Abstract
The Arabidopsis MADS box transcription factor APETALA1 (AP1) was identified as a substrate for farnesyltransferase and shown to be farnesylated efficiently both in vitro and in vivo. AP1 regulates the transition from inflorescence shoot to floral meristems and the development of sepals and petals. AP1 fused to green fluorescent protein (GFP) retained transcription factor activity and directed the expected terminal flower phenotype when ectopically expressed in transgenic Arabidopsis. However, ap1mS, a farnesyl cysteine–acceptor mutant of AP1, as well as the GFP–ap1mS fusion protein failed to direct the development of compound terminal flowers but instead induced novel phenotypes when ectopically expressed in Arabidopsis. Similarly, compound terminal flowers did not develop in era1-2 transformants that ectopically expressed AP1. Together, the results demonstrate that AP1 is a target of farnesyltransferase and suggest that farnesylation alters the function and perhaps specificity of the transcription factor.
INTRODUCTION
Protein prenylation has been reported for several proteins that participate in growth regulation and signal transduction, cell cycle regulation, nuclear architecture, and membrane trafficking. These proteins include the Ras super family of small GTP binding proteins, the γ subunit of trimeric GTP binding proteins, yeast a mating factor, cyclic GMP phosphodiesterase, and type I inositol-(1,4,5)-trisphosphate 5′-phosphatase (Anderegg et al., 1988; Hancock et al., 1989; Vorburger et al., 1989; Finegold et al., 1990; Anant et al., 1992; De Smedt et al., 1996). Prenylation results in the covalent attachment of either farnesyl diphosphate (FPP) or geranylgeranyl diphosphate (GGPP) to a conserved cysteine near the C terminus of proteins by farnesyltransferase (FTase) or geranylgeranyltransferase-I (GGTase-I). The enzymes recognize a conserved CaaX box sequence motif in which C is a cysteine residue, a is usually an aliphatic amino acid, and X is any amino acid modified by FTase or, with only a few exceptions, leucine modified by GGTase-I (Schafer and Rine, 1992).
Recent studies have shown that plant FTase is structurally and functionally conserved (Yalovsky et al., 1997; Rodríguez-Concepción et al., 1999b) and is required in signal transduction by abscisic acid in stomatal guard cells (Pei et al., 1998). Lack of FTase in Arabidopsis affects the control of cell division in the shoot apical meristem, resulting in developmental alterations (Yalovsky et al., 2000, this issue). The FTase mutant era1-2 (for enhanced response to abscisic acid), in which the gene for the FTase β subunit (AtFTB) is deleted (Cutler et al., 1996), has enlarged shoot apical and floral meristems that produce larger leaves and inflorescences and an increased number of floral organs than does the wild type. Moreover, era1-2 plants are late flowering, and flowers often fail to develop (Yalovsky et al., 2000, this issue). Treatment of tobacco tissue culture cells with FTase inhibitors causes cell cycle arrest (Morehead et al., 1995), indicating that the enzyme is required for normal cell cycle progression in plants as well. Despite the emerging importance of protein farnesylation in plant growth and development, however, only ANJ1, the plant homolog of the bacterial chaperone DnaJ, has been identified as a substrate for FTase (Zhu et al., 1993).
The amino acid sequence of the floral MADS box transcription factor APETALA1 (AP1) (Mandel et al., 1992) terminates in CFAA (Figure 1), a typical CaaX box recognition motif for FTase. AP1 regulates the transition from inflorescence shoot to floral meristems and the development of sepals and petals (Irish and Sussex, 1990; Mandel et al., 1992; Bowman et al., 1993; Gustafson-Brown et al., 1994; Mandel and Yanofsky, 1995a; Hempel et al., 1997). ap1 mutants develop leaflike bracts instead of sepals, lack or have fewer petals than the wild type, and develop axillary flowers in the axis of the first whorl organs (Irish and Sussex, 1990; Bowman et al., 1993). In wild-type plants, AP1 is expressed at high levels early in the ontogeny of flower primordia, indicating a direct role for the gene in specifying floral fate (Mandel et al., 1992; Gustafson-Brown et al., 1994; Hempel et al., 1997). Ectopic expression of AP1 in wild-type Arabidopsis directs the conversion of vegetative and inflorescence meristems into floral meristems and the development of compound terminal flowers (Mandel and Yanofsky, 1995b; Lijegren et al., 1999).
Figure 1.
The CaaX Box Motif Is Conserved between AP1 and Homologs from Distantly Related Plant Species.
Amino acid sequence alignment of the C-terminal 20 amino acids of APETALA1 (GenBank accession number Z16421) and selected homologs: SQUAMOSA (X63701) from Antirrhinum majus; AP1H (AJ000759) from Malus domestica (apple); AP1H.2 (U67452) and AP1H.1 (Z37968) from Brassica oleracea; CAULIFLOWER (L36925), AGL8 (Q38876), and AGAMOUS (X53579) from Arabidopsis thaliana; MADS3 (X99653) from Betula pendula; MADS2 (AF068726) from Nicotiana sylvestris; AP1H (AJ251300) from Brassica rapa; and AP1H (AF034093) from Populus tremuloides. The putative CaaX box sequences are shaded; dots indicate residues identical to AP1; dashes indicate gaps formed by the alignment program; the underscoring indicates a conserved amino acid substitution between AP1 and a given homolog; and boxes highlight substitutions of the CaaX box cysteines.
Homologs of AP1 from Arabidopsis (Bowman et al., 1993; Gustafson-Brown et al., 1994; Kempin et al., 1995) and other plant species contain similar CaaX box sequences (Figure 1), suggesting that a subfamily of proteins exists within the plant MADS box superfamily of transcription factors that may be prenylated. We show here that AP1 is a novel substrate for FTase, which efficiently farnesylates the MADS box transcription factor both in vitro and in vivo. Ectopic expression of an AP1 cysteine-acceptor mutant protein that cannot be farnesylated failed to direct the development of compound terminal flowers but induced other novel phenotypes.
RESULTS
AP1 Is Prenylated in Vitro and in Vivo
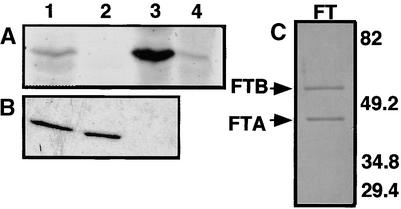
Incubation of purified AP1 fused with glutathione S-transferase (GST–AP1) with 3H-FPP and recombinant plant FTase prepared from either yeast or baculovirus-infected Sf9 cells (Yalovsky et al., 1997) showed that the fusion protein was labeled by FTase. The protein was not labeled in an extract prepared from yeast ram1Δ cells, which lack FTase activity (Trueblood et al., 1993; Yalovsky et al., 1997) (Figure 2A). Labeling was also substantially reduced in a protein extract from mock-infected Sf9 cells containing only endogenous FTase. To confirm that prenylation was dependent on the CFAA sequence motif, we constructed a GST–AP1 fusion protein in which the farnesyl cysteine acceptor was replaced with serine (GST–ap1mS), thereby creating a nonfunctional farnesylation motif. We also predicted that mutation of the C-terminal alanine to leucine (GST–ap1mL) would convert the AP1 CaaX box sequence to a preferred substrate for GGTase-I. Incubation of the three fusion proteins with purified FTase confirmed that farnesylation required the conserved cysteine (Figures 2B and 2C). GST–ap1mL was also labeled weakly with 3H-FPP but not with 3H-GGPP, consistent with earlier reports that plant FTase can farnesylate GGTase-I–like substrates but cannot utilize GGPP (Yalovsky et al., 1997). Such complementation would be similar to the promiscuity of GGTase-I observed in yeast and mammalian cells (Trueblood et al., 1993; Dalton et al., 1995; James et al., 1995).
Figure 2.
In Vitro Prenylation of AP1.
(A) GST–AP1 is a protein substrate for prenylation. Reactions in lanes 1 and 3 contained plant FTase extracts from yeast or baculovirus-infected Sf9 cells, respectively, coexpressing the two subunits of tomato FTase. Control extracts were from ram1Δ yeast cells (lane 2) or mock-infected Sf9 cells (lane 4).
(B) Farnesylation of GST–AP1 requires a functional CaaX box. Purified plant Ftase, shown in (C), was incubated with GST–AP1 (lane 1), GST–ap1mL (lane 2), and GST–ap1mS (lane 3).
(C) Purified recombinant plant FTase prepared from baculovirus-infected Sf9 cells. Positions of protein bands for FTase subunits α and β (FTA and FTB, respectively) are indicated. Numbers at right indicate positions of molecular-weight markers (Bio-Rad) in kilodaltons.
To confirm that AP1 is prenylated in vivo, we expressed full-length AP1 or ap1mS in Nicotiana benthamiana plants, using a potato virus X (PVX) vector (Figure 3). Proteins were prepared from infected leaves labeled with 3H-mevalonic acid and separated on SDS–polyacrylamide gels, which were then used either for immunoblot analysis with polyclonal AP1 antibodies (Figure 3A) or for fluorography to detect labeled AP1 (Figure 3B). AP1 wild-type and mutant proteins were expressed to a similar extent in leaves infected with PVX–AP1 or PVX–ap1mS. A labeled protein the same size as AP1 was detected only in extracts prepared from PVX–AP1—infected leaves but not in those prepared from PVX–ap1mS—infected or PVX mock-infected leaves. Together, these results establish that both the AP1 CFAA CaaX box sequence and the full-length transcription factor are substrates for FTase.
Figure 3.
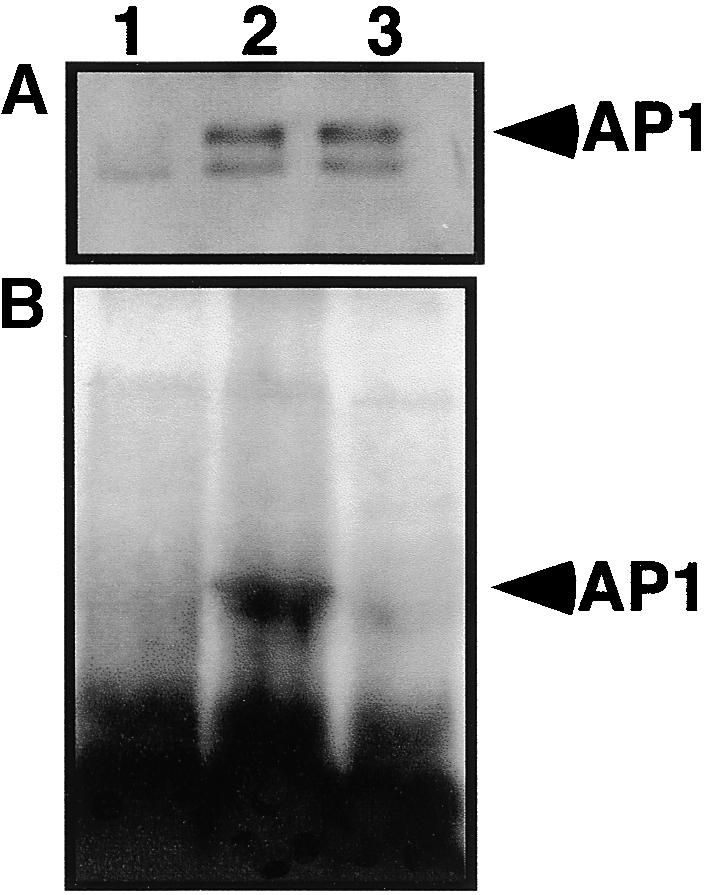
AP1 Is Prenylated in Vivo.
(A) Immunoblots of protein extracts from PVX-infected N. benthamiana leaves labeled with 3H-mevalonic acid. After infection with control PVX (lane 1), PVX–AP1 (lane 2), or PVX–ap1mS (lane 3) viruses, protein extracts were subjected to immunoblot analysis with polyclonal antibodies prepared against the C-terminal third of AP1.
(B) In vivo labeling of AP1. Fluorography of gel with the same protein extracts as shown in (A).
Given that shoot apices and floral primordia have FTase activity (Cutler et al., 1996; Schmitt et al., 1996; Zhou et al., 1997), AP1 is probably farnesylated in cells of the floral meristem. Kinetic studies have also shown that the CFAA sequence was farnesylated with a Km of 3.4 μM (0.9 ng/μL), which is similar to the Km of 5 μM found for farnesylation of H-ras by rat FTase (Reiss et al., 1991). In contrast, based on prenylation assays with purified plant FTase, the Km for farnesylation of the CAULIFLOWER CaaX box CYAA (Figure 1) was calculated to exceed 100 μM (data not shown). Thus, the CAULIFLOWER MADS box transcription factor is unlikely to be farnesylated in vivo. Together, these experiments show that AP1 is a substrate for FTase, thus making AP1 the first reported prenylated transcription factor.
Nonfarnesylated AP1 Does Not Direct Development of Compound Terminal Flowers
To investigate the function of AP1 prenylation in Arabidopsis, we ectopically expressed both AP1 and the farnesyl cysteine–acceptor mutant ap1mS in Columbia (Col-0) plants under regulation of the cauliflower mosaic virus (CaMV) 35S promoter (Figure 4). Thirty T1 kanamycin-resistant (KanR) transformants were initially selected for either 35S::AP1 or 35::ap1mS. The plants were allowed to self-pollinate, and two 35S::AP1 and three 35S::ap1mS T2 lines that showed both Mendelian segregation for the KanR marker and the strongest phenotypes were characterized further. Transformants expressing the 35S::AP1 gene developed the typical terminal flower gain-of-function phenotype (Mandel and Yanofsky, 1995a; Lijegren et al., 1999) (Figure 4A, 35S::AP1), but plants transformed with 35S::ap1mS did not develop compound terminal flowers in either primary or secondary shoot inflorescences (Figure 4A, 35S::ap1mS). Instead, these transformants flowered early and had curled leaves.
Figure 4.
Ectopic Expression of AP1 and ap1mS in Wild-Type and era1-2 Plants.
(A) Transgenic Arabidopsis Col-0 plants expressing either wild-type AP1 or farnesyl cysteine–acceptor mutant ap1mS under the control of the CaMV 35S promoter (35S::AP1 and 35S::ap1mS, respectively). cl, curled leaf; tf, compound terminal flowers.
(B) Transgenic Arabidopsis era1-2 mutant plants expressing wild-type AP1 under control of the CaMV 35S promoter (35S::AP1).
To confirm that lack of farnesylation affects AP1 activity, we also ectopically expressed the 35S::AP1 gene in era1-2 (Figure 4B). Twenty T1 KanR transformants were selected, eight of which showed an early-flowering phenotype. Interestingly, none of the primary shoot inflorescences of transgenic 35S::AP1 era1-2 transformants terminated in a compound terminal flower, not even in transformants that flowered early. Together, these results suggest that in era1-2, the nonfarnesylated AP1 protein is active and can confer an early-flowering phenotype when ectopically expressed, but its ectopic function differs from that of the wild-type protein with respect to induction of terminal flower development.
In many transformants, ectopic expression of 35S::ap1mS caused development of unexpanded leaves that often lacked chlorophyll. In some cases, leaf expansion ceased and a colorless callus-like tissue developed instead (data not shown). These results suggest that relative to AP1, the nonfarnesylated transcription factor had novel activities that impaired normal leaf development.
Ectopic Expression of Green Fluorescent Protein–ap1mS Induced New Phenotypes but Failed to Direct Development of Compound Terminal Flowers
To further substantiate that lack of farnesylation affects the function or localization of ectopically expressed AP1, we constructed fusion proteins between green fluorescent protein (GFP) and the N terminus of the transcription factor. GFP–AP1 and GFP–ap1mS were expressed in transgenic Arabidopsis under the control of the CaMV 35S promoter to correlate ectopic protein expression assessed by GFP fluorescence with the observed phenotypes. Of 78 transgenic KanR GFP–AP1 plants, 15 showed the dominant gain-of-function phenotype typical of ectopic AP1 expression. As Figure 5 shows, these plants developed only a few rosette leaves, and their very short primary inflorescence stem bore only one or two flowers (Figure 5A). GFP fluorescence was detected in the nuclei of both floral and leaf tissues (Figure 5B), demonstrating that GFP–AP1 is correctly localized and active. Only one of the transformants that showed GFP fluorescence did not develop a terminal flower phenotype. In this plant, GFP fluorescence was detected in the nuclei of stomatal guard cells but not in meristem or flower tissues (data not shown), suggesting that the transgene had inserted into a locus that altered the activity of the CaMV 35S promoter. This result supports an earlier conclusion (Mandel and Yanofsky, 1995a) that AP1 must be active in meristem or leaf tissues (or both) to direct the terminal flower phenotype. No GFP fluorescence was detected in the remaining 62 KanR plants, which either appeared to be wild-type plants or developed alterations in flower morphology reminiscent of the ap1 phenotype. This phenotype probably resulted from inactivation of the AP1 functions by cosuppression of the GFP–AP1 transgene and the endogenous AP1 gene (data not shown).
Figure 5.
A GFP–AP1 Fusion Protein Retains Ectopic AP1 Transcription Factor Activity.
(A) Transgenic Arabidopsis Col-0 plants expressing a chimeric gene for the GFP–AP1 fusion protein under the control of the CaMV 35S promoter. tf, compound terminal flowers.
(B) Confocal laser scanning microscope images of a gynoecium (left) and a petal area (right) demonstrating the ectopic expression and nuclear localization of GFP–AP1 in transgenic plants shown in (A). 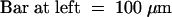 ;
; 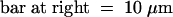 .
.
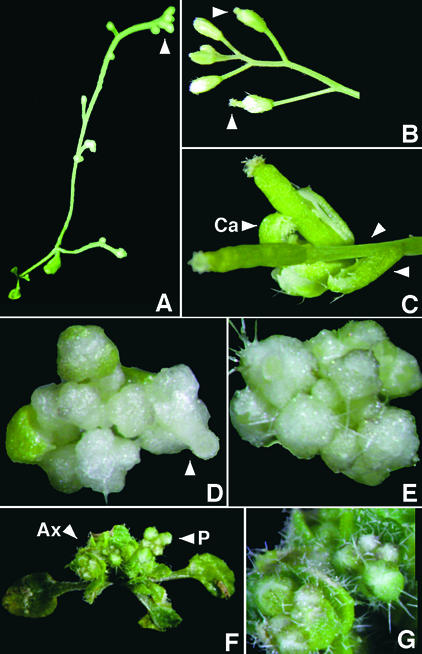
The transgenic GFP–ap1mS plants flowered early, but their primary inflorescence stem was indeterminate and developed numerous flowers rather than a single compound terminal flower (Figures 6A, 6B, and 6D to 6F). In some plants, the axillary inflorescence terminated in compound terminal flowers with an elongated stem between the outer whorl organs (Figure 6C), thus resembling 35S::AP1 leafy (lfy) plants (Lijegren et al., 1999). Interestingly, some flowers also had carpelloid leaves with stigmatic papillae (Figure 6C), resembling lfy mutants (Weigel et al., 1992) and short-day-grown era1-2 plants (Yalovsky et al., 2000, this issue). In other plants, both the primary and the axillary rosette inflorescences were indeterminate, and both rosette leaves and sepals were very hairy (Figures 6F and 6G), closely resembling the 35S::ap1mS plants described above. In approximately half of the GFP–ap1mS plants, the inflorescences were colorless (Figures 6D and 6E); in many flowers, the gynoecium protruded from the closed flowers (Figures 6B and 6D), which resembled the era1-2 plants (Yalovsky et al., 2000, this issue). Together, these results demonstrate that unlike that of GFP–AP1, ectopic expression of GFP–ap1mS did not direct the development of terminal flowers but instead produced new phenotypes not observed with AP1 or GFP–AP1.
Figure 6.
Ectopic Expression of GFP–ap1mS Did Not Induce Development of Compound Terminal Flowers.
Shown are four plants that expressed a high level of the farnesyl cysteine–acceptor mutant ap1mS fused to GFP (GFP–ap1mS) under the control of CaMV 35S promoter.
(A) A six-week-old plant with an elongated stem that produced numerous flowers. The primary stem inflorescence (arrowhead) did not terminate in a compound flower.
(B) Primary stem inflorescence of the plant shown in (A). Elongated gynoecia, which protruded from closed flowers, are visible (arrowheads).
(C) An axillary inflorescence from the plant shown in (A). An elongated stem between the first whorl organs and the rest of the flower (arrowheads) and a carpeloid leaf (Ca) with stigmatic papillae are clearly visible.
(D) Primary stem inflorescence composed of numerous colorless flowers and a flower with an elongated gynoecium (arrowhead).
(E) Primary stem inflorescence consisting of numerous colorless flowers. Many trichomes are visible.
(F) A plant with nonterminal primary (P) and axillary (Ax) inflorescences.
(G) The axillary inflorescences from the same plant shown in (F) in more detail. An inflorescence with multiple flowers and the increased number of hair cells on both leaves and sepals are visible.
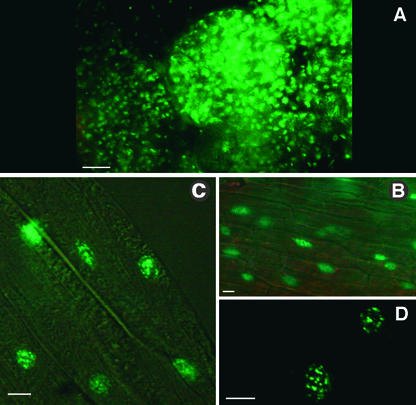
Of 106 transgenic KanR GFP–ap1mS plants, 52 showed GFP fluorescence in nuclei of all tissues examined. Figure 7 shows examples of fluorescing nuclei from two of the plants shown in Figure 6. The strong GFP fluorescence, which was visible in nearly every cell, suggests a direct correlation between expression of the GFP–ap1mS fusion protein and the resulting novel phenotypes caused by the lack of farnesylation of the transcription factor.
Figure 7.
Nuclear Localization of the GFP–ap1mS Fusion Proteins.
(A) A flower bud from the plant shown in Figures 6F and 6G.
(B) and (C) GFP fluorescence is localized exclusively to nuclei. Images of stamen (B) and sepal (C) tissues were taken from the plant shown in Figures 6A to 6C. Images were produced under dim light and UV illumination (to induce GFP fluorescence) by using Nomarsky differential interference contrast optics to visualize cell boundaries.
(D) High-magnification image of two of the nuclei shown in (C). The GFP–ap1mS produces speckles, which are dispersed throughout the nuclear matrix.
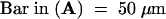 ;
; 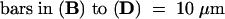 .
.
DISCUSSION
We have identified AP1 as a substrate for FTase and demonstrated that farnesylation is required for the established ectopic function of the transcription factor. Ectopic expression of the farnesyl cysteine–acceptor mutant protein ap1mS in wild-type plants or AP1 in the era1-2 FTase null mutant directs the development of novel phenotypes that differ from the terminal flower phenotype reported for the ectopic expression of AP1, suggesting that farnesylation alters the function of the transcription factor.
What Is the Biochemical Role of AP1 Farnesylation?
The functional role of the AP1 C-terminal domain is not well understood. However, the C-terminal sequence similarities between AP1 and its homologs from distantly related plant species (Figure 1) suggest that the C-terminal domain has conserved functions. In only three of the reported AP1 homologs was the conserved CaaX box cysteine converted into serine, tyrosine, or arginine by single-base changes (Figure 1). Interestingly, Brassica oleracea has at least two AP1 homologs; AP1H2 has a conserved CFAA CaaX box, but the cysteine in AP1H1 is replaced by serine (Figure 1). In view of the conservation of the CFAA CaaX box sequence in many AP1 homologs and the efficient farnesylation of the cysteine acceptor, perhaps we should reevaluate the DNA sequence in those plants in which single-base changes have apparently converted the farnesyl cysteine acceptor to other amino acids. Identification of additional CaaX box–containing AP1 homologs in other species, such as Populus tremuloides, will provide further information on the extent to which the AP1 transcription factor may be modified by farnesylation in flowering plants.
It is unlikely that farnesylation of AP1 is required for the DNA binding activity of the transcription factor. In vitro, AP1 binds to target promoter DNA sequences as a homodimer (Riechmann et al., 1996). Only the MADS box domain and part of the L-loop amino acid sequence are required for homodimerization of AP1 and binding to CArG box DNA sequences (Riechmann et al., 1996). However, a truncated AP1 protein consisting of only the MADS box and L domains was inactive in vivo (Krizek and Meyerowitz, 1996), demonstrating that homodimerization and DNA binding are not sufficient for AP1 to function as an active transcription factor. The above results are consistent with the genetic analysis of the ap1-4 and ap1-8 alleles, which encode mutant AP1 proteins in which C-terminal sequences, including the CFAA FTase recognition sequence, are deleted (M. Yanofsky, personal communication). Both alleles produce intermediate ap1 mutant phenotypes (Bowman et al., 1993), confirming that the C-terminal domain is required for correct AP1 function.
Farnesylation of AP1 might also be important in interactions of the transcription factor with other proteins. In Antirrhinum, the AP1 ortholog SQUAMOSA forms a tetramer with DEFICIENS and GLOBOSA, orthologs of the Arabidopsis B-type proteins AP3 and PISTILLATA. Assembly of the heterotetrameric transcription factor complex requires the C-terminal domain of SQUAMOSA (Egea-Cortines et al., 1999). In addition, conservation of the CaaX box farnesylation motif in several other MADS box and homeobox plant transcription factors (Figure 1) (Nambara and McCourt, 1999; Yalovsky et al., 1999) supports the view that farnesylation may have structural or regulatory roles in the assembly of certain types of plant transcription factor complexes. For example, farnesylation of Ras in mammalian cells is required for efficient recruitment of the GDP/GTP exchange factor after receptor signaling but not for Ras function per se (Zhang and Casey, 1996).
Our results are difficult to reconcile with a published report in which a chimeric protein consisting of the MADS box and L domains of AP1 and the K-domain and C-terminal domain of AGAMOUS (which does not terminate in a CaaX motif) could induce the AP1 ectopic phenotype when expressed in wild-type plants (Krizek and Meyerowitz, 1996). In contrast, the farnesyl cysteine–acceptor mutant protein ap1mS produced a different phenotype (Figures 4 and 6). The conserved C-terminal sequence of AP1 is highly divergent from the C-terminal domain of AGAMOUS (Figure 1). It is possible, therefore, that in the chimeric protein, other amino acid sequences in the AGAMOUS domain could substitute for certain functions in the AP1 C-terminal domain that would be affected by the farnesyl cysteine–acceptor mutation in ap1mS. Because the interactions of AP1 and AGAMOUS are currently best understood only at the genetic level, it is difficult to explain how the AP1–AGAMOUS chimeric protein differs functionally from the single amino acid mutation in the farnesyl cysteine–acceptor mutant ap1mS.
The biochemical and genetic analyses of AP1 prenylation confirmed that the C-terminal CaaX box of AP1 is a functional farnesylation site that can be modified by FTase with a Km comparable to that of mammalian H-Ras (Reiss et al., 1990) and that the nonfarnesylated AP1 protein has unusual activity. The reduction in shoot-to-flower transition in 35S::ap1mS and 35S::GFP–ap1mS plants (Figures 4 and 6) and the suppression of shoot-to-flower transition in transgenic 35S::AP1 era1-2 (Figure 4) suggest that the nonfarnesylated AP1 has a dominant-negative effect on flower development. Taken together, these data suggest that farnesylation of AP1 is likely to direct specific, novel biochemical activities of the transcription factor, although additional studies are required to test this hypothesis and to define the biochemical function of farnesylation.
Why Does Lack of FTase in era1-2 Fail to Produce a Strong ap1 Phenotype?
The pleiotropic phenotype of the era1-2 mutant suggests that several specific developmental and signaling processes rely on protein farnesylation (Yalovsky et al., 2000, this issue). The late-flowering phenotype of era1-2, considered with the apparent homeotic transformation of floral organs and partial inhibition of floral development (Yalovsky et al., 2000, this issue), indicates that protein farnesylation is required both during the transition to reproductive growth and at specific times during flower development. The phenotypic changes in era1-2 become more severe when mutant plants are grown in short-day conditions (Yalovsky et al., 2000, this issue). A similar overlapping pleiotropy is also observed in ap1 mutants, although no ap1 allele has been reported in which only the farnesyl cysteine acceptor is mutated. Because the lack of FTase activity in era1-2 would be expected to affect the functions of other proteins during reproductive growth as well, it is perhaps not surprising that the mutant does not show a strong ap1 floral phenotype. Instead, an altered function of a nonfarnesylated AP1 transcription factor in era1-2 probably would be partially compensated or modified by changes in other pathways.
Another possibility is that a small amount of AP1 in era1-2 might be modified by GGTase-I, thereby partially restoring its specific function, although our experiments have confirmed that AP1 is not an efficient substrate for GGTase-I (data not shown). Moreover, in era1-2, the high Km of AP1 for GGTase-I could be reduced by increased activities of GGTase-I, although immunological analysis of era1-2 protein extracts with an antibody against the β subunit of AtGGTase did not reveal any difference in AtGGTase protein concentrations (S. Yalovsky and W. Gruissem, unpublished results).
To explain the absence of an apparent ap1 phenotype in Arabidopsis FTase mutants, Nambara and McCourt (1999) suggested that other genes might exist for the β subunit of AtFTase. Considering the conservation of prenyltransferases surveyed in eukaryotic cells (Yalovsky et al., 1997, 1999; Rodríguez-Concepción et al., 1999b), however, this scenario seems unlikely and is not supported by biochemical data, which confirm that era1-2 lacks FTase activity (Cutler et al., 1996). Also, polyclonal antibodies against AtFTB failed to detect cross-reacting protein bands (Yalovsky et al., 2000, this issue), and low-stringency hybridization with genomic DNA from era1-2 and AtFTB cDNA as a probe did not produce any cross-reacting DNA bands that would indicate the presence of a divergent gene (S. Yalovsky and W. Gruissem, unpublished results).
Lipid Modifications of Specific Transcription Factors and Growth Regulators Have Been Identified
Although AP1 is the first transcription factor to be identified that is efficiently farnesylated in vivo and in vitro, lipid and cholesterol modifications of specific transcription factors and growth regulators have already been reported. SREBP, a transcription factor involved in the regulation of cholesterol metabolism in mammals, is synthesized as a plasma membrane–bound precursor. A lipid modification sensitive to changes in blood cholesterol concentrations initiates proteolytic cleavage to release the mature SREBP from the plasma membrane, followed by transport of transcription factor to the nucleus (Brown and Goldstein, 1997). Similarly, modification of hedgehog proteins by cholesterol appears to be critical for their function, because a low-cholesterol diet perturbs embryo development (Porter et al., 1996).
The precise role of farnesylation for AP1 function remains to be established, although we speculate it may provide a functional link between a key transcription factor controlling flower development and the activity of the isoprenoid pathway in meristems during reproductive growth. As such, farnesylation may facilitate interaction of AP1 with other developmentally regulated transcription factors or nuclear envelope complexes that coordinate cell division with the temporal regulation of developmental gene expression in response to the nutritional status of the cell. Several other prenylated proteins have been identified in yeast and mammalian cells that regulate aspects of signal transduction, cell division, and chromosome segregation and development; homologs of these proteins exist in plants as well (Nambara and McCourt, 1999; Rodríguez-Concepción et al., 1999b; Yalovsky et al., 1999). Together, the characterization of prenylated proteins involved in various aspects of plant signal transduction and development implicated by the era1 mutants will provide important clues into the mechanisms by which plants integrate key cellular biochemical and regulatory processes.
METHODS
Construction of Expression Vectors
To construct the pGST-AP1 plasmid, we cloned the cDNA encoding the C-terminal 146–amino acid residues of AP1 in frame with the glutathione S-transferase (GST) coding region into plasmid pGEX-3X (Pharmacia). Plasmid pGST-AP1 was modified by polymerase chain reaction (PCR) with primers APF (5′-GTCAAGGATCCCATTATCTTGGGGAAGACTTGC-3′) and APL (5′-CTATTGAATTCATAAGGCGAAGCAGCCAAGGTTGC-3′) to produce pGST-ap1mL (i.e., a fusion plasmid in which the AP1 C-terminal alanine is mutated to leucine) and with APF and APS (5′-CTATTGAATTCATGCGGCGAAGGAGCCAAGGTTGC-3′) to produce pGST-ap1mS. The corresponding 43-kD fusion proteins (GST–AP1, GST–ap1mL, and GST–ap1mS) were expressed in Escherichia coli XL-1 Blue and purified on glutathione Sepharose 4B beads (Pharmacia) according to the manufacturer's instructions. The PCR products were sequenced to verify that the correct fragments were amplified.
Full-length cDNAs of AP1 and ap1mS were prepared for subcloning into potato virus X (PVX) expression vectors. pPVXAP1 (pSY503) and pPVXap1mS (pSY504) were constructed by ligating the cDNAs encoding AP1 or ap1mS into pPVX. Plasmids were linearized and then transcribed to infect Nicotiana benthamiana plants (Chapman et al., 1992). AP1 and ap1mS were subcloned into pGFP-MRC (Rodríguez-Concepción et al., 1999a), a modified version of pRTL-sGFP, to produce a C-terminal in-frame fusion between GFP and the AP1 derivatives, creating plasmids pGFP-AP1 (pSY505) and pGFP-ap1mS (pSY506). For ectopic expression in Arabidopsis thaliana of GFP–AP1 and GFP–ap1mS, pSY505 and pSY506 were digested with HindIII to isolate 35S::GFP–AP1–NOS or 35S::GFP–ap1m–NOS fragments. These fragments were subcloned into the plant binary vectors SLJ7292 (Jones et al., 1992) or pCAMBIA2300, which were digested with the same enzyme to create pSY507 and pSY527. For ectopic expression in Arabidopsis of AP1 and ap1mS, full-length cDNA constructs were subcloned into pMDI to create pSY509 and pSY510.
Protein Expression
The cDNAs encoding LeFTB and AtFTA were subcloned into pFastBacHTb (Gibco BRL) (LeFTB) (pSY105) and pFastBacHTc (AtFTA) (pSY302). Recombinant baculoviruses expressing the genes were prepared in the Bac-to-Bac system (Gibco BRL) according to the manufacturer's instructions. Recombinant proteins were purified on 1-mL Ni-NTA columns (Qiagen, Chatsworth, CA) according to the manufacturer's instructions. Fractions containing purified proteins were pooled and dialyzed/concentrated under vacuum against a solution of 50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 50 μM ZnCl2, and 1 mM DTT, on ice, by using a Collodion apparatus (Schleicher and Schuell) with a dialysis membrane having a 25-kD cutoff.
In Vitro Prenylation Assays
Prenylation assays were performed as previously described (Yalovsky et al., 1997) except for the following modifications. Fusion proteins were used as substrates instead of synthetic peptides, and in some reactions, purified FTase was used at 0.5 μmol.
In Vivo Labeling of AP1
For labeling experiments, we excised leaves with the intact petiole from PVX-infected plants and then incubated them in a ventilated hood to facilitate transpiration in buffer A (0.5 × Murashige-Skoog salt mixture [Gibco BRL] and 20 μM lovastatin [Merck]). After 2 to 5 hr, leaves were transferred to new tubes containing 50 μCi of 3H-mevalonic acid (60 Ci/mmol; American Radiolabeled Chemicals, St. Louis, MO) in buffer A. After the mevalonic acid was absorbed, leaves were transferred to buffer A. After 12 to 15 hr, petioles were cut off and proteins were extracted by grinding equal weights of lamina tissues in SDS buffer (Laemmli, 1970). To precipitate nonsoluble material, we centrifuged the extracts for 5 min at 1000g and then for 5 min at 15,000g. Equal volumes from each extract were loaded on 10% SDS–polyacrylamide gels. After electrophoresis, the gel was fixed, fluorographed (Amplify; Amersham), and exposed to XAR film (Kodak) for 14 days.
Immunoblots
Nitrocellulose membranes were first blocked with 5% nonfat milk and subsequently incubated for 12 hr at 4°C with the AP1 antibody (diluted 1:1,000), washed with Tris-buffered saline containing Tween-20, and incubated for 1 hr with 20,000-fold-diluted goat anti–rabbit secondary antibody conjugated to horseradish peroxidase for developing with a Super Signal Substrate kit (Pierce).
Plant Growth and Analysis
Arabidopsis seeds were sown in Sunshine mix (Fison Horticulture, Bellevue, WA) and kept for 5 days at 4°C before being moved into the greenhouse. Plants were grown at 22°C under long-day (16-hr-light/8-hr-dark) growth conditions. Transgenic plants were grown under continuous light or long-day growth conditions.
Confocal and Fluorescence Microscopy
Scans were made with a confocal argon laser scanning microscope (Molecular Dynamics, Sunnyvale, CA) at both 488 and 514 nm. Filters and detectors were set up in the following order: (1) neutral density filter attenuating light to 10% transmittance; (2) dichroic long-pass filter (DRLP) at 520 nm; (3) sample; (4) DRLP at 520 nm; (5) long-pass barrier filter at 530 nm; (6) beam splitter at 595 nm; (7) detector 1, either not active or with a barrier filter at 610 nm used to detect red (chlorophyll) fluorescence; and (8) detector 2, with a barrier filter at 530 nm used to detect GFP fluorescence. Fluorescence images were prepared using a Zeiss (Zeiss, Inc., Thornwood, NY) Axioplan 2 equipped with Nikon (Tokyo, Japan) Coolpix 950 digital camera.
Construction and Analysis of Transgenic Arabidopsis
All plasmids were transformed into Agrobacterium tumefaciens strain GV3101/mp90 by electroporation. This strain was used to transform Arabidopsis ecotype Columbia (Col-0) plants by vacuum infiltration (Bechtold et al., 1993; Bent et al., 1994), with 0.005% Silwet L-77 (Lehle Seeds, Round Rock, TX) added to the infiltration media or by floral dip (Clough and Bent, 1998). Transgenic plants were selected by plating seeds on growth media with 50 μg/mL kanamycin added. All kanamycin-resistant (KanR) plants were transferred to soil and examined for GFP fluorescence at 520 nm by using a Zeiss Axioplan fluorescence microscope.
Acknowledgments
We thank Drs. Martin Yanofsky, Cristina Ferrandiz, Yuval Cohen, Gail McLean, Nir Ohad, Yuki Mizukami, and Teresa Rubio for materials and advice; Dr. Peter McCourt for providing era1-2 seeds; and Dr. Martin Yanofsky for communicating unpublished data. We are especially indebted to Anita Kulukian for maintenance of the plant material, other members of the Gruissem laboratory for their help and advice, and Dr. Nir Ohad for technical support. The research was supported by the U.S. Department of Energy Grant No. 85ER13375 to W.G. and the Israel Science Foundation Grant No. 571/99 to S.Y. M.R.-C. was supported by a fellowship from the Ministerio de Educación y Cultura (Spain).
References
- Anant, J.S., Ong, O.C., Xie, H., Clarke, S., O'Brein, P.J., and Fung, B.K.K. (1992). In vivo differential prenylation of retinal cyclic GMP phosphodiesterase catalytic subunits. J. Biol. Chem. 267, 687–690. [PubMed] [Google Scholar]
- Anderegg, R.J., Betz, R., Carr, S.A., Crabb, J.W., and Duntze, W. (1988). Structure of the Saccharomyces cerevisiae mating hormone a-factor: Identification of S-farnesyl cysteine as a structural component. J. Biol. Chem. 263, 18236–18240. [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium–mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Ser. III 316, 1194–1199. [Google Scholar]
- Bent, A.F., Kunkel, B.N., Dahlbeck, D., Brown, K.L., Schmidt, R., Giraudat, J., Leung, J., and Staskawicz, B.J. (1994). RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 265, 1856–1860. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Alvarez, J., Weigel, D., Meyerowitz, E.M., and Smyth, D.R. (1993). Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119, 721–743. [Google Scholar]
- Brown, M.S., and Goldstein, J.L. (1997). The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340. [DOI] [PubMed] [Google Scholar]
- Chapman, S., Kavanagh, T., and Baulcombe, D. (1992). Potato virus X as a vector for gene expression in plants. Plant J. 2, 549–557. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cutler, S., Ghassemian, M., Bonetta, D., Cooney, S., and McCourt, P. (1996). A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science 273, 1239–1241. [DOI] [PubMed] [Google Scholar]
- Dalton, M.B., Fantle, K.S., Bechtold, H.A., Demaio, L., Evans, R.M., Krystosek, A., and Sinensky, M. (1995). The farnesyl protein transferase inhibitor BZA-5B blocks farnesylation of nuclear lamins and p21ras but does not affect their function or localization. Cancer Res. 55, 3295–3304. [PubMed] [Google Scholar]
- De Smedt, F., Boom, A., Pesesse, X., Schiffmann, S.N., and Erneux, C. (1996). Post-translational modification of human brain type I inositol-1,4,5-trisphosphate 5-phosphatase by farnesylation. J. Biol. Chem. 271, 10419–10424. [DOI] [PubMed] [Google Scholar]
- Egea-Cortines, M., Saedler, H., and Somer, H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18, 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegold, A.A., Schafer, W.R., Rine, J., Whiteway, M., and Tamanoi, F. (1990). Common modifications of trimeric G proteins and Ras protein: Involvement of polyisoprenylation. Science 249, 165–169. [DOI] [PubMed] [Google Scholar]
- Gustafson-Brown, C., Savidge, B., and Yanofsky, M.F. (1994). Regulation of the Arabidopsis floral homeotic gene APETALA1. Cell 76, 131–143. [DOI] [PubMed] [Google Scholar]
- Hancock, J.F., Magee, A.I., Childs, J.E., and Marshall, C.J. (1989). All Ras proteins are polyisoprenylated but only some are palmitoylated. Cell 57, 1167–1177. [DOI] [PubMed] [Google Scholar]
- Hempel, F.D., Weigel, D., Mandel, M.A., Ditta, G., Zambryski, P.C., Feldman, L.J., and Yanofsky, M.F. (1997). Floral determination and expression of floral regulatory genes in Arabidopsis. Development 124, 3845–3853. [DOI] [PubMed] [Google Scholar]
- Irish, V.F., and Sussex, I.M. (1990). Function of the APETALA1 gene during Arabidopsis floral development. Plant Cell 2, 741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, G.L., Goldstein, J.E., and Brown, S.K. (1995). Polylysine and CVIM sequences of K-RasB dictate specificity of prenylation and confer resistance to benzodiazepine peptidomimetic in vitro. J. Biol. Chem. 270, 6221–6226. [DOI] [PubMed] [Google Scholar]
- Jones, D.G., Shlumukov, L., Carland, F., English, J., Scofield, S.R., Bishop, G.J., and Harrison, K. (1992). Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Res. 1, 285–297. [DOI] [PubMed] [Google Scholar]
- Kempin, S.A., Savidge, B., and Yanofsky, M. (1995). Molecular basis of the cauliflower phenotype in Arabidopsis. Science 267, 522–525. [DOI] [PubMed] [Google Scholar]
- Krizek, B.A., and Meyerowitz, E.M. (1996). Mapping the protein regions responsible for the functional specificities of the Arabidopsis MADS domain organ identity proteins. Proc. Natl. Acad. Sci. USA 93, 4063–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lijegren, S.J., Gustafson-Brown, C., Pinyopich, A., Ditta, G.S., and Yanofsky, M.F. (1999). Interactions among APETALA1, LEAFY, and TERMINAL FLOWER specify meristem fate. Plant Cell 11, 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel, A.M., and Yanofsky, M.F. (1995. a). A gene triggering flower formation in Arabidopsis. Nature 377, 522–524. [DOI] [PubMed] [Google Scholar]
- Mandel, A.M., and Yanofsky, M.F. (1995. b). The Arabidopsis AGL8 MADS box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1. Plant Cell 7, 1763–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel, M.A., Gustafson-Brown, C., Savidge, B., and Yanofsky, M.F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. [DOI] [PubMed] [Google Scholar]
- Morehead, T.A., Biermann, B.J., Crowell, D.N., and Randall, S.K. (1995). Changes in protein isoprenylation during growth of suspension-cultured tobacco cells. Plant Physiol. 109, 277–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara, E., and McCourt, P. (1999). Protein farnesylation in plants: A greasy tale. Curr. Opin. Plant Biol. 2, 388–392. [DOI] [PubMed] [Google Scholar]
- Pei, Z.-M., Ghassemian, M., Kwak, C.M., McCourt, P., and Schroeder, J.I. (1998). Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282, 287–290. [DOI] [PubMed] [Google Scholar]
- Porter, J.A., Young, K.E., and Beachy, P.A. (1996). Cholesterol modification of hedgehog signaling proteins in animal development. Science 274, 255–259. [DOI] [PubMed] [Google Scholar]
- Reiss, Y., Goldstein, J.L., Seabra, M.C., and Casey, P.J., and Brown, M.S. (1990). Inhibition of purified p21 ras farnesyl transferase by Cys-AAX tetrapeptides. Cell 62, 81–88. [DOI] [PubMed] [Google Scholar]
- Reiss, Y., Stradley, S.J., Gierach, L.M., Brown, M.S., and Goldstein, J.L. (1991). Sequence requirement for peptide recognition by rat brain p21ras protein farnesyltransferase. Proc. Natl. Acad. Sci. USA 88, 732–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., Krizek, B.A., and Meyerowitz, E.M. (1996). Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 93, 4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Concepción, M., Yalovsky, S., Zik, M., Fromm, H., and Gruissem, W. (1999. a). The prenylation status of a novel plant calmodulin directs plasma membrane or nuclear localization of the protein. EMBO J. 18, 1996–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Concepción, M., Yalovsky, S., and Gruissem, W. (1999. b). Protein prenylation in plants: Old friends and new targets. Plant Mol. Biol. 39, 865–870. [DOI] [PubMed] [Google Scholar]
- Schafer, W.R., and Rine, J. (1992). Protein prenylation: Genes, enzymes, targets and functions. Annu. Rev. Genet. 30, 209–237. [DOI] [PubMed] [Google Scholar]
- Schmitt, D., Callan, K., Kim, S.-H., and Gruissem, W. (1996). Molecular and biochemical characterization of tomato farnesyl-protein transferase. Plant Physiol. 112, 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueblood, C.E., Ohya, Y., and Rine, J. (1993). Genetic evidence for in vivo cross-specificity of the CaaX box protein prenyltransferases farnesyltransferase and geranylgeranyltransferase-I in Saccharomyces cerevisiae. Mol. Cell. Biol. 13, 4260–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorburger, K., Kitten, G.T., and Nigg, E.A. (1989). Modification of nuclear lamin proteins by a mevalonic acid derivative occurs in reticulocyte lysates and requires the cysteine residue of the C-terminal CXXM motif. EMBO J. 8, 4007–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., Alvarez, J., Smyth, D.R., Yanofsky, M.F., and Meyerowitz, E.M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. [DOI] [PubMed] [Google Scholar]
- Yalovsky, S., Trueblood, C.E., Callan, K.L., Narita, J.O., Jenkins, S.M., Rine, J., and Gruissem, W. (1997). Plant farnesyltransferase can restore yeast Ras signaling and mating. Mol. Cell. Biol. 17, 1986–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky, S., Rodríguez-Concepción, M., and Gruissem, W. (1999). Lipid modification of proteins—Slipping in and out of membranes. Trends Plant Sci. 4, 429–438. [DOI] [PubMed] [Google Scholar]
- Yalovsky, S., Kulukian, A., Rodríguez-Concepción, M., Young, C.A., and Gruissem, W. (2000). Functional requirement of plant farnesyltransferase during development in Arabidopsis. Plant Cell 12, 1267–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F.L., and Casey, P.J. (1996). Protein prenylation: Molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65, 241–269. [DOI] [PubMed] [Google Scholar]
- Zhou, D., Qian, D., Carmer, C.L., and Yang, Z. (1997). Developmental and environmental regulation of tissue- and cell-specific expression for a pea protein farnesyltransferase gene in transgenic plants. Plant J. 12, 921–930. [DOI] [PubMed] [Google Scholar]
- Zhu, J.-K., Bressan, R.A., and Hasegawa, P.M. (1993). Isoprenylation of the plant molecular chaperone ANJ1 facilitates membrane association and function at high temperature. Proc. Natl. Acad. Sci. USA 90, 8557–8561. [DOI] [PMC free article] [PubMed] [Google Scholar]