Abstract
An activation tagging screen in which the cauliflower mosaic virus 35S enhancer was inserted randomly into an Arabidopsis genome homozygous for the floral homeotic mutation apetala2-1 (ap2-1) resulted in a line (28-5) with extraordinarily wide, heart-shaped ovaries. The ovary of the 28-5 ap2-1 mutant shows an oval shape because of increased numbers of enlarged cells. When the ap2-1 mutation is crossed out of the genetic background, more elongated rather than wider fruits are obtained. Normally, Arabidopsis fruits will develop to a normal size only when the ovules are present and fertilized. In the 28-5 single mutant, the siliques keep growing despite failure of fertilization and can reach nearly normal size. When wild-type pollen was used to pollinate the mutant pistil, the pollinated 28-5 silique became >10% longer and 40% wider than a wild-type silique, although producing very few seeds. The enhancer insertion in line 28-5 acts by hyperactivating a cytochrome P450 gene, CYP78A9. The pistil of 28-5 ap2-1 mutant flowers shows a structure similar to that of Capsella bursa-pastoris, a distant mustard relative of Arabidopsis, suggesting that the processes regulated by the CYP78A9-encoded protein may be involved in evolutionary control of carpel shape.
INTRODUCTION
Many fruit crops have fruits that develop only when ovules are present and fertilized. Pollination and fertilization increase the amounts of phytohormones in the ovary, stimulate cell division, and lead to fruit set and growth (Gillaspy et al., 1993). To produce seedless fruits, plant hormones such as auxin analogs (in the case of strawberries) or gibberellins (in cases such as apples, cucumbers, and eggplant), mutants capable of parthenocarpic development, or plants altered in their ploidy are typically used (Bukovac and Nakagawa, 1967; Robinson et al., 1971; Mapelli et al., 1978; Mazzucato et al., 1998). Other possible methods for generating seedless fruits require inducing an enzyme in a plant hormone biosynthetic pathway. The iaaM gene, driven by an ovule-specific promoter, induces seedless fruits, but only when flowers are emasculated (Rotino et al., 1997).
In Arabidopsis, recent studies have identified numerous mutants that display abnormal gynoecium and fruit development (reviewed in Bowman et al., 1999). Several parthenocarpic mutants have been isolated and analyzed intensively (Ohad et al., 1996, 1999; Chaudhury et al., 1997; Luo et al., 1999). However, only a few parthenocarpic plants suitable as crop cultivars are available (Mazzucato et al., 1998, 1999).
Fruit-bearing domesticated plants have larger fruits than do their wild-type progenitors (Alpert and Tanksley, 1996). In many fruit crops, large fruit traits have been selected over the many years since the beginning of cultivation. However, the genes controlling crop fruit size and shape have not been cloned so far. In Arabidopsis, mutations in the MADS-box gene FRUITFULL result in inappropriate maturation of the cells of the valves, leading to short fruits (Gu et al., 1998). However, ectopic expression experiments in which FUL is constitutively expressed suggest that the FUL function is to specify valve identity, not to control cellular elongation (Liljegren et al., 1998).
Here, we report the discovery of a gene for which overexpression changes the shape and size of the fruit of the mustard Arabidopsis. The large fruit phenotype results from overexpression of a cytochrome P450 gene. The large group of cytochrome P450 enzymes are involved in oxidation reactions (Nebert and Gonzalez, 1987). The >400 plant P450 genes sequenced so far have been divided into 85 subfamilies, called CYP (Nelson, 1999). The cytochrome P450 cDNA overexpressed in the mutant line of Arabidopsis encodes a protein most similar to those encoded by members of the CYP78A subfamily and has been designated CYP78A9. Although the function of the proteins encoded by the CYP78A subfamily is not known, other CYP78A genes have been isolated as coding for floral- or meristem-specific transcripts. CYP78A2 from Phalaenopsis and maize CYP78A1 are expressed specifically in the pollen tube and tassel primordia, respectively (Larkin, 1994; Nadeau et al., 1996). CYP78A5 from Arabidopsis is expressed in the peripheral regions of the vegetative and reproductive shoot meristems (Zondlo and Irish, 1999). We show that the gene CYP78A9 is expressed in floral organs. In the wild-type Arabidopsis flower, a relatively high expression of CYP78A9 RNA was observed in the funiculus of developing ovules. In the mutant line, the gene is highly expressed in the carpel wall, especially in the inner side. The strong effects of the overexpression of CYP78A9 as well as its specific expression patterns suggest a role for the CYP78A9 gene in fruit development.
RESULTS
Characterization of the Arabidopsis Activation Tagging Line 28-5 apetala2-1
To screen for genes involved in cellular differentiation and growth of floral organs, we initiated a T-DNA activation tagging screen in an apetala2-1 (ap2-1) homozygous genetic background (Weigel et al., 2000). A mutant with a pistil wider than normal was identified from T1 plants and named 28-5 ap2-1. As shown in Figure 1, the pistil of 28-5 ap2-1 flowers was three to five times wider in the lateral direction than that of the ap2-1 mutant. The enlarged region of the pistil was empty, and most of the ovules were underdeveloped (Figure 1C). A medial view showed that the pistil was flat (Figure 1D). After anthesis, silique lengths of 28-5 ap2-1 and ap2-1 mutants were almost identical (Figures 1E and 1J). In contrast, petals and stamens of 28-5 ap2-1 flowers were much shorter than those of ap2-1 (Figures 1A, 1B, and 1D). Because of the short stamens and reduced pollen production (see below), no self-pollination was observed in 28-5 ap2-1 flowers. Fully elongated 28-5 ap2-1 mutant siliques were as much as 1.9-fold longer and 3.1-fold wider than those of emasculated ap2-1 flowers (Table 1). Epidermal cells of 28-5 ap2-1 carpel valves were 20 to 30% wider than those of the ap2-1 mutant (Figures 1E to 1N). The cells of 28-5 ap2-1 carpel valves were distorted in comparison with those of ap2-1 (Figures 1G and 1L, 1H and 1M, 1I and 1N). The distortion of the cell shape was more severe near the replum than at the edge of the valves. Near the replum, some of the valve cells were abnormally wide (Figure 1L). The number of cells in 28-5 ap2-1 carpel valves was greatly increased over that in ap2-1 homozygotes (Figures 1F and 1K). Thus, the abnormal enlargement of 28-5 ap2-1 pistils seems to result from both increased cell number and larger cell size. In addition to the flower phenotype, 28-5 ap2-1 plants showed dark green leaves, stout stems, and late flowering (data not shown).
Figure 1.
Phenotypes of ap2-1 and 28-5 ap2-1 Mutant Flowers.
(A) ap2-1 mutant flower.
(B) and (C) 28-5 ap2-1 mutant flowers. The pistils of 28-5 ap2-1 mutant flowers are three to five times wider laterally than those of ap2-1 (B), and the enlarged region is empty (C).
(D) Scanning electron micrograph (SEM) of a 28-5 ap2-1 flower viewed from above.
(E) to (I) SEM of ap2-1 pistil.
(J) to (N) SEM of 28-5 ap2-1 pistil.
(E) and (J) View of stage 16 pistil, showing lateral expansion of the mutant ovary. (F) and (K) Close-up view toward the top of the pistils. (G) and (L) Close-up view near the centers of the pistils. Re, replum; Va, valve. (H) and (M) Close-up view of the edges of the pistils. The carpel cells of 28-5 ap2-1 are distorted and larger than those of ap2-1. (I) and (N) Higher magnification of the cells at the edge of (H) and (M), respectively.  ; b
; b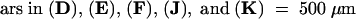 ;
;  ;
;  .
.
Table 1.
Dimensions of Fully Elongated Siliques of Emasculated ap2-1, 28-5 ap2-1 Mutant, Emasculated Wild Type, 28-5 Single Mutant, Self-Pollinated Wild Type, and Pollinated 28-5 Single Mutant with Wild-Type Pollen or 28-5 Pollen
| Genotype | Length (mm) | Width (mm) | Length/Width |
|---|---|---|---|
| Emasculated ap2-1 (n = 10)a |
2.7 ± 0.6b | 0.7 ± 0.1 | 3.9 |
| 28-5 ap2-1 (n = 31) | 5.2 ± 0.5 | 2.2 ± 0.2 | 2.4 |
| Emasculated wild type (n = 10) | 3.1 ± 0.2 | 0.6 ± 0.1 | 5.2 |
| 28-5 single (n = 45) | 7.8 ± 1.2 | 1.0 ± 0.1 | 7.8 |
| Wild type (self- pollinated) (n = 16) | 12.0 ± 1.0 | 1.0 ± 0.0 | 12.0 |
| Pollinated 28-5 single with wild-type pollen (n = 16) | 14.0 ± 0.3 | 1.8 ± 0.4 | 7.7 |
| Pollinated 28-5 single with 28-5 pollen (n = 8) |
9.8 ± 0.2 | 1.7 ± 0.1 | 5.8 |
n is the number of siliques tested.
Values are means with standard deviations.
The original mutant line 28-5 ap2-1 was almost sterile. Almost all ovules were shriveled, and the dehiscence of 28-5 ap2-1 anthers was much delayed (Figures 1C and 1D). In the flowers produced later in development, some pollen was produced; however, this pollen was unable to fertilize wild-type ovules (data not shown). Our initial attempts to get seeds by crossing wild-type pollen to the 28-5 ap2-1 mutant failed. Therefore, we maintained the original 28-5 ap2-1 line in tissue culture.
The 28-5 Single Mutant Exhibited Elongated Carpels in the Absence of Pollination
Using tissue culture methods, we propagated many 28-5 ap2-1 mutant plants. All of the plants exhibited the mutant phenotype, and we ultimately obtained a few seeds by extensive application of wild-type pollen to 28-5 ap2-1 mutant pistils. As shown in Figure 2, when the ap2-1 mutation was crossed out of the genetic background, more elongated rather than wider fruits were obtained. In 28-5 single mutant flowers, short stamens and reduced fertility were also observed. The elongation of petals was only delayed, however, and they finally reached the same size as those of the wild type. The length of the sepals and petals of the 28-5 flowers at anthesis was almost identical to that of the wild type (Figure 2A). In contrast, stamens of 28-5 flowers were ∼50% shorter than those of the wild type (Figures 2B and 2E). At anthesis, the 28-5 mutant pistil was ∼15% longer and twice as wide as that of the wild type (Figure 2B). The silique of the 28-5 mutant continued to elongate after anthesis, despite the absence of mature ovules and the failure of fertilization (Figures 2C and 2D). Fully elongated 28-5 single mutant siliques were as much as 2.5-fold longer and 1.7-fold wider than those of emasculated wild-type flowers (Figure 2C and Table 1). The average length of wild-type emasculated siliques was ∼3.1 mm. In contrast, the average length of the 28-5 single mutant siliques was 7.8 mm, and the largest 28-5 mutant siliques produced in older plants reached 10 mm (Figure 2D and Table 1). By scanning electron microscopy, most of the ovules of 28-5 flowers were observed to be shriveled, with no apparent embryo sac (Figures 2E to 2H and data not shown). A few ovules showed the normal oval shape (Figure 2H). Applying wild-type pollen to 28-5 mutant pistils produced a few fertilized seeds.
Figure 2.
Phenotypes of Wild-Type and 28-5 Single Mutant Flowers.
(A) and (B) Wild-type (Landsberg erecta) (left) and 28-5 single mutant flowers (right). The length of the sepals and petals of 28-5 flowers did not differ from those of the wild type. However, stamens of 28-5 mutants were ∼50% shorter than those of the wild type. The 28-5 single mutant pistils were longer and wider than those of the wild type.
(C) Wild-type silique ∼14 days after emasculation (left), 28-5 single mutant siliques ∼3 days (middle), and 5 days (right) after anthesis. The siliques of 28-5 mutants continued to elongate without fertilization. Stigmatic papillae of elongating 28-5 siliques were still intact.
(D) Unpollinated dried 28-5 single mutant silique. The silique of the 28-5 single mutant showed a parthenocarpic phenotype. One carpel was removed to view the inside (right). No seeds were produced.
(E) SEM of 28-5 single mutant pistil. Part of one carpel was removed to view the ovules inside.
(F) to (H) Close-up view of the ovules. Most of the ovules were shriveled around the region where the embryo sac would be in the wild type (G). However, a few ovules showed normal morphology (H).
(I) Self-pollinated wild-type silique (left) and pollinated 28-5 single mutant silique with wild-type pollen (middle). Pollinated 28-5 silique elongated to as much as 18 mm in one extreme case (right).
 ;
;  ;
;  ;
;  .
.
The 28-5 Single Mutant Exhibited Enlarged Siliques When Pollinated
When wild-type pollen was applied to 28-5 single mutant pistils, the siliques became further enlarged. The pollinated 28-5 siliques were 10 to 20% longer on average than those of self-pollinated wild type (Table 1). The average length of wild-type pollinated siliques was ∼12 mm (left side of Figure 2I and Table 1), whereas the average length of pollinated 28-5 single mutant siliques was 14 mm (middle of Figure 2I and Table 1). Pollinated 28-5 siliques as long as 18 mm were found in an extreme case (right side of Figure 2I). The pollinated 28-5 siliques were also 40 to 80% wider than those of the wild type (Figure 2I and Table 1). The enlarged siliques were more frequently observed on the latest-arising flowers of old plants. In general, the enlarged 28-5 siliques contained very few seeds, ranging from two or three to ∼30.
Anthers of 28-5 flowers showed delayed dehiscence, with reduced pollen production. In addition, the stamens were very short, which resulted in lack of self-pollination. After hand pollination with 28-5 pollen, the 28-5 siliques elongated. The average length of the 28-5 siliques pollinated with 28-5 pollen was 9.8 mm (Table 1), but these siliques contained no seed. The 28-5 pollen seemed to function to stimulate carpel elongation without resulting in fertilization.
Molecular Cloning of the 28-5 Gene, CYP78A9
Because the 28-5 mutant was male sterile and exhibited reduced female fertility, we maintained the line by applying wild-type or ap2-1 pollen to 28-5 mutant pistils. In the T2 generation, all of the lines with the 28-5 mutant phenotype contained T-DNA insertions, whereas the plants that appeared wild type did not (data not shown), suggesting that T-DNA was linked with the mutation. Genomic DNA gel blot analysis using the 35S enhancer region as a probe showed that the 28-5 mutant genome contained only one T-DNA copy. We cloned the flanking regions of the T-DNA by plasmid rescue. Figure 3A shows the map of the T-DNA insertion site in the 28-5 line and the rescued clones. Using the EcoRI-rescued clone as a probe, a genomic DNA gel blot was performed on wild-type and 28-5 mutant genomic DNA digested with EcoRI. In the wild-type DNA lane, a single band (a 3.1-kb EcoRI fragment) was observed. In the 28-5 mutant DNA lane, a 6.1-kb EcoRI fragment was observed in addition to the wild-type band (3.1-kb EcoRI fragment, Figure 3B), indicating that 28-5 was heterozygous for the T-DNA insertion and therefore that the 28-5 mutation functioned in a dominant manner. The locus was mapped to chromosome 3 at ∼83 centimorgans by hybridization to IGF bacterial artificial chromosome (BAC) library filters.
Figure 3.
Physical Map of the 28-5 Locus and Overexpression of 28-5 Gene 1 in the 28-5 Mutant.
(A) Schematic map of the T-DNA activation-tagged 28-5 gene. The regions of the T-DNA activation tagging vector pSKI015 (http://biosun.salk.edu/LABS/pbio-w/) are shown: Bastar, bar gene; pB KS, pBluescript KS+; triangles, cauliflower mosaic virus 35S enhancers. Recognition sites for restriction endonucleases BamHI (B), EcoRI (E), KpnI (K), and XhoI (X) are shown on the map. Rescued plasmids, designated by lines, are shown below. 28-5 genomic DNA was digested with EcoRI, XhoI, and KpnI to clone the sequences adjacent to the right border and with BamHI and SpeI to clone the sequences adjacent to the left border. The right border clones revealed two genes at that locus (shown in schematic). 28-5 gene 1 was identified ∼2 kb from the right border of the T-DNA insertion site. 28-5 gene 2, for which transcription starts in the opposite orientation to gene 1, was found 8 kb from the right border.
(B) DNA gel blot genomic DNA hybridization analysis of the wild type (Wt) and the 28-5 mutant. Genomic DNA was digested with EcoRI, separated by electrophoresis, and blotted. The blot was probed with the EcoRI-rescued plasmid. Arrows indicate DNA size markers in kilobases.
(C) RNA gel blot hybridization analysis of wild-type and 28-5 mutant callus tissue. Ten micrograms of total RNA was hybridized with the EcoRI fragment shown in (A) as a probe. The result indicated that the 28-5 gene 1 was overexpressed in the 28-5 callus and that the transcribed RNA was 1.9 kb long.
One gene (28-5 gene 1) was found ∼2 kb from the right border of the T-DNA insertion site (Figure 3A, bottom). By RNA gel blot analysis of RNA isolated from the 28-5 ap2-1 mutant and from wild-type calli, this gene was found to be overexpressed in 28-5 ap2-1 mutant callus (Figure 3C). The same result was observed by reverse transcription–polymerase chain reaction (RT-PCR) with RNA isolated from 28-5 single mutant and wild-type inflorescences, as shown in Figure 4B. Downstream of 28-5 gene 1 is the 3′ region of another gene (28-5 gene 2), the transcription of which starts ∼8 kb from the T-DNA right border (Figure 3A). RNA gel blot analysis showed the expression of the gene 2 in 28-5 ap2-1 mutant callus to be slightly more than that of the wild type (data not shown). No gene flanking the T-DNA left border was found by RNA gel blot analysis to be overexpressed in 28-5, as determined by using a left border flanking region of ∼7 kb as a probe (data not shown).
Figure 4.
Overexpression of 28-5 Gene 1 Reproduced the 28-5 Mutant Phenotypes.
(A) Diagram of the transformation construct. The KpnI fragment, containing four tandemly arrayed cauliflower mosaic virus 35S enhancer and the 28-5 gene 1 (CYP78A9), was cloned into Agrobacterium binary vector pPZP211, and the construct was transformed into ap2-1 or wild-type plants by Agrobacterium-mediated transformation. Bastar, bar gene; LB, left border; RB, right border; pB KS, pBluescript KS+.
(B) Amplification by RT-PCR of 28-5 gene 1 cDNA with primers 28-5-1F and 28-5-1R. Total RNA was isolated from inflorescences of wild-type plants (Wt), 28-5 single mutants (28-5), a transgenic plant showing wild-type phenotypes (T.G.−), and a transgenic plant showing strong mutant phenotypes (T.G.+). The arrow indicates the 580-bp PCR product amplified from 28-5 gene 1. As a control, ubiquitin extension protein (UBQ5) (Callis et al., 1990) was amplified.
(C) Transgenic plant in the wild-type background. A pistil continued to elongate without fertilization. Stamens were very short and were not visible before removal of the outer floral organs.
(D) Silique of a transgenic plant in the wild-type background showing parthenocarpy. One carpel was removed to view the ovules inside.
(E) Flower of a transgenic plant in the ap2-1 mutant background. A very wide pistil was observed. Some sepals and petals were removed to show the stamens, which were very short.
(F) ap2-1 flower at anthesis.
(G) Capsella bursa-pastoris flower at anthesis.
(H) C. bursa-pastoris fruit.
 .
.
To demonstrate that the 28-5 mutant phenotype was a consequence of overexpression of 28-5 gene 1, a DNA fragment containing 35S enhancers and the entire gene 1 coding region (a KpnI fragment) was cloned into the binary vector pPZP211 and transformed into wild-type and ap2-1 mutant Arabidopsis plants by Agrobacterium-mediated transformation (Figure 4A). The construct contained neither the 28-5 gene 2 coding region nor the region flanking the T-DNA left border in the original 28-5 line. Approximately half (78 of 158) of the T1 plants in the wild-type background showed 28-5 mutant phenotypes, such as short stamens and enlarged carpels (Figures 4C and 4D). Of these 78 lines, 34 showed the strong phenotype and 44 lines showed a somewhat milder phenotype. The other 80 lines appeared wild type. Using RT-PCR, we checked the expression levels of the 28-5 gene 1 in inflorescences from wild-type plants, 28-5 single mutants, a transgenic line with the mutant phenotype, and a transgenic line without the mutant phenotype. We confirmed that the mutant phenotypic severity was correlated with the relative amounts of expression of 28-5 gene 1 (Figure 4B). The expression level of 28-5 gene 1 in plants with a wild-type phenotype was almost identical to that of the wild-type control. In contrast, the expression level in the transgenic line with a strong mutant phenotype was almost the same as that in the 28-5 single mutant.
When we transformed ap2-1 mutant plants with the construct, five of 18 transgenic plants reproduced the mutant phenotypes of short stamens and a very wide flat pistil (Figure 4E). In addition, these lines were male-sterile and showed reduced female fertility, as had been seen in the original 28-5 ap2-1 mutant. Thus, the 28-5 mutant phenotype was induced by overexpression of the 28-5 gene 1. Moreover, the difference in silique shape between the 28-5 ap2-1 mutant and 28-5 single mutant is presumably attributable to the altered AP2 activity.
28-5 gene 1 spans ∼3 kb of genomic DNA and consists of three exons with consensus splice sites at the exon–intron boundaries (Figure 3A). The predicted 28-5 protein has 534 amino acids and a calculated molecular mass of 60.0 kD. The deduced amino acid sequence of this protein is similar to the conserved anchor region, the proline-rich hinge region, and the oxygen and heme binding domains of microsomal cytochrome P450, as defined by Nebert and Gonzalez (1987). Therefore, the 28-5 protein appears to include all of the functionally important domains of a P450 monooxygenase (Pan et al., 1995). Accordingly, the 28-5 protein was designated CYP78A9 by the P450 Nomenclature Committee (GenBank accession number AB036059). CYP78A9 exhibits the greatest similarity to the CYP78A3 protein of Glycine max (66% amino acid identity; GenBank AF022463) and strong similarity to Pinus radiata CYP78A4 (54% identity; GenBank AF049067), Arabidopsis CYP78A5 (54% identity [Zondlo and Irish, 1999]), Phalaenopsis sp CYP78A2 (54% identity [Nadeau et al., 1996]), and Zea mays CYP78A1 (48% identity [Larkin, 1994]). In addition, the Arabidopsis genome sequencing project has uncovered additional CYP78A subfamily members highly similar to CYP78A9: CYP78A6 (81% identity; GenBank AC005819), CYP78A8 (66% identity; GenBank AC007323), and CYP78A7 (52% identity; GenBank AC016893). CYP78As are all group A cytochrome P450s, which are phylogenetically more closely related to each other than to non–group A cytochrome P450s, and they appear to catalyze plant-specific reactions.
CYP78A9 Expression
To examine the organ specificity of CYP78A9 expression in wild-type Arabidopsis, RT-PCR was performed with total RNA from various tissues with CYP78A9-specific primers. CYP78A9 was expressed only in floral organs, not in vegetative tissue (data not shown). In contrast, the closest Arabidopsis homolog, CYP78A6 (81% amino acid identity with CYP78A9), was ubiquitously expressed, as shown by a RT-PCR experiment (data not shown). To define the expression pattern of the CYP78A9 gene in the floral meristem, in situ hybridization experiments were performed. Figure 5A shows that in the wild-type Arabidopsis flower, the CYP78A9 transcript was not detected in flower buds up to stage 13. At stage 14, CYP78A9 was expressed in funiculi (the stalks of developing ovules) in relatively high amounts (Figure 5B, cross-section; Figure 5C, longitudinal section). In contrast, the gene is ectopically overexpressed in all floral organs of 28-5 ap2-1. The strongest signal was observed in the carpel valves of 28-5 ap2-1, especially on the inner side of the carpels (Figure 5D).
Figure 5.
In Situ Hybridization of CYP78A9 to Arabidopsis Flowers.
(A) Longitudinal section of the flowers at stage 6 and stage 12. In the wild-type Arabidopsis flower, the strong localization of the CYP78A9 transcript was not detected in the floral buds up to stage 13.
(B) Cross-section of the pistil of a stage 14 flower. The CYP78A9 gene was expressed in the funiculi (stalks of the developing ovules) in stage 14 flowers (arrowheads).
(C) Longitudinal section of the pistil of a stage 14 flower. Arrowheads show funiculi.
(D) Longitudinal medial section of the 28-5 ap2-1 mutant pistil. The CYP78A9 RNA is ectopically overexpressed in the carpel valves, especially in the inner side of the carpels.
 .
.
DISCUSSION
Most types of fruit crops have fruits that will develop only when ovules are present and fertilized (Gillaspy et al., 1993). Fruit development is roughly divided into four steps: pollination, fertilization, seed development, and ovary enlargement. Pollination and fertilization increase the concentrations of phytohormones in the ovary, stimulate cell division, and lead to fruit set and growth (Bukovac and Nakagawa, 1967; Rodrigo and García-Martínez, 1998). Positive growth stimuli for inducing fruit development are expected to be produced by pollen or ovules after fertilization (Gillaspy et al., 1993). Some lines of evidence suggest that increased concentrations of auxin and gibberellin in the ovary induce parthenocarpic fruit development (Bukovac and Nakagawa, 1967; Robinson et al., 1971; Rodrigo and García-Martínez, 1998; Rebers et al., 1999). Overexpression of CYP78A9 allows fruit growth independently of fertilization in 28-5 and 28-5 ap2-1 mutant pistils. As shown by a RT-PCR experiment, CYP78A9 is expressed only in floral organs, not in vegetative organs. In situ hybridization experiments show that the CYP78A9 transcript is specifically localized in the funiculus at anthesis. So far, no known plant hormone treatments mimic the entire 28-5 mutant phenotype. Thus, CYP78A9 might be involved in a process that is independent of the actions of known hormones in Arabidopsis. One of the functions of CYP78A9 may be production of a signal that activates or enhances fruit development. That cytochrome P450 proteins are often involved in synthesis or degradation of plant secondary products indicates that the function of the CYP78A9 gene and its relatives might be the production of an undiscovered plant growth substance that can affect cell size and cell division patterns in developing carpels.
Some members of the cytochrome P450 family are known to be involved in important biochemical pathways, such as the biosynthesis of brassinosteroids, flavonoids, and lignin (Holton et al., 1993; Toguri et al., 1993; Holton, 1995; Werck-Reichhart, 1995; Szekeres et al., 1996; Choe et al., 1998; Kim et al., 1998; Mathur et al., 1998; Salchert et al., 1998; de Vetten et al., 1999; Kaltenbach et al., 1999; Neff et al., 1999; Osakabe et al., 1999). Although the function of the CYP78A subfamily is not known, most of the other CYP78A subfamily member genes have been isolated as flower or meristem specific in their expression (Larkin, 1994; Nadeau et al., 1996; Zondlo and Irish, 1999). CYP78A2 from Phalaenopsis and maize CYP78A1 are expressed only in the pollen tube and tassel primordia, respectively (Larkin, 1994; Nadeau et al., 1996). CYP78A5 from Arabidopsis is expressed in the peripheral regions of the vegetative and reproductive shoot meristems (Zondlo and Irish, 1999). Overexpression of CYP78A5 causes twisting and kinking of the stem as well as defects in floral development (Zondlo and Irish, 1999). In contrast, the 28-5 mutant does not show twisting stems; instead, stems of the 28-5 line were rather stout. Similar floral defects, such as short stamens and reduced aborted ovules, were observed in the lines overexpressing CYP78A5 or CYP78A9. Overexpression of these genes might affect the same or related pathways, even though the gene products are not highly similar (51% amino acid identity).
The Arabidopsis genome sequencing project has revealed additional genes that are very similar to CYP78A9. CYP78A6 and CYP78A8 have 81 and 66% amino acid identity with CYP78A9, respectively. We recently isolated an insertional mutant in the CYP78A6 gene. The homozygotes show no mutant phenotype (data not shown). Possibly these gene products have redundant or related functions. Future work to identify insertional mutants in CYP78A9 and CYP78A8 and to generate double and triple mutants will help elucidate the developmental functions of the members of the CYP78A subfamily.
Arabidopsis wild-type fruits are 12-fold longer than they are wide (Table 1). In contrast, the 28-5 ap2-1 mutant fruits are only 2.4-fold longer than wide (Table 1). The architecture of the 28-5 ap2-1 pistil is similar to that of Capsella bursa-pastoris, which is in the same family (the mustard family, Brassicaceae) as Arabidopsis (Figures 1B, 1J, 4G, and 4H). Thus, changes in CYP78A9 expression or activity may be one of the critical factors involved in specification of carpel morphology in the mustard family. Functional analyses of cytochrome P450 (CYP78A9) in Capsella flowers might elucidate the molecular basis for some of the variations in fruit shape that have occurred in the evolution of the mustards.
Moreover, introducing CYP78A9 to crop plants and causing its expression in the tissues of developing fruits might provide a method for controlling fruit shape and size and facilitating parthenogenetic fruit development, as it does in Arabidopsis. Eventually, the elucidation of the biochemical function of the gene product may lead to the discovery of one or more new plant growth substances with use in regulation of fruit development.
METHODS
Generation of Activation-Tagged Lines
Seeds of genotype apetala 2-1 (ap2-1; ecotype Landsberg erecta of Arabidopsis thaliana) (Okamuro et al., 1997) were planted in pots and transformed with the Agrobacterium tumefaciens strain GV3101 pMP90RK, which carries the binary vector pSKI015 (Weigel et al., 2000), by the vacuum-infiltration method (Bechtold et al., 1993). T1 plants were selected by using the herbicide Basta and screened by observing flower phenotype.
Tissue Culture
The original 28-5 mutant was sterile and was maintained by tissue culture by following the procedure described by Akama et al. (1992). Flowers from the 28-5 mutant were excised and placed on plates containing callus-inducing medium for 10 to 20 days. Callus was efficiently propagated from the peduncle of the flowers. The callus was transferred onto plates containing shoot-inducing medium, and by changing the plates containing shoot-inducing medium every week, the regenerated shoots were obtained. The shoots were transferred to plates containing root-inducing medium. After roots were generated, the plants were transplanted into soil.
Cloning and Sequencing the 28-5 Gene
The flanking region of the T-DNA insertion of the 28-5 mutant was isolated by plasmid rescue. Genomic DNA was prepared by using buffer containing 3% hexadecyltrimethylammonium bromide, 100 mM Tris-HCl, pH 8.0, 20 mM EDTA, 1.4 M NaCl, and 0.2% β-mercaptoethanol. Approximately 0.5 μg of the genomic DNA was digested with EcoRI, KpnI, and XhoI to rescue the region flanking the T-DNA right border. To rescue the flanking region of the T-DNA left border, the DNA was digested with BamHI and SpeI. The DNA was self-ligated and transformed into competent cells (Electro-Max DH10B cells; Gibco BRL) by using the Gene Pulser II electroporator (Bio-Rad Laboratories). DNA sequencing was performed by using the ABI PRISM Dye Terminator method (Perkin-Elmer).
RNA Gel Blot Analysis
RNA was prepared by using TRI reagent (Molecular Research Center Inc., Cincinnati, OH). For each sample, 10 μg of total RNA was combined with loading buffer, heated at 65°C for 10 min, and loaded onto an RNA gel (260 mL of water and 30 mL of 10 × MES buffer [1 × MES buffer is 20 mM MOPS, 5 mM sodium acetate, and 1 mM EDTA buffer], 3.6 g of agarose, and 9.0 mL of 37% formaldehyde). After electrophoresis, the RNA was transferred to Biodyne-A membranes (Pall BioSupport, East Hills, NY) and hybridized with each probe.
Retransformation of 28-5 Gene 1 (CYP78A9)
The clone rescued by use of KpnI-digested DNA contained the 35S enhancer and the entire 28-5 gene 1 (shown in Figure 4A). This clone was linearized by digestion with KpnI and ligated into the KpnI site of Agrobacterium binary vector pPZP211 (Hajdukiewicz et al., 1994). The construct was transformed first into the Agrobacterium strain ASE by electroporation and then into wild-type Arabidopsis or ap2-1 mutant plants by the vacuum infiltration method (Bechtold et al., 1993). Seeds collected from the vacuum-infiltrated plants were sterilized and plated onto selection plates containing B5 medium (Sigma Chemical, St. Louis, MO), 0.8% Bacto-agar, and 50 μg/mL kanamycin.
Mapping of 28-5 Gene 1 (CYP78A9)
The chromosomal locus of the 28-5 gene was determined by using a bacterial artificial chromosome (BAC) filter (IGF library) obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus). The 28-5 EcoRI genomic fragment (shown in Figure 3A) was labeled with random primers and used as a probe. The membrane was washed under high-stringency conditions (0.2 × SSC [1 × SSC is 0.15 M NaCl, 0.015 sodium citrate] and 0.1% SDS at 65°C). Positive signals were obtained from the BAC clones 10E6, 7E18, 12G4, 19H10, 9C8, 8P1, 21F14, and 18J21, all of which are located at 83 centimorgans on chromosome 3.
Reverse Transcription–Polymerase Chain Reaction
Approximately 2 μg of total RNA was used for cDNA synthesis using the Thermoscript reverse transcription–polymerase chain reaction (RT-PCR) system (Gibco BRL). The conditions for PCR amplification were as follows: 1 cycle at 94°C for 5 min and then 12, 24, or 28 cycles at 94°C for 40 sec, 58°C for 30 sec, and 72°C for 1 min. The following oligonucleotides were used as primers for RT-PCR: p28-5-1F (5′-CGCGAATCAAATCGTGAAGTGTCTCA-3′) and p28-5-1R (5′-GACTCTTCGACAGCCCTTGATCGT-3′). For use as a positive control, a ubiquitin extension protein (UBQ-5) (Callis et al., 1990) was amplified under the same conditions except for a 52°C annealing temperature with the following two primers: Nubq (5′-GGTGCTAAGAAGAGGAAGAAT-3′) and Cubq (5′-CTCCTTCTTTCTGGTAAACGT-3′).
In Situ Hybridization
In situ hybridization was performed by following the procedure previously described (Sakai et al., 1995). To generate a 28-5 gene 1 (CYP78A9)–specific probe, the 5′ region of the CYP78A9 was amplified by PCR, cloned into pBluescript SK+ (Stratagene), and used as an in vitro transcription template. The following oligonucleotides were used as primers for PCR of genomic DNA: pE1S1 (5′-GTCACGTGCAATCCTGATGTAGCT-3′) and pE2A1 (5′-CGACGACCGTCGATGATCGTG-3′). After hybridization, the samples were treated with 20 μg/mL RNase A for 30 min at 37°C, washed with 5 mM DTT and 0.1 × SSC at 57°C, exposed to NTB-2 emulsion (Kodak) for 4 weeks at 4°C, and developed with D-19 developer (Kodak).
Acknowledgments
We thank Detlef Weigel (The Salk Institute for Biological Studies, La Jolla, CA) for supplying the activation tagging vector pSKI015. We are grateful to Caroline Lim, Marjorie L. James, and Giao Hang for their help with making transgenic plants and their molecular analysis. We also thank Caroline Lim, Carolyn Ohno, Catherine Baker, Chiou-Fen Chuang, Doris Wagner, Eva Ziegelhoffer, Frank Wellmer, Giao Hang, Michael Vishnevetsky, and Venugopala Reddy Gonehal for critical reading of the manuscript. This work was supported by National Institutes of Health Grant No. GM45697 to E.M.M. T.I. was supported by a Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowship for Research Abroad from 1997 through 1999.
References
- Akama, K., Okada, K., and Shimura, Y. (1992). Efficient transformation of Arabidopsis thaliana: Comparison of the efficiencies with various organs, plant ecotypes and Agrobacterium strains. Plant Cell Rep. 12, 7–11. [DOI] [PubMed] [Google Scholar]
- Alpert, K.B., and Tanksley, S.D. (1996). High-resolution mapping and isolation of a yeast artificial chromosome contig containing fw2.2: A major fruit weight quantitative trait locus in tomato. Proc. Natl. Acad. Sci. USA 93, 15503–15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. 316, 1194–1199. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Baum, S.F., Eshed, Y., Putterill, J., and Alvarez, J. (1999). Molecular genetics of gynoecium development in Arabidopsis. Curr. Top. Dev. Biol. 45, 155–205. [DOI] [PubMed] [Google Scholar]
- Bukovac, M.J., and Nakagawa, S. (1967). Comparative potency of gibberellins in inducing parthenocarpic fruit growth in Malus sylvestris Mill. Experientia 23, 865. [DOI] [PubMed] [Google Scholar]
- Callis, J., Raasch, J.A., and Vierstra, R.D. (1990). Ubiquitin extension proteins of Arabidopsis thaliana: Structure, localization, and expression of their promoters in transgenic tobacco. J. Biol. Chem. 265, 12486–12493. [PubMed] [Google Scholar]
- Chaudhury, A.M., Ming, L., Miller, C., Craig, S., Dennis, E.S., and Peacock, W.J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94, 4223–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, S., Dilkes, B.P., Fujioka, S., Takatsuto, S., Sakurai, A., and Feldmann, K.A. (1998). The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10, 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vetten, N., ter Horst, J., van Schaik, H.P., de Boer, A., Mol, J., and Koes, R. (1999). A cytochrome b5 is required for full activity of flavonoid 3′,5′-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc. Natl. Acad. Sci. USA 96, 778–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillaspy, G., Ben-David, H., and Gruissem, W. (1993). Fruits: A developmental perspective. Plant Cell 5, 1439–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Q., Ferrandiz, C., Yanofsky, M.F., and Martienssen, R. (1998). The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125, 1509–1517. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Holton, T.A. (1995). Modification of flower colour via manipulation of P450 gene expression in transgenic plants. Drug Metab. Drug Interact. 12, 359–368. [DOI] [PubMed] [Google Scholar]
- Holton, T.A., Brugliera, F., Lester, D.R., Tanaka, Y., Hyland, C.D., Menting, J.G., Lu, C.Y., Farcy, E., Stevenson, T.W., and Cornish, E.C. (1993). Cloning and expression of cytochrome P450 genes controlling flower colour. Nature 366, 276–279. [DOI] [PubMed] [Google Scholar]
- Kaltenbach, M., Schröder, G., Schmelzer, E., Lutz, V., and Schröder, J. (1999). Flavonoid hydroxylase from Catharanthus roseus: cDNA, heterologous expression, enzyme properties and cell-type specific expression in plants. Plant J. 19, 183–193. [DOI] [PubMed] [Google Scholar]
- Kim, G.T., Tsukaya, H., and Uchimiya, H. (1998). The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 12, 2381–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, J.C. (1994). Isolation of a cytochrome P450 homologue preferentially expressed in developing inflorescences of Zea mays. Plant Mol. Biol. 25, 343–353. [DOI] [PubMed] [Google Scholar]
- Liljegren, S.J., Ferrándiz, C., Alvarez-Buylla, E.R., Pelaz, S., and Yanofsky, M.F. (1998). Arabidopsis MADS-box genes involved in fruit dehiscence. Flowering Newsl. 25, 9–19. [Google Scholar]
- Luo, M., Bilodeau, P., Koltunow, A., Dennis, E.S., Peacock, W.J., and Chaudhury, A.M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli, S., Frova, C., Torti, G., and Soressi, G.P. (1978). Relationship between set, development and activities of growth regulators in tomato berries. Plant Cell Physiol. 19, 1281–1288. [Google Scholar]
- Mathur, J., et al. (1998). Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 14, 593–602. [DOI] [PubMed] [Google Scholar]
- Mazzucato, A., Taddei, A.R., and Soressi, G.P. (1998). The parthenocarpic fruit (pat) mutant of tomato (Lycopersicon esculentum Mill.) sets seedless fruits and has aberrant anther and ovule development. Development 125, 107–114. [DOI] [PubMed] [Google Scholar]
- Mazzucato, A., Testa, G., Biancari, T., and Soressi, G.P. (1999). Effect of gibberellic acid treatments, environmental conditions, and genetic background on the expression of the parthenocarpic fruit mutation in tomato. Protoplasma 208, 18–25. [Google Scholar]
- Nadeau, J.A., Zhang, X.S., Li, J., and O'Neill, S.D. (1996). Ovule development: Identification of stage-specific and tissue-specific cDNAs. Plant Cell 8, 213–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert, D.W., and Gonzalez, F.J. (1987). P450 genes: Structure, evolution, and regulation. Annu. Rev. Biochem. 56, 945–993. [DOI] [PubMed] [Google Scholar]
- Neff, M.M., Nguyen, S.M., Malancharuvil, E.J., Fujioka, S., Noguchi, T., Seto, H., Tsubuki, M., Honda, T., Takatsuto, S., Yoshida, S., and Chory, J. (1999). BAS1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc. Natl. Acad. Sci. USA 96, 15316–15323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D.R. (1999). Cytochrome P450 and the individuality of species. Arch. Biochem. Biophys. 369, 1–10. [DOI] [PubMed] [Google Scholar]
- Ohad, N., Margossian, L., Hus, Y., Williams, C., Repetti, P., and Fischer, R.L. (1996). A mutation that allows endosperm development without fertilization. Proc. Natl. Acad. Sci. USA 93, 5319–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad, N., Yadegari, R., Margossian, L., Hannon, M., Michaeli, D., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11, 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro, J.K., Szeto, W., Lotys-Prass, C., and Jofuku, K.D. (1997). Photo and hormonal control of meristem identity in the Arabidopsis flower mutants apetala2 and apetala1. Plant Cell 9, 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe, K., Tsao, C.C., Li, L., Popko, J.L., Umezawa, T., Carraway, D.T., Smeltzer, R.H., Joshi, C.P., and Chiang, V.L. (1999). Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc. Natl. Acad. Sci. USA 96, 8955–8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Z., Durst, F., Werck-Reichart, D., Gardner, H.W., Camara, B., Comish, C., and Backhus, R.A. (1995). The major protein of guayule rubber particles is a cytochrome P450. J. Biol. Chem. 270, 8487–8494. [DOI] [PubMed] [Google Scholar]
- Rebers, M., Kaneta, T., Kawaide, H., Yamaguchi, S., Yang, Y.Y., Imai, R., Sekimoto, H., and Kamiya, Y. (1999). Regulation of gibberellin biosynthesis genes during flower and early fruit development of tomato. Plant J. 17, 241–250. [DOI] [PubMed] [Google Scholar]
- Robinson, R., Cantliffe, D., and Shannon, S. (1971). Morphactin-induced parthenocarpy in cucumber. Science 171, 1251–1252. [DOI] [PubMed] [Google Scholar]
- Rodrigo, M.J., and García-Martínez, J.L. (1998). Hormonal control of parthenocarpic ovary growth by the apical shoot in pea. Plant Physiol. 116, 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotino, G.L., Perri, E., Zottini, M., Sommer, H., and Spena, A. (1997). Genetic engineering of parthenocarpic plants. Natl. Biotechnol. 15, 1398–1401. [DOI] [PubMed] [Google Scholar]
- Sakai, H., Medrano, L.J., and Meyerowitz, E.M. (1995). Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378, 199–203. [DOI] [PubMed] [Google Scholar]
- Salchert, K., Bhalerao, R., Koncz-Kalman, Z., and Koncz, C. (1998). Control of cell elongation and stress responses by steroid hormones and carbon catabolic repression in plants. Philos. Trans. R. Soc. London Ser. B 353, 1517–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres, M., Németh, K., Koncz-Kálmán, Z., Mathur, J., Kauschmann, A., Altmann, T., Rédei, G.P., Nagy, F., Schell, J., and Koncz, C. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182. [DOI] [PubMed] [Google Scholar]
- Toguri, T., Umemoto, N., Kobayashi, O., and Ohtani, T. (1993). Activation of anthocyanin synthesis genes by white light in eggplant hypocotyl tissues, and identification of an inducible P-450 cDNA. Plant Mol. Biol. 23, 933–946. [DOI] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart, D. (1995). Cytochromes P450 in phenylpropanoid metabolism. Drug Metab. Drug Interact. 12, 221–243. [DOI] [PubMed] [Google Scholar]
- Zondlo, S.C., and Irish, V.F. (1999). CYP78A5 encodes a cytochrome P450 that marks the shoot apical meristem boundary in Arabidopsis. Plant J. 19, 259–268. [DOI] [PubMed] [Google Scholar]







