Abstract
The petunia loci anthocyanin1 (an1), an2, an4, and an11 are required for the transcription of anthocyanin biosynthetic genes in floral organs. The an2 and an11 loci were recently cloned and shown to encode a MYB-domain transcriptional activator and a cytosolic WD40 protein, respectively. Here, we report the isolation of an1 by transposon tagging. an1 encodes a new member of the basic helix-loop-helix family of transcription factors that is functionally and evolutionarily distinct from JAF13, the apparent petunia ortholog of maize RED1 and snapdragon DELILA. We provide genetic evidence that the transcription factors encoded by an1, an2, and an4 operate in an unexpectedly complex regulatory hierarchy. In leaves, ectopic expression of AN2 induces an1 expression, whereas in anthers, an1 expression depends on an4, encoding (or controlling) a MYB protein that is paralogous to AN2. Experiments with transgenic plants expressing a post-translationally controlled AN1–GLUCOCORTICOID RECEPTOR fusion protein indicated that independent of protein synthesis, AN1 directly activates the expression of the dfrA gene encoding the enzyme dihydroflavonol 4-reductase and of Pmyb27 encoding a MYB-domain protein of unknown function.
INTRODUCTION
Pigmentation is a phenotypic trait that provides an excellent system for studying how groups of genes are coordinately expressed in a tissue-specific manner (reviewed in Mol et al., 1995, 1998). Synthesis of anthocyanin pigments from malonyl-CoA and 4-coumaroyl CoA requires some 10 to 15 enzymes, and many of the structural genes encoding these enzymes have been isolated (Holton and Cornish, 1995). Regulatory genes that control the tissue-specific expression of structural anthocyanin genes have been identified by mutation in several species.
In maize, a MYB domain and a basic helix-loop-helix (bHLH)-type transcription factor, encoded by the colorless1 (c1)/purple leaf1 (pl1) and the red1 (r1)/booster1 (b1) gene family, respectively, are required for activation of structural anthocyanin genes (reviewed in Mol et al., 1998; Weisshaar and Jenkins, 1998). At least one other factor, encoded by pale aleurone color1 (pac1), is required for activating the structural genes in the maize kernel (Selinger and Chandler, 1999). The nature of the PAC1 protein, however, is not known. In two-hybrid assays in yeast and maize cells, B1 interacts with C1, which suggests that both proteins are in the same transcription complex (Goff et al., 1992). In vitro, C1 binds with low affinity to functional cis-acting elements in the promoters of structural genes (Sainz et al., 1997), but thus far, no DNA binding activity has been detected for R1. Whether this is because R1 needs to dimerize with an as yet unknown partner or because the C1/R1 complex interacts in vivo with the promoter of an unknown “intermediate” regulator is unclear at this stage. The gene intensifier1 (in1) is an inhibitor of anthocyanin synthesis because loss-of-function mutations in this gene result in a more intense pigmentation of the kernel. A small fraction of the in1 transcripts encode a bHLH protein homologous to R1. However, the large majority of transcripts are misspliced and encode truncated proteins, which may be responsible for the inhibitory character of this locus (Burr et al., 1996).
In flowers and seeds of dicotyledonous species, expression of the structural genes encoding the “early” enzymes of the pathway, which are required for the synthesis of all flavonoid classes, and those encoding the “late” anthocyanin-specific enzymes is controlled by distinct genes (Martin et al., 1991; Quattrocchio et al., 1993, 1998; Shirley, 1996). For example, in petunia flowers, the loci anthocyanin1 (an1), an2, and an11 are required for transcription of anthocyanin-specific genes such as dfrA (encoding dihydroflavonol 4-reductase; Huits et al., 1994a), rt (encoding anthocyanin rhamnosyl-transferase; Kroon et al., 1994), and an9 (encoding a glutathione S-transferase; Alfenito et al., 1998) but not for expression of chsA (encoding chalcone synthase), chi (encoding chalcone-flavanone isomerase), or an3 (encoding flavanone 3-β-hydroxylase) (Quattrocchio et al., 1993).
The an2 locus encodes a MYB-domain protein that is functionally interchangeable with C1 from maize (Quattrocchio et al., 1998, 1999). jaf13 of petunia and delila from snapdragon encode highly similar bHLH proteins thought to be orthologs of R1 from maize (Goodrich et al., 1992; Quattrocchio et al., 1998). Ectopic JAF13 expression induces anthocyanin accumulation and transcription of the dfrA gene; however, no loss-of-function mutants are known for jaf13. The an11 locus encodes a highly conserved WD40 repeat protein that is localized in the cytosol. Because an11− mutants can be rescued, at least partially, by overexpression of AN2, we previously suggested that AN11 may post-translationally regulate the activity of the anthocyanin transcription factors by an as yet unknown mechanism (de Vetten et al., 1997). The observation that mutation of the Arabidopsis transparent testa glabra (ttg) gene, encoding a homologous WD40 protein (Walker et al., 1999), can be complemented by (over)expression of R1 from maize points in the same direction (Lloyd et al., 1992). The function of an1 in this regulatory system remains, thus far, unclear because of the elusive nature of the gene product. Recently, Kubo et al. (1999) showed that anthocyaninless2 (a2) of Arabidopsis encodes a protein with similarity to homeodomain transcription factors. However, whether ANL2 coregulates the anthocyanin-specific genes together with the mentioned MYB, bHLH, and WD40 proteins or is an activator of the early structural genes is unclear.
In this article, we report the isolation and molecular characterization of the an1 locus. an1 encodes a bHLH protein that is most similar to IN, and less to R1 and JAF13. Analysis of an1 expression in an4− mutants and in transgenic plants ectopically expressing AN2 shows that AN1 operates downstream of AN2 and AN4. Induction experiments in transgenic plants in which AN1 was placed under post-translational control showed that AN1 directly activated transcription of a structural anthocyanin gene, independent of protein synthesis. These data indicate that anthocyanin transcription factors operate in a regulatory hierarchy.
RESULTS
Isolation of the an1 Gene
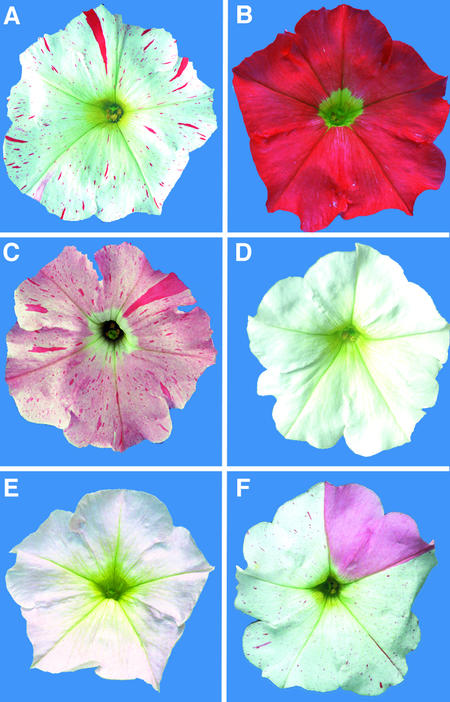
The petunia line W138 contains an an1 allele, initially designated an1s/+-p but later renamed an1-W138, that is somatically and germinally unstable (Doodeman et al., 1984; Huits et al., 1994b). Consequently, an1-W138 flowers are white with red (full revertant) and pink (partial revertant) spots (Figure 1A). In germinal cells, instability of an1-W138 results in a fraction of the progeny having completely red, pink, or white flowers (Figures 1B to 1D).
Figure 1.
Phenotypes of Flowers Harboring Different an1 Alleles.
(A) Flower of line W138, homozygous for the allele an1-W138.
(B) Flower of a plant heterozygous for an1-W138 and a full revertant An1+ allele.
(C) Flower of a plant heterozygous for an1-W138 (evident from the red spots) and an excision allele with partial activity (evident from the pink background color).
(D) Flower of a plant homozygous for a stable recessive null allele derived from an1-W138.
(E) Flower of a homozygote for an1-A2255, a stable allele with low activity.
(F) Flower of a homozygote for the unstable an1-X2191 allele.
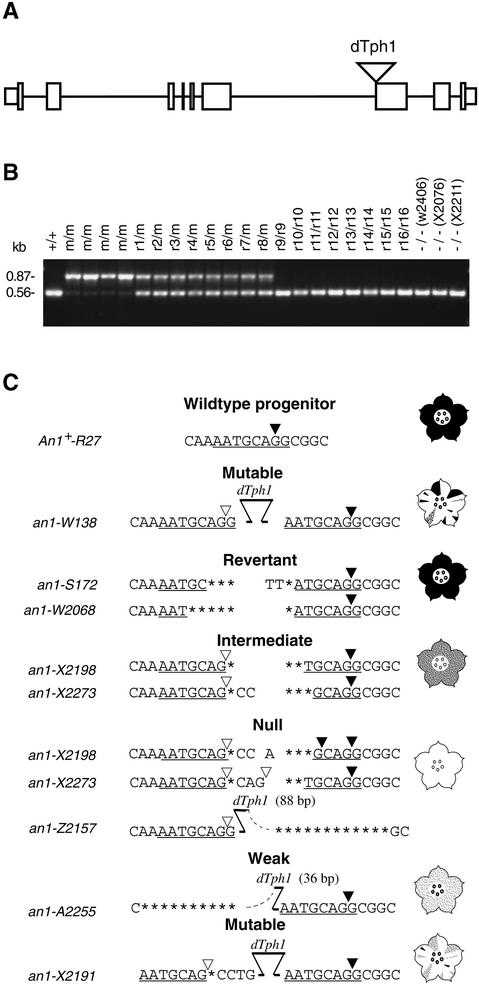
Because somatic instability of the an1-W138 allele responded to the same genetic element (Act1) that is required for transposition of dTph1 elements (Huits et al., 1994b), we anticipated that an1-W138 harbored a dTph1 insertion. To isolate a fragment of the an1 locus, we cloned dTph1 flanking sequences unique for an1-W138 plants by a combination of inverse polymerase chain reaction (PCR) and differential screening of amplification products (Souer et al., 1995). This yielded four genomic fragments that contained a dTph1 insertion in six different an1-W138 plants but not in plants homozygous for two independent An1+ reversion alleles. DNA gel blot analysis of a larger set of plants showed that fragment 65 hybridized to a 0.9-kb RsaI fragment in all an1-W138 homozygotes and to a 0.6-kb fragment in all homozygotes for revertant or stable recessive alleles tested, whereas heterozygotes for an1-W138 and a revertant allele contained both fragments (data not shown). Because these results indicated that fragment 65 originated from the an1 locus, we isolated the gene and corresponding petal cDNAs from libraries made from an An1+ line. Comparison of the genomic sequence with the cDNA and the original dTph1 flanking fragment showed that it contained a gene composed of eight exons, which in petunia line W138 contained a dTph1 insertion at the border of intron 6 and exon 7 (Figure 2A).
Figure 2.
Isolation of the an1 Gene.
(A) Map of the an1 gene. Boxes represent exon sequences, separated by thin lines representing introns. Protein coding sequences are indicated by boxes of double height. The dTph1 insertion in the an1-W138 allele is represented by a triangle.
(B) Ethidium bromide–stained PCR products amplified from plants harboring the parental An1-R27 allele (+), the mutable an1-W138 allele (m), 16 independently isolated full-revertant alleles (r1 to r16), or three independently isolated stable recessive (−) an1 alleles (W2406, X2076, and X2211). The genotypes indicated above the lanes were determined by phenotypic analysis of progeny obtained by self-pollination. The size of the PCR products is indicated at left.
(C) Sequences of the parental An1-R27 allele, the unstable an1-W138 allele, and derived excision alleles. The flower diagrams indicate the petal color specified by each group of alleles. The 8-bp target site duplication caused by the dTph1 insertion in an1-W138 is underlined. Nucleotides that were lost during the attempted dTph1 excision are indicated by asterisks. The open and closed triangles in An1-R27 and an1-X2198 indicate confirmed acceptor splice sites. Triangles in other alleles indicate putative acceptor splice sites that were inferred by using a consensus sequence (Brown et al., 1996). Closed triangles indicate (putative) splice sites that generate an an1 transcript with an intact reading frame, whereas open triangles indicate (putative) splice sites that generate an1 transcripts with a frameshift.
Subsequently, we amplified the region containing the dTph1 insertion from a large number of plants harboring an1-W138 or derived stable alleles with full (revertant) or null activity. Figure 2B shows that the presence of the an1-W138 allele resulted in the amplification of a 0.87-kb fragment, consistent with the presence of a dTph1 insertion, whereas all of the 17 derived stable alleles yielded a 0.58-bp fragment, indicating that these alleles resulted from excisions of that dTph1 element.
Sequencing of PCR amplification products of an1-W138 showed that a 284-bp dTph1 element had inserted at the border of intron 6 and exon 7 and duplicated 8 bp of the target sequence, including the splice acceptor site, whereas the derived stable alleles contained different transposon footprints at this position (Figure 2C). In the full-revertant An1+ excision alleles, the footprint sequence contained only one acceptor splice site that fit the consensus sequence (Brown et al., 1996), indicating that exons 6 and 7 were correctly spliced in all transcripts. The two intermediate alleles contain two potential acceptor splice sites. Splicing to the downstream site would produce a functional transcript with a wild-type sequence, whereas splicing on the upstream site would generate a transcript with a frameshift. Sequence analysis of an1 fragments that were amplified by reverse transcription (RT)–PCR from RNA of X2198 flowers showed that in vivo, both splice sites indeed were used (data not shown), thus explaining the intermediate phenotype. The stable null alleles X2198 and X2273 contain three potential splice sites. Splicing to the most downstream of these would produce a functional transcript, but splicing to either of the two upstream sites would produce transcripts with a frameshift. The stable an1− phenotypes specified by these alleles suggest that the majority of transcripts are (mis)spliced to one or both of the two upstream sites. In a third null allele, an1-Z2157, 195 bp of the right dTph1 end and 10 bp of flanking sequence, including the splice site, were deleted and only 88 bp from the left end of dTph1 remained.
One rare excision event produced an an1 allele, an1-A2255, that specifies flowers with a very pale rather than completely white color, indicating that anthocyanin synthesis has been strongly reduced but not completely blocked (Figure 1E). In an1-A2255, 248 bp at the left end of dTph1 and 10 bp of the an1 flanking (intron) sequence were lost, whereas 36 bp of the right dTph1 end remained present (Figure 2C); as a result, exon 6 was spliced inframe to a cryptic splice site located ∼200 bp downstream in the middle of exon 7 (C. Spelt, F. Quattrocchio, J.N.M. Mol, and R. Koes, unpublished data).
Among W138 progeny, a new unstable an1 allele was found that specified white flowers in which the revertant spots were predominantly pink and only occasionally red (Figure 1F). Sequencing showed that the dTph1 element remained in its original position but now flanked by a footprint sequence on its left side (Figure 2C). Presumably, an1-X2191 arose by a faulty transposition attempt in which only the left end of dTph1 was cleaved and was repaired again without excision of the transposon, similar to the an11-G5543 allele (de Vetten et al., 1997).
These results show that genetic alterations of an1-W138 correlate without exception with (attempted) excisions of the dTph1 element. Therefore, we conclude that the isolated DNA is derived from an1.
an1 Encodes a Novel bHLH Protein
RNA gel blot and RT-PCR experiments showed that the An1+ line R27 expresses a major an1 transcript mRNA of ∼2.5 kb. To determine the nature of the AN1 protein, we sequenced two independently isolated cDNA clones. The longest cDNA measured 2454 bp and contained a 2004-bp open reading frame specifying a 668–amino acid protein (Figure 3A). The second cDNA represented a partial cDNA derived from an incompletely spliced an1 transcript because its 5′ end started 1023 bp downstream from the translation start codon and because it still contained the 473-bp intron located between exons 7 and 8 (data not shown).
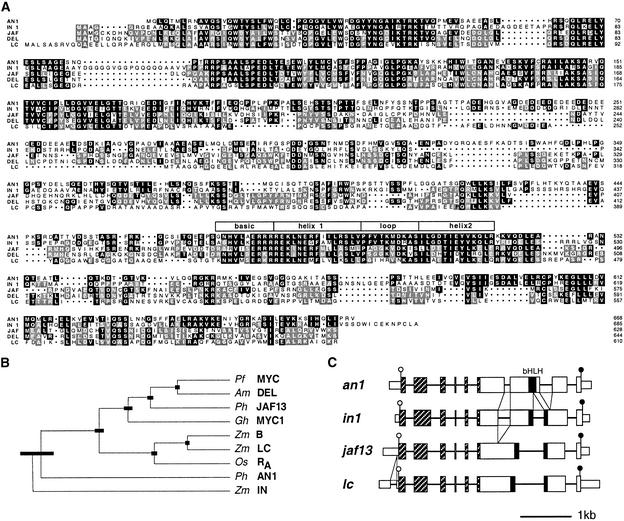
Figure 3.
Molecular Analysis of the an1 Gene.
(A) Alignment of the proteins encoded by an1 and jaf13 (JAF) from petunia, in1 and lc1 (LC) from maize, and del from snapdragon. Sequence identity between AN1 and any one of the other proteins is indicated by black shading. Identical amino acids that are not conserved in AN1 are indicated by gray shading. Numbering of the protein sequences is shown at right, and the position of the bHLH domain is indicated by the box overlying this region. Dots represent gaps introduced to improve the alignment.
(B) Phylogenetic tree of bHLH proteins implicated in pigmentation constructed by the unweighted pair method using arithmetic averages (UPGMA) algorithm (Sokal and Michener, 1958). The sequences are from the following sources: snapdragon (Am DEL, Goodrich et al., 1992; GenBank accession number M84913), Perilla frutescens (Pf MYC, Gong et al., 1999; accession number AB0204051), Gerbera hybrida (Gh MYC1, Elomaa et al., 1998; accession number AJ007709), rice (Os RA,, Hu et al., 1996; accession number U39860), maize (Zm B, Radicella et al., 1991; accession number X57276; Zm LC, Ludwig et al., 1989; accession number M26227; Zm IN, Burr et al., 1996; accession number U57899), and petunia (Ph JAF13, Quattrocchio et al., 1998; accession number AF020543; and Ph AN1, this paper, accession numbers AF260918 and AF260919). The thick bars indicate the standard error in the positions of the branch point.
(C) Intron–exon structures of an1, in1, jaf13, and lc1 (LC). Exons are drawn to scale, and coding sequences are indicated by double height. Introns are not drawn to scale but are presented in such a way that the conserved positions of 3′ acceptor splice sites are aligned. The positions of additional introns that are not conserved in all genes are indicated by the triangles between the diagrams. The regions encoding the conserved N-terminal domains and the bHLH domain are indicated by striped and black boxes, respectively. The open and closed circles denote the start and stop codons, respectively, of the protein coding region.
Database searches showed that the predicted AN1 protein had structural similarities with several plant bHLH proteins, including those encoded by the r1 family (R-S, LC1, SN1, and B1) and the in1 locus of maize, delila of snapdragon, and jaf13 of petunia. Figure 3A shows that AN1 shares a highly conserved N-terminal domain of ∼170 amino acids with the other bHLH proteins. The bHLH domain of AN1, located between positions 474 and 522, is virtually identical to that of IN, with somewhat less similarity to the bHLH domains of DELILA, LC1, and JAF13. The C-terminal region downstream of the bHLH domain is in general less conserved between these proteins. In this region, again AN1 shares the most similarity with IN. The middle domain of the bHLH proteins is the least conserved; the JAF13–DEL pair displays the strongest conservation in this region.
To visualize the quantitative similarity between bHLH proteins implicated in anthocyanin synthesis, we constructed the phylogenetic tree shown in Figure 3B, which indicates that these bHLH proteins fall into different classes. One class includes JAF13, DELILA, GMYC, and MYC1, all of dicot origin, and also RA and the R1 family proteins LC1 and B1, all of monocot origin; AN1 and IN, however, seem to represent at least one and possibly two separate classes of proteins. This classification of bHLH proteins is supported by the intron–exon structures of the corresponding genes, as shown in Figure 3C. The protein coding sequences of b1, lc1, jaf13, and ra contain seven introns in conserved positions (Ludwig et al., 1989; Hu et al., 1996; Quattrocchio et al., 1998), whereas in1 contains an additional intron that splits the region encoding the weakly conserved middle domain (Burr et al., 1996). This extra intron seems to be conserved in an1, although the low sequence similarity in this region makes it impossible to assess whether both introns are in precisely the same position. In addition, an1 lacks the intron that splits the region encoding the bHLH domain but has a unique intron slightly downstream.
Together, these data show that an1 encodes a bHLH protein that is not orthologous to R1, DEL, or JAF13, implying that AN1 is a novel regulator of the anthocyanin pathway.
Spatiotemporal Control of an1 Expression
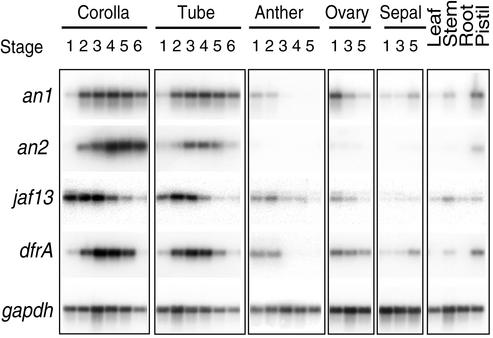
To analyze the expression pattern of the an1 gene, we compared the amount of an1, dfr (an an1-controlled structural anthocyanin gene encoding dihydroflavonol 4-reductase), an2, jaf13, and gapdh (encoding the glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase) mRNA in various tissues. Figure 4 shows that the limb and tube of developing flower corollas contain relatively large amounts of an1 and dfr mRNA, reaching a maximum around stage 4, when the corolla starts to unfold. an1 and dfr mRNA are also clearly detectable in the pistil, consistent with the presence of anthocyanins in the style. In the petunia line that we used (V30), green tissues such as stem, leaf, and sepal can become partially pigmented, especially in older plants, which coincides with the presence of low amounts of an1 and dfr transcripts. The ovary also contains an1 mRNA, consistent with the observation that ovaries express dfr in an an1-dependent manner, even though they do not contain anthocyanins (Huits et al., 1994a). We did not, however, detect an1 transcripts in root, a tissue that never expresses structural anthocyanin genes.
Figure 4.
Spatiotemporal Expression Pattern of an1.
Transcripts of an1, an2, jaf13, dfrA, and gapdh were detected by quantitative RT-PCR in the corolla and tube, anthers, ovaries, and sepals from flowers at various developmental stages and from leaves, stems, roots, and pistils of the wild-type line V30, as indicated above the lanes.
Expression of an2 is essentially limited to the limb, with somewhat less expression in the tube and the pistil, which is consistent with the finding that mutation of an2 affects only pigmentation in the petal limb. These results suggest that an2 function is redundant with and taken over by other loci in distinct tissues (see Quattrocchio et al., 1998, 1999). In contrast, an1 is expressed in all tissues that express dfr and jaf13 (Figure 4). This, combined with the observation that mutation of an1 blocks pigmentation in all tissues (de Vlaming et al., 1984; Quattrocchio et al., 1993), indicates that an1 function is only barely, if at all, redundant.
Forced Expression of AN1 and AN2 Causes Ectopic dfr Transcription
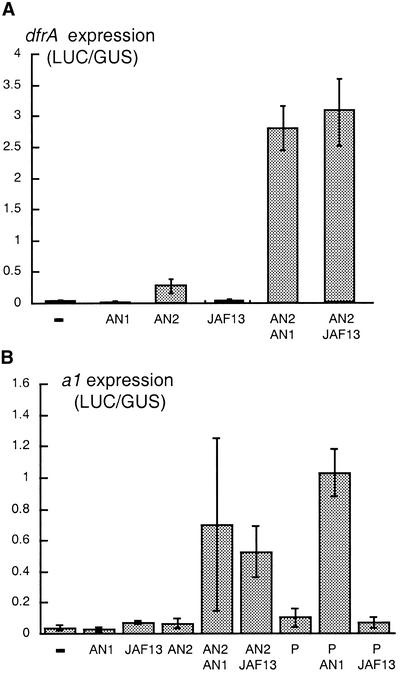
To test whether AN1 is a transcription activator or a repressor (as IN is) and to study how it regulates the tissue-specific expression pattern of dfr, we expressed AN1 ectopically, either alone or in combination with AN2, and examined the activation of the dfr promoter. To this end, the an1 cDNA containing the full 2004-bp open reading frame was fused to the cauliflower mosaic virus 35S promoter, 35S-an1. Similar constructs, 35S-an2 and 35S-jaf13, were used to express AN2 and JAF13 (see Quattrocchio et al., 1998). Different combinations of these regulators were introduced into petunia leaf cells by particle bombardment; their activity was measured by monitoring the expression of a cobombarded luciferase reporter gene that was driven by the dfrA promoter, dfr-luc. Figure 5A shows that expression of either AN1 or JAF13 alone did not activate the dfr promoter, and expression of AN2 caused only weak induction. However, coexpression of AN2 plus AN1 or AN2 plus JAF13 strongly induced the dfr promoter, although coexpression of all three regulators did not further upregulate dfr promoter activity (data not shown).
Figure 5.
Activation of dfr and a1 by Ectopically Expressed AN1, AN2, JAF13, and P.
(A) Activation of dfr-luc by AN1, AN2 and JAF13.
(B) Activation of a1-luc by AN1, AN2, JAF13 and P.
The columns and error bars in (A) and (B) denote the mean and standard error of the activity of the dfr-luc or a1-luc reporter gene after bombardment with 35S-an1, 35S-an2, 35S-jaf13, or 35S-p alone or in combination. Reporter gene activity, measured as luciferase (LUC) enzyme activity, is expressed in arbitrary units and was normalized to GUS enzyme activity expressed from a cobombarded reference gene, 35S-gus, which consisted of the β-glucuronidase gene driven by the 35S promoter.
In maize kernels, the anthocyaninless1 gene (a1) encoding DFR is activated by C1 and R1 in the aleurone, whereas in the pericarp, the activator is P1, a MYB-type transcription factor. In maize suspension cells, forced expression of P1 alone is sufficient to induce transcription of a1 (Grotewold et al., 1994; Bruce et al., 2000), and this induction cannot be enhanced by coexpression of R1; this indicates that P1, unlike C1, functions independent of R1-like bHLH factors (Grotewold, 1995; Sainz et al., 1997).
Combined expression of AN2 and JAF13 could also activate the a1 promoter in particle-bombarded petunia cells (Quattrocchio et al., 1998). Figure 5B shows that the combined expression of AN2 and AN1 or of AN2 and JAF13 activates the a1 promoter with similar efficiency. In these petunia cells, the a1 promoter could also be (weakly) activated by expression of P1 from a cobombarded 35S-p1 transgene. Strikingly, the activation by P1 was enhanced ∼10-fold by coexpression of AN1, whereas JAF13 had no effect (Figure 5B), indicating that P1 responds differently to AN1 and JAF13.
We conclude from these experiments that AN1 is a transcription-activating protein functionally different from JAF13 (and R1) and that it is an important determinant of the tissue-specific expression pattern of dfr and other structural anthocyanin genes.
AN1 Is Directly Involved in Transcription Activation of dfr and myb27
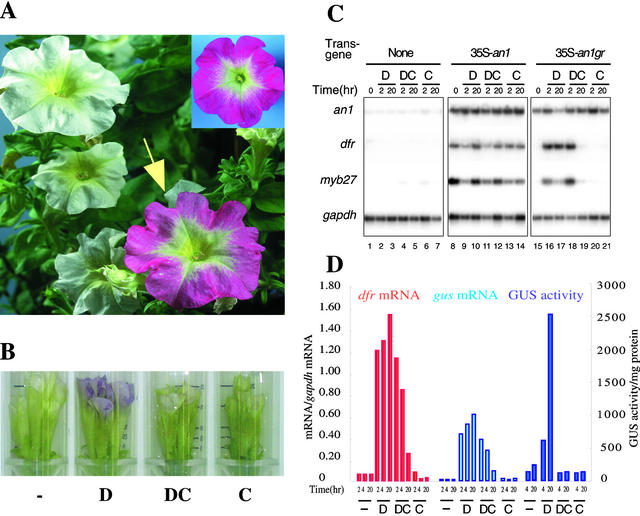
Previous experiments had shown that an1− petals lack transcripts of at least nine structural anthocyanin genes (see, e.g., Quattrocchio et al., 1993; Kroon et al., 1994; de Vetten et al., 1999) and of one potential regulatory gene, Pmyb27, which encodes a protein with similarity to MYB-domain transcription factors (Mur, 1995). Thus, AN1 might induce dfr transcription directly (e.g., as part of the transcription complex on the dfr promoter) or indirectly (by inducing the expression of an intermediate regulator such as MYB27). To distinguish between these possibilities, we constructed the 35S-an1 gr gene, in which the 35S promoter drives the expression of a chimeric protein consisting of the complete AN1 sequence and the ligand binding domain of the rat glucocorticoid receptor (GR), and introduced this gene into the an1− line W242. Treatment with dexamethasone (DEX), a synthetic steroid, was expected to induce AN1GR activity by releasing the protein from an inhibitory cytoplasmic complex rather than by stimulating de novo synthesis. In case protein synthesis were blocked, DEX treatment would still activate transcription of primary target genes but not of the secondary target genes controlled indirectly by AN1. In addition, we generated transgenic W242 plants containing a 35S-an1 transgene or the empty vector.
We obtained 10 transformants harboring 35S-an1, three of which had full-colored flowers indistinguishable from those of the isogenic An1+ line M87, indicating that the transgene fully complemented the an1− mutation. The other seven transformants had white, pale, or spotted flowers, apparently because of poor or irregular expression of the transgene. All 20 primary 35S-an1 gr transformants had white (an1−) flowers similar to those of transgenic plants containing the empty vector. However, dipping single 35S-an1 gr flowers in a solution of DEX led in ∼16 hr to full restoration of anthocyanin synthesis in six of the transgenics (Figure 6A). In the other 14 transformants, DEX treatment had little or no effect on pigmentation, presumably because of poor expression of the 35S-an1 gr transgene.
Figure 6.
Direct Activation of dfr and myb27 by an AN1GR Fusion Protein.
(A) A plant of the an1− line W242 harboring 35S-an1, 20 hr after one floral bud (arrow) had been dipped in 10 μM DEX. For comparison, the inset in the top right corner shows a flower of a transgenic W242 plant complemented with the 35S-an1 transgene.
(B) Phenotype of a transgenic W242 flower harboring 35S-an1 gr after 20 hr of incubation in a test tube containing water (−), 10 μM DEX (D), 10 μM DEX plus 100 μM CHX (DC), or 100 μM CHX (C).
(C) Detection of an1, dfr, myb27, and gapdh transcripts in petal limbs of W242 flowers containing 35S-an1 gr, 35S-an1, or the empty vector after incubation in 10 μM DEX (D), 10 μM DEX plus 100 μM CHX (DC), or 100 μM CHX (C) for 2 or 20 hr. The numbering of the lanes is indicated at the bottom.
(D) dfr-GUS expression in an1− mutant plants harboring 35S-an1 gr. Flowers treated for various periods with DEX or CHX or both, as indicated in (B) and (C), were assayed for gus mRNA, dfr mRNA, and GUS enzyme activity. gus and dfr mRNA amounts, expressed in arbitrary units, were determined by hybridization of RT-PCR products, quantification of the radioactivity by PhosphorImaging, and normalization to the amount of gapdh mRNA.
To study in further detail how AN1 induces pigmentation, we switched to a more easily controlled induction system in which detached flower buds were incubated with specific solutions in a test tube (Figure 6B); we monitored expression of the AN1 target genes by quantitative RT-PCR. Figure 6C shows that in buds containing 35S-an1 gr, the dfr and myb27 mRNAs became detectable within 2 hr after DEX treatment and that after 20 hr, the quantities of these transcripts were comparable to those in fully colored buds harboring the constitutive 35S-an1 transgene. No induction of dfr and myb27 mRNA was seen in buds of transformants harboring the empty vector, confirming that DEX induction requires the AN1GR protein.
Pilot experiments showed that in the presence of 100 μM cycloheximide (CHX), DEX could no longer induce pigment synthesis, indicating that translation was effectively blocked (Figure 6B), whereas at 50 μM or less CHX, DEX-induced pigmentation was still detectable. RT-PCR analysis showed that the presence of 100 μM CHX did not reduce DEX-induced dfr and myb27 mRNA amounts within the first 2 hr (Figure 6C, lane 18). Treatment with CHX alone had no clear effect on dfrA expression but did cause a weak induction of the myb27 mRNA. After 20 hr of treatment with DEX and CHX, the dfr and myb27 mRNAs decreased to barely detectable amounts in 35S-an1 gr flowers (lane 20) but remained high in 35S-an1 flowers (lane 12). We believe that this difference is due to the relatively low stability of the AN1GR fusion protein compared with AN1, combined with the continuous turnover of dfrA and myb27 transcripts during the course of the experiment.
Because it was crucial to assess whether translation was completely blocked under the experimental conditions used, we analyzed the expression of a β-glucuronidase (gus) reporter gene driven by the dfr promoter. We backcrossed an An1+ dfr-gus transgenic plant (Huits et al., 1994a) with a 35S-an1 gr plant (as the recurrent parent) and selected an1− 35S-an1 gr dfr-gus plants from the progeny. Flower buds from these plants were analyzed for induction of dfr and gus expression, as described above. Figure 6D shows that DEX induced dfr and gus mRNA to a similar extent, irrespective of whether CHX was present or not. However, in the presence of CHX, no appreciable GUS enzyme activity was found, confirming that translation was completely blocked.
We conclude from these results that AN1 is a direct activator of dfrA transcription and does not operate by inducing the transcription or translation of a hitherto unknown intermediary regulator.
Regulation of an1 Expression by an2 and an4
Because an1 functions relatively late in the regulatory hierarchy, the question arose whether an2, an4, or an11 also controls pigmentation directly or by regulating an1 expression. To address this question, we examined by RT-PCR the expression of an1, an2, jaf13, and an11 mRNA in various mutants.
The an11 gene is, like an1, required for pigmentation of all tissues, including the petal limb (de Vlaming et al., 1984; Quattrocchio et al., 1993). RT-PCR analysis showed that petal limbs of the isogenic lines R27 (An11+) and W134 (an11−) contained similar amounts of an1 transcripts (data not shown). Because an11-W134 homozygotes lack detectable AN11 protein (de Vetten et al., 1997), this suggests that AN11 is not required for an1 transcription (data not shown).
RT-PCR analysis showed that petal limbs from line W82, which harbors the unstable dTph1 insertion allele an2-W82 (Quattrocchio et al., 1999), and those from derived An2+ germinal revertants contained similar amounts of an1 transcripts (data not shown). However, because an2-W82 is not a null allele, this result is difficult to interpret. Therefore, we analyzed an1 transcripts in an F1 hybrid of the lines W115 and W59, both of which contain frameshift an2 alleles that have completely lost the ability to activate dfrA (Quattrocchio et al., 1999), and compared them with an1 mRNA in complemented W115/W59 plants harboring a 35S-an2 transgene. Figure 7 shows that the 35S-an2 complementants express high amounts of an2 mRNA, which restores dfr expression (lanes 4 to 6) and pigmentation (see Quattrocchio et al., 1998). However, complementation does not affect the amount of an1 (or an11 or jaf13) mRNA, suggesting that an2 is not essential for an1 expression in the petal limb.
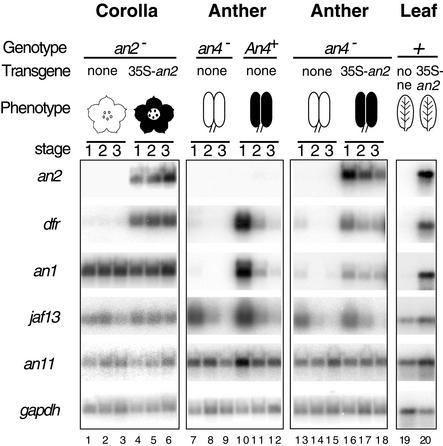
Figure 7.
Genetic Control of an1 Expression.
Transcripts of an2, dfr, an1, jaf13, an11, and gapdh were detected by RT-PCR in different developmental stages of an2− corollas from the hybrid W115/W59 and complementants harboring 35S-an2 (lanes 1 to 6), in anthers from selected an4− and An4+ progeny of the backcross (W138 × V30) × W138 (lanes 7 to 12), in an4− anthers from the hybrid W115/W59 and complementants harboring 35S-an2 (lanes 13 to 18), and in leaves of W115/W59 and W115/W59 plants harboring 35S-an2 (lanes 19 and 20). The presence of anthocyanin pigments in the various tissues is indicated by cartoons. The numbering of the lanes is indicated at bottom.
Mutation of an4 blocks pigmentation of the anthers but not of the corolla limb. Initial experiments showed that anthers of line R27 (an4−) lacked an1 transcripts (data not shown), whereas the transcripts were detectable in anthers of line V30 (An4+) (Figure 4). To “normalize” for differences in the R27 and V30 genetic backgrounds, we analyzed anthers pooled from multiple An1+An4+ and An1+an4− plants obtained by the backcross (W138 × V30) × W138. Figure 7 shows that An4+ anthers express dfr and an1 mRNA, as expected (lanes 10 to 12), whereas both transcripts are downregulated in an4− anthers (lanes 7 to 9). The an4− genotype did not, however, affect expression of an11, jaf13, or gapdh mRNA.
Previous experiments showed that the introduction of a 35S-an2 gene in the an4− line W115 and the hybrid W115/W59 restored pigmentation in the corolla (complementation of an2) as well as the anthers, indicating that ectopically expressed AN2 complements an4 (Quattrocchio et al., 1998). Figure 7 shows that an4− anthers of W115/W59 contain very small amounts of dfr and an1 mRNA (lanes 13 to 15). As expected, dfr mRNA quantities are upregulated again in anthers of the 35S-an2 complementants, although the amount of dfrA mRNA remains less than that in the An4+ V30/W138 plants, possibly because of differences in the genetic background. Surprisingly, the amounts of an1 mRNA in these complemented anthers were restored to the same extent as was dfr (lanes 16 to 18). Moreover, whereas dfrA, an1, and jaf13 mRNAs disappear simultaneously in An4+ anthers when ageing, the expression of dfrA and an1 is prolonged relative to jaf13 in an4− 35S-an2 anthers, apparently because 35S-an2 expression persists during anther development (lanes 16 to 18).
In leaves of transgenic plants, ectopic expression of AN2 activated the expression of dfrA, but not of the early structural genes chsA, chi, and f3h, and increased pigmentation in the veins (Quattrocchio et al., 1998). Figure 7 shows that AN2 also induces the expression of an1 mRNA in this tissue, up to amounts ordinarily found in floral tissues only, whereas AN2 has little or no effect on the expression of jaf13 and an11.
Together, these data indicate that expression of an1 is controlled by an2 and an4 (see also Discussion).
DISCUSSION
AN1 Represents a bHLH Protein That Is Distinct from JAF13, DEL, and R1
The AN1 protein has high similarity with R1 from maize and DEL from snapdragon, particularly in the ∼170–amino acid N-terminal domain and in the bHLH domain. Given the similar phenotypes of an1−, r1−, and del− mutants, the question arises whether an1, r1, and del are orthologous genes. If so, this may imply that jaf13, previously identified by homology to r1 and del and by gain-of-function studies, is another member of the same gene family, possibly with little or no activity. However, numerous findings indicate that this idea is incorrect; rather, they suggest that an1 is a novel regulator and cannot be considered the ortholog of r1 and del. For example, if the many insertions and deletions are ignored, AN1 is more similar to IN than to R1 or DEL (Figure 3). Moreover, R1 is more similar to petunia JAF13 than it is to AN1, and the intron–exon structure of the r1 gene is more similar to that of jaf13 than that of an1. Functional assays also reveal differences between AN1 and JAF13 or R1. First, AN2 can induce expression of an1 but not of jaf13 (Figure 7). Second, activity of the P1 protein can be enhanced by AN1 but not by JAF13 (Figure 5B) or R1 (Grotewold, 1995; Sainz et al., 1997). Third, using two-hybrid screens in yeast, we identified a protein that interacts with the bHLH domain of AN1 but fails to recognize JAF13 in yeast (A. Kroon and R. Koes, unpublished results).
Collectively, these findings argue strongly against an1 being the petunia ortholog of r1 and del; they are better explained by assuming that the common ancestor of monocot and dicot plants had two genes coding for bHLH proteins, one of which is the ancestor of today's r1, delila, and jaf13 and the other of an1.
The interaction between P1 and AN1 in activating the a1 expression in petunia cells may seem surprising, given that P1 controls the synthesis of phlobaphenes, a type of flavonoid pigment not present in petunia flowers. However, the pigment responsible for the brown color of petunia seeds is unknown, although it can be converted into an anthocyanin (delphinidin) by treatment with mineral acid. That its synthesis requires some of the flower pigmentation loci, including an1, an11, and an6 (an6 contains the dfrA gene) (Koes et al., 1990), suggests that it may a be flavonoid polymer such as, for instance, condensed tannins or a phlobaphene. Thus, the interaction of AN1 with P1 may reflect interaction with an unknown MYB protein from petunia involved in pigmentation of the seed coat.
Regulation of an1 Expression
In maize, transcription of c1 is controlled by VIVIPAROUS1 (VP1), a transcriptional regulator that controls multiple aspects of seed maturation, including anthocyanin synthesis, seed dormancy, and expression of amylase genes (Hoecker et al., 1995; Suzuki et al., 1997). Our data on an1 indicate that the anthocyanin-specific regulators also operate in some sort of transcriptional hierarchy.
AN4 is required for expression of an1 in anthers (Figure 7). Several observations indicate that an4 encodes a paralog of AN2 or controls its expression. an2 and an4 mutants have complementary phenotypes; they result in loss of pigmentation in either the petal limb (an2−) or the anthers (an4−) with little or no effect on pigmentation of the seed coat or the flower stem (de Vlaming et al., 1984; Quattrocchio et al., 1993). Moreover, an4 mutants are complemented by ectopic expression of AN2, as judged by the restoration of pigmentation (Quattrocchio et al., 1998) and expression of dfrA and an1 (Figure 7). Finally, we recently identified two new MYB-type proteins that appear, based on sequence homology and expression patterns, to be paralogs of AN2. One of these genes is strongly expressed in An4+ anthers but not in an4− anthers (A. Kroon, C. Spelt, and R. Koes, unpublished data).
In gain-of-function experiments, AN2 (re)activates an1 expression in an4− anthers (complementation of an4) and in leaves, a tissue that normally does not express the anthocyanin pathway. This effect of AN2 appears to be specific for an1, because structural anthocyanin genes such as chs, chi, and an3, which are regulated independent of an1, an2, an4 and an11, are not activated in 35S-an2 leaves (Quattrocchio et al., 1998); moreover, AN2 does not have a clear effect on the expression of jaf13 or an11 (Figure 7).
The conclusion that like an4, an2 is a regulator of an1 expression seems inconsistent with the finding that an1 transcripts are still expressed in an2− petal limbs. This discrepancy can be explained in at least two ways. First, the an2− alleles of W115 and W59 express roughly wild-type amounts of mRNA and encode a protein that is truncated immediately after the MYB domain (Quattrocchio et al., 1999). Although these an2− alleles fail to activate dfrA transcription, even when expressed from the strong 35S promoter (Quattrocchio et al., 1999), the truncated AN2 protein might still be stable and capable of activating an1 expression. Second, the function of an2 seems redundant, even in the petal limb, because an2− petal limbs are pale rather than completely white and continue to express small amounts of structural anthocyanin mRNAs. The residual transcription activity presumably is provided by one of the above-mentioned AN2 paralogs, which may be sufficient for full activation of an1 and only partial activation of the structural anthocyanin genes (which are expressed much more strongly than an1).
Despite several attempts, we have not been able to solve this apparent discrepancy. Experiments to test whether AN2 induces dfrA transcription indirectly (by way of an1) through expressing a DEX-regulated AN2GR protein in transgenic plants were unsuccessful because the AN2GR fusion protein failed to complement an2 and an4 mutations (apparently AN2GR is unstable or nonfunctional). Also, transient assays designed to test whether a truncated AN2 protein could activate an1 transcription failed because a luc reporter gene fused to an 1.5-kb fragment upstream from the an1 was transcriptionally inactive. Whether this is because important cis-acting regulatory elements of an1 are located outside the promoter region or because this region of an1 represents a large intron rather than the promoter is unclear at this stage.
Activation of Structural Anthocyanin Genes
Although the first regulatory anthocyanin genes, c1 and r1, were isolated >10 years ago, we still know very little about the mechanism by which they activate the structural anthocyanin genes. In vitro DNA binding studies showed that C1 can bind to functional cis-acting elements in the promoters of the a1 and a2 genes (Sainz et al., 1997; Lesnick and Chandler, 1998). However, because C1 binds with relatively low affinity to a1 (Sainz et al., 1997) and because multiple MYB-domain proteins may be involved in the activation of structural anthocyanin genes (see Solano et al., 1995; Quattrocchio et al., 1999), it is difficult to judge whether the DNA–protein interactions observed in vitro are relevant in vivo. For R1, neither direct DNA binding nor an enhancement of the DNA binding capacity of C1 could be demonstrated, even though R1 is known to be required for transcriptional activation by C1. Because nearly all available data are from genetic experiments, it is unclear whether the identified regulators activate the structural anthocyanin genes directly or are part of a more complex regulatory hierarchy in which they regulate the structural genes in an indirect manner. Obviously, such information is important for the correct interpretation of the in vitro analyses of DNA–protein interactions.
Our results show that AN1 is both an activator of transcription (not an inhibitor as IN is) and a key factor in determining the expression domain of dfr (Figure 5). In transient assays, expression of AN1 together with AN2 is sufficient to activate the dfrA promoter in leaf cells. Surprisingly, the function of AN1 can be fully replaced by JAF13 in such assays, even though the phenotype of an1− petals—which still normally express jaf13 mRNA (Quattrocchio et al., 1998)—indicates that ordinarily JAF13 cannot replace AN1. Most likely, AN1 and JAF13 have different affinities for partner proteins or target DNA sequences, or both—subtle differences that are not recognizable when these proteins are produced in excess from a transgene driven by the strong 35S promoter.
In transient assays, expression of AN2 causes only weak activation of dfrA-luc or dfrA-gus reporter genes, and coexpression of a bHLH protein such as JAF13, AN1, or R1 is required for full activation of dfrA (Figure 4; Quattrocchio et al., 1998). In stable transformants, however, ectopic expression of AN2 alone is sufficient for a strong induction of dfr mRNA in leaves, up to amounts that ordinarily are seen in pigmented floral tissues only (Figure 7), and the effect of AN2 is enhanced only slightly by coexpression of JAF13 (Quattrocchio et al., 1998). These seemingly conflicting results are probably attributable to the different time spans of the experiments. In stable transformants, ectopic expression of AN2 induces the expression of AN1 (Figure 7) and possibly of other regulatory proteins required for subsequent activation of dfr. In transient assays, a period of only 20 hr elapses between the introduction of DNAs and the assay of reporter gene activity, which may be too little time for AN2 to induce expression of sufficient AN1 for activation of dfr. However, if 35S-an1 is cointroduced, AN1 starts to accumulate immediately, resulting in a stronger expression of the dfr reporter gene during the 20 hr of the experiment.
The experiments in which the localization and activity of an AN1GR fusion protein could be controlled post-translationally strongly indicate that AN1 is a direct regulator of dfr and does not operate by activating the transcription/translation of an intermediate regulatory gene (Figure 7). The same strategy was used to demonstrate the direct regulation of apetala1 by LEAFY and of nap (nam-homolog activated by apetala3) by APETALA3 in floral meristems of Arabidopsis (Sablowski and Meyerowitz, 1998; Wagner et al., 1999). However, our results do not necessarily imply that AN1 contacts the dfr promoter directly. AN1 may well be recruited to the transcription complex by protein–protein interactions only (as an adapter or coactivator), particularly because the putative DNA binding “basic” region upstream of the HLH domain in AN1 (as well as in JAF13, R1, DEL, and homologous genes from other plants) contains only a few basic amino acids in comparison with mammalian bHLH proteins with a proven DNA binding capacity.
METHODS
Petunia Lines and Mutants
The petunia lines R27 (an4−), W138 (an1-W138, an4−), and W137 (an1-W137, an4−) are all from the same genetic background (R27). The line W242 contains a null allele of an1, which was obtained by the insertion and excision of a dTph1 element in progeny of the An1+ line M87 (C. Spelt, F. Quattrocchio, J.N.M. Mol, and R. Koes, unpublished data). Details on line W82 harboring the unstable an2-W82 allele, the derived An2+ revertants, and the an2−an4− lines W115 and W59 have been described by Quattrocchio et al. (1999). Lines V30, V23, M87 (all wild type for all an genes), W82, W115, W59, and R27 are all from different genetic backgrounds. To obtain An4+ and an4− anther tissue in a normalized genetic background, we used the backcross (W138 × V30) × W138. B1 plants that were An1+ (scored from coloration of the petals) and Hf1+ (determined by thin-layer chromatographic analysis of petal anthocyanins) were selected, and anthers from multiple An4+ (blue/purple anthers) or an4− (yellow anthers) plants were pooled for RNA isolation. All plants were grown in a normal greenhouse.
Molecular Analysis of the an1 Locus
DNA prepared from an an1-W138 homozygous plant was digested with RsaI, diluted, and circularized with T4 ligase. dTph1 flanking sequences were amplified by inverse polymerase chain reaction (PCR), using dTph1 specific primers and cloned into M13 phage to obtain a library of dTph1 flanking sequences (Souer et al., 1995). Duplicate plaque lifts, taken on Hybond-N membranes (Amersham), were hybridized to 32P-labeled dTph1 flanking sequences generated by inverse PCR amplification of circularized RsaI-digested DNA from a second an1-W138 homozygote (+ probe) or a plant homozygous for an An1+ revertant allele (− probe). Two hundred differentially hybridizing plaques were isolated and grouped in ∼20 classes by cross-hybridization and sizing of the insert. Representative clones of each class were hybridized to dot blots containing inverse PCR–amplified dTph1 flanking sequences from four an1-W138 unstable plants and from plants homozygous for four independent An1+ revertant alleles. Clone 65 produced the hybridization pattern expected for an an1 fragment, and further DNA gel blot and PCR analyses confirmed that it contained part of the an1 locus (Figure 2).
Genomic and cDNA clones representing the an1 locus and mRNA were isolated by hybridization of clone 65 to a cDNA library prepared from petal RNA and a genomic library of an An1+ line (R27). Nested deletions of cDNA and genomic clones were sequenced by the dideoxychain termination method, using fluorescent primers complementary to the cloning vector and an Applied Biosystems (Foster City, CA) DNA sequencer (model 370A). Sequences were analyzed with the program Geneworks (Intelligenetics, Mountain View, CA), and alignments were optimized by hand.
Construction of Transgenes and Plant Transformation
The 35S-an1 gene was constructed by ligation of the 2.45-kb an1 cDNA between the 35S promoter of cauliflower mosaic virus and the 3′ end of the nopaline synthase gene of plasmid PIP M1. For construction of 35S-an1 gr, primers 1.20 and 1.34 were used to amplify the complete coding region (without stop codon) of an1; this fragment was inserted into PIP M1, together with a fragment encoding the ligand binding domain (amino acids 508 to 795) of the glucocorticoid receptor (GR) obtained from the plasmid pBI-ΔGR (Lloyd et al., 1994). Details of the genes 35S-jaf13, 35S-an2, dfrA-gus, dfrA-luc, and 35S-P1 can be found elsewhere (Grotewold et al., 1994; Huits et al., 1994a; de Vetten et al., 1997; Quattrocchio et al., 1998).
For plant transformation, 35S-an1 and 35S-an1 gr were inserted into the binary T-DNA vector pBIV M2 and introduced into the an1− line W242 by leaf-disc transformation. Transgenic plants containing 35S-an2 were generated by Agrobacterium-mediated leaf-disc transformation of F1 plants from the cross W115 × W59.
Transient transformation of petunia leaf cells was performed by particle bombardment, as described previously (de Vetten et al., 1997; Quattrocchio et al., 1998).
Dexamethasone Induction Experiments
Transgenic plants expressing the AN1GR fusion protein were selected by dipping stage 4 floral buds in 10 μM dexamethasone (DEX) and visually inspecting for pigment synthesis after ∼16 to 20 hr, when the flower had opened.
For treatments with DEX or cycloheximide (CHX), or both, stage 4 floral buds were taken from the plants and incubated in transparent 50-mL test tubes with 10 mL of water containing 10 μM DEX or 100 μM CHX (or both) on a rocking and rolling test tube mixer. This ensured that the tissues were continuously exposed to the solution without being completely submerged.
Expression Analyses
RNA isolation, RNA gel blot analyses, and reverse transcription (RT)–PCR were performed as described previously (Quattrocchio et al., 1993; de Vetten et al., 1997). In comparing mRNA quantities in different genotypes, care was taken that the analyzed tissues were harvested simultaneously. Radioactive hybridization products were detected and quantified by using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Transient expression assays were performed by particle bombardment of petunia W115 leaves, as described previously (de Vetten et al., 1997; Quattrocchio et al., 1998)
Acknowledgments
We thank Jan Kooter for commenting on the manuscript, Erich Grotewold for a gift of the 35S-p1 construct, Alan Lloyd for a gift of pBI-ΔGR, and Daisy Kloos, Pieter Hoogeveen, and Martina Meesters for their care of the plants. This research was sponsored in part by the Netherlands Organisation for Scientific Research and by a grant from the European Union (No. BIO4-CT98-0432).
References
- Alfenito, M.R., Souer, E., Goodman, C.D., Buell, R., Mol, J., Koes, R., and Walbot, V. (1998). Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell 10, 1135–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.W., Smith, P., and Simpson, C.G. (1996). Arabidopsis consensus intron sequences. Plant Mol. Biol. 32, 531–535. [DOI] [PubMed] [Google Scholar]
- Bruce, W., Folkerts, O., Garnaat, C., Crasta, O., Roth, B., and Bowen, B. (2000). Expression profiling of the maize flavonoid pathway genes controlled by estradiol-inducible transcription factors CRC and P. Plant Cell 12, 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr, F.A., Burr, B., Scheffler, B.E., Blewitt, M., Wienand, U., and Matz, E.C. (1996). The maize repressor-like gene intensifier1 shares homology with the r1/b1 multigene family of transcription factors and exhibits missplicing. Plant Cell 8, 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vetten, N., Quattrocchio, F., Mol, J., and Koes, R. (1997). The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants and animals. Genes Dev. 11, 1422–1434. [DOI] [PubMed] [Google Scholar]
- de Vetten, N., ter Horst, J., van Schaik, H.-P., den Boer, B., Mol, J., and Koes, R. (1999). A cytochome b5 is required for full activity of flavonoid 3′5′-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc. Natl. Acad. Sci. USA 96, 778–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vlaming, P., Cornu, A., Farcy, E., Gerats, A.G.M., Maizonnier, D., Wiering, H., and Wijsman, H.J.W. (1984). Petunia hybrida: A short description of the action of 91 genes, their origin and their map location. Plant Mol. Biol. 2, 21–42. [Google Scholar]
- Doodeman, M., Boersma, E.A., Koomen, W., and Bianchi, F. (1984). Genetic analysis of instability in Petunia hybrida 1. A highly unstable mutation induced by a transposable element inserted at the An1 locus for flower colour. Theor. Appl. Genet. 67, 345–355. [DOI] [PubMed] [Google Scholar]
- Elomaa, P., Mehto, M., Kotilainen, M., Helariutta, Y., Nevalainen, L., and Teeri, T.H. (1998). A bHLH transcription factor mediates organ, region and flower type specific signals on dihydroflavonol-4-reductase (dfr) gene expression in the inflorescence of Gerbera hybrida (Asteraceae). Plant J. 16, 93–99. [DOI] [PubMed] [Google Scholar]
- Goff, S.A., Cone, K.C., and Chandler, V.L. (1992). Functional analysis of the transcription activator encoded by the maize B-gene: Evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 6, 864–875. [DOI] [PubMed] [Google Scholar]
- Gong, Z.Z., Yamagishi, E., Yamazaki, M., and Saito, K. (1999). A constitutively expressed Myc-like gene involved in anthocyanin biosynthesis from Perilla frutescens: Molecular characterization, heterologous expression in transgenic plants and transactivation in yeast cells. Plant Mol. Biol. 41, 33–44. [DOI] [PubMed] [Google Scholar]
- Goodrich, J., Carpenter, R., and Coen, E.S. (1992). A common gene regulates pigmentation pattern in diverse plant species. Cell 68, 955–964. [DOI] [PubMed] [Google Scholar]
- Grotewold, E. (1995). Does P protein require a partner as C1 protein does? Maize Genet. Coop. Newsl. 69, 32. [Google Scholar]
- Grotewold, E., Drummond, B.J., Bowen, B., and Peterson, T. (1994). The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76, 543–553. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., Vasil, I.K., and McCarty, D.R. (1995). Integrated control of seed maturation and germination programs by activator and repressor functions of Viviparous-1 of maize. Genes Dev. 9, 2459–2469. [DOI] [PubMed] [Google Scholar]
- Holton, T.A., and Cornish, E.C. (1995). Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7, 1071–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J., Anderson, B., and Wessler, S. (1996). Isolation and characterisation of rice genes: Evidence for distinct evolutionary paths in rice and maize. Genetics 142, 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huits, H.S.M., Gerats, A.G.M., Kreike, M.M., Mol, J.N.M., and Koes, R.E. (1994. a). Genetic control of dihydroflavonol 4-reductase gene expression in Petunia hybrida. Plant J. 6, 295–310. [DOI] [PubMed] [Google Scholar]
- Huits, H.S.M., Koes, R.E., Wijsman, H.J.W., and Gerats, A.G.M. (1994. b). Genetic characterization of Act1, the activator of a non-autonomous transposable element from Petunia hybrida. Theor. Appl. Genet. 91, 110–117. [DOI] [PubMed] [Google Scholar]
- Koes, R.E., Van Blokland, R., Quattrocchio, F., Van Tunen, A.J., and Mol, J.N.M. (1990). Chalcone synthase promoters in petunia are active in pigmented and unpigmented cell types. Plant Cell 2, 379–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon, J., Souer, E., de Graaff, A., Xue, Y., Mol, J., and Koes, R. (1994). Cloning and structural analysis of the anthocyanin pigmentation locus Rt of Petunia hybrida: Characterization of insertion sequences in two mutant alleles. Plant J. 5, 69–80. [DOI] [PubMed] [Google Scholar]
- Kubo, H., Peeters, A.J., Aarts, M.G., Pereira, A., and Koornneef, M. (1999). ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell 11, 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesnick, M.L., and Chandler, V.L. (1998). Activation of the maize anthocyanin gene a2 is mediated by an element conserved in many anthocyanin promoters. Plant Physiol. 117, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, A.M., Walbot, V., and Davis, R.W. (1992). Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258, 1773–1775. [DOI] [PubMed] [Google Scholar]
- Lloyd, A.M., Schena, M., Walbot, V., and Davis, R. (1994). Epidermal cell fate determination in Arabidopsis: Patterns defined by a steroid-inducible regulator. Science 266, 436–439. [DOI] [PubMed] [Google Scholar]
- Ludwig, S.R., Habera, L.F., Dellaporte, S.L., and Wessler, S.R. (1989). Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc. Natl. Acad. Sci. USA 86, 7092–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, C., Prescott, A., Mackay, S., Bartlett, J., and Vrijlandt, E. (1991). Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant J. 1, 37–49. [DOI] [PubMed] [Google Scholar]
- Mol, J.N.M., Holton, T.A., and Koes, R.E. (1995). Floriculture: Genetic engineering of commercial traits. Trends Biotechnol. 13, 350–355. [Google Scholar]
- Mol, J., Grotewold, E., and Koes, R. (1998). How genes paint flowers and seeds. Trends Plant Sci. 3, 212–217. [Google Scholar]
- Mur, L. (1995). Characterization of Members of the myb Gene Family of Transcription Factors from Petunia hybrida. PhD Dissertation (Amsterdam, The Netherlands: Vrije Universiteit).
- Quattrocchio, F., Wing, J.F., Leppen, H.T.C., Mol, J.N.M., and Koes, R.E. (1993). Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 5, 1497–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio, F., Wing, J.F., van der Woude, K., Mol, J.N.M., and Koes, R. (1998). Analysis of bHLH and MYB-domain proteins: Species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 13, 475–488. [DOI] [PubMed] [Google Scholar]
- Quattrocchio, F., Wing, J., van der Woude, K., Souer, E., de Vetten, N., Mol, J., and Koes, R. (1999). Molecular analysis of the anthocyanin2 gene of Petunia and its role in the evolution of flower color. Plant Cell 11, 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radicella, J.P., Turks, D., and Chandler, V.L. (1991). Cloning and nucleotide sequence of a cDNA encoding B-Peru, a regulatory protein of the anthocyanin pathway from maize. Plant Mol. Biol. 17, 127–130. [DOI] [PubMed] [Google Scholar]
- Sablowski, R.W., and Meyerowitz, E.M. (1998). A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92, 93–103. [DOI] [PubMed] [Google Scholar]
- Sainz, M., Grotewold, E., and Chandler, V. (1997). Evidence for direct activation on an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb-domain proteins. Plant Cell 9, 611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger, D.A., and Chandler, V.L. (1999). A mutation in the pale aleurone color1 gene identifies a novel regulator of the maize anthocyanin pathway. Plant Cell 11, 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley, B.W. (1996). Flavonoid biosynthesis: “New” functions for an “old” pathway. Trends Plant Sci. 1, 363–402. [Google Scholar]
- Sokal, R.R., and Michener, C.D. (1958). A statistical method for evaluating systematic relationships. Univ. Kans. Sci. Bull. 38, 1409–1438. [Google Scholar]
- Solano, R., Nieto, C., Avila, J., Cañas, L., Diaz, I., and Paz-Ares, J. (1995). Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (Myb.Ph3) from Petunia hybrida. EMBO J. 14, 1773–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer, E., Quattrocchio, F., de Vetten, N., Mol, J.N.M., and Koes, R.E. (1995). A general method to isolate genes tagged by a high copy number transposable element. Plant J. 7, 677–685. [DOI] [PubMed] [Google Scholar]
- Suzuki, M., Kao, C.Y., and McCarty, D.R. (1997). The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, D., Sablowski, R.W., and Meyerowitz, E.M. (1999). Transcriptional activation of APETALA1 by LEAFY. Science 285, 582–584. [DOI] [PubMed] [Google Scholar]
- Walker, A.R., Davison, P.A., Bolognesi-Winfield, A.C., James, C.M., Srinivasan, N., Blundell, T.L., Esch, J.J., Marks, M.D., and Gray, J.C. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11, 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshaar, B., and Jenkins, G.I. (1998). Phenylpropanoid biosynthesis and its regulation. Curr. Opin. Plant Biol. 1, 251–257. [DOI] [PubMed] [Google Scholar]