Abstract
CONSTANS (CO) promotes flowering of Arabidopsis in response to long photoperiods. Transgenic plants carrying CO fused with the cauliflower mosaic virus 35S promoter (35S::CO) flowered earlier than did the wild type and were almost completely insensitive to length of day. Genes required for CO to promote flowering were identified by screening for mutations that suppress the effect of 35S::CO. Four mutations were identified that partially suppressed the early-flowering phenotype caused by 35S::CO. One of these mutations, suppressor of overexpression of CO 1 (soc1), defines a new locus, demonstrating that the mutagenesis approach is effective in identifying novel flowering-time mutations. The other three suppressor mutations are allelic with previously described mutations that cause late flowering. Two of them are alleles of ft, indicating that FT is required for CO to promote early flowering and most likely acts after CO in the hierarchy of flowering-time genes. The fourth suppressor mutation is an allele of fwa, and fwa soc1 35S::CO plants flowered at approximately the same time as co mutants, suggesting that a combination of fwa and soc1 abolishes the promotion of flowering by CO. Besides delaying flowering, fwa acted synergistically with 35S::CO to repress floral development after bolting. The latter phenotype was not shown by any of the progenitors and was most probably caused by a reduction in the function of LEAFY. These genetic interactions suggest models for how CO, FWA, FT, and SOC1 interact during the transition to flowering.
INTRODUCTION
Flowering time of Arabidopsis is influenced by both environmental conditions, which include length of day, temperature, light quality, and nutrient deprivation, and developmental factors associated with the age of the plant (Martínez-Zapater et al., 1994). This complexity is mediated by several genetic pathways that control the response to different environmental signals (Koornneef et al., 1998a; Levy and Dean, 1998). These pathways were originally demonstrated by analyzing the phenotypes of late-flowering mutants (Martínez-Zapater and Somerville, 1990; Koornneef et al., 1991). The mutations were classified on the basis of their effects on environmental responses, particularly those involving temperature and photoperiod, and their genetic interactions. These experiments led to the proposal that three general pathways promote flowering of Arabidopsis.
One of these pathways, called the long-day pathway, controls the promotion of flowering by day length. Arabidopsis flowers rapidly under long-day conditions consisting of 16 hr of light but flowers later during short days with 8 or 10 hr of light. The mutations that define the long-day pathway (constans [co], gigantea [gi], lhy, ft, fwa, and fha) delay flowering under long-day but not short-day conditions (Redei, 1962; Koornneef et al., 1991).
Mutants of the second class (e.g., fca, fve, luminidependens [ld], and fy) flower later than those of the wild type under both long and short days and respond strongly to vernalization—an extended exposure to low temperatures soon after germination (Martínez-Zapater and Somerville, 1990; Koornneef et al., 1991). This class of mutations is assigned to the autonomous flowering pathway, which is active under all day lengths. This pathway was recently shown to act, at least in part, by reducing the abundance of the mRNA encoded by the FLOWERING LOCUS C (FLC) gene (Michaels and Amasino, 1999; Sheldon et al., 1999).
Mutations of the third class reduce synthesis or response to the growth regulator gibberellin (GA). These have their most severe effect under short-day conditions, and mutations that have a strong effect on GA biosynthesis can prevent flowering in plants growing under short days (Wilson et al., 1992).
Further evidence that these phenotypic classes represent genetic pathways resulted from analyses of double mutants. Generally, double mutants containing two mutations assigned to one of these classes do not flower later than the more extreme parental mutant, whereas combining mutations from different classes causes a greater delay in flowering than does either single mutation (Koornneef et al., 1998b). Because the three pathways are partially redundant, inactivation of two pathways results in a more severe phenotype than inactivation of one.
Several of these flowering-time genes have now been cloned and generally encode regulatory proteins. CO, LD, FLC, LHY, and CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) encode putative transcription factors (Lee et al., 1994; Putterill et al., 1995; Schaffer et al., 1998; Wang and Tobin, 1998; Michaels and Amasino, 1999; Sheldon et al., 1999), whereas FCA encodes a probable RNA binding protein (Macknight et al., 1997). The FT gene encodes a protein with similarity to phosphatidylethanolamine binding proteins (Kardailsky et al., 1999; Kobayashi et al., 1999) and is a homolog of TERMINAL FLOWER 1 (Bradley et al., 1997; Oshima et al., 1997), whereas the GI gene encodes a putative membrane protein of unknown function (Fowler et al., 1999; Park et al., 1999).
The order in which genes act within a single flowering-time pathway has been difficult to assess genetically because each mutation causes a similar phenotype. The order of gene action was determined genetically in other plant signaling pathways, such as the ethylene signaling pathway in which repressor and promotive steps occur within the same pathway, and mutations affecting these distinct functions have opposite phenotypes (Roman et al., 1995). Some information on the order in which flowering-time genes act has come from analyzing the expression of cloned genes in mutant backgrounds. Such analyses have demonstrated, for example, that the autonomous pathway genes and genes for vernalization act upstream of FLC to repress its expression (Michaels and Amasino, 1999; Sheldon et al., 1999). Similarly, in the long-day pathway, CO expression is reduced in a cryptochrome 2 (cry2) mutant, and FT expression is reduced by co mutations, suggesting that these genes act in the order CRY2-CO-FT (Guo et al., 1998; Kardailsky et al., 1999; Kobayashi et al., 1999).
The long-day, autonomous, and GA pathways must eventually converge to regulate the expression and function of the same floral meristem identity genes that control flower development (Simon et al., 1996; Blazquez et al., 1998; Nilsson et al., 1998; reviewed in Piñeiro and Coupland, 1998), and the effects of flowering-time genes on the expression or function of floral meristem identity genes have also been informative. Inducing the function of CO by using a steroid-inducible construct increased LEAFY (LFY) expression within 24 hr after steroids were applied (Simon et al., 1996). These results suggest that LFY acts downstream of CO.
The relationship between flowering time and floral meristem identity genes has also been examined by using transgenic plants in which the LFY gene was overexpressed from the cauliflower mosaic virus (CaMV) 35S promoter. Broadly, the effect of the 35S::LFY transgene on flowering time was not greatly affected by most flowering-time mutations—again suggesting that the flowering-time genes act to promote LFY transcription (Nilsson et al., 1998). However, ft and fwa were exceptions among the flowering-time mutations because they did delay flowering of 35S::LFY plants (Nilsson et al., 1998). These mutations may therefore act to influence the activity of LFY protein. Ruiz-Garcia et al. (1997) came to a similar conclusion by examining the effects of combining ft and fwa mutations with lfy and concluded that these flowering-time genes do not act to regulate LFY transcription but rather regulate transcription of another floral meristem identity gene, APETALA1 (AP1). Overexpression of FT, however, did cause premature expression of LFY (Kardailsky et al., 1999), probably as an indirect effect of activation of AP1 by FT.
Our interest is in the long-day pathway, and we have previously described the CO gene (Putterill et al., 1995). Transgenic plants containing increased copy numbers of CO flower earlier than the wild type under long- and short-day conditions, suggesting that the amount of CO expression in wild-type plants limits flowering time. This was confirmed by expressing the steroid-inducible CO fusion protein from the strong CaMV 35S promoter (Simon et al., 1996). In the presence of the steroid dexamethasone, overexpression of the fusion protein was sufficient to promote very early flowering under both long and short days. Transcriptional regulation of CO is therefore an important determinant of the photoperiodic regulation of flowering time. Here, we describe transgenic plants expressing the wild-type CO protein from the CaMV 35S promoter. The transgene causes early flowering and therefore provides a phenotype opposite to that of the co loss-of-function mutations. Suppressor mutagenesis of these plants was used to identify genes that are required for CO function. This approach identified mutations in the previously identified flowering-time genes ft and fwa as well as in a previously unidentified gene that we designate SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1). The roles of these genes in the control of flowering time are discussed.
RESULTS
Effects of Expression of the CO Gene from the CaMV 35S Promoter
The CaMV 35S promoter was fused to genomic DNA encoding the CO open reading frame (see Methods). The gene fusion (35S::CO) was introduced into a Landsberg erecta (Ler) line homozygous for the co-2 mutation. Plants homozygous for the transgene at a single locus were identified from among the progeny of five independent transformants. RNA was extracted from all five lines, and gel blots were made and hybridized with a CO probe. As expected from previous experiments, no hybridization was detected in wild-type plants because of the very low expression of CO (Putterill et al., 1995). However, mRNA of the expected size was recognized in all five transformants (Figures 1A and 1B), indicating that transcription of the gene was considerably increased in all of the transgenic lines. The phenotypes of these five lines were then studied in detail.
Figure 1.
Detection of CO mRNA in Wild-Type and Transgenic Plants.
(A) Total RNA extracted from wild-type Ler (WT) and transgenic plants carrying 35S::CO:GR (CO:GR) or five transformants carrying 35S::CO (lanes 1 to 5) was transferred to a filter and hybridized with the CO cDNA.
(B) As a control for loading, the filter was stripped and hybridized with a fragment derived from the tubulin gene (see Methods).
All five lines flowered earlier than did wild-type plants under both long- and short-day conditions (Figure 2 and Table 1). Earlier flowering was apparent both in the time between sowing until floral buds were visible in the center of the rosette and in the fewer leaves formed by the shoot meristem before the initiation of flower development. Overexpression of CO in these lines, therefore, causes early flowering under long- and short-day conditions, although the effect is most dramatic under short-day conditions (Table 1).
Figure 2.
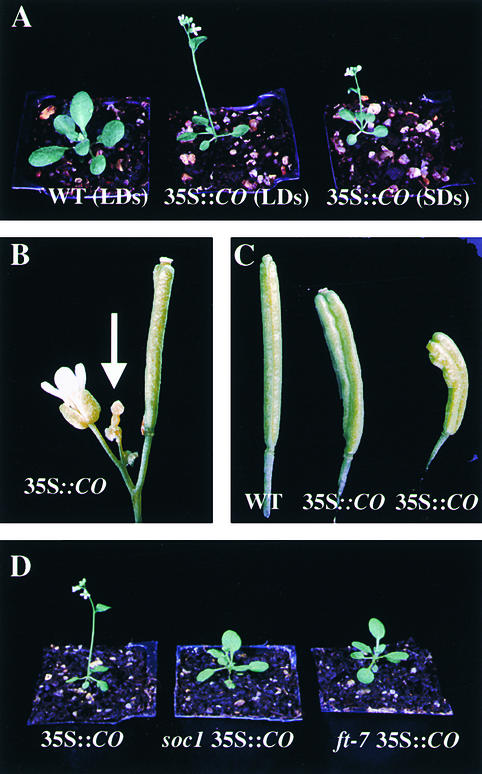
Phenotype of 35S::CO and 35S::CO Plants Carrying Suppressor Mutations.
(A) A 3-week-old wild-type (WT) plant grown under long days (LDs) (left), a 35S::CO plant grown under long days (center), and a 35S::CO plant grown under short days (SDs) (right).
(B) The inflorescence apex and the pistil-like structure formed at the apex of the shoot of a 35S::CO plant (arrow).
(C) Siliques of wild-type (left) and 35S::CO (center and right) plants.
(D) A 3-week-old 35S::CO plant (left), a soc1 35S::CO plant (center), and a ft-7 35S::CO plant (originally designated suppressor mutant 1) (right) grown under long-day conditions.
Table 1.
Effect of the Suppressor Mutations and fca-1 on Flowering Time in the Presence of 35S::CO
| Genotype | No. of Rosette Leaves | No. of Cauline Leaves | Total No. of Leaves | No. of Days to Flowering |
|---|---|---|---|---|
| ft-7 35S::COa | ||||
| Long day | 5.7 ± 0.6 | 3.1 ± 0.8 | 8.8 ± 0.8 | 16.5 ± 0.8 |
| Short day | 8.9 ± 0.9 | 3.2 ± 0.5 | 12.1 ± 1.1 | 22.0 ± 1.3 |
| fwa-100 35S::COb | ||||
| Long day | 7.6 ± 0.5 | 7.2 ± 0.9 | 14.8 ± 1.0 | NDc |
| Short day | 8.8 ± 1.1 | 5.9 ± 1.1 | 14.7 ± 1.7 | ND |
| soc1 35S::COd | ||||
| Long day | 4.4 ± 0.6 | 2.5 ± 0.5 | 6.9 ± 0.6 | 15.4 ± 1.5 |
| Short day | 5.0 ± 0.4 | 1.5 ± 0.5 | 6.5 ± 0.8 | 18.0 ± 0.7 |
| ft-8 35S::COe | ||||
| Long day | 6.3 ± 0.6 | 3.2 ± 0.6 | 9.6 ± 0.8 | ND |
| Short day | 10.4 ± 0.7 | 2.9 ± 0.4 | 13.3 ± 0.7 | ND |
| fca-1 35S::CO | ||||
| Long day | 4.8 ± 0.4 | 2.1 ± 0.4 | 6.8 ± 0.5 | ND |
| ft-8 fca-1 35S::COe | ||||
| Long day | 15.4 ± 1.2 | 5.7 ± 0.7 | 21.1 ± 1.3 | ND |
| ft-1 35S::CO | ||||
| Long day | 4.1 ± 0.1 | 2.5 ± 0.3 | 6.6 ± 0.3 | ND |
| Short day | 7.1 ± 0.2 | 3.2 ± 0.3 | 10.2 ± 0.5 | ND |
| 35S::CO | ||||
| Long day | 2.5 ± 0.5 | 2.0 ± 0.4 | 4.5 ± 0.8 | 13.5 ± 0.9 |
| Short day | 2.4 ± 0.5 | 2.0 ± 0.6 | 4.5 ± 0.5 | 15.0 ± 1.2 |
| Ler | ||||
| Long day | 5.5 ± 0.5 | 3.0 ± 0.4 | 8.5 ± 0.7 | 17.0 ± 0.7 |
| Short day | 31.4 ± 1.8 | 12.2 ± 1.0 | 43.6 ± 2.0 | 46.7 ± 1.6 |
ft-7 was originally designated suppressor mutation 1.
fwa-100 was originally designated suppressor mutation 2.
ND, not determined.
soc1 was originally designated suppressor mutation 3.
ft-8 was originally designated suppressor mutation 4.
The 35S::CO plants were almost insensitive to the length of day, and their total leaf number was the same under both long- and short-day conditions (Table 1). In contrast, wild-type plants flowered ∼30 days earlier under long-day conditions than under short days (Table 1). Occasionally, 35S::CO plants grown during short days formed more rosette leaves and fewer cauline leaves than did long-day-grown plants, suggesting that in these individuals, day length affected the node at which internode elongation first occurred.
In wild-type plants, the distribution of trichomes alters around the time of the transition to flowering (Chien and Sussex, 1996; Telfer et al., 1997); therefore, we tested whether 35S::CO caused trichome distribution to change earlier than it did in wild-type plants. The rosette leaves that form early during the development of wild-type plants form numerous trichomes on their upper (adaxial) sides and no trichomes on their lower (abaxial) sides, whereas leaves that develop later form fewer adaxial trichomes and many abaxial trichomes. This effect is influenced by day length, because abaxial trichomes are formed later in shoot development under short-day conditions (Chien and Sussex, 1996; Telfer et al., 1997). Wild-type plants growing under our long-day conditions usually produced their first abaxial trichomes on leaf 5, which was often the first cauline leaf, although ∼20% of the plants formed a few abaxial trichomes on leaf 4. After their initial appearance, abaxial trichomes were always more frequent on successive leaves. Abaxial trichomes appeared earlier on 35S::CO plants growing under the same conditions: they were present on leaf 3 of ∼60% of the plants and on leaf 4 of all plants. Under short-day conditions, 35S::CO plants always had abaxial trichomes on leaf 4, but they were present on the third leaf of only ∼20% of plants. Abaxial trichomes, therefore, appear earlier on 35S::CO plants than on the wild type, and their distribution is slightly influenced by day length.
35S::CO plants also showed characteristic modifications in flower and shoot morphology. The transformants formed fewer flowers than did the wild type. For example, under long-day conditions, 35S::CO plants formed 11 ± 1.5 flowers compared with 29.3 ± 1.6 for Ler. Also, the shoot of 35S::CO plants terminated in the formation of a pistil-like structure (Figure 2). The shoot of 35S::CO plants is therefore determinate, in contrast to the indeterminate shoot of wild-type plants. The flowers of 35S::CO plants also show alterations in morphology, particularly to the gynoecium, which often comprises three or four carpels rather than the two that are present in wild-type flowers (Figure 2).
Isolation of Mutations That Suppress the Extreme Early-Flowering Phenotype of 35S::CO Plants
To identify mutations that suppress the effect of overexpression of CO, we performed two screens for mutations that delay the extreme early-flowering phenotype of 35S::CO plants. In the first experiment, seeds carrying 35S::CO were mutagenized with ethyl methanesulfonate (EMS), and individuals that flowered later than the 35S::CO progenitor were identified in an M2 population of ∼20,000 plants grown under short-day conditions (see Methods). Five M2 plants that flowered substantially later than 35S::CO were identified. Three of these mutations were reliably transmitted to subsequent generations and were designated suppressor mutations 1 to 3.
In the second screen, fca-1 35S::CO seeds were mutagenized with EMS. The flowering-time gene FCA acts in the autonomous flowering-time pathway (Koornneef et al., 1998b; Levy and Dean, 1998), which is distinct from the long-day pathway in which CO acts. The fca-1 mutation strongly delays flowering under long-day conditions, whereas 35S::CO almost entirely overcomes this effect, although fca-1 35S::CO plants flower slightly later than do 35S::CO plants (Table 1). To reduce the likelihood of isolating mutations, such as fca-1, that affect the autonomous flowering-time pathway and therefore to focus the screen on genes more closely related to CO, fca-1 35S::CO plants in which this pathway was already inactivated were used for the mutagenesis. An M2 population of ∼5000 plants, generated by EMS mutagenesis, was screened under long-day conditions, and one plant flowering later than the fca-1 35S::CO progenitor was identified. The mutation in this plant was designated suppressor mutation 4.
Genetic Characterization of the Suppressor Mutations
All four suppressor mutations in the 35S::CO background were crossed to wild-type Ler, and F2 populations were generated. Individuals that flowered as early as 35S::CO were present in each F2 population, indicating that all of the suppressor mutations are extragenic suppressors of 35S::CO and none is closely linked to 35S::CO. Furthermore, in these populations, plants that carried each of the suppressor mutations but lacked 35S::CO were identified. All of the suppressor mutations caused a late-flowering phenotype under long days in the absence of 35S::CO (Table 2).
Table 2.
Effect of the Suppressor Mutations and fca-1 on Flowering Time in Wild-Type Background
| Genotype | No. of Rosette Leaves | No. of Cauline Leaves | Total No. of Leaves | No. of Days to Flowering |
|---|---|---|---|---|
| ft-7a | ||||
| Long day | 16.4 ± 1.0 | 7.1 ± 0.7 | 23.5 ± 1.6 | 31.5 ± 0.9 |
| Short day | 31.7 ± 1.8 | 12.8 ± 1.0 | 44.5 ± 1.7 | 54.6 ± 3.1 |
| fwa-100b | ||||
| Long day | 16.5 ± 0.7 | 11.6 ± 0.8 | 28.1 ± 1.1 | 29.9 ± 1.4 |
| Short day | 36.5 ± 2.4 | 18.8 ± 1.4 | 55.3 ± 2.5 | 53.7 ± 2.2 |
| soc1c | ||||
| Long day | 9.2 ± 0.4 | 3.5 ± 0.5 | 12.7 ± 0.6 | 19.1 ± 0.5 |
| Short day | 38.5 ± 2.9 | 15.3 ± 1.6 | 53.9 ± 2.7 | 56.3 ± 1.6 |
| ft-8d | ||||
| Long day | 15.6 ± 0.8 | 7.5 ± 0.7 | 23.1 ± 1.0 | NDe |
| ft-1 | ||||
| Long day | 9.8 ± 0.5 | 4.1 ± 0.5 | 13.8 ± 0.4 | ND |
| Short day | 32.1 ± 1.6 | 15.4 ± 0.7 | 47.4 ± 1.8 | 51.3 ± 1.9 |
| fca-1 | ||||
| Long day | 21.2 ± 2.7 | 6.8 ± 0.9 | 28.0 ± 3.4 | ND |
| Ler | ||||
| Long day | 5.5 ± 0.5 | 3.0 ± 0.4 | 8.5 ± 0.7 | 17.0 ± 0.7 |
| Short day | 31.4 ± 1.8 | 12.2 ± 1.0 | 43.6 ± 2.0 | 46.7 ± 1.6 |
ft-7 was originally designated suppressor mutation 1.
fwa-100 was originally designated suppressor mutation 2.
soc1 was originally designated suppressor mutation 3.
ft-8 was originally designated suppressor mutation 4.
ND, not determined.
Suppressor Mutations 1 and 4 Are Alleles of ft, and Suppressor Mutation 2 Is an Allele of fwa
Suppressor mutations 1 and 4 were mapped between the simple sequence length polymorphism markers nga280 and nga111 on chromosome 1 (see Methods), in the vicinity of the ft locus. To test whether these mutations are alleles of ft, we crossed them to ft-1 (Koornneef et al., 1991), which causes a less severe delay in flowering time than do both suppressor mutations. The F1 progeny of a cross between suppressor 1 or 4 and ft-1 flowered later than did wild-type plants and ft-1 mutants but earlier than did the parental suppressor mutant. The late-flowering phenotype of the F1 plants suggested that both suppressor mutations 1 and 4 are alleles of ft. FT was cloned recently (Kardailsky et al., 1999; Kobayashi et al., 1999). The FT gene from suppressor mutants 1 and 4 was therefore sequenced, and in both cases, mutations were identified. Suppressor mutation 1 is predicted to introduce a premature stop codon that results in a 137–amino acid protein rather than the 175–amino acid wild-type protein (see Methods). The truncated 137–amino acid protein is identical to that predicted to be formed from the ft-2 mutant allele (Kardailsky et al., 1999; Kobayashi et al., 1999). In suppressor mutant 4, a mutation was found in the 3′ splice acceptor site of the third intron of the FT gene (see Methods). Suppressor mutations 1 and 4 are therefore alleles of ft and are referred to as ft-7 and ft-8, respectively.
Suppressor mutation 2 showed genetic linkage to the cleaved amplified polymorphic sequence marker g8300 on chromosome 4. The marker mapped at ∼17.4 centimorgans (cM) from suppressor mutation 2 (see Methods). The fwa mutation, which causes late flowering and, like suppressor mutation 2, is semidominant (see below), is also located on chromosome 4 around the position of suppressor mutation 2. To examine whether suppressor mutation 2 is an allele of fwa, we performed segregation analysis. All 694 F2 progeny from a cross between suppressor mutant 2 and fwa-1 showed late flowering, indicating strong linkage between them. These observations, together with the effects of fwa on floral meristem identity described below, indicate that suppressor mutant 2 is an allele of fwa, and we refer to it as fwa-100.
Demonstration That Suppressor Mutation 3 Defines a Novel Flowering-Time Gene
Suppressor mutation 3 is recessive and is located in a region ∼16 cM from simple sequence length polymorphism marker nga168 on chromosome 2 (see Methods). Further mapping demonstrated that the mutation cosegregated with marker 40E1T7, ∼10 cM from marker AthBio2 (see Methods). The fpa mutation, which causes late flowering, is the only flowering-time locus previously shown to be located in this region. In an allelism test between suppressor mutation 3 and fpa-2, the F1 progeny flowered at a time similar to that of the wild type, demonstrating that suppressor mutation 3 is not an allele of fpa. These experiments demonstrate that suppressor mutation 3 defines a novel flowering-time gene, which we designated SOC1.
The soc1 mutation caused 35S::CO plants to produce an extra two or three leaves under both long and short days (Table 1). The soc1 35S::CO mutants were insensitive to day length, flowering at the same time under long and short days; therefore, the SOC1 gene seems to be required equally under both conditions for the phenotype of the 35S::CO plants. In the absence of 35S::CO, the soc1 mutation also delayed flowering under both long and short days (Table 2).
The Delay in Flowering of 35S::CO Caused by ft and Its Enhancement by fca-1
Unlike the 35S::CO progenitor, the ft-7 35S::CO plants responded to length of day, flowering later under short-day conditions. This suggests that the FT gene is more strongly required for 35S::CO function under short days. In the absence of 35S::CO, however, the ft-7 and ft-1 mutations had no effect or caused only a slight increase in leaf number under short-day conditions. This confirms previous observations indicating that FT is required only weakly for flowering under short-day conditions (Koornneef et al., 1991, 1998b).
The fca-1 mutation strongly enhances the effect of ft-8. For example, fca-1 ft-8 35S::CO produced ∼21 leaves, compared with 10 for ft-8 35S::CO and seven for fca-1 35S::CO (Table 1). Similarly, in greenhouse-grown plants, ft-8 fca-1 produced 53 leaves compared with ∼30 for both ft-8 and fca-1. These experiments suggest that fca-1 and ft-8 have additive effects on flowering time, which is consistent with their acting in different flowering-time pathways.
The Delay in Flowering of 35S::CO Caused by fwa
The fwa-100 35S::CO mutants formed ∼10 more leaves than did 35S::CO plants (Table 1). However, approximately half of the extra leaves were cauline leaves, and given that fwa-100 35S::CO also shows severe defects in floral development, the increase in cauline leaf number may reflect in part a loss of floral identity rather than a delay in the transition to reproductive development. Nevertheless, the increase in rosette leaf number caused by fwa-100 in a 35S::CO background under long days is more than that caused by soc1 or ft mutations, suggesting that fwa has the most severe effect on flowering time of 35S::CO under these conditions (Table 1).
The Combination of soc1 and fwa-1 Strongly Suppresses the Promotion of Flowering by 35S::CO
None of the suppressor mutations completely suppressed the early-flowering phenotype of 35S::CO plants. This may reflect genetic redundancy between the genes that act after CO in the long-day pathway. The effects of combining ft-1 and fwa-1 on flowering were tested previously, and the double mutants flowered at a time similar to the single mutants, suggesting that FT and FWA act in the same pathway (Koornneef et al., 1998a). To address whether SOC1 acts in the same pathway as FT and FWA or in a different pathway, the flowering time of an F2 population made from a cross between soc1 35S::CO and fwa-1/+ 35S::CO was examined. Eight of 138 F2 plants produced markedly more rosette leaves than did fwa-1 35S::CO or soc1 35S::CO (Figure 3). These plants showed severe floral meristem identity defects, as seen in fwa/fwa 35S::CO plants (see below), but their rosette leaf number was twice the average leaf number of fwa-1/ fwa-1 35S::CO plants and similar to that of co-2 mutants (Figure 3). This is a greater difference than was expected from a simply additive interaction between soc1 and fwa-1 and suggests that they affect separate flowering pathways downstream of CO.
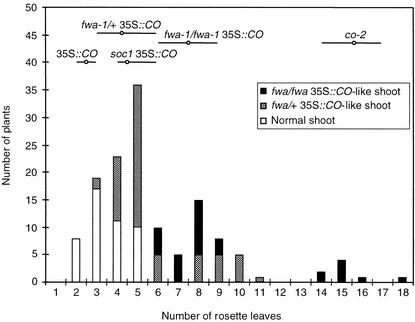
Figure 3.
Segregation of the Late-Flowering Phenotype in the F2 Population of a Cross between soc1 35S::CO and fwa-1/+ 35S::CO Plants.
Plants were grown under long-day conditions. Plants showing severe (fwa-1/fwa-1 35S::CO–like) and weak (fwa-1/+ 35S::CO–like) floral meristem identity defect segregated in the population and are indicated by the black and cross-hatched bars, respectively. Plants showing a normal 35S::CO–like shoot are indicated by the open bars. The open circles and horizontal lines indicate the average and the range, respectively, of rosette leaf numbers of defined genotypes grown under the same conditions. The plants flowering with between 14 and 18 rosette leaves are presumed to have the genotype soc1/soc1 fwa-1/fwa-1 35S::CO and represent approximately one in 17 of the total population.
Analysis of the Floral Meristem Identity Phenotype of fwa 35S::CO
The fwa-100 mutation is semidominant, and fwa-100 35S::CO plants showed a defect in floral meristem identity (see below) in addition to later flowering. The progeny derived from self-pollination of plants heterozygous for fwa-100 and homozygous for 35S::CO segregated in a ratio of ∼1:2:1; that is, 24.7% showed the normal 35S::CO phenotype and had not inherited fwa-100; 51.3% showed a phenotype similar to that of the parental plants; and 24.0% showed an extreme loss of floral meristem identity and flowered later than did the parents. The flower and shoot morphology phenotype and the late-flowering phenotype cosegregated, suggesting that both result from a single mutation. The fwa-100 mutation is therefore semidominant, and in the presence of 35S::CO it affects flowering time and floral meristem identity.
The floral meristem identity phenotype observed in fwa-100 35S::CO plants was not present in fwa-100 single mutants, suggesting that it occurs only in the presence of 35S::CO. To confirm this notion, the fwa-100 mutant was backcrossed to 35S::CO plants; in the F1 progeny, the floral meristem identity phenotype reappeared. Therefore, this phenotype is present only in plants that carry both fwa-100 and 35S::CO.
To test whether fwa-1 caused the same floral meristem identity phenotype in the presence of 35S::CO that was observed in fwa-100 35S::CO plants, we crossed fwa-1 to 35S::CO plants. The fwa-1 35S::CO plants produced in the F1 and F2 generations of this cross showed flowering-time and floral meristem identity phenotypes similar to those observed for fwa-100 35S::CO (Table 3).
Table 3.
Effects of fwa on Vegetative and Floral Organ Number in a 35S::CO Background
| Genotype | Rosette Leaf |
Cauline Leaf |
Secondary Shoot |
Whorl 1 LeaflikeSepala |
Sepala | Whorl 1 or 2 Sepaloid/ Petaloid Organa |
Whorl 2 Petala |
Secondary Flowera |
Whorl 3 Stamena |
Whorl 4Carpela |
|---|---|---|---|---|---|---|---|---|---|---|
| Ler | 5.5 ± 0.5 | 3.0 ± 0.4 | 3.0 ± 0.4 | 0 ± 0 | 4.0 ± 0 | 0 ± 0 | 4.0 ± 0 | 0 ± 0 | 5.8 ± 0.4 | 2.0 ± 0 |
| 35S::CO co-2 | 2.5 ± 0.5 | 2.0 ± 0.4 | 2.0 ± 0.4 | 0 ± 0 | 4.0 ± 0 | 0 ± 0 | 4.0 ± 0 | 0 ± 0 | 6.1 ± 0.4 | 2.6 ± 0.4 |
| fwa-100 | 16.5 ± 0.7 | 12.5 ± 0.9 | 12.7 ± 1.0 | 0 ± 0 | 4.9 ± 0.8 | 0.8 ± 0.9 | 2.6 ± 1.0 | 0 ± 0.1 | 6.0 ± 0.2 | 2.0 ± 0 |
| fwa-1 | 12.9 ± 0.7 | 8.9 ± 0.8 | 9.0 ± 0.7 | 0 ± 0 | 4.3 ± 0.6 | 0 ± 0.2 | 4.0 ± 0.4 | 0 ± 0 | 6.0 ± 0.1 | 2.0 ± 0.1 |
|
fwa-100/+ 35S::CO co-2 |
4.2 ± 0.5 | 3.1 ± 0.7 | 6.4 ± 2.3 | 1.3 ± 1.6 | 2.4 ± 1.7 | 0.4 ± 0.9 | 2.8 ± 1.7 | 0.4 ± 0.8 | 5.0 ± 0.9 | 2.0 ± 0 |
|
fwa-1/+ 35S::CO co-2 |
3.8 ± 0.7 | 3.2 ± 0.7 | 6.6 ± 1.7 | 1.1 ± 1.3 | 2.2 ± 1.6 | 0.5 ± 0.9 | 2.9 ± 1.5 | 0.3 ± 0.6 | 6.4 ± 1.3 | 2.2 ± 0.4 |
|
fwa-1/+ 35S::CO/+ co-2/+ |
5.0 ± 0.6 | 3.1 ± 1.0 | 4.8 ± 1.7 | 1.1 ± 1.4 | 2.8 ± 1.6 | 0.4 ± 0.9 | 3.5 ± 1.2 | 0.2 ± 0.5 | 5.8 ± 0.4 | 2.0 ± 0.2 |
|
fwa-1/+ 35S::CO/+ 35S::AP1/+ co-2/+ |
4.1 ± 0.7b | 3.4 ± 1.3b | 4.2 ± 1.4b | 1.2 ± 1.5 | 2.3 ± 1.5 | 0.4 ± 0.9 | 3.3 ± 1.3 | 0.1 ± 0.3 | 5.7 ± 0.6 | 2.0 ± 0 |
|
fwa-100/fwa-100 35S::CO co-2 |
7.6 ± 0.5 | 7.2 ± 0.9 | >30c | NDd | ND | ND | ND | ND | ND | ND |
|
fwa-1/fwa-1 35S::CO co-2 |
6.7 ± 0.5e | 6.1 ± 0.6e | >30c,e | ND | ND | ND | ND | ND | ND | ND |
In each case, >15 plants were analyzed, except where indicated.
Only the first to 10th flowers produced by the main shoot were scored in each genotype.
Seven plants were analyzed.
Exact number was difficult to determine because the main shoot of these plants produced secondary shoots indefinitely.
ND, not determined. Floral organ numbers for these plants were not scored because the main shoot never produced flowers.
Ten plants were analyzed.
Under long days, fwa-100 35S::CO double homozygotes never formed floral organs, except for carpel-like structures observed at the apex of some lateral shoots after 2 months of growth. They produced more lateral shoots subtended by cauline leaves than did wild-type plants and then formed >30 lateral shoots that were not subtended by cauline leaves (Table 3) until shoot growth terminated in a cluster of leaves (Figure 4). Under short days, these plants produced flowers at the apex of some lateral shoots, and these flowers contained carpels, stamens, petals, and leaflike sepals (Figure 4). They also formed fewer cauline leaves than they did under long-day conditions (Table 1). These observations suggest that the floral meristem identity defect of fwa-100 35S::CO plants is influenced by day length and is weaker under short-day conditions.
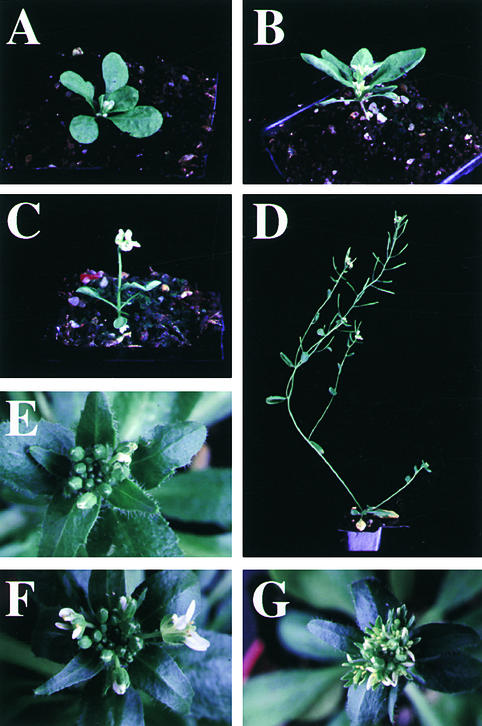
Figure 4.
Shoot and Flower Phenotype of fwa 35S::CO Plants.
All plants were grown under long-day conditions except for the plant shown in (F).
(A) A 35S::CO plant (left), an fwa-100/+ 35S::CO plant (center), and an fwa-100/fwa-100 35S::CO plant (right). All plants are 1 month old.
(B) A 5-week-old fwa-1/fwa-1 35S::CO plant.
(C) A 2-month-old fwa-1/fwa-1 35S::CO plant showing no obvious floral organs, except occasional carpelloid structures at the apex of lateral shoots.
(D) Top view of the apex of the primary inflorescence of a 1-month-old fwa-1/fwa-1 35S::CO plant.
(E) Structure of the apex of a lateral branch of a 2-month-old fwa-100/fwa-100 35S::CO plant. A few carpel-like structures are visible among the leaves.
(F) Structure of the apex of a lateral branch of a 2-month-old fwa-100/fwa-100 35S::CO plant grown under short-day conditions.
(G) A fwa-100/+ 35S::CO flower with leaflike sepals and a secondary flower.
(H) A fwa-100/fwa-100 flower with sepal/petal chimeric organs (arrows).
Plants heterozygous for fwa-100 and homozygous for 35S::CO exhibited weaker floral meristem identity defects than did the double homozygotes. After production of several coflorescences, the main stems formed flowers that often had inflorescence-like characters, such as leaflike sepals, leaflike or sepaloid petals, and secondary flowers that formed within flowers (Table 3). Although flower phenotypes were highly variable, the first and second whorls seemed to be more affected than the third and fourth whorls (Table 3). These plants also often formed coflorescences on the main inflorescence after nodes at which solitary flowers had formed.
In fwa-100 single mutants, severe effects on floral structure were observed very rarely (Table 3), although chimeric organs containing both sepal and petal tissue were often observed and occasionally secondary flowers were formed within flowers. 35S::CO plants never showed such floral meristem identity phenotypes (Table 3).
The Floral Meristem Identity Phenotype of fwa-1 35S::CO Plants Can Be Corrected by 35S::LFY but Not by 35S::AP1
The lfy fwa double mutant exhibits an extreme floral meristem identity defect, suggesting that fwa controls floral meristem identity genes redundantly with LFY (Ruiz-Garcia et al., 1997). This enhancement is similar to that observed when fwa is combined with 35S::CO. However, this phenotypic similarity cannot be readily explained, because 35S::CO is expected to activate LFY expression (Simon et al. 1996; Nilsson et al., 1998) and therefore would not be expected to have the same effect on fwa as an lfy mutation would.
To investigate the cause of the floral meristem identity defect of fwa 35S::CO, we tested whether overexpression of the floral meristem identity genes LFY and AP1 could suppress the phenotype of fwa 35S::CO. Plants homozygous for 35S::LFY were crossed to fwa-1/+ 35S::CO plants. All of the 40 F1 progeny showed the characteristic 35S::LFY–like shoot phenotype and formed a terminal flower (Figure 5). Half of the F1 progeny were fwa-1/+ 35S::CO/+ 35S::LFY/+, but they did not show the floral meristem identity phenotype described for fwa-1/+ 35S::CO/+ plants. Therefore, overexpression of LFY overcomes the floral meristem identity defect of fwa-1 35S::CO plants. However, 35S::LFY did not affect the delay in flowering time of 35S::CO caused by fwa-1, which is consistent with previous reports that fwa substantially delayed flowering of 35S::LFY plants (Nilsson et al., 1998).
Figure 5.
Effects of 35S::LFY and 35S::AP1 on the fwa 35S::CO Phenotype and the Effects of Dexamethasone Applications on 35S::CO:GR fwa Plants.
All plants were grown under long-day conditions.
(A) An 18-day-old 35S::LFY plant showing a terminal flower phenotype.
(B) An 18-day-old fwa-1/+ 35S::CO/+ 35S::LFY/+ plant showing a terminal flower phenotype.
(C) An 18-day-old 35S::AP1 plant showing a terminal flower phenotype.
(D) A 5-week-old fwa-1/+ 35S::CO/+ 35S::AP1/+ plant.
(E) Top view of a 6-week-old fwa-1/+ co-2/+ plant treated with dexamethasone 3 weeks after sowing.
(F) Top view of a 6-week-old fwa-1/+ 35S::CO:GR co-2/+ plant not treated with dexamethasone.
(G) Top view of a 6-week-old fwa-1/+ 35S::CO:GR co-2/+ plant treated with dexamethasone 3 weeks after sowing.
In contrast, when plants heterozygous for 35S::AP1 were crossed to a fwa-1/+ 35S::CO, 18 of 38 F1 progeny showed a shoot and flower phenotype similar to that of fwa-1/+ 35S::CO plants (Figure 5). The presence of 35S::AP1 in seven of these plants was confirmed by polymerase chain reaction (PCR); despite the presence of 35S::AP1, those plants were indistinguishable from fwa-1/+ 35S::CO/+ plants (Table 3). Therefore, unlike 35S::LFY, the 35S::AP1 transgene did not suppress the floral meristem identity defect of fwa-1 35S::CO plants.
In an fwa Background, the Effect of 35S::CO on Floral Meristem Identity Can Be Separated from Its Effect on Flowering Time
The floral meristem identity defect shown by 35S::CO fwa plants may be an indirect consequence of 35S::CO driving early flowering in the presence of fwa or of a more direct interaction between 35S::CO and fwa that occurs independent of flowering time. To distinguish between these possibilities, we used a fusion of CO to the ligand binding domain of the rat glucocorticoid receptor (35S::CO:GR). This fusion enables CO overexpression to be induced by application of the steroid dexamethasone (Simon et al., 1996). The 35S::CO:GR transgenic plants were crossed to fwa-1 mu-tants, and the F1 plants were treated with dexamethasone at various times after germination. When plants were treated repeatedly with dexamethasone from germination onwards, the phenotype was very similar to that of fwa-1/+ 35S::CO/+ plants, demonstrating that when continually induced, CO:GR has an effect similar to that of 35S::CO.
When plants were treated once with dexamethasone at 7, 14, or 21 days after sowing, flowering occurred in response to the application of dexamethasone, and floral defects similar to those described for fwa-1/+ 35S::CO plants (Figure 5) were present. The plants treated 21 days after sowing produced as many leaves as fwa-1/+ 35S::CO:GR/+ co-2/+ plants not treated with dexamethasone or as fwa-1/+ co-2/+ plants; however, these plants still had floral defects. In contrast, such flower defects were not observed in fwa-1/+ 35S::CO:GR/+ co-2/+ plants not treated with dexamethasone or in dexamethasone-treated fwa-1/+ co-2/+ plants (Figure 5). This result suggests that the floral meristem identity defect in 35S::CO fwa plants indicates a direct effect on floral development and not an indirect consequence of their delayed flowering time.
DISCUSSION
Effect of 35S::CO on Flowering Time
35S::CO caused early flowering in both long- and short-day conditions, so that the transgenic plants were almost completely insensitive to day length. A similar phenotype was reported previously for 35S::CO:GR plants treated with dexamethasone early in development (Simon et al., 1996). Together with previous observations that showed a correlation between copy number of the CO gene and flowering time (Koornneef et al., 1991; Putterill et al., 1995), this suggests that transcriptional control of CO expression is an important determinant in the photoperiodic control of flowering. All five 35S::CO lines contained the CO mRNA in much greater abundance than did wild-type plants, but the amount of the mRNA varied between lines. Nevertheless, all five lines showed similar phenotypes, suggesting that beyond the expression level of the weakest line, increasing the abundance of CO mRNA does not have a further effect on phenotype. Above that amount, therefore, other factors may limit CO function.
35S: :CO Can Overcome the Effect of Mutations in the Autonomous Pathway
The fca-1 mutation strongly delays flowering in an Ler background but only slightly delays flowering of 35S::CO plants. FCA acts in the autonomous flowering-time pathway, whereas CO acts in the long-day pathway (Koornneef et al., 1998a, 1998b; Levy and Dean, 1998). Nevertheless, increasing the activity of the long-day pathway by overexpressing the CO gene can almost completely overcome the effect of strong mutations in the autonomous pathway. This suggests that flowering time is controlled by the balance of activity of these two pathways and that when the long-day pathway is superactivated, the autonomous pathway is not required for early flowering.
Nevertheless, the fca-1 mutation does slightly delay flowering of 35S::CO plants. This effect is strongly enhanced in the presence of the ft-8 mutation so that ft-8 fca-1 35S::CO plants flower much later than either fca-1 35S::CO or ft-8 35S::CO plants. These observations suggest that the autonomous pathway is required for 35S::CO to have its full effect. The marked enhancement of the effect of fca-1 by ft-8 suggests that the contribution of the autonomous pathway is partially redundant with FT.
Identification of Suppressor Mutations of 35S::CO and the Role of the Novel Flowering-Time Gene SOC1
Suppressor mutagenesis of a gain-of-function allele has been used in model organisms such as Drosophila and Caenorhabditis elegans to identify factors that act downstream of a gene of interest (Stern and DeVore, 1994; Karim et al., 1996). In plants, screens for extragenic suppressors of a loss-of-function allele have been described (e.g., Pepper and Chory, 1997; Reed et al., 1998), as have mutations that suppress the effect of constitutive activation of ethylene signaling (Roman et al., 1995). To identify genes that act downstream of CO to control flowering, we performed suppressor mutagenesis of 35S::CO.
Three loci were identified as suppressors of the early-flowering phenotype of 35S::CO. One of them, soc1, defines a novel flower-promoting gene; its position on chromosome 2 distinguishes it from all other flowering-time loci described to date. The soc1 mutation had not been previously identified by mutagenesis of wild-type Arabidopsis, demonstrating that using suppressor mutagenesis of transgenic Arabidopsis plants overexpressing a gene is an effective way to identify novel loci likely to be functionally associated with the gene of interest. The other three suppressor mutations were alleles of ft and fwa.
The soc1 mutation delays flowering under both long- and short-day conditions. This is a characteristic of mutations that affect the autonomous flowering pathway rather than the long-day pathway in which CO acts. If SOC1 acts in the autonomous pathway, then the delay in flowering of 35S::CO plants caused by soc1 would be similar to the effect of fca-1. Alternatively, SOC1 may act both downstream of CO in the long-day pathway and in the autonomous pathway. In this case, soc1 mutations may show effects similar to those of mutations in the autonomous pathway but nevertheless act in the long-day pathway downstream of CO. We favor this latter possibility because of the relative severity of the effect of soc1 on 35S::CO. The delay caused by soc1 on the flowering time of wild-type plants is very much weaker than that of ft, fwa, and fca (Table 2), but its effect on 35S::CO is at least as strong as that of fca-1 (Table 1). The relatively strong effect of soc1 on the phenotype of 35S::CO may argue that SOC1 acts more directly downstream of CO than does FCA. Furthermore, soc1 has an effect similar to that of ft-1 in both wild-type and 35S::CO backgrounds (Tables 1 and 2), and extensive genetic and molecular data now place FT downstream of CO (see below).
FT and FWA Act Downstream of CO
The suppressor mutations 1 and 4 are alleles of ft, and suppressor mutation 2 is an allele of fwa. FT and FWA were proposed to act in the same pathway as CO, based on analyses of double mutants and their response to environmental conditions (Koornneef et al., 1998b). These observations are extended by our findings that the ft and fwa mutations partially suppress 35S::CO, which suggests that FT and FWA act downstream of CO (Figure 6A). Furthermore, Kardailsky et al. (1999) and Kobayashi et al. (1999) have recently cloned the FT gene and found that not only is FT mRNA abundance reduced in the co-2 mutant background and rapidly increased by activation of CO but also that 35S::FT suppresses co-2 mutations. Together with our genetic data, this strongly suggests that FT acts downstream of CO to promote flowering; however, because strong mutant alleles of ft only partially suppress 35S::CO (Table 1), CO must also activate other downstream genes (see below). In addition, fwa largely suppresses the early-flowering phenotype caused by the overexpression of FT (Kardailsky et al., 1999; Kobayashi et al., 1999), which would again be consistent with the idea that fwa affects the long-day pathway downstream of CO.
Figure 6.
Schematic Model for the Interaction of Genes in the Control of Flowering Time and Floral Meristem Identity.
(A) Model for control of flowering time.
(B) Model for control of floral meristem identity (FMI). The thin numbered lines represent two possibilities, described in the text, for how the interaction between 35S::CO and fwa might inhibit LFY function. In both (A) and (B), arrows represent promotive interactions and T-bars represent inhibitory interactions. The proposed inhibitory function of FWA is based on the assumption that fwa mutations are gain-of-function mutations.
FT is most strongly required for the phenotype of 35S::CO plants under short days. Whereas 35S::CO plants flowered at the same time under long and short days, ft 35S::CO plants flowered later under short days than under long days. FT is either not required or only minimally required for flowering of wild-type plants under short days (Table 2; see also Koornneef et al., 1998a), and it is probably strongly required under short days in 35S::CO plants because 35S::CO activates transcription of FT under these conditions. The weaker effect of ft on 35S::CO under long days suggests that under these conditions, other genes may be activated that can partially compensate for ft. This could occur through post-translational activation of CO under long-day conditions, or it might occur independently of CO.
Genetic Evidence for Parallel Flowering-Time Pathways Downstream of CO
No single mutation that completely suppresses early flowering of 35S::CO was identified from our screens. 35S::CO could still accelerate flowering of any of the suppressor mutants. Such partial suppression could be explained by genetic redundancy between genes that act downstream of CO. Of the three suppressor mutations, ft and fwa were previously proposed to affect the same pathway and not to act redundantly (Koornneef et al., 1998a). Therefore, 35S::CO must promote flowering of the ft/fwa mutants by way of another pathway. Analysis of an F2 population generated by intercrossing soc1 35S::CO and fwa-1 35S::CO suggested that SOC1 may define such a pathway (Figure 6A), because combining both soc1 and fwa-1 in a 35S::CO background largely suppressed early flowering of 35S::CO, and the number of rosette leaves produced by these plants was similar to that produced by co-2 mutants. Previous genetic and molecular experiments suggested that CO does not promote flowering by way of FT in a simple linear pathway (Simon et al., 1996; Kardailsky et al., 1999; Kobayashi et al., 1999), and our data indicate that CO promotes flowering via at least two genetic pathways, one involving SOC1 and a second involving FT/FWA.
Effect of fwa on Floral Meristem Identity
Both fwa-100 35S::CO and fwa-1 35S::CO showed an extreme floral meristem identity defect that was not shown by any of the parents (Figure 4) and was similar to that of lfy fwa double mutants (Ruiz-Garcia et al., 1997). CO was also overexpressed late in the development of fwa mutants using 35S::CO:GR, and the floral meristem identity defect was induced irrespective of flowering time. Therefore, the effect of 35S::CO on floral meristem identity in fwa is not an indirect effect of early flowering. Suppression of this defect by 35S::LFY suggests that LFY expression or activity may be decreased in fwa 35S::CO plants. Also petal/sepal chimeric organs observed in fwa-100 single mutant flowers are reminiscent of the flower phenotype caused by weak lfy alleles (Figure 4; Weigel et al.. 1992). These observations suggested that fwa may slightly affect LFY and that 35S::CO may strongly enhance the effect. No difference between fwa mutants and wild-type plants regarding LFY mRNA abundance was detected in previous experiments (Ruiz-Garcia et al., 1997; Nilsson et al., 1998), indicating that the slight effect of fwa may not be easily detectable or that the repression of LFY does not occur by decreasing the amount of its mRNA.
In contrast to 35S::LFY, 35S::AP1 did not suppress the floral meristem identity defect of fwa 35S::CO. The AP1 gene is directly activated by LFY (Parcy et al., 1998; Wagner et al., 1999), and 35S::AP1 can largely but not completely suppress lfy (Mandel and Yanofsky, 1995; Liljegren et al., 1999). The observation that 35S::LFY suppresses fwa-1 35S::CO but 35S::AP1 does not suggests that the floral meristem identity phenotype of fwa-1 35S::CO plants is largely caused by reductions in LFY activity affecting the expression of genes other than AP1. In addition to its effect on AP1, LFY has also been proposed to activate the AGAMOUS and AP3 genes by interaction with different coactivators (Parcy et al., 1998). Similarly, LFY may activate other floral meristem identity genes, which could be repressed by the combination of fwa and 35S::CO. Consistent with this idea, Ruiz-Garcia et al. (1997) suggested that LFY and FWA could control other floral meristem identity genes as well as AP1, because the floral meristem identity defect in lfy fwa was more severe than in ap1 fwa, ap1 lfy, or ap1 cauliflower lfy.
The fwa mutant alleles may be gain-of-function mutations caused by hypomethylation, because all known fwa mutations are dominant, and Kakutani (1997) reported that a dominant late-flowering mutation induced in a decreased DNA methylation 1 background was likely to be an allele of fwa. In addition, some fwa mutants showed other epigenetic features. For example, an epigenetic allele of superman (clark kent-1) was identified from the fwa-1 mutant line (Jacobsen and Meyerowitz, 1997). The fwa-100 mutant line that we identified as suppressor mutation 2 showed phenotypic instability, such as variation in the severity of the floral meristem identity defect between branches (data not shown).
If the fwa mutant allele results from increased expression of the FWA gene, then overexpression of CO can somehow be correlated with increased expression of FWA to inhibit LFY activity. This appears to be specific to the fwa 35S::CO interaction and was not shown by fwa 35S::FT plants (Kardailsky et al., 1999; Kobayashi et al., 1999). The effect on floral meristem identity of combining 35S::CO and fwa may be explained by two general models. The first model is based on that of Ruiz-Garcia et al. (1997), who proposed that the severe enhancement of the phenotype of lfy mutants caused by fwa occurs because the fwa mutation represses a floral meristem identity gene, the function of which is redundant with LFY. The fwa 35S::CO phenotype can be explained by this model if the overexpression of CO in combination with fwa also acts to repress the function of LFY or of a gene downstream of LFY. That CO may activate genes that repress flowering was shown previously for the activation of TFL1 by 35S::CO:GR (Simon et al., 1996) and proposed because co-1 35S::FT plants flowered slightly earlier than did 35S::FT (Kobayashi et al., 1999), although a similar effect was not seen with co-2 (Kardailsky et al., 1999). A second model proposes that the FWA product is present at increased amounts in the fwa mutant and that 35S::CO causes the amount of overexpression to be increased further or increases the activity of FWA protein. In this case, if FWA represses floral meristem identity gene expression, and the severity of this effect is proportional to the degree of overexpression of FWA, then 35S::CO would enhance the effect of fwa on floral meristem identity.
In conclusion, the genetic approach that we have used to identify genes required for CO function demonstrated that CO controls several distinct activities that regulate flowering. One of these somehow acts together with FWA to repress flower development. In addition, two parallel pathways act downstream of CO to promote flowering; one of these pathways involves FT/FWA, and the other includes SOC1. The isolation of SOC1 will shed further light on the function of this second pathway.
METHODS
Plant Materials and Growth Conditions
Wild-type plants were Arabidopsis thaliana in the wild-type Landsberg erecta (Ler) ecotype. All mutant and transgenic lines were in the Ler background. constans-2 (co-2) mutants carried the transparent testa4 (tt4) mutation (Putterill et al., 1995), which causes a white seed color phenotype and was used as a visible marker to indicate the presence of the co-2 mutation. Seeds of late-flowering mutants fca-1, ft-1, fpa-2, and fwa-1 were kindly provided by M. Koornneef (Koornneef et al., 1991). 35S::LEAFY (LFY) and 35S::APETALA1 (AP1) transgenic seeds were gifts of D. Weigel (Weigel and Nilsson, 1995) and M. Yanofsky (Mandel and Yanofsky, 1995), respectively. The fusion of CO to the ligand binding domain of the rat glucocorticoid receptor (35S::CO:GR) was as described previously (Simon et al., 1996). Seeds were placed on moist filter paper at 4°C for 4 or 5 days, planted in soil, and germinated in growth cabinets or greenhouses.
Measurement of Flowering Time and Floral Organ Number
Flowering time and floral organ analyses were performed with plants grown in controlled-environment cabinets under either short-day (10-hr-light/14-hr-dark) or long-day (10-hr-light/6-hr-day extension/ 8-hr-dark) conditions as described in Putterill et al. (1995). Flowering time was measured by scoring the number of leaves on the main stems or the number of days from sowing until flower buds were visible by eye at the center of the rosette. Floral organ number was scored under a stereoscopic microscope. Data from one experiment are presented as mean ±sd, where  plants or 50 to 80 flowers, except where indicated.
plants or 50 to 80 flowers, except where indicated.
Construction of 35S::CO
To make the 35S::CO fusion, we inserted the 35S promoter upstream of a genomic clone of the CO gene. The entire CO gene had previously been inserted into the EcoRV site of pBluescript (Stratagene, La Jolla, CA) as a 4.2-kb EcoRV-PvuII fragment in plasmid d178. The EcoRV site is located ∼2450 bp upstream of the translational start site of CO, and the PvuII site is located ∼400 bp downstream of the translational stop codon. The fragment was inserted into the EcoRV site of pBluescript such that the EcoRV site of the EcoRV-PvuII fragment was adjacent to the HindIII site of the polylinker. The 35S promoter was removed from vector pJIT62 (P. Mullineaux, John Innes Centre) as a 400-bp KpnI-HindIII fragment. Plasmid d178 was cleaved with KpnI and HindIII, and the largest fragment generated was ligated to the KpnI-HindIII fragment containing the 35S promoter. This ligation generated a fusion of the 35S promoter to the CO gene at a HindIII site 60 bp upstream of the CO translational start codon. To enable easy transfer of this fragment to the binary vector, the KpnI site was converted to a ClaI site by using the adapter 5′-TATCGATAGTAC-3′. The fusion was then removed as a ClaI-BamHI fragment and inserted into vector SLJ1711 (Jones et al., 1992). The 35S::CO construct was introduced into co-2 tt4 mutants by the root transformation procedure (Valvekens et al., 1988). Transgenic line 5 was used for further study.
RNA Gel Blot Analysis
RNA gel blot analysis was performed as previously described (Schaffer et al., 1998). RNA was extracted from soil-grown plants with 3 to 4 visible leaves.
Mutagenesis and Phenotypic Screens for Mutations That Delay Flowering of 35S::CO
Approximately 20,000 35S::CO co-2 seeds in three independent batches and 10,000 35S::CO fca-1 seeds were mutagenized by imbibition in 0.3% ethyl methanesulfonate (EMS; Sigma) for 8 to 9 hr, followed by washing with 0.1 M Na2SO3 and distilled water (five times). M2 seeds were collected in pools, with each pool containing seeds of ∼50 M1 plants. Approximately 20,000 M2 seeds representing ∼4000 M1 plants after mutagenesis of 35S::CO co-2 seeds were sown on soil and screened for late-flowering mutants under short-day conditions in a greenhouse. Approximately 5000 M2 seeds representing ∼500 M1 plants from the mutagenesis of 35S::CO fca-1 seeds were sown on soil and screened for a late-flowering phenotype under long-day conditions in a greenhouse.
Genetic Analysis
The suppressor mutations in 35S::CO co-2 background were backcrossed to 35S::CO co-2 plants twice before phenotypic analysis. Detailed information on the construction of lines and genetic segregation ratios is available from the authors.
Molecular Determination of the Presence of co-2, 35S::CO, and 35S::AP1
For detection of the co-2 mutation, the primers CO41 (5′-GGTCCCAACGAAGAAGTGC-3′) and CO52 (5′-GGAACAGCCACGAAGCAA-3′) were used. After amplification, the polymerase chain reaction (PCR) products were digested with BspLU11I (Boehringer Mannheim), which produces 324-, 292-, and 114-bp fragments for the wild-type allele and 616- and 114-bp fragments for the co-2 allele.
For detection of the 35S::CO transgene, a set of primers 35S01 5′-TATCCTTCGCAAGACCCTTC-3′ and CO52 was used. This primer pair amplifies an ∼600-bp fragment.
For detection of the 35S::AP1 transgene, the primers 5′-GAAGAGGATAGAGAACAAGAT-3′ and 5′-ATTGACGTCGGACTCAGG-TG-3′ were used. This primer pair amplifies a 241-bp fragment from the AP1 cDNA and a 1583-bp from the genomic copy.
Template DNA for PCR reactions was extracted from leaves of each genotype, as described by Sawa et al. (1997).
Genetic Mapping
Suppressor mutants 1, 3, or 4 were crossed to wild-type Columbia to generate mapping populations. For mapping suppressor 1, 3, or 4, DNA was isolated (Sawa et al., 1997) from 32, 30, and 48 F2 plants that showed a late-flowering phenotype, respectively, and analyzed with simple sequence length polymorphism (Bell and Ecker, 1994) and cleaved amplified polymorphic sequence (CAPS) markers (Konieczny and Ausubel, 1993). soc1 showed strong linkage to a CAPS marker 40E1T7 (no recombinants were found in analysis of 58 chromosomes) and weaker linkage to a simple sequence length polymorphism marker, AthBIO2 (seven recombinants were found in analysis of 60 chromosomes), on chromosome 2.
To facilitate restriction fragment length polymorphism mapping of the suppressor mutations that were recovered in the 35S::CO co-2 plants in the Ler background, 35S::CO was introgressed into Columbia. The introgression was performed by crossing 35S::CO co-2 Ler to wild-type Columbia four times. Plants inheriting 35S::CO were selected by their early-flowering phenotype and the results of a PCR test (see above) performed after each cross. Plants homozygous for 35S::CO were selected after the fourth backcross. 35S::CO Columbia plants were then used to map suppressor 2 by crossing 35S::CO Ler carrying this mutation to 35S::CO Columbia plants, and DNA was isolated from plants homozygous for suppressor mutant 2 and carrying 35S::CO (Sawa et al., 1997). A total of 15 F2 plants showing the severe floral meristem identity defect phenotype were analyzed with simple sequence length polymorphism and CAPS markers. Suppressor mutation 2 showed linkage to CAPS marker g8300 (five recombinants in analysis of 30 chromosomes) on chromosome 4.
Map distance between the markers and the suppressor mutations was determined by using the Kosambi mapping function (Koornneef and Stam, 1992).
DNA Sequencing
The FT gene was amplified by PCR from genomic DNA of suppressor mutants 1 and 4 with two sets of primers: 5′-CAGAAACAATCAACACAGAGAAACCACCTG-3′ and 5′-TCAATTTAGCTTGGGTGTGG-3′, and 5′-TTATGTTGTGGTGCCATAGC-3′ and 5′-ACTATAGGCATCATCACCGTTCGTTCGTTACTCG-3′. These pairs of primers were used to amplify 1406- and 571-bp fragments, respectively, that covered all of the FT coding region. PCR products were purified with the Quiaquick PCR purification kit (Qiagen, Chatsworth, CA) and were sequenced on one strand with a DNA sequencing kit (Perkin Elmer) by using the same primers used to amplify the FT coding regions. Once mutations were found, another strand of an independently amplified PCR product was sequenced to verify that the DNA sequence alteration was not a PCR-derived error or a sequencing error. A mutation in ft-7 changed the 138th amino acid residue Trp (TGG) to a stop codon (TGA). ft-8 contained a mutation in the 3′ splice acceptor site of the third intron (AG changed to AA.).
Application of Dexamethasone
Dexamethasone (10 μM; Sigma) was prepared and applied to plants as described previously (Simon et al.,1996).
Acknowledgments
We are grateful to Detlef Weigel for supplying the primers for the sequencing of FT prior to publication and to Detlef Weigel and Takashi Araki for communicating results prior to publication. Many thanks to Xiahoe Hu for help with plant care. Alon Samach and Paula Suarez-Lopez made many useful comments on the manuscript. This work was supported by grants from the European Community and Human Frontiers Science Program Organisation to G.C.; by fellowships from the Novartis Foundation, Japan, to H.O., and from the European Community to C.P.; and by a Sainsbury studentship to K.G. from the Gatsby Trust.
References
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Blazquez, M.A., Green, R., Nilsson, O., Sussman, M.R., and Weigel, D. (1998). Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell 10, 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley, D., Ratcliffe, O., Vincent, C., Carpenter, R., and Coen, E. (1997). Inflorescence commitment and architecture in Arabidopsis. Science 275, 80–83. [DOI] [PubMed] [Google Scholar]
- Chien, J.C., and Sussex, I.M.. (1996). Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 111, 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler, S., Lee, K., Onouchi, H., Richardson, K., Samach, A., Morris, B., Coupland, G., and Putterill, J. (1999). Molecular characterisation of GIGANTEA: A circadian clock–controlled gene that regulates photoperiodic flowering in Arabidopsis. EMBO J. 18, 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Yang, H., Mockler, T.C., and Lin, C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Jacobsen, S.E., and Meyerowitz, E.M. (1997). Hypermethylated SUPERMAN epigenetic alleles in Arabidopsis. Science 277, 1100–1103. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G., Shlumukov, L., Carland, F., English, J., Scofield, S.R., Bishop, G.J., and Harrisson, K. (1992). Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Res. 1, 285–297. [DOI] [PubMed] [Google Scholar]
- Kakutani, T. (1997). Genetic characterization of late-flowering traits induced by DNA hypomethylation mutation in Arabidopsis thaliana. Plant J. 12, 1447–1451. [DOI] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V.K., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Karim, F.D., Chang, H.C., Therrien, M., Wasserman, D.A., Laverty, T., and Rubin, G.M. (1996). A screen for genes that function downstream of Ras1 during Drosophila eye development. Genetics 143, 315–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., and Stam, P. (1992). Genetic analysis. In Methods in Arabidopsis Research, C. Koncz, N.-H. Chua, and J. Schell, eds (Singapore: World Scientific), pp. 83–99.
- Koornneef, M., Hanhart, C.J., and van der Veen, J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Alonso-Blanco, C., Peeters, A.J.M., and Soppe, W. (1998. a). Genetic control of flowering time in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 345–370. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Alonso-Blanco, C., de Vries, H.B., Hanhart, C.J., and Peeters, A.J. (1998. b). Genetic interactions among late-flowering mutants of Arabidopsis. Genetics 148, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I., Aukerman, M.J., Gore, S.L., Lohman, K.N., Michaels, S.D., Weaver, L.M., John, M.C., Feldmann, K.A., and Amasino, R.M. (1994). Isolation of LUMINIDEPENDENS: A gene involved in the control of flowering time in Arabidopsis. Plant Cell 6, 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, Y.Y., and Dean, C. (1998). The transition to flowering. Plant Cell 10, 1973–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren, S.J., Gustafson-Brown, C., Pinyopich, A., Ditta, G.S., and Yanofsky M.F. (1999). Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell 11, 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macknight, R., Bancroft, I., Page, T., Lister, C., Schmidt, R., Love, K., Westphal, L., Murphy, G., Sherson, S., Cobbet, C., and Dean, C. (1997). FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89, 737–745. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., and Yanofsky, M.F. (1995). A gene triggering flower formation in Arabidopsis. Nature 377, 522–524. [DOI] [PubMed] [Google Scholar]
- Martínez-Zapater, J.M., and Somerville, C.R. (1990). Effect of light quality and vernalization on late flowering mutants of Arabidopsis thaliana. Plant Physiol. 92, 770–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Zapater, J.M., Coupland, G., Dean, C., and Koornneef, M. (1994). The transition to flowering in Arabidopsis. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 403–433.
- Michaels, S.D., and Amasino, R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, O., Lee, I., Blazquez, M.A., and Weigel, D. (1998). Flowering-time genes modulate the response to LEAFY activity. Genetics 150, 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima, S., Murata, M., Sakamoto, W., Ogura, Y., and Motoyoshi, F. (1997). Cloning and molecular analysis of the Arabidopsis gene TERMINAL FLOWER 1. Mol. Gen. Genet. 254, 186–194. [DOI] [PubMed] [Google Scholar]
- Parcy, F., Nilsson, O., Busch, M.A., Lee, I., and Weigel, D. (1998). A genetic framework for floral patterning. Nature 395, 561–566. [DOI] [PubMed] [Google Scholar]
- Park, D.H., Somers, D.E., Kim, Y.S., Choy, Y.H., Lim, H.K., Soh, M.S., Kim, H.J., Kay, S.A., and Nam, H.G. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285, 1579–1582. [DOI] [PubMed] [Google Scholar]
- Pepper, A.E., and Chory, J. (1997). Extragenic suppressors of the Arabidopsis det1 mutant identify elements of flowering-time and light-response regulatory pathways. Genetics 145, 1125–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeiro, M., and Coupland, G. (1998). The control of flowering time and floral identity in Arabidopsis. Plant Physiol. 117, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. [DOI] [PubMed] [Google Scholar]
- Redei, G.P. (1962). Supervital mutants of Arabidopsis. Genetics 47, 443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W., Elumalai, R.P., and Chory, J. (1998). Suppressors of an Arabidopsis thaliana phyB mutation identify genes that control light signaling and hypocotyl elongation. Genetics 148, 1295–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman, G., Lubarsky, B., Kieber, J.J., Rothenberg, M., and Ecker, J.R. (1995). Genetic-analysis of ethylene signal-transduction in Arabidopsis thaliana: Five novel mutant loci integrated into a stress-response pathway. Genetics 139,1393–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Garcia, L., Madueno, F., Wilkinson, M., Haughn, G., Salinas, J., and Martinez-Zapater, J.M. (1997). Different roles of flowering-time genes in the activation of floral initiation genes in Arabidopsis. Plant Cell 9, 1921–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, S., Ito, T., and Okada, K. (1997). A rapid method for detection of single base changes in Arabidopsis thaliana using the polymerase chain reaction. Plant Mol. Biol. Rep. 15, 179–185. [Google Scholar]
- Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carre, I., and Coupland, G. (1998). The late elongated hypocotyl mutation in Arabidopisis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229. [DOI] [PubMed] [Google Scholar]
- Sheldon, C.C., Burn, J.E., Perez, P.P., Metzger, J., Edwards, J.A., Peacock, W.J., and Dennis, E.S. (1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, R., Igeno, I.M., and Coupland, G. (1996). Activation of floral meristem identity genes in Arabidopsis. Nature 384, 59–62. [DOI] [PubMed] [Google Scholar]
- Stern, M.J., and DeVore, D.L. (1994). Extending and connecting signalling pathways in C. elegans. Dev. Biol. 166, 443–459. [DOI] [PubMed] [Google Scholar]
- Telfer, A., Bollman, K.M., and Poethig, R.S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124, 645–654. [DOI] [PubMed] [Google Scholar]
- Valvekens, D., Van Montagu, M., and Van Lijsebettens, M. (1988). Agrobacterium tumefaciens–mediated transformation of Arabidopsis root explants using kanamycin selection. Proc. Natl. Acad. Sci. USA 85, 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, D., Sablowski, R.W., and Meyerowitz, E.M. (1999). Transcriptional activation of APETALA1 by LEAFY. Science 285, 582–584. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Nilsson, O. (1995). A developmental switch sufficient for flower initiation in diverse plants. Nature 377, 495–500. [DOI] [PubMed] [Google Scholar]
- Weigel, D., Alvarez, J., Smyth, D.R., Yanofsky, M.F., and Meyerowitz, E.M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. [DOI] [PubMed] [Google Scholar]
- Wilson, R.N., Heckman, J.W., and Somerville, C.R. (1992). Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 100, 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]