Abstract
Cell expansion, a developmental process regulated by both endogenous programs and environmental stimuli, is critically important for plant growth. Here, we report the isolation and characterization of RSG (for repression of shoot growth), a transcriptional activator with a basic leucine zipper (bZIP) domain. To examine the role of RSG in plant development, we generated transgenic tobacco plants expressing a dominant-negative form of RSG, which repressed the activity of full-length RSG. In transgenic plants, this expression severely inhibited stem internode growth, specifically cell elongation. These plants also had less endogenous amounts of the major active gibberellin (GA) in tobacco, GA1. Applying GAs restored the dwarf phenotypes of transgenic tobacco plants that expressed the dominant-negative form of RSG. To investigate the function of RSG in the regulation of the endogenous amounts of GAs, we identified a target for RSG. RSG bound and activated the promoter of Arabidopsis GA3, one of the genes encoding enzymes involved in GA biosynthesis. Moreover, the dominant-negative form of RSG decreased expression of the GA3 homolog in transgenic tobacco plants. Our results show that RSG, a bZIP transcriptional activator, regulates the morphology of plants by controlling the endogenous amounts of GAs.
INTRODUCTION
The growth of multicellular organisms is accomplished by orderly cell division and regulated cell expansion. In plants, the contribution of cell expansion to growth and development is much more important than in most other organisms, the direction and extent of cell expansion determining the final shapes and sizes of all plant organs. In the phase of controlled expansion that generally follows cell division in meristems, the daughter cells may often increase in volume by ⩾50-fold (Steeves and Sussex, 1989). Phytohormones such as auxins, gibberellins (GAs), and brassinolides induce the elongation of cells along the longitudinal axis, whereas cytokinins and ethylene cause expansion of cells along the transverse axis (Shibaoka, 1994).
GAs, which are tetracyclic diterpenoid growth factors, are essential regulators in many aspects of plant development, including stem elongation, seed germination, and flowering. The control of polarized cell expansion is thought to involve cortical microtubules, characteristically found beneath the cell membrane in plant cells (Giddings and Staehelin, 1988; Cyr, 1994). GAs promote the orientation of cortical microtubules perpendicularly to the growing axis of the cell; the corresponding cellulose deposition that results allows the cells to expand only in the growing axis, thereby producing thin, elongated plants (Shibaoka, 1994). GA-deficient Arabidopsis mutants display characteristic phenotypes that include dark green leaves and stunted growth attributable to the inhibition of stem elongation, especially during the cell elongation phase. Investigators using these mutants have isolated several genes that encode gibberellin biosynthetic enzymes (Sun and Kamiya, 1994; Chiang et al., 1995; Xu et al., 1995; Yamaguchi et al., 1996; Helliwell et al., 1998). Both endogenous developmental programs and environmental stimuli affect the expression of these enzymes. Therefore, elucidating the transcriptional regulation of GA biosynthetic enzymes is crucial to identifying the molecular mechanisms involved in plant development and to understanding how these mechanisms help plants adapt to changes in their environment. However, transcription factors that regulate GA biosynthetic genes have not been identified.
In this report, we describe a new basic leucine zipper (bZIP) transcriptional activator, designated RSG (for repression of shoot growth), and its ability to regulate cell elongation by controlling the amounts of GAs present. A standard approach in plant development research is the analysis of morphological mutants. However, when a regulatory gene belongs to a multigene family and has functions that are the same as those of other members of the family, isolation of the loss-of-function mutant becomes difficult. We used a strategy to repress the function of the wild-type gene by using a dominant mutant protein. This strategy has been used successfully with yeast (Hope and Struhl, 1986), mammalian (Lloyd et al., 1991), and plant cells (Unger et al., 1993; Rieping et al., 1994) to investigate the in vivo functions of bZIP transcriptional factors. Expression of a dominant-negative form of RSG severely inhibited the process of cell elongation in stems and reduced the endogenous amount of GA1, which is the major, active GA in tobacco, in transgenic plants. To investigate the function of RSG in the regulation of GA content, we identified a target gene of RSG. This gene encodes ent-kaurene oxidase in the GA biosynthetic pathway. These results indicate that RSG regulates plant morphology through the transcriptional control of a GA biosynthetic enzyme.
RESULTS
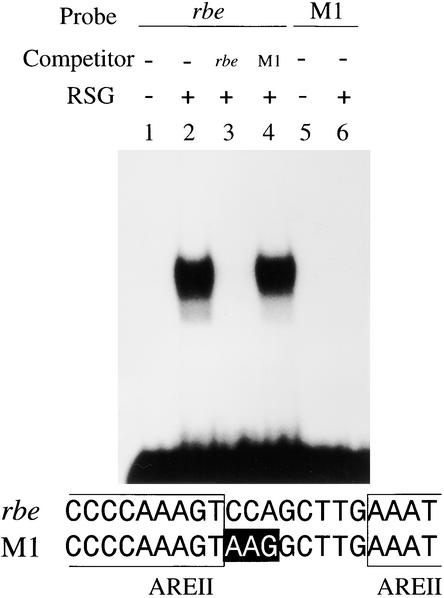
To identify a DNA binding protein for the auxin-responsive element II (AREII) cis regulatory element, which is involved in the auxin responsiveness of the parB gene from tobacco (Takahashi et al., 1995), we performed a yeast one-hybrid screen with a 2 × AREII construct to isolate a tobacco cDNA for RSG (see Methods). The 2 × AREII construct was strongly activated by RSG, whereas a single-copy AREII construct was not markedly activated by RSG in yeasts (data not shown). Gel mobility shift experiments showed that recombinant RSG bound specifically to an artificial sequence, named rbe (for RSG binding element), that had been generated in the process of dimerizing AREII but was not able to bind to the mutated version of rbe, named M1, in which the sequence TCCAGCTTGA had been changed to TAAGGCTTGA (Figure 1). Thus, although RSG is not an AREII binding protein, it seems to play a crucial role in regulating plant morphogenesis, based on the results presented here.
Figure 1.
Sequence-Specific Binding of RSG.
Recombinant RSG was used in a gel retardation assay. rbe was used as a probe for lanes 1 to 4, and M1 was used as a probe for lanes 5 and 6. The DNA sequences of oligonucleotides used as probes are shown. The mutated bases in M1 are highlighted. rbe was generated in the process of dimerizing AREII. The sequences from AREII are boxed. (+), addition to the reaction mixtures; (−), omission from the reaction mixtures.
Structure of RSG
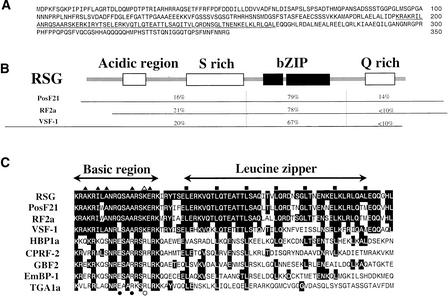
Nucleotide sequence analysis revealed an open reading frame that was predicted to encode a protein of 350 amino acids (Figure 2A). A search for homologous sequences in the GenBank database identified a bZIP domain in RSG between residues 194 and 262. An acidic region, preceded by a phenylalanine cluster near the N terminus (Uesugi et al., 1997), a serine-rich region, and a glutamine-rich region in the C-terminal region might serve as transcriptional activation domains. The amino acid sequence of the predicted RSG product exhibited strong similarities in the bZIP region to PosF21 from Arabidopsis (Aeschbacher et al., 1991), RF2a from rice, which is involved in leaf development (Yin et al., 1997), and vs-1 binding factor (VSF-1) from tomato (Torres-Schumann et al., 1996). In contrast, similarities of amino acid sequences were limited in regions outside the bZIP domain for these sequences (Figure 2B). Unlike other plant bZIP proteins, which have a conserved arginine residue at position −10 relative to the first leucine residue in the leucine-zipper region, RSG, PosF21, RF2a, and VSF-1 have a lysine residue at this position (Figure 2C). This arginine residue at position −10 is important for the DNA binding specificity of bZIP proteins (Aukerman et al., 1991; Suckow et al., 1994). Thus, RSG, PosF21, RF2a, and VSF-1 seem to form a distinct class among bZIP proteins.
Figure 2.
Predicted Amino Acid Sequence and Domain Structure of RSG.
(A) Deduced amino acid sequence of RSG. The bZIP domain is underlined. The GenBank accession number of RSG cDNA is AB040471.
(B) Schematic domain structure of RSG. The amino acid sequence identities with PosF21, RF2a, and VSF-1 are also indicated.
(C) Alignment of amino acid sequences in the bZIP region. Sequences of the bZIP domain of RSG were compared with those of PosF21 (Aeschbacher et al., 1991), RF2a (Yin et al., 1997), VSF-1 (Torres-Schumann et al., 1996), HBP1a (Tabata et al., 1989), CPRF-2 (Weisshaar et al., 1991), GBF2 (Schindler et al., 1992b), EmBP-1 (Guiltinan et al., 1990), and TGA1a (Katagiri et al., 1989). Highlighted residues indicate amino acids that are identical to those of RSG. Squares indicate the position of leucine residues conserved in the bZIP proteins. Triangles indicate the amino acid residues specifically conserved among RSG, PosF21, RF2a, and VSF-1 in the basic region. Circles indicate the amino acid residues conserved among other plant bZIP proteins in the basic region. The open triangles and circles indicate the amino acid residues at position −10 relative to the first leucine residue in the leucine zipper region, which are important for DNA binding specificity.
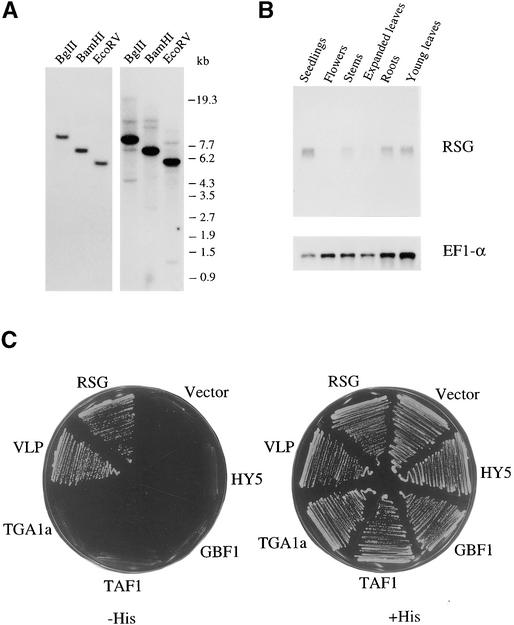
Genomic DNA gel blot hybridization using the DNA region that encodes the bZIP domain of RSG as a probe under both stringent and less stringent conditions (Figure 3A) showed that RSG seems to be encoded by a single-copy gene with a few related genes in the tobacco genome. In fact, we isolated a few RSG-related cDNAs, including the one encoding VLP (for VSF-1–like protein; see below). RNA gel blot analysis showed that the mRNAs for RSG are expressed in seedlings, stems, roots, and young leaves and are barely detectable in expanded leaves and flowers (Figure 3B). A faint signal is visible below the RSG band in the lane for the stems sample; however, the meaning of this signal is currently unknown.
Figure 3.
Genomic Organization and Expression of RSG.
(A) DNA gel blot analysis of RSG. Ten micrograms of genomic DNA was digested with the indicated enzymes and probed with a 32P-labeled DNA fragment corresponding to the bZIP domain of RSG. At left, under stringent hybridization conditions; at right, under less stringent hybridization conditions. Length markers are indicated at right in kilobases.
(B) RNA gel blot analysis of RSG mRNA. One microgram of poly(A)+ RNA isolated from the indicated organs was used for gel blot analysis under stringent hybridization conditions. The blot was hybridized with radiolabeled RSG cDNA and then reprobed with EF1-α cDNA as a loading control.
(C) Specificity of dimerization of RSG. Interactions between RSG and other plant bZIP proteins were assayed by using the yeast two-hybrid system. Yeast cells HF7c were simultaneously transformed with a plasmid expressing a GAL4 DNA binding domain fused to the bZIP domain of RSG and with plasmids expressing the GAL4 activation domain fused to the bZIP domains of the indicated RSG, VLP (a RSG-related protein), TGA1a, TAF1, GBF1, HY5, or a control vector. Cells containing both plasmids were streaked on the plates with (+His) or without (−His) histidine but with 0.2 mM 3-aminotriazole.
The bZIP proteins generally function as either homodimers or heterodimers. Dimerization specificity depends on the amino acid sequences of the two zipper regions. Thus, each bZIP protein in the cell can form dimers with only a small set of other bZIP proteins. For example, the bZIP class of proteins that includes TGA1a (Katagiri et al., 1989) can form dimers with other members of this class but cannot heterodimerize with another class of bZIP proteins called G-box binding factors (GBFs; Schindler et al., 1992a). To investigate the dimerization specificity of RSG, we tested the interactions of RSG with other bZIP proteins by using the yeast two-hybrid assay (Figure 3C). The bZIP domain of RSG did not interact with those of other classes of bZIP proteins, namely, GBF1 (Schindler et al., 1992b), HY5 (for LONG HYPOCOTYL5; Oyama et al., 1997), TGA1a, or TAF-1 (Oeda et al., 1991), but did interact with those of RSG and the RSG-related protein VLP. This result indicates that RSG selectively forms a dimer with itself and its related protein. VLP was isolated as an RSG-interacting factor by a yeast two-hybrid screen (to be described in a forthcoming manuscript). In the bZIP region, the deduced amino acid sequence of VLP showed 83% identity to that of RSG.
Dominant-Negative Form of RSG
Because RSG is a member of a small gene family, isolation of the loss-of-function mutant and suppression of RSG function by an antisense RNA would be difficult. Indeed, transgenic tobacco plants in which the antisense construct of RSG was expressed did not show remarkable morphological alteration (data not shown). Thus, to investigate the role of RSG in the development of tobacco, we tried to repress its function by introducing a dominant-negative transgene. Removal of either the activation domain or the DNA binding domain, whether occurring naturally or constructed in vitro, can produce a dysfunctional transcription factor. Such mutants can inhibit the function of wild-type factors in a dominant-negative fashion (Lloyd et al., 1991; Nakabeppu and Nathans, 1991; Deng and Karin, 1993). Myb-like transcriptional activators from Arabidopsis, GLABRA1 (GL1), and WEREWOLF (WER), which regulate the differentiation of epidermal cells, are perturbed by another Myb-like protein, CAPRICE (CPC), which lacks a transcriptional activation domain (Wada et al., 1997; Lee and Schiefelbein, 1999). A dominant-negative form of PG13 (the gene product of g13), a member of the TGA1a bZIP family, suppresses the activity of wild-type TGA1a/PG13 in transgenic tobacco plants (Rieping et al., 1994). These findings suggest that RSG without activation domains could repress the activity of full-length RSG and its related bZIP proteins.
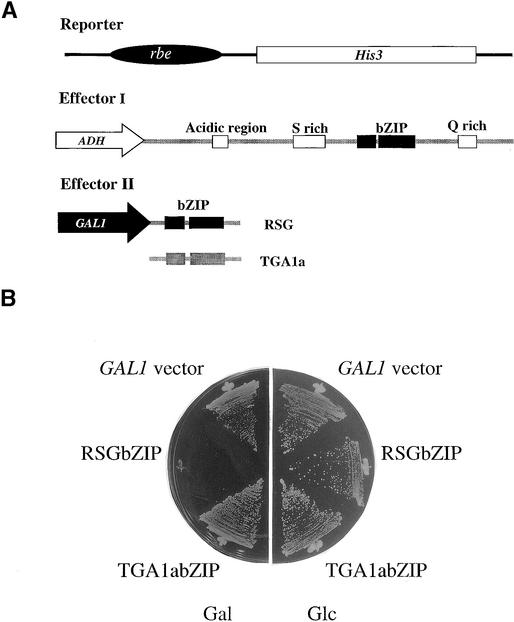
The full-length RSG protein exhibits the function of a transcriptional activator in yeast and plant cells. In yeast, expression of the bZIP domain of RSG under the control of the GAL1 promoter can inhibit transcriptional activation by full-length RSG, whereas the bZIP domain of TGA1a, another class of the tobacco bZIP protein, or the empty vector does not (Figures 4A and 4B). When the GAL1 promoter was not induced, no inhibition of transcriptional activation was observed. These results show that the transcriptional activity of full-length RSG was selectively repressed by the bZIP domain of RSG, at least in yeast.
Figure 4.
Dominant-Negative Effect of the bZIP Domain of RSG in Yeasts.
(A) Schematic representation of the structures of the reporter and effectors. Effector I expresses full-length RSG. Effector II expresses the bZIP domain of RSG or TGA1a.
(B) Inhibition of full-length RSG by the bZIP domain of RSG in yeasts. The plasmids pLysGAL1bZIPRSG (encoding the bZIP domain of RSG), pLysGAL1bZIPTGA1a (encoding the bZIP domain of TGA1a), or pLysGAL1 (for a control vector) were transformed with pLeuGADHRSG, which expressed the full-length RSG under the control of an ADH promoter, and selected on a medium without lysine or leucine. Yeast strain YPH499 carrying AREII × 2-HIS3 was used as a host. The transformants were streaked on media without histidine, lysine, or leucine but with 0.2 mM 3-aminotriazole containing galactose (Gal; left) or glucose (Glc; right).
Phenotypes of 35S:RSGbZIP–Transformed Tobacco Plants
To repress endogenous RSG, we generated transgenic tobacco plants in which the bZIP domain of RSG was expressed under the control of the 35S promoter of the cauliflower mosaic virus. Eleven of 22 independent transformants exhibited a marked decrease in plant height (Figure 5A). The number of leaves produced before flowering was 18.0 ± 1.4 (mean ±se of four plants) in the control SR1 tobacco plants and 21.0 ± 1.3 in 35S:RSGbZIP–transformed tobacco plants. Because 35S:RSGbZIP plants and the SR1 controls had a comparable number of leaves, the decrease in height was the result of a reduction in the internode length. The leaves of 35S:RSGbZIP–transformed plants were smaller than those of control SR1 tobacco plants, had a wrinkled surface, and were slightly dark green. Although no striking change in morphology was found in flowers or roots, fewer flowers were produced and flowering time was delayed.
Figure 5.
Phenotypes of Transgenic Tobacco Plants Expressing the bZIP Domain of RSG.
(A) Comparison of SR1 tobacco (left) and the transgenic tobacco harboring the 35S:RSGbZIP construct (right).
(B) Transverse section of the eighth internode of control SR1 tobacco.
(C) Transverse section of the eighth internode of the transgenic tobacco harboring the 35S:RSGbZIP construct.
(D) Morphology of epidermal cells of the seventh internode of SR1 tobacco.
(E) Morphology of epidermal cells of the seventh internode of the transgenic tobacco harboring the 35S:RSGbZIP construct.
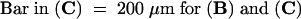 ;
; 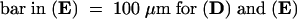 .
.
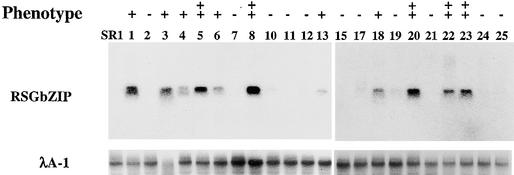
RNA gel blot analysis showed that the transgene was expressed in all the transgenic tobacco plants that had altered morphology (Figure 6), indicating that the morphological changes of 35S:RSGbZIP–transformed plants were the result of expression of the transgene. Some of the 35S:RSGbZIP–transformed tobacco plants were fertile, and their offspring inherited the dwarf phenotypes with the expression of the transgene; however, the germination frequency was reduced. On the other hand, in plants expressing the dominant-negative forms of PG13 and TGA1a, another class of bZIP proteins, there were no morphological alterations in the transgenic tobacco plants (Rieping et al., 1994; Miao and Lam, 1995). Thus, the altered phenotypes of the transgenic tobacco plants used in this study are specific to the dominant-negative form of RSG. Because RSG was selectively repressed by its own bZIP domain in yeast (Figure 4) and formed a dimer only with RSG and an RSG-related protein among the few that we tested (Figure 3C), the morphological changes of 35S:RSGbZIP–transformed plants more likely resulted from the suppression of a few RSG-related proteins rather than the cross-suppression of other classes of bZIP transcriptional factors.
Figure 6.
RNA Gel Blot Analysis of Transgenic Tobacco Plants Harboring the 35S:RSGbZIP Construct.
Ten micrograms of total RNA extracted from an expanded leaf was subjected to electrophoresis, transferred to Biodyne B (Pall, East Hills, NY) membrane, and hybridized with the DNA corresponding to the bZIP domain (amino acids 164 to 286) and λA-1 encoding one of the rRNAs as a control. +, transgenic tobacco plants showing altered morphology; ++, transgenic tobacco plants showing severe phenotypes (i.e., internode length <20% of control SR1); − , transgenic tobacco plants showing no altered morphology.
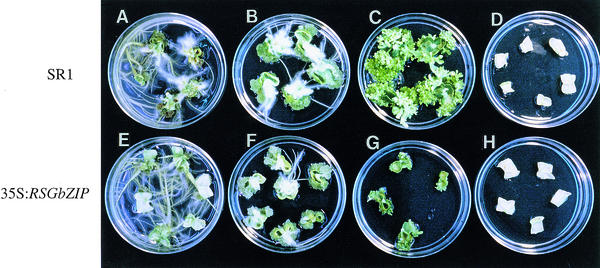
To examine the effects of the transgene on the growth of other parts of the plant, the root, callus, and shoot were induced from leaf squares in a medium containing various concentrations of auxin and cytokinin. The auxin-to-cytokinin ratio controls the formation of roots, shoots, and callus tissue in vitro (Skoog and Miller, 1957). Whereas the growth of both root (Figures 7A and 7E) and callus (Figures 7B and 7F) from R1 35S:RSGbZIP–transformed tobacco plants was comparable to that of the control SR1 tobacco plants, shoot growth was severely inhibited (Figures 7C and 7G). No differentiation or growth was observed without hormones (Figures 7D and 7H). These results show that the dominant-negative form of RSG did not inhibit the overall growth of tobacco but specifically inhibited the growth of the shoot. Furthermore, in relation to induction of organs, expression of the dominant-negative form of RSG did not affect the responsiveness of the cells to auxin and cytokinin (Figure 7).
Figure 7.
Growth of Organs of Transgenic Tobacco Plants Expressing the bZIP Domain of RSG.
The squares of leaves from the R1 35S:RSGbZIP transgenic tobacco (line 8) and SR1 tobacco were cut into 10 × 10-mm squares and cultured for 4 weeks on Murashige and Skoog medium (Murashige and Skoog, 1962).
(A) and (E) Plates containing 2 mg/L IAA and 0.02 mg/L kinetin for the root.
(B) and (F) Plates containing 2 mg/L IAA and 0.20 mg/L kinetin for the callus.
(C) and (G) Plates containing 0.02 mg/L IAA and 2 mg/L kinetin for the shoot.
(D) and (H) Plates containing no hormone as a control.
Plates (A) to (D) were used for leaves from control SR1 tobacco plants. Plates (E) to (H) were used for leaves from the transgenic tobacco expressing the bZIP domain of RSG. All plates contained 250 mg/L vancomycin and 100 mg/L carbenicillin.
We next investigated the morphology of the stem cells to determine whether inhibition of cell division or cell elongation was the cause of growth repression in the shoot, especially in the internode. The epidermal cells in stems of 35S:RSGbZIP–transformed tobacco plants were shorter than those in the control SR1 plants (Figures 5D and 5E). The cell shape and cell file of stem epidermis were irregular in 35S:RSGbZIP transformants. However, differences, if any, in the transverse sections of stems were not conspicuous (Figures 5B and 5C). Therefore, the repression of internode elongation in 35S:RSGbZIP–transformed tobacco plants could be accounted for mostly by the inhibition of cell elongation in the growing axis.
GAs Are Decreased in 35S:RSGbZIP–Transformed Tobacco Plants
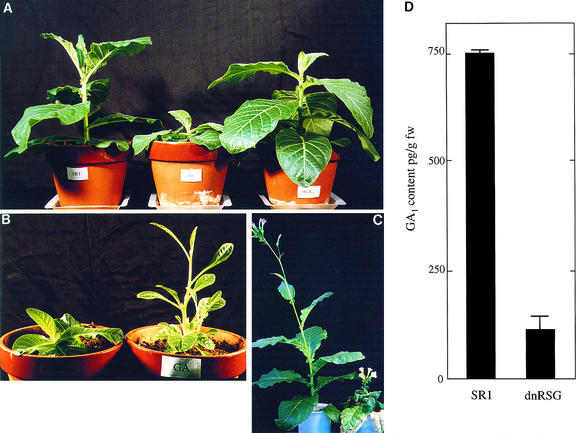
GA-deficient Arabidopsis mutants display characteristic phenotypes, including dark green leaves and a dwarf growth habit attributable to reduced stem elongation (Sun and Kamiya, 1994; Chiang et al., 1995; Xu et al., 1995; Yamaguchi et al., 1996; Helliwell et al., 1998). In this study, because the dwarf phenotypes of 35S:RSGbZIP–transformed tobacco plants seemed similar to those of GA-deficient mutants, we examined the effect of applying GAs to the 35S:RSGbZIP–transformed tobacco plants. The 35S:RSGbZIP–carrying plants were sprayed with solutions of GA3, GA9, or GA18. As shown in Figure 8A, the GAs restored internode length as well as the size and surface of 35S:RSGbZIP tobacco plant leaves to the same as those of control SR1 tobacco plants. GA3 was the most effective among the GAs tested such that overdosing with GA3 changed the shape of 35S:RSGbZIP–carrying tobacco plants to that of the elongated slender plants (Figure 8B). The plant hormones brassinolide and indole-3-acetic acid (IAA) also promote cell elongation; however, applying either of these to the affected plants failed to restore the phenotypes of the transgenic tobacco plants (data not shown).
Figure 8.
Decrease in Endogenous GAs in the 35S:RSGbZIP Transformants.
(A) GAs restored the phenotype of 35S:RSGbZIP–transformed tobacco plants. The control SR1 tobacco is at left, the 35S:RSGbZIP transformant is center, and the GA-treated 35S:RSGbZIP plant is at right. The 35S:RSGbZIP–transformed tobacco plant at right was sprayed with a solution of 10−4 M GA9, and the plant at center was sprayed with water once weekly for 4 weeks.
(B) 35S:RSGbZIP plants overdosed with GA3. After the 35S:RSGbZIP plant at left was sprayed with water and the 35S:RSGbZIP plant at right with 10−4 M GA3 every day for 2 weeks, plants were grown without spraying for 1 week.
(C) The effect of uniconazole P on the growth of tobacco. The SR1 tobacco plant at right received 250 mL of 10 mg/L uniconazole P; the SR1 tobacco plant at left received only water.
(D) GA1 contents in 35S:RSGbZIP plants. Each column represents the mean ±se of four plants. fw, fresh weight.
An inhibitor for GA biosynthesis, uniconazole P, was applied to control SR1 tobacco plants to determine the effect of decreasing the GA content on the plant morphology. Uniconazole P altered the morphology of control SR1 tobacco plants so that they resembled 35S:RSGbZIP transformants but in a more pronounced manner (Figure 8C). GA perception and signal transduction pathways appeared to be normal in the transgenic tobacco plants because GAs restored stem elongation and overdosing produced thin, elongated plants (Figure 8B). These observations suggest that the dwarf phenotypes of 35S:RSGbZIP–carrying plants were due to a decrease in the endogenous amount of GAs. We identified GA1, the major active GA in tobaccos, by gas chromatography–selected ion monitoring (GC-SIM) with an internal standard and quantified the amount present by ELISA. The endogenous amount of GA1 in the 35S:RSGbZIP plants was only 15% of that in the control SR1 plants (Figure 8D). These results indicate that the dominant-negative form of RSG inhibited cell elongation through decreasing the amount of GAs present.
RSG Activates the GA3 Promoter
One problem with the strategy of using dominant-negative mutations is the potential for influencing other, unrelated regulatory proteins. Although RSG does not interact with those known plant bZIP proteins that we have tested, we cannot completely rule out the possibility that the morphological changes of 35S:RSGbZIP–transformed plants could reflect the cross-inhibition of other transcriptional factors. Identifying the target gene of RSG would provide the most direct evidence that RSG regulates the endogenous amount of GAs. Our results using the dominant-negative form of RSG suggest that RSG might regulate one of the genes that encode enzymes for biosynthesis of GA. Substantial progress has been made in isolating the genes encoding the enzymes of the GA biosynthetic pathway (Hedden and Kamiya, 1997). We found GA18 to be effective in promoting cell elongation in 35S:RSGbZIP plants (data not shown). GA18 is not biologically active by itself but becomes active by sequential conversion to GA1 by a GA 20-oxidase (Hedden and Kamiya, 1997). This suggested that the lesion in the transgenic tobaccos could involve the enzyme upstream of GA 20-oxidase in the GA biosynthetic pathway.
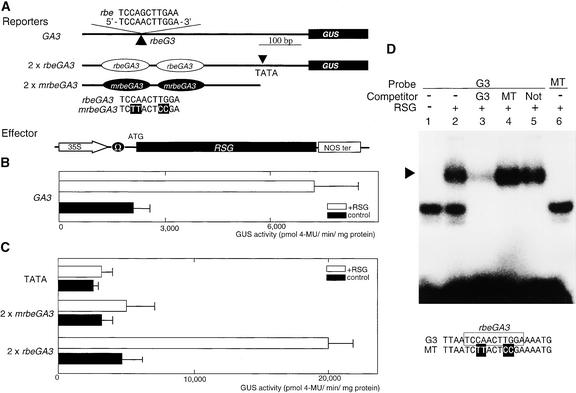
Because RSG binds to a characteristic sequence, rbe, that contains TCCAGCTTGA, we examined whether similar sequences exist in the promoter region of genes encoding the GA biosynthetic pathway. A sequence closely related to rbe, TCCAACTTGG, was found 416 bp upstream of the tentative TATA box of the Arabidopsis GA3 gene that encodes a cytochrome P-450 enzyme having the activity of an ent-kaurene oxidase (Figure 9A). The ent-kaurene oxidase protein catalyzes ent-kaurene to ent-kaurenoic acid in the GA biosynthetic pathway (Helliwell et al., 1999) and is upstream of GA 20-oxidase. We next investigated whether the expression of GA3 was regulated by RSG. Figure 9B shows that Arabidopsis GA3 promoter–β-glucuronidase (GUS) was activated by RSG in the transient assay system with tobacco mesophyll protoplasts. The transcription of the cauliflower mosaic virus 35S promoter was increased only 1.1-fold by RSG. Furthermore, an 11-bp dimer of the rbe-related sequence in the GA3 gene (rbeGA3) was sufficient for transcriptional activation by RSG. The mutation in rbeGA3 eliminated RSG-dependent GUS activity (Figure 9C). Gel retardation experiments showed that recombinant RSG specifically bound to rbeGA3 but was not able to bind to the mutated rbeGA3, which abolishes RSG-dependent transcriptional activation (Figure 9D). Furthermore, the formation of a complex of rbeGA3 with RSG was not inhibited in the presence of an excess amount of the mutated sequence or unrelated DNA sequences.
Figure 9.
RSG Regulates the Arabidopsis GA3 ent-Kaurene Oxidase Gene.
(A) Schematic representation of the reporters and the effector. The 528 bp of the Arabidopsis GA3 promoter was fused to the GUS gene (GA3). The 11-bp dimers of the rbe-related sequence in the GA3 gene (2 × rbeGA3) and of the mutated sequence (2 × mrbeGA3) were fused to the TATA box of parB driving GUS. The mutagenized nucleotides in the mrbeGA3 are highlighted. The effector plasmid expressed the full-length RSG under the control of 35S promoter with a viral translation enhancer (Ω). The empty effector vector was used as the control. NOS ter indicates the polyadenylation signal of the gene for nopaline synthetase.
(B) Activation of GA3 promoter by RSG. The reporter construct (GA3, 7 μg) and the effector construct (35S-RSG, 3 μg) were cotransfected to tobacco mesophyll protoplasts. The open bar represents GUS activity of protoplasts transfected with the effector expressing RSG; the closed bar represents GUS activity of protoplasts transfected with the control vector. Each assay was done in triplicate. The error bars indicate sd.
(C) Activation of rbeGA3 by RSG. Transfections were performed with tobacco mesophyll protoplasts, with 7 μg of the reporter constructs and 3 μg of the effector construct. TATA indicates the parB TATA box–GUS construct. The open bars represent GUS activity of protoplasts transfected with the effector expressing RSG; the closed bars represent GUS activity of protoplasts transfected with the control vector. Each assay was done in triplicate. The error bars indicate sd.
(D) Gel retardation assay using recombinant RSG. The specific RSG–DNA complexes are indicated by an arrowhead. Oligonucleotides containing rbeGA3 (G3, lanes 1 to 5) or mrbeGA3 (MT, lane 6) were used as the probes. The rbeGA3 sequence is boxed, and the mutated bases are highlighted. For lane 3, G3 was the competitor. For lane 4, MT, a mutated version of G3, was the competitor. For lane 5, the NotI linker (Not) was used as an unrelated competitor. The lower shifted bands in lanes 1, 2, and 6 are nonspecific DNA–protein complexes. (+), present; (−), absent.
4-MU, 4-methylumbelliferone.
These results strongly suggest that the transcriptional suppression of the ent-kaurene oxidase gene was responsible for the decrease in the GA content and consequently induced the dwarf phenotypes in 35S:RSGbZIP–transformed tobacco plants. To confirm whether the expression of the ent-kaurene oxidase gene was repressed in the transgenic tobacco plants, we used reverse transcription–polymerase chain reaction (RT-PCR) with degenerate primers and isolated from tobacco a cDNA that encoded putative ent-kaurene oxidase. The amino acid sequence of the predicted product of this 894-bp fragment of cDNA (NtKO, for Nicotiana tabacum ent-kaurene oxidase; Figure 10A) was 64% identical and 93% similar to that of Arabidopsis GA3. However, this region was outside the heme binding domain, which is conserved among members of the P-450 family. The amino acid sequence identity between NtKO and other plant P-450 enzymes in the databases was <25%. Because we could not detect the NtKO transcript, even when we used poly(A)+ RNA in the gel blot analysis, we conducted RT-PCR with total RNA from the young leaves of control SR1 and 35S:RSGbZIP–transformed plants. To quantify the NtKO transcript, we executed amplification by PCR with increasing numbers of cycles and using NtKO-specific primers and then detected the products by DNA gel blot hybridization. Tobacco arcA, which encodes a WD-40 protein (Ishida et al., 1993), was used as an internal control for RT-PCR. The amount of NtKO transcript in 35S:RSGbZIP plants was decreased to <23% of that in control SR1 plants (Figure 10B).
Figure 10.
Predicted Amino Acid Sequence and the Expression of NtKO.
(A) Comparison of the partial amino acid sequences of NtKO (tobacco) and GA3 (Arabidopsis). Identical amino acids are highlighted, and similar amino acids are marked with gray boxes. Numbers at right indicate amino acid positions from the first methionine of GA3 (Arabidopsis). The regions corresponding to the primer sequences used for RT-PCR are shown by lines above the sequence. GenBank accession number of NtKO is AB040485.
(B) Comparison of NtKO mRNA contents by RT-PCR. Amplifications were performed for five, seven, nine, or 11 cycles, and the products were detected by DNA gel blot hybridization. Tobacco arcA was amplified in the same reaction and used as an internal control of RT-PCR. SR1, control SR1 tobacco plants; TG, 35S:RSGbZIP transgenic tobacco plants.
DISCUSSION
Our study focuses on the functional analysis of a bZIP transcriptional activator, RSG. The dominant-negative form of RSG was expressed to inhibit endogenous RSG, which resulted in the transcriptional repression of the gene encoding ent-kaurene oxidase, a GA biosynthetic enzyme. This downregulation reduced the endogenous amounts of GAs and repressed cell elongation in stems in 35S:RSGbZIP–transformed tobacco plants. Thus, RSG regulates plant morphology through transcriptional control of a GA biosynthetic enzyme.
RSG Is a Member of a Distinct Class of bZIP Proteins
Many bZIP proteins have been isolated from various plants. Whereas RSG is a typical bZIP protein, with PosF21, VSF-1, and RF2a it forms a distinct class of bZIP proteins based on highly conserved bZIP domains and the presence of lysine rather than arginine at position −10 relative to the first leucine residue in the leucine zipper region (Figure 2C). The arginine residue at position −10 is conserved in all other bZIP proteins of both animals and plants. In many cases, the bZIP proteins bind to DNA containing the palindromic ACGT core sequence; however, the proposed binding site of RF2a is CCA(N)nTGG (Yin et al., 1997), and that of VSF-1 is CCTCCGTTG (Ringli and Keller, 1998). In this study, RSG specifically bound to the rbe sequence, which contained TCC-AGCTTGA (Figure 1), and to rbeGA3, which contained TCC-AACTTGG (Figure 9D). These two sequences are similar to the proposed binding site of RF2a. Substitution for an arginine residue by lysine at position −10 changes the optimal binding site of the yeast bZIP protein GCN4 (Suckow et al., 1994) from the palindromic ATF/CRE (for activating transcription factor 1/cAMP response element) site (GACGTC) to the pseudopalindromic AP-1 (for activator protein 1) site (TGACTCA). On the other hand, the mutant VSF-1, with a lysine-to-arginine substitution at this position, binds more efficiently to the palindromic G-box element (GCCACG-TGGC) than does the wild-type protein (Ringli and Keller, 1998). Moreover, RSG binds more efficiently to rbe than to the G-box (Y. Takahashi, unpublished results). The presence of lysine rather than arginine at position −10 of this class of bZIP proteins, including RSG, may contribute to binding to the target sequences that do not contain a palindromic ACGT core sequence.
Dominant-Negative Effects
In the case of functionally redundant genes, loss-of-function mutations and expression of antisense RNA might not result in a phenotype. A clear example of this is provided by studies of the ethylene receptors of Arabidopsis. Single loss-of-function mutation in ETHYLENE RESPONSE1 (ETR1)–related genes does not exhibit defects in ethylene response (Hua and Meyerowitz, 1998). Mice that bear loss-of-function mutations in a wide variety of important genes, including those encoding c-Src (Soriano et al., 1991), the nerve growth factor receptor (Lee et al., 1992), and MyoD (Rudnicki et al., 1992), are viable and have no obvious change in phenotype. Functional redundancy may represent a widespread feature in some regulatory networks controlling complex developmental processes in multicellular organisms. Five RSG-related genes are currently documented in the Arabidopsis genome database. An Arabidopsis mutant in which one of these genes was interrupted by a T-DNA insertion did not show an obvious change in phenotype (Babiychuk et al., 1997).
For this reason, we used a dominant-negative strategy to address the in vivo function of RSG. The expression of dominant mutant RSG protein decreased the endogenous levels of GAs probably through the repression of RSG and a few related proteins. However, one must be aware of a potential influence on other, unrelated regulatory proteins by the dominant-negative mutated protein. In this study, to demonstrate the role of RSG in the regulation of the endogenous amount of GAs, we identified a target gene of RSG, which encodes ent-kaurene oxidase in the GA biosynthetic pathway. Because the observed morphological changes of 35S:RSGbZIP plants were essentially limited to GA-deficient phenotypes, that is, reduced stem elongation, dark green leaves, and decrease in germination efficiency, the effects of the dominant-negative form of RSG on other regulatory factors were small, if any. A similar approach would be applicable to other functionally redundant transcriptional regulators to study their functions in vivo.
No remarkable change was found in morphology or growth of roots in 35S:RSGbZIP–transformed tobacco plants. Various GA-deficient plants, which are characterized by dwarf shoot habits, exhibit nearly normal root elongation (Baluška et al., 1993). This phenomenon has been explained by the proposition that GAs control the growth of root cells at a lower concentration range than that required to control the growth of shoots (Tanimoto, 1991).
Role of RSG in the GA Synthetic Pathway
Recently, molecular genetic studies using morphological mutants have revealed many proteins that exhibit amino acid sequences similar to those of transcription factors involved in plant development (Oyama et al., 1997; Wada et al., 1997; Walsh et al., 1997; Lotan et al., 1998). To understand how these transcription factors affect morphogenesis and differentiation, their target genes must be identified. However, only a few of such target genes have been identified in plants (Sablowski and Meyerowitz, 1998). Our study, using a dominant-negative mutant, suggests that RSG regulates one of the genes that encode enzymes for GA biosynthesis. At this time, the GA biosynthetic pathway has been established, and several genes encoding GA biosynthetic enzymes have been cloned. This information has helped in our search for target genes of RSG. Our results show that the ent-kaurene oxidase gene is an important target gene of RSG and that its regulation might be direct. A transcription factor usually regulates the expression of several target genes. The possibility that RSG potentially regulates the genes for other proteins involved in GA biosynthesis in addition to the ent-kaurene oxidase gene should not be overlooked.
We have generated transgenic tobaccos that express full-length RSG under the control of the 35S promoter; however, these transgenic plants exhibited no obvious morphological changes (data not shown). Because many enzymes are involved in GA biosynthesis and because some of their genes, such as those encoding GA 3β-hydroxylase and GA 20-oxidase, are downregulated by GAs through feedback suppression (Chiang et al., 1995; Phillips et al., 1995), the increase of only one enzyme could not have resulted in a drastic elevation of GA levels. Indeed, the overexpression of the GA1 gene encoding copalyl diphosphate synthase did not alter the morphology of plants (Sun and Kamiya, 1994). Another possibility is that other transcription factors in addition to RSG might also be required for full activation of the ent-kaurene oxidase gene. In this regard, using a two-hybrid system, we have identified a few DNA binding proteins, including VLP, that interact with RSG (D. Igarashi and Y. Takahashi, unpublished results).
The sequence of the bZIP domain of RSG exhibited considerable similarity to that of RF2a, which was isolated as a DNA binding protein for the phloem-specific cis element of the rice tungro bacilliform virus. Overexpression of RF2a in transgenic rice plants did not result in any obvious changes, as with RSG in tobacco plants; however, antisense suppression of RF2a caused leaf twisting and a stunting of shoots without alterations in the roots (Yin et al., 1997). Because these phenotypes seemed to be similar to those of 35S:RSGbZIP plants, we think RF2a might also play an important role in the regulation of GA contents. The expression of genes encoding GA biosynthetic enzymes is regulated by both an endogenous developmental program and environmental stimuli (Silverstone et al., 1997; Helliwell et al., 1998; Yamaguchi et al., 1998). Further investigations of how the transcriptional activity of RSG is controlled by both internal and external signals will help unravel the molecular mechanisms for the fine regulation of the endogenous amounts of this plant hormone that controls many aspects of plant development.
METHODS
cDNA Cloning
The reporter plasmid pAREII × 2-HIS3, containing two tandem copies of the auxin-responsive element II (AREII), was constructed by the insertion of the HIS3 gene into the EcoRI-SalI site and of the AREII dimer into the SmaI site of pRS304. The reporter plasmid pAREII × 2-LacZ was constructed by the insertion of the LacZ gene with a TATA box into the HindIII-BamHI site and of the AREII dimer into the filled-in SphI site of pRS306. Tobacco cDNA was generated from poly(A)+ RNA isolated from 2-day-old BY-2 cells and cloned into yeast expression vector pGAD10 by using a cDNA construction kit (Clontech, Palo Alto, CA). The number of transformants was 5 × 106 with an average insert size of 1.3 kb. A purified plasmid library DNA was used to transform the YPH499::AREII × 2 strain. Of 6 × 107 yeast transformants, 113 colonies grew on plates without histidine but containing 3-aminotriazole (1 μM), and these were screened for LacZ activity. The library plasmids were isolated from LacZ-positive clones and transformed back into yeasts. Clones that restored the activities of reporters were sequenced, and four clones encoding RSG (for repression of shoot growth) were isolated. A full-length cDNA clone for RSG was obtained from a λgt10 cDNA library of BY-2 cells by using a partial cDNA for RSG.
Gel Mobility Shift Assay
The coding region of RSG was cloned into the EcoRI site and the XhoI site of expression vector pET-30a (Novagen, Madison, WI) using synthetic oligonucleotides. The plasmid was transformed into Escherichia coli BL21 (DE3)pLysE. Cells were grown at 37°C and induced by the addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 1 mM.
Nucleotide sequences of the double-stranded oligonucleotides used for the gel mobility shift assays are rbe (5′-GAGCCC-CAAAGTCCAGCTTGAAAT-3′ and 5′-GTGATTTCAAGCTGGACT-TTGGGG-3′), M1 (5′-GAGCCCCAAAGTAAGGCTTGAAAT-3′ and 5′-GTGATTTCAAGCCTTACTTTGGGG-3′), G3 (5′-GAGTTAATCCAA-CTTGGAAAATG-3′ and 5′-GTGCATTTTCCAAGTTGGATTAA-3′), and MT (5′-GAGTTAATCTTACTCCGAAAATG-3′ and 5′-GTGCATTTTCGG-AGTAAGATTAA-3′). The oligonucleotides were annealed and then labeled with α-32P-dCTP and the Klenow fragment of DNA polymerase I. Binding mixtures contained 50 fmol of a labeled probe, 1 μg of a sonicated extract of E. coli expressing RSG or of control E. coli, and 2 μg of poly(dA-dT). DNA competitors were used at 1000-fold molar excess. The binding buffer consisted of 25 mM Hepes-KOH, pH 7.9, 10 mM MgCl2, 50 mM KCl, 0.5 mM EDTA, 10% glycerol, and 0.5 mM DTT. Reactions were incubated at 4°C for 30 min and loaded onto 4% polyacrylamide gels containing 6.7 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 3.3 mM sodium acetate.
DNA and RNA Gel Blot Hybridization
The DNA region encoding the basic leucine zipper (bZIP) domain (amino acids 164 to 286) was used as a probe. Hybridization conditions were 6 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS for 18 hr at 65°C for stringent conditions or at 55°C for less stringent conditions. Under the stringent conditions, after hybridization, the membrane was washed at 65°C in 2 × SSC containing 0.1% SDS and then autoradiographed for 1 day. For the less stringent conditions, the membrane was washed at 55°C and then autoradiographed for 4 days.
Generation of Transgenic Tobacco Plants
The coding region of the bZIP domain of RSG (amino acids 159 to 287) was cloned into the vector pBI121 between the BamHI and EcoRI sites and introduced into tobacco (Nicotiana tabacum cv Petit Havana SR1) by way of Agrobacterium tumefaciens–mediated transformation.
Dominant-Negative Effect and Two-Hybrid Assay in Yeast
The GAL1 expression plasmid pLysGAL1 was constructed by the insertion of the GAL1 promoter and terminator from pYO773 into the SacI and SalI sites of pRS317. The coding regions of the bZIP domains of RSG (amino acids 159 to 287) and TGA1a (amino acids 42 to 151) were isolated by polymerase chain reaction (PCR) and cloned into the site of pLysGAL1. The resulting plasmids were named pLysGAL1bZIPRSG and pLysGAL1bZIPTGA1a, respectively. The ADH (for alcohol dehydrogenase) expression plasmid pGADH was constructed by self-ligation of the filled-in KpnI and EcoRI fragment from pGAD424 (Clontech). The PCR-amplified cDNA fragment encoding full-length RSG was cloned into the regenerated EcoRI and BamHI of pGADH, which was named pLeuGADHRSG. Plasmids pLysGAL1bZIPRSG, pLysGAL1bZIPTGA1a, and pLysGAL1 were transformed into YPH499::AREII × 2 with pLeuGADHRSG and selected on a medium containing neither lysine nor leucine. For the induction of the GAL1 promoter, the transformants were grown once on a medium with neither leucine nor lysine but containing galactose. They were next streaked onto a medium that contained 0.2 mM 3-aminotriazole and galactose but not leucine, lysine, or histidine.
The cDNA fragments encoding the bZIP domain of RSG (amino acids 159 to 287), LONG HYPOCOTYL5 (HY5; amino acids 68 to 148), G-box binding factor1 (GBF1; amino acids 206 to 297), TAF1 (amino acids 161 to 265), TGA1a (amino acids 42 to 151), and vs-1 binding factor (VSF-1)–like protein (VLP; amino acids 157 to 346) were PCR-amplified, sequenced, and cloned into the yeast expression vector pGAD424 (Clontech). The coding region of the bZIP domain of RSG (amino acids 159 to 287) was cloned into the yeast expression vector pGBT9 (Clontech), which was named pGBT9-RSGbZIP. Yeast cells (HF7c) were simultaneously transformed with pGBT9-RSGbZIP and with plasmids expressing the activation domain of GAL4 fused to the bZIP domain of RSG, HY5, GBF1, TAF1, TGA1a, and VLP. The transformants were streaked onto a medium that contained 0.2 mM 3-aminotriazole and galactose but not leucine, lysine, or histidine.
Application of Plant Growth Regulators
The responsiveness to gibberellins (GAs), brassinolide, and indole-3-acetic acid (IAA) was examined with 35S:RSGbZIP–transformed tobacco plants 1.5 to 2 cm tall (∼2 months after germination). These tobacco plants were sprayed once a week with a solution of 10−4 M GA3, GA9, GA18, or IAA or 10−6 M brassinolide. Control plants were sprayed with water. For overdosing experiments, plants were sprayed daily with 10−4 M GA3.
The effect of uniconazole P was examined on plants grown individually in 1-liter beakers. SR1 tobacco plants received 250 mL of 10 mg/L uniconazole P (Wako, Osaka, Japan). Control plants received 250 mL of water.
Identification and Quantification of GA1
The occurrence of GA1 in the SR1 control plants was also confirmed by gas chromatography–selected ion monitoring (GC-SIM), in which deuterium-labeled GA1 (2H5-GA1) was used as an internal standard. The plants were homogenized and extracted in 80% acetone after addition of 2H5-GA1 (50 ng) and submitted to solvent partitioning to obtain an acidic ethyl acetate–soluble (AE) fraction. The AE fraction was prepurified by HPLC on an octadecylsilane column eluted with a mixture of CH3CN, H2O, and acetic acid (0.5%), increasing the concentration of CH3CN from 5 to 80% in 50 min. The fraction containing GA1 (retention time 33 to 34 min) was determined by ELISA and by using an anti-GA4 antibody (Yamaguchi et al., 1990). The GA1 fraction was concentrated by evaporation, trimethylsilylated in N-methyl-N-trimethylsilyl trifluoroacetamide, and analyzed by GC-SIM in which mass-to-charge ratios (m/z) of 565 and 452 were monitored for the internal standard and 560 and 447 for endogenous GA1. The occurrence of endogenous GA1 was evident from the observation of the ion peaks of m/z 560 and 447 at the same retention time as the peaks of m/z 565 and 452. New control SR1 and transgenic tobacco leaves were processed in the same manner as described above for the quantification of GA1 by ELISA. Use of ELISA to analyze fractions obtained from octadecylsilane-HPLC at retention times ranging from 31 to 36 min showed clear immunoreactivity in the fraction collected at the retention time of 33 to 34 min, and the GA1 in the fraction was quantified.
Transient Assay
The promoter of Arabidopsis thaliana GA3 from positions −624 to −97 (+1, initiation codon) was isolated by PCR and joined to the parB minimal promoter–GUS (for β-glucuronidase). The 5′ end of the parB minimal promoter was 4 bp upstream of the TATA box of parB. The 2 × rbeGA3 oligonucleotides (containing two tandem copies of rbeGA3) and 2 × mrbeGA3 oligonucleotides (containing two tandem copies of mrbeGA3) were cloned into the parB minimal promoter–GUS. The sequences of 2 × rbeGA3 were 5′-AGCTTCCAACTTGGATCCAACTTGGA-3′ and 5′-AGCTTCCAAGTTGGATCCAAGTTGGA-3′. The sequences of 2 × mrbeGA3 were 5′-AGCTTCTTACTCCGATCTTACTCCGA-3′ and 5′-AGCTTCGGAGTAAGATCGGAGTAAGA-3′. The effector plasmid expressed the full-length RSG cDNA under the control of the cauliflower mosaic virus 35S promoter with a viral translation enhancer (Ω). The control effector was the empty vector.
Protoplasts were prepared from mesophyll tissues of tobacco (cv Xanthi-nc), essentially as described by Nagy and Maliga (1976), but with enzymes at 0.5% cellulase Onozuka RS (Seishin Pharmaceutical Co., Tokyo, Japan) and 0.1% macerozyme (Yakult Pharmaceutical Co., Tokyo, Japan). DNA transfection was performed according to Krens et al. (1982). Protoplasts (0.3 mL of 3 × 105) in a MaMg medium (0.54 M mannitol, 15 mM MgCl2, and 4.7 mM Mes, pH 5.6) were incubated for 20 min at room temperature with 20% (w/v) polyethylene glycol 4000, 3 μg of effector plasmids, and 7 μg of reporter plasmids. Cells were diluted with 4.5 mL of a K3 medium (Nagy and Maliga, 1976) and cultured at 28°C for 30 hr. Immediately after the addition of 4.5 mL of a W5 medium (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 5 mM glucose, pH 5.8), cells were collected and GUS activity was determined.
Isolation and Detection of NtKO
To isolate the cDNA clone encoding tobacco ent-kaurene oxidase, we performed reverse transcription (RT)–PCR using total RNA isolated from young tobacco leaves and degenerate primers (5′-GAYTAYGAYGAYTTYCAYAAR-3′ and 5′-YTTYTTRTCCATRTTRCANCC-3′). Samples were heated to 94°C for 5 min and then subjected to 30 cycles of 94°C for 30 sec, 53°C for 60 sec, and 74°C for 60 sec. DNA fragments of ∼900 bp were isolated after gel electrophoresis and used for secondary PCR reaction. The resulting product was cloned into pUC18.
The yield of PCR product is proportional to the starting amount of the template only under conditions in which PCR amplification proceeds exponentially at a constant efficiency. For the detection of N. tabacum ent-kaurine oxidase (NtKO) transcripts, PCR was performed with cDNA derived from 0.5 μg of total RNA for five, seven, nine, or 11 cycles at 94°C for 20 sec, 58°C for 30 sec, and 74°C for 75 sec. The primer sequences were 5′-CACTTTGATAGAAAATGT-ATCTAAG-3′ and 5′-GCCCATTCTGTGCTCACTACTGTGG-3′ for NtKO and 5′-ATTCTAGAACCATGGCGCAAGAATCACTAGTACTC-3′ and 5′-ATGGATCCATAACGGCCAATACCCCA-3′ for arcA, an internal control of RT-PCR. The PCR products of 0.5 kb for NtKO and 1.0 kb for arcA were then detected by using DNA gel blot analysis with NtKO and arcA as probes. The intensity of the radioactive bands was quantified by Scion Image (Scion Corp., Frederick, MD).
Acknowledgments
We are grateful to Daisuke Igarashi, Mineko Konishi, and Dr. Toshiyuki Nagata for technical assistance and helpful discussions during the course of this work. We also thank Drs. Makiko Chono and Hiroshi Kawaide for assistance in the quantification of GA. This study was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, and Culture (Japan) and a grant from the Ministry of Agriculture, Forestry, and Fisheries (Japan) to Y.T.
References
- Aeschbacher, R.A., Schrott, M., Potrykas, I., and Saul, M.W. (1991). Isolation and molecular characterization of PosF21, an Arabidopsis thaliana gene which shows characteristics of a bZIP class transcription factor. Plant J. 1, 303–316. [PubMed] [Google Scholar]
- Aukerman, M.J., Schmidt, R.J., Burr, B., and Burr, F.A. (1991). An arginine to lysine substitution in the bZIP domain of an opaque2 mutant in maize abolishes specific DNA binding. Genes Dev. 5, 310–320. [DOI] [PubMed] [Google Scholar]
- Babiychuk, E., Fuangthong, M., Van Montagu, M., Inzé, D., and Kushnir, S. (1997). Efficient gene tagging in Arabidopsis thaliana using a gene trap approach. Proc. Natl. Acad. Sci. USA 94, 12722–12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluška, F., Parker, J.S., and Barlow, P.W. (1993). A role for gibberellic acid in orienting microtubules and regulating cell growth polarity in the maize root cortex. Planta 191, 149–157. [Google Scholar]
- Chiang, H.-H., Hwang, I., and Goodman, H.M. (1995). Isolation of the Arabidopsis GA4 locus. Plant Cell 7, 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr, R.J. (1994). Microtubules in plant morphogenesis: Role of the cortical array. Annu. Rev. Cell Biol. 10, 153–180. [DOI] [PubMed] [Google Scholar]
- Deng, T., and Karin, M. (1993). JunB differs from c-Jun in its DNA-binding and dimerization domains, and represses c-Jun by formation of inactive heterodimers. Genes Dev. 7, 479–490. [DOI] [PubMed] [Google Scholar]
- Giddings, T.H., and Staehelin, L.A. (1988). Spatial relationship between microtubules and plasma-membrane rosettes during the deposition of primary wall microfibrils in Closterium sp. Planta 173, 22–30. [DOI] [PubMed] [Google Scholar]
- Guiltinan, M.J., Marcotte, W.R., Jr., and Quatrano, R.S. (1990). A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250, 267–271. [DOI] [PubMed] [Google Scholar]
- Hedden, P., and Kamiya, Y. (1997). Gibberellin biosynthesis: Enzymes, genes and their regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 431–460. [DOI] [PubMed] [Google Scholar]
- Helliwell, C.A., Sheldon, C.C., Olive, M.R., Walker, A.R., Zeevaart, J.A.D., Peacock, W.J., and Dennis, E.S. (1998). Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc. Natl. Acad. Sci. USA 95, 9019–9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, C.A., Poole, A., Peacock, W.J., and Dennis, E.S. (1999). Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol. 119, 507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope, I.A., and Struhl, K. (1986). Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell 46, 885–894. [DOI] [PubMed] [Google Scholar]
- Hua, J., and Meyerowitz, E.M. (1998). Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94, 261–271. [DOI] [PubMed] [Google Scholar]
- Ishida, S., Takahashi, Y., and Nagata, T. (1993). Isolation of cDNA of an auxin-regulated gene encoding a G protein β subunit–like protein from tobacco BY-2 cells. Proc. Natl. Acad. Sci. USA 90, 11152–11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri, F., Lam, E., and Chua, N.-H. (1989). Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature 340, 727–730. [DOI] [PubMed] [Google Scholar]
- Krens, F.A., Molendijk, L., Wullems, G.J., and Schilperoort, R.A. (1982). In vitro transformation of plant protoplasts with Ti-plasmid DNA. Nature 296, 72–74. [Google Scholar]
- Lee, K.-F., Li, E., Huber, L.J., Landis, S.C., Sharpe, A.H., Chao, M.V., and Jaenisch, R. (1992). Targeted mutation of the gene encoding the low affinity NGF receptor p75 leads to deficits in the peripheral sensory nervous system. Cell 69, 737–749. [DOI] [PubMed] [Google Scholar]
- Lee, M.M., and Schiefelbein, J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473–483. [DOI] [PubMed] [Google Scholar]
- Lloyd, A., Yancheva, N., and Wasylyk, B. (1991). Transformation suppressor activity of a Jun transcription factor lacking its activation domain. Nature 352, 635–638. [DOI] [PubMed] [Google Scholar]
- Lotan, T., Ohto, M., Yee, K.M., West, M.A.L., Lo, R., Kwong, R.W., Yamagishi, K., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (1998). Arabidopsis LEAFY COTYLEDON is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. [DOI] [PubMed] [Google Scholar]
- Miao, Z.-H., and Lam, E. (1995). Construction of a trans-dominant inhibitor for members of the TGA family of transcription factors conserved in higher plants. Plant J. 7, 887–896. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nagy, J.I., and Maliga, P. (1976). Callus induction and plant regeneration from mesophyll protoplasts of Nicotiana sylvestris. Z. Pflanzenphysiol. 78, 453–455. [Google Scholar]
- Nakabeppu, Y., and Nathans, D. (1991). A naturally occurring truncated form of FosB that inhibits Fos/Jun transcriptional activity. Cell 64, 751–759. [DOI] [PubMed] [Google Scholar]
- Oeda, K., Salinas, J., and Chua, N.-H. (1991). A tobacco bZIP transcription activator (TAF-1) binds to a G-box–like motif conserved in plant genes. EMBO J. 10, 1793–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama, T., Shimura, Y., and Okada, K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11, 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, A.L., Ward, D.A., Uknes, S., Appleford, N.E.L., Lange, T., Huttly, A.K., Gaskin, P., Graebe, J.E., and Hedden, P. (1995). Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 108, 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieping, M., Fritz, M., Prat, S., and Gatz, C. (1994). A dominant-negative mutant of PG13 suppresses transcription from a cauliflower mosaic virus 35S truncated promoter in transgenic tobacco plants. Plant Cell 6, 1087–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringli, C., and Keller, B. (1998). Specific interaction of the tomato bZIP transcription factor VSF-1 with a non-palindromic DNA sequence that controls vascular gene expression. Plant Mol. Biol. 37, 977–988. [DOI] [PubMed] [Google Scholar]
- Rudnicki, M.A., Braun, T., Hinuma, S., and Jaenisch, R. (1992). Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell 71, 383–390. [DOI] [PubMed] [Google Scholar]
- Sablowski, R.W.M., and Meyerowitz, E.M. (1998). A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92, 93–103. [DOI] [PubMed] [Google Scholar]
- Schindler, U., Beckman, H., and Cashmore, A.R. (1992. a). TGA1 and G-box binding factors: Two distinct classes of Arabidopsis leucine zipper proteins compete for the G-box–like element TGACGTGG. Plant Cell 4, 1309–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler, U., Menkens, A.E., Beckman, H., Ecker, J.R., and Cashmore, A.R. (1992. b). Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO J. 11, 1261–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibaoka, H. (1994). Plant hormone–induced changes in the orientation of cortical microtubules: Alterations in the cross-linking between microtubules and the plasma membrane. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45, 527–544. [Google Scholar]
- Silverstone, A.L., Chang, C.-W., Krol, E., and Sun, T.-P. (1997). Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J. 12, 9–19. [DOI] [PubMed] [Google Scholar]
- Skoog, F., and Miller, C. (1957). Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 11, 118–131. [PubMed] [Google Scholar]
- Soriano, P., Montgomery, C., Geske, R., and Bradley, A. (1991). Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell 64, 693–702. [DOI] [PubMed] [Google Scholar]
- Steeves, T.A., and Sussex, I.M. (1989). The development of the shoot system. In Patterns in Plant Development (New York: Cambridge University Press), pp. 203–228.
- Suckow, M., Schwamborn, K., Kisters-Woike, B., von Wilcken-Bergmann, B., and Müller-Hill, B. (1994). Replacement of invariant bZIP residues within the basic region of the yeast transcription activator GCN4 can change its DNA binding specificity. Nucleic Acids Res. 22, 4395–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T.-P., and Kamiya, Y. (1994). The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6, 1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata, T., Takase, H., Takayama, S., Mikami, K., Nakatsuka, A., Kawata, T., Nakayama, T., and Iwabuchi, M. (1989). A protein that binds to a cis-acting element of wheat histone genes has a leucine zipper motif. Science 245, 965–967. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., Sakai, T., Ishida, S., and Nagata, T. (1995). Identification of auxin-responsive elements of parB and their expression in apices of shoot and root. Proc. Natl. Acad. Sci. USA 92, 6359–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto, E. (1991). Gibberellin requirement for the normal growth of roots. In Gibberellins, N. Takahashi, B.O. Phinney, and J. MacMillan, eds (New York: Springer-Verlag), pp. 229–240.
- Torres-Schumann, S., Ringli, C., Heierli, D., Amrhein, N., and Keller, B. (1996). In vitro binding of the tomato bZIP transcriptional activator VSF-1 to a regulatory element that controls xylem-specific gene expression. Plant J. 9, 283–296. [DOI] [PubMed] [Google Scholar]
- Uesugi, M., Nyanguile, O., Lu, H., Levine, A.J., and Verdine, G.L. (1997). Induced α helix in the VP16 activation domain upon binding to a human TAF. Science 277, 1310–1313. [DOI] [PubMed] [Google Scholar]
- Unger, E., Parsons, R.L., Schmidt, R.J., Bowen, B., and Roth, B.A. (1993). Dominant-negative mutants of Opaque2 suppress transactivation of a 22-kD Zein promoter by Opaque2 in maize endosperm cells. Plant Cell 5, 831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, T., Tachibana, T., Shimura, Y., and Okada, K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277, 1113–1116. [DOI] [PubMed] [Google Scholar]
- Walsh, J., Waters, C.A., and Freeling, M. (1997). The maize gene liguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade–sheath boundary. Genes Dev. 11, 208–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshaar, B., Armstrong, G.A., Block, A., da Costa e Silva, O., and Hahlbrock, K. (1991). Light-inducible and constitutively expressed DNA-binding proteins recognizing a plant promoter element with functional relevance in light responsiveness. EMBO J. 7, 1777–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y.-L., Li, L., Wu, K., Peters, A.J.M., Gage, D.A., and Zeevaart, J.A.D. (1995). The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: Molecular cloning and functional expression. Proc. Natl. Acad. Sci. USA 92, 6640–6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, I., Nakazawa, H., Nakagawa, R., Suzuki, Y., Kurogochi, S., Murofushi, N., Takahashi, N., and Weiler, E.W. (1990). Identification and semi-quantification of gibberellins from the pollen and anthers of Zea mays by immunoassay and GC/MS. Plant Cell Physiol. 31, 1063–1069. [Google Scholar]
- Yamaguchi, S., Saito, T., Abe, H., Yamane, H., Murofushi, N., and Kamiya, Y. (1996). Molecular cloning and characterization of a cDNA encoding the gibberellin biosynthetic enzyme ent-kaurene synthase B from pumpkin (Cucurbita maxima L.). Plant J. 10, 203–213. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S., Smith, M.W., Brown, R.G.S., Kamiya, Y., and Sun, T.-P. (1998). Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10, 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y., Zhu, Q., Dai, S., Lamb, C., and Beachy, R. (1997). RF2a, a bZIP transcriptional activator of the phloem-specific rice tungro bacilliform virus promoter, functions in vascular development. EMBO J. 16, 5247–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]