Abstract
Brassinosteroids (BRs) are plant growth–promoting natural products required for plant growth and development. Physiological studies have demonstrated that exogenous BR, alone or in combination with auxin, enhance bending of the lamina joint of rice. However, little is known about the function of endogenous BR in rice or other grass species. We report here the phenotypical and molecular characterization of a rice dwarf mutant, d61, that is less sensitive to BR compared to the wild type. We cloned a rice gene, OsBRI1, with extensive sequence similarity to that of the Arabidopsis BRI gene, which encodes a putative BR receptor kinase. Linkage analysis showed that the OsBRI1 gene is closely linked to the d61 locus. Single nucleotide substitutions found at different sites of the d61 alleles would give rise to amino acid changes in the corresponding polypeptides. Furthermore, introduction of the entire OsBRI1 coding region, including the 5′ and 3′ flanking sequences, into d61 plants complemented the mutation to display the wild-type phenotype. Transgenic plants carrying the antisense strand of the OsBRI1 transcript showed similar or even more severe phenotypes than those of the d61 mutants. Our results show that OsBRI1 functions in various growth and developmental processes in rice, including (1) internode elongation, by inducing the formation of the intercalary meristem and the longitudinal elongation of internode cells; (2) bending of the lamina joint; and (3) skotomorphogenesis.
INTRODUCTION
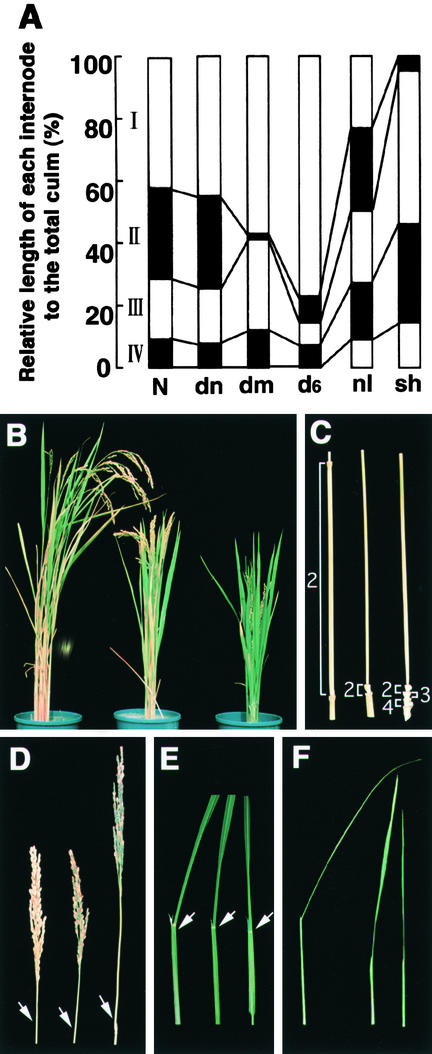
Numerous dwarf mutants of rice have been accumulated and characterized because of their agronomic importance. These dwarf mutants are categorized into six groups, based on the elongation pattern of the upper four or five internodes. Figure 1A (redrawn from Takeda, 1977) shows schematically the relative length of each internode to the total culm. In rice, each internode is numbered from top to bottom such that the uppermost internode just below the panicle is the first. As this representation shows, in the dn-type mutants the length of each internode is almost uniformly shortened, resulting in an elongation pattern similar to that of the wild-type plant. In contrast, the dm-type mutants show a specific shortening of the second internode. Similar shortening of specific internodes is also observed in the sh- and d6-type mutants, in which only the uppermost internode or the internodes below it are shortened, respectively.
Figure 1.
Phenotype of the d61 Mutants.
(A) Schematic representation of the various elongation patterns of internodes in the wild type (N) and various rice dwarf mutants (dn-, dm-, d6-, nl- and sh-types), adapted from Takeda (1977).
(B) Gross morphology of a wild-type plant (left); d61-1 mutant (center), a weak allele; and d61-2 mutant (right), a strong allele.
(C) Elongation pattern of internodes. The wild-type plant (left) shows the N-type of the elongation pattern, whereas the d61-1 (center) and d61-2 (right) mutants show typical dm- and d6-type patterns, respectively. The number of each internode is indicated.
(D) Panicle structure. The wild-type plant (left) has a short panicle; the d61-1 (center) and d61-2 (right) mutants have longer panicles. The arrows indicate the nodes.
(E) Leaf morphology. The leaf of the wild-type plant (left) is bent at the lamina joint indicated by the white arrow, whereas the leaves of d61-1 (center) and d61-2 (right) mutants are more erect.
(F) Leaf sheath morphology. The leaf sheath in the d61-1 (center) and d61-2 (right) mutants is shorter than in the wild-type plant (left).
The mechanism of internode elongation of rice culm has not been clarified well; however, we do know that the transition of the shoot apical meristem (SAM) from the vegetative to the reproductive stage induces internode elongation. Although a similar phenomenon is also seen in Arabidopsis, there is an important difference between the two species: the elongated internodes in rice are derived from the vegetative SAM, whereas those in Arabidopsis come from the reproductive SAM. In rice, the uppermost four or five internodes develop from the vegetative SAM and initially are indistinguishable from the lower unelongated internodes. When the SAM shifts to the reproductive phase, the uppermost four or five internodes differentiate in response to the development of intercalary meristems in the internodes. This synchronicity between the phase change of the SAM and development of the intercalary meristem leads to the hypothesis that these processes might be linked by one or more signals coming from the SAM to the uppermost four or five phytomers when its phase change occurs.
Recent molecular genetic approaches have revealed that plant dwarfism is often caused by defects in the biosynthesis and perception of plant hormones such as gibberellin (GA) and brassinosteroids (BRs). In Arabidopsis, for example, several mutants are BR-related mutants that have a distinctive dwarf phenotype with dark green rugose leaves. When grown in the dark, these mutants show a deetiolated (DET) phenotype with less hypocotyl elongation (Chory et al., 1991; Kauschmann et al., 1996). Through the characterization of these Arabidopsis mutants, BRs are shown to play important roles in normal growth and also in light and dark development during morphogenesis.
In contrast to studies with Arabidopsis and some other dicot plants, only a few reports have been published on the physiological effect of BRs in the growth and development of monocot plants. The most well-known effect is the stimulation of lamina inclination in rice, which has been used for a sensitive bioassay of BRs (Maeda, 1965; Wada et al., 1981; Takeno and Pharis, 1982). Treatment with low or high concentrations of BRs, respectively, promotes or inhibits the growth of roots in rice (Radi and Maeda, 1988). In etiolated wheat seedlings, treatment with brassinolide (BL) or its derivative, castasterone, stimulates unrolling of the leaf blades (Wada et al., 1985). Although these effects of exogenously supplied BRs do suggest that growth and developmental processes in grass plants should be responsive to endogenous BRs, the exact functions of endogenous BRs in these plants remain unclear. In addition, the literature indicates some apparent disagreement as to whether BRs induce cell elongation in grass plants. Yokota and Takahashi (1986) reported that BL did not induce elongation of the leaf sheath of rice, whereas He et al. (1991) reported that BL induced elongation of the coleoptile and mesocotyl in maize. Recently, we isolated a novel dwarf mutant, d61, that shows specific shortening of internodes. Mutants carrying the weak allele specifically fail to elongate the second internode (dm-type mutants), whereas those with the strong allele fail to elongate all of the internodes except the uppermost one (d6-type mutants) (Wu et al., 1999). We have also isolated the D61 gene, which encodes a putative protein kinase with a high similarity to BRI1, a putative BR receptor in Arabidopsis. Through characterizing the d61 mutants, we have demonstrated that BR plays important roles in internode elongation and bending of the lamina joint in rice.
RESULTS
Characterization of Rice d61 Dwarf Mutants
Rice dwarf mutants d61-1 and d61-2 were obtained independently by mutagenesis with N-methyl-N-nitrosourea (Figure 1B). These were initially characterized as two independent mutants, because the culm of d61-2 is much shorter than that of d61-1. In addition, d61-1 shows a typical dm-type pattern of internode elongation, whereas d61-2 shows the d6-type (Figure 1C). However, crosses between these mutants demonstrated that they are alleles of a single locus. To our knowledge, this is the only case of rice mutants of a single locus that show different, specific patterns of inhibition of internode elongation.
The phenotypes of these mutants are abnormal in other respects. For example, the neck internode of the mutants is longer than that of wild-type plants (Figure 1D). The neck internode length shows an inverse relationship to the length of culm in these plants, such that wild-type plants have the longest culm and shortest neck internode, the d61-1 mutant has intermediate length culm and neck internodes, and d61-2 has the shortest culm and the longest neck internode. Another abnormal phenotype of these mutants is erect leaves (Figure 1E). In wild-type plants, the leaf blade bends away from the vertical axis of the leaf sheath toward the abaxial side. When the leaf blades and sheaths are fully elongated, cells at the adaxial side of the lamina joint (indicated by arrows in Figure 1E) start to elongate, causing the leaf blade to bend away from the leaf sheath. In the d61-1 mutant, some leaves still show slight bending, but in the d61-2 mutant almost all of the leaves are completely erect (Figure 1E). The lack of bending of the mutant leaves is caused by impaired development of the lamina joint, because these are abnormally developed even in the d61-1 mutant with the mild phenotype. The mutants show a further abnormal phenotype in having shorter leaf sheaths than do the wild-type plants (Figure 1F).
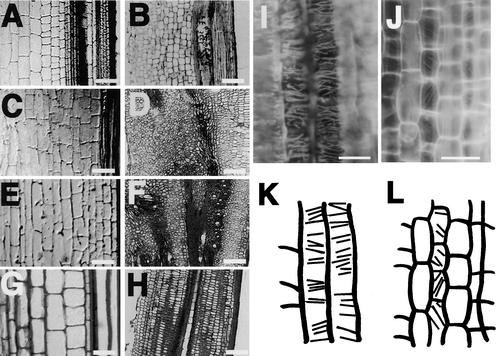
Internode elongation is caused by cell division in the intercalary meristem, followed by cell elongation in the elongation zone (Hoshikawa, 1989). Therefore, dwarfing of the culms could be the result of a defect in one or both of these processes. To distinguish between these possibilities, we examined sections of each internode from adult plants under the microscope. Figures 2A to 2H show the cell morphology of the upper four internodes in a wild-type plant and in the d61-2 mutant. In the wild-type plant, cells in all internodes were longitudinally elongated and well organized with longitudinal files (Figures 2A, 2C, 2E, and 2G). Similar longitudinal cell files were also seen in the first elongated internodes of the mutant plants, although the cells were a little shorter than those in the wild-type plant (Figure 2B). In the nonelongated internodes of the mutant, however, such as the second and third internodes, the arrangement of cells was disorganized with no organized cell files apparent (Figures 2D and 2F). Disorganization of the internodal cells indicates that the intercalary meristems of the mutants, which ordinarily generate the longitudinal files of elongated cells, are not developed in the nonelongated internodes. In the fourth internode, organized cell files were present but the cells were much shorter than in the wild-type plant (cf. Figures 2G and 2H). This suggests that intercalary meristems did develop in these internodes but the cells failed to elongate.
Figure 2.
Structure of Well-Developed Internodes from Wild-Type and d61-2 Rice Plants and Orientation of Microtubules in Elongating Cells in the First Internode of Wild-Type and d61-2 Plants.
(A) and (B) Longitudinal sections of the first internode from the wild type and d61-2, respectively.
(C) and (D) Longitudinal sections of the second internode from the wild type and d61-2, respectively.
(E) and (F) Longitudinal sections of the third internode from the wild type and d61-2, respectively.
(G) and (H) Longitudinal sections of the fourth internode from the wild type and d61-2, respectively.
(I) and (J) Immunofluorescence images of the microtubule arrangement in internodal parenchyma cells of the first internode from the wild type and d61-2, respectively.
(K) and (L) Schematic presentation of the microtubule arrangement in internodal parenchyma cells of the first internode from the wild type and d61-2, respectively.
Bars in (A) to (H) = 100 μm; bars in (I) to (L) = 50 μm.
Cell elongation depends on the orientation of microfibrils (Green, 1962; Ledbetter and Porter, 1963); therefore, we also examined the arrangement of microtubules in the internodal cells of wild-type and d61-2 mutant plants by immunofluorescence microscopy. In wild-type plants, the microtubules in cells of elongating internodes were arranged in an orderly manner at right angles to the direction of elongation (Figures 2I and 2K). In the d61-2 mutant, however, the microtubules in cells of the first elongating internode were arranged in different directions in each cell, apparently at random (Figures 2J and 2L). Moreover, the microtubules in the mutant appeared to be thinner and less distinct relative to those of the wild-type plant (cf. Figures 2I and 2J). We saw no organized microtubule arrangement in cells from nonelongated internodes of the mutant plants. Taken together, these results show that nonelongated internodes in the mutants fail to develop an intercalary meristem and that the cells lack an organized arrangement of microtubules. In those internodes that do elongate in the mutants, albeit less than in the wild-type plants, an intercalary meristem does develop but the cells lack a well-organized microtubule arrangement.
The d61 Mutant Is Less Sensitive to BL than Wild-Type Plants
The degree of bending between the leaf blade and leaf sheath in rice is known to be sensitive to the concentration of BL or its related compounds. This unusual character of rice leaves is the basis for a quantitative bioassay for BRs, known as the lamina joint test (Wada et al., 1981)—even though the biological function of endogenous BRs in monocotyledonous plants, including rice, remains unknown.
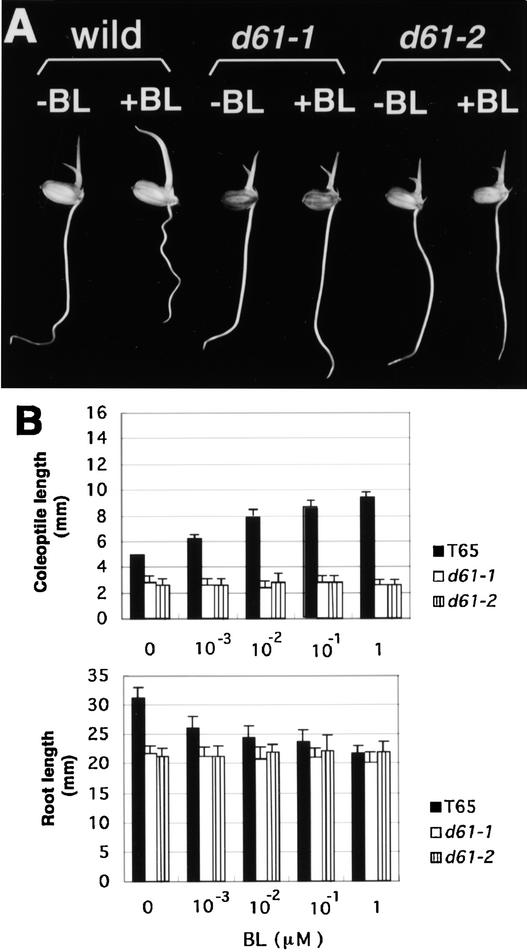
The dwarf phenotype and erect leaves of the d61 mutant led us to speculate that the D61 gene product could be involved in either the biosynthesis or signal transduction of BRs. To test these possibilities, we performed the following experiments. First, seeds of the wild-type and mutant plants were germinated on agar plates containing 1 μM BL. When wild-type seeds were germinated on BL plates, the coleoptiles elongated abnormally in the presence of exogenous BRs, resulting in a twisted shape; the foliage leaves grew poorly and did not break through the coleoptile (Figure 3A). Furthermore, root elongation was inhibited, the roots being not straight but developing in a wavy form. The wild-type plants grew normally in the absence of exogenously supplied BL: coleoptile elongation stopped at an early stage of germination and the foliage leaves elongated to break out of the coleoptile. In contrast to the wild-type plants, the d61 mutants showed normal growth patterns in both the presence and absence of exogenously supplied BRs (Figure 3A). We measured the effects of a range of BL concentrations on the length of coleoptiles and roots of seedlings (Figure 3B). Neither root nor coleoptile length was affected in the mutants by BL, whereas in the wild plants, the coleoptile length was increased and the root length was decreased after treatment with a higher concentration of BL. These results suggest that the mutant plants are less sensitive to exogenous BL than are the wild-type plants.
Figure 3.
Response of Seedlings to BL.
(A) Seeds of the wild type (left) and the dwarf mutants d61-1 (center) and d61-2 (right) were germinated on agar plates in the presence (+) or absence (−) of 1 μM BL. Seedlings were examined 1 day after germination.
(B) Effect of BL on the coleoptile and root elongation in wild-type and d61 seedlings. The plants were germinated in the same conditions as (A) with the indicated concentration of BL. Data presented are the means of results from five plants. Bars indicate sd.
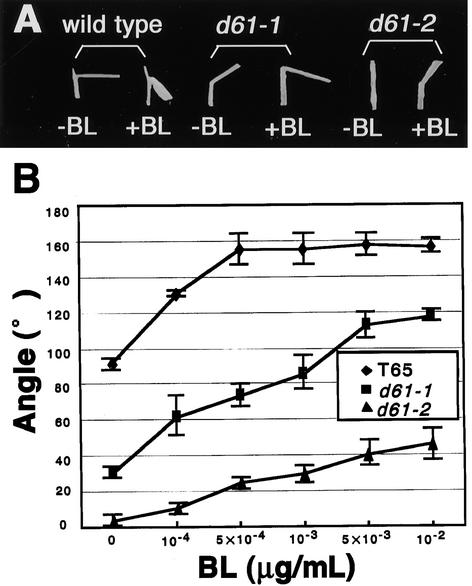
We also tested the sensitivity of the mutants to BL with a more quantitative method. As previously mentioned, rice leaves can be used for the quantitative bioassay of BRs. If the mutants are less sensitive to BRs, the degree of bending between the leaf blade and sheath of the mutant plants should be less than that of the wild-type plants. When wild-type T65 plants (the background strain of the mutants) were used for the lamina joint test, the leaf blade was bent almost at right angles to the axis of the leaf sheath in the absence of exogenous BL, becoming even more bent in the presence of increasing concentrations of BL (Figure 4A). The bending axis plateaued at ∼160° at the BL concentration of 5 ×10−4 μg mL−1 (Figure 4B). When the mutants were so tested, the degree of bending also increased with higher concentrations of BL, as in the wild-type plants (Figures 4A and 4B). However, the extent of bending of the leaves from the mutants was much less than that of the wild-type plants under the same conditions, and the bending axis did not plateau even at a BL concentration of 10−2 μg mL−1 BL (Figure 4B). The results of the lamina joint test thus confirm that the sensitivity of the mutants to BRs is less than that of the wild-type plants.
Figure 4.
Effect of BL on the Degree of Inclination of Etiolated Leaf Lamina in Wild-Type and d61 Plants.
(A) Typical response of the second leaf lamina joint from wild-type, d61-1, and d61-2 plants to BL at 10−3 μg mL−1.
(B) The dose response to BL of the bending angle in the wild type and d61 mutants. Data presented are the means of results from six plants. Bars indicate sd.
Quantitative Analysis of BRs in the d61-2 Mutant
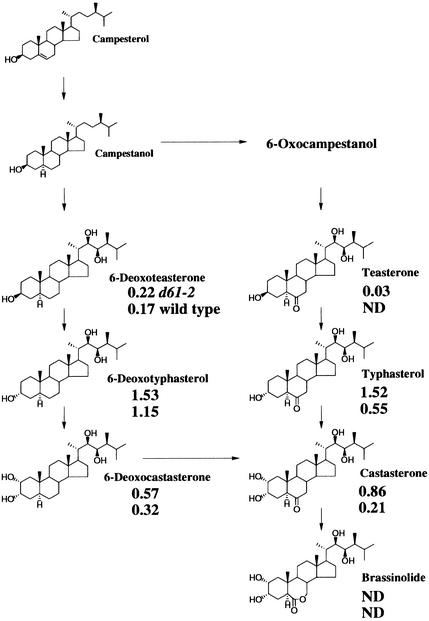
The above experiments demonstrate that the d61 mutants have less sensitivity than wild types to BRs. As a consequence, we conjectured, perhaps they try to compensate for this by synthesizing more BRs. Thus, we measured the concentration of BRs in both the d61-2 mutant and wild-type plants by using gas chromatography–selected ion monitoring analyses. BL was not detected in shoots from either the mutant or wild-type plants, suggesting that this compound is a minor component of the total BR pool (Figure 5). However, all of the other known BRs were detected in both plant types, except for teasterone, which was not found in the wild-type plants. The concentrations of all of the BR compounds detected were greater in the mutant plants, most notably, castasterone, which was four times higher in the mutant than in the wild type. The greater amounts of BRs in the d61-2 mutant are consistent with an attempt by the plants to compensate for their diminished sensitivity to BRs.
Figure 5.
Endogenous BRs in Wild-Type and d61-2 Rice Plants.
The amounts (ng g−1 fresh weight) of BL and its biosynthetic precursors in d61-2 (upper) and wild-type (lower) rice plants are shown. ND, not detected.
d61 Mutants Show a DET Phenotype
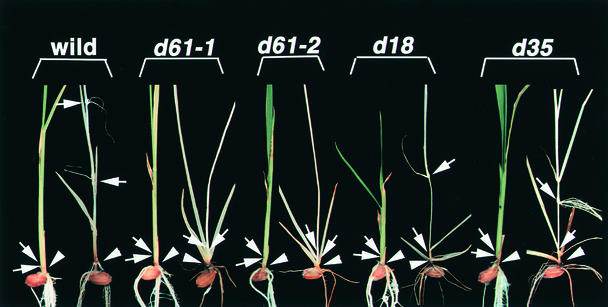
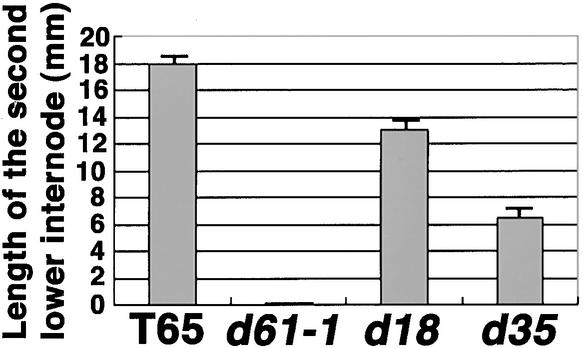
Hypocotyl elongation, opening of cotyledons, and emergence of primary leaves are all impaired in Arabidopsis mutants having deficiencies in BR biosynthesis or BR sensitivity when grown in complete darkness (Chory, 1993; Kauschmann et al., 1996; Szekeres et al., 1996). To test whether the d61 mutants also showed such skotomorphogenic characteristics, we grew them in the dark. Wild-type plants germinated in the dark showed unusual elongation of the mesocotyl and internodes in comparison with light-grown seedlings (Figures 6 and 7). Such elongation of the mesocotyl and internodes did not occur in the mutants, however, even in the dark. This failure of the mesocotyl or internodes to elongate in the dark is not a common characteristic of rice dwarf mutants. For example, two other dwarf mutants, d18 and d35, which are deficient in GA biosynthesis, showed elongated mesocotyls and internodes when grown in the dark. Thus, the reduced elongation of the mesocotyl and internodes appears to be a specific feature of the d61 mutants.
Figure 6.
DET Phenotype of the Wild Type, d61, and Two Gibberellin-Deficient Rice Mutants (d18 and d35) in the Dark.
The left- and right-hand seedlings of each pair were grown for 2 weeks in the light and the dark, respectively. Arrows indicate the nodes and arrowheads indicate the mesocotyls.
Figure 7.
Length of the Second Lower Internodes of the Wild Type and Mutants Grown in the Dark.
Data presented are the means of the results from five plants. Bars indicate sd.
The d61 Locus Is Closely Linked to a Homolog of the Arabidopsis BRI1 Gene
To map the D61 locus, we crossed the d61-2 mutant with an indica rice cultivar, Kasarath. The linkage analysis between the mutant phenotype and restriction fragment length polymorphism (RFLP) markers released from the Rice Genome Project (Tsukuba, Japan) revealed that the D61 locus is mapped to the long arm of chromosome 1, with tight linkage to the RFLP marker C1370.
We also tested the linkage between the mutant phenotype and a rice gene homologous to the Arabidopsis BRI1 gene. The Arabidopsis BRI1 gene is the only gene thus far that has been described to be involved in BR signal transduction (Li and Chory, 1997). The reduced sensitivity to BL of the rice d61 mutants suggested that a rice homolog of the Arabidopsis BRI1 gene might be a good candidate for the genetic lesion in these mutants. Therefore, we performed a BLAST search, which revealed one clone of a rice expressed sequence tag, S1676, with a high sequence similarity to the Arabidopsis BRI1 gene. An RFLP between japonica (12.5 kb) and indica (17.5 kb) rice was observed when genomic DNAs were digested with EcoRI and probed with the cDNA clone. All F2 plants with the mutant phenotype were homozygous for the japonica allele (12.5 kb), whereas the F2 plants with the wild-type phenotype were either homozygous for the indica allele (17.5 kb) or heterozygous with both the japonica and indica alleles (data not shown). This result demonstrates that the D61 locus is closely linked to the position of the rice gene that is homologous to the Arabidopsis BRI1.
Identification of the d61 Gene
Linkage analysis strongly suggested that the d61 mutation is caused by loss of function of the rice homolog of the Arabidopsis BRI1 gene. To test this, we isolated the rice BRI1 homologous gene, OsBRI1 (Oryza sativa BRI1). The structure of the OsBRI1 gene is quite similar to the Arabidopsis BRI1 gene throughout its length (Figure 8). The predicted OsBRI1 polypeptide contains several domains that are also present in Arabidopsis BRI1, the functions of which were discussed by Li and Chory (1997). These include a putative signal peptide, two conservatively spaced cysteine pairs, a leucine-rich repeat (LRR) domain, a transmembrane domain, and a kinase domain. The N terminus of the predicted OsBRI1 polypeptide has a hydrophobic segment that is predicted to act as a signal peptide to transport the protein to the plasma membrane. In the BRI1 protein, Li and Chory (1997) predicted a potential four-heptad-amphipathic leucine zipper motif after the signal peptide, but OsBRI1 does not have such a typical leucine zipper motif in the corresponding region.
Figure 8.
Deduced Amino Acid Sequences of OsBRI1 and Arabidopsis BRI1.
Identical residues are shaded. The underlined regions indicate (1) a putative signal peptide, (2) a leucine zipper motif, (3) N-side, and (4) C-side of a cysteine pair.
A putative extracellular domain (from Met-1 to Leu-670), consisting of 22 tandem copies of a LRR of ∼24 amino acids with 12 potential N-glycosylation sites (Asn-X-Ser/Thr), is flanked by pairs of conservatively spaced cysteines. The LRR has been implicated to function in protein–protein interactions (Kobe and Deisenhofer, 1994).
Compared with the BRI1 sequence, OsBRI1 is missing three LRR domains, which correspond to the third to fifth repeats of the Arabidopsis BRI1. The two LRRs before this deletion are less conserved except for the consensus residues found in other LRR proteins, but the LRRs of both proteins are well conserved after the deletion in both the LRR consensus and non-LRR consensus residues. An unusual feature of the LRR region of BRI1 is the presence of a 70–amino acid island between the 21st and 22nd LRRs (Li and Chory, 1997). A highly similar feature is also present in OsBRI1, which has the same number of amino acids between the 18th and 19th LRRs, corresponding to the site of the island in BRI1. Li and Chory (1997) showed that exchange of an amino acid residue in this island resulted in the loss of function of BRI1 and discussed whether this motif is important for direct interaction with BRs or for maintaining the structure of the BR binding domain.
The protein kinase domain of OsBRI1 has all 11 conserved subdomains of eukaryotic protein kinases, retaining the invariant amino acid residues in their proper positions (Hanks and Quinn, 1991). The protein kinase domain of OsBRI1 is highly related to that of BRI1 (44% identity over the entire region). It is also related to the kinase domains of other receptor-like protein kinases in higher plants such as ERECTA (Torii et al., 1996), CLV1 (Clark et al., 1997), and RLK5 (Walker, 1993) from Arabidopsis and Xa21 from rice (Song et al., 1995). The highly conserved structure of OsBRI1 and these receptor-like protein kinases, especially in subdomains VIb and VIII, suggests that OsBRI1 is a serine/threonine kinase rather than a tyrosine kinase (Hanks and Quinn, 1991).
We also compared the sequences of OsBRI1 in d61-1 and d61-2 mutants with that of the wild type. We identified a single nucleotide substitution in each mutant allele (C to T in d61-1 and G to A in d61-2) at different sites. The mutation in d61-1 results in the change from threonine to isoleucine at residue 989 in subdomain IX of the kinase domain, which is conserved between OsBRI1 and BRI1. The mutation in d61-2 will change valine to methionine at residue 491 in the 17th LRR, just before the unusual 70–amino acid interrupting region. These mutations in the OsBRI1 genes from the d61 mutants provide strong evidence that OsBRI1 encodes the D61 locus.
Molecular Complementation of the d61 Mutation by the Wild-Type OsBRI1 Gene
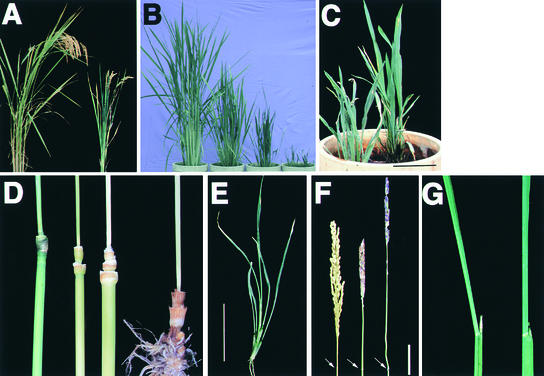
To confirm that OsBRI1 corresponds to the d61 locus, we performed complementation analysis of the d61-1 mutant by introduction of the wild-type OsBRI1 gene. Transformation of d61-1 with a control vector that carried no rice genomic DNA had no apparent effect on the culm length or the structure of leaves (Figure 9A). However, when a 10.5-kb DNA fragment containing the entire wild-type OsBRI1 gene was introduced, the normal phenotype was recovered in almost all plants that were resistant to hygromycin (Figure 9A). This result confirms that the d61 mutant phenotype is caused by the loss-of-function mutation in the OsBRI1 gene.
Figure 9.
Complementation and Antisense Phenotype of OsBRI1.
(A) d61 plant with the wild-type OsBRI1gene (left) and d61 plant introduced with a vector only (right).
(B) Dwarf phenotype of OsBRI1 antisense plants with intermediate and severe phenotypes (right) in comparison with a wild-type plant (left).
(C) Close-up view of OsBRI1 antisense plants with the severe phenotype.
(D) Naked culm internodes of transgenic plant. From left to right, wild-type plant with normal elongation pattern of internodes, and OsBRI1 antisense plants with the dm, dm–d6, and d6 phenotypes, respectively.
(E) Abnormal leaf morphology of an OsBRI1 antisense plant with a severe phenotype, showing lack of developed sheath organs.
(F) Panicle morphology in wild-type (left) and OsBRI1 antisense plants with the mild (center) and intermediate (right) phenotypes. The arrows indicate the nodes.
(G) Leaf morphology of wild-type (left) and OsBRI1 antisense plants with mild phenotype (right), showing the erect leaves in the latter.
Bar in (C) = 5 cm; bar in (E) = 10 cm; bar in (F) = 2 cm.
Organ-Specific Expression of OsBRI1
Very little is known about the function of endogenous BRs in monocotyledonous plants. Therefore, to investigate this we determined by RNA gel blot analysis the expression pattern of the OsBRI1 gene in various rice organs. We used a partial cDNA fragment, corresponding to the region from Ser-740 to Asp-1116 in the kinase domain, as a probe that specifically hybridized to the genomic DNA fragment encoding OsBRI1 (data not shown). A single, strongly hybridizing band was detected in RNA from the vegetative shoot apices (Figure 10A). The size of the band was ∼3.5 kb, almost the same as that of the longest cDNA clone. Weaker hybridization was also observed in RNA from flowers, rachis, roots, and expanded leaf sheaths, whereas no hybridization was observed in RNA from expanded leaf blades or seeds. Thus, the expression of OsBRI1 varies markedly between organs, suggesting that the sensitivity to BRs also differs among these organs.
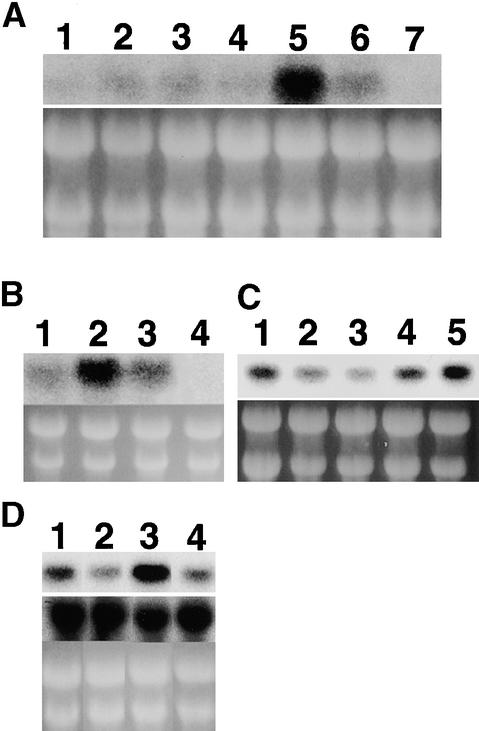
Figure 10.
Expression Pattern of OsBRI1 in Various Organs.
Total RNA (10 μg) from various organs of wild-type plants was probed by hybridization with an OsBRI1 cDNA clone.
(A) Organ-specific expression of OsBRI1. Lane 1, leaf blade; lane 2, leaf sheath; lane 3, developed flower; lane 4, rachis; lane 5, shoot apex; lane 6, root; and lane 7, seed.
(B) Region-specific expression of OsBRI1 in developing first internodes. Lane 1, node; lane 2, divisional zone; lane 3, elongation zone; and lane 4, elongated zone.
(C) Differential expression of OsBRI1 in each elongating internode. Lanes 1 to 4, the divisional and elongation zones of the first to fourth internodes, respectively, at the actively elongating stage for each internode; lane 5, the unelongated stem at the vegetative phase.
(D) Light-dependent and BL-dependent expression of OsBRI1 (upper panel). Rice seedlings were grown for 10 days in the light (lanes 1 and 2) or dark (lanes 3 and 4) on agar plates in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of 1 μM BL. The middle panel shows the amount of the actin mRNA probed with an expressed sequence tag clone, S14002, from Rice Genome Project as a control.
We also tested the expression of OsBRI1 in different parts of elongating culms (Figure 10B). We divided elongating culms into four parts: the node and the division, elongation, and elongated zones of the internode. The strongest hybridization was found in RNA from the division zone. RNA from the elongation zone also gave a strong signal. RNA from the node gave only a weak signal, and that from the elongated internode gave no signal at all. That is, each region of the elongating culm may have different sensitivities to BR, with the most sensitive parts of the elongating culm being the division and elongation zones, where cells are actively dividing and elongating.
We examined the expression of OsBRI1 in the elongation zones of the upper four internodes at the stage when each internode was actively elongating. A strongly hybridizing band was found in RNA from the elongation zone of the uppermost (first) and the lowest (fourth) internodes, whereas hybridization was relatively weak in the second and third internodes (Figure 10C). Apparently, therefore, the internodes also differ in sensitivity to BRs, such that the second and third internodes are the least sensitive. Interestingly, expression of OsBRI1 was also high in the stem at the vegetative stage, in which the internodes do not elongate, indicating that high expression of OsBRI1 in the culm does not necessarily coincide with internode elongation.
We also tested the effects of exogenously applied BL and light on the amount of OsBRI1 mRNA. Germinating seeds were placed on 0.9% agar plates with or without 1 μM BL and were grown for 6 days in the light or dark. OsBRI1 expression was greater in seedlings grown in the dark than in light (Figure 10D). The expression of OsBRI1 was also regulated by exogenously supplied BL, its expression being decreased in the presence of exogenous BL in both light- and dark-grown seedlings, whereas the actin expression did not differ among the RNAs from these seedlings.
Phenotypic Analysis of Transgenic Plants Expressing the Antisense Strand of OsBRI1
The above phenotypic analyses of the d61 mutants and the single-base exchange in the OsBRI1 genes in each mutant suggest that these might not be null alleles but perhaps could retain some partial function. To investigate further the function of BRs in rice, we attempted to generate other mutants with more severe phenotypes by overexpressing the antisense strand of the OsBRI1 transcript under the control of the rice Actin1 gene promoter (Zhang et al., 1991). Almost all of the resulting transgenic plants (18 of 20) produced erect leaves (Figure 9G and Table 1). All of the transformants showed a dwarfed phenotype varying in severity (Figure 9B and Table 1), with less amount of the transcript in the shoot apex region relative to that in the wild-type plant (data not shown). In the plants with the weakest phenotype, the length of each internode was partially and uniformly shortened, resulting in an elongation pattern similar to that of the wild plant; these plants thus resemble dn-type mutants. Plants with intermediate phenotypes had the typical internode elongation patterns of dm- (specific reduction of the second internode) or d6-type (specific reduction of the second to fourth internodes) mutants or a mixed dm–d6-type phenotype (specific reduction of the second and third internodes) (Figure 9D). The inverse relationship between the neck internode and culm length was also seen in these antisense plants (Figure 9F). Plants with the severe phenotype formed only abnormal leaves without developed sheath organs, and the internodes did not elongate (Figure 9E). These plants were <15 cm tall, even after >1 year, and did not produce seeds (Figure 9C). The other phenotypes were inherited in subsequent generations and cosegregated with hygromycin resistance. The cosegregation between the abnormal phenotypes and hygromycin resistance, and the similarity between the intermediate phenotype of the antisense plants and the d61 mutants, demonstrate that the antisense strand acts to suppress the function of OsBRI1 in the transgenic plants.
Table 1.
Phenotype of OsBRI1 Antisense Plants
| Transgenic Plant | Erect Leaves | Internode Elongation Patterna |
|---|---|---|
| 1 | Yes | dm |
| 2 | Yes | Severe dwarfb |
| 3 | Yes | dm |
| 4 | Yes | Severe dwarf |
| 5 | Yes | Mixed dm–d6 |
| 6 | Yes | Severe dwarf |
| 7 | No | dn |
| 8 | Yes | dn |
| 9 | No | dn |
| 10 | Yes | Severe dwarf |
| 11 | Yes | Mixed dm–d6 |
| 12 | Yes | dn |
| 13 | Yes | Mixed dm–d6 |
| 14 | Yes | Severe dwarf |
| 15 | Yes | dn |
| 16 | Yes | dn |
| 17 | Yes | Severe dwarf |
| 18 | Yes | dm |
| 19 | Yes | Mixed dm–d6 |
| 20 | Yes | Mixed dm–d6 |
The elongation pattern of rice internode was represented in Fig-ure 1A.
Plants with the severe dwarf only formed abnormal leaves without internode elongation (see Figure 9C).
DISCUSSION
D61 Encodes a Putative BR Receptor
The following observations from the phenotypic analyses of the d61 mutants revealed that the plants are insensitive or show reduced sensitivity to BL. First, unlike wild-type seedlings, d61 seedlings did not show abnormal growth on agar plates in the presence of BL (Figure 3). Second, the bending response of the leaf blades from the d61 plants was less than that of the wild-type plants (Figure 4). Finally, the content of BRs in the d61-2 mutant exceeded that in the wild-type plants (Figure 5). All these results strongly suggest that the lesion in the d61 mutants affects the perception of BRs. The mutations at the D61 locus were found to be in the gene OsBRI1, which is homologous to the Arabidopsis BRI1 gene that encodes a putative BR receptor (Li and Chory, 1997). The predicted OsBRI1 polypeptide shares several domains with the BRI1 polypeptide highly conservatively, including a putative signal peptide, two conservatively spaced cysteine pairs, a LRR domain, and a protein kinase domain (Figure 8).
Function of BRs in Rice
The phenotypes of the d61 mutants and the antisense plants with decreased expression of OsBRI1 gave us direct indication of the physiological roles of endogenous BRs in rice plants. The d61 mutants had straight leaves with little or no inclination of the lamina in comparison with wild-type plants. This phenotype thus demonstrates that endogenous BRs do have an important role in leaf development—as might be expected from the effects of exogenous treatment with BRs. The degree of lamina inclination correlated to the severity of the dwarfism, with the mildly dwarfed mutant d61-1 having slightly inclined leaves and the more highly dwarfed mutant d61-2 having straight leaves (Figure 1E). The continuity of the severity in lamina inclination suggests that the longitudinal elongation of the surface cells on the adaxial side of lamina, which causes the lamina inclination, responds continuously to the BR signal. This correlation is consistent with the observation that the degree of lamina inclination is a good indicator of the concentration of BRs in vivo.
The BR signal is also necessary for elongation of internodes in the stem. In contrast to lamina inclination, internode elongation seems to be an all-or-nothing event. In d61-1 mutants, which show relatively weak phenotypes, the second internodes are completely stunted but the other internodes are elongated. In the OsBRI1 antisense plants, with an intermediate phenotype, both the second and third internodes are stunted; and in those plants with the most severe phenotype, only the uppermost internode is elongated. Whether internode elongation occurs or not depends essentially on the development of an intercalary meristem in each internode. These observations therefore indicate that a BR signal is essential for the development of intercalary meristems.
d61 Mutants Are Defective in Skotomorphogenesis
The differences in morphology between Arabidopsis mutants that are defective in BR biosynthesis or BR signal transduction and the wild-type plants are more apparent when the plants are grown in the dark. The mutants have a shortened hypocotyl and open cotyledons and develop primary leaves, whereas wild-type Arabidopsis plants develop a long hypocotyl and small, closed cotyledons (Chory et al., 1991; Kauschmann et al., 1996). This DET or constitutive photomorphogenesis (COP) phenotype in darkness is a common feature of these Arabidopsis BR-related mutants. A similar DET or COP phenotype is also observed in a tomato extreme dwarf (dx) mutant that shows a short hypocotyl, lack of apical hook, and expansion of cotyledons (Bishop et al., 1996). In contrast to these observations, a pea BR-defective mutant, lkb, does not show such a DET phenotype (Nomura et al., 1997). Because the d61 rice mutants are also defective in BR signaling, this led us to investigate their development in the dark to determine whether such DET or COP phenotypes are also found in monocotyledonous plants. The mesocotyl and internodes were elongated in wild-type plants grown in the dark, but elongation was arrested in the mutants. This arrested elongation of the mesocotyl and internodes are an uncommon phenotype among rice dwarf mutants. Thus, the DET phenotype of the d61 mutants differentiates them from GA-related dwarf mutants. The results also indicate that BR signals are important for skotomorphogenesis in both dicot- and monocotyledonous plants, with defects in BR signal giving rise to similar DET phenotypes.
More OsBRI1 was expressed in dark-grown seedlings than in light-grown seedlings (Figure 10D), which suggests that the dark-grown rice seedlings are more sensitive to BRs than are the light-grown plants (Worley and Mitchell, 1971). High sensitivity to BRs in dark-grown plants should cause the elongation of internode cells in situations where the cells in light-grown plants do not respond to BRs. The correlation between high expression of OsBRI1 and internode elongation is seen not only in the dark-grown plants but also in culms at the flowering stage under normal growth conditions. High amounts of OsBRI1 mRNA were also observed in elongating internodes (Figure 10C). These results suggest that the high expression of OsBRI1 in the culm is important for active internode elongation and that internode elongation correlates to the amount of expression of OsBRI1 (see below). In contrast to that in rice, the extent of BRI1 expression in Arabidopsis is little changed between dark- and light-grown seedlings (Li and Chory, 1997). The reason for this difference between the expression patterns of the rice OsBRI1 and the Arabidopsis BRI1 is not known but could be related to the different photoperiod responses of these two species, rice being a short-day plant and Arabidopsis a long-day plant.
Internodes Differ in Their Sensitivity to BRs
RNA gel blot analysis demonstrated that the amount of OsBRI1 expression differed between internodes, expression being greater in the uppermost and fourth internodes than in the second and third internodes (Figure 10C). The suggestion that the uppermost and fourth internodes are more sensitive to BRs than are the second and third internodes is supported by transgenic plants expressing the antisense strand of OsBRI1 with the intermediate, dm–d6-type, phenotype. Those plants show specific reduction of the second and third internodes as well as elongation of the uppermost and fourth internodes. Presumably, the greater OsBRI1 expression in the uppermost and fourth internodes allows these internodes to respond to the BR signal by inducing elongation.
This greater expression of OsBRI1 in the uppermost and fourth internodes can explain the unusual internode elongation pattern of the dm–d6-type, but it cannot account for the d6 or dm types. The d6 type, in which all of the internodes except the uppermost are reduced, could indicate that the uppermost internode is exposed to more BRs than is the fourth internode. The timing of the elongation of the uppermost internode corresponds to the development of anthers in the flowers, organs that in many plants have been observed to contain high amounts of BRs (Grove et al., 1979; Plattner et al., 1986; Suzuki et al., 1986; Ikekawa et al., 1988; Takatsuto et al., 1989; Gamoh et al., 1990). Presumably, large quantities of BRs could move down from the anthers to lower organs, such as the uppermost internode, and there induce internode elongation. However, elongation of the fourth internode is completed before the flowers develop, which means that this internode does not receive the high-BR signal from the flowers at the time of its active elongation.
The differential sensitivity of the internodes to BRs, as related to the extent of OsBRI1 expression, and differences in the strength of the BR signal alone cannot explain the specific retardation of the second internode observed in the dm-type mutants. More likely, elongation of the second internode is regulated by several factors, because several independent dwarf mutants have the dm-type phenotype, including d1, d2, d11, and d61. Very recently, the D1 gene was isolated and found to encode a protein similar in structure to the α subunit of a G protein (Gα) (Ashikari et al., 1999; Fujisawa et al., 1999). The D1 Gα-like protein is now thought to be involved in the GA signal transduction pathway, because the d1 mutant alleles show little or no sensitivity to active GA (M. Tanaka-Ueguchi, Y. Fujisawa, M. Kobayashi, M. Ashikari, Y. Iwasaki, H. Kitano, and M. Matsuoka, unpublished data). Thus, perhaps a specific mechanism links BR and GA signal transduction in the second internode and is responsible for induction of elongation there.
METHODS
Plant Growth Conditions
Rice plants (Oryza sativa) were grown in the field or in the greenhouse at 30°C (day) and 24°C (night).
Lamina Joint Inclination Bioassay
Rice seeds were soaked in distilled water and imbibed for 48 hr at 30°C. After the imbibed seeds were washed with distilled water several times, they were germinated in a dark chamber at 30°C for 8 days. The second leaf lamina joint was used for the inclination test as described by Wada et al. (1981).
Isolation and Sequencing of the OsBRI1 Gene
A rice cDNA clone, S1676, with high homology to the Arabidopsis thaliana BRI1 gene (Li and Chory, 1997) was found using a BLAST search of the expressed sequence tag database. The S1676 clone was used to probe a rice genomic DNA library to isolate the genomic DNA clone encoding OsBRI1. Hybridization was performed as described in Church and Gilbert (1984), except that membranes were hybridized at greater stringency (68°C).
Genomic DNA Isolation and DNA Gel Blot Analysis
Rice genomic DNA was isolated from leaf tissue by using an ISOPLANT DNA isolation kit (Nippon GENE Co., Toyama, Japan). One microgram of the genomic DNA was digested with appropriate restriction enzymes, and after agarose gel electrophoresis the separated digests were transferred onto Hybond N+ membranes (Amersham) under alkaline conditions. The membrane was probed with the partial cDNA fragment (corresponding to the region from Ser-740 to Asp-1116 in the kinase domain) that specifically hybridized with the genomic DNA fragment encoding OsBRI1. Hybridization was performed at high stringency (68°C).
RNA Isolation and RNA Gel Blot Analysis
RNA was isolated from various rice tissues as described by Chomczynski and Sacchi (1987). Ten micrograms of total RNA was electrophoresed in 1% agarose gel and then transferred to a Hybond N+ membrane for analysis by hybridization with the same probe and hybridization conditions as described above for DNA gel blot analysis.
Quantification of Endogenous Brassinosteroids
Wild-type and d61-2 plants were grown in a greenhouse with a 16-hr day and 8-hr night. Shoots from 2-month-old plants were harvested and lyophilized immediately after harvest. Lyophilized shoots (equivalent to 50 g fresh weight) were extracted twice with 250 mL of methanol:CHCl3 (4:1 [v:v]), and deuterium-labeled internal standards (2 ng/g fresh weight) were added. Purification and gas chromatographic–mass spectroscopic analyses were performed as described in Noguchi et al. (1999).
OsBRI1 Antisense Gene Construct and Rice Transformation
A promoter–terminator cassette (pBIAct1nos) containing the Act1 promoter (Zhang et al., 1991) and the NOS terminator was prepared by substituting the Act1 promoter for the 35S promoter in the hygromycin-resistance binary vector, pBI35Snos (Sato et al., 1999), between the HindIII and XbaI sites. The cDNA clone encoding the entire OsBRI1 coding region was introduced between the XbaI and SmaI sites of pBIAct1nos. Vector pBI-cont, containing no insert, was used as a control vector. The OsBRI1-antisense and control constructs were introduced into the rice cultivar Nipponbare by Agrobacterium tumefaciens–mediated transformation, as described by Hiei et al. (1994).
Complementation of the d61 Mutant
A genomic OsBRI1 clone, including the entire coding region and the 5′ and 3′ flanking regions, was inserted as a 10.5-kb restriction fragment between the XbaI and SmaI sites of the hygromycin-resistance binary vector pBI101-Hm3 (Sato et al., 1998). The OsBRI1 gene construct was introduced into the d61-1 rice mutant as described above. Control plants were transformed with the pBI-cont vector.
Preparation of Sections for Microscopic Analysis
Developing or developed culms at various stages were fixed in formalin:glacial acetic acid:70% ethanol (1:1:18 [v:v:v]) and dehydrated in a graded ethanol series. The samples were embedded in a Technovit 7100 resin (Kurzer, Wehrheim, Germany), polymerized at 45°C, and cut into 3- to 5-μm-thick sections that were stained with Toluidine Blue and examined under the light microscope.
Observation of Microtubule Arrangement
Internodal parenchyma tissue was prefixed for 45 min at room temperature in 3.7% (w/v) paraformaldehyde in microtubule-stabilizing buffer: 0.1 M piperazine–diethanolsulfonic acid, 1 mM MgCl2, 5 mM EGTA, 0.2% (v/v) Triton X-100, and 1% (w/v) glycerol, pH 6.9. Longitudinal sections were cut with a fresh razor blade, collected in the fixation medium, and after 40 min in the fixation medium, washed in microtubule-stabilizing buffer. The sections were then incubated for 1 hr at 37°C with a rabbit monoclonal antibody raised against α-tubulin, diluted 1:500 in phosphate-buffered saline containing 0.1% (v/v) Triton X-100 (PBST). The sections were washed three times in PBST and then incubated overnight at 4°C with fluorescein isothiocyanate–conjugated mouse anti–rabbit IgG antibody diluted 1:50 in PBST. After three washes in the same buffer, the sections were mounted in antifading solution (Fluoro Guard Antifade reagent; Bio-Rad, Hercules, CA) and viewed under a fluorescence microscope.
Acknowledgments
We are grateful to Dr. Kenzo Nakamura (Nagoya University) for providing the vector for rice transformation and Dr. Kouich Mizuno (Osaka University) for providing a rabbit monoclonal antibody raised against α-tubulin. We thank Ms. Tomoyo Asano for her advice and help on the lamina joint inclination bioassay. This work was supported in part by a Grant-in-Aid from the Ministry of Agriculture, Forestry and Fishery and by a Special Coordination Fund of the Science and Technology Agency.
References
- Ashikari, M., Wu, J., Yano, M., Sakaki, T., and Yoshimura, A. (1999). Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the α-subunit of GTP-binding protein. Proc. Natl. Acad. Sci. USA 96, 10284–10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, G.J., Harrison, K., and Jones, J.D.G. (1996). The tomato dwarf gene isolated by heterologous transposon tagging encodes the first member of a new cytochrome P450 family. Plant Cell 8, 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Chory, J. (1993). Out of darkness: Mutants reveal pathways controlling light-regulated development in plants. Trends Genet. 9, 167–172. [DOI] [PubMed] [Google Scholar]
- Chory, J., Nagpal, P., and Peto, C.A. (1991). Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3, 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 3, 575–585. [DOI] [PubMed] [Google Scholar]
- Fujisawa, Y., Kato, T., Ohki, S., Ishikawa, A., Kitano, H., Sasaki, T., Asahi, T., and Iwasaki, Y. (1999). Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc. Natl. Acad. Sci. USA 96, 7575–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamoh, K., Okamoto, N., Takatsuto, S., and Tejima, I. (1990). Determination of traces of natural brassinosteroids as dansylaminophenylboronates by liquid chromatography with fluorimetric detection. Anal. Chim. Acta 228, 101–105. [Google Scholar]
- Green, P.B. (1962). Mechanism for plant cellular morphogenesis. Science 138, 1404–1405. [DOI] [PubMed] [Google Scholar]
- Grove, M.D., Spencer, G.F., Rohwedder, W.K., Mandava, N., Worley, J.F., Warthen, J.D., Jr., Steffens, G.L., Flippen-Anderson, J.L., and Cook, J.C., Jr. (1979). Brassinolide, a plant growth–promoting steroid isolated from Brassica napus pollen. Nature 281, 216–217. [Google Scholar]
- Hanks, S.K., and Quinn, A.M. (1991). Protein kinase catalytic domain sequence database: Identification of conserved features of primary structure and classification of family members. Methods Enzymol. 200, 38–62. [DOI] [PubMed] [Google Scholar]
- He, R.-Y., Wang, G.-J., and Wang, X.-S. (1991). Effects of brassinolide on growth and chilling resistance of maize seedlings. In Brassinosteroids—Chemistry, Bioactivity and Applications (ACS Symp. Ser. 474), H.G.C. Cutler, T. Yokota, and G. Adam, eds (Washington, DC: American Chemical Society), pp. 220–230.
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hoshikawa, K. (1989). Stem. In The Growing Rice Plant (Tokyo: Nobunkyo), pp. 123–148.
- Ikekawa, N., Nishiyama, F., and Fujimoto, Y. (1988). Identification of 24-epibrassinolide in bee pollen of the broad bean, Vicia faba L. Chem. Pharm. Bull. 36, 405–407. [Google Scholar]
- Kauschmann, A., Jessop, A., Koncz, C., Szekeres, M., Willmitzer, L., and Altmann, T. (1996). Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 9, 701–703. [Google Scholar]
- Kobe, B., and Deisenhofer, J. (1994). The leucine-rich repeat; a versatile binding motif. Trends Biochem. Sci. 19, 415–421. [DOI] [PubMed] [Google Scholar]
- Ledbetter, M.C., and Porter, K.R. (1963). A “microtubule” in plant cell fine structure. J. Cell Biol. 19, 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- Maeda, E. (1965). Rate of lamina inclination in excised rice leaves. Physiol. Plant. 18, 813–827. [Google Scholar]
- Noguchi, T., Fujioka, S., Choe, S., Takatsuto, S., Yoshida, S., Yuan, H., Feldmann, K.A., and Tax, F.E. (1999). Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 121, 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, T., Nakayama, M., Reid, J.B., Takeuch, Y., and Yokota, T. (1997). Blockage of brassinosteroid biosynthesis and sensitivity causes dwarfism in garden pea. Plant Physiol. 93, 572–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plattner, D., Taylor, S.L., and Grove, M.D. (1986). Detection of brassinolide and castasterone in Alnus glutinosa (European alder) pollen by mass spectrometry/mass spectrometry. J. Natl. Prod. 49, 540–545. [Google Scholar]
- Radi, S.H., and Maeda, E. (1988). Effect of brassinolide on the cultured rice root growth as modified by Figaron and gibberellic acid. Jpn. J. Crop Sci. 57, 191–198. [Google Scholar]
- Sato, Y., Sentoku, N., Nagato, Y., and Matsuoka, M. (1998). Isolation and characterization of a rice homeobox gene, OSH15. Plant Mol. Biol. 38, 983–998. [DOI] [PubMed] [Google Scholar]
- Sato, Y., Sentoku, N., Miura, Y., Hirochika, H., Kitano, H., and Matsuoka, M. (1999). Loss of function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J. 18, 992–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W.-Y., Wang, G.-L., Chen, L.-L., Kim, H.-S., Pi, L.-Y., Holsten, T., Gardner, J., Wang, B., Zhao, W.-X., Zhu, L.-H., Fauquet, C., and Ronald, P. (1995). A receptor kinase–like protein encoded by the rice disease resistance gene Xa21. Science 270, 1804–1806. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., Yamaguchi, I., Yokota, T., and Takahashi, N. (1986). Identification of castasterone, typhasterol and teasterone from the pollen of Zea mays. Agric. Biol. Chem. 50, 3133–3138. [Google Scholar]
- Szekeres, M., Németh, K., Koncz-Kálmán, Z., Mathur, J., Kauschmann, A., Altmann, T., Rédei, G.P., Nagy, F., Schell, J., and Koncz, C. (1996). Brassinosteroids rescue the deficiency of CY90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182. [DOI] [PubMed] [Google Scholar]
- Takatsuto, S., Yokota, T., Omote, K., Gamoh, K., and Takahashi, N. (1989). Identification of brassinolide, castasterone and norcastasterone (brassinone) in sunflower (Helianthus annuus L.) pollen. Agric. Biol. Chem. 53, 2177–2180. [Google Scholar]
- Takeda, K. (1977). Internode elongation and dwarfism in some gramineous plants. Gamma Field Symp. 16, 1–18. [Google Scholar]
- Takeno, K., and Pharis, R.P. (1982). Brassinosteroid-induced bending of the leaf lamina of dwarf rice seedlings: An auxin-mediated phenomenon. Plant Cell Physiol. 23, 1275–1281. [Google Scholar]
- Torii, K.U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R.F., and Komeda, Y. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, K., Marumo, S., Ikekawa, N., Morisaki, M., and Mori, K. (1981). Brassinolide and homobrassinolide promotion of lamina inclination of rice seedlings. Plant Cell Physiol. 22, 323–325. [Google Scholar]
- Wada, K., Kondo, H., and Marumo, S. (1985). A simple bioassay for brassinosteroids: A wheat leaf-unrolling test. Agric. Biol. Chem. 49, 2249–2251. [Google Scholar]
- Walker, J.C. (1993). Receptor-like protein kinase gene of Arabidopsis thaliana. Plant J. 3, 451–456. [DOI] [PubMed] [Google Scholar]
- Worley, J.F., and Mitchell, J.W. (1971). Growth responses induced by brassins (fatty plant hormones) in bean plants. J. Am. Soc. Hort. Sci. 96, 270–273. [Google Scholar]
- Wu, X., Ihara, Y., Takeda, K., and Kitano, H. (1999). New dm-type dwarf mutants varying in internode elongation pattern are controlled by different mutant genes at the same locus in rice (Oryza sativa L.). Breed. Sci. 49, 147–153. [Google Scholar]
- Yokota, T., and Takahashi, N. (1986). Chemistry, physiology and agricultural application of brassinolide and related steroids. In Plant Growth Substances 1985, M. Bopp, ed (Berlin: Springer-Verlag), pp.129–138.
- Zhang, W., McElroy, D., and Wu, R. (1991). Analysis of rice Act1 5′ region activity in transgenic rice plants. Plant Cell 3, 1155–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]