Abstract
Cells modulate the expression of nuclear genes in response to changes in the functional state of mitochondria, an interorganelle communication pathway called retrograde regulation. In yeast, expression of the CIT2 gene shows a typical retrograde response in that its expression is dramatically increased in cells with dysfunctional mitochondria, such as in ρo petites. Three genes control this signaling pathway: RTG1 and RTG3, which encode basic helix-loop-helix leucine zipper transcription factors that bind as heterodimer to the CIT2 upstream activation site, and RTG2, which encodes a protein of unknown function. We show that in respiratory-competent (ρ+) cells in which CIT2 expression is low, Rtg1p and Rtg3p exist as a complex largely in the cytoplasm, and in ρo petites in which CIT2 expression is high, they exist as a complex predominantly localized in the nucleus. Cytoplasmic Rtg3p is multiply phosphorylated and becomes partially dephosphorylated when localized in the nucleus. Rtg2p, which is cytoplasmic in both ρ+ and ρo cells, is required for the dephosphorylation and nuclear localization of Rtg3p. Interaction of Rtg3p with Rtg1p is required to retain Rtg3p in the cytoplasm of ρ+ cells; in the absence of such interaction, nuclear localization and dephosphorylation of Rtg3p is independent of Rtg2p. Our data show that Rtg1p acts as both a positive and negative regulator of the retrograde response and that Rtg2p acts to transduce mitochondrial signals affecting the phosphorylation state and subcellular localization of Rtg3p.
INTRODUCTION
Cells can monitor and respond to changes in the state of their organelles. In the endoplasmic reticulum (ER), for example, there is a stress-related signal transduction pathway that responds to the accumulation of unfolded proteins in the lumen of the ER, activating expression of genes encoding some ER-resident proteins, such as the chaperone BiP (for review, see Kaufman, 1999). Similarly, cells can modulate the expression of nuclear genes in response to alterations in mitochondrial function, a response termed retrograde regulation (Parikh et al., 1987; Liao and Butow, 1993). In animal cells, interference of mitochondrial gene expression and loss of mitochondrial DNA result in changes in the level of a subpopulation of nuclear-derived mRNAs (Poyton and McEwen, 1996). Biswas et al. (1999) showed recently that in mouse C2C12 cells, decreases in mitochondrial DNA content or the addition of mitochondrial poisons resulted in elevated cytosolic Ca2+ levels. This was accompanied by an activation of the calcineurin and c-Jun N-terminal kinase pathways, a reduction in the level of nuclear factor-κB, and increased transcription of the sarcoplasmic reticular ryanodine receptor-1 Ca2+ release channel and the cytochrome oxidase subunit Vb gene. These effects were attributed to changes in the mitochondrial membrane potential, ΔΨm, and an attendant reduction in ATP levels.
In yeast, the retrograde signaling pathway functions as a homeostatic or stress response mechanism to adjust various biosynthetic and metabolic activities to the alterations in the mitochondrial state (Liao et al., 1991; Shyjan and Butow, 1993; Small et al., 1995; Liu and Butow, 1999). One member of the retrograde responsive set of genes is CIT2, which encodes a peroxisomal isoform of citrate synthase that catalyzes the first step in the glyoxylate cycle, a metabolic pathway responsible for the conversion of two carbon compounds (generated, for example, from the oxidation of long-chain fatty acids) into intermediates such as succinate that can enter the mitochondrial tricarboxylic acid (TCA) cycle. This metabolic interaction between the glyoxylate and TCA cycles enables cells to use two carbon compounds for anabolic pathways, because the glyoxylate cycle bypasses the steps in the TCA cycle at which two equivalents of CO2 are released. Thus, activation of the CIT2 retrograde response allows for a more efficient use of carbon for biosynthetic processes.
In wild-type, respiratory-competent cells (ρ+), CIT2 expression is low. But in cells with compromised mitochondrial function, such as those with mutations in one or more genes encoding enzymes of the TCA cycle, or those that are respiratory deficient because they lack mitochondrial DNA (ρo petites), CIT2 expression is high (Liao et al., 1991; Liao and Butow, 1993; Chelstowska and Butow, 1995; Kos et al., 1995; Small et al., 1995). Depending on the severity or number of different mitochondrial lesions, CIT2 expression can be elevated as much as 30- to 40-fold. Both basal and elevated levels of CIT2 expression are dependent on three genes, RTG1, RTG2, and RTG3. RTG1 and RTG3 encode basic helix-loop-helix leucine zipper (bHLH-Zip) transcription factors (Liao and Butow, 1993; Jia et al., 1997). Most members of this family bind to canonical E box target sites, CANNTG. Rtg1p (18 kDa) and Rtg3p (54 kDa) are unusual, however, because they do not recognize E boxes, but, rather, activate transcription by binding as a heterodimer to a novel site called an R box (GTCAC), two of which are located in the CIT2 promoter (Jia et al., 1997). Neither protein alone is able to bind to a target R box site. Although transcriptional activation requires both Rtg1p and Rtg3p, only Rtg3p has been found to contain transcriptional activation domains (Rothermel et al., 1997). The basic domain of Rtg3p strongly resembles that of many other bHLH transcription factors containing conserved amino acid residues (e.g., histidine, glutamic acid, and arginine) with conserved spacing that have been shown to be important for contacting target site DNA (Ferré-D'Amaréet al., 1993; Ellenberger, 1994; Ellenberger et al., 1994). In contrast, Rtg1p, although essential for CIT2 expression, is a novel member of the bHLH family, because its truncated basic domain lacks the conserved amino acid residues noted above, and it has no discernable transactivation activity (Rothermel et al., 1997). We have suggested that Rtg1p may facilitate the binding of Rtg3p to R box sites through its interaction with Rtg3p (Jia et al., 1997; Rothermel et al., 1997).
How Rtg2p functions in the regulation of gene expression is less clear. Rtg2p is a novel protein with an N-terminal ATP binding motif similar to that found in hsp70 homologues, actin, and sugar kinases (Bork et al., 1992). In addition, Rtg2p shares some sequence similarity with bacterial polyphosphatases and phosphatases that hydrolyze the transcriptional regulators guanosine penta- and tetraphosphate (Koonin, 1994). Genetic and transactivation studies suggested that Rtg2p acts upstream of the Rtg1p–Rtg3p complex in the regulation of CIT2 expression (Rothermel et al., 1997). Finally, although none of the RTG genes is essential for viability, null alleles of any one of them result in pleiotropic phenotypes, including not only a loss of CIT2 expression but an inability of cells to grow on acetate as a sole carbon source and a growth requirement for glutamate or aspartate. These phenotypes are characteristic of cells with defects in the TCA and glyoxylate cycles.
The RTG genes are also involved in the retrograde control of expression of a cytosolic d-lactate dehydrogenase activity encoded by a previously uncharacterized gene, YEL071, now named DLD3 (Chelstowska et al., 1999), and in a novel, dual regulation of expression of the TCA cycle genes CIT1, ACO1, IDH1, and IDH2 (Liu and Butow, 1999). These latter genes encode proteins responsible for catalyzing the first three steps of the TCA cycle leading to the synthesis of α-ketoglutarate. Their expression in cells with robust mitochondrial function is largely dependent on the Hap2,3,4,5p transcription complex, but as mitochondrial respiratory function becomes more compromised, their expression becomes more dependent—and, in some cases, entirely dependent—on the RTG genes. We have suggested that the HAP-to-RTG switch ensures that sufficient glutamate is synthesized from α-ketoglutarate for biosynthetic processes in cells with dysfunctional mitochondria. Collectively, these findings suggest that diverse metabolic activities may be under the control of the RTG genes.
The major objective of the present study was to understand how these RTG-dependent pathways of gene expression are activated in cells with compromised mitochondrial function. We show that in cells with robust mitochondrial function, in which expression of the RTG-dependent indicator gene CIT2 is low, Rtg1p and Rtg3p exist as a complex in the cytoplasm, and in cells with dysfunctional mitochondria, in which CIT2 expression is greatly elevated, these transcription factors accumulate in the nucleus. Nuclear translocation of Rtg3p correlates with (incomplete) dephosphorylation of the protein. Rtg2p is exclusively a cytoplasmic protein and is required for the nuclear localization and dephosphorylation of Rtg3p. Surprisingly, in addition to its requirement for transcriptional activation as a heterodimer with Rtg3p at target gene R box sites when the retrograde response is turned on, Rtg1p also functions to retain Rtg3p in a phosphorylated state in the cytoplasm when the retrograde response is off. These findings suggest a novel role for a component of a transcriptional activation complex and offer the first mechanistic view of the control of signaling between mitochondria and the nucleus in yeast.
MATERIALS AND METHODS
Yeast Strains, Plasmids, and Growth Conditions
The S. cerevisiae strains used in this study are derivatives of strain PSY142 (MATα, leu2 lys2 ura3 ρ+). The ρo derivatives were obtained by several passages of ρ+ cells through YPD medium (1% yeast extract, 2% bacto peptone, and 2% dextrose) containing 25 μg of ethidium bromide/ml. Gene disruptions of RTG1, RTG2, or RTG3 in PSY142 were carried out as described (Liao and Butow, 1993; Rothermel et al., 1995; Jia et al., 1997). Cells were grown at 30°C in YPR medium (YP plus 2% raffinose) or YNBR medium (0.67% yeast nitrogen base containing 1% casamino acids [+cas] and 20 mg/l uracil as required).
pRS416Rtg3-GFP was constructed by PCR amplification of the RTG3 coding and 5′ flanking regions from −745 to +1458 using the oligonucleotides 5′-GTCCTGTCTAGATACAGGCAAC-3′ and 5′-AAACTACTCGAGACCCCGAACC-3′ (restriction sites used for cloning are underlined). The oligonucleotides 5′-GGTTCGGGGGGTACCTAGTTTATG-3′ and 5′-TCATTTTCCGGATCCACTTTAT-AG-3′ were used to PCR amplify 881 bp of the 3′ untranslated region (UTR) of RTG3. The PCR products were cleaved with the appropriate restriction enzymes, and a 727-bp XhoI–KpnI fragment containing the coding region of a bright green version of green fluorescent protein (bGFP; see below) was cloned into the XbaI–BamHI site of the yeast centromere plasmid pRS416. Truncated versions of Rtg3-GFP (pRS416Rtg3Δ376–486-GFP, pRS416Rtg3Δ2–279-GFP, pRS416Rtg3Δ345–486-GFP, pRS416Rtg3Δ314–344-GFP, and pRS416Rtg3Δ280–298-GFP) were constructed by amplifying the appropriate DNA fragments by PCR using pRS416Rtg3-GFP as template and ligating the resulting fragments into pRS416 or a 6.5-kb XbaI–XhoI fragment of pRS416Rtg3-GFP. Further information on construction of p416Rtg3-GFP derivatives is available upon request. The bGFP contains three amino acid substitutions, F99S, M153T, and V163A (Okamoto et al., 1998).
To construct pRS416Rtg1-GFP, the RTG1 coding and 5′ flanking region from −720 to +531 was amplified by PCR using the oligonucleotides 5′-TTGTCTAGAAATTCGGATACGCAAAA-3′ and 5′-AGTCTCGAGCGCTACCATTACCGTACTCAC-3′. The oligonucleotides 5′-AGTGGTACCAAGTACTTCTGACTCTCAC-3′ and 5′-CCTGGATCCTTCCCGAGGATACAA-3′ were used to PCR amplify 288 bp of the 3′ UTR of RTG1. These fragments together with the 727-bp XhoI–KpnI fragment of bGFP were cloned into the XbaI–BamHI site of pRS416. Similarly, for pRS416Rtg2-GFP, the RTG2 coding and 5′ flanking region from −396 to +1766 was amplified by PCR using the oligonucleotides 5′-ATAAAGCTTCACCCCAATCCTTTCTGTTATT-3′ and 5′-CTTTATTCTCGAGAAAATTGCACGCC-3′. The oligonucleotides 5′-TGGCGTGGTACCTTATGAAGAATAAAGA-3′ and 5′-TCAGGATCCTGGATATGAGACATGC-3′ were used to PCR amplify 2988 bp of the 3′ UTR of RTG2. These fragments together with the 727-bp XhoI–KpnI fragment bGFP fragment were cloned into pRS416.
Transplacements of the RTG3 gene by the various Rtg3-GFP derivatives were carried out using linear XbaI–BamHI fragments of the full-length or truncated version of Rtg3-GFP from the different plasmids described above that were transformed by standard procedures into an rtg3::URA3 recipient strain of PSY142 (Jia et al., 1997) and selecting for Ura− transformants by plating on solid YNB+cas medium containing 3% glycerol, 0.1% 5-fluoroorotic acid, and 20 mg/ml uracil. All transformants were verified by Southern hybridization. Similarly, transplacement of RTG1 or RTG2 to Rtg1-GFP or Rtg2-GFP, respectively, was done by transforming an XbaI–BamHI fragment of Rtg1-GFP including 720-bp upstream and 288-bp downstream sequence of RTG1 from pRS416Rtg1-GFP or a HindIII fragment of Rtg2-GFP including 396-bp upstream and 1377-bp downstream sequence of RTG2 from pRS416Rtg2-GFP, respectively, into rtg1::URA3 or rtg2::URA3 derivatives of PSY142.
Microscopy
Yeast strains containing Rtg3-GFP fusions were grown to logarithmic phase (OD600, 0.7–1.0) in YNBR+cas medium. Samples were observed using a Leica (Deerfield, IL) microscope (model DMRXE) equipped for an HBO 100 W/2 mercury arc lamp, an X100 Plan-Apochromat objective, and epifluorescence with the following filter set: 450 to 490-nm bandpass excitation filter, 510-nm dichroic reflector, and >515-nm long-pass emission filter for GFP. Images were collected with a charge-coupled device camera (model C5810; Hamamatsu, Hamamatsu City, Japan), and processed in Adobe (Mountain View, CA) Photoshop 5.0.
RNA Isolation and Northern Blot Analysis
Cells were grown in YPR medium to an OD600 of 0.7–1.0. Total yeast RNA was prepared using the hot phenol method as described (Schmitt et al., 1990). Northern blot analysis was performed as described (Jia et al., 1997).
Western Blot Analysis
Trichloroacetic acid precipitates of total yeast cell proteins were prepared by pelleting cells from OD 0.7–1.0 culture as described before (Rothermel et al., 1995). For SDS-PAGE, equal volumes of extract dissolved in SDS-PAGE sample buffer (100 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 0.002% bromphenol blue, and 0.1 M dithiothreitol) were loaded onto an SDS-PAGE gel with the appropriate concentration of acrylamide and separated using the Ready Gel system (Bio-Rad, Hercules, CA). Proteins were transferred to nitrocellurose membranes (Schleicher & Schuell, Keene, NH) by semidry transfer units (Hoefer Scientific, San Francisco, CA). Immunodetection of proteins was carried out using primary rabbit anti-Rtg1p and anti-Rtg3p polyclonal antibodies. Anti-Rtg1p polyclonal antibody was raised as described by Rothermel et al. (1995). Anti-Rtg3p polyclonal antibody was raised against a purified maltose-binding protein–tagged version of the protein. Anti-rabbit immunoglobulin G-coupled HRP (Bio-Rad) was used as the second antibody and was visualized using the ECL system (Amersham, Arlington Heights, IL).
Immunoprecipitation
Cells were grown to OD600 0.7–1.0 in 50 ml of YNBR+cas, pelleted, and resuspended in 500 μl of solution A (50 mM Tris-HCl, pH 7.4, 50 mM NaCl, and 0.2% Triton X-100 containing 10 μg/ml protease inhibitors aprotinin, pepstatin A, and leupeptin and 1 mM PMSF) and 0.5 g of glass beads (0.5 mm). In some cases, 0.5 mM NaF and 5 mM sodium pyrophosphate were included in solution A to block phosphatase activity. Cells were broken by vortexing for 4 min (eight times for 30 s, with 30 s on ice between each vortex). The lysate was transferred to chilled 2-ml Eppendorf tubes and centrifuged at 21,000 × g for 30 min. Protein in the supernatant was adjusted to 3 μg/μl, and a 500-μl aliquot was incubated with polyclonal antiserum against Rtg3 or polyclonal antiserum against GFP (1 μl/100 μl of extract) at 4°C for 2 h. Then 150 μl of a slurry of protein G-Sepharose (Boehringer Mannheim, Indianapolis, IN) were added to the reaction mixture. The immune complexes were released by boiling in SDS-PAGE sample buffer after washing five times with solution A. The released immune complexes were analyzed by Western blotting as described above.
RESULTS
Interaction between Rtg1p and Rtg3p Is Independent of the Retrograde Response
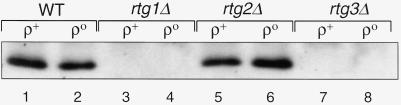
To determine whether the intrinsic interaction between Rtg1p and Rtg3p might be modulated to control CIT2 expression, Rtg3p was immunoprecipitated with Rtg3p-specific anitserum from whole-cell extracts of wild-type ρ+ and ρo cells and various mutant derivatives of these strains, and the immunoprecipitates were analyzed by Western blotting with antiserum specific for Rtg1p. In extracts of ρ+ and ρo wild-type cells, similar amounts of Rtg1p were coprecipitated with anti-Rtg3p antiserum (Figure 1, lanes 1 and 2). As expected, no Rtg1p was detected in extracts prepared from rtg1Δ (lanes 3 and 4) or rtg3Δ (lanes 7 and 8) mutant derivatives of these strains. Thus, Rtg1p and Rtg3p appear to interact comparably in ρ+ and ρo cells despite the large difference in CIT2 expression between these strains. Moreover, in rtg2Δ ρ+ and ρo cells, we detected the same complex between Rtg3p and Rtg1p (lanes 5 and 6). These data are consistent with previous yeast two-hybrid experiments showing that the interaction between Rtg1p and Rtg3p is independent of Rtg2p and that these proteins are similarly expressed in ρ+ and ρo cells (Rothermel et al., 1997).
Figure 1.
Rtg1p and Rtg3p interact in both ρ+ and ρo petite cells. Whole-cell extracts were prepared from wild-type (WT) ρ+ and ρo cells and rtg1Δ, rtg2Δ, or rtg3Δ mutant derivatives of these strains. Extracts were adjusted to 3 mg/ml protein and incubated with 5 μl of antiserum raised against recombinant Rtg3p. The immunoprecipitates were then analyzed by Western blotting with antiserum raised against recombinant Rtg1p.
Activation of the Retrograde Response Correlates with the Translocation of Rtg1p and Rtg3p from the Cytoplasm to the Nucleus
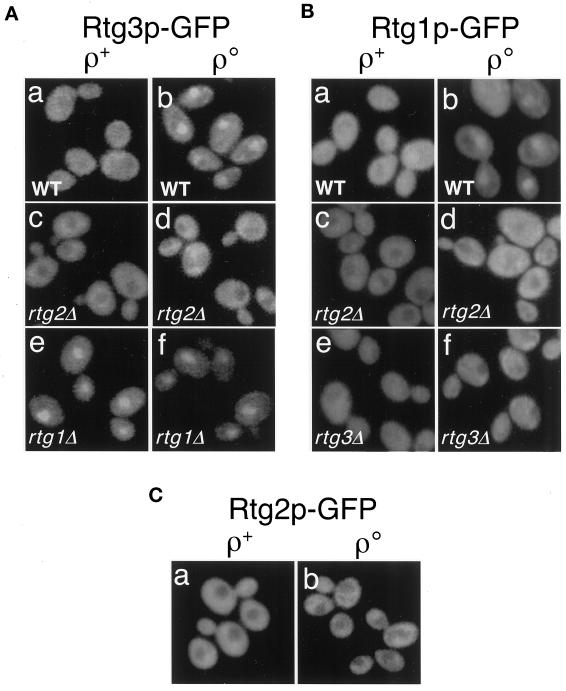
To investigate the possibility that the retrograde response is controlled by regulation of the subcellular localization of Rtg1p and Rtg3p, we constructed integrating vectors encoding in-frame fusions between the C termini of full-length Rtg1p and Rtg3p and GFP. Expression of each of the Rtg-GFP fusion proteins was placed under the control of its natural promoter. These constructs were used in integrative transformations to replace the respective chromosomal copies of RTG1 and RTG3, and the expression of the GFP fusion proteins was examined in ρ+ and ρo wild-type and various mutant derivatives of these strains. Preliminary experiments verified that each GFP fusion protein expressed from an integrated single-copy gene was functional in vivo, because each could complement their respective rtg null allele as determined by restoration of CIT2 expression and the activation of the retrograde response in ρo cells (our unpublished results).
In wild-type ρ+ cells in which CIT2 expression is low, Rtg3p-GFP is predominantly cytoplasmic (Figure 2A, a). By contrast, Rtg3p-GFP shows a predominantly nuclear localization in ρo cells in which CIT2 expression is high (Figure 2A, b). This same pattern of cytoplasmic versus nuclear localization was also observed for Rtg1p-GFP expressed in wild-type ρ+ and ρo cells (Figure 2B, a and b). Thus, one level of control of the retrograde response is by regulation of the subcellular localization of the Rtg1p–Rtg3p complex.
Figure 2.
Subcellular localization of Rtg3p (A), Rtg1p (B), and Rtg2p (C) in wild-type (WT) and in various rtgΔ mutant derivatives of ρ+ and ρo cells. Constructs encoding C-terminal-tagged GFP derivatives of full-length Rtg1p, Rtg2p, and Rtg3p were transplaced into their respective chromosomal loci and expressed under the control of the each of the native promoters. Cells were grown in YNBR+cas medium. In wild-type ρ+ cells, Rtg3p-GFP (A, a) and Rtg1p-GFP (B, a) are largely cytoplasmic. In otherwise wild-type ρo cells, however, both Rtg3p-GFP (A, b) and Rtg1p-GFP (B, b) are concentrated in the nucleus. Rtg2p-GFP expressed from a single-copy gene transplaced into the RTG2 locus appears strictly cytoplasmic both in ρ+ and ρo cells (C, a and b). The effects of rtg1Δ, rtg2Δ, or rtg3Δ mutations on the subcellular localization of these GFP fusion proteins are shown. Localization of the GFP fusion proteins was determined by epifluorescence microscopy as described in MATERIALS AND METHODS.
Rtg2p Is a Cytoplasmic Protein and Is Required for Nuclear Localization of Rtg1p and Rtg3p
Although Rtg2p is essential for CIT2 expression in both ρ+ and ρo cells, it lacks any obvious DNA binding motifs and shows no activity as a transcriptional activator (Rothermel et al., 1995). It was of interest, therefore, to determine the subcellular localization of this protein as well. As was done for Rtg1p and Rtg3p, we constructed a full-length fusion protein between the C terminus of Rtg2p and GFP and transplaced it into the RTG2 locus under control of the RTG2 promoter. Complementation experiments also indicated that Rtg2p-GFP is functional in vivo (our unpublished results). In both ρ+ and ρo cells, Rtg2p-GFP is strictly cytoplasmic and appears to be excluded from the nucleus in those cells (Figure 2C, a and b).
Given these findings and previous genetic data that Rtg2p acts upstream of the Rtg1p–Rtg3p complex (Rothermel et al., 1997), how might Rtg2p function as a cytoplasmic protein in the regulation of expression of retrograde responsive genes? One obvious possibility is that Rtg2p controls the subcellular localization of Rtg1p and Rtg3p. To test this, we deleted the RTG2 gene in strains harboring the transplaced copies of Rtg3-GFP and Rtg1-GFP and determined the effect on the localization of the GFP fusion proteins. These experiments show that the rtg2Δ mutation had no obvious effect on the cytoplasmic localization of either Rtg3p-GFP or Rtg1p-GFP in ρ+ cells (Figure 2, A, c, and B, c, respectively) but blocked their nuclear accumulation in ρo cells (Figure 2, A, d, and B, d). These data suggest that Rtg2p regulates RTG-dependent gene expression by controlling the nuclear localization of Rtg1p and Rtg3p.
Rtg1p Is Required to Retain Rtg3p in the Cytoplasm in ρ+ Cells but Not Vice Versa
Because Rtg1p and Rtg3p interact in the cytoplasm in ρ+ cells as well as in the nucleus when bound to R box target sites, we asked whether the subcellular localization of either protein might be affected by the absence of the other. To this end, we examined the localization of Rtg3p-GFP expressed from the transplaced gene in rtg1Δ ρ+ and ρo cells and, similarly, the localization of Rtg1-GFP in rtg3Δ ρ+ and ρo cells. Surprisingly, we observed that the absence of Rtg1p in ρ+ cells resulted in a predominantly nuclear localization of Rtg3p-GFP (Figure 2A, e), comparable with that observed in otherwise wild-type ρo petite cells (Figure 2A, b). The nuclear localization of Rtg3p-GFP in ρo cells was unaffected by the rtg1Δ mutation (Figure 2A, f). In sharp contrast to these observations, Rtg1p-GFP remained cytoplasmic in both ρ+ and ρo rtg3Δ cells (Figure 2B, e and f). These observations suggest that nuclear retention of Rtg1p requires that Rtg3p also be present in the nucleus. They suggest further that Rtg1p functions not only as a positive regulator in the transcriptional activation of retrograde responsive genes via its interaction with Rtg3p at R box target sites but also as a negative regulator by contributing to the sequestration of Rtg3p in the cytoplasm in ρ+ cells when the retrograde response is off and the level of target gene expression is low.
Nuclear Localization of Rtg1p and Rtg3p Correlates with Dephosphorylation of Rtg3p
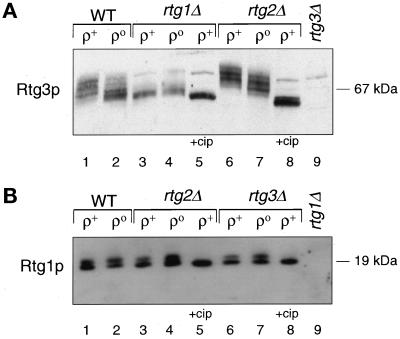
The subcellular localization of some transcription factors has been shown to be regulated by phosphorylation (for reviews, see Jans and Hubner, 1996; Nigg, 1997; Hopper, 1999). To address whether Rtg1p and Rtg3p are phosphoproteins and, if so, whether phosphorylation correlates with their subcellular localization, we examined the electrophoretic mobility of these proteins by Western blotting of extracts from wild-type ρ+ and ρo cells and different rtg mutant derivatives using Rtg3p- and Rtg1p-specific antiserum. In wild-type ρ+ cells, different electrophoretic mobility forms of Rtg3p are detected (Figure 3A, lane 1), suggesting that in ρ+ cells, Rtg3p is multiply phosphorylated (also see below). In extracts from wild-type ρo cells (Figure 3A, lane 2), there is a distinct shift in the mobility of Rtg3p to faster-migrating species, indicating a substantial but incomplete dephosphorylation of the protein compared with ρ+ cells. In rtg1Δ ρ+ and ρo cells, Rtg3p is predominately unphosphorylated (Figure 3A, lanes 3 and 4), and treatment of the extracts with calf intestinal alkaline phosphatase (Figure 3A, lane 5), resulted in the appearance of a single species whose electrophoretic mobility is close to that expected for a 54-kDa protein. In contrast, Rtg3p becomes hyperphosphorylated in rtg2Δ ρ+ and ρo cells, evident by a dramatic shift to much slower-migrating species (Figure 3A, lanes 6 and 7); these species also are converted to a faster-migrating form upon treatment with calf intestinal alkaline phosphatase (Figure 3A, lane 8). From these data we conclude that the nuclear accumulation of Rtg3p correlates with incomplete dephosphorylation of the protein and that both its subcellular localization and phosphorylation state are controlled by Rtg2p.
Figure 3.
Effects of the functional state of mitochondria and rtgΔ mutations on the phosphorylation state of Rtg3p and Rtg1p. Logarithmic phase cultures of wild-type (WT) ρ+ and ρo cells and rtgΔ mutant derivatives of these strains were grown in YPR medium, and cell-free extracts were prepared as described in MATERIALS AND METHODS. Aliquots of these extracts were analyzed by Western blotting with antisera raised against recombinant Rtg3p (A) or Rtg1p (B). In some cases the extracts were treated with 5 U of calf intestinal alkaline phosphatase (cip) before Western blot analysis, as indicated.
Rtg1p is also a phosphoprotein, because we generally observe two bands both in ρ+ and ρo cells (Figure 3B) that collapse to a single band after alkaline phosphatase treatment of the extract (Figure 3B, lanes 5 and 8). However, in contrast to the results with Rtg3p, we have not observed any significant difference in the distribution of these forms of the protein in ρ+ and ρo cells (Figure 3B, lanes 1 and 2) or in rtg2Δ (Figure 3B, lanes 3 and 4) or rtg3Δ (Figure 3B, lanes 6 and 7) mutant derivatives of these cells. Thus, the phosphorylation state of Rtg1p does not appear to play a role in its subcellular localization.
An rtg1Δ Mutation Suppresses the Effects of an rtg2Δ Mutation on Rtg3p Localization and Phosphorylation States in ρ+ and ρo Cells
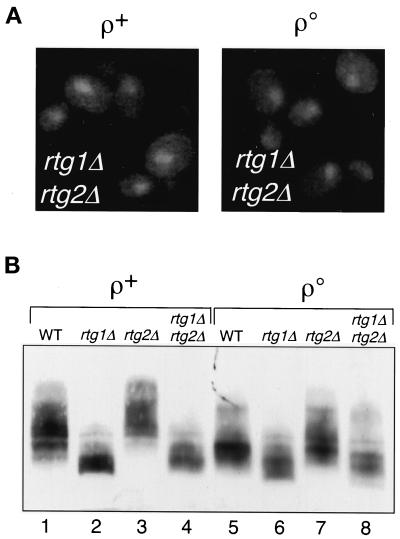
It is clear from the data presented above that the rtg1Δ and rtg2Δ mutations have entirely opposite effects on the nuclear localization and phosphorylation state of Rtg3p. It was of obvious interest therefore to examine the behavior of Rtg3p in rtg1Δ rtg2Δ double-mutant ρ+ and ρo strains. As shown in Figure 4A, left panel, the nuclear localization of Rtg3p-GFP in ρ+ cells induced by the rtg1Δ mutation (Figure 2A, e) is unaffected in the rtg1Δ rtg2Δ double mutant. However, in ρo cells (Figure 4A, right panel), the rtg1Δ mutation reversed the block in nuclear localization of Rtg3p-GFP caused by the rtg2Δ mutation (Figure 2A, d). In a similar manner, we compared the phosphorylation state of Rtg3p in ρ+ rtg1Δ rtg2Δ cells with that in the rtg1Δ and rtg2Δ single mutants and in otherwise wild-type ρo cells (Figure 4B). In both ρ+ and ρo cells, the hyperphosphorylation of Rtg3p caused by the rtg2Δ mutation (lanes 3 and 7) is reversed in the rtg1Δ rtg2Δ double mutant (lanes 4 and 8), resulting in the same dephosphorylated Rtg3p species as observed in rtg1Δ single-mutant cells (lanes 2 and 6). Collectively, these data show that rtg1Δ is epistatic to rtg2Δ in affecting the subcellular localization and phosphorylation state of Rtg3p.
Figure 4.
An rtg1Δ mutation is epistatic to an rtg2Δ mutation. (A) Subcellular localization of Rtg3p-GFP in ρ+ or ρo rtg1Δ rtg2Δ double-mutant cells was determined by epifluorescence microscopy. (B) Extracts were prepared from wild-type ρ+ and ρo cells and the indicated rtg mutant derivatives of these strains and analyzed by Western blotting with antiserum specific for Rtg3p.
Functional Domains of Rtg3p
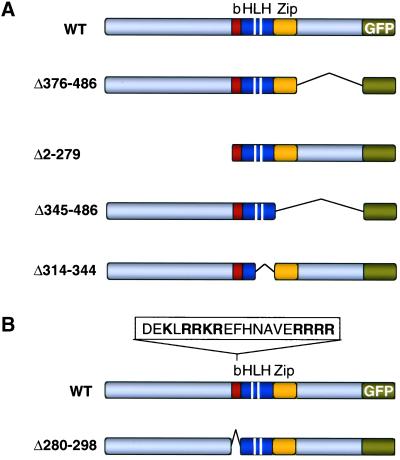
To identify domains of Rtg3p that are important determinants in its subcellular localization, we constructed several Rtg3 deletion mutants tagged at their C terminus with GFP (Figure 5A) and expressed these from the natural RTG3 promoter as single-copy genes transplaced into the RTG3 locus. In these experiments we were particularly interested in knowing how these mutations affected not only the subcellular localization of Rtg3p but also its phosphorylation state and ability to interact with Rtg1p in ρ+ and ρo cells. For some of the mutants, we also determined whether their localization and phosphorylation states were affected in rtg1Δ or rtg2Δ mutant cells and whether the mutants affected CIT2 retrograde expression. The results of these experiments are shown in Figure 6. Of the constructs shown in Figure 5A, only the C-terminal deletion mutant, Rtg3Δ376–486-GFP, behaved in all of the assays essentially identical to that of wild-type Rtg3p. Thus, Rtg3Δ376–486-GFP binds Rtg1p (Figure 6A, lane 2) and is a phosphorylated protein (Figure 6B, lane 1) that exists predominantly in the cytoplasm of ρ+ cells (Figure 6C) and in the nucleus in ρo cells (Figure 6C), partially dephosphorylated (Figure 6B, lane 2). From these observations, it is not surprising that Rtg3Δ376–486-GFP supports a robust CIT2 retrograde response (Figure 6D, lanes 5 and 6) requiring Rtg1p and Rtg2p (Figure 6D, lanes 7 and 8). Moreover, phosphorylation and retention of Rtg3Δ376–486-GFP in the cytoplasm of ρ+ cells require Rtg1p (Figure 6, B, lane 3, and C, respectively), and its dephosphorylation and nuclear accumulation are dependent on Rtg2p (Figure 6, B, lane 4, and C, respectively).
Figure 5.
Deletion mutants of Rtg3p-GFP. (A) Shown in order below that of wild-type (WT) Rtg3p-GFP are representations of mutants with deletion of the C or N terminus, the Zip, and loop-helix 2 (LH) domains of Rtg3p-GFP. The white bars separate the helix 1-loop-helix 2 domains. (B) A portion of the amino acid sequence in the basic region (b) of wild-type Rtg3p is shown and below it a representation of the deletion mutant lacking the basic region. A consensus bipartite NLS is indicated in bold.
Figure 6.
Effects of domain deletions on various properties of Rtg3p-GFP. Constructs encoding the deletion mutants of Rtg3p-GFP indicated in Figure 5 were transplaced into the RTG3 locus of wild-type and various mutant derivatives of ρ+ cells. ρo derivatives of those strains were obtained by ethidium bromide mutagenesis. The various ρ+ and ρo strains and rtg mutant derivatives were analyzed in A for their ability to interact with Rtg1p by Western blotting with anti-Rtg1p antiserum of immunoprecipitates obtained by incubation of whole-cell extracts with anti-GFP antiserum, in B for their phosphorylation state by Western blot analysis using anti-Rtg3p antiserum, in C for their subcellular localization by epifluorescence microscopy, and in D and E for their ability to support CIT2 expression as determined by Northern blotting with a CIT2-specific probe as described in MATERIALS AND METHODS. RNA loads were normalized to the level of ACT1 mRNA using an ACT1-specific probe.
In contrast to the results with Rtg3Δ376–486-GFP, the N-terminal deletion mutant Rtg3Δ2–279-GFP (Figure 5A), although still able to interact with Rtg1p (Figure 6A, lane 5), is dephosphorylated and predominantly nuclear in ρ+ as well as in ρo rtg2Δ cells (Figure 6, B, lanes 1 and 2, and C, respectively). Nevertheless, this mutant protein is unable to support CIT2 expression in either ρ+ or ρo cells (Figure 6E, lanes 7 and 8). These results are consistent with the presence of a transactivation domain in the N terminus of Rtg3p (Rothermel et al., 1997; Massari et al., 1999). Most of the potential phosphorylation sites of Rtg3p are located in the N-terminal domain, which contains 80% of the total serine and threonine residues of the protein. This would account for the observations that only single band is observed for Rtg3Δ2–279-GFP in the various strains indicated in Figure 6B, lanes 1–4, all of which have the same mobility as an alkaline phosphatase-treated extract from Rtg3Δ2–279-GFP rtg2Δ cells (Figure 6B, lane 5). These findings suggest that phosphorylation of Rtg3p within its N-terminal domain is important for its retention in the cytoplasm.
Most bHLH-Zip proteins function as homo- or heterodimers through interactions that include the Zip and HLH domains (Ferré-D'Amaréet al., 1993; Ellenberger, 1994). To investigate further the notion that interaction of Rtg1p with Rtg3p is important for retaining Rtg3p in the cytoplasm in ρ+ cells, we examined two Rtg3p-GFP deletion mutants, Rtg3Δ345–486-GFP, which lacks the C-terminal and Zip domain, and Rtg3Δ314–344-GFP, which lacks the loop-helix 2 domain (Figure 5A). Both mutant proteins are predicated to be compromised in their ability to interact with Rtg1p. Immunoprecipitation experiments confirm these predictions, showing that neither of these deletion mutants forms a stable complex with Rtg1p (Figure 6A, lanes 3 and 6), and neither could support CIT2 expression (Figure 6E, lanes 3–6). Moreover, like Rtg3Δ2–279-GFP, both of the deletion mutants were largely dephosphorylated (Figure 6B, lanes 1–3) even in rtg2Δ cells (Figure 6B, lane 4) and were localized in the nucleus in ρ+ and in ρo rtg2Δ cells (Figure 6C). The similarity in the localization and phosphorylation states of these deletion mutants to that of wild-type Rtg3p in rtg1Δ cells further supports the notion that Rtg1p functions to sequester Rtg3p in the cytoplasm of ρ+ cells.
Rtg3p contains a putative bipartite nuclear localization sequence (NLS) in the basic domain of the bHLH motif (Figure 5B). To test whether it is a functional NLS, that sequence was deleted in the mutant protein, Rtg3pΔ280–298 (Figure 5B). Immunoprecipitation experiments showed that the absence of the putative NLS region in Rtg3pΔ280–298 did not impair the ability of this mutant protein to interact with Rtg1p (Figure 6A, lane 4). Importantly, Rtg3pΔ280–298-GFP failed to localize to the nucleus in ρo rtg1Δ cells (Figure 6C), a genetic background that we have shown is optimal for nuclear accumulation of wild-type Rtg3p. These findings are consistent with the absence of CIT2 expression in ρ+ and ρo cells expressing Rtg3pΔ280–298-GFP (Figure 6E, lanes 9 and 10). From these experiments, we conclude that the bipartite sequence in Rtg3p functions as an NLS.
The finding that Rtg3pΔ280–298-GFP fails to localize to the nucleus but can still interact with Rtg1p allowed us to assess whether the conversion of Rtg3p to more dephosphorylated forms in ρo or in rtg1Δ ρ+ cells is a cytoplasmic or nuclear activity. As shown in Figure 6B, lanes 1–3, Rtg3pΔ280–298-GFP, despite being cytoplasmic, is largely unphosphorylated. In rtg2Δ ρo cells, Rtg3pΔ280–298-GFP appears slightly phosphorylated (Figure 6B, lane 4), although not nearly to the same extent as seen for wild-type Rtg3p-GFP in rtg2Δ ρo cells. Considered together, these results suggest that cytoplasmic Rtg3p is a substrate for a regulated kinase or phosphatase activity or both.
The Phosphorylation State of Rtg3p Is Subject to a Feedback Control
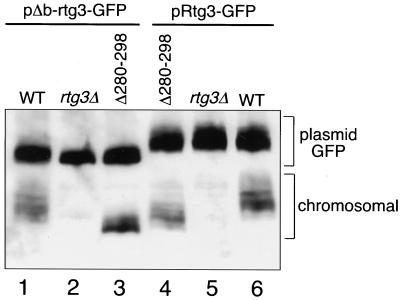
It was somewhat surprising to us that Rtg3pΔ280–298-GFP was largely unphosphorylated in ρ+ or in rtg2Δ ρo cells, despite being complexed with Rtg1p in the cytoplasm. The analysis of this deletion mutant, as well as the other deletion mutants described above, was carried out in cells in which the wild-type, chromosomal copy of RTG3 was replaced with those genes encoding the deletion mutant variants. Although none of the RTG genes is essential for viability, the RTG system appears to influence the expression of a broad spectrum of genes (C. Epstein, unpublished observations). Moreover, even in ρ+ cells with robust mitochondrial function, there is some RTG-dependent gene expression. Thus, the absence of a functional Rtg3p might itself be stressful to cells and, like mitochondrial dysfunctions, might lead to an activation of events resulting in the dephosphorylation and nuclear translocation of cytosolic Rtg3p. To examine this possibility, the chromosomal copy of RTG3 in ρ+ cells was replaced with the NLS deletion mutant rtg3Δ280–298 (which lacks the C-terminal GFP tag), and Rtg3Δ280–298-GFP or wild-type Rtg3-GFP was coexpressed in those cells from the centromeric plasmid pΔb-rtg3-GFP or pRtg3-GFP, respectively. Western blot analysis using Rtg3p-specific antiserum was then performed to assess the phosphorylation state of the chromosomally expressed Rtg3pΔ280–298, which could be readily distinguished from the plasmid-expressed proteins because it lacks the 27-kDa GFP extension. Analysis of controls with either the chromosomal wild-type RTG3 or rtg3Δ alleles was carried out in parallel. As shown in Figure 7, lane 3, chromosomally expressed Rtg3pΔ280–298 is largely dephosphorylated in cells expressing Rtg3Δ280–298-GFP from pΔb-rtg3-GFP, whereas it is significantly more phosphorylated in cells expressing wild-type Rtg3-GFP from pRtg3-GFP (Figure 7, lane 4). Control experiments show that chromosomally expressed wild-type Rtg3p is phosphorylated in these ρ+ cells regardless of whether those cells are coexpressing Rtg3Δ280–298-GFP or wild-type Rtg3-GFP (Figure 7, lanes 1 and 6, respectively). From these data we conclude that Rtg3pΔ280–298 is capable of being phosphorylated and that both phosphorylation and dephosphorylation activities of Rtg3p are cytoplasmic. The data also support the conclusion that the phosphorylation state of cytoplasmic Rtg3p is subject to modulation by stress responses, which include not only mitochondrial dysfunction but also the absence of a functional Rtg3p.
Figure 7.
Rtg3p phosphorylation is subject to a feedback control. ρ+ wild-type (WT), rtg3Δ, and rtg3Δ280–298 deletion mutant allele transplaced into the chromosomal RTG3 locus, each transformed with a centromeric plasmid, pΔb-rtg3-GFP encoding the Rtg3pΔ280–298-GFP deletion mutant or with pRtg3-GFP encoding wild-type Rtg3p-GFP, were grown in YNBR+cas medium to midlogarithmic phase. Whole-cell extracts were prepared and analyzed by Western blotting using Rtg3p-specific antiserum. The positions of the plasmid-encoded GFP-tagged and chromosomally expressed Rtg3ps are indicated.
DISCUSSION
The Functional State of Mitochondria Determines the Subcellular Localization and Phosphorylation State of Rtg3p
We have shown that a key factor in the control of mitochondria-to-nuclear signaling is the regulated nuclear localization of the bHLH-Zip transcription factor Rtg3p. In ρ+ cells in which CIT2 expression is low, Rtg3p is localized largely in the cytoplasm bound to its heterodimeric partner Rtg1p. In ρo petite cells in which CIT2 expression is high, both Rtg3p and Rtg1p are predominantly nuclear. These changes in the subcellular localization of Rtg3p are accompanied by changes in its phosphorylation state: When present in the cytoplasm, wild-type Rtg3p is multiply phosphorylated, and when localized in the nucleus in ρo cells, it is partially dephosphorylated. Rtg1p is also a phosphoprotein, but we have not detected any difference in its phosphorylation state between ρ+ and ρo cells. Although Rtg1p and Rtg3p are both required for expression of CIT2, only Rtg3p appears to have a direct transactivation function (Rothermel et al., 1997), and only its nuclear localization is regulated. Unlike Rtg3p, the nuclear accumulation of the 18-kDa Rtg1p (plus 27 kDa of GFP) appears to be passive (in general, proteins smaller than 45–50 kDa can freely diffuse into the nucleus), requiring only that Rtg3p be present in the nucleus, presumably to anchor Rtg1p there in an active transcription complex. It is also possible that Rtg1p is transported to the nucleus as a complex with Rtg3p. A tentative model summarizing the findings of the present work is shown in Figure 8.
Figure 8.
Model of the control of mitochondria-to-nuclear signaling. In cells with dysfunctional mitochondria, one or more signals, one of which is possibly the level of glutamate produced from the TCA cycle, are transmitted from mitochondria (bold, dashed arrow) via Rtg2p to a cytoplasmic complex between Rtg1p and a highly phosphorylated form of Rtg3p. This complex, which may include other factors not indicated, becomes transiently dissociated along with a dephosphorylation of Rtg3p. Rtg1p and Rtg3p then translocate to the nucleus and assemble for transcriptional activation at target gene R box sites, GTCAC. The phosphorylation state of cytoplasmic Rtg3p is sensitive to a feedback response, indicated by the light green arrow, in that the absence of Rtg1p–Rtg3p-dependent transcription in the nucleus activates further dephosphorylation and nuclear translocation of cytoplasmic Rtg3p. It is not known whether dephosphorylation of cytoplasmic Rtg3p is caused by inactivation of a kinase or activation of a phosphatase.
It is now clear that phosphorylation plays an important role in regulating the distribution of some proteins between the cytosol and nucleus in response to nutritional or other environmental signals (Jans and Hubner, 1996; Nigg, 1997; Hopper, 1999). For instance, several phosphorylation events control the subcellular localization of Pho4, a transcriptional activator required in a pathway of gene regulation that monitors changes in the external concentration of phosphate (Oshima, 1997; Kaffman et al., 1998b; Komeili and O'Shea, 1999). In phosphate-poor medium, Pho4 is dephosphorylated, and in the nucleus, when cells are exposed to a phosphate-rich medium, a subset of serine residues in Pho4 are phosphorylated by the Pho80-Pho85 cyclin–cyclin-dependent kinase complex (Kaffman et al., 1994) effecting Pho4 nuclear export (Kaffman et al., 1998a) through interaction with Msn5p, a member of the importin-β family of nuclear receptors (Fornerod et al., 1997; Gorlich et al., 1997). Phosphorylation of Pho4 has also been shown to be important in preventing Pho4 nuclear import by blocking its interaction with the nuclear import receptor Pse1p/Kap121, another member of the importin-β nuclear receptor family (Kaffman et al., 1998b). Nuclear import of the transcriptional repressor Mig1p is also regulated by phosphorylation (De Vit et al., 1997). In high-glucose medium, a dephosphorylated form of Mig1p is rapidly imported into the nucleus, where it acts as a negative regulator of gene expression. In low-glucose medium, nuclear Mig1p is phosphorylated by the protein kinase Snf1p and, like Pho4, is exported to the cytoplasm via the Msn5p pathway (De Vit and Johnston, 1999). Preliminary experiments suggest that Msn5p may also function in regulating Rtg3p nuclear export (T. Sekito, unpublished observations). The relevant kinase and phosphatase activities affecting the phosphorylation state of Rtg3p, as well as the phosphorylation sites critical for its regulated subcellular localization, remain to be identified. Experiments are presently under way to resolve these issues. Most of the potential phosphorylation sites in Rtg3p are located in the N-terminal region, which contains 80% of the total serine and threonine residues of the protein. Deletion of this N-terminal domain in the mutant Rtg3Δ2–279-GFP did not affect its interaction with Rtg1p, but the mutant protein was nevertheless constitutively localized in the nucleus, implicating one or more of the N-terminal phosphorylated residues in regulating the subcellular location of Rtg3p.
In addition to containing most of the potential phosphorylation sites, the N-terminal domain of Rtg3p has an important transactivation function. Recently, Massari et al. (1999) identified a novel amino acid motif (LDFS) at the extreme N terminus of Rtg3p located within a conserved α-helical activation domain, termed AD1 (Aronheim et al., 1993; Quong et al., 1993; Massari et al., 1996). The LDFS motif has been found in class I HLH proteins such as E2A, HEB, and E2-2 (Massari et al., 1999), and appears to be unique among yeast bHLH proteins. The LDFS motif has been suggested to function in transactivation by interacting with components of the SAGA complex allowing chromatin remodeling through histone modification in and around target genes (Grant et al., 1997, 1998). Mutants lacking Gcn5p, a histone acetylase and a component of the SAGA complex, were shown to have a ∼50% reduction in CIT2 expression (www.wi.mit.edu/young/expression.html), suggesting that the AD1 domain is important but not essential for CIT2 expression.
Identification of the Rtg3p NLS
The basic region of Rtg3p contains a bipartite sequence similar to many NLS's. When that sequence was deleted in the mutant, Rtg3pΔ280–298-GFP, the protein remained cytoplasmic in ρo cells and in cells lacking Rtg1p, genetic backgrounds in which wild-type Rtg3p was predominantly nuclear. Moreover, just the basic HLH domain of Rtg3p can accumulate in the nucleus (our unpublished results), further supporting the notion that the bipartite element functions as an NLS. Although deletion of the basic region in Rtg3pΔ280–298-GFP did not appear to compromise the protein's ability to interact with Rtg1p, it was surprising to find that, despite its cytoplasmic location, Rtg3pΔ280–298-GFP was largely unphosphorylated in ρ+ cells and in cells lacking Rtg2p; in the latter, wild-type Rtg3p is hyperphosphorylated. Because the gene encoding the deletion mutant was transplaced into the chromosomal copy of RTG3, there was no functional form of Rtg3p present in those cells. Although the level of expression of genes such as CIT2 and DLD3 is much lower in ρ+ than in ρo cells (Liao and Butow, 1993; Chelstowska and Butow, 1995; Chelstowska et al., 1999), that (low) level of expression is nevertheless dependent on the RTG genes. We reasoned, therefore, that the loss of expression of one or more RTG-dependent genes in ρ+ cells attributable to the absence of the functional form of Rtg3p might itself trigger a stress or feedback response, similar to the ρo state, initiating nuclear translocation of Rtg3p; for Rtg3pΔ280–298-GFP, which can bind Rtg1p but cannot translocate to the nucleus, this response might be manifest as an unphosphorylated form of the protein. This notion was supported by the finding that when a wild-type form of Rtg3p was coexpressed from a plasmid in cells expressing Rtg3pΔ280–298 from a chromosomal gene transplaced into RTG3 locus, the deletion mutant protein became phosphorylated. In addition to suggesting a feedback mechanism for Rtg3p subcellular localization, these experiments suggest that both phosphorylation and dephosphorylation of Rtg3p are cytoplasmic activities. They do not, however, exclude the possibility that nuclear dephosphorylation and phosphorylation activities may also function in nuclear import and export of Rtg3p associated, respectively, with activation or repression of the retrograde response.
Dual Function of Rtg1p
Two complementary lines of evidence suggest that when RTG-dependent gene expression is low, as in ρ+ cells, Rtg1p functions as a negative regulator by sequestering Rtg3p in the cytoplasm. First, simply deleting the RTG1 gene resulted in nuclear accumulation and dephosphorylation of Rtg3p, effects associated with activation of the retrograde response. Second, deletion of two domains of Rtg3p that, by analogy with other bHLH-Zip homo- or heterodimeric interactions, should be important interfaces for interaction with Rtg1p—the Zip domain (Rtg3Δ345–486-GFP) and the loop-helix 2 domain (Rtg3Δ314–344-GFP)—gave the same result as observed with full-length Rtg3p-GFP in a rtg1Δ background, namely, unphosphorylated proteins that are constitutively localized in the nucleus. Indeed, immunoprecipitation experiments showed that these Rtg3p deletion mutants do not interact with Rtg1p. Because Rtg1p also functions as a positive effector, being required together with Rtg3p for transcriptional activation of target gene expression (Jia et al., 1997; Rothermel et al., 1997), Rtg1p would thus have a novel dual activity in the regulation of the retrograde response. Rtg1p alone has no ability to activate gene expression when bound to a promoter as a fusion protein with a heterologous DNA binding domain (Rothermel et al., 1997). Rtg3p, by contrast, which cannot bind to an R box in the absence of Rtg1p (Jia et al., 1997), is a robust transactivator of gene expression in cells lacking Rtg1p when expressed as a fusion protein containing a heterologous DNA binding domain (Rothermel et al., 1997). We speculated that the role of Rtg1p in transcriptional activation is as an accessory protein to Rtg3p, whereby the heterodimer allows for binding and correct positioning of Rtg3p at R box target sites (Jia et al., 1997).
Transcription factors and other regulatory proteins may be sequestered in the cytoplasm by a variety of mechanisms, including interaction with other proteins (retention factors) that prevent nuclear translocation until the appropriate signals effect their release. In ρ+ cells, Rtg1p would function like the cytoplasmic retention factors IκB and Cactus, two negative regulatory proteins that down-regulate gene expression by sequestering the Rel homology transcription factors nuclear factor-κB and Dorsal, respectively, in the cytoplasm (Sen and Baltimore, 1986; Baeuerle and Baltimore, 1988; Roth et al., 1991; Geisler et al., 1992; Kidd, 1992). Under appropriate signals, IκB and Cactus are targeted for degradation by their phosphorylation, allowing access of the transcription factors to the nuclear import machinery (Brown et al., 1995; Reach et al., 1996).
An alternative explanation for the finding that interaction with Rtg1p is necessary for retention of Rtg3p in the cytoplasm in ρ+ cells is activation of the feedback response caused by the absence of RTG-dependent gene expression, suggested from the finding that the constitutively cytoplasmic deletion mutant Rtg3pΔ280–298 is phosphorylated as long as a functional Rtg3p is coexpressed. In that scenario, Rtg1p would not be functioning in ρ+ cells strictly as a cytoplasmic anchor for Rtg3p, but rather, the loss of RTG-dependent gene expression caused by the absence of Rtg1p initiates the feedback dephosphorylation and nuclear import of Rtg3p. However, this interpretation is not easily reconciled with the findings that Rtg3p is nuclear and dephosphorylated in the rtg1Δ rtg2Δ double mutant, whereas in the rtg2Δ single mutant, in which RTG-dependent gene expression is also blocked, Rtg3p is cytoplasmic and hyperphosphorylated. Moreover, we have observed that Rtg3pΔ280–298-GFP is substantially phosphorylated in ρ+ rtg2Δ cells (our unpublished results), suggesting that the dephosphorylation attributable to the feedback response cannot completely overcome the absence of Rtg2p. The feedback response could effect a destabilization of this tethering complex via a change in the phosphorylation state of Rtg3p. Attempts to override the constitutive cytoplasmic localization of the NLS deletion protein Rtg3pΔ280–298-GFP by introducing ectopic NLS sequences within the C- or N-terminal regions of the protein were unsuccessful, suggesting that the mechanism of retention of Rtg3p in the cytoplasm by Rtg1p binding may not be by simple occlusion of the Rtg3p NLS. Further experiments will be required to explore this possibility in greater detail.
Function of Rtg2p
Previous genetic data indicated that Rtg2p acts upstream of Rtg1p and Rtg3p (Rothermel et al., 1997). The current data support and extend those findings and suggest that Rtg2p may act as a proximal sensor of mitochondrial dysfunction by promoting the dephosphorylation and nuclear accumulation of Rtg3p when the retrograde response is activated in ρo cells. Although the biochemical function of Rtg2p remains to be established, its absence results in a hyperphosphorylated, constitutively cytoplasmic form of Rtg3p. Because the requirement for Rtg2p in the dephosphorylation and nuclear localization of Rtg3p can be bypassed in rtg1Δ rtg2Δ mutant cells, one plausible mechanism for the action of Rtg2p would be to effect the dissociation of a cytoplasmic Rtg1p–Rtg3p complex, allowing accessibility of Rtg3p to a phosphatase activity or preventing accessibility to a kinase. The effects of Rtg2p on the Rtg1p–Rtg3p complex may be indirect, however, because neither yeast two-hybrid (Rothermel et al., 1997) nor coimmunoprecipitation experiments hint at any interaction between Rtg2p and the Rtg1p–Rtg3p complex.
ACKNOWLEDGMENTS
We are grateful to Zhengchang Liu and other members of the Butow laboratory for many helpful discussions and also for sharing many of the reagents and strains used in these studies. This work was supported by grants from the National Institutes of Health (GM-22525) and from The Robert A. Welch Foundation (I-0642).
REFERENCES
- Aronheim A, Shiran R, Rosen A, Walker MD. The E2A gene product contains two separable and functionally distinct transcription activation domains. Proc Natl Acad Sci USA. 1993;90:8063–8067. doi: 10.1073/pnas.90.17.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeuerle PA, Baltimore D. I kappa B: a specific inhibitor of the NF-kappa B transcription factor. Science. 1988;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- Biswas G, Adebanjo OA, Freedman BD, Anandatheerthavarada HK, Vijayasarathy C, Zaidi M, Kotlikoff M, Avadhani NG. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 1999;18:522–533. doi: 10.1093/emboj/18.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin and hsp70 heat shock proteins. Proc Natl Acad Sci USA. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of I kappa B-alpha proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- Chelstowska A, Butow RA. RTG genes in yeast that function in communication between mitochondria and the nucleus are also required for expression of genes encoding peroxisomal proteins. J Biol Chem. 1995;270:18141–18146. doi: 10.1074/jbc.270.30.18141. [DOI] [PubMed] [Google Scholar]
- Chelstowska A, Liu Z, Jia Y, Amberg D, Butow RA. Signaling between mitochondria and the nucleus regulates the expression of a new d-lactate dehydrogenase activity in yeast. Yeast. 1999;15:1377–1391. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1377::AID-YEA473>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- De Vit MJ, Johnston M. Regulated export of the S. cerevisiae glucose repressor Mig1 requires its phosphorylation and the exportin Msn5. Curr Biol. 1999;9:1231–1241. doi: 10.1016/s0960-9822(99)80503-x. [DOI] [PubMed] [Google Scholar]
- De Vit MJ, Waddle JA, Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger T. Getting a grip on DNA recognition: structures of the basic region leucine zipper, and the basic region helix-loop-helix DNA-binding domains. Curr Opin Struct Biol. 1994;4:12–21. [Google Scholar]
- Ellenberger T, Fass D, Arnaud M, Harrison SC. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994;8:970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR, Prendergast GC, Ziff EB, Burley SK. Recognition by MAX of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals (see comments) Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Geisler R, Bergmann A, Hiromi Y, Nusslein-Volhard C. cactus, a gene involved in dorsoventral pattern formation of Drosophila, is related to the I kappa B gene family of vertebrates. Cell. 1992;71:613–621. doi: 10.1016/0092-8674(92)90595-4. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Grant PA, Sterner DE, Duggan LJ, Workman JL, Berger SL. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 1998;8:193–197. doi: 10.1016/s0962-8924(98)01263-x. [DOI] [PubMed] [Google Scholar]
- Hopper AK. Nucleocytoplasmic transport: inside out regulation. Curr Biol. 1999;9:R803–R806. doi: 10.1016/s0960-9822(99)80494-1. [DOI] [PubMed] [Google Scholar]
- Jans DA, Hubner S. Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol Rev. 1996;76:651–685. doi: 10.1152/physrev.1996.76.3.651. [DOI] [PubMed] [Google Scholar]
- Jia Y, Rothermel B, Thornton J, Butow RA. A basic helix-loop-helix zipper transcription complex functions in a signaling pathway from mitochondria to the nucleus. Mol Cell Biol. 1997;17:1110–1117. doi: 10.1128/mcb.17.3.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A, Herskowitz I, Tjian R, O'Shea EK. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- Kaffman A, Rank NM, O'Neill EM, Huang LS, O'Shea EK. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 1998a;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- Kaffman A, Rank NM, O'Shea EK. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 1998b;12:2673–2683. doi: 10.1101/gad.12.17.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- Kidd S. Characterization of the Drosophila cactus locus and analysis of interactions between cactus and dorsal proteins. Cell. 1992;71:623–635. doi: 10.1016/0092-8674(92)90596-5. [DOI] [PubMed] [Google Scholar]
- Komeili A, O'Shea EK. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Yeast protein controlling inter-organelle communication is related to bacterial phosphatases containing the Hsp70-type ATP-binding domain. Trends Biochem Sci. 1994;19:156–157. doi: 10.1016/0968-0004(94)90275-5. [DOI] [PubMed] [Google Scholar]
- Kos W, Kal AJ, Vanwilpe S, Tabak HF. Expression of genes encoding peroxisomal proteins in Saccharomyces cerevisiae is regulated by different circuits of transcriptional control. Biochem Biophys Acta. 1995;1264:79–86. doi: 10.1016/0167-4781(95)00127-3. [DOI] [PubMed] [Google Scholar]
- Liao X, Butow RA. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- Liao X, Small WC, Srere PA, Butow RA. Intramitochondrial functions regulate nonmitochondrial citrate synthase (CIT2) expression in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:38–46. doi: 10.1128/mcb.11.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Butow RA. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol Cell Biol. 1999;19:6720–6728. doi: 10.1128/mcb.19.10.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari ME, Grant PA, Pray-Grant MG, Berger SL, Workman JL, Murre C. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol Cell. 1999;4:63–73. doi: 10.1016/s1097-2765(00)80188-4. [DOI] [PubMed] [Google Scholar]
- Massari ME, Jennings PA, Murre C. The AD1 transactivation domain of E2A contains a highly conserved helix which is required for its activity in both Saccharomyces cerevisiae and mammalian cells. Mol Cell Biol. 1996;16:121–129. doi: 10.1128/mcb.16.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Perlman PS, Butow RA. The sorting of mitochondrial DNA and mitochondrial proteins in zygotes: preferential transmission of mitochondrial DNA to the medial bud. J Cell Biol. 1998;142:613–623. doi: 10.1083/jcb.142.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y. The phosphatase system in Saccharomyces cerevisiae. Genes Genet Syst. 1997;72:323–334. doi: 10.1266/ggs.72.323. [DOI] [PubMed] [Google Scholar]
- Parikh VS, Morgan MM, Scott R, Clements LS, Butow RA. The mitochondrial genotype can influence nuclear gene expression in yeast. Science. 1987;235:576–580. doi: 10.1126/science.3027892. [DOI] [PubMed] [Google Scholar]
- Poyton RO, McEwen JE. Crosstalk between nuclear and mitochondrial genomes. Annu Rev Biochem. 1996;65:563–607. doi: 10.1146/annurev.bi.65.070196.003023. [DOI] [PubMed] [Google Scholar]
- Quong MW, Massari ME, Zwart R, Murre C. A new transcriptional-activation motif restricted to a class of helix-loop-helix proteins is functionally conserved in both yeast and mammalian cells. Mol Cell Biol. 1993;13:792–800. doi: 10.1128/mcb.13.2.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reach M, Galindo RL, Towb P, Allen JL, Karin M, Wasserman SA. A gradient of cactus protein degradation establishes dorsoventral polarity in the Drosophila embryo. Dev Biol. 1996;180:353–364. doi: 10.1006/dbio.1996.0308. [DOI] [PubMed] [Google Scholar]
- Roth S, Hiromi Y, Godt D, Nusslein-Volhard C. cactus, a maternal gene required for proper formation for the dorsoventral pattern in the Drosophila embryo. Development. 1991;112:371–388. doi: 10.1242/dev.112.2.371. [DOI] [PubMed] [Google Scholar]
- Rothermel B, Thornton J, Butow RA. Rtg3p, a basic helix-loop-helix/leucine zipper protein functions in mitochondrial-induced changes in gene expression, contains independent activation domains. J Biol Chem. 1997;272:19801–19807. doi: 10.1074/jbc.272.32.19801. [DOI] [PubMed] [Google Scholar]
- Rothermel BA, Shyjan AW, Etheredge JL, Butow RA. Transactivation by Rtg1p, a basic helix-loop-helix protein that functions in communication between mitochondria and the nucleus in yeast. J Biol Chem. 1995;49:29476–29482. doi: 10.1074/jbc.270.49.29476. [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from S. cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R, Baltimore D. Inducibility of κ immunoglobulin enhancer-binding protein NF-κB by a posttranslational mechanism. Cell. 1986;47:921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- Shyjan AW, Butow RA. Intracellular dialogue. Curr Biol. 1993;3:398–400. doi: 10.1016/0960-9822(93)90212-7. [DOI] [PubMed] [Google Scholar]
- Small WC, Brodeur RD, Sandor A, Fedorova N, Li G, Butow RA, Srere PA. Enzymatic and metabolic studies on retrograde regulation mutants in yeast. Biochemistry. 1995;16:5569–5576. doi: 10.1021/bi00016a031. [DOI] [PubMed] [Google Scholar]