Abstract
The kinases responsible for phosphorylation of inositol-containing lipids are essential for many aspects of normal eukaryotic cell function. Genetic and biochemical studies have established that the phosphatidylinositol (PtdIns) 3-kinase encoded by the yeast VPS34 gene is essential for the efficient sorting and delivery of proteins to the vacuole; the kinase encoded by the human VPS34 homolog has been equally implicated in the control of intracellular vesicle traffic. The plant VPS34 homolog also is required for normal growth and development, and although a role for PtdIns 3-kinase in vesicle trafficking is likely, it has not been established. In this study, we have shown that considerable PtdIns 3-kinase activity is associated with the internal matrix of nuclei isolated from carrot suspension cells. Immunocytochemical and confocal laser scanning microscopy studies using the monoclonal antibody JIM135 (John Innes Monoclonal 135), raised against a truncated version of the soybean PtdIns 3-kinase, SPI3K-5p, revealed that this kinase appears to have a distinct and punctate distribution within the plant nucleus and nucleolus. Dual probing of root sections with JIM135 and anti–bromo-UTP antibodies, after in vitro transcription had been allowed to proceed in the presence of bromo-UTP, showed that SPI3K-5p associates with active nuclear and nucleolar transcription sites. These findings suggest a possible link between PtdIns 3-kinase activity and nuclear transcription in plants.
INTRODUCTION
Since their discovery in the early to mid-1980s, the enzymes responsible for D-3 phosphorylation of phosphoinositides, that is, the phosphatidylinositol (PtdIns) 3-kinases, have been the subject of intense study (reviewed in Carpenter and Cantley, 1996). The PtdIns 3-kinases are now known to be involved in a plethora of cellular processes, ranging from mitogenesis, membrane trafficking and ruffling to glucose uptake, oxidative burst responses, chemotaxis, and apoptosis.
Several distinct PtdIns 3-kinase isoforms are known to exist in eukaryotic cells. The currently identified PtdIns 3-kinases can be divided broadly into four families based on sequence homology and their preferred inositol lipid substrate or substrates. However, only one family has been found in all the eukaryotic cells studied so far, namely, the PtdIns-specific Vps34p-related 3-kinases. The other families presumably are more organism specific and have evolved later from a common primordial PtdIns 3-kinase family. Despite the high degree of sequence homology between members of this family and the other PtdIns 3-kinases, the Vps34p-related kinases all have the distinguishing biochemical feature that they phosphorylate only PtdIns, both in vivo and in vitro, and are unable to utilize PtdIns 4-monophosphate (PtdIns(4)P) or PtdIns 4,5-bisphosphate as substrates. The VPS34 gene product, Vps34p, was first identified in a screen of yeast mutants defective in vesicle sorting (Stack and Emr, 1994). Subsequently, a human homolog was identified, and Vps34p-related PtdIns 3-kinases now are known to exist in a range of other eukaryotes, including Dictyostelium (Zhou et al., 1995), Drosophila (Linassier et al., 1997), and plants (Hong and Verma, 1994; Welters et al., 1994). Studies of yeast have shown that Vps34p—or perhaps more precisely its product, PtdIns 3-phosphate (PtdIns(3)P)—plays an essential role in vesicle-mediated delivery of vacuolar enzymes, a finding that has led to the general view that the primary function of Vps34p-related 3-kinases is to control specific reactions during vesicle trafficking events. This view is supported by two studies of the plant PtdIns 3-kinases showing that specific induction and repression of PtdIns 3-kinase gene expression during root nodule formation are correlated directly with the degree of membrane proliferation (Hong and Verma, 1994) and that the PtdIns 3-kinase inhibitor, wortmannin, also inhibits at least one type of vacuolar sorting (Matsuoka et al., 1995). That the PtdIns 3-kinase is essential for normal plant growth was demonstrated by Welters et al. (1994), who showed that the expression of the Arabidopsis PtdIns 3-kinase (AtVPS34) antisense constructs led to second-generation transformed plants with very severely affected phenotypes. Although most current data thus lend support to the hypothesis that PtdIns 3-kinase is involved in vesicle trafficking in plant cells, we previously demonstrated that a substantial proportion of PtdIns 3-kinase activity in plant cells is associated with a detergent-resistant nucleocytoskeletal compartment (Dove et al., 1994). However, the methods for preparing plant nucleocytoskeletal fractions involve manipulations known to greatly upregulate the plant phosphoinositide signaling system (Drøbak and Watkins, 1994), so the potential exists for signal-induced translocation of the PtdIns 3-kinase to the cytoskeleton/nucleus in such studies.
To further investigate the subcellular distribution and possible function of the plant PtdIns 3-kinase, we expressed a truncated version of the soybean PtdIns 3-kinase, SPI3K-5, in Escherichia coli and raised monoclonal antibodies against the recombinant enzyme. Using several approaches, including immunocytochemistry and confocal microscopy, we found that the plant PtdIns 3-kinase (SPI3K-5p) is catalytically active in plant nuclei and appears to be intimately associated with active nuclear/nucleolar transcription sites. This suggests that PtdIns 3-kinases, at least in plant cells, may play a novel and hitherto unsuspected role in the transcriptional process.
RESULTS
To investigate the possible presence and localization of PtdIns 3-kinases in plant nuclei, we developed a new method for isolating the plant cell nuclei without using detergents. Figure 1 shows an autoradiogram obtained from thin-layer chromatography (TLC) separation of radiolabeled phospholipids isolated from nuclei incubated in the presence of γ-33P-ATP, in either the absence or the presence of exogenously added PtdIns liposomal lipid substrate. Using the borate system of Walsh et al. (1991), one can separate PtdIns(3)P and PtdIns(4)P by one-dimensional TLC because of the formation of PtdIns(4)P–borate complexes. The data in Figure 1 (lane 1) show that both PtdIns(3)P and PtdIns(4)P can be synthesized in roughly equal quantities in non-detergent-treated plant nuclei and that sufficient endogenous nuclear PtdIns is present to sustain both PtdIns 3- and PtdIns 4-kinase activity. Addition of exogenous lipid substrate (PtdIns) results in a marked increase in the formation of radiolabeled PtdIns monophosphates, with PtdIns(4)P showing the greatest relative increase in rate of synthesis (lane 2).
Figure 1.
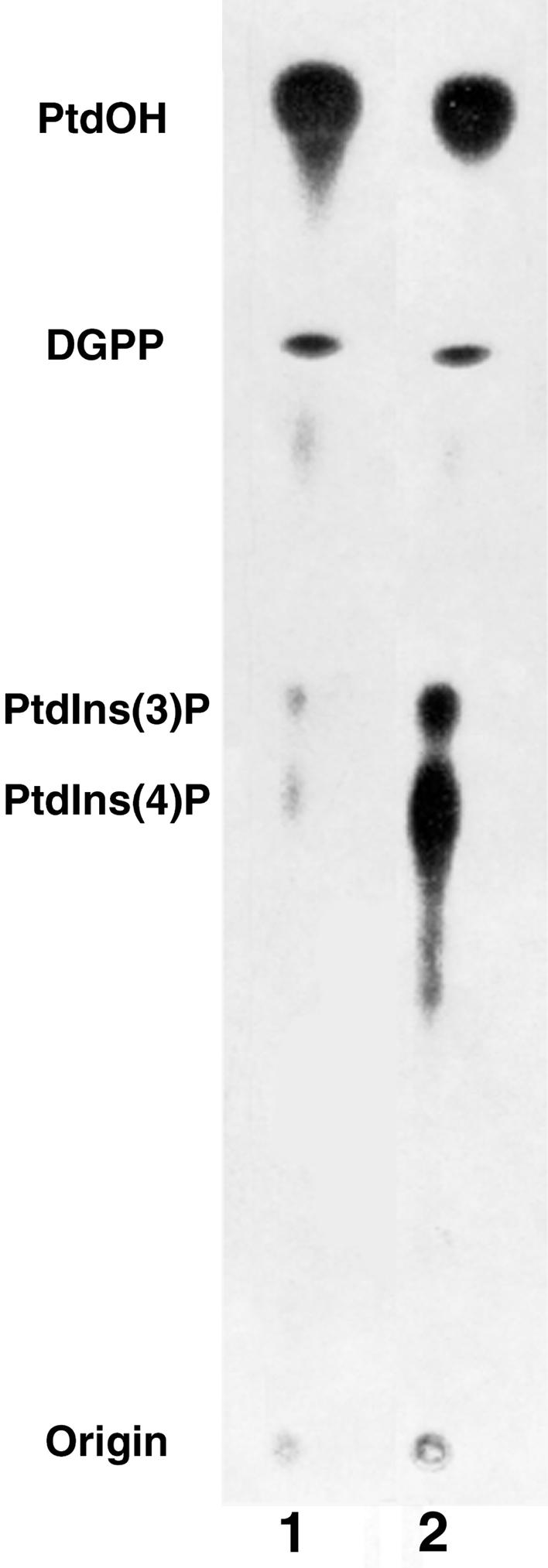
Autoradiograph of 33P-Labeled Lipids Isolated from Plant Nuclei and Separated by One-Dimensional TLC.
A borate-based separation system was used to allow separation of PtdInsP isomers. Nuclei were incubated with γ-33P-ATP either in the absence of exogenous PtdIns (lane 1) or in the presence of added exogenous liposomal PtdIns (lane 2). Kinase assays were terminated after 10 min at 25°C. DGPP, diacylglycerolpyrophosphate.
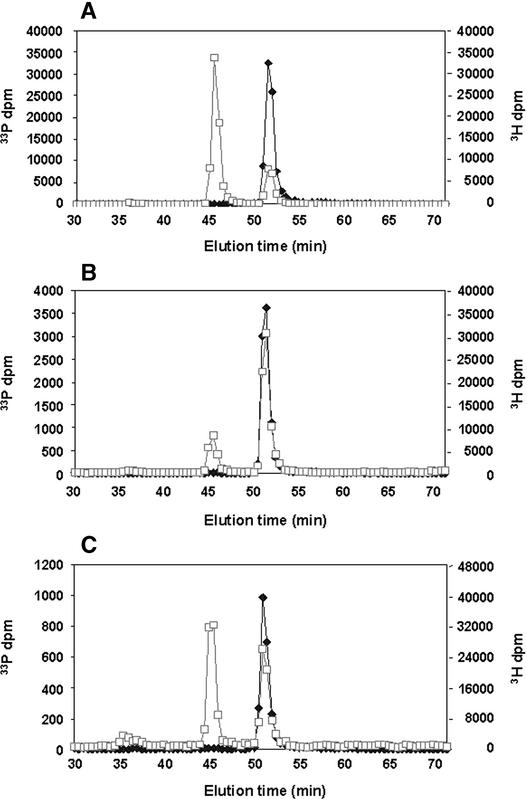
To verify the isomeric configuration of the two radiolabeled lipid products tentatively identified as PtdIns(4)P and PtdIns(3)P in Figure 1, and to gain additional information about their relative abundance, radiolabeled lipids were recovered from the TLC plate and deacylated, and the resulting glyceroPtdIns (GroPtdIns) phosphate products were analyzed by HPLC. The results of these experiments are shown in Figure 2. Because radiolabeled GroPtdIns(3)P was not commercially available at the time of this study, we prepared 33P-labeled authentic GroPtdIns(3)P by performing lipid kinase assays with exogenous PtdIns as the substrate and homogenates from Sf21 cells expressing SPI3K-5p as the enzyme source. Figure 2A shows the elution profile of authentic 33P-GroPtdIns(3)P and 3H-GroPtdIns(4)P. Note that a small peak of 33P-GroPtdIns(4)P is also present, reflecting native PtdIns 4-kinase activity in Sf21 cells.
Figure 2.
HPLC Separation of 33P-Labeled GroPtdInsP Derived from PtdIns 3-Kinase Assays Performed in the Presence of Exogenous PtdIns.
γ-33P-ATP–labeled lipids were separated by TLC. The bands corresponding to PtdIns monophosphates were recovered and subjected to deacylation and HPLC analysis.
(A) HPLC profiles for deacylated products obtained from lysates of insect cells expressing the soybean PtdIns 3-kinase, SPI3K-5p, in a baculovirus expression construct and authentic GroPtdIns(4)P.
(B) and (C) Separations of GroPtdInsP derivatives obtained from nuclei isolated in the absence of detergent (B) and from nuclei pretreated with 0.4% (v/v) Triton X-100 (C). Authentic 3H-GroPtdIns(4)P was included as an internal standard in all separations. 33P, squares; 3H, diamonds.
Figure 2B illustrates the HPLC profile of GroPtdInsP derivatives obtained from nuclei isolated in the absence of detergent and assayed in the presence of exogenous PtdIns. Peaks corresponding to both GroPtdIns(3)P and GroPtdIns(4)P are evident. The radiolabeling of GroPtdIns(4)P is approximately three- to fourfold higher than in GroPtdIns(3)P, indicating that the 4-kinase activity in intact nuclei dominates over 3-kinase activity when assayed with exogenous substrates. In contrast, Figure 2C shows the HPLC profile of various GroPtdInsP forms obtained from nuclei treated with 0.4% (v/v) Triton X-100 and kinase activity assayed in the presence of exogenous PtdIns. Here, the radiolabeling of GroPtdIns(3)P now supersedes that of GroPtdIns(4)P. These results suggest that most PtdIns 4-kinase activity in plant nuclei is present in the nuclear membranes; this view agrees with studies of mammalian cells by Payrastre et al. (1992), who also demonstrated that PtdIns 4-kinase activity was found almost exclusively in the nuclear membranes. The fact that the amounts of GroPtdIns(3)P present remain almost unchanged by the detergent treatment suggests that PtdIns 3-kinase is associated with detergent-resistant structures or with internal regions of the nucleus that are inaccessible to detergents. Simple detergent effects on enzyme activity can be ruled out because Triton X-100 was removed before the kinase assays were performed. Irvine and Divecha (1992) established that mammalian nuclei possess a partially autonomous phosphoinositide system; components of the plant phosphoinositide system also have been demonstrated in the nucleus. That plant nuclei possess PtdIns 4-kinase activity has been shown by Hendrix et al. (1989), and that a small proportion of this activity is detergent resistant also has been demonstrated (Xu et al., 1992). Although we previously found a detergent-resistant PtdIns 3-kinase associated with cytoskeletal complexes, which also contain nuclear components, the results shown in Figure 2 directly demonstrate the presence of a catalytically active detergent-resistant PtdIns 3-kinase in isolated plant nuclei.
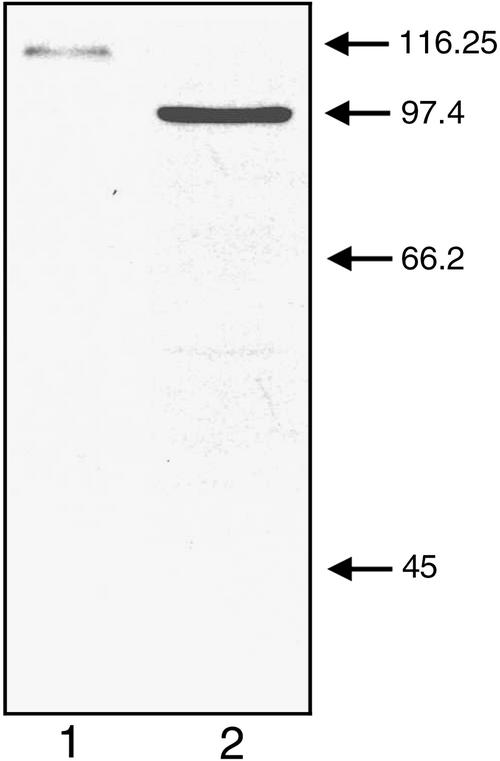
To further study the subnuclear distribution of the PtdIns 3-kinase enzyme, a monoclonal antibody (named John Innes Monoclonal 135, or JIM135) was raised against a truncated form of the soybean PtdIns 3-kinase expressed in E. coli. This antibody recognizes the truncated E. coli–expressed peptide (data not shown). The protein gel blot seen in Figure 3 shows that JIM135 also recognizes the full-length PtdIns 3-kinase (SPI3K-5p) expressed in Sf21 cells (lane 2) as well as a single peptide (of 111 kD) obtained from a whole-cell protein extract from soybean roots (lane 1). The observed difference in relative electrophoretic mobility of the Sf21-expressed and the native soybean SPI3K-5p may have several explanations. As pointed out by King and Possee (1992), the extent of post-translational modification of foreign proteins expressed in Sf21 cells can be difficult to predict and may in many cases give rise to apparent differences between the relative electrophoretic mobility of native and Sf21-expressed proteins when analyzed by SDS-PAGE. We also have preliminary evidence suggesting that SPI3K-5p can be phosphorylated (data not shown), whereas the potential for in vivo phosphorylation in insect cells remains questionable.
Figure 3.
Gel Blot of Proteins Extracted from Soybean Root Tissue and Sf21 Cells Expressing the Full-Length Soybean PtdIns 3-Kinase (SPI3K-5p).
Lane 1 contains proteins isolated from soybean root tissue; lane 2 shows protein isolated from Sf21 cells expressing full-length SPI3K-5p. Blots were probed with the anti-PtdIns 3-kinase antibody JIM135 as described in the text. Arrows indicate the position of authentic relative molecular mass markers (in kilodaltons).
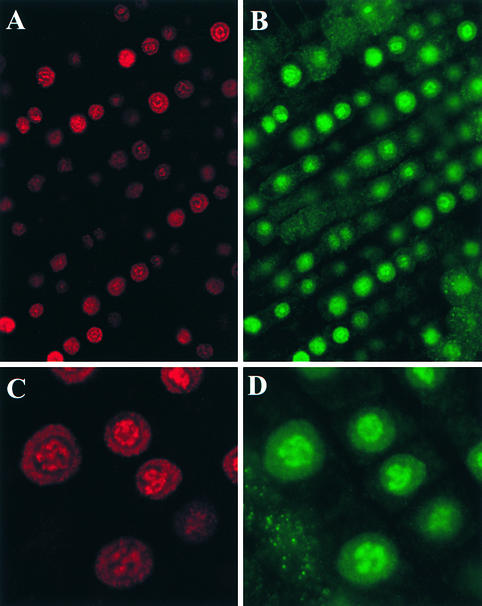
This antibody subsequently was used to locate the SPI3K-5p PtdIns 3-kinase epitope in fixed root sections of soybean. Figure 4 illustrates data obtained by confocal microscopy and shows that the SPI3K-5p epitope has a distinct and punctate distribution within the nucleus. Labeling can be seen throughout the nucleus but predominantly in the nucleolus. Because the widespread punctate distribution of the JIM135 labeling appeared very similar to that previously shown for transcription sites (Thompson et al., 1997; Abranches et al., 1998), we decided to investigate further whether the PtdIns 3-kinase distribution could be correlated to this pattern and so performed a set of double-labeling experiments. Nuclei in root sections first were permitted to transcribe in the presence of bromo-UTP and subsequently were fixed. The sections then were probed with JIM135 and an anti–bromo-UTP antibody. The results of the double-labeling experiment are shown in Figure 5; BrUTP incorporation is shown in red (Figures 5A and 5C) and JIM135 labeling in green (Figures 5B and 5D). The BrUTP labeling shows a bright, punctate distribution of transcription sites in the nucleolus, which Thompson et al. (1997) showed is located within the nucleolar dense fibrillar component, together with other punctate transcription sites within the nucleoplasm, as was demonstrated in plant cells by Abranches et al. (1998). In control experiments omitting either the anti-BrUTP or the JIM135 antibodies, the signal was abolished in the red or the green channel, respectively. The double-labeled images shown in Figure 5 were collected sequentially on the confocal microscope so that the excitation laser line, the dichroic mirror, and the emission filter were specific for each fluorochrome (488-nm excitation for fluorescein isothiocyanate and 568 nm for Cy3 [Amersham, Buckinghamshire, UK]). Under these conditions, cross-talk between the red and green channels was negligible.
Figure 4.
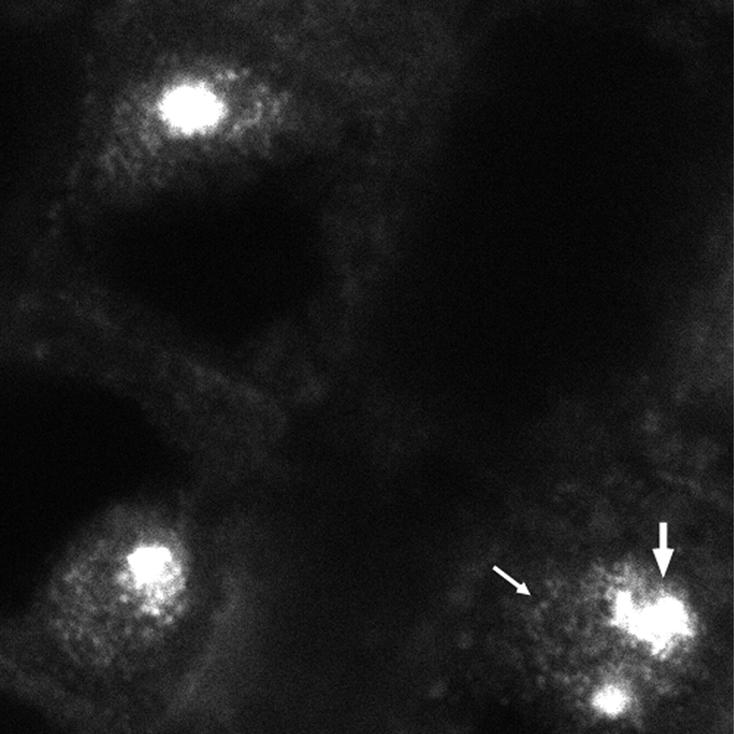
Immunolocalization of Soybean PtdIns 3-Kinase in Root Tissue.
Single confocal image from a vibratome section of soybean root labeled with the JIM135 antibody. Many foci of labeling are seen in both the nucleoplasm (small arrow) and the nucleolus (large arrow).
Figure 5.
Dual Labeling of PtdIns 3-Kinase and Active Transcription Sites in the Nucleus and Nucleolus.
(A) and (C) BrUTP incorporation shown in red.
(B) and (D) JIM135 labeling shown in green.
Both the nucleoplasmic and nucleolar labeling patterns coincide, indicating that the localization of JIM135 is at, or closely associated with, active transcription sites.
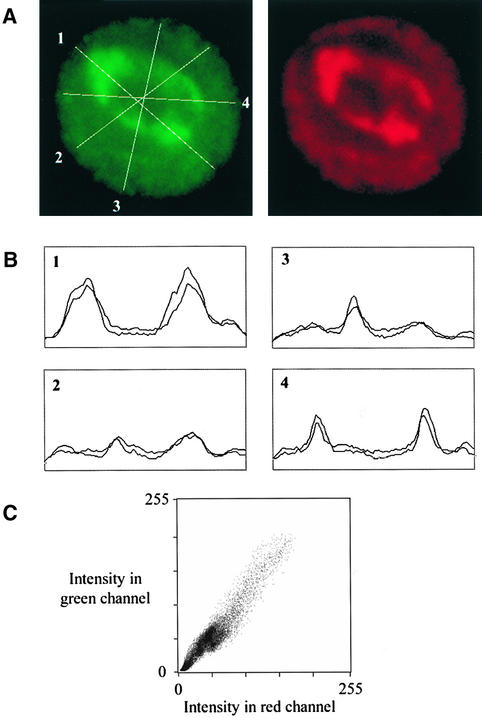
The labeling in the red and green channels shown in Figure 5 is clearly very similar. To quantify the correlation between the BrUTP transcription labeling and the JIM135 labeling more precisely, we performed detailed analysis on individual nuclei as shown in Figure 6. A single nucleus selected from a larger field is shown in Figure 6A, probed with JIM135 (green) and anti-BrUTP (red). First, we plotted line intensity profiles in each channel with a series of lines through the nucleus. Comparing the line profiles from the two channels showed that the positions of the maxima and minima in the two channels coincided precisely. Second, we calculated image correlation plots to show how closely the overall labeling patterns obtained with the two antibodies correlated. In this plot, the intensity for each pixel in the green (JIM135) channel was plotted against the intensity for the corresponding pixel in the red (BrUTP) channel as a scatter graph. In this type of plot, the same intensity of labeling in the two channels produces a point lying on the diagonal axis of the graph, whereas different intensities produce a point not on the diagonal; accordingly, the scatter of the points around the diagonal indicates the degree of correlation between the two channels. The two image channels we tested were highly correlated, as shown by the points being closely localized around the diagonal axis. The labeling patterns in Figure 5 and those in Figure 6 thus demonstrate that much of the JIM135 antigen, that is, the plant PtdIns 3-kinase, is located at or near active transcription sites, both in the nucleolus (pol I transcription) and in the nucleoplasm (pol II and/or pol III transcription).
Figure 6.
Confocal Microscopy Imaging of a Single Nucleus Chosen from a Larger Field Probed with Either JIM135 or an Anti-BrUTP Antibody.
(A) Nucleus probed with JIM135 is shown in green (left), and nucleus probed with an anti-BrUTP antibody is shown in red (right).
(B) Line intensity profiles of the green and the red channels across the four randomly selected lines through the nucleus shown in (A).
(C) Image correlation plot (Z1–Z2 plot) in which the intensity in the green channel (JIM135) is plotted as a function of the intensity in the red channel (anti-BrUTP). Details are given in the text.
DISCUSSION
That catalytically active PtdIns 3-kinases are associated with the plant nuclear matrix and that the localization of these enzymes appears to coincide with sites of active transcription obviously suggest that the plant PtdIns 3-kinase may have roles other than (or perhaps more likely, in addition to) those associated with vacuolar sorting and membrane trafficking. Because a role for the Vps34p-related PtdIns 3-kinases in processes related to transcriptional activity has not been identified previously in other cell types, our findings raise several questions. We cannot, of course, categorically rule out the possibility that JIM135 has an alternative in situ target that is not identified in protein gel blot analyses. However, the biochemical demonstration of detergent-resistant PtdIns 3-kinase activity in nuclei and the protein gel blot results obtained with both native and heterologously expressed proteins make this scenario highly implausible. Additional studies now are needed to clarify the potential roles of the plant nuclear PtdIns 3-kinase; several hypotheses, however, deserve brief consideration.
Until recently, comparatively little was known about the functional domains of Vps34p-related PtdIns 3-kinases except that they all contain a conserved catalytic lipid kinase domain (phosphoinositide kinase [PIK] domain) and an unidentified motif involved in the interaction with one or more adaptor proteins. Most Vps34p-related PtdIns 3-kinases also possess intrinsic protein kinase activity, but the physiological relevance of this remains to be determined. Several studies, however, have begun to shed light on the molecular mechanism involved in the function of the PtdIns 3-kinase product, PtdIns(3)P. For some time, vesicle targeting events have been known in many cases to depend on small GTP binding proteins. Thus, endocytosis requires the small GTP binding protein Rab5. Several studies have established a direct link between Rab5, PtdIns 3-kinase activity (i.e., PtdIns(3)P formation), and the early endosomal autoantigen (EEA1) associated with early endosomes in mammalian cells. A domain responsible for PtdIns(3)P binding was identified recently in EEA1. This domain, christened the FYVE finger, is ∼70 amino acids long, has eight potential Zn2+-coordinating cysteine residues, and binds two Zn2+ ions (overview in Wiedermann and Cockroft, 1998). The interaction between the FYVE finger domain and PtdIns(3)P now has been shown to be essential for proper EEA1 function; in addition, EEA1 needs to bind to Rab5-GTP. Thus, the concomitant presence of PtdIns(3)P and Rab5-GTP in a membrane domain ensures a unique recognition site for EEA1. FYVE finger–containing proteins now also have been identified in plant cells (Wiedermann and Cockroft, 1998; S. DeVos and B.K. Drøbak, unpublished data), but their potential function remains to be elucidated. We currently are investigating whether any of the FYVE finger–containing plant proteins have a nuclear localization and whether they have the potential to interact directly or indirectly with PtdIns 3-kinase. The presence of several small GTP binding proteins in plant nuclei has been reported already (Drøbak et al., 1995).
In mammalian cells, some evidence links inositol-containing lipids to the regulation of nuclear transcriptional activity. Santi et al. (1994) thus suggested that inositol lipid metabolism may play a role in the nuclear modifications accompanying steroid hormone induction of transcriptional activity, and in another study, Maraldi et al. (1997) showed how the generation of 3-phosphorylated lipids accompanied cell responses to interleukin-1α. Furthermore, saos-2 cells have been shown to possess a nuclear PtdIns 3-kinase activity that is upregulated by interleukin-1α treatment (Maraldi et al., 1997). In a more recent study, Marchisio et al. (1998) showed that nuclei of HL-60 cells have a constitutive PtdIns 3-kinase activity that is upregulated during granulocytic differentiation. However, before any parallels can be drawn between these results and the findings in our study, note that the studies on mammalian cells mentioned above investigated PtdIns 3-kinases belonging to the type I family of PtdIns 3-kinases, which utilize PtdIns 4,5-bisphosphate as a substrate. As we pointed out previously, eukaryotic PtdIns 3-kinases are likely to have evolved from a common ancestral 3-kinase family, and the possibility of some substantial species-specific functional overlap between members of these families should not be ruled out at present.
Thus, several members of a group of proteins closely related to both the PtdIns 3- and PtdIns 4-kinases have been found to be directly involved in regulation of transcriptional activity. This group of proteins, known as the PIK-related kinases, forms a distinct family of proteins (reviewed in Keith and Schreiber, 1995). Despite their obvious similarity to PIKs, their relevant substrates have not been identified, and in vitro experiments thus far have been unsuccessful in demonstrating any ability to phosphorylate inositol lipids.
Whatever the precise roles of the PtdIns 3-kinases and 3-phosphorylated inositol lipids turn out to be in plant cells, their functions are almost certain to be considerably more diverse and multifaceted than is assumed currently. Our present findings suggest that the nucleus and nucleolus are among the subcellular locations at which at least some of these enigmatic lipid/protein kinases and their products exert their functions.
METHODS
Chemicals
3H-l-α-phosphatidylinositol 4-phosphate (specific activity 185 Gbq/mmol) and γ-33P-ATP (specific activity 37 Tbq/mmol) were from Dupont-NEN (Zaventem, Belgium). All other chemicals, unless otherwise stated, were from Sigma (Poole, Dorset, UK).
Cell Culture and Isolation of Nuclei
Nonembryonic carrot (Daucus carota) cell cultures were maintained in full-strength Murashige and Skoog medium as described by Drøbak et al. (1995). For the isolation of nuclei, cells were concentrated by centrifugation in an MSE swing-out centrifuge (Denley, UK) at 100g for 10 min at room temperature, after which the supernatant was removed. The cell pellet was added to 50 mL of fresh culture medium: 0.5 M mannitol, 0.01% (w/v) Onezuka Y-23, and 0.1% (w/v) RS cellulase (Yakult Honsha Co. Ltd., Tokyo, Japan). Cells were incubated (at 25°C and 120 rpm) in the protoplast medium overnight. Protoplasts were harvested by centrifugation (100g for 10 min), and the protoplast pellet was gently resuspended in nuclear isolation buffer (NIB), which consisted of 0.5 M sucrose, 10 mM NaCl, and 10 mM Mes, pH 5.3. The protoplast suspension was transferred to 100-mL volumetric flasks and centrifuged for 10 min at 150g. Intact protoplasts were collected from the neck of the flask. Then, 10 mL of NIB with the sucrose content reduced to 0.4 M was added per 400 mg of the protoplast suspension remaining and equilibrated for 5 min on ice. Protease inhibitors (1 mM benzamidine-HCl, 0.1 mg/mL phenanthroline, 1 mg/mL aprotinin, 1 mg/mL leupeptin, 1 mg/mL pepstatin A, and 1 mM phenylmethylsulfonyl fluoride) in a solution of 0.1 mM DTT and 5 mM EDTA were added, and the protoplast suspension was passed three times through a 25-gauge needle; the resulting homogenate was layered onto a crude sucrose gradient (from 1.6 to 2.3 M sucrose in NIB) overlying a cushion of 0.5 mL of Maxidens (Nycomed, Oslo, Norway). The nuclear preparation was centrifuged at 100,000g for 2 hr, and nuclei were collected at the sucrose/Maxidens interphase. Nuclei were resuspended in NIB containing 0.4 M sucrose and protease inhibitors.
Lipid Kinase Assay
Lipid kinase assays were performed in a total volume of 120 μL in a medium containing 50 mM Hepes/KOH, pH 7.5, 5 mM MgCl2, 30 mM DTT, 0.33 μg/μL PtdIns, and 50 μM ATP (containing γ-33P-ATP, specific activity 75 kBq/nmol). The kinase reaction was started by adding 20 mg of protein, unless otherwise stated. Assays were performed at 25°C and terminated after 10 min. Thin-layer chromatography (TLC) analysis was performed as described by Dove et al. (1994). Deacylation of radiolabeled phosphoinositides was performed as described by Drøbak and Roberts (1992). Protein was determined by the dye binding method, with BSA as standard (Bradford, 1970).
HPLC
All HPLC separations were performed with a Partisil 10 SAX column (250 × 4.6 mm; Whatman, Maidstone, UK). Deacylated 33P-labeled lipid derivatives were analyzed using the gradient of Dove et al. (1994), with minor modifications: solvent A (Analar water; Gibco BRL, Paisley, UK) and solvent B (1 M [NH4]2HPO4, pH 3.8); and percentage of solvent B: 0 to 5 min, 0%; 5 to 60 min, 0 to 20%; 60 to 65 min, 20%; 65 to 68 min, 20 to 0%; and 68 to 75 min, 0%. The flow rate was maintained at 1.0 mL/min, and fractions were collected at 30-sec intervals from 30 to 71.5 min. Radioactivity was determined by liquid scintillation spectrophotometry (Rack-Beta; LKB, Bromma, Sweden) using a high-ionic scintillation cocktail (Canberra-Packard, Pangbourne, UK) and a dual 33P/3H scintillation program. Authentic standards and samples were coinjected with nonradioactive ADP for each HPLC analysis, and the absorbance at 256 nm was monitored to ensure reproducibility between separations.
Expression of Soybean PtdIns 3-Kinase and Production of JIM135 Monoclonal Antibody
Because expression of the full-length SPI3K-5 was found to be toxic to Escherichia coli, the SPI3K-5 cDNA (GenBank accession number L27265) contained within plasmid pRSETc (Invitrogen, La Jolla, CA) was digested with NdeI, and the 5′-terminal 1.3-kb fragment was ligated into plasmid pET15b (Novogen, Madison, WI). This was transformed into E. coli strain BL21(DE3) pLYSs by using the rubidium chloride method of Ano and Shoda (1992). The bacteria were grown to mid-log phase in Luria-Bertani broth (LB) supplemented with 100 μg/mL carbenicillin and 2% (w/v) glucose, after which the cells were pelleted. Cells were washed in LB containing 100 μg/mL carbenicillin, repelleted, and suspended in fresh LB containing 100 μg/mL carbenicillin; isopropyl-β-d-thiogalactopyranoside then was added to a final concentration of 1 mM. Cells were incubated at 37°C for 2 hr on an orbital shaker and finally pelleted at 6000g for 10 min at 4°C. Recombinant protein was recovered from inclusion bodies essentially as described by Nguyen et al. (1993). The protein was purified from inclusion bodies after solubilization and separation on SDS-PAGE curtain gels. A 2-cm strip of the gel stained with Coomassie Brilliant Blue R 250 was used to locate the protein band of interest. The region of the gel containing the protein was excised and frozen at ∼20°C; the protein was recovered by electroelution (model 422 electroeluter; Bio-Rad) according to the manufacturer's instructions. Expression of catalytically active PtdIns 3-kinase (SPI3K-5p, root form; Hong and Verma, 1994) was achieved with use of the Gibco BRL Bac-to-Bac system. The SPI3K-5 gene was removed from pRSETc by restriction enzyme digestion and ligated into pFastbac1 (Gibco BRL) downstream of the polyhedrin promoter. Recombinant bacmids were constructed by transposing the mini-Tn7 flanking the promoter and inserting the bacmid mini-attTn7 attachment site by using the pMON7124 helper plasmid. Bacmid DNA was grown overnight in E. coli and isolated by miniprep procedures. High molecular mass bacmid DNA was used to transfect Sf21 cells. Viral stocks (>107 plaque-forming units/mL) were harvested from transfected cells and used to infect additional Sf21 cells. Cells were harvested after 96 hr, and the manufacturer's instructions were followed throughout. Fisher rats (Charles River Laboratories Ltd., Margate, UK) were used for production of polyclonal antibodies according to the methods of Köhler and Milstein (1975) and Galfré and Milstein (1981) and following the normal practices of the John Innes Centre Monoclonal Antibody Unit, using the rat myeloma IR983F (Bazin, 1982) and MRC-5 feeder cells (Jacobs et al., 1970). Positive hybridomas were identified by protein gel blot analysis and in situ hybridization on soybean roots. JIM135, chosen after cloning by limited diffusion because of its superior antigenicity, was determined to belong to the IgM family by the method of Ouchterlony (1949) and using a rat monoclonal isotyping kit (Serotec, Kidlington, UK).
Electrophoresis and Immunoblotting Analysis
SDS-PAGE and electroblotting were performed as described by Drøbak et al. (1995) except that no DTT was added to the gel loading buffer, and samples were heated to 65°C before separation. The protein gel blots were incubated overnight with JIM135 at 1:10 (v/v) dilution in blocking solution at 4°C and processed as described by Drøbak et al. (1995).
Immunofluorescence on Tissue Sections
The terminal 3 to 4 mm of 48- to 96-hr-old soybean roots were excised and fixed for 1 hr at room temperature in 4% (w/v) formaldehyde in 50 mM Pipes-NaOH, pH 6.9, 5 mM EGTA, and 5 mM MgSO4 (PEM) buffer. After three washes in PBS (14 mM Na2HPO4, 3 mM NaH2PO4, and 150 mM NaCl, adjusted to pH 7.4 with 1 M HCl), 20- to 30-μm sections were cut using a vibratome (series 1000; TAAB Laboratory Equipment Ltd., Berkshire, UK). The sections were dried onto multiwell slides coated with glutaraldehyde-activated aminopropyltriethoxysilane. Sections were permeabilized with 2% (w/v) cellulase Onozuka R10 (Yakult Pharmaceuticals) in PBS and incubated for 2 hr at room temperature with JIM135 diluted 1:100 (v/v) in PBS containing 3% (w/v) BSA. After washing, the sections then were incubated with an anti-rat Cy3 (Amersham, Buckinghamshire, UK) antibody. After a final rinse, the nuclei were counterstained with the DNA dye 4,6-diamidino-2-phenylindole.
In Vitro Transcription on Vibratome Sections
The procedure of Thompson et al. (1997) was used with minor adjustments. The terminal 3 to 4 mm of 48- to 96-hr-old soybean roots were excised, and 30- to 40-μm-thick sections were cut with the vibratome in modified physiological buffer (MPB)—100 mM KCH3COO, 20 mM KCl, 20 mM Hepes-KOH, pH 7.4, 1 mM MgCl2, 1 mM ATP, 1% (v/v) thiodiglycol, 2 μg/mL aprotenin, and 0.5 mM phenylmethylsulfonyl fluoride—containing 1 M hexylene glycol. Sections were transferred to a tissue-handling device, treated with MPB containing 0.05% (v/v) Triton X-100 for 1 min, washed in MPB, incubated for 5 min in transcription buffer—MPB containing 50 μM CTP, 50 μM GTP, 25 μM bromo-UTP, 125 μM MgCl2 100 U/mL RNA guard—and finally washed in MPB. Sections were fixed in 4% (w/v) formaldehyde in PEM for 1 hr, washed, dried onto multiwell slides coated with glutaraldehyde-activated aminopropyltriethoxysilane, and permeabilized with 2% (w/v) cellulase Onozuka R10 in PBS. Incorporated BrUTP was detected by incubation with mouse anti–bromo-UTP antibodies (Boehringer Mannheim; diluted 1:20 [v/v] in PBS containing 3% [w/v] BSA), followed by incubation with donkey anti–mouse cy3 (Jackson Laboratories, Bar Harbor, ME; diluted 1:100 [v/v] in PBS containing 3% [w/v] BSA). Nuclei then were counterstained with the DNA dye 4′,6-diamidino-2-phenylindole. All images were obtained with a Bio-Rad MRC-1000 laser scanning confocal microscope, and image processing was performed with National Institutes of Health image or Scion software (Scion, Frederick, MD).
Acknowledgments
T.D.B. gratefully acknowledges a John Innes Foundation postgraduate studentship. L.E.H. was supported by European Community Human and Capital Mobility Grant No. CHRX CT940699. Work in the B.K.D. and P.J.S. laboratories was supported by the Biotechnology and Biological Sciences Research Council Intracellular Signaling Initiative. We thank Z. Hong and D.P.S. Verma for the generous gift of SPI3K-5 cDNA (L27265) and Carole Thomas for her valuable help and advice on Baculovirus/Sf21 cell expression. We also gratefully acknowledge the help and support received from Anna Cullingford during the antibody preparation and the help of Phil Taylor and Grant Calder in preparation of the figures.
References
- Abranches, R., Beven, A.F., Wells, B., and Shaw, P.J. (1998). Transcription sites are not correlated with chromosome territories in wheat nuclei. J. Cell Biol. 143, 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ano, T., and Shoda, M. (1992). Ultra-rapid transformation of Escherichia coli by an alkali cation. Biosci. Biotechnol. Biochem. 56, 1505. [DOI] [PubMed] [Google Scholar]
- Bazin, H. (1982). Production of rat monoclonal antibodies with the LOU rat non-secreting IR983F myeloma cell line. In Peptides of the Biological Fluids. H. Peeters, ed (New York: Pergamon Press), pp. 615–618.
- Bradford, M.M. (1970). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Carpenter, C.L., and Cantley, L.C. (1996). Phosphoinositide kinases. Curr. Opin. Cell Biol. 8, 153–158. [DOI] [PubMed] [Google Scholar]
- Dove, S.K., Lloyd, C.W., and Drøbak, B.K. (1994). Identification of a PtdIns 3-hydroxy kinase in plant cells: Association with the cytoskeleton. Biochem. J. 303, 347–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drøbak, B.K., and Roberts, K. (1992). Analysis of components of the plant phosphoinositide system. In Molecular Plant Pathology, Vol. 2, S.J. Gurr, M.J. McPherson, and D.J. Bowles, eds (Oxford, UK: IRL Press), pp. 195–221.
- Drøbak, B.K., and Watkins, P.A.C. (1994). Inositol(1,4,5)trisphosphate production in plant cells: Stimulation by the venom peptides, mellitin and mastoparan. Biochem. Biophys. Res. Commun. 205, 739–745. [DOI] [PubMed] [Google Scholar]
- Drøbak, B.K. Watkins, P.A.C., Bunney, T.D., Dove, S.K., Shaw, P.J., White, I.R., and Millner, P.A. (1995). Association of multiple GTP-binding proteins with the plant cytoskeleton and nuclear matrix. Biochem. Biophys. Res. Commun. 210, 7–13. [DOI] [PubMed] [Google Scholar]
- Galfré, G., and Milstein, C. (1981). Preparation of monoclonal antibodies: Strategies and procedures. Methods Enzymol. 73, 3–46. [DOI] [PubMed] [Google Scholar]
- Hendrix, K.W., Assefa, H., and Boss, W.F. (1989). The polyphosphoinositides, PtdIns monophosphate and PtdIns bisphosphate, are present in nuclei isolated from carrot protoplast. Protoplasma 151, 62–72. [Google Scholar]
- Hong, Z., and Verma, D.P.S. (1994). A PtdIns 3-kinase is induced during soybean nodule organogenesis and is associated with membrane proliferation. Proc. Natl. Acad. Sci. USA 91, 9617–9621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine, R.F., and Divecha, N. (1992). Phospholipids in the nucleus—Metabolism and possible functions. Semin. Cell Biol. 3, 225–235. [DOI] [PubMed] [Google Scholar]
- Jacobs, J.P., Jones, C.M., and Baille, J.P. (1970). Characteristics of a human diploid cell designated MRC-5. Nature 227, 168–170. [DOI] [PubMed] [Google Scholar]
- Keith, C.T., and Schreiber, S.L. (1995). PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science 270, 50–51. [DOI] [PubMed] [Google Scholar]
- King, L.A., and Possee, R.D. (1992). The Baculovirus Expression System. (London: Chapman and Hall).
- Köhler, G., and Milstein, C. (1975). Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256, 495–497. [DOI] [PubMed] [Google Scholar]
- Linassier, C., MacDougall, L.K., Domin, J., and Waterfield, M.D. (1997). Molecular cloning and biochemical characterization of a Drosophila PtdIns-specific phosphoinositide 3-kinase. Biochem. J. 321, 849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraldi, N.M., Marmiroli, S., Cocco, L., Capitani, S., Barnabei, O., and Manzoli, F.A. (1997). Nuclear lipid-dependent signal transduction in human osteosarcoma cells. Adv. Enzyme Regul. 37, 351–375. [DOI] [PubMed] [Google Scholar]
- Marchisio, M., Bertagnolo, V., Colamussi, M.L., Capitani, S., and Neri, L.M. (1998). PtdIns 3-kinase in HL-60 nuclei is bound to the nuclear matrix and increases during granulocytic differentiation. Biochem. Biophys. Res. Commun. 253, 346–351. [DOI] [PubMed] [Google Scholar]
- Matsuoka, K., Bassham, D.C., Raikhel, N.V., and Nakamura, K. (1995). Different sensitivity to wortmannin of two vacuolar sorting signals indicates the presence of distinct sorting machineries in tobacco cells. J. Cell Biol. 130, 1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, L.H., Jensen, D.B., and Burgess, R.R. (1993). Overproduction and purification of σ32, the Escherichia coli heat shock transcription factor. Protein Expression Purif. 4, 425–433. [DOI] [PubMed] [Google Scholar]
- Ouchterlony, O. (1949). Antigen–antibody reaction in gels. Ark. Kemi Miner. Geol. 26B, 1–9. [Google Scholar]
- Payrastre, B., Nievers, M., Boonstra, J., Breton, M., and Verkleij, A.J. and van Bergenen Henegouwen, P.M.P. (1992). A differential location of phosphoinositide kinases, diacylglycerol kinase and phospholipase C in the nuclear matrix. J. Biol. Chem. 267, 5078–5084. [PubMed] [Google Scholar]
- Santi, P., Marmiroli, S., Falcieri, E., Bertagnolo, V., and Capitani, S. (1994). Inositol lipid phosphorylation and breakdown in rat liver nuclei is affected by hydrocortisone blood levels. Cell Biochem. Funct. 12, 201–207. [DOI] [PubMed] [Google Scholar]
- Stack, J.H., and Emr, S.D. (1994). Vps34p required for yeast vacuolar protein sorting is a multiple specificity kinase that exhibits both protein kinase and PtdIns-specific PI 3-kinase activities. J. Biol. Chem. 269, 31552–31562. [PubMed] [Google Scholar]
- Thompson, W.F., Beven, A.F., Wells, B., and Shaw, P.J. (1997). Sites of rDNA transcription are widely dispersed through the nucleolus in Pisum sativum and can comprise single genes. Plant J. 12, 571–582. [DOI] [PubMed] [Google Scholar]
- Walsh, J.P., Caldwell, K.K., and Majerus, P.W. (1991). Formation of PtdIns 3-phosphate by isomerization from PtdIns. Proc. Natl. Acad. Sci. USA 88, 9184–9187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welters, P., Takegawa, K., Emr, S.D., and Chrispeels, M.J. (1994). ATVPS34, a PtdIns 3-kinase of Arabidopsis thaliana is an essential protein with homology to a calcium-dependent lipid-binding domain. Proc. Natl. Acad. Sci. USA 91, 11398–11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedermann, C., and Cockroft, S. (1998). Sticky fingers grab a lipid. Nature 394, 426–427. [DOI] [PubMed] [Google Scholar]
- Xu, P., Lloyd, C.W., Staiger, C.J., and Drøbak, B.K. (1992). Association of PtdIns 4-kinase with the plant cytoskeleton. Plant Cell 4, 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, K., Takegawa, K., Emr, S.D., and Firtel, R.A. (1995). A PtdIns (PI) kinase gene family in Dictyostelium discoideum: Biological roles of putative mammalian p110 and yeast Vps34p PI3-kinase homologues during growth and development. Mol. Cell. Biol. 15, 5645–5651. [DOI] [PMC free article] [PubMed] [Google Scholar]