Abstract
Serine carboxypeptidases contain a conserved catalytic triad of serine, histidine, and aspartic acid active-site residues. These enzymes cleave the peptide bond between the penultimate and C-terminal amino acid residues of their protein or peptide substrates. The Arabidopsis Genome Initiative has revealed that the Arabidopsis genome encodes numerous proteins with homology to serine carboxypeptidases. Although many of these proteins may be involved in protein turnover or processing, the role of virtually all of these serine carboxypeptidase-like (SCPL) proteins in plant metabolism is unknown. We previously identified an Arabidopsis mutant, sng1 (sinapoylglucose accumulator 1), that is defective in synthesis of sinapoylmalate, one of the major phenylpropanoid secondary metabolites accumulated by Arabidopsis and some other members of the Brassicaceae. We have cloned the gene that is defective in sng1 and have found that it encodes a SCPL protein. Expression of SNG1 in Escherichia coli demonstrates that it encodes sinapoylglucose:malate sinapoyltransferase, an enzyme that catalyzes a transesterification instead of functioning like a hydrolase, as do the other carboxypeptidases. This finding suggests that SCPL proteins have acquired novel functions in plant metabolism and provides an insight into the evolution of secondary metabolic pathways in plants.
INTRODUCTION
Plants produce thousands of unique molecules that are collectively referred to as secondary metabolites. Even within the angiosperms, many of these compounds are unique to specific taxa, indicating that the pathways that produce them may have evolved within the last 100,000 years. A central question in the study of plant secondary metabolism concerns how the catalytic diversity of plant secondary metabolism has arisen. What classes of genes and proteins have been co-opted, presumably from their ancestral roles in primary metabolism, to serve as catalysts in the synthesis of secondary metabolites?
In Arabidopsis, the phenylpropanoid pathway leads to the production of sinapic acid esters, a group of fluorescent UV-protective secondary metabolites derived from phenylalanine (Figure 1). These compounds are dispensable under laboratory conditions and thus provide targets for the genetic dissection of phenylpropanoid metabolism. The analysis of these compounds is facilitated by their blue fluorescence under UV light both in vivo and after thin-layer chromatography (TLC) (Chapple et al., 1992; Ruegger et al., 1999). Arabidopsis and some other members of the Brassicaceae accumulate three major sinapic acid esters. In the biosynthetic pathway leading to these compounds, sinapoylglucose is the immediate precursor of sinapoylcholine and sinapoylmalate, which accumulate in seeds and leaves, respectively. 1-O-Sinapoylglucose is a β-acetal ester with a high free energy of hydrolysis (Mock and Strack, 1993); it provides the necessary free energy for the transacylation reaction catalyzed by sinapoylglucose:malate sinapoyltransferase (SMT; EC 2.3.1.92) (Strack, 1982), which generates sinapoylmalate in vegetative tissues (Sharma and Strack, 1985). During seed maturation, however, sinapoylglucose is converted to sinapoylcholine by sinapoylglucose:choline sinapoyltransferase (SCT; EC 2.3.1.91) (Strack et al., 1983). Despite the detailed biochemical understanding of this pathway, none of the genes involved has been cloned, and relatively little is known about its regulation.
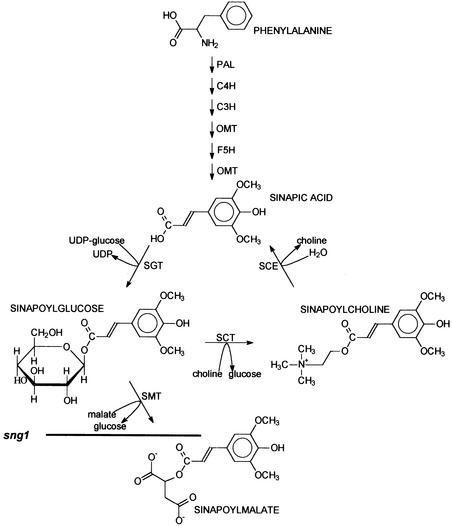
Figure 1.
The Pathway of Sinapate Ester Biosynthesis.
The enzymes required for the conversion of phenylalanine to sinapic acid are phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), p-coumarate 3-hydroxylase (C3H), caffeic acid/5-hydroxyferulic acid O-methyltransferase (OMT), and ferulate 5-hydroxylase (F5H). The enzymes unique to sinapate ester biosynthesis are UDP-glucose: sinapic acid glucosyltransferase (SGT), sinapoylglucose:malate sinapoyltransferase (SMT), sinapoylglucose:choline sinapoyltransferase (SCT), and sinapoylcholinesterase (SCE). The biochemical block in the sng1 mutant is indicated with a horizontal line across the step catalyzed by SMT.
A screen of 7600 ethyl methanesulfonate (EMS)–mutagenized plants by TLC analysis of methanolic leaf extracts identified two allelic mutants that lacked sinapoylmalate and instead accumulated its biosynthetic precursor, sinapoylglucose (Lorenzen et al., 1996). On the basis of their biochemical phenotypes, these mutants were named sinapoylglucose accumulator 1 (sng1-1) and sinapoylglucose accumulator 2 (sng1-2). Although SMT activity was readily detectable in wild-type leaf extracts, it was undetectable in extracts of mutant leaf tissue, suggesting that the defect in the sng1 mutant is in the gene encoding SMT or in a gene required for SMT expression.
Here, we show that the SNG1 locus encodes SMT and that SMT exhibits homology with serine carboxypeptidases (EC 3.4.16.1), hydrolases that use proteins or peptides as substrates. These findings indicate that this class of proteins has been recruited to function in plant secondary metabolic pathways and that their catalytic repertoire has broadened through evolution to include transacylation reactions such as that catalyzed by SMT.
RESULTS
sng1 Mutants Can Be Identified by Their Appearance under UV Light
The fluorescence of sinapoylmalate accumulated in the epidermis of Arabidopsis leaves can be visualized in vivo when wild-type plants are observed under UV light (Chapple et al., 1992; Ruegger et al., 1999). Mature sng1 mutants contain sinapoylglucose concentrations comparable to those found in wild-type plants; for unknown reasons, however, they are less fluorescent than are their wild-type counterparts (Figure 2). We used the diminished fluorescence phenotype of the sng1 mutant to identify two independent sng1 alleles (sng1-3 and sng1-4) among the T-DNA–tagged Arabidopsis lines available from the Arabidopsis Biological Resource Center (The Ohio State University, Columbus, OH). Backcrosses to the wild-type ecotype Wassilewskija, followed by tests of cosegregation, demonstrated that the kanamycin-resistant phenotype engendered by the T-DNA cosegregated with the mutant phenotype of sng1-4 but not with sng1-3.
Figure 2.

Fluorescence of Wild-Type and sng1 Plants under UV Light.
Because of a defect in sinapoylmalate biosynthesis, sng1 plants appear slightly darker than the wild type under UV light.
To provide additional resources for cloning the SNG1 gene, we identified several sng1 alleles (sng1-5 through sng1-8) from fast neutron–mutagenized populations of Arabidopsis. Because fast neutrons are known to generate deletions, lines that have a sng1 phenotype would be likely to carry restriction fragment-length polymorphisms, which would be helpful in the map-based cloning of SNG1. From a screen of 42,000 plants representing 12 parental groups, four independent mutants were identified with UV and TLC phenotypes similar to sng1. The biochemical phenotype of these mutants was verified by HPLC analysis, and all of the mutants failed to complement sng1-1, indicating that these plants carry new sng1 alleles (data not shown).
The SNG1 Gene Encodes a Serine Carboxypeptidase-like Protein
Inverse polymerase chain reaction (PCR) was used to amplify the genomic DNA adjacent to the T-DNA insertion in sng1-4 by using primers designed against the known sequence of the T-DNA vector. The resulting fragment was used to screen a cosmid library constructed in the transformation-competent binary vector pBIC20 (Meyer et al., 1996). Three classes of cosmids were recovered by this screening, as determined by digestion with HindIII. All three classes shared a common 3.9-kb fragment that hybridized with the inverse PCR product in DNA gel blot analysis (data not shown).
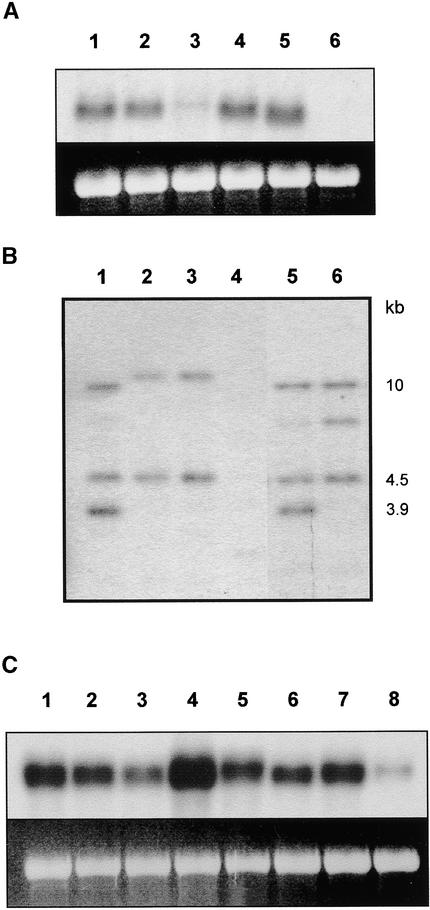
Before attempting to complement the sng1 mutant, we used two independent approaches to determine whether the 3.9-kb fragment shared by these cosmids was likely to carry at least a portion of the SNG1 gene. First, we used the 3.9-kb fragment to identify potential SNG1 transcripts and to compare their abundance in plants homozygous for each of four sng1 alleles (Figure 3A). RNA gel blot hybridization analysis identified a potential SNG1 transcript that was present in similar quantities in the leaf tissues of Columbia and Wassilewskija ecotypes. Transcript was present in wild-type quantities in the lines homozygous for sng1-2 and sng1-3, although the transcript in sng1-3 is possibly truncated. Transcript abundance was substantially decreased in the EMS-induced sng1-1 mutant and was undetectable in the T-DNA–tagged line, sng1-4. Considering that EMS-induced missense mutations and insertional mutagenesis often lead to mRNA destabilization, these data provided correlative evidence that we had cloned the SNG1 gene. Next, we used the fast neutron–induced sng1 alleles to determine whether these lines exhibited DNA polymorphisms associated with the putative SNG1 locus (Figure 3B). These experiments demonstrated that three of the four mutant lines carried deletions large enough to be detected by DNA gel blot analysis, and of those three, all had deletions that affected or eliminated hybridization of the 3.9-kb HindIII fragment to their genomic DNA. These data provide additional support that the SNG1 protein is at least partially encoded by this DNA. Based on the sequence data described below, one cosmid (hereafter referred to as pBIC20-SNG1; see Figure 4) was characterized further.
Figure 3.
Expression Analysis of the Putative SNG1 Gene and Its Structure in sng1 Deletion Mutants.
(A) Gel blot hybridization of RNA isolated from leaves of wild type and sng1 mutants probed with the 3.9-kb fragment of pBIC20-SNG1. Lane 1, Columbia wild type; lane 2, Wassilewskija wild type; lanes 3 to 6, sng1-1 through sng1-4.
(B) DNA gel blot analysis of fast neutron–induced sng1 alleles. Genomic DNA was prepared from M2 plants, digested with HindIII, electrophoresed, blotted, and probed with the 9.3-, 3.9-, and 4.5-kb HindIII fragments indicated in Figure 4. Lane 1, Columbia wild type; lanes 2 and 3, two isolates of sng1-5 from a single parental group; lanes 4 to 6, sng1-6, sng1-7, and sng1-8, respectively.
(C) Gel blot hybridization analysis of SNG1 expression in wild-type Arabidopsis. RNA was prepared from various tissues and probed with the 3.9-kb fragment of pBIC20-SNG1. Lane 1, young leaves; lane 2, mature leaves; lane 3, senescent leaves; lane 4, 10-day-old seedlings; lane 5, stems; lane 6, siliques; lane 7, flowers; lane 8, roots.
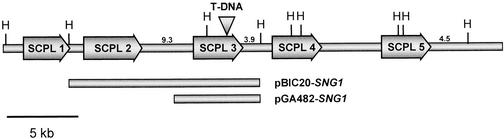
Figure 4.
The Region of the Arabidopsis Genome Surrounding the SNG1 Locus.
The bacterial artificial chromosome (BAC) clone F21P24 was found to include the putative SNG1 gene (SCPL 3) as well as four additional SCPL genes, each of which is indicated with arrows. The sixth SCPL gene, which is upstream of SCPL 1, is not indicated because it is thought to be a pseudogene. Shown are the position of the T-DNA insertion in the sng1-4 allele and the regions of Arabidopsis genomic DNA carried by the pBIC20-SNG1 and pGA482-SNG1 complementation constructs.
The 3.9-kb HindIII restriction fragment of pBIC20-SNG1 (Figure 4) was subcloned and sequenced. BLASTX analysis (Altschul et al., 1990) indicated that this region of the genome was likely to encode a protein with homology to serine carboxypeptidase proteins (score 48; E value 5 × e−5; closest homolog, carboxypeptidase I precursor from Hordeum vulgare; GenBank accession number J03897). To examine the expression of this putative carboxypeptidase, we probed an RNA gel blot with the 3.9-kb fragment from pBIC20-SNG1 (Figure 3C). Although sinapoylmalate is accumulated primarily in leaves of Arabidopsis and related crucifers, the putative SNG1 gene was expressed in almost all of the tissues examined. The greatest amount of message was observed in 10-day-old seedlings; only a low amount of SNG1 mRNA was found in roots.
To further characterize pBIC20-SNG1, we subcloned and partially sequenced the 9.3-kb HindIII fragment upstream of the 3.9-kb fragment (Figure 4). As expected, BLASTX analysis of the 3′ end of the fragment (relative to the direction of the putative SNG1 open reading frame) demonstrated homology to serine carboxypeptidase proteins. This was consistent with the previous analysis of the 5′ end of the downstream 3.9-kb fragment, which showed homology to internal sequences of serine carboxypeptidase proteins. Surprisingly, analysis of the 5′ end of the 9.3-kb fragment also indicated that this region encodes a serine carboxypeptidase-like (SCPL) protein. These data provided the first suggestion that at least two SCPL proteins are encoded near the SNG1 locus.
pBIC20-SNG1 Complements the sng1 Mutant Phenotype
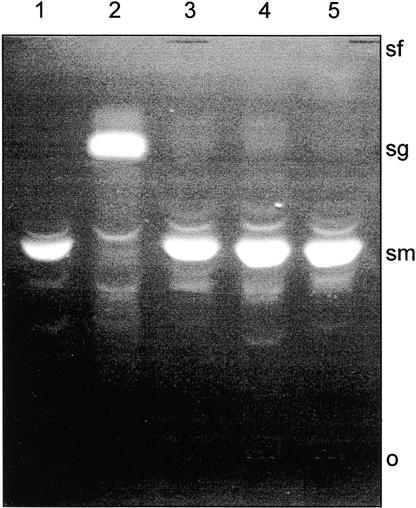
To provide definitive proof that pBIC20-SNG1 carries the SNG1 genomic sequence, we introduced this cosmid into Agrobacterium tumefaciens C58 pGV3850 by electroporation, and cultures harboring the binary vector were used to transform the sng1-1 mutant. Thirty-four kanamycin-resistant seedlings representing 19 independent transformation events were transferred to soil and tested for their profile of sinapate esters by TLC. All plants contained sinapoylmalate instead of, or in addition to, sinapoylglucose, indicating a total or partial complementation of the mutant phenotype (Figure 5). These data unequivocally demonstrate that the gene that is defective in the sng1 mutant is encoded on the pBIC20-SNG1 cosmid.
Figure 5.
Analysis of Sinapate Ester Content in Wild-Type, Mutant, and Transgenic Lines.
Leaf extracts were prepared from Columbia wild type (lane 1), sng1-1 (lane 2), and three sng1-1 transformants carrying the pBIC20-SNG1 transgene (lanes 3 to 5). Extracts were analyzed by TLC on silica gel plates with the mobile phase n-butanol:acetic acid:water, 5:2:3 (v/v/v). Sinapoylmalate (sm) and sinapoylglucose (sg) were visualized under 312 nm UV light. o, origin; sf, solvent front.
At the time the initial sequence data for pBIC20-SNG1 were obtained, TAMU BAC F21P24 was being sequenced by the Arabidopsis Genome Initiative. When the complete BAC sequence was released, it revealed that BAC F21P24 carries the SNG1 locus and five SCPL genes surrounding the SNG1 gene (Figure 4). One of these genes (not shown in Figure 4) has been annotated in the database as a pseudogene because the region corresponding to its first exon is flanked by sequences highly similar to ATPases, suggesting that this SCPL gene lacks a promoter. All of the SCPL proteins encoded by this region of the genome are highly similar to one another, their deduced amino acid sequences sharing between 69 and 78% amino acid identity. Their similarity and tandem arrangement suggest that they may be the result of relatively recent gene duplication events. RNA gel blot hybridization experiments indicate that these genes are expressed only in very low quantities in all tissues examined previously for SNG1 expression (data not shown). These data also indicate that the widespread expression previously observed for SNG1 is not an artifactual result of cross-hybridization to mRNAs of these other SCPL genes.
Our partial sequence data and the release of the sequence of BAC F21P24 demonstrate that pBIC20-SNG1 encodes two SCPL proteins (Figure 4). To prove unambiguously which gene is defective in the sng1 mutant, we generated a new construct (pGA482-SNG1) that contains only the downstream SCPL gene under the control of 1.1 kb of its upstream regulatory sequence and used this construct to transform the sng1 mutant. Like pBIC20-SNG1, the genomic sequence carried on pGA482-SNG1 complements the sng1 phenotype (data not shown), indicating that we have identified the SNG1 gene (SCPL 3 in Figure 4).
SMT Is a SCPL Protein
We used the 3.9-kb fragment of pBIC20-SNG1 to screen a cDNA library prepared from 10-day-old Arabidopsis seedlings (Meyer et al., 1994) and retrieved several clones, of which we sequenced the longest (GenBank accession number AF275313). Analyzing the predicted N-terminal sequence by using the algorithm described by Nielsen et al. (1997) and available at the SignalP website (www.cbs.dtu dk/services/SignalP/) indicated the presence of a signal peptide, which is probably cleaved after residue S19. If this prediction is correct, then the inferred translation product of 49.4 kD would give rise to a mature protein with a mass of 47.2 kD. Analysis of the SMT sequence by the PSORT algorithm (psort.nibb.ac.jp/) predicted six possible glycosylation sites and indicated that the protein is most likely localized in the vacuole. These predictions are consistent with previous research that demonstrated SMT to be a vacuolar protein (Strack and Sharma, 1985). Most importantly, the sequence ASIVKFLPGFEGPLPFE was found immediately after the predicted cleavage site. This peptide matched at 16 of 17 residues the N-terminal sequence obtained when SMT was purified from Brassica napus, as described by Gräwe et al. (1992), blotted onto polyvinylidene difluoride membrane, and sequenced with a model 120A liquid phase protein sequencer (data not shown).
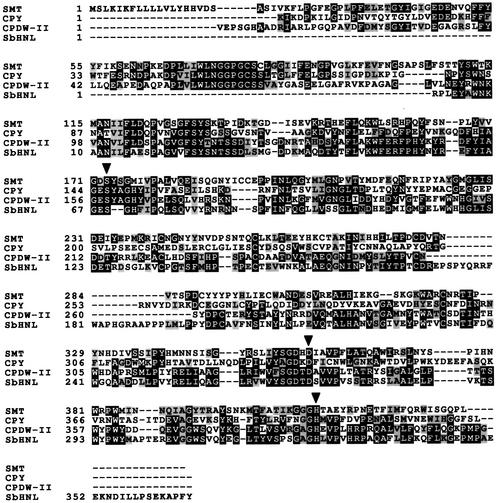
Comparison of the inferred amino acid sequence of the cDNA with those in the database indicated substantial similarities with SCPL proteins from plants, animals, and yeast. The inferred amino acid sequence of the putative SMT cDNA shares 18% identity with carboxypeptidase Y from Saccharomyces cerevisiae and 23% identity with the wheat carboxypeptidase for which the crystal structure has been determined (Liao and Remington, 1990; Liao et al., 1992) (Figure 6). The putative SMT sequence shares the conserved serine, aspartic acid, and histidine residues (S173, D358, and H411 in the SMT sequence) that have been demonstrated through inhibitor studies and site-directed mutagenesis (Hayashi et al., 1973, 1975; Bech and Breddam, 1989), as well as crystallographic analysis (Liao and Remington, 1990; Liao et al., 1992), to make up the catalytic triad that is essential for enzymatic activity. The involvement of an active-site serine residue in the SMT protein is supported by the observation that preincubation with phenylmethylsulfonyl fluoride inhibited by 30% the activity of SMT extracted from Arabidopsis leaves.
Figure 6.
Alignment of the SNG1 Gene Product with Serine Carboxypeptidases and SCPL Proteins.
An alignment of SMT with the yeast carboxypeptidase Y (CPY), wheat carboxypeptidase (CPDW-II), and the hydroxynitrile lyase from Sorghum bicolor (SbHNL) (only an incomplete sequence was available in the database) was prepared by using the ClustalW algorithm. Amino acids that are identical in two or more proteins are shaded in black, and conservative amino acid substitutions are shaded in gray. Putative active residues in SMT (S-173, D-358, and H-411) are designated with black arrowheads, based on their alignment with the carboxypeptidase Y catalytic triad. Dashes denote gaps introduced to optimize the amino acid alignment.
Although these data provided strong evidence that the SNG1 gene encodes SMT, we could not exclude the possibility that SNG1 is a serine carboxypeptidase required for the proteolytic activation of one or more vacuolar proproteins that include SMT. Indeed, this interpretation could be supported by our findings that SNG1 transcript is expressed in tissues other than those known to accumulate sinapoylmalate. In addition, the five SCPL genes clustered at the SNG1 locus on chromosome 2 encode proteins having amino acid sequences in their N-terminal regions that are highly similar (only one to three amino acid substitutions) to the N-terminal sequence of SMT purified from B. napus. Consequently, this amino acid sequence is not necessarily diagnostic for SMT. Finally, although carboxypeptidases have been shown to catalyze acyltransferase reactions under nonphysiological conditions (Widmer and Johansen, 1979; Widmer et al., 1980), the literature contains no precedents for SCPL proteins acting as acyltransferases in vivo.
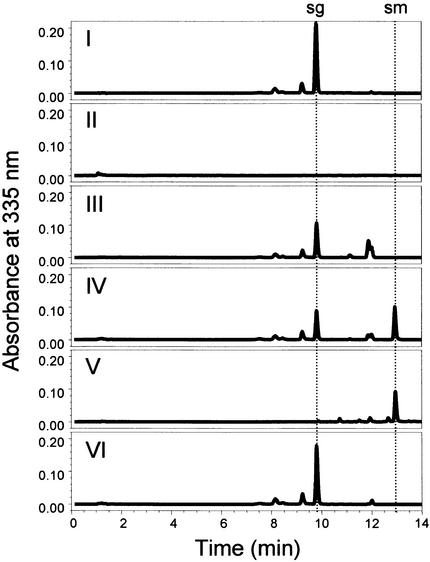
To determine unequivocally whether the SNG1 gene encodes SMT, we expressed the SNG1 cDNA in Escherichia coli (Figure 7). For these experiments, the portion of the SNG1 open reading frame corresponding to the mature N-terminally trimmed polypeptide was subcloned into pET28A under the control of the T7 promoter. As analyzed by SDS-PAGE, no obvious differences were observed between the soluble proteins extracted from cells carrying pET28A and pET28A-SNG1 grown at 14°C in either the presence or absence of isopropyl-β-d-thiogalactopyranoside (IPTG). In contrast, a distinct novel band with a molecular mass of ∼44 kD was visible in uninduced and induced cells carrying the pET28A-SNG1 vector. The size of this band was somewhat less than, but reasonably consistent with, the expected size of the SNG1 protein. Although these data indicated that the bulk of the SNG1 protein was present in inclusion bodies, we assayed samples of the supernatant protein for SMT activity. We anticipated that if only a small percentage of the protein was correctly folded and soluble, then its enzymatic activity could be readily measured even if the protein were undetectable by SDS-PAGE analysis. HPLC analysis demonstrated that sinapoylmalate was formed by extracts of cells harboring the pET28A-SNG1 construct when incubated in the presence of sinapoylglucose and malate (Figure 8). Omission of enzyme, sinapoylglucose, or malate eliminated the production of sinapoylmalate, as did the use of extracts of cells harboring only the pET28A vector. This experiment provides conclusive proof that the SNG1 gene encodes SMT.
Figure 7.
Analysis of SNG1 Expressed in E. coli.
(A) SDS-PAGE analysis of soluble fractions of E. coli harboring pET28A (lanes 1 and 2) and the SNG1 expression vector pET28A-SNG1 (lane 3 and 4) grown in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of 0.8 mM IPTG.
(B) The same analysis as given for (A) but using insoluble fractions.
Figure 8.
Analysis of SMT Activity in E. coli Expressing the SNG1 Gene.
Enzyme assays and leaf extracts were analyzed by HPLC with UV detection at 335 nm. Assay I contained all assay components except the E. coli protein extract. Assays II to IV contained 100 μg of soluble protein from E. coli harboring pET28A-SNG1; assay II lacked sinapoylglucose (sg); assay III lacked malate; assay IV contained all assay components. HPLC run V represents a methanolic extract of wild-type Arabidopsis leaves containing sinapoylmalate (sm). Assay VI included all assay components incubated with 100 μg of soluble protein of E. coli harboring the original pET28A vector. All protein extracts were obtained from cultures that had not been induced with IPTG. All assays were incubated at 30°C for 14 hr. The identity of the SMT reaction product was confirmed by liquid chromatography–mass spectrometry on a Micromass Quattro Ultima (Micromass; Manchester, UK) triple quadrupole instrument in negative ion electrospray mode (m/z− for sinapoylmalate 339.15).
sng1 Deletion Mutants Are Not Defective in Other Aspects of Sinapate Ester Metabolism
The reaction catalyzed by SMT is biochemically analogous to two other reactions of sinapate ester biosynthesis and turnover. Like SMT, SCT catalyzes an acyltransfer reaction which uses sinapoylglucose as a sinapate donor, but choline instead of malate is the sinapate acceptor. Sinapoylcholinesterase (SCE; EC 3.1.1.49; Nurmann and Strack, 1979; Strack et al., 1980) also uses a sinapate ester as a substrate but catalyzes a hydrolysis reaction in which water can be considered to act as the sinapate acceptor. In light of the biochemical similarity of these reactions, we tested the hypotheses that SCT and SCE are also SCPL proteins and that these enzymes are encoded by the SCPL genes flanking the SNG1 locus.
The deletion in the sng1-6 mutant spans the SNG1 locus and deletes all or part of each of the surrounding SCPL genes. When the sng1-6 mutant was examined for phenotypes that would be expected to be associated with the loss of the activities of SCT (failure to accumulate sinapoylcholine in seeds) and SCE (failure to hydrolyze sinapoylcholine during early seedling development), no abnormalities in sinapate ester metabolism were detected (data not shown). These results indicate that either the genes flanking SNG1 are not involved in sinapate ester metabolism, or their functions are redundant with proteins encoded elsewhere in the genome.
DISCUSSION
Serine carboxypeptidases have been identified in a wide array of organisms. They catalyze the hydrolysis of the C-terminal peptide bond in proteins or peptides and are usually thought of as being involved in protein degradation and processing. The best studied of these is serine carboxypeptidase Y from S. cerevisiae, a vacuolar protein that is initially synthesized as a preproenzyme. This enzyme has been used extensively in studies of protein transport, targeting, and processing (for examples, see Valls et al., 1990; Ramos et al., 1994; Ramos and Winther, 1996). Alkylation by suicide inhibitors and subsequent mutagenesis experiments have identified the active-site serine and histidine residues (Hayashi et al., 1973, 1975; Bech and Breddam, 1989), and crystallization of the enzyme has led to identification of the other amino acid residues that make up the pocket in which substrate is bound (Endrizzi et al., 1994).
In plants, serine carboxypeptidases and proteins that share amino acid sequence homology with them (SCPL proteins) have been isolated from several species, and SCPL genes have been identified in expressed sequence tag and genomic sequencing projects. The proteins from wheat and barley have been particularly well studied because of their inferred role in mobilization of seed storage reserves (Baulcombe et al., 1987; Doan and Fincher, 1988; Degan et al., 1994), and the homodimeric wheat serine carboxypeptidase II has been crystallized (Liao and Remington, 1990; Liao et al., 1992). SCPL proteins from cauliflower, rice, and tomato have also been purified and characterized (Doi et al., 1980; Walker-Simmons and Ryan, 1980; Kim and Hayashi, 1983; Mehta and Mattoo, 1996; Mehta et al., 1996). SCPL genes have been isolated from Arabidopsis, pea, and rice on the basis of their homology to SCPL cDNAs from wheat and barley (Bradley, 1992; Washio and Ishikawa, 1994; Jones et al., 1996). SCPL enzymes also play a role in herbicide metabolism, in which a SCPL has been shown to catalyze the first step in the catabolism of an alachlor glutathione S-conjugate by removing the terminal glycine residue of the glutathione moiety (Wolf et al., 1996). On the basis of these and other studies, investigators have suggested various functions for SCPL enzymes, ranging from protein turnover and C-terminal processing to roles in wound responses and xenobiotic metabolism.
Although plant SCPL enzymes and genes have been the subject of numerous publications, their natural substrates are largely unknown. Virtually all SCPL enzymes have been purified from plants based on their ability to degrade artificial peptide substrates. In many cases, their role in proteolysis has been implied or assumed because the enzymes have been isolated from tissues actively engaged in protein turnover and show little apparent substrate specificity. Although some of these enzymes may be carboxypeptidases, no genetic proof has demonstrated their in vivo function.
SNG1 Encodes SMT, a SCPL Protein
The Arabidopsis Genome Initiative has provided evidence that the Arabidopsis genome probably contains at least 40 SCPL genes. Although some of these proteins may indeed function in proteolysis, our data demonstrate that these proteins can have unexpected functions in secondary metabolism in plants. Thus, the provisional annotation of these genes as carboxypeptidase-like is reasonable but may be misleading. We have cloned the SNG1 gene and found that it encodes SMT, a SCPL protein. Our working model is that although carboxypeptidases hydrolyze a peptide bond directly or indirectly by using water as a nucleophile, SMT cleaves an ester bond by using the hydroxyl of malate as a nucleophile, possibly by way of a “sinapoylated” enzyme intermediate. Consequently, SMT catalyzes a transesterification reaction rather than a hydrolysis. This model is consistent with previous research demonstrating that carboxypeptidase Y can catalyze transpeptidation and transesterification reactions under alkaline nonphysiological conditions (Widmer and Johansen, 1979; Widmer et al., 1980). The activity of SCPL proteins toward both esters and peptides is also consistent with Grimm's rules for hydride displacement (Grimm, 1925) and the classification of esters and amides as bioisosteres (Burger, 1970).
The fact that SMT is a SCPL protein may indicate that plants have recruited the catalytic abilities of these enzymes to act in other capacities. Although sinapate ester biosynthesis is limited as to taxonomic distribution, the study of SCPL genes in this pathway has shed light on the catalytic diversity of this class of enzymes and may implicate enzymes of this class in other important pathways. For example, glucose ester transesterification reactions are found in pathways leading to numerous other plant secondary metabolites, including the synthesis of chlorogenic acid in sweet potato (Villegas and Kojima, 1986) and gallotannins in oak (Gross, 1983). Similarly, indoleacetic acid (IAA)–glucose serves as an activated form of IAA, which is used in a transesterification reaction to form IAA-inositol (Michalczuk and Bandurski, 1982). These reactions are analogous to sinapoylmalate biosynthesis and may be catalyzed by a SCPL enzyme. SCPL proteins may also be important in vacuolar targeting. The most abundant anthocyanin in wild carrot, Daucus carota, is a sinapoylated cyanidin glycoside (Harborne et al., 1983; Glässgen et al., 1992), and the enzyme catalyzing the “sinapoylation” of the nonacylated anthocyanin uses sinapoylglucose as the activated sinapate donor (Glässgen and Seitz, 1992). Because isolated Daucus vacuoles actively take up the sinapoylated anthocyanin but not the nonacylated form (Hopp and Seitz, 1987), the sinapate moiety may function as a vacuolar uptake tag. Alternatively, the sinapate moiety may be required for, or be the site of, glutathione derivatization by a glutathione S-transferase analogous to that encoded by the maize Bronze2 gene (Marrs et al., 1995).
Two other pathways of plant secondary metabolism are known to involve SCPL proteins. The wild tomato, Lycopersicon pennellii, accumulates 2,3,4-tri-O-acylglucoses in its trichomes (Ghangas and Steffens, 1993, 1995). The gene encoding one of these enzymes (A.X. Li and J.C. Steffens, unpublished results; GenBank accession number AF248647) and three related acyltransferase genes from Solanum berthaultii (T. Wei and J.C. Steffens, unpublished results; GenBank accession numbers AF006078, AF006079, and AF006080) have been cloned and shown to encode SCPL proteins. SMT shares considerable amino acid identity with these acyltransferases (∼42%), suggesting that SCPL proteins that function as acyltransferases may be more similar to one another than they are to actual carboxypeptidases. The gene encoding the hydroxynitrile lyase involved in cyanogenic glycoside degradation in S. bicolor has also been cloned and shown to encode a SCPL enzyme (Wajant et al., 1994). The available amino acid sequence of the sorghum hydroxynitrile lyase is 18% identical to SMT and also shares the conserved catalytic triad found in SCPL proteins (Figure 6). The fact that this enzyme is a lyase, rather than a hydrolase or acyltransferase, is another demonstration of the catalytic diversity of these proteins and of their widespread occurrence in plant metabolism.
SCT and SCE Are Not Clustered with SNG1
The reactions catalyzed by SMT and SCT are similar in that they both involve transesterification of sinapate from sinapoylglucose onto the hydoxyl group of an acceptor molecule. The SCE reaction is also similar, but in this case, the acceptor molecule is water. Given that SMT is a SCPL protein, we postulate that SCT and SCE may also be enzymes of this type. This hypothesis is supported by limited sequence data available from previous work on SCT (Vogt et al., 1993). Although relatively little is known about SCE, carboxypeptidase Y and tomato leaf carboxypeptidase have esterase activity (Walker-Simmons and Ryan, 1980; Christensen, 1994), and other esterases are known to use the same catalytic triad as is found in SCPL proteins (Dodson and Wlodawer, 1998). Taken together, these data suggest that all three of the enzymes involved in the final stages of sinapate ester biosynthesis in Arabidopsis may be SCPL proteins.
The sng1 deletion mutants allowed us to test the hypothesis that the SCPL genes encoding SMT, SCT, and SCE are clustered at the SNG1 locus on chromosome 2. Given the restricted taxonomic distribution of sinapate esters in the plant kingdom, it was tempting to speculate that a series of SCPL gene duplication events early in the evolutionary history of the Brassicaceae led to the development of the sinapate ester biosynthetic and interconversion pathways. If the sinapate ester biosynthetic genes had remained clustered, then some of the sng1 deletion mutants would be expected to have multiple defects in sinapate ester biosynthesis. Instead, these mutants showed no defects in sinapoylcholine synthesis or turnover, indicating either that these genes are not involved in sinapate ester biosynthesis or that their function is genetically redundant. Identification of a mutant that accumulates sinapoylglucose in its seeds instead of sina-poylcholine (C. McMichael and C. Chapple, unpublished results) suggests that function of SCT, at least, is not redundant.
Conclusions
SCPL proteins may function in a broad range of biochemical pathways, including those of secondary metabolite biosynthesis, herbicide conjugation, and germination-associated degradation of seed protein reserves. Thus, these proteins may be vital for normal plant growth and development, for the synthesis of compounds that protect plants against pathogens and UV light, and for resistance to natural and manmade xenobiotics. The identification of SMT as a SCPL protein has cast new light on the potential of these enzymes to serve as participants in diverse biochemical pathways.
METHODS
Plant Material
Arabidopsis thaliana Heynh. ecotypes Columbia or Landsberg erecta were cultivated at a light intensity of 100 μE m−2 sec−1 at 23°C under a photoperiod of 16-hr light/8-hr dark in ProMix potting mixture (Premier Horticulture, Red Hill, PA). For seedling plant material to be used in the analysis of SNG1 mRNA accumulation, seeds were surface sterilized for 30 min in a 2:1 (v/v) mixture of 0.1% Triton X-100 and household bleach. Seeds were rinsed thoroughly with sterile water and plated on Miracloth (Calbiochem) discs on modified Murashige and Skoog (1962) medium (ammonia-free medium to which an additional 20.6 mM potassium nitrate was added instead of ammonium nitrate) containing 0.7% agar.
Secondary Metabolite Analysis
Leaf extracts were prepared from 100-mg samples of fresh leaf tissue suspended in 1 mL of 50% methanol. Samples were ground briefly, then centrifuged at 12,000g for 5 min. Sinapate ester content was qualitatively determined by UV fluorescence after chromatography of the extracts on silica gel thin-layer chromatography (TLC) plates in a mobile phase of n-butanol:acetic acid:water, 5:2:3 (v/v/v) or quantitatively determined by HPLC.
HPLC Analysis
Plant extracts and sinapoylglucose:malate sinapoyltransferase (SMT) assays were analyzed by HPLC on a Nova-Pak (Waters Associates; Milford, MA) C18 column (60 Å pore size, 4 μM particle size) by using a 15-min gradient at 1 mL min−1 from 6% acetonitrile:1.5% phosphoric acid to 48% acetonitrile:1.5% phosphoric acid. Absorbance of the effluent at 335 nm was measured.
Analysis of Nucleic Acids
For DNA gel blot analyses, DNA was extracted from leaf material (Rogers and Bendich, 1985), digested with restriction endonucleases, electrophoretically separated, transferred to Hybond N+ membrane (Amersham), and hybridized with cDNA probes according to standard protocols (Sambrook et al., 1989). RNA was extracted from tissues (Goldsbrough and Cullis, 1981), electrophoretically separated, transferred to Hybond N+ membrane (Amersham), and hybridized with radiolabeled probes prepared from genomic clones according to standard protocols. The genomic and cDNA clones were sequenced on a Pharmacia (Uppsala, Sweden) ALFexpress automated DNA sequencer with the use of standard primers.
Inverse Polymerase Chain Reaction and Identification of cDNA and Genomic Clones
To isolate regions flanking the T-DNA insert in the sng1-4 mutant, we extracted genomic DNA as described above. Genomic DNA was digested with BclI and circularized with T4 DNA ligase. Inverse polymerase chain reaction (PCR) was performed with the primers 5′-GATGCACTCGAAATCAGCCA-3′ and 5′-GCGCGGAGTCATTAC-AGTTA-3′ and using 35 1-min cycles and a primer annealing temperature of 54°C . These reactions amplified a single 768-bp fragment. SNG1 cDNA and genomic clones were identified by standard techniques (Sambrook et al., 1989), with the inverse PCR fragment amplified from sng1-4 used as a probe. The SNG1 cDNA clone was identified in a library prepared from mRNA from 10-day-old abi1 seedlings (Meyer et al., 1994). The SNG1 genomic clones were identified in an A. thaliana (Landsberg erecta) library generated in the binary cosmid vector pBIC20 (Meyer et al., 1996). For generation of the pGA482-SNG1 construct, a region corresponding to the SNG1 promoter was amplified by PCR using the upstream primer 5′-CGG-GTACCAGCAAAACGCATCAACCATAAAC-3′ and the downstream primer 5′-GAGGGCCGGGACAATCATA-3′. The upstream primer introduced a new KpnI site into the sequence, and the downstream primer bound downstream of the HindIII site that is internal to the SNG1 gene. The amplification product was subcloned into pGEMT-Easy (Promega) for sequencing and then liberated with KpnI and HindIII for subcloning into similarly digested pGA482 (An, 1987). The resulting vector was then digested with HindIII, and the 3.9-kb HindIII fragment from pBIC20-SNG1 was inserted and checked for orientation by PCR to generate pGA482-SNG1.
Plant Transformation
Constructs for plant transformation were introduced into Agrobacterium tumefaciens C58 pGV3850 (Zambrisky et al., 1983) by electroporation. Plant transformation was performed by vacuum infiltration (Bent et al., 1994), with minor modifications (Bell-Lelong et al., 1997). Transformed seedlings (T1) identified by selection on Murashige–Skoog medium containing 50 mg L−1 kanamycin and 200 mg L−1 timentin were transferred to soil.
Constructs for Expression of SNG1 in Escherichia coli
Two oligonucleotides designed to amplify a fragment of the SNG1 cDNA encoding a protein lacking the predicted signal peptide were used to create a fragment suitable for cloning into the pET28A expression vector (Novagen; Madison, WI). The N-terminal oligonucleotide 5′-TCATGACCTCTATCGTCAAGTTTCTTCC-3′ incorporated a start codon and the restriction site PagI (TCATGA) and altered the N-terminal alanine codon (GCC) to a threonine codon (ACC). The C-terminal oligonucleotide, 5′-GTCGACTTACAGGGGTTGGCCACTG-3′, incorporated a SalI restriction site after the stop codon. The SNG1 gene was amplified by PCR, subcloned, and sequenced. The SNG1 gene was excised by PagI-SalI digestion and cloned into the NcoI-SalI–digested pET28A vector to yield pET28A-SNG1. For analysis of SNG1 expression and activity, the E. coli host BL21DE3 was transformed with the empty pET28A vector and pET28A-SNG1.
E. coli Growth Conditions and Preparation of E. coli Extracts
For heterologous expression of SNG1, an overnight culture of bacteria grown at 37°C was diluted 200-fold into fresh Luria-Bertani medium and grown at 18°C to an OD600nm of 0.6. Cells were subsequently induced with 0.8 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and grown for 48 hr at 14°C. Cells were harvested and lysed in 2.5 mL of 20 mM Tris-HCl, pH 8.0, plus 500 mM NaCl using a french press. The cell lysate was cleared by centrifugation at 14,000g at 4°C for 30 min. Supernatant (soluble protein fraction) and pellet (insoluble protein fraction) were analyzed by SDS-PAGE. The protein concentration of the soluble fraction was determined by using the Bradford assay (Bradford, 1976).
Enzyme Assays
SMT assays were performed with 12.5 μL of 0.5 mM sinapoylglucose in 100 mM potassium phosphate buffer, pH 7.5, 5 μL of 100 mM potassium phosphate buffer, pH 6.0, 5 μL of 1 M malic acid in potassium phosphate buffer, pH 6.0, and 5 μL of E. coli extract corresponding to 100 μg of protein. Assays were incubated for 14 hr at 30°C, stopped by addition of 30 μL of methanol, and stored at −70°C before analysis by HPLC. Sinapoylglucose for use in enzyme assays was purified from the sng1 mutant of Arabidopsis (Lorenzen et al., 1996).
The impact of phenylmethylsulfonyl fluoride on SMT activity was determined by using enzyme prepared from Arabidopsis leaf tissue, as described previously (Lorenzen et al., 1996). Assays were conducted essentially as described above after 30 min preincubation of the enzyme extract in the presence of 30 mM of the inhibitor. Percentage inhibition was determined by measuring the amount of product generated after a further 60-min incubation in the presence of substrates compared with control reactions.
NOTE ADDED IN PROOF
It should be noted that since the submission of this manuscript, a paper describing the SCPL acyltransferases from wild tomato has been published (Li, A.X., and Steffens, J.C. [2000]. An acyltransferase catalyzing the formation of diacylglucose is a serine carboxypeptidase-like protein. Proc. Natl. Acad. Sci. USA 97, 6902–6907).
Acknowledgments
This work was supported by grants from the Division of Energy Biosciences, United States Department of Energy, and the Purdue University Office of International Programs in Agriculture to C.C. and a grant from the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie to D.S. We are grateful to Maike Lorenzen (Halle) for SMT purification, Dr. A. Otto (Max-Delbrück-Zentrum, Berlin, Germany) for amino acid sequencing, and John E. Buckholz and Dr. Barbara S. Larsen (DuPont) for liquid chromatographic–mass spectrometric analysis. This is journal paper number 16327 of the Purdue University Agricultural Experiment Station.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- An, G. (1987). Binary Ti vectors for plant transformation and promoter analysis. Methods Enzymol. 153, 292–305. [Google Scholar]
- Baulcombe, D.C., Barker, R.F., and Jarvis, M.G. (1987). A gibberellin responsive wheat gene has homology to yeast carboxypeptidase Y. J. Biol. Chem. 262, 13726–13735. [PubMed] [Google Scholar]
- Bech, L.M., and Breddam, K. (1989). Inactivation of carboxypeptidase Y by mutational removal of the putative essential histidyl residue. Carlsberg Res. Commun. 54, 165–171. [DOI] [PubMed] [Google Scholar]
- Bell-Lelong, D.A., Cusumano, J.C., Meyer, K., and Chapple, C. (1997). Cinnamate-4-hydroxylase expression in Arabidopsis: Regulation in response to development and the environment. Plant Physiol. 113, 729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A.F., Kunkel, B.N., Dahlbeck, D., Brown, K.L., Schmidt, R., Giraudat, J., Leung, J., and Staskawicz, B.J. (1994). RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 265, 1856–1860. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Bradley, D. (1992). Isolation and characterization of a gene encoding a carboxypeptidase Y-like protein from Arabidopsis thaliana. Plant Physiol. 98, 1526–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger, A. (1970). In Medicinal Chemistry, A. Burger, ed, 3rd ed. (New York: Wiley).
- Chapple, C.C.S., Vogt, T., Ellis, B.E., and Somerville, C.R. (1992). An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4, 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, U. (1994). Effects of pH on carboxypeptidase-Y–catalyzed hydrolysis and aminolysis reactions. Eur. J. Biochem. 220, 149–153. [DOI] [PubMed] [Google Scholar]
- Degan, F.D., Rocher, A., Cameron-Mills, V., and von Wettstein, D. (1994). The expression of serine carboxypeptidases during maturation and germination of barley grain. Proc. Natl. Acad. Sci. USA 91, 8209–8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan, N.P., and Fincher, G.B. (1988). The A- and B-chains of carboxypeptidase I from germinated barley originate from a single precursor polypeptide. J. Biol. Chem. 263, 11106–11110. [PubMed] [Google Scholar]
- Dodson, G., and Wlodawer, A. (1998). Catalytic triads and their relatives. Trends Biochem. Sci. 23, 347–352. [DOI] [PubMed] [Google Scholar]
- Doi, E., Komori, N., Matoba, T., and Morita, Y. (1980). Purification and some properties of a carboxypeptidase in rice bran. Agric. Biol. Chem. 44, 85–92. [Google Scholar]
- Endrizzi, J.A., Breddam, K., and Remington, S.J. (1994). 2.8-Å structure of yeast serine carboxypeptidase. Biochemistry 33, 11106–11120. [DOI] [PubMed] [Google Scholar]
- Ghangas, G.S., and Steffens, J.C. (1993). UDPglucose:fatty acid transglucosylation and transacylation in triacylglucose biosynthesis. Proc. Natl. Acad. Sci. USA 90, 9911–9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghangas, G.S., and Steffens, J.C. (1995). 1-O-Acyl-β-d-glucoses as fatty acid donors in transacylation reactions. Arch. Biochem. Biophys. 316, 370–377. [DOI] [PubMed] [Google Scholar]
- Glässgen, W.E., and Seitz, H.U. (1992). Acylation of anthocyanins with hydroxycinnamic acids via 1-O-acylglucosides by protein preparations from cell cultures of Daucus carota L. Planta 186, 582–585. [DOI] [PubMed] [Google Scholar]
- Glässgen, W.E., Wray, V., Strack, D., Metzger, J.W., and Seitz, H.U. (1992). Anthocyanins from cell suspension cultures of Daucus carota. Phytochemistry 31, 1593–1601. [DOI] [PubMed] [Google Scholar]
- Goldsbrough, P.B., and Cullis, C.A. (1981). Characterization of the genes for ribosomal RNA in flax. Nucleic Acids Res. 9, 1301–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräwe, W., Bachhuber, P., Mock, H.-P., and Strack, D. (1992). Purification and characterization of sinapoylglucose:malate sinapoyltransferase from Raphanus sativus L. Planta 187, 236–241. [DOI] [PubMed] [Google Scholar]
- Grimm, H.G. (1925). Über Bau und Grösse der Nichtmetallhydride. Z. Elektrochem. 31, 474–480. [Google Scholar]
- Gross, G.G. (1983). Synthesis of mono-, di- and trigalloyl-β-d-glucose by β-glucogallin-dependent galloyltransferases from oak leaves. Z. Naturforsch. 38c, 519–523. [Google Scholar]
- Harborne, J.B., Mayer, A.M., and Bar-Nun, N. (1983). Identification of the major anthocyanin of carrot cells in tissue culture as cyanidin 3-(sinapoylxylosylglucosylgalactoside). Z. Naturforsch. 38c, 1055–1056. [Google Scholar]
- Hayashi, R., Moore, S., and Stein, W.H. (1973). Serine at the active center of yeast carboxypeptidase. J. Biol. Chem. 248, 8366–8369. [PubMed] [Google Scholar]
- Hayashi, R., Bai, Y., and Hata, T. (1975). Evidence for an essential histidine in carboxypeptidase Y. Reaction with the chloromethyl ketone derivative of benzyloxycarbonyl-l-phenylalanine. J. Biol. Chem. 250, 5221–5226. [PubMed] [Google Scholar]
- Hopp, W., and Seitz, H.U. (1987). The uptake of acylated anthocyanin into isolated vacuoles from a cell suspension culture of Daucus carota. Planta 170, 74–85. [DOI] [PubMed] [Google Scholar]
- Jones, C.G., Lycett, G.W., and Tucker, G.A. (1996). Protease inhibitor studies and cloning of a serine carboxypeptidase cDNA from germinating seeds of pea (Pisum sativum L.). Eur. J. Biochem. 235, 574–578. [DOI] [PubMed] [Google Scholar]
- Kim, Y., and Hayashi, R. (1983). Properties of a serine carboxypeptidase in cauliflower. Agric. Biol. Chem. 47, 2655–2667. [Google Scholar]
- Liao, D.-I., and Remington, S.J. (1990). Structure of wheat serine carboxypeptidase II at 3.5-Å resolution. A new class of serine proteinase. J. Biol. Chem. 265, 6528–6531. [DOI] [PubMed] [Google Scholar]
- Liao, D.-I., Breddam, K., Sweet, R.M., Bullock, T., and Remington, S.J. (1992). Refined atomic model of wheat serine carboxypeptidase II at 2.2-Å resolution. Biochemistry 31, 9796–9812. [DOI] [PubMed] [Google Scholar]
- Lorenzen, M., Racicot, V., Strack, D., and Chapple, C. (1996). Sinapic acid ester metabolism in wild type and a sinapoylglucose-accumulating mutant of Arabidopsis. Plant Physiol. 112, 1625–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs, K.A., Alfenito, M.R., Lloyd, A.M., and Walbot, V. (1995). A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 375, 397–400. [DOI] [PubMed] [Google Scholar]
- Mehta, R.A., and Mattoo, A.K. (1996). Isolation and identification of ripening-related tomato fruit carboxypeptidase. Plant Physiol. 110, 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, R.A., Warmbardt, R.D., and Mattoo, A.K. (1996). Tomato fruit carboxypeptidase. Properties, induction upon wounding, and immunocytochemical localization. Plant Physiol. 110, 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, K., Leube, M.P., and Grill, E. (1994). A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264, 1452–1455. [DOI] [PubMed] [Google Scholar]
- Meyer, K., Benning, G., and Grill, E. (1996). Cloning of plant genes based on genetic map location. In Genome Mapping in Plants, A.H. Paterson, ed (New York: Academic Press, and Austin, TX: Landes Bioscience Publishers), pp. 137–154.
- Michalczuk, L., and Bandurski, R.S. (1982). Enzymic synthesis of 1-O-indol-3-ylacetyl-β-D-glucose and indol-3-ylacetyl-myo-inositol. Biochem. J. 207, 273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock, H.-P., and Strack, D. (1993). Energetics of the uridine 5′-diphosphoglucose:hydroxycinnamic acid acyl-glucosyltransferase reaction. Phytochemistry 32, 575–579. [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Nurmann, G., and Strack, D. (1979). Sinapine esterase. I. Characterization of sinapine esterase from cotyledons of Raphanus sativus. Z. Naturforsch. 34c, 715–720. [Google Scholar]
- Ramos, C., and Winther, J.R. (1996). Exchange of regions of the carboxypeptidase Y propeptide. Sequence specificity and function in folding in vivo. Eur. J. Biochem. 242, 29–35. [DOI] [PubMed] [Google Scholar]
- Ramos, C., Winther, J.R., and Kielland-Brandt, M.C. (1994). Requirement of the propeptide for in vivo formation of active yeast carboxypeptidase Y. J. Biol. Chem. 269, 7006–7012. [PubMed] [Google Scholar]
- Rogers, S.O., and Bendich, A.J. (1985). Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 5, 69–76. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Meyer, K., Cusumano, J.C., and Chapple, C. (1999). Regulation of ferulate-5-hydroxylase expression in Arabidopsis in the context of sinapate ester biosynthesis. Plant Physiol. 119, 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Plainview, NY: Cold Spring Harbor Laboratory Press).
- Sharma, V., and Strack, D. (1985). Vacuolar localization of 1-sinapoylglucose: l-malate sinapoyltransferase in protoplasts from cotyledons of Raphanus sativus. Planta 163, 563–568. [DOI] [PubMed] [Google Scholar]
- Strack, D. (1982). Development of 1-O-sinapoyl-β-d-glucose:l-malate sinapoyltransferase activity in cotyledons of red radish (Raphanus sativus L. var. sativus). Planta 155, 31–36. [DOI] [PubMed] [Google Scholar]
- Strack, D., and Sharma, V. (1985). Vacuolar localization of the enzymatic synthesis of hydroxycinnamic acid esters of malic acid in protoplasts from Raphanus sativus leaves. Physiol. Plant. 65, 45–50. [Google Scholar]
- Strack, D., Nurmann, G., and Sachs, G. (1980). Sinapine esterase. II. Specificity and change of sinapine esterase activity during germination of Raphnaus sativus. Z. Naturforsch. 35c, 963–966. [Google Scholar]
- Strack, D., Knogge, W., and Dahlbender, B. (1983). Enzymatic synthesis of sinapine from 1-O-sinapoyl-β-d-glucose and choline by a cell-free system from developing seeds of red radish (Raphanus sativus L. var. sativus). Z. Naturforsch. 38c, 21–27. [Google Scholar]
- Valls, L.A., Winther, J.R., and Stevens, T.H. (1990). Yeast carboxypeptidase Y vacuolar targeting signal is defined by four propeptide amino acids. J. Cell Biol. 111, 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas, R.J.A., and Kojima, M. (1986). Purification and characterization of hydroxycinnamoyl d-glucose quinate hydroxycinnamoyl transferase in the root of sweet potato, Ipomoea batatas Lam. J. Biol. Chem. 261, 8729–8733. [PubMed] [Google Scholar]
- Vogt, T., Aebershold, R., and Ellis, B. (1993). Purification and characterization of sinapine synthase from seeds of Brassica napus. Arch. Biochem. Biophys. 300, 622–628. [DOI] [PubMed] [Google Scholar]
- Wajant, H., Mundry, K.-W., and Pfizenmaier, K. (1994). Molecular cloning of hydroxynitrile lyase from Sorghum bicolor (L.). Homologies to serine carboxypeptidases. Plant Mol. Biol. 26, 735–746. [DOI] [PubMed] [Google Scholar]
- Walker-Simmons, M., and Ryan, C.A. (1980). Isolation and properties of carboxypeptidase from leaves of wounded tomato plants. Phytochemistry 19, 43–47. [Google Scholar]
- Washio, K., and Ishikawa, K. (1994). Cloning and sequencing of the gene for type I carboxypeptidase in rice. Biochim. Biophys. Acta 1199, 311–314. [DOI] [PubMed] [Google Scholar]
- Widmer, F., and Johansen, J.T. (1979). Enzymatic peptide synthesis. Carboxypeptidase Y catalyzed formation of peptide bonds. Carlsberg Res. Commun. 44, 37–46. [Google Scholar]
- Widmer, F., Breddam, K., and Johansen, J.T. (1980). Carboxypeptidase Y catalyzed peptide synthesis using amino acid alkyl esters as amine components. Carlsberg Res. Commun. 45, 453–463. [Google Scholar]
- Wolf, A.E., Dietz, K.-J., and Schröder, P. (1996). Degradation of glutathione S-conjugates by a carboxypeptidase in the plant vacuole. FEBS Lett. 384, 31–34. [DOI] [PubMed] [Google Scholar]
- Zambrisky, P., Joos, H., Genetello, C., Leemans, J., van Montagu, M., and Schell, J. (1983). Ti plasmid vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity. EMBO J. 2, 2143–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]