independently (Letourneur and Klausner, 1992). IgG Fc receptor, for example, contains two tyrosine- and one LL-based motifs at its cytosolic tail. However, neither motif is absolutely required for basolateral targeting; instead, all three signals function as determinants for basolateral targeting as well as endocytosis (Hunziker and Fumey, 1994; Matter et al., 1994). KOR1 seems to use a similar mechanism, because both m6- and m7-GFP accumulated at a greater concentration at the cell plate than at the plasma membrane. The utilization of multiple signals may increase the targeting specificity by a cooperative action among the signals or may ensure that a protein is properly transported even in the event that one of the signals is impaired. In pea, BP80, a type I transmembrane protein, contains a YXXΦ motif in its cytoplasmic tail. Deletion of the cytoplasmic tail and the TMD resulted in secretion of the truncated protein (Paris et al., 1997). A subsequent study, however, revealed that the TMD alone was sufficient for lytic vacuole targeting of the protein, and the entire cytoplasmic tail appeared to be dispensable (Jiang and Rogers, 1998). Whereas the involvement of the TMD, along with the YXXΦ and LL motifs in polarized targeting, has been reported in both yeast and animal cells (Marks et al., 1997; Munro, 1998), it would be interesting to determine whether the KOR1 TMD also plays a role in its targeting.
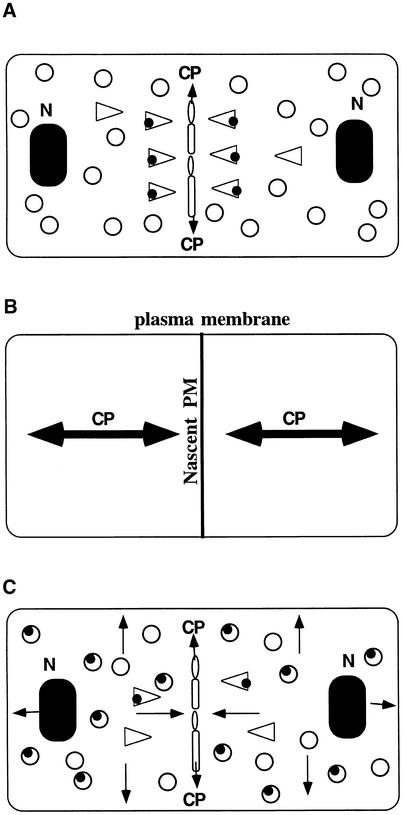
On the basis of our localization data, the phenotypes of the three kor1 alleles, and the phenotypes of the kor1 substitution mutants, we propose that KOR1 localizes to the cell plate by polarized targeting and is involved in cell plate maturation (Figure 9A). After the completion of cytokinesis, the protein remains in the plasma membrane to modify primary cell walls in subsequent expansive growth. Once the surface areas of the plasma membrane have expanded by cell growth, the KOR1 concentration is gradually reduced. Accordingly, KOR1 accumulates at the highest level in the cell plate, followed by the nascent plasma membrane, whereas fully expanded cells contain the lowest concentrations of the protein (Figure 9B). Functionally, this concentration gradient is consistent with the metabolic status of cell walls in that forming new walls at the cell plate requires the most activity, whereas fully expanded cells require the least. This model predicts that mutations in the polarized targeting signals will cause mislocalization of the protein (Figure 9C) and affect cytokinesis but may have negligible effects on cell elongation. Indeed, all these predictions were experimentally verified in this study. In m6 and m7 plants, expression of the mutant transgenes at the normal level fully restored the impaired expansive growth but not the defective cytokinesis. This indicates that whereas the mislocalized EGase suffices the requirement for proper cell elongation, a decreased concentration of the protein at the cell plate is incapable of maintaining normal cytokinesis. Consistent with this notion, overexpression of m6 or m7 was able to completely rescue defects in cytokinesis as well as cell elongation (data not shown). Unlike m6 and m7, the kor1-1 mutation caused a reduced expression of the wild-type gene and appeared to affect cell elongation but had no detectable effects on cytokinesis (Nicol et al., 1998). Presumably, a decrease in the gene expression, as in kor1-1, may not be particularly detrimental for cell divisions, because the cell plate is able to “concentrate” the wild-type protein during cytokinesis. Conversely, a decrease in expression to a similar extent may substantially affect cell elongation because the protein will be serially diluted in an expanding cell. To verify this explanation, it would be interesting to express m6 or m7 in kor1-1, which should be able to fully complement the kor1-1 phenotype.
Figure 9.
A Model of KOR1 Localization by Polarized Targeting.
(A) Golgi-derived and the cell plate–targeting vesicles (triangles) carrying KOR1 (filled circles) localize to the cell plate during cytokinesis.
(B) Concentration gradient of KOR1 in dividing cells. The thickness of the lines indicates the relative concentration of the protein in different locales.
(C) Depolarized targeting of m6 or m7 proteins.
Open circles indicate nonpolarized targeting vesicles, and open ovals indicate fusing vesicles. Single arrows indicate the direction of the KOR1 targeting and double arrows the direction of the cell plate growth. See text for details. CP, cell plate; N, nucleus; PM, plasma membrane.
Activities of the secreted plant EGases have long been associated with cell wall modifications during organ abscission, fruit ripening, and expansive growth. Here, we present evidence showing that the membrane-anchored EGase KOR1 plays a crucial role in cytokinesis. We also show that cytokinetic plant cells use a polarized targeting mechanism to selectively transport KOR1 to the cell plate. Whereas these findings provide a fresh view for our current understanding of cytokinesis and protein targeting in plant cells, our future challenge is to identify the substrate and products of KOR1 and the receptor or receptors that interact with the EGase.
METHODS
Plant Materials and Genetic Analysis
Unless otherwise indicated, the Arabidopsis thaliana C24 ecotype was used, and plants were grown at 22°C in 16-hr-light/8-hr-dark conditions. T-DNA mutagenesis was performed by transforming roots with Agrobacteria LBA4404 cells carrying the binary vector pGE9 (C. Dong, G. Xia, and N.-H. Chua, unpublished data). The kor1-2 mutant was identified from a screen of ∼2000 lines by visual inspection. T3 kor1-2/+ plants were crossed with wild-type C24, Columbia, and Landsberg plants. The F2 seeds were sown on kanamycin-selective or nonselective A medium (1 × Murashige and Skoog [Murashige and Skoog, 1962] salts and 3% sucrose). On selective medium, 2423 of the germinated seedlings were resistant to kanamycin, whereas the remaining 797 seedlings appeared to be sensitive (3.04:1.00), indicating a single T-DNA insertion in the mutant genome. When germinated on nonselective medium, 885 of 3489 seedlings showed the mutant phenotype (2.94:1.00), indicating that the mutation was recessive in a single Mendelian locus. Mutant plants were then transferred to kanamycin-selective medium 7 to 10 days after germination. All of the 885 transferred mutant plants were resistant to kanamycin, suggesting that the mutation was tightly linked to the T-DNA.
Light and Electron Microscopy
Light microscopic analysis of Arabidopsis embryos was performed essentially as described by Mayer et al. (1993). Other methods have been previously described (Takahashi et al., 1995).
Molecular Cloning of the KOR1 Gene
All molecular manipulations were performed by standard methods (Sambrook et al., 1989; Altschul et al., 1990). DNA gel blot analysis with pGE9 as a probe revealed that HindIII cleavage of kor1-2 genomic DNA yielded two bands of ∼4.5 and 6.6 kb. Based on HindIII fragmentation of kor1-2 genomic DNA, two partial libraries, H4.5 and H6.6, were constructed. The libraries were screened with the NPTII coding sequence and the nos polyadenylation sequence as probes. Four identical clones were isolated from the H4.5 library, which contained a 2.7-kb pGE9 sequence flanked by 2.1 kb of the 3′ end of the ORE gene. The ORE sequence was used as a probe to screen a genomic library (CD4-8) obtained from the Arabidopsis Biological Resources Center (ABRC). Two identical clones, containing an insert of ∼13 kb, were isolated. This genomic DNA fragment contained both the ORE and KOR1 genes. The KOR1 cDNA was obtained from an expressed sequence tag (EST) clone, G1C11T7 (accession number N96075; provided by ABRC), which was identical to OR16pep except for lacking the first 8 bp.
Some discrepancies on the KOR1 genomic sequences are apparent between our data and those of Nicol et al. (1998). Most notably, both the number and position of introns are different (cf. Figure 3A of this article with Figure 4A of Nicol et al., 1998). Our data showed that KOR1 contains five introns instead of the four reported by Nicol et al. (1998). In addition, the positions of other four introns were different, according to restriction mapping. We have discussed these discrepancies with Nicol et al. (1998), who later published in the database a corrected KOR1 sequence (accession number AF073875; from the Columbia strain) identical to our sequence (from the Landsberg strain) except for a 1-bp mismatch in intron 4. This mismatch is unlikely to reflect polymorphism between two ecotypes, because our sequence is completely identical to the one recently published by the Kazusa group (accession number AB025613; from the Columbia strain).
RNA Gel Blot Analysis and Reverse Transcription–Polymerase Chain Reaction
Total RNA was prepared using the Plant RNA Prep Kit (Qiagen) according to the manufacturer's instructions. RNA gel blot analysis was performed according to Sambrook et al. (1989). For quantitative reverse transcription–polymerase chain reaction (RT-PCR), the first-strand cDNA was synthesized by using 1 μg of total RNA primed by oligo-dT. One-tenth of the reaction mixture (treated with RNase A) was used for PCR amplification with two pairs of primers that anchored within the coding sequences of the KOR1 and actin genes, respectively. The reaction was run for 15 cycles, and the product was used for DNA gel blot analyses with the KOR1 and actin probes. The data were collected with a PhosphorImage scanner (Molecular Dynamics), and the amount of KOR1 transcript was normalized with that of actin. Under such assay conditions (or after amplification for 25 cycles in the PCR), the KOR1 transcript was not detectable in 7- or 14-day-old kor1-2 plants. At these stages, the amount of KOR1 transcript in kor1-1 was 12.7% of that of wild-type plants (after PCR amplification for 15 cycles).
Molecular Complementation and Expression of the kor1 Substitution Mutant Genes in kor1-2 Plants
Wild-type genomic DNA fragments containing the putative ORE (a ClaI-BglII fragment), KOR1 (an NsiI-SalI fragment; see Figure 3A), or ORE-KOR1 (a ClaI-SalI fragment) transcription units were cloned into p72-1, a binary vector carrying a hygromycin selection marker. The KOR1::KOR1-cDNA fusion gene vector pHD2 was made by using the PstI site upstream of intron 1 (see Figure 3A). kor1-2/+ plants were transformed by vacuum infiltration with Agrobacteria ABI cells carrying these constructs (Bechtold et al., 1993) or by coculture with roots (Koncz et al., 1989). Fifty-two pORE lines were analyzed. All displayed a phenotype similar to that of the original mutant, and all pORE lines tested (n = 18) had an ORE expression pattern similar to that of wild type. For pKOR1 (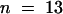 ), pORE-KOR1 (
), pORE-KOR1 (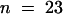 ), and pHD2 (
), and pHD2 (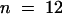 ) transgenic lines, complementation was confirmed by genetic analysis. One pKOR1, three pORE-KOR1, and one pHD2 lines were further analyzed by genomic DNA gel blot hybridization to confirm that they were homozygous for kor1-2.
) transgenic lines, complementation was confirmed by genetic analysis. One pKOR1, three pORE-KOR1, and one pHD2 lines were further analyzed by genomic DNA gel blot hybridization to confirm that they were homozygous for kor1-2.
The point mutations (Figure 5) were introduced into the KOR1 cDNA by PCR based on pHD2. The KOR1–HA fusion genes were made by inserting one (KOR1-HA1) or four (KOE1-HA4) copies of a 27-bp double-stranded DNA fragment into the NcoI site (blunted) of the KOR1 cDNA (codon 618). The inserted DNA fragment encoded a HA tag peptide (9 amino acid residues), and the open reading frames of the fusion genes were terminated by the stop codon of KOR1. The mutant or fusion genes were introduced into kor1-2/+ plants by vacuum infiltration. Whereas KOR1-HA1 is fully functional, KOR1-HA4 only partially complements the mutant phenotype. Identical results were obtained by protein gel blot analyses and immunocytology for both types of transgenic plants.
Preparation of Anti-KOR1 Antibody and Protein Gel Blot Analysis
A KOR1 cDNA fragment encoding the first 71 amino acid residues was cloned into the pMAL-C2 vector (Bio-Labs) to generate an MBP-KOR1 fusion gene. Production and purification of the MBP-KOR1 recombinant protein were performed according to the manufacturer's instructions. The purified fusion protein was used to immunize rabbits (Harlow and Lane, 1988). Antisera were affinity-purified by using the purified glutathione S-transferase (GST)-KOR1 (residues 1 to 71) recombinant protein as a ligand. The purified anti-KOR1 antibody, diluted 1:1000 to 1:3000, was used in protein gel blot analyses (Zuo et al., 1995).
Expression of Green Fluorescent Protein Fusion Proteins in Tobacco BY2 Cells
To construct KOR1-GFP, an XhoI-NcoI (blunted with Klenow) fragment containing a part of the 5′-untranslated region (UTR) and the sequence encoding the first 618 amino acids of KOR1 was inserted at the XhoI-NcoI (blunted) sites of pX–green fluorescent protein (GFP) (Kost et al., 1998). Substitution mutations were made by replacing the KOR1 sequence of the fusion gene with mutated sequences. All fusion genes were cloned into the XVE vector pER10, which carried a kanamycin-selective marker (J. Zuo, Q.-W. Niu, and N.-H. Chua, unpublished data). In this vector, expression of the target genes is strictly regulated by estradiol. Transformation of BY-2 cells was performed as described by Gu and Verma (1997). Transgenic cells subcultured for 5 to 7 days were transferred to the inductive medium, which contained 2 μM 17-β-estradiol, and incubated for 16 to 18 hr. GFP fluorescence was analyzed by confocal microscopy (Kost et al., 1998). At least 10 independent lines were tested for each construct.
Acknowledgments
We thank the Arabidopsis Biological Resources Center for providing seeds and DNAs, and Drs. R. Fisher, S. Misera, L. Zhou, D.W. Meinke, V. Sundaresan, and H. Hofte for providing seeds. We are grateful to Dr. V. Sundaresan for sharing unpublished data and to Drs. G. Jedd, U. Klahre, and C. Li for discussions and critical reading of the manuscript. We thank Ms. Yang Sun Chan for assistance with scanning electron microscopy. B.K. was supported by a postdoctoral fellowship from the Swiss National Science Foundation (No. 823A-046686). This work was supported by a grant from the National Science Foundation (Grant No. IBN 9420038).
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Aoyama, T., and Chua, N.-H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Arioli, T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720. [DOI] [PubMed] [Google Scholar]
- Asada, T., Kuriyama, R., and Shibaoka, H. (1997). TKRP125, a kinesin-related protein involved in the centrosome-independent organization of the cytokinetic apparatus in tobacco BY-2 cells. J. Cell Sci. 110, 179–189. [DOI] [PubMed] [Google Scholar]
- Assaad, F., Mayer, U., Wanner, G., and Jurgens, G. (1996). The KEULE gene is involved in cytokinesis in Arabidopsis. Mol. Gen. Genet. 253, 267–277. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Ser. III Sci. Vie 316, 1194–1199. [Google Scholar]
- Beguin, P. (1990). Molecular biology of cellulose degradation. Annu. Rev. Microbiol. 44, 219–248. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Bogre, L., Calderini, O., Binarova, P., Mattauch, M., Till, S., Kiegerl, S., Jonak, C., Pollaschek, C., Barker, P., Huskisson, N.S., Hirt, H., and Heberle-Bors, E. (1999). A MAP kinase is activated late in plant mitosis and becomes localized to the plane of cell division. Plant Cell 11, 101–114. [PMC free article] [PubMed] [Google Scholar]
- Bowser, J., and Reddy, A.S. (1997). Localization of a kinesin-like calmodulin-binding protein in dividing cells of Arabidopsis and tobacco. Plant J. 12, 1429–1437. [DOI] [PubMed] [Google Scholar]
- Brummell, D.A., Lashbrook, C.C., and Bennett, A.B. (1994). Plant endo-1,4-β-d-glucanases. Structure, properties, and physiological function. Am. Chem. Soc. Symp. Ser. 566, 100–129. [Google Scholar]
- Brummell, D.A., Catala, C., Lashbrook, C.C., and Bennett, A.B. (1997). A membrane-anchored E-type endo-1,4-β-glucanase is localized on Golgi and plasma membranes of higher plants. Proc. Natl. Acad. Sci. USA 94, 4794–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderini, O., Bogre, L., Vicente, O., Binarova, P., Heberle-Bors, E., and Wilson, C. (1998). A cell cycle regulated MAP kinase with a possible role in cytokinesis in tobacco cells. J. Cell Sci. 111, 3091–3100. [DOI] [PubMed] [Google Scholar]
- Carpita, N.C., McCann, M., and Griffing, L.R. (1996). The plant extracellular matrix: News from the cell's frontier. Plant Cell 8, 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.C., and McCormick, S. (1996). sidecar pollen, an Arabidopsis thaliana male gametophytic mutant with aberrant cell divisions during pollen development. Development 122, 3243–3253. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (1997). Relaxation in a high-stress environment: The molecular bases of extensible cell walls and cell enlargement. Plant Cell 9, 1031–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove, D.J. (1999). Enzymes and other agents that enhance cell wall extensibility. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 391–417. [DOI] [PubMed] [Google Scholar]
- Farkas, V., and Maclachlan, G. (1988). Stimulation of pea 1,4-glucanase activity by oligosaccharides derived from xyloglucan. Carbohydr. Res. 184, 213–219. [Google Scholar]
- Field, C., Li, R., and Oegema, K. (1999). Cytokinesis in eukaryotes: A mechanistic comparison. Curr. Opin. Cell Biol. 11, 68–80. [DOI] [PubMed] [Google Scholar]
- Fry, S.C.. (1989). Cellulases, hemicellulases and auxin-stimulated growth: A possible relationship. Physiol. Plant. 75, 532–536. [Google Scholar]
- Galweiler, L., Guan, C., Muller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230. [DOI] [PubMed] [Google Scholar]
- Geffen, I., Fuhrer, C., Leitinger, B., Weiss, M., Huggel, K., Griffiths, G., and Spiess, M. (1993). Related signals for endocytosis and basolateral sorting of the asialogycoprotein receptor. J. Biol. Chem. 268, 20772–20777. [PubMed] [Google Scholar]
- Glotzer, M. (1997). The mechanism and control of cytokinesis. Curr. Opin. Cell Biol. 9, 815–823. [DOI] [PubMed] [Google Scholar]
- Gu, X., and Verma, D.P.S. (1996). Phragmoplastin, a dynamin-like protein associated with cell plate formation in plants. EMBO J. 15, 695–704. [PMC free article] [PubMed] [Google Scholar]
- Gu, X., and Verma, D.P.S. (1997). Dynamics of phragmoplastin in living cells during cell plate formation and uncoupling of cell elongation from the plane of cell division. Plant Cell 9, 157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Heese, M., Mayer, U., and Jurgens, G. (1998). Cytokinesis in flowering plants: Cellular process and developmental integration. Curr. Opin. Plant Biol. 1, 486–491. [DOI] [PubMed] [Google Scholar]
- Henrissat, B. (1991). A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat, B., Claeyssens, M., Tomme, P., Lemesle, L., and Mornon, J.P. (1989). Cellulase families revealed by hydrophobic cluster analysis. Gene 81, 83–95. [DOI] [PubMed] [Google Scholar]
- Hoson, T. (1993). Regulation of polysaccharide breakdown during auxin-induced cell wall loosening. J. Plant Res. 103, 369–381. [Google Scholar]
- Hulskamp, M., Parekh, N.S., Grini, P., Schneitz, K., Zimmermann, I., Lolle, S.J., and Pruitt, R.E. (1997). The STUD gene is required for male-specific cytokinesis after telophase II of meiosis in Arabidopsis thaliana. Dev. Biol. 187, 114–124. [DOI] [PubMed] [Google Scholar]
- Hunziker, W., and Fumey, C. (1994). A di-leucine motif mediates endocytosis and basolateral sorting of macrophage IgG Fc receptors in MDCK cells. EMBO J. 13, 2963–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L., and Rogers, J.C. (1998). Integral membrane protein sorting to vacuoles in plant cells: Evidence for two pathways. J. Cell Biol. 143, 1183–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen, T., Bonifacino, J.S., and Riezman, H. (1997). Linking cargo to vesicle formation: Receptor tail interactions with coat proteins. Curr. Opin. Cell Biol. 9, 488–495. [DOI] [PubMed] [Google Scholar]
- Koncz, C., Martini, N., Mayerhofer, R., Koncz-Kalman, Z., Korber, H., Redei, G.P., and Schell, J. (1989). High-frequency T-DNA–mediated gene tagging in plants. Proc. Natl. Acad. Sci. USA 86, 8467–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.-H. (1998). A GFP–mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16, 393–401. [DOI] [PubMed] [Google Scholar]
- Lauber, M.H., Waizenegger, I., Steinmann, T., Schwarz, H., Mayer, U., Hwang, I., Lukowitz, W., and Jurgens, G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139, 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneur, F., and Klausner, R.D. (1992). A novel di-leucine motif and a tyrosine-based motif independently mediate lysosomal targeting and endocytosis of CD3 chains. Cell 69, 1143–1157. [DOI] [PubMed] [Google Scholar]
- Liu, B., Cyr, R.J., and Palevitz, B.A. (1996). A kinesin-like protein, KatAp, in the cells of Arabidopsis and other plants. Plant Cell 8, 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.-M., Johnson, S., and Wang, T.L. (1995). cyd, a mutant of pea that alters embryo morphology, is defective in cytokinesis. Dev. Genet. 16, 321–331. [Google Scholar]
- Liu, Z., Running, M.P., and Meyerowitz, E.M. (1997). TSO1 functions in cell division during Arabidopsis flower development. Development 124, 665–672. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., Mayer, U., and Jurgens, G. (1996). Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84, 61–71. [DOI] [PubMed] [Google Scholar]
- Marks, M.S., Hiroshi, H., Kirchhausen, T., and Bonifacino, J.S. (1997). Protein sorting by tyrosine-based signals: Adapting to the Ys and wherefores. Trends Cell Biol. 7, 124–128. [DOI] [PubMed] [Google Scholar]
- Matter, K., Hunziker, W., and Mellman, I. (1992). Basolateral sorting of LDL receptor in MDCK cells: The cytoplasmic domain contains two tyrosine-dependent targeting determinants. Cell 71, 741–753. [DOI] [PubMed] [Google Scholar]
- Matter, K., Yamamoto, E.M., and Mellman, I. (1994). Structural requirements and sequence motifs for polarized sorting and endocytosis of LDL and Fc receptors in MDCK cells. J. Cell Biol. 126, 991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse, A.G., White, S., and Lightfoot, R. (1995). Genes required for cellulose synthesis in Agrobacterium tumefaciens. J. Bacteriol. 177, 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, U., Buttner, G., and Jurgens, G. (1993). Apical-basal pattern formation in the Arabidopsis embryo: Studies on the role of the GNOM gene. Development 117, 149–162. [Google Scholar]
- Mayer, U., Herzog, U., Berger, F., Inze, D., and Jurgens, G. (1999). Mutations in the pilz group genes disrupt the microtubule cytoskeleton and uncouple cell cycle progression from cell division in Arabidopsis embryo and endosperm. Eur. J. Cell Biol. 78, 100–108. [DOI] [PubMed] [Google Scholar]
- Mellman, I. (1996). Endocytosis and molecular sorting. Annu. Rev. Cell Dev. Biol. 12, 575–625. [DOI] [PubMed] [Google Scholar]
- Mellman, I., Yamamoto, E., Whitney, J.A., Kim, M., Hunziker, W., and Matter, K. (1993). Molecular sorting in polarized and non-polarized cells: Common problems, common solutions. J. Cell Sci. 17, 1–7. [DOI] [PubMed] [Google Scholar]
- Mineyuki, Y., and Gunning, B.E.S. (1990). A role for the preprophase band of microtubules in maturation of new cell walls, and a general proposal on the function of preprophase band sites in cell division of higher plants. J. Cell Sci. 97, 527–537. [Google Scholar]
- Muller, A., Guan, C., Galweiler, L., Tanzler, P., Huijser, P., Marchant, A., Parry, G., Bennett, M., Wisman, E., and Palme, K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17, 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro, S. (1998). Localization of proteins to the Golgi apparatus. Trends Cell Biol. 8, 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nickle, T.C., and Meinke, D.W. (1998). A cytokinesis-defective mutant of Arabidopsis (cyt1) characterized by embryonic lethality, incomplete cell walls, and excessive callose accumulation. Plant J. 15, 321–332. [DOI] [PubMed] [Google Scholar]
- Nicol, F., His, I., Jauneau, A., Vernhettes, S., Canut, H., and Hofte, H. (1998). A plasma membrane–bound putative endo-1,4-β-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17, 5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris, N., Rogers, S.W., Jiang, L., Kirsch, T., Beevers, L., Phillips, T.E., and Rogers, J.C. (1997). Molecular cloning and further characterization of a probable plant vacuolar sorting receptor. Plant Physiol. 115, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell, R. (1998). Cell walls: Structures and signals. Curr. Opin. Plant Biol. 1, 504–510. [DOI] [PubMed] [Google Scholar]
- Potikha, T., and Delmer, D.P. (1995). A mutant of Arabidopsis thaliana displaying altered patterns of cellulose deposition. Plant J. 7, 453–460. [Google Scholar]
- Reiter, W.D.. (1998). The molecular analysis of cell wall components. Trends Plant Sci. 3, 27–32. [Google Scholar]
- Rose, J.K.C., and Bennett, A.B. (1999). Cooperative disassembly of the cellulose–xyloglucan network of plant cell walls: Parallels between cell expansion and fruit ripening. Trends Plant Sci. 4, 176–183. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Samuels, A.L., Giddings, T.H., Jr., and Staehelin, L.A. (1995). Cytokinesis in tobacco BY-2 and root tip cells: A new model of cell plate formation in higher plants. J. Cell Biol. 130, 1345–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman, M., Preuss, D., Li, F.L., Browne, W.E., Scott, R.J., and Dickinson, H.G. (1997). TETRASPORE is required for male meiotic cytokinesis in Arabidopsis thaliana. Development 124, 2645–2657. [DOI] [PubMed] [Google Scholar]
- Staehelin, L.A., and Hepler, P.K. (1996). Cytokinesis in higher plants. Cell 84, 821–824. [DOI] [PubMed] [Google Scholar]
- Takahashi, T., Gasch, A., Nishizawa, N., and Chua, N.-H. (1995). The DIMINUTO gene of Arabidopsis is involved in regulating cell elongation. Genes Dev. 9, 97–107. [DOI] [PubMed] [Google Scholar]
- Tang, B.L., and Hong, W. (1999). A possible role of di-leucine–based motifs in targeting and sorting of the syntaxin family of proteins. FEBS Lett. 446, 211–212. [DOI] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W.R., Cutler, S., Somerville, C.R., and Turner, S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11, 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainotti, L., Spolaore, S., Pavanello, A., Baldan, B., and Casadoro, G. (1999). A novel E-type endo-β-1,4-glucanase with a putative cellulose-binding domain is highly expressed in ripening strawberry fruits. Plant Mol. Biol. 40, 323–332. [DOI] [PubMed] [Google Scholar]
- Turner, S.R., and Somerville, C.R. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9, 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn, K.C., Hoffman, J.C., Hahn, M.G., and Staehelin, L.A. (1996). The herbicide dichlobenil disrupts cell plate formation: Immunogold characterization. Protoplasma 194, 117–132. [Google Scholar]
- von Heijine, G. (1992). Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225, 487–494. [DOI] [PubMed] [Google Scholar]
- Zuo, J., Rungger, D., and Voellmy, R. (1995). Multiple layers of regulation of human heat shock transcription factor 1. Mol. Cell. Biol. 15, 4319–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]