Abstract
4′-O-Methylation of an isoflavonoid intermediate is a key reaction in the biosynthesis of the phytoalexin medicarpin in legumes. However, isoflavone O-methyltransferase (IOMT) from alfalfa converts the isoflavone daidzein to 7-O-methyl daidzein (isoformononetin) in vitro as well as in vivo in unchallenged leaves of transgenic alfalfa ectopically expressing IOMT. In contrast, elicitation of IOMT-overexpressing plants with CuCl2 or infecting these plants with Phoma medicaginis leads to greater accumulation of formononetin (4′-O-methyl daidzein) and medicarpin in the leaves than does elicitation or infection of control plants, and no isoformononetin is detected. Overexpression of IOMT results in increased induction of phenylpropanoid/isoflavonoid pathway gene transcripts after infection but has little effect on basal expression of these genes. IOMT-overexpressing plants display resistance to P. medicaginis. The apparently different regiospecificities of IOMT in vivo and in vitro are discussed in relation to potential metabolic channeling at the entry point into the isoflavonoid pathway.
INTRODUCTION
The isoflavonoids of the Leguminosae are phenylpropanoid-derived plant natural products exhibiting a range of biological activities. Simple isoflavones such as daidzein, genistein, and biochanin A exhibit estrogenic, antiangiogenic, antioxidant, and anticancer activities (Dixon, 1999), and the health-promoting activity of high-soy diets is believed to reside in their isoflavone components (Barnes et al., 1990; Adlercreutz et al., 1991; Adlercreutz, 1998; Lee et al., 1991). Daidzein and genistein induce the nodulation genes of Bradyrhizobium japonicum as an early step in establishing the nitrogen-fixing symbiosis in soybean (Kosslak et al., 1987), and they are precursors in the biosynthesis of antimicrobial isoflavonoid phytoalexins in a wide variety of legumes (Dixon and Paiva, 1995). Furthermore, because of their estrogenic activity, high concentrations of isoflavonoids can affect the reproductive capacity of sheep that graze forage legumes (Shutt, 1976). Genetic manipulation of isoflavonoid biosynthesis in transgenic plants therefore has potential for a positive impact on plant, animal, and human health (Dixon et al., 1999).
Although phytoalexins exhibit antimicrobial activity in vitro and are induced to potentially antimicrobial concentrations in plant tissues in response to infection (Dixon, 1986; Kuć, 1995), there is little direct evidence of their function as resistance factors in vivo. A large body of literature reports temporal and spatial correlations between phytoalexin accumulation and resistance (Dixon et al., 1983), and fungal detoxification of phytoalexins has in some cases been an important factor in virulence (VanEtten et al., 1989), suggesting a role for phytoalexins in resistance. However, at present, knowledge is lacking as to which enzymes control phytoalexin accumulation in most plant species and might therefore be targets for gain (or loss)-of-function genetic approaches to address more critically the roles of phytoalexins in resistance.
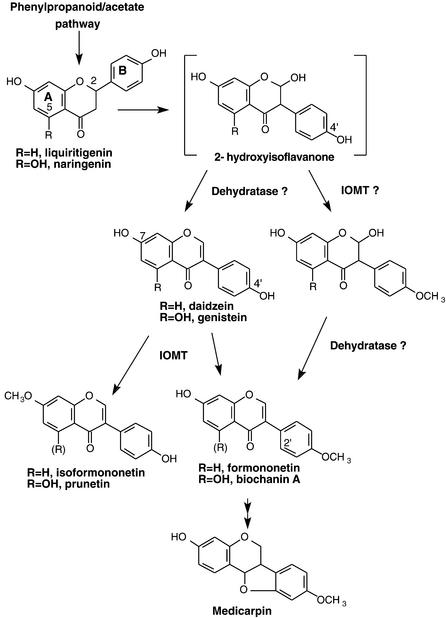
The biosynthetic branch pathway leading to isoflavones in leguminous plants starts with a cytochrome P450–mediated 2-hydroxylation/aryl migration of a flavanone intermediate (Kochs and Grisebach, 1986; Hakamatsuka et al., 1991; Steele et al., 1999; Jung et al., 2000), as shown in Figure 1. After aryl migration, the 2-hydroxyisoflavanone intermediate undergoes dehydration to yield the corresponding isoflavone (Hakamatsuka et al., 1998). Genistein (4′,5,7-trihydroxyisoflavone) is the product of aryl migration/dehydration of naringenin, whereas daidzein (4′,7-dihydroxyisoflavone) is formed in a similar manner from liquiritigenin (4′,7-dihydroxyflavanone). 4′-O-Methylation of daidzein yields formononetin, whereas 4′-O-methylation of genistein yields biochanin A, an anticancer compound found in chickpea (Yanagihara et al., 1993).
Figure 1.
Formation and O-Methylation of Isoflavones.
The flavanones naringenin and liquiritigenin are formed by the action of chalcone synthase and chalcone isomerase, with loss of the 5-hydroxyl in liquiritigenin occurring by coaction of chalcone reductase with chalcone synthase. Aryl migration of the B-ring is catalyzed by 2-hydroxyisoflavanone synthase, leading (after dehydration) to daidzein or genistein. 4′-O-Methylation could hypothetically occur at two stages: after dehydration to daidzein or before dehydration, at the level of the 2-hydroxyisoflavanone. Conversion of formononetin to medicarpin proceeds by way of 2′-hydroxylation, two successive reductions (catalyzed by isoflavone reductase and vestitone reductase), and a ring closure step (Dixon, 1999).
In alfalfa and many other legumes, including clover, pea, and chickpea, methylation of the 4′-hydroxyl is a prerequisite for further substitutions of the isoflavonoid nucleus that can lead to the pterocarpan phytoalexins such as medicarpin (Figure 1) (Dixon, 1999). However, the exact nature of this O-methylation reaction is currently unclear. In light of radiolabeling studies in copper-induced alfalfa seedlings in which formononetin (but, surprisingly, not daidzein) was incorporated into the pterocarpan medicarpin, Dewick and Martin (1979b) proposed that 4′-O-methylation might be an integral part of the aryl migration reaction of isoflavone biosynthesis. However, aryl migration can occur in vitro and in vivo in the absence of methylation (Kochs and Grisebach, 1986; Kessmann et al., 1990; Steele et al., 1999; Jung et al., 2000). Nonetheless, no S-adenosyl-l-methionine (SAM)–dependent O-methyltransferase (OMT) has been characterized that can catalyze 4′-O-methylation of daidzein. Instead, the SAM-dependent isoflavone OMT (IOMT) from alfalfa (Edwards and Dixon, 1991; He and Dixon, 1996) and chickpea (Barz and Welle, 1992) produces 7-O-methyl daidzein (isoformononetin) in vitro, as shown in Figure 1; to our knowledge, this rarely occurring plant natural product has not previously been reported from this species.
Alfalfa IOMT also converts daidzein to isoformononetin when expressed in Escherichia coli (He et al., 1998). Because IOMT enzyme activity and transcripts are strongly induced in elicited alfalfa cell cultures in coordination with the accumulation of medicarpin, formononetin, and the other enzyme activities involved in medicarpin biosynthesis (Dalkin et al., 1990; He et al., 1998), this suggests that IOMT might somehow be involved in medicarpin biosynthesis in vivo, despite its apparent seven-position specificity. We now confirm this hypothesis by using reverse-genetic approaches in transgenic alfalfa.
The role of the indole phytoalexin camalexin in disease resistance of Arabidopsis has been addressed by genetic loss-of-function experiments. Two phytoalexin-deficient (pad) mutants exhibit increased susceptibility to virulent pathogens but are unaffected in their response to avirulent pathogens (Glazebrook and Ausubel, 1994). The pad-3 mutant, which appears to be deficient in a camalexin biosynthetic cytochrome P450 monooxygenase (Zhou et al., 1999), is unaffected in its response to Pseudomonas syringae and to two obligate fungal pathogens but exhibits increased susceptibility to the necrotrophic fungus Alternaria brassicola (Thomma et al., 1998, 1999), thus suggesting a role for camalexin in resistance to this fungus. We here confirm the role of the isoflavonoid phytoalexin medicarpin in disease resistance in alfalfa by using a gain-of-function approach based on genetic manipulation of IOMT.
RESULTS
Generation of Transgenic Plants with Altered Expression of IOMT
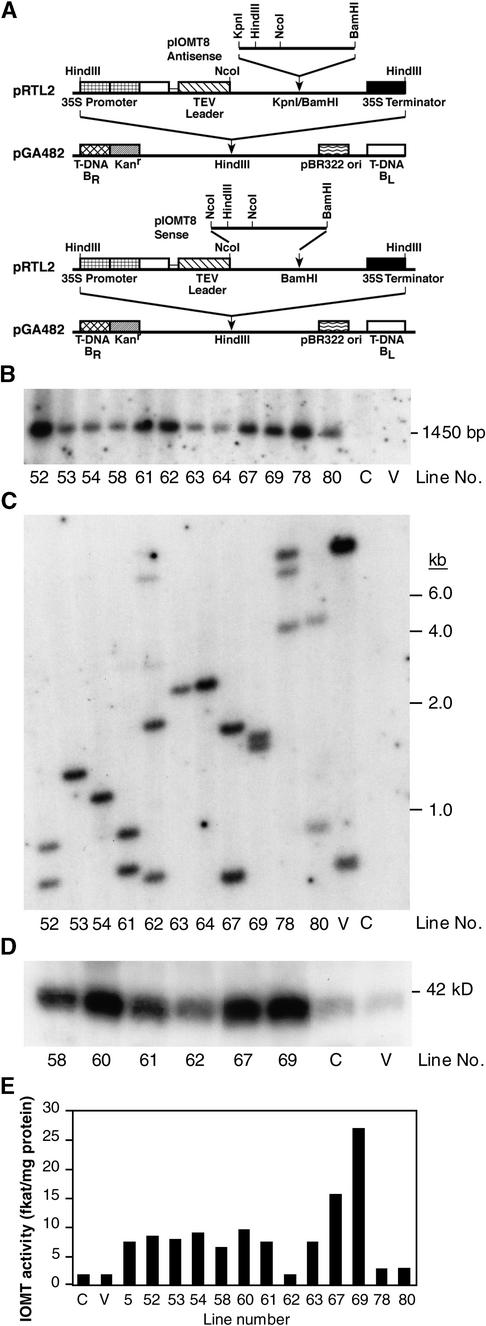
Gene constructs containing the full-length alfalfa IOMT8 cDNA in the sense and antisense orientations under control of the constitutive 35S promoter are shown in Figure 2A. These were introduced into alfalfa by leaf disc transformation. After plant regeneration by way of somatic embryogenesis, transformants (47 sense and 44 antisense) were analyzed by polymerase chain reaction (PCR) for genomic insertion of IOMT transgene sequences. Of the transformants analyzed, >70% (33/47 sense, 32/44 antisense) contained the full-length IOMT cDNA insert (data not shown). Genomic integration of the transgene construct was confirmed in a subset of the transformants by DNA gel blot analysis. The data presented in Figure 2B show that the 1.45-kb EcoRI fragment containing the IOMT transgene is absent from untransformed and empty vector–transformed lines. Genomic DNA from a range of transformants was then cut with HindIII, and the blots were probed with the T-DNA left border, as shown in Figure 2C. This analysis indicated that all the transformants analyzed arose from independent integration events.
Figure 2.
Generation and Molecular Analysis of Transgenic Alfalfa Altered in Expression of IOMT.
(A) Binary vector constructs that harbor IOMT sequences in sense and antisense orientations (see Methods for details). BL, left border; BR, right border; Kanr, kanamycin resistance gene; ori, origin of replication; TEV, tobacco etch virus leader sequence.
(B) DNA gel blot analysis of empty vector (V), untransformed control (C), and a range of IOMT sense transformants. Genomic DNA was digested with EcoRI, which cuts twice within the sense construct, and hybridized with an 800-bp IOMT8 probe.
(C) DNA gel blot border analysis of the same DNA samples shown in (B) but after digestion of DNA with HindIII and hybridization with the T-DNA left border sequence.
(D) Protein gel blot analysis of leaf extracts from a range of IOMT sense transgene-expressing, empty vector (V), and untransformed control (C) alfalfa plants. Blots were probed with anti-(alfalfa IOMT) polyclonal antiserum.
(E) Enzymatic activity of IOMT in leaf tissue from untransformed (C) and empty vector–transformed (V) controls and from a range of IOMT8 sense transformants.
Healthy alfalfa plants do not express the isoflavonoid pathway in leaves. Leaf tissue extracts from sense transformants were therefore subjected to protein gel blot analysis to assess the amounts of ectopic expression of the IOMT transgene product in leaves, as shown in Figure 2D. Lines 67 and 69 expressed the greatest concentrations of IOMT protein. Finally, IOMT enzyme activity was measured in extracts from leaves of greenhouse-grown plants. The data presented in Figure 2E indicate that several independent lines harboring the IOMT sense construct expressed high IOMT activity, with the greatest activities in lines 67 and 69. However, lines 62 and 78, although harboring at least three copies of the transgene, had very low IOMT activity. Thin-layer chromatographic and HPLC analysis of the labeled product formed from daidzein and S-[3H-methyl]adenosyl-l-methionine showed that only isoformononetin (Figure 1) was produced in vitro by the enzyme extracted from healthy, IOMT-overexpressing alfalfa leaves (data not shown).
IOMT Expression Affects the Biosynthesis of 4′-O-Methylated Isoflavonoids in Vivo
Alfalfa roots accumulate formononetin glucosyl malonate (FGM) and medicarpin glucosyl malonate (MGM) (Sumner et al., 1996). The amounts of these compounds, and of free medicarpin, were quite variable in root extracts from greenhouse-grown plants, presumably because of the various degrees of stress encountered as the roots pushed through the soil medium. For analysis of potential downregulation of isoflavonoid contents in antisense IOMT transgenic lines, plants selected initially on the basis of metabolite concentrations were vegetatively propagated, and the replicated rooted cuttings were grown under growth chamber conditions to yield enough samples for statistical analysis of results. Root tissue from cuttings of three independent antisense lines exhibited markedly reduced concentrations of the three compounds in comparison with the following average values (950 ± 340 nmol/g fresh weight for FGM, 85 ± 40 nmol/g fresh weight for medicarpin, and 117 ± 35 nmol/g fresh weight for MGM) from a population of 46 independently propagated empty-vector control plants. The overall reductions for the independent cuttings from the three lines (line 42,  ; line 54,
; line 54, 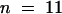 ; line 60,
; line 60, 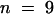 ) were 26 to 46% for FGM, 29 to 75% for medicarpin, and 18 to 46% for MGM. However, although this suggests that IOMT may be involved in the biosynthesis of formononetin and medicarpin, IOMT activity was very low (close to assay baseline) in control root extracts, and we were therefore unable to demonstrate a relationship between enzyme activity and metabolite concentrations in the antisense lines.
) were 26 to 46% for FGM, 29 to 75% for medicarpin, and 18 to 46% for MGM. However, although this suggests that IOMT may be involved in the biosynthesis of formononetin and medicarpin, IOMT activity was very low (close to assay baseline) in control root extracts, and we were therefore unable to demonstrate a relationship between enzyme activity and metabolite concentrations in the antisense lines.
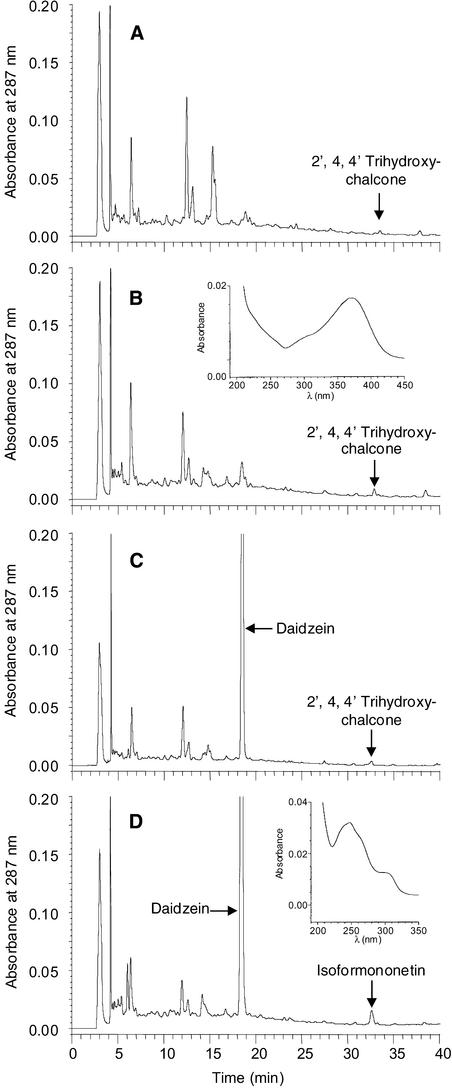
We next examined the effects of overexpression of IOMT in healthy alfalfa leaves. Figures 3A and 3B show HPLC profiles of leaf extracts from growth chamber–grown cuttings of IOMT-overexpressing (line 69) and vector control plants, respectively, after infiltration with Pi buffer. The profiles were qualitatively very similar. No formononetin, isoformononetin, or medicarpin (free or conjugated) was detected. The isoflavonoid pathway is expressed substantially in alfalfa leaves only after stress, such as infection or elicitation (Higgins, 1972; Paiva et al., 1994), and there would therefore be no isoflavonoid substrates present in unstressed leaves for IOMT to act on. A compound with very similar retention time to isoformononetin was identified by UV spectroscopy as 2′,4,4′-trihydroxychalcone, a precursor of daidzein, as seen in the inset in Figure 3B. However, isoformononetin was clearly produced when unlabeled daidzein in Pi buffer was infiltrated into leaves of IOMT-overexpressing transgenic alfalfa, as shown in Figure 3D and inset, whereas the daidzein remained unconverted when fed to leaves of control plants, as shown in Figure 3C. Thus, IOMT exhibits seven-position regiospecificity when ectopically expressed in planta, identical to the specificity displayed in vitro, if the substrate is exogenously supplied to unchallenged tissues.
Figure 3.
Metabolism of Daidzein after Infiltration into Untreated Alfalfa Leaves.
(A) and (B) HPLC profiles of phenolic compounds from leaves of vector control plant line 62C (A) and IOMT sense transgenic line 69 (B) 24 hr after infiltration with Pi buffer.
(C) and (D) HPLC profiles of leaf extracts from the same vector control (C) and sense transgenic (D) lines 24 hr after infiltration with daidzein in Pi buffer.
The insets show UV spectra of 2′,4,4′-trihydroxychalcone (B) and isoformononetin (D).
Trifoliate leaves were elicited by placing cuttings in 3 mM CuCl2, a treatment previously used as a reliable method for induction of isoflavonoid accumulation in alfalfa (Dewick and Martin, 1979a); other cuttings were exposed only to water (control). Copper-induced IOMT activities were 9.0 ± 0.3 μkat/kg protein in control lines and 37.6 ± 1.2 μkat/kg protein in the overexpressing lines. Quantities of isoflavonoids were determined by HPLC. Metabolite profiles of leaves from control (empty vector) plants and IOMT-overexpressing line 67 exposed to CuCl2 or water are shown in Figures 4A to 4D. No formononetin, isoformononetin, or medicarpin was observed in water-treated control or IOMT-overexpressing plants, as seen in the HPLC traces shown in Figures 4C and 4D. Copper treatment led to accumulation of free and conjugated apigenin, 7,4′-dihydroxyflavone, 7,4′-dihydroxyflavanone, and low amounts of formononetin and medicarpin in the control line (Figure 4A); however, induction of the latter two compounds was much stronger in the IOMT-overexpressing line (Figure 4B). Importantly, no isoformononetin accumulated in response to elicitation.
Figure 4.
Abiotic Elicitation of Isoflavonoid Compounds in Control and IOMT-Overexpressing Transgenic Alfalfa.
(A) and (B) HPLC profiles of leaf extracts from CuCl2-treated plants of vector control line 9C (A) and IOMT-overexpressor line 67 (B) 32 hr after exposure to 3 mM CuCl2. Peaks 1 to 4 were identified as apigenin conjugate, 7,4′-dihydroxyflavone, 7,4′-dihydroxyflavanone, and apigenin aglycone, respectively. Formononetin and medicarpin peaks are marked.
(C) and (D) HPLC profiles of leaf extracts from plants of vector control line 9C (C) and IOMT-overexpressor line 67 (D) 32 hr after transfer of seedlings to water.
(E) to (H) Amounts of formononetin ([E] and [G]) or medicarpin ([F] and [H]) in leaves of replicate vegetatively propagated vector control lines 9C and 62C (control lines) and IOMT-overexpressing lines 67 and 69 assessed 32 hr after exposure to water or 3 mM CuCl2.
Analysis of replicate rooted cuttings from multiple empty-vector control lines and two independent IOMT-overexpressing lines (67 and 69) confirmed that overexpression of IOMT in elicited alfalfa leaves led to formation of substantially more formononetin and medicarpin than in the control lines after exposure to CuCl2 (Figures 4E to 4H). The amounts of induced 7,4′-dihydroxyflavanone and apigenin were quite variable in leaves of CuCl2-induced alfalfa and did not appear to correlate with IOMT activity (data not shown).
When leaves of IOMT-overexpressing alfalfa that had been previously elicited with CuCl2 were fed unlabeled daidzein, isoformononetin was produced in the unelicited leaves but was not detected in the elicited leaves. Instead, formononetin was detected as a result of elicitation, as shown in Figure 4, but feeding daidzein did not increase the amount of formononetin (data not shown). When the in vitro IOMT activity was measured in extracts from control or CuCl2-elicited leaves of the various lines, the product was always isoformononetin, indicating that CuCl2 itself does not affect the regiospecificity of the enzyme.
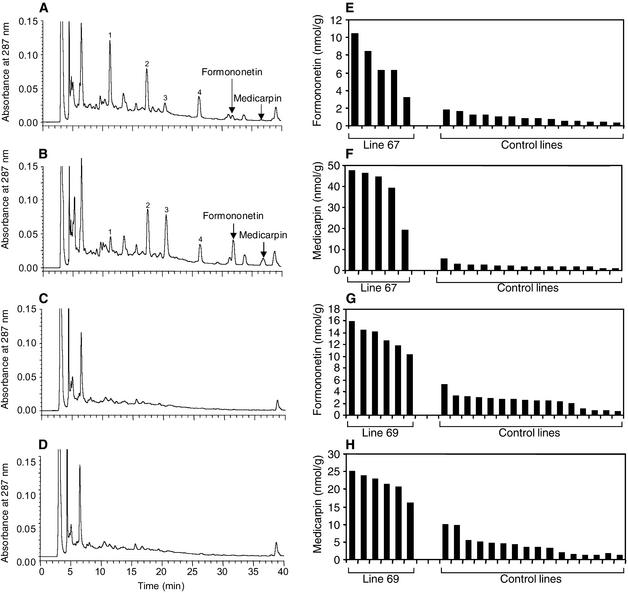
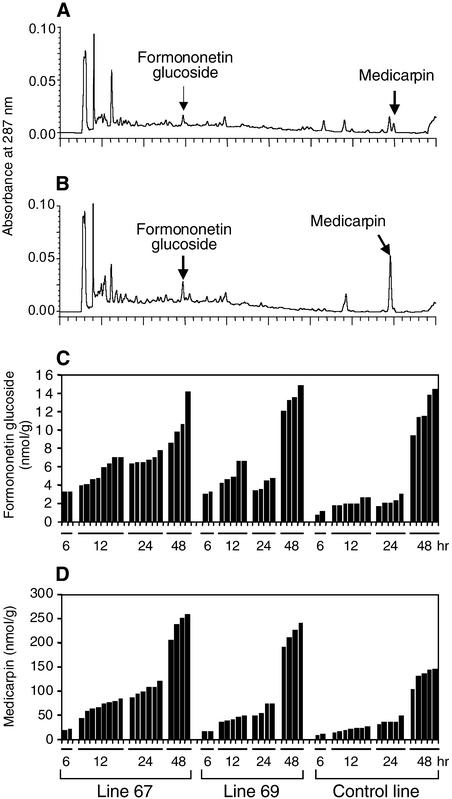
The elicitation experiments were repeated on trifoliate leaves, this time using, instead of abiotic elicitation, infection by spores of Phoma medicaginis, another treatment that induces the enzymes of the isoflavonoid pathway and consequent medicarpin accumulation in alfalfa leaves (Paiva et al., 1994). The data illustrated in Figures 5A and 5B show that fungal infection led to the appearance of formononetin glucoside and medicarpin in the empty-vector control line, but even greater amounts were produced in the IOMT-overexpressing line. As before, no medicarpin or formononetin conjugate was detected in the uninfected control or IOMT-overexpressing line. No isoformononetin was observed in infected leaves. The results shown in Figures 5C and 5D show a time-course study with replicate rooted cuttings from the empty-vector control and two independent IOMT-overexpressing lines (67 and 69) harvested at various times after fungal infection. The data clearly confirm that overexpression of IOMT in infected alfalfa leaves leads to earlier induction and greater amounts of formononetin glucoside and medicarpin than in the control plants.
Figure 5.
Accumulation of Isoflavonoid Compounds in Response to Fungal Infection in Control and IOMT-Overexpressing Transgenic Alfalfa.
(A) and (B) HPLC profiles of extracts from P. medicaginis–infected leaves of empty-vector control line 64C (A) and IOMT-overexpressor line 67 (B) 12 hr after inoculation. Peaks of formononetin glucoside and medicarpin are marked.
(C) and (D) Amounts of formononetin glucoside (C) or medicarpin (D) in leaves of replicate vegetatively propagated vector control (line 64C) and IOMT-overexpressing plants (lines 67 and 69) at various times after inoculation with P. medicaginis.
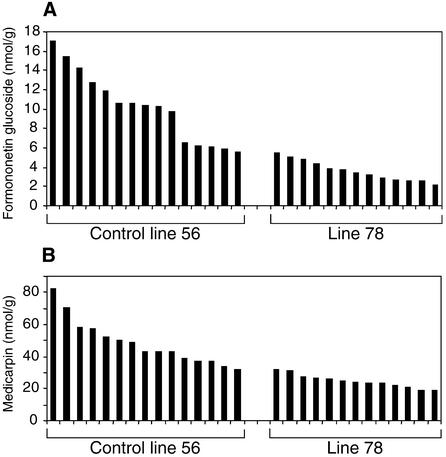
Sense transformant 78 has a high transgene copy number but only weakly expressed IOMT protein (data not shown) or activity in the leaves (Figure 2). This line may therefore exhibit epigenetic gene silencing (Matzke and Matzke, 1995). Parallel analysis of replicate cuttings of line 78 and empty-vector control line 56C indicated that, after infection with P. medicaginis, the induction of formononetin glucoside and medicarpin in line 78 was markedly less than in the control line, as shown in Figure 6. Overall, the formononetin glucoside was reduced by 65% and medicarpin by 58%. This reflected the relative IOMT activities in plants of the two lines; Phoma-induced activities were 38.8 μkat/kg for line 56C and 18.1 μkat/kg for line 78. By 20 hr after infection, the amounts of formononetin glucoside were quite high in the independent cuttings of control line 56C (cf. Figures 5 and 6). The experiments whose results are shown in Figures 5 and 6 were performed at different times, and although all experiments were performed in a growth chamber under apparently identical conditions, the amounts of induced isoflavonoids can vary with the developmental state of the plant and the rate at which the infection develops. We are therefore not able to conclude whether downregulation of IOMT in Phoma-infected leaves has a similar effect on both formononetin glucoside and medicarpin or whether the major effect is on medicarpin alone.
Figure 6.
Accumulation of Isoflavonoid Compounds in Control and Putatively IOMT Gene-Silenced Transgenic Alfalfa.
(A) The amounts of formononetin glucoside were measured in leaves from replicate cuttings of vector control line 56C and IOMT sense transformant 78 20 hr after inoculation with spores of P. medicaginis.
(B) As in (A), but showing medicarpin.
Taken together, these elicitation and infection studies indicate that genetic manipulation of IOMT, an enzyme ascribed activity as an isoflavone 7-O-methyltransferase in vitro, leads to modulation of 4′-O-methylated isoflavonoids in vivo in transgenic alfalfa.
Overexpression of IOMT Leads to Increased Disease Resistance to P. medicaginis
P. medicaginis is a successful pathogen of alfalfa, causing brown disease lesions on leaves of the cultivar (Regen SY) we used for the genetic transformation. To determine whether the more rapid accumulation of the phytoalexin medicarpin leads to resistance to Phoma in plants overexpressing IOMT, we inoculated the IOMT-overexpressing and empty-vector control plants on the leaves by using a pinwheel to produce a line of small wounds into which fungal spores would enter to initiate infection on the susceptible alfalfa cultivar. The sizes of the brown Phoma lesions were then measured 5 days after infection. The data presented in Figure 7A clearly show that the lesion size was much less in IOMT-overexpressing line 69 than in the control. Measurement of multiple lesions 5 days after infection gave an average lesion size of 0.96 mm (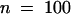 ) for control plants and 0.41 mm (
) for control plants and 0.41 mm (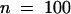 ) for line 69 (a 57.3% decrease) and also 0.46 mm (
) for line 69 (a 57.3% decrease) and also 0.46 mm (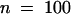 ) for IOMT-overexpressing line 67 compared with 1.17 mm (
) for IOMT-overexpressing line 67 compared with 1.17 mm (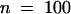 ) for the corresponding control line grown and infected in parallel (a 60.7% decrease), as shown in Figure 7B. These reductions in lesion size correlated with 12-hr-postinoculation increases in medicarpin concentrations of 260% for line 69 and 370% for line 67 in comparison with the parallel control line (see Figure 5). At 48 hr after inoculation, the increases were 168% for line 69 and 184% for line 67. That there was virtually no overlap in the values for lesion size between the control and IOMT-overexpressing lesion populations clearly demonstrates the relationship between medicarpin content and resistance to P. medicaginis.
) for the corresponding control line grown and infected in parallel (a 60.7% decrease), as shown in Figure 7B. These reductions in lesion size correlated with 12-hr-postinoculation increases in medicarpin concentrations of 260% for line 69 and 370% for line 67 in comparison with the parallel control line (see Figure 5). At 48 hr after inoculation, the increases were 168% for line 69 and 184% for line 67. That there was virtually no overlap in the values for lesion size between the control and IOMT-overexpressing lesion populations clearly demonstrates the relationship between medicarpin content and resistance to P. medicaginis.
Figure 7.
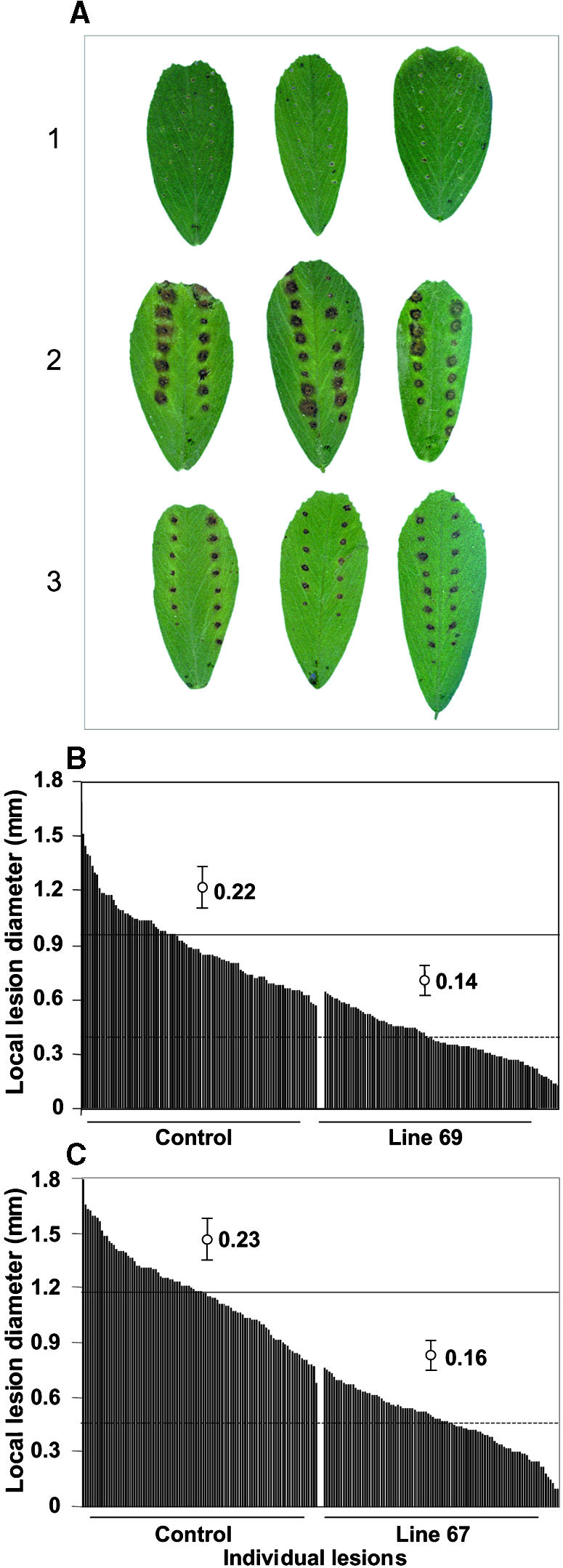
Disease Resistance of Transgenic Alfalfa Modified in Expression of Isoflavone O-Methyltransferase.
(A) Leaves of control vector–transformed line 64C (line 2, middle) and of IOMT-overexpressing line 69 (line 3, bottom) were wounded on the leaves with a tracing wheel and then sprayed with a spore suspension of P. medicaginis (5 × 105 spores/mL). Leaves of line 64C were also sprayed with water alone (line 1, top). The inoculated plants were then incubated in a growth chamber for 5 days under conditions described in Methods; typical disease symptoms are shown.
(B) Sizes of 100 individual lesions on leaves of empty-vector control line 64C and IOMT-overexpressing line 69 measured 5 days after inoculation. The average value for the size of the wounds produced by the pinwheel alone for a parallel series of 100 wound sites has been subtracted. The upper horizontal solid line shows the mean for the control line, and the lower dashed horizontal line shows the means for the IOMT-overexpressing line. The bars show the standard deviations.
(C) As in (B), but showing lesions on IOMT-overexpressing line 67 and the empty-vector control line 64C infected in parallel.
Comparison of Phoma lesion size in line 78 with that in control lines 5 days after infection indicated that the decrease in medicarpin concentrations resulting from downregulation of IOMT did not increase disease severity in this line compared with the control lines. In fact, there was no substantial difference in lesion size between lines 56C and 78 (data not shown).
Expression of Isoflavonoid Pathway Genes as a Consequence of IOMT Overexpression
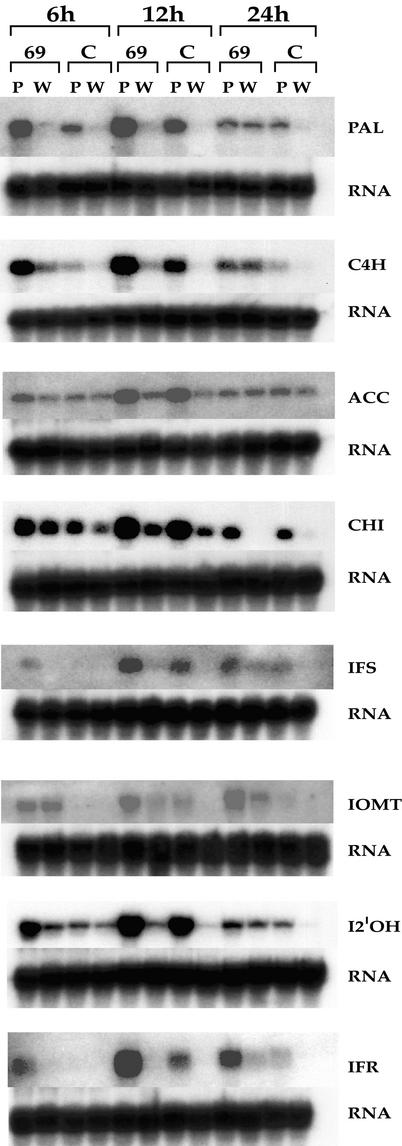
These data suggest that increased production of 4′-O-methylated isoflavonoids and the resulting disease resistance in IOMT-overexpressing plants might be the direct result of increased concentrations of IOMT and therefore point to a direct involvement of IOMT in the synthesis of these compounds. However, because the 4′-O-methylated compounds are stress-induced phytoalexins in alfalfa, it is possible that lines with increased IOMT activity have some kind of metabolic stress that leads to increased phytoalexin production independent of flux through IOMT. This is unlikely, however, in view of the lack of detectable phytoalexins in unelicited or uninfected IOMT-overexpressing plants (Figures 4 and 5). To further rule out this possibility and to determine whether other enzymes of the isoflavonoid pathway might be modulated after expression of IOMT, we examined the amounts of transcripts encoding eight of the enzymes involved in the conversion of phenylalanine to medicarpin in an empty-vector control and in an IOMT-overexpressing line as a function of time after infection with P. medicaginis. The results shown in Figure 8 confirm that IOMT transcripts were constitutively high in line 69 in the absence of Phoma infection, consistent with the transgene being under control of the 35S promoter. In uninfected leaves ectopically expressing IOMT, the amounts of transcripts of the other enzymes of the phenylpropanoid/isoflavonoid pathway were not markedly greater than those in the empty-vector control. Thus, overexpression of IOMT does not, of itself, induce the isoflavonoid pathway as a stress response, a conclusion consistent with the metabolic profile data shown in Figures 4 and 5.
Figure 8.
RNA Gel Blot Analysis of Isoflavonoid Pathway Transcripts in Phoma-Infected Leaves of Empty-Vector Control (Line 64C) and IOMT-Overexpressing (Line 69) Alfalfa.
Total RNA was isolated from leaves at 6, 12, and 24 hr (h) after infection with P. medicaginis (P) or treatment with water (W) and probed with labeled cDNA probes for alfalfa PAL, alfalfa C4H, alfalfa acetyl CoA carboxylase (ACC), alfalfa chalcone isomerase (CHI), Medicago truncatula IFS, alfalfa IOMT, M. truncatula I2′OH, and alfalfa IFR. Blots were stripped and reprobed with an rRNA probe to check loading and transfer efficiency. C, control.
After infection with P. medicaginis, empty-vector control alfalfa leaves had a weak, relatively slow induction of phytoalexin pathway enzyme transcripts, as shown in Figure 8. This response is typical of that found in compatible interactions (Lamb et al., 1992). However, the amounts of the transcripts encoding phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), isoflavone synthase (IFS), isoflavone 2′-hydroxylase (I2′OH), and isoflavone reductase (IFR) were much greater in IOMT-overexpressing lines than in the control line as early as 6 hr after infection. IFS and IFR transcripts remained uninduced at this time in the control line. PAL, C4H, IFS, I2′OH, and IFR transcripts were also present in somewhat greater amounts in line 69 than in the control line at 12 hr after infection. Chalcone isomerase transcripts were only slightly greater in line 69 than in the control line after infection. However, the transcripts encoding cytosolic acetyl CoA carboxylase, the enzyme that provides malonyl CoA for the chalcone synthase reaction and that is coinduced as part of the phytoalexin response in elicited alfalfa cells (Shorrosh et al., 1994), were not particularly greater in line 69 than in the empty-vector control line in any of the treatments. Thus, overexpression of IOMT leads to markedly increased induction of most of the other enzymes directly involved in isoflavonoid biosynthesis but does not appear to affect expression of these enzymes in the absence of the inducing agent.
DISCUSSION
Regiospecificity of Isoflavone 7-O-Methyltransferase in Vivo
Many plant OMT sequences are now available in the databases (Ibrahim et al., 1998; Joshi and Chiang, 1998). In many cases, the enzymes appear to exhibit strict regiospecificity; for example, a series of distinct, position-specific OMTs is involved in the synthesis of polymethylated flavonols in Chrysosplenium americanum (Ibrahim et al., 1987; Gauthier et al., 1996). However, OMTs may be regiospecific but act on multiple structurally related substrates, such as the well-studied OMTs of lignin biosynthesis (Bugos et al., 1991; Li et al., 1997, 2000), and in some cases, they have been shown to be more promiscuous, acting on different classes of plant natural products (Gauthier et al., 1998; Frick and Kutchan, 1999). Database searches reveal that alfalfa IOMT has greatest sequence identity (50.5%) to 6a-hydroxymaackiain 3-OMT from pea, an enzyme also involved in biosynthesis of isoflavonoid phytoalexins (Wu et al., 1997). The reaction catalyzed by this enzyme is functionally analogous to the 7-O-methylation of daidzein, the 3-position of pterocarpans being equivalent to the 7-position of isoflavones.
Modulation of IOMT expression in transgenic alfalfa has corresponding effects on the amounts of 4′-O-methylated isoflavonoids present, suggesting that IOMT is somehow involved in the synthesis of these compounds in alfalfa, despite its specificity for the A-ring hydroxyl group of daidzein in vitro. Given our present findings, the apparent lack of isoformononetin accumulation in alfalfa, and the paradoxical in vivo labeling data concerning isoflavone O-methylation (Dewick and Martin, 1979b), we suggest two different hypotheses to explain the involvement of isoflavone 7-OMT in the biosynthesis of 4′-O-methylated isoflavonoids. In the first model, IOMT is part of a complex or metabolic channel in elicited or infected cells, in close association with the membrane-bound P450 that catalyzes the aryl migration of the B-ring (Figure 1). Such an association could lead to altered regiospecificity by facilitating direct presentation of daidzein into the active site of IOMT in a specific orientation with respect to the enzyme. Such channeling of daidzein is supported by the lack of any substantial pool of this compound in alfalfa (Dalkin et al., 1990), the lack of incorporation of daidzein into medicarpin in radiotracer studies (Dewick and Martin, 1979b), and the lack of isoformononetin production in elicited alfalfa leaves that overexpress IOMT. It is also supported by recent studies indicating that alfalfa IOMT, although an operationally soluble enzyme, associates with endoplasmic reticulum membranes after elicitation (C.-J. Liu, X.-Z. He, and R.A. Dixon, unpublished results).
In the second model, the unstable 2-hydroxyisoflavanone intermediate of the IFS reaction shown in Figure 1 would be the true substrate for 4′-O-methylation. We have not been able to test this model directly, because the recombinant 2-hydroxyisoflavanone synthase we recently cloned from soybean acts to convert liquiritigenin directly to daidzein in the insect cell expression system used, with no apparent accumulation of 2-hydroxyisoflavanone intermediate (Steele et al., 1999). Even if the second model is correct, metabolic channeling between 2-hydroxyisoflavanone synthase and IOMT may still be necessary to prevent spontaneous dehydration before methylation, an essential feature of the model if daidzein itself is not a substrate for 4′-O-methylation.
The increased amounts of PAL, IFR, and IFS transcripts in infected plants overexpressing IOMT make it difficult to conclude that the IOMT, rather than other enzymes (particularly IFS), is rate limiting for medicarpin biosynthesis in alfalfa. However, the lack of increased expression of any enzymes of isoflavonoid biosynthesis except IOMT in unchallenged leaves overexpressing the enzyme indicates that ectopic expression of IOMT does not lead to increased phytoalexin accumulation through some kind of stress response. Rather, the increased amounts of phenylpropanoid/isoflavonoid pathway gene transcripts in Phoma-infected IOMT-overexpressing plants could simply reflect the fact that the rapid accumulation of medicarpin resulting from IOMT overexpression protects the plant cells in the vicinity of the infection site from succumbing to the necrotrophic pathogen, thereby allowing them to continue producing defense gene transcripts. Alternatively, if the enzymes of the isoflavonoid pathway are organized in a metabolic channel, then mechanisms may exist to maintain stoichiometric concentrations of members of the complex based on perception of expression of other members of the complex; this would explain the increased expression of IFS, which may be the true rate-limiting step in the isoflavonoid pathway. Interestingly, inhibiting flux through IOMT by blocking cellular SAM synthesis through inhibition of S-adenosyl-l-homocysteine hydrolase with tubericidin causes accumulation of flavone derivatives in association with a reduction in isoflavonoid phytoalexin biosynthesis in alfalfa (Daniell et al., 1997). Thus, blocking isoflavone methylation chemically also blocks aryl migration in vivo, suggesting that the two processes are somehow linked mechanistically.
Genetic Manipulation of Isoflavonoid Phytoalexin Production and Disease Resistance
In recent years, the complete biosynthetic pathway to the phytoalexin medicarpin has been elucidated, and most of the genes encoding the biosynthetic enzymes have been cloned (Dixon, 1999). Thus far, however, there have been no reports of attempts to manipulate the isoflavonoid pathway by genetically modifying expression of biosynthetic pathway genes. Some of the intermediates of the medicarpin pathway retain substantial antifungal activity against some alfalfa pathogens, which might make it difficult to interpret assumed loss-of-function experiments (Blount et al., 1993), and targets for gain-of-function experiments have not, until now, been identified.
Introducing novel phytoalexins into plants can increase disease resistance. For example, production of the stilbene resveratrol in transgenic tobacco or alfalfa expressing a grapevine stilbene synthase gene leads to increased resistance to Botrytis cinerea (Hain et al., 1993) and P. medicaginis (Hipskind and Paiva, 2000), respectively. However, such studies do not address the functions of the endogenous phytoalexins of the host plant. In the case of IOMT-overexpressing alfalfa, the host's natural phytoalexin is overproduced, allowing assessment of the potential role of that compound in disease resistance in the natural host. IOMT-overexpressing alfalfa plants also accumulate increased amounts of formononetin glucoside after infection. This should not complicate interpretation of the results, however, because formononetin, in contrast to medicarpin, exerts no inhibitory effects on the growth of P. medicaginis, at least in vitro (Blount et al., 1993).
Clearly, increasing medicarpin production leads to effective resistance of alfalfa to the leaf spot pathogen P. medicaginis, providing direct evidence for the involvement of an isoflavonoid compound in disease resistance. Failure to demonstrate increased susceptibility in plants with decreased amounts of medicarpin may simply reflect the incomplete inhibition of accumulation of the phytoalexin in these studies. In contrast, in Arabidopsis phytoalexin-deficient mutants that exhibited increased susceptibility to virulent A. brassicola, camalexin was totally absent (Thomma et al., 1999).
Phytoalexins are often as phytotoxic as they are fungitoxic (Skipp et al., 1977), thus obviating constitutive overexpression of the pathway end products as a feasible strategy for engineering defense. However, constitutive IOMT overexpression leads to an increased phytoalexin response only after microbial infection, making this a viable approach for engineering disease resistance in legumes that accumulate 4′-O-methylated isoflavonoid phytoalexins. Such species include other Medicago spp, chickpea, clovers, and pea (Ingham, 1982). Future studies are now needed to evaluate the effects of the enhanced phytoalexin response in IOMT-overexpressing plants on other pathogens, and symbionts, of alfalfa.
METHODS
Plant Material
Alfalfa (Medicago sativa cv Regen SY) was maintained under standard greenhouse conditions or grown in a growth chamber (16 hr of light at 25°C, 8 hr of darkness at 19°C, 80 to 90% relative humidity).
Chemicals
Flavonoids were purchased from Indofine Chemicals (Somerville, NJ), except for isoformononetin (Apin Chemicals, Oxfordshire, UK). S-[3H-Methyl]adenosyl-l-methionine (radiolabeled SAM; 2.2 to 3.1 TBq/mmol) was obtained from Amersham (Arlington Heights, IL). Medicarpin was from our laboratory collection.
Elicitation and Infection of Alfalfa Leaves
For abiotic elicitation, rooted alfalfa cuttings were incubated in 3 mM CuCl2 for 8 hr at room temperature. Mature trifoliate leaves were placed on two layers of moist filter paper in a Petri dish and incubated for another 24 hr. To produce inoculum for fungal infection, Phoma medicaginis was cultured on potato dextrose agar for 1 month at 25°C. Spores were harvested by agitation in water containing 0.025% (v/v) Tween 20. After small wounds were generated on trifoliate leaves with a pinwheel, the leaves were dip-inoculated in ∼5 × 105 spores/mL and incubated in a moist chamber at room temperature. CuCl2-treated and infected leaves for isoflavonoid analysis were frozen in liquid N2 and stored at −80°C.
Vector Construction and Transformation of Alfalfa
All recombinant DNA techniques were performed essentially as described elsewhere (Sambrook et al., 1989). For sense vector construction, the expression plasmid pET15b/IOMT8, which contains the full-length IOMT8 cDNA (He et al., 1998), was digested with BamHI and NcoI. A 1.2-kb fragment was isolated and subcloned into the NcoI and BamHI sites of plasmid pRTL2 (Restrepo et al., 1990). For antisense vector construction, the full-length IOMT8 cDNA was amplified by polymerase chain reaction (PCR) using primers corresponding to the 5′ and 3′ ends of the cDNA clone with added KpnI and BamHI restriction sites; the PCR product was then digested with KpnI and BamHI before being cloned into pRTL2. Plasmids containing isoflavone O-methyltransferase (IOMT) sequences in pRTL2 were digested with HindIII, and 2.2-kb fragments containing the cauliflower mosaic virus (CaMV) 35S promoter, tobacco etch virus 5′-untranslated sequence, IOMT sequence, and CaMV terminator were isolated and ligated into the HindIII site of the binary vector pGA482 (An, 1986) (Figure 2A). The constructs were mobilized into Agrobacterium tumefaciens strain LBA4404 by electroporation (Cell-porator; Gibco BRL, Gaithersburg, MD). Alfalfa was transformed and regenerated through somatic embryogenesis with kanamycin (25 mg/L) as the selection marker, as described elsewhere (Thomas et al., 1990). All transformations were performed on clonally propagated cuttings of a single, highly regenerable line.
PCR Analysis of Alfalfa Transformants
Kanamycin-resistant alfalfa transformants were tested for the presence of IOMT sequences by PCR. Genomic DNA was extracted according to Edwards et al. (1991). DNA was resuspended in 10 mM Tris-HCl, pH 8.0. PCR reactions were performed in 50-μL volumes containing 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.25 mM MgCl2, 200 μM dNTPs, 0.5 units of Taq polymerase, 0.4 μM oligonucleotide primers, and 200 ng of plant DNA. Amplification of IOMT sequences was performed with 5′ (5′-GGCCATATGGCTTCATCAATTATGGC-3′) and 3′ (5′-CGGGATCCTTATGGATAGATCTCAA-3′) sequences of IOMT8 as primers, using a Perkin-Elmer Cetus 480 thermocycler with 35 cycles of denaturation (94°C for 1 min), annealing (55°C for 1 min), and extension (72°C for 1 min, with a 5-min extension at 72°C for the last cycle). The amplified endogenous IOMT band was larger than the amplified transgene band because of the presence of an intron in the endogenous gene.
DNA Gel Blot Analysis
Genomic DNA was isolated from leaf tissues with a Nucleon Phytopure plant DNA extraction kit (Amersham, Piscataway, NJ), digested with EcoRI, subjected to electrophoresis through an 0.8% agarose gel, and transferred to a Hybond-N membrane (Amersham) by capillary blotting. The membrane was hybridized with 32P-labeled IOMT probe (from nucleotides 249 to 1035) and washed once in 0.2 × SSC (1 × SSC is 0.15 NaCl and 0.015 M sodium citrate) containing 0.1% SDS at 42°C for 20 min and then three times for 30 min each at 65°C.
RNA Gel Blot Analysis
Total RNA was prepared with TRI Reagent (Molecular Research Center, Cincinnati, OH). RNA (30 μg/lane) was denatured and separated by electrophoresis through a 1% agarose/formaldehyde gel and transferred to a Hybond-N membrane by capillary blotting. Membranes were hybridized with probes prepared as follows. The phenylalanine ammonia-lyase (PAL) probe was from an alfalfa partial cDNA clone for PAL (Gowri et al., 1991), which was digested with HindIII, and a 700-bp fragment obtained was gel-purified. Cinnamate 4-hydroxylase (C4H) and acetyl CoA carboxylase probes were prepared by PCR with the alfalfa cDNA clones (Fahrendorf and Dixon, 1993; Shorrosh et al., 1994) as templates. The chalcone isomerase probe was prepared from the alfalfa cDNA clone (McKhann and Hirsch, 1994), which was digested with EcoRI, and a fragment of ∼1 kb was gel-purified. Isoflavone synthase (IFS) and isoflavone 2′-hydroxylase (I2′OH) probes were prepared from corresponding M. truncatula cDNA clones (C.L. Steele and R.A. Dixon, unpublished results), which were digested with EcoRI and XhoI. The IOMT probe was the nucleotide 249 to 1035 fragment of alfalfa IOMT8 (He et al., 1998). The isoflavone reductase (IFR) probe was prepared from the alfalfa cDNA clone for IFR (Paiva et al., 1991), which was digested with EcoRI. Probes were labeled with 32P by using the random priming procedure. After hybridization, the membranes were washed under high-stringency conditions, as described above for DNA gel blot analysis. RNA loading and transfer were checked by reprobing the blots with a soybean 18S rRNA probe.
Enzyme Assays
IOMT activity was analyzed as previously described (He and Dixon, 1996). Reaction mixtures were extracted with 0.5 mL of ethyl acetate, the ethyl acetate was evaporated under N2, and the residues were resuspended in methanol. Product analysis by thin-layer chromatography was performed as described previously (Edwards and Dixon, 1991). For product analysis by HPLC, 20 μL of resuspended residue was applied to a C18 column (5-μm particle size, 4.6 × 250 mm) and eluted with a gradient of increasing solvent B in solvent A (solvent A, 1% phosphoric acid in water; solvent B, acetonitrile: from 0 to 40 min, the gradient increased from 20 to 45% solvent B; from 40 to 42 min, solvent B increased from 45 to 95%). The eluate was monitored at 287 nm. Identity of products was confirmed by coelution and diode array analysis of UV spectra with authentic samples of formononetin and isoformononetin.
Protein Gel Blot Analysis
Polyclonal antiserum against IOMT was produced by immunizing rabbits (Covance Research Products., Denver, PA) with the protein antigen expressed from Escherichia coli (He et al., 1998). Protein extracts from leaf tissues were solubilized in sample buffer (25 mM Tris-HCl, pH 6.8, 1% SDS, 2.5% 2-mercaptoethanol, and 5% glycerol). Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes (Amersham) by electrophoretic blotting in transfer buffer (25 mM Tris-HCl, pH 8.3, 150 mM glycine, and 20% [v/v] methanol). Blots were probed with polyclonal antiserum raised against purified alfalfa IOMT as the primary antibody and anti–(rabbit IgG)–peroxidase conjugate as the secondary antibody. The IOMT signal was detected by exposing the blots to x-ray film shortly after incubation with ECL reagent (Amersham).
Feeding Studies and Phenolic Analysis
For feeding studies, 1 mg of daidzein was dissolved in a minimum amount of 0.5 M NaOH, and 1 mL of 0.1 M phosphate buffer, pH 7.2, was added (Dewick and Martin, 1979b). The daidzein suspension was infiltrated into alfalfa leaves with a 1-mL syringe. Extraction and HPLC analysis of phenolic compounds was performed as described previously (Dalkin et al., 1990). Twenty microliters of the final methanolic extract was applied to a C18 HPLC column for analysis, as described above. Retention times and calibration curves for formononetin, isoformononetin, medicarpin, and isoflavonoid conjugates were established with authentic samples. The concentrations of the compounds are expressed as nanomoles per gram (fresh weight) of sample.
Acknowledgments
We thank Darla Boydston, Cuc Ly, and Paul Horton for artwork and Drs. Nancy Paiva and Christopher Steele for helpful discussions and critical reading of the manuscript. This work was supported by the Samuel Roberts Noble Foundation.
References
- Adlercreutz, H., Honjo, H., Higashi, A., Fotsis, T., Hämäläinen, E., Hasegawa, T., and Okada, H. (1991). Urinary excretion of lignans and isoflavonoid phytoestrogens in Japanese men and women consuming a traditional Japanese diet. Am. J. Clin. Nutr. 54, 1093–1100. [DOI] [PubMed] [Google Scholar]
- Adlercreutz, M. (1998). Epidemiology of phytoestrogens. Baillieres Clin. Endocrinol. Metab. 12, 605–623. [DOI] [PubMed] [Google Scholar]
- An, G. (1986). Development of plant promoter expression vectors and their use for analysis of differential activity of nopaline synthase promoter in transformed tobacco cells. Plant Physiol. 81, 86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, S., Grubbs, C., Setchell, K.D.R., and Carlson, J. (1990). Soybeans inhibit mammary tumors in models of breast cancer. In Mutagens and Carcinogens in the Diet. M.W. Pariza, ed (New York: Wiley-Liss), pp. 239–253. [PubMed]
- Barz, W., and Welle, R. (1992). Biosynthesis and metabolism of isoflavones and pterocarpan phytoalexins in chickpea, soybean and phytopathogenic fungi. Recent Adv. Phytochem. 26, 139–164. [Google Scholar]
- Blount, J.W., Dixon, R.A., and Paiva, N.L. (1993). Stress responses in alfalfa (Medicago sativa L.). XVI. Antifungal activity of medicarpin and its biosynthetic precursors; implications for the genetic manipulation of stress metabolites. Physiol. Mol. Plant Pathol. 41, 333–349. [Google Scholar]
- Bugos, R.C., Chiang, V.L.C., and Campbell, W.H. (1991). cDNA cloning, sequence analysis and seasonal expression of lignin-bispecific caffeic acid/5-hydroxyferulic acid O-methyltransferase of aspen. Plant Mol. Biol. 17, 1203–1215. [DOI] [PubMed] [Google Scholar]
- Dalkin, K., Edwards, R., Edington, B., and Dixon, R.A. (1990). Stress responses in alfalfa (Medicago sativa L.). I. Elicitor-induction of phenylpropanoid biosynthesis and hydrolytic enzymes in cell suspension cultures. Plant Physiol. 92, 440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell, T., O'Hagan, D., and Edwards, R. (1997). Alfalfa cell cultures treated with a fungal elicitor accumulate flavone metabolites rather than isoflavones in the presence of the methylation inhibitor tubericidin. Phytochemistry 44, 285–291. [Google Scholar]
- Dewick, P.M., and Martin, M. (1979. a). Biosynthesis of pterocarpan and isoflavan phytoalexins in Medicago sativa: The biochemical interconversion of pterocarpans and 2′-hydroxyisoflavans. Phytochemistry 18, 591–596. [Google Scholar]
- Dewick, P.M., and Martin, M. (1979. b). Biosynthesis of pterocarpan, isoflavan and coumestan metabolites of Medicago sativa: Chalcone, isoflavone and isoflavanone precursors. Phytochemistry 18, 597–602. [Google Scholar]
- Dixon, R.A. (1986). The phytoalexin response: Elicitation, signaling and the control of host gene expression. Biol. Rev. 61, 239–291. [Google Scholar]
- Dixon, R.A. (1999). Isoflavonoids: Biochemistry, molecular biology and biological functions. In Comprehensive Natural Products Chemistry, Vol. 1, U. Sankawa, ed (Amsterdam: Elsevier), pp. 773–823.
- Dixon, R.A., and Paiva, N.L. (1995). Stress-induced phenylpropanoid metabolism. Plant Cell 7, 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, R.A., Dey, P.M., and Lamb, C.J. (1983). Phytoalexins: Enzymology and molecular biology. Adv. Enzymol. Relat. Areas Mol. Biol. 55, 1–136. [DOI] [PubMed] [Google Scholar]
- Dixon, R.A., Canovas, P., Guo, Z.-J., He, X.-Z., Lamb, C., and McAlister, F. (1999). Molecular controls for isoflavonoid biosynthesis in relation to plant and human health. Recent Adv. Phytochem. 33, 133–160. [Google Scholar]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19, 1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, R., and Dixon, R.A. (1991). Isoflavone O-methyltransferase activities in elicitor-treated cell suspension cultures of Medicago sativa. Phytochemistry 30, 2597–2606. [Google Scholar]
- Fahrendorf, T., and Dixon, R.A. (1993). Stress responses in alfalfa (Medicago sativa L.). XVIII: Molecular cloning of the elicitor-inducible cinnamic acid 4-hydroxylase cytochrome P450 from alfalfa. Arch. Biochem. Biophys. 305, 509–515. [DOI] [PubMed] [Google Scholar]
- Frick, S., and Kutchan, T.M. (1999). Molecular cloning and functional expression of O-methyltransferases common to isoquinoline alkaloid and phenylpropanoid biosynthesis. Plant J. 17, 329–339. [DOI] [PubMed] [Google Scholar]
- Gauthier, A., Gulick, P., and Ibrahim, R.K. (1996). cDNA cloning and characterization of a 3′/5′-O-methyltransferase for partially methylated flavonols from Chrysosplenium americanum. Plant Mol. Biol. 32, 1163–1169. [DOI] [PubMed] [Google Scholar]
- Gauthier, A., Gulick, P.J., and Ibrahim, R.K. (1998). Characterization of two cDNA clones which encode O-methyltransferases for the methylation of both flavonoid and phenylpropanoid compounds. Arch. Biochem. Biophys. 351, 243–249. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., and Ausubel, F.M. (1994). Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. USA 91, 8955–8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowri, G., Paiva, N.L., and Dixon, R.A. (1991). Stress responses in alfalfa (Medicago sativa L.). 12. Sequence analysis of phenylalanine ammonia-lyase (PAL) cDNA clones and appearance of PAL transcripts in elicitor-treated cell cultures and developing plants. Plant Mol. Biol. 17, 415–429. [DOI] [PubMed] [Google Scholar]
- Hain, R., Reif, H.-J., Krause, E., Langebartels, R., Kindl, H., Vornam, B., Wiese, W., Schmelzer, E., Schrier, P.H., Stocker, R.H., and Stenzel, K. (1993). Disease resistance results from foreign phytoalexin expression in a novel plant. Nature 361, 153–156. [DOI] [PubMed] [Google Scholar]
- Hakamatsuka, T., Hashim, M.F., Ebizuka, Y., and Sankawa, U. (1991). P-450–dependent oxidative rearrangement in isoflavone biosynthesis: Reconstitution of P-450 and NADPH:P450 reductase. Tetrahedron 47, 5969–5978. [Google Scholar]
- Hakamatsuka, T., Mori, K., Ishida, S., Ebizuka, Y., and Sankawa, U. (1998). Purification of 2-hydroxyisoflavanone dehydratase from the cell cultures of Pueraria lobata. Phytochemistry 49, 497–505. [Google Scholar]
- He, X.-Z., and Dixon, R.A. (1996). Affinity chromatography, substrate/product specificity and amino acid sequence analysis of an isoflavone O-methyltransferase from alfalfa (Medicago sativa L.). Arch. Biochem. Biophys. 336, 121–129. [DOI] [PubMed] [Google Scholar]
- He, X.-Z., Reddy, J.T., and Dixon, R.A. (1998). Stress responses in alfalfa (Medicago sativa L.). XXII. cDNA cloning and characterization of an elicitor-inducible isoflavone 7-O-methyltransferase. Plant Mol. Biol 36, 43–54. [DOI] [PubMed] [Google Scholar]
- Higgins, V.J. (1972). Role of the phytoalexin medicarpin in three leaf spot diseases of alfalfa. Physiol. Plant Pathol. 2, 289–300. [Google Scholar]
- Hipskind, J.D., and Paiva, N.L. (2000). Constitutive accumulation of a resveratrol-glucoside in transgenic alfalfa increases resistance to Phoma medicaginis. Mol. Plant-Microbe Interact. 13, 551–562. [DOI] [PubMed] [Google Scholar]
- Ibrahim, R.K., De Luca, V., Khouri, H., Latchinian, L., Brisson, L., and Charest, P.M. (1987). Enzymology and compartmentation of polymethylated flavonol glucosides in Chrysosplenium americanum. Phytochemistry 26, 1237–1245. [Google Scholar]
- Ibrahim, R.K., Bruneau, A., and Bantignies, B. (1998). Plant O-methyltransferases: Molecular analysis, common signature and classification. Plant Mol. Biol. 36, 1–10. [DOI] [PubMed] [Google Scholar]
- Ingham, J. (1982). Phytoalexins from the Leguminosae. In Phytoalexins, J.A. Bailey and J.W. Mansfield, eds (New York: Halsted Press), pp. 21–80.
- Joshi, C.P., and Chiang, V.L. (1998). Conserved sequence motifs in plant S-adenosyl-l-methionine–dependent methyltransferases. Plant Mol. Biol. 37, 663–674. [DOI] [PubMed] [Google Scholar]
- Jung, W., Yu, O., Lau, S.-M.C., O'Keefe, D.P., Odell, J., Fader, G., and McGonigle, B. (2000). Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat. Biotechnol. 18, 208–212. [DOI] [PubMed] [Google Scholar]
- Kessmann, H., Choudhary, A.D., and Dixon, R.A. (1990). Stress responses in alfalfa (Medicago sativa L.). III. Induction of medicarpin and cytochrome P450 enzyme activities in elicitor-treated cell suspension cultures and protoplasts. Plant Cell Rep. 9, 38–41. [DOI] [PubMed] [Google Scholar]
- Kochs, G., and Grisebach, H. (1986). Enzymic synthesis of isoflavones. Eur. J. Biochem. 155, 311–318. [DOI] [PubMed] [Google Scholar]
- Kosslak, R.M., Bookland, R., Barkei, J., Paaren, H.E., and Applebaum, E.R. (1987). Induction of Bradyrhizobium japonicum common nod genes by isoflavones isolated from Glycine max. Proc. Natl. Acad. Sci. USA 84, 7428–7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuć, J. (1995). Phytoalexins, stress metabolism, and disease resistance in plants. Annu. Rev. Phytopathol. 33, 275–297. [DOI] [PubMed] [Google Scholar]
- Lamb, C.J., Ryals, J.A., Ward, E.R., and Dixon, R.A. (1992). Emerging strategies for enhancing crop resistance to microbial pathogens. Bio/Technology 10, 1436–1445. [DOI] [PubMed] [Google Scholar]
- Lee, H.P., Gourley, L., Duffy, S.W., Esteve, J., Lee, J., and Day, N.E. (1991). Dietary effects on breast-cancer risk in Singapore. Lancet 337, 1197–1200. [DOI] [PubMed] [Google Scholar]
- Li, L., Popko, J.L., Zhang, X.-H., Osakabe, K., Tsai, C.-J., Joshi, C.P., and Chiang, V.L. (1997). A novel multifunctional O-methyltransferase implicated in a dual methylation pathway associated with lignin biosynthesis in loblolly pine. Proc. Natl. Acad. Sci. USA 94, 5461–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Popko, J.L., Umezawa, T., and Chiang, V.L. (2000). 5-Hydroxyconiferyl aldehyde modulates enzymatic methylation for syringyl monolignol formation, a new view of monolignol biosynthesis in angiosperms. J. Biol. Chem. 275, 6537–6545. [DOI] [PubMed] [Google Scholar]
- Matzke, M.A., and Matzke, A.J.M. (1995). How and why do plants inactivate homologous (trans)genes? Plant Physiol. 107, 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann, H.I., and Hirsch, A.M. (1994). Isolation of chalcone synthase and chalcone isomerase cDNAs from alfalfa (Medicago sativa L.): Highest transcript levels occur in young roots and root tips. Plant Mol. Biol. 24, 767–777. [DOI] [PubMed] [Google Scholar]
- Paiva, N.L., Edwards, R., Sun, Y., Hrazdina, G., and Dixon, R.A. (1991). Stress responses in alfalfa (Medicago sativa L.). XI. Molecular cloning and expression of alfalfa isoflavone reductase, a key enzyme of isoflavonoid phytoalexin biosynthesis. Plant Mol. Biol. 17, 653–667. [DOI] [PubMed] [Google Scholar]
- Paiva, N.L., Oommen, A., Harrison, M.J., and Dixon, R.A. (1994). Regulation of isoflavonoid metabolism in alfalfa. Plant Cell Tissue Organ Cult. 38, 213–220. [Google Scholar]
- Restrepo, M.A., Freed, D.D., and Carrington, J.C. (1990). Nuclear transport of plant potyviral proteins. Plant Cell 2, 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular cloning. In Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Shorrosh, B.S., Dixon, R.A., and Ohlrogge, J.B. (1994). Molecular cloning, characterization and elicitation of acetyl CoA carboxylase from alfalfa. Proc. Natl. Acad. Sci. USA 91, 4323–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutt, D.A. (1976). The effects of plant oestrogens on animal reproduction. Endeavour 75, 110–113. [DOI] [PubMed] [Google Scholar]
- Skipp, R.A., Selby, C., and Bailey, J.A. (1977). Toxic effects of phaseollin on plant cells. Physiol. Plant Pathol. 10, 221–227. [Google Scholar]
- Steele, C.L., Gijzen, M., Qutob, D., and Dixon, R.A. (1999). Molecular characterization of the enzyme catalyzing the aryl migration reaction of isoflavonoid biosynthesis in soybean. Arch. Biochem. Biophys. 367, 147–150. [DOI] [PubMed] [Google Scholar]
- Sumner, L., Paiva, N.L., Dixon, R.A., and Geno, P.W. (1996). High-performance liquid chromatography/continuous-flow liquid secondary ion mass spectrometry of flavonoid glucosides in leguminous plant extracts. J. Mass Spectrom. 31, 472–485. [DOI] [PubMed] [Google Scholar]
- Thomas, M.R., Johnson, L.B., and White, F.F. (1990). Selection of interspecific somatic hybrids of Medicago by using Agrobacterium-transformed tissue. Plant Sci. 69, 189–198. [Google Scholar]
- Thomma, B.P.H.J., Eggermont, K., Penninckx, I.A.M.A., Mauch-Mani, B., Vogelsang, R., Cammue, B.P.A., and Broekaert, W.F. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95, 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P.H.J., Nelissen, I., Eggermont, K., and Broekaert, W.F. (1999). Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicola. Plant J. 19, 163–171. [DOI] [PubMed] [Google Scholar]
- VanEtten, H.D., Matthews, D.E., and Matthews, P.S. (1989). Phytoalexin detoxification: Importance for pathogenicity and practical implications. Annu. Rev. Phytopathol. 27, 143–164. [DOI] [PubMed] [Google Scholar]
- Wu, Q., Presig, C.L., and VanEtten, H.D. (1997). Isolation of the cDNAs encoding (+)6-hydroxymaackiain 3-O-methyltransferase, the terminal step for the synthesis of the phytoalexin pisatin in Pisum sativum. Plant Mol. Biol. 35, 551–560. [DOI] [PubMed] [Google Scholar]
- Yanagihara, K., Ito, A., Toge, T., and Numoto, M. (1993). Antiproliferative effects of isoflavones on human cancer cell lines established from the gastrointestinal tract. Cancer Res. 53, 5815–5821. [PubMed] [Google Scholar]
- Zhou, N., Tootle, T.L., and Glazebrook, J. (1999). Arabidopsis pad3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11, 2419–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]