Abstract
A new subfamily of sucrose transporters from Arabidopsis (AtSUT4), tomato (LeSUT4), and potato (StSUT4) was isolated, demonstrating only 47% similarity to the previously characterized SUT1. SUT4 from two plant species conferred sucrose uptake activity when expressed in yeast. The Km for sucrose uptake by AtSUT4 of 11.6 ± 0.6 mM was ∼10-fold greater than for all other plant sucrose transporters characterized to date. An ortholog from potato had similar kinetic properties. Thus, SUT4 corresponds to the low-affinity/high-capacity saturable component of sucrose uptake found in leaves. In contrast to SUT1, SUT4 is expressed predominantly in minor veins in source leaves, where high-capacity sucrose transport is needed for phloem loading. In potato and tomato, SUT4 was immunolocalized specifically to enucleate sieve elements, indicating that like SUT1, macromolecular trafficking is required to transport the mRNA or the protein from companion cells through plasmodesmata into the sieve elements.
INTRODUCTION
The reduced carbon produced through photosynthesis in mature leaves is distributed by the vascular system, mainly in the form of sucrose, to support the growth of heterotrophic (sink) tissues such as developing leaves, the shoot apex, roots, and reproductive organs. Within the vascular tissue, the sieve elements in the phloem form the conduits for long-distance transport. Sieve elements are highly specialized, lacking many organelles (including a nucleus and vacuole) at maturity, and hence depend on tightly associated companion cells for metabolic support (Sjölund, 1997). The loading of sucrose into the sieve element/companion cell (SE/CC) complex in many plants requires the active uptake of sucrose from the extracellular space. Because of variability in the rate of photosynthesis according to changes in environmental conditions, and because sink demands change depending on development and external factors, we can reasonably assume that the rate of phloem loading of sucrose is regulated. In fact, the phenotype of transgenic plants overexpressing pyruvate decarboxylase indicates that sugar export from potato leaves can be upregulated by as much as 10-fold (Tadege et al., 1998). The increase in sucrose transport activity caused by modification of a conserved histidine in the first external loop (Lu and Bush, 1998) indicates that sucrose transporters may be directly regulated at the protein level. In addition, the amounts of mRNA for sucrose transporter SUT1 from potato are developmentally controlled and hormonally regulated (Riesmeier et al., 1993; Harms et al., 1994).
Clearly, multiple kinetic components of sucrose uptake are present in leaves (Delrot and Bonnemain, 1981; Maynard and Lucas, 1982). As demonstrated by autoradiography, 14C-sucrose, externally applied to source leaves of Vicia faba or Beta vulgaris, is taken up by mesophyll cells and phloem (Fondy and Geiger, 1977; Giaquinta, 1977; Delrot, 1981). The overall Km for sucrose uptake into leaves is pH dependent, with greater affinity being measured at pH 5.0 (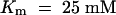 ) than at pH 8.0 (
) than at pH 8.0 (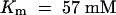 ) (Giaquinta, 1977). In V. faba leaves, at least two saturable sucrose uptake components have been described: one high-affinity/low-capacity (HALC) system with a Km of 2.7 mM and a Vmax of 0.71 nmol cm−2 min−1 and one low-affinity/high-capacity (LAHC) system with a Km of 25.8 mM and a Vmax of 3.6 nmol cm−2 min−1 (Delrot and Bonnemain, 1981). The properties of SUT1 fit with those of the HALC system, whereas transporters representing the LAHC system have not been identified.
) (Giaquinta, 1977). In V. faba leaves, at least two saturable sucrose uptake components have been described: one high-affinity/low-capacity (HALC) system with a Km of 2.7 mM and a Vmax of 0.71 nmol cm−2 min−1 and one low-affinity/high-capacity (LAHC) system with a Km of 25.8 mM and a Vmax of 3.6 nmol cm−2 min−1 (Delrot and Bonnemain, 1981). The properties of SUT1 fit with those of the HALC system, whereas transporters representing the LAHC system have not been identified.
All plant sucrose transporters isolated thus far are encoded by the SUT gene family (reviewed in Lalonde et al., 1999). The first SUT cDNA clones were isolated from spinach and potato by expression cloning in yeast (Riesmeier et al., 1992, 1993). SUT1 and closely related homologs from various plants encode high-affinity proton-coupled sucrose transporters with a Km in the range of 0.3 to 2 mM (Riesmeier et al., 1992, 1993; Gahrtz et al., 1994; Sauer and Stolz, 1994; Boorer et al., 1996; Weig and Komor, 1996; Hirose et al., 1997; Zhou et al., 1997; Shakya and Sturm, 1998). Its expression in Xenopus oocytes showed StSUT1 to have a 1:1 transport stoichiometry for sucrose:H+ (Boorer et al., 1996) and a slow transport rate with a rate-limiting step of ∼5 sec−1 (Boorer et al., 1996). The Km for sucrose of StSUT1 and AtSUC1 is pH dependent with higher affinity at more acidic external pH (Boorer et al., 1996; Zhou et al., 1997). This is consistent with the pH dependence of the Km for whole-leaf sucrose uptake (Giaquinta, 1977). However, the measurement of both low- and high-affinity saturable sucrose transport in leaf discs under conditions in which the external pH is buffered (Delrot and Bonnemain, 1981) indicates the presence of two or more distinct transport activities.
In tobacco and potato, the severe phenotype of plants expressing SUT1 genes in antisense orientation has been interpreted to indicate that high-affinity sucrose uptake into the SE/CC complex is required for phloem loading of sucrose (Riesmeier et al., 1994; Kühn et al., 1996; Lemoine et al., 1996; Bürkle et al., 1998). However, phloem transport requires loading sucrose into minor veins as well as the operation of retrieval mechanisms along the transport pathway. SUT1, which is expressed all along this pathway and has high affinity for sucrose, might be essential for maintaining the sucrose gradient and thereby controlling the rate of phloem translocation. High-affinity sucrose transport is especially important for retrieval, as suggested by Maynard and Lucas (1982). In contrast, one would expect the LAHC transporter to be present only in zones with high rates of phloem loading, that is, in minor veins. This process may be similar to glucose uptake in kidney, in which high-capacity uptake is mediated by the Na+ glucose cotransporter (SGLT2) and high-affinity transport is mediated by SGLT1 with an ∼10-fold lower Km for glucose compared to SGLT2 (Mackenzie et al., 1996).
In this study, a novel low-affinity sucrose transporter from Arabidopsis and its orthologs from tomato and potato were isolated. In agreement with the transporter having a function in phloem loading, expression of this transporter in source leaves was restricted to minor veins of Arabidopsis, indicating functional differentiation of the phloem. Expression was also found in sink tissues, where SUT4 may have a role in high-capacity sucrose uptake. By immunofluorescence, SUT4 protein was localized specifically to sieve elements in tomato and potato. Because mature sieve elements do not contain nuclei, transcription must take place in companion cells, and macromolecular trafficking is required to incorporate the sucrose transporter into the plasma membrane of the sieve element. Depending on the sucrose concentrations in the extracellular space as well as within the SE/CC complex, transporters of different affinities may be expressed to optimize the capacity and affinity of sucrose uptake into the sieve elements, both in the loading zone and along the path, as proposed by Lalonde et al. (1999).
RESULTS
Isolation of Sucrose Transporter cDNAs
Physiological studies indicate that plants contain multiple sucrose transport activities. HALC sucrose transporters have been identified from a broad range of plants; except for sucrose binding protein (Overvoorde et al., 1996), however, genes encoding potential LAHC sucrose transporters have not been identified. To determine whether more distant homologs of high-affinity sucrose transporters, with distinct functions, exist in plants, a tomato cDNA library was screened with a fragment of the genomic clone of NtSUT3, a tobacco sucrose transporter expressed specifically in pollen (Lemoine et al., 1999). Eighteen clones were isolated, 15 corresponding to LeSUT1 (GenBank accession number X82275) and three representing full-length cDNAs from a novel gene subsequently named LeSUT4 (GenBank accession number AF176950). The deduced amino acid sequence of LeSUT4 is 47% identical to that of LeSUT1. Therefore, LeSUT4 belongs to a new subfamily within the SUT family of sucrose transporters (reviewed in Ward et al., 1998). The LeSUT4 sequence (GenBank accession number AF176950) was also used to isolate an ortholog from potato: StSUT4 (GenBank accession number AF237780) is 97.8% identical at the amino acid level to LeSUT4 (Figure 1). Database analysis revealed a closely related Arabidopsis gene (AtSUT4) on chromosome 1 (GenBank accession number AC000132) with 82% homology (67% identity) to LeSUT4 at the amino acid level. Corresponding cDNA clones were isolated from Arabidopsis by the polymerase chain reaction (PCR). All three proteins—LeSUT4, StSUT4, and AtSUT4—are highly related and contain 12 predicted transmembrane domains with highest sequence identity within the membrane spans.
Figure 1.
Alignment and Structural Prediction of SUT4 Proteins from Tomato, Potato, and Arabidopsis.
The deduced amino acid sequences for AtSUT4 from Landsberg erecta ecotype (GenBank accession number AF175321), LeSUT4 (AF176950), and StSUT4 (AF237780) were aligned by using the Megalign program (DNASTAR, Madison, WI). Amino acids identical to the LeSUT4 sequence are shaded. Locations of 12 potential membrane-spanning domains (TM1 to TM12) as predicted by the TM-Pred program (Hofmann and Stoffel, 1993) are indicated.
Functional Analysis of AtSUT4 in Yeast
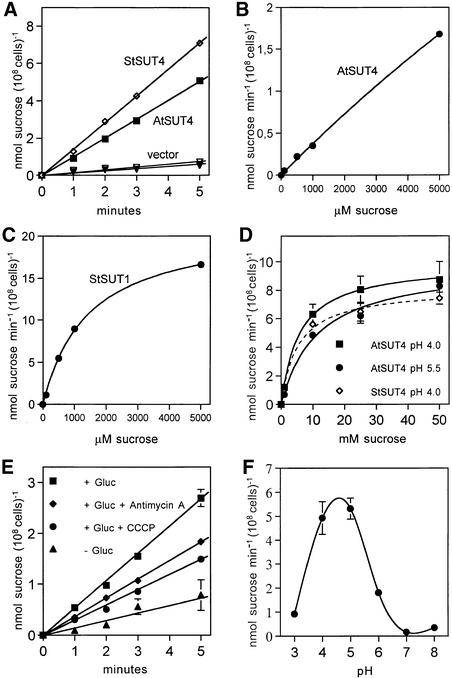
For testing the function of AtSUT4, a modified version of the SUSY7 yeast strain (Riesmeier et al., 1992) allowing selection for URA3-containing plasmids (SUSY7/ura3) was used (Barker et al., 2000). On media containing sucrose as the sole carbon source, SUSY7/ura3 transformed with StSUT1 grows better (Figure 2A; Riesmeier et al., 1994) than does yeast transformed with the empty pDR196 vector (Figure 2C). Expression of AtSUT4 allowed yeast growth on sucrose (Figure 2B), providing the first indication that AtSUT4 encodes a functional sucrose transporter. Uptake experiments with yeast expressing each of the three transporters (AtSUT4, StSUT4, or LeSUT4) revealed that both AtSUT4 and StSUT4 were functional sucrose transporters, whereas LeSUT4 was not functional. The time course of 14C-sucrose uptake by SUSY7/ura3 expressing AtSUT4 or StSUT4 is shown in Figure 3A. The uptake rate of 14C-sucrose was linear and markedly higher than that of the vector controls. Sucrose-uptake rates for AtSUT4 did not saturate at low sucrose concentrations (Figure 3B) in contrast to the rates for the high-affinity sucrose transporter StSUT1 (Figure 3C). Kinetic analysis of 14C-sucrose uptake by SUSY7/ura3 expressing AtSUT4 revealed a Km for sucrose of 11.6 ± 0.6 mM (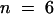 ) at pH 5.5 and 5.9 ± 0.8 mM (
) at pH 5.5 and 5.9 ± 0.8 mM (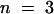 ) at pH 4.0 (Figure 3D). StSUT4 had a similar low affinity, with a Km of 6.0 ± 1.2 mM (
) at pH 4.0 (Figure 3D). StSUT4 had a similar low affinity, with a Km of 6.0 ± 1.2 mM (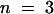 ) at pH 4.0. In comparison, the Km values for sucrose of AtSUC1 and AtSUC2 from Arabidopsis, when expressed in yeast and assayed at pH 5.5, are 0.5 and 0.7 mM, respectively (Sauer and Stolz, 1994). The Km for sucrose of StSUT1 from potato expressed in Xenopus oocytes is also pH dependent (Boorer et al., 1996), giving lower Km values at more acidic pH. At pH 5.0, the Km of StSUT1 for sucrose is 0.5 mM. 14C-sucrose uptake via AtSUT4 was stimulated by glucose and inhibited by the electron transport inhibitor antimycin A and the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP) (Figure 3E). These data are consistent with a proton-coupled sucrose uptake mechanism. Uptake of 14C-sucrose by way of AtSUT4 is highly pH dependent, with an optimum at pH 4.0 to 5.0 (Figure 3F).
) at pH 4.0. In comparison, the Km values for sucrose of AtSUC1 and AtSUC2 from Arabidopsis, when expressed in yeast and assayed at pH 5.5, are 0.5 and 0.7 mM, respectively (Sauer and Stolz, 1994). The Km for sucrose of StSUT1 from potato expressed in Xenopus oocytes is also pH dependent (Boorer et al., 1996), giving lower Km values at more acidic pH. At pH 5.0, the Km of StSUT1 for sucrose is 0.5 mM. 14C-sucrose uptake via AtSUT4 was stimulated by glucose and inhibited by the electron transport inhibitor antimycin A and the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP) (Figure 3E). These data are consistent with a proton-coupled sucrose uptake mechanism. Uptake of 14C-sucrose by way of AtSUT4 is highly pH dependent, with an optimum at pH 4.0 to 5.0 (Figure 3F).
Figure 2.
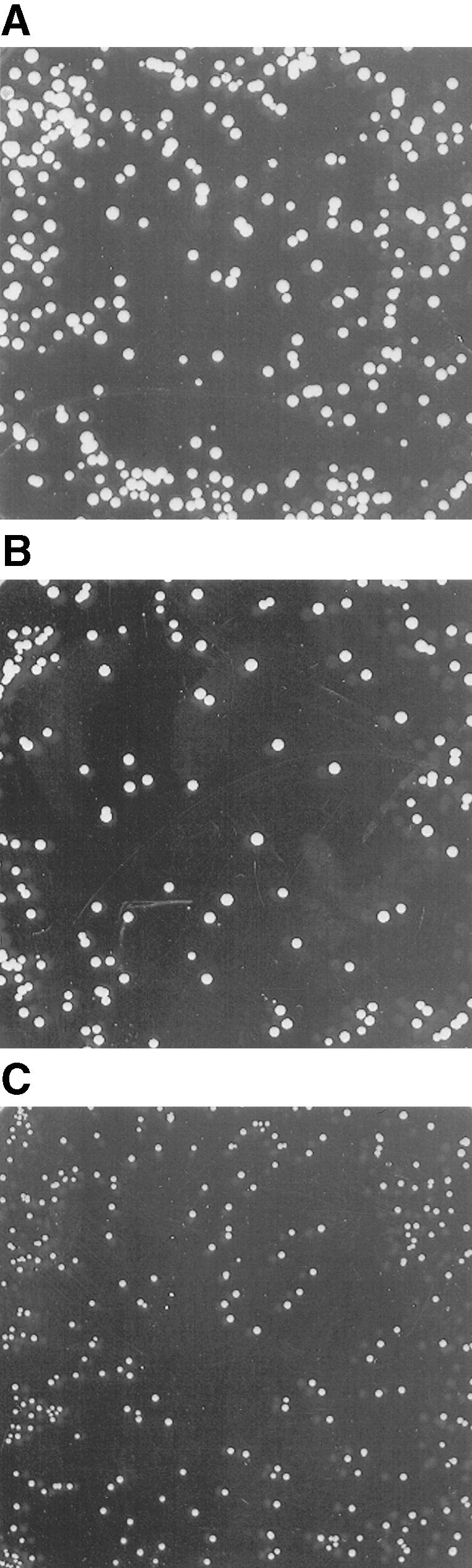
Expression of AtSUT4 in the Yeast Strain SUSY7/ura3 Allows Growth on Sucrose as the Sole Carbon Source.
(A) SUSY7/ura3 yeast transformed with StSUT1 in the vector pDR195.
(B) SUSY7/ura3 yeast transformed with AtSUT4 in the vector pDR196.
(C) SUSY7/ura3 yeast transformed with the empty pDR196 vector.
In (A) to (C), yeast were plated on synthetic minimal media containing 2% (w/v) sucrose at pH 4.0 and 20 mg L−1 tryptophan and grown for 10 days at 28°C.
Figure 3.
Kinetic Analysis of 14C-Sucrose Uptake by Yeast Strain SUSY7/ura3 Expressing AtSUT4.
(A) Time course of sucrose uptake. SUSY7/ura3 yeast transformed with StSUT4 in pDR195, AtSUT4 in pDR196, or empty pDR195/pDR196 vector were assayed for 14C-sucrose uptake at 1 mM sucrose and pH 4.0.
(B) Rate of sucrose uptake by SUSY7/ura3 expressing AtSUT4. Transport assays were performed at pH 5.5, and uptake rates are plotted against final sucrose concentrations in the assays. Background uptake rates (empty vector) were subtracted.
(C) Sucrose-uptake rate of SUSY7/ura3 expressing StSUT1, performed as in (B).
(D) Sucrose-uptake kinetics of SUSY7/ura3 expressing AtSUT4 or StSUT4 from experiments as described in (A). Data are presented as the mean ±se for three independent transformants. A nonlinear regression fit of the Michaelis–Menten equation is shown: for AtSUT4 at pH 5.5, 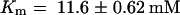 ,
, 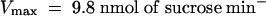 1
1 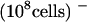 1 (
1 (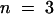 ); at pH 4.0,
); at pH 4.0, 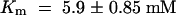 ,
, 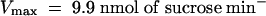 1
1  1 (
1 ( ). For StSUT4 at pH 4.0,
). For StSUT4 at pH 4.0, 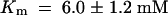 ,
, 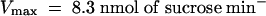 1
1  1 (
1 ( ).
).
(E) Uptake of 14C-sucrose by SUSY7/ura3 expressing AtSUT4 is inhibited by addition of 10 μM antimycin A or 2.5 μM CCCP. Inhibitors were added 1 min before the addition of sucrose to the assay. (+) or (−) Gluc, with or without glucose. Data are presented as mean ±se, 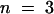 .
.
(F) pH dependence of sucrose uptake by SUSY7/ura3 expressing AtSUT4. Experiments were performed at 1 mM sucrose; rates are presented as means ±se ( ).
).
Competition by other sugars with 14C-sucrose uptake by AtSUT4 indicated that like SoSUT1 (Riesmeier et al., 1992), AtSUC1, and AtSUC2 (Sauer and Stolz, 1994), the only physiological sugar to compete effectively was maltose (data not shown). To test directly whether maltose was a substrate for transport, the kinetics of 14C-maltose uptake were measured. The Km for maltose uptake at pH 4.0 was 13.1 mM and the Vmax was 4.6 nmol sucrose min−1 (108 cells)−1, which is approximately half (46%) of the transport rate for sucrose (Figure 3D).
Analysis of AtSUT4 Expression in Arabidopsis
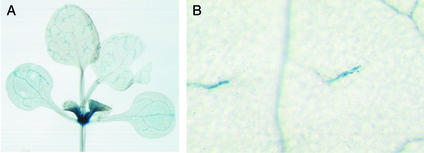
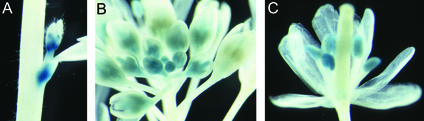
Promoter fragments of 2.2 and 3.1 kb, upstream of the translation start of AtSUT4 from Arabidopsis, were fused transcriptionally to the β-glucuronidase (GUS) gene. The GUS expression pattern for both constructs was identical, with predominant staining in sink tissues (Figure 4). GUS activity was greatest in the sink leaves (Figure 4A); after transition to source leaves, its expression was restricted to minor veins (Figures 4A and 4B). These data indicate a functional difference between phloem in the minor veins and in other veins in the leaf. In contrast to SUT1 from solanaceous species and AtSUC2 from Arabidopsis (expressed in all veins in source leaves), AtSUT4 is expressed only in the loading zone (minor veins) of source leaves. In the inflorescence, GUS activity was found in axils of branches (Figure 5A) and within developing flowers in the anther and pistil (Figures 5B and 5C). At anthesis, staining in the flowers was restricted to anthers (Figure 5C).
Figure 4.
Arabidopsis Plants Transformed with AtSUT4 Promoter GUS Constructs and Stained with 5-Bromo-4-Chloro-3-Indolyl β-d-Glucopyranoside.
(A) Eighteen-day-old plant showing minor vein staining in source leaves and strong sink leaf staining.
(B) Minor vein staining in 18-day-old plant stained with 5-bromo-4-chloro-3-indolyl β-d-glucopyranoside and ferricyanide.
Plants were transformed with a 2.2-kb promoter construct (see Methods) and grown in soil in a greenhouse.
Figure 5.
Arabidopsis Plants Expressing GUS Driven by an ATSUT4 Promoter.
(A) Staining of an axil of a branch in the inflorescence.
(B) An inflorescence showing staining in younger flowers.
(C) Older flowers showed staining in anthers and less in the pistil and filaments.
Plants were transformed with a 3.1-kb promoter construct (see Methods) and grown in the greenhouse for 4 to 5 weeks.
Expression of SUT Genes in Tomato
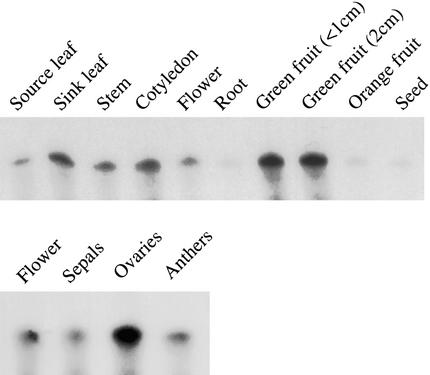
RNA gel blot analysis was used to investigate the organ-specific expression of LeSUT4. Although the signal was at the limits of detection, LeSUT4 expression nonetheless appeared to be greatest in sink leaves, stems, and green fruit (data not shown). To increase the sensitivity of detection, RNase protection assays (RPA) were performed. Consistent with the RNA gel blots, LeSUT4 expression was greatest in sink leaves, stems, cotyledons, and immature fruit (Figure 6). These data correlate with the expression pattern of AtSUT4 in Arabidopsis, as determined by promoter GUS analysis (Figures 4 and 5). As in Arabidopsis, expression was relatively strong in cotyledons and sink leaves of tomato and low in source leaves (Figure 6).
Figure 6.
Analysis of LeSUT4 Expression in Tomato by RNase Protection Analysis of the Organ-Specific Expression of the LeSUT4 Gene.
Twenty micrograms of total RNA from each organ was hybridized overnight at 45°C with a 32P-labeled probe. The protected 340-base RNA fragment is shown.
Immunolocalization of SUT4 in Potato and Tomato
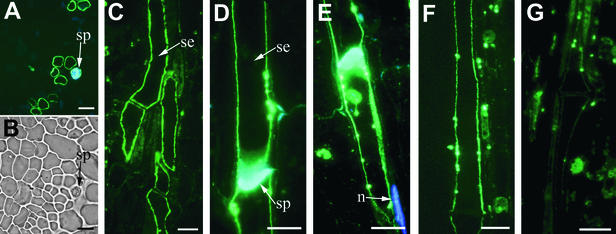
Polyclonal antisera were raised in rabbits by using synthetic peptides corresponding to the sequence within the LeSUT4 central loop. The corresponding region in StSUT4 is 90% identical to LeSUT4, and the anti-LeSUT4 antisera cross-reacted with potato. Immunofluorescent detection of StSUT4 was performed on stem tissue from potato. StSUT4 was detected only in sieve elements (Figures 7A and 7C), recognizable by the presence of sieve plates (Figures 7A and 7B). To be able to compare this localization with the data obtained for LeSUT1 and LeSUT2 from tomato (Kühn et al., 1997; Barker et al., 2000), immunolocalization studies were also performed with the tomato ortholog LeSUT4 (Figures 7D to 7F). LeSUT4 was detected in sieve elements in tomato stem (Figures 7D and 7E) and identified by counterstaining with aniline blue. 4′,6-Diamidino-2-phenylindole (DAPI) staining further differentiated enucleate sieve elements from companion cells (Figure 7E). LeSUT4 was also detected in sieve elements of petioles of source leaves and in the midvein of sink leaves (data not shown). Preimmune serum, purified by using protein A–Sepharose columns and used at the same protein concentration as the affinity-purified anti-LeSUT4 antisera, gave no immunofluorescence signal (data not shown). Incubation with the secondary antibody alone also gave no detectable signal (data not shown). Peptide competition experiments were performed to test antibody specificity (see Methods). Preincubation of anti-LeSUT4 (central loop) antibodies with the peptide used for immunization greatly reduced the fluorescence signal (Figure 7G), whereas use of equal concentrations of an unrelated control peptide had no effect (Figure 7F).
Figure 7.
Immunocytochemical Localization of SUT4 Protein in Sieve Elements of Potato and Tomato.
Sections (1 μm thick) of stem were treated with anti-SUT4 polyclonal antiserum; antibody binding was detected with an anti–rabbit IgG–fluorescein isothiocyanate conjugate.
(A) Cross-section of unicryl-embedded potato stem immunostained with anti-SUT4 and counterstained with aniline blue. sp, sieve plate.
(B) Phase-contrast image of the section in (A).
(C) Longitudinal section of potato stem immunostained with anti-SUT4. se, sieve element.
(D) Longitudinal section of tomato stem immunostained with anti-SUT4 and counterstained with aniline blue.
(E) Longitudinal section of tomato stem immunostained with anti-SUT4 and counterstained with aniline blue and DAPI. Companion cell adjacent to the sieve element contains a DAPI-stained nucleus (n).
(F) Longitudinal section of tomato stem preincubated with nonspecific control peptide and immunostained with anti-SUT4.
(G) Longitudinal section of tomato stem preincubated with specific central loop peptide used to generate the antisera and then immunostained with anti-SUT4.
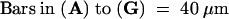 .
.
Specificity of the antisera was verified with yeast cells expressing LeSUT1 or LeSUT4. In protein blots, LeSUT4 was detected as a single specific band at a molecular mass of 46 kD (data not shown). No cross-reaction of anti-LeSUT4 sera with LeSUT1 was detected, and no bands were detected by protein blots of membranes isolated from yeast transformed with empty vector. Protein blot also detected no reaction with membranes isolated from tomato leaves, probably as a result of low and cell-specific expression, consistent with results from RPA analysis. The ability of the anti-LeSUT4 sera to recognize the protein after fixation was confirmed by immunostaining yeast spheroplasts that expressed LeSUT4 (data not shown).
DISCUSSION
Different Functions for Members of the SUT Family
Long-distance transport of sucrose from source to sink tissue is a key determinant for crop yield. However, the mechanism and regulation of sucrose loading in source leaves and unloading in sink tissues are not fully understood. Several different sucrose transport activities are thought to be required for phloem function (reviewed in Lalonde et al., 1999), including sucrose exporters, uptake transporters, and vacuolar transporters. To date, only members of the SUT family have been isolated, and these have been characterized as proton-coupled sucrose uptake transporters (Riesmeier et al., 1992). Despite having highly similar transport activities, members of the SUT family have diverse functional roles, including the loading of sucrose into phloem at the sieve elements (Kühn et al., 1997) or in companion cells (Stadler and Sauer, 1996), as well as sucrose uptake into sink cells such as seeds (Weber et al., 1997; Tegeder et al., 1998) and pollen (Lemoine et al., 1999; Stadler et al., 1999). All members of the SUT family characterized thus far catalyze high-affinity sucrose uptake with measured Km values for sucrose between 0.3 and 2 mM (Riesmeier et al., 1992, 1993; Gahrtz et al., 1994, 1996; Sauer and Stolz, 1994; Boorer et al., 1996; Weig and Komor, 1996; Weber et al., 1997; Zhou et al., 1997; Shakya and Sturm, 1998). Thus, SUT1 corresponds to the HALC sucrose transport system described in leaves of V. faba (Delrot and Bonnemain, 1981) and B. vulgaris (Maynard and Lucas, 1982). The high-affinity transporters SUT1 and AtSUC2 are expressed not only in minor veins but also in the efferent veins, in the stem, and even in the sieve elements of sink tissues (Truernit and Sauer, 1995; A. Weise, J.M. Ward, and W.B. Frommer, unpublished data). Blocking the HALC system inhibits long-distance transport of sucrose and allocation of assimilate (Riesmeier et al., 1994; Bürkle et al., 1998).
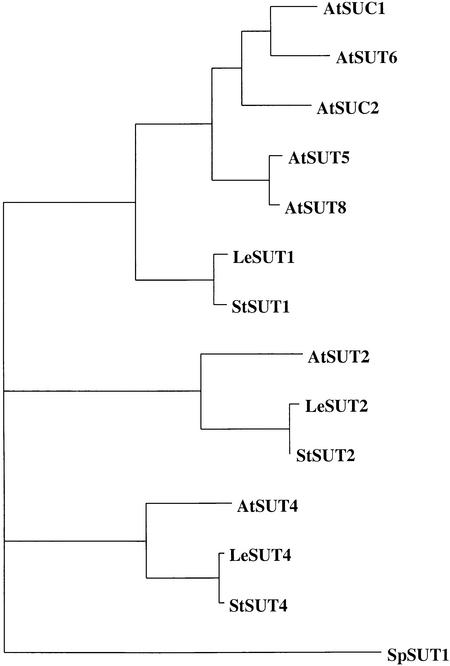
A saturable, LAHC sucrose uptake system has been studied in leaves (Delrot and Bonnemain, 1981), but no gene has been assigned to this function. In this study, representatives of a novel and only distantly related subfamily of sucrose transporters, SUT4, were identified from Arabidopsis, potato, and tomato. A phylogenetic tree of all sucrose transporter homologs from Arabidopsis, tomato, and potato identified to date is presented in Figure 8. Because expression of AtSUT4 and StSUT4 in yeast conferred low-affinity saturable sucrose uptake activity, SUT4 appears to represent a LAHC sucrose transport system. Direct evidence links a low affinity for sucrose with high-capacity transport within the SUT family; that is, all mutations in histidine-65 of AtSUC1, a high-affinity sucrose transporter, that caused an increase in Km for sucrose also resulted in higher Vmax (Lu and Bush, 1998). However, the transport rate of SUT4 relative to SUT1 is not known. One way to test this hypothesis would be to compare particle density (determined by freeze-fracture electron microscopy) with transport rates after expression in Xenopus oocytes (Eskandari et al., 1998).
Figure 8.
Phylogenetic Analysis of Sucrose Transporter Homologs from Arabidopsis, Tomato, and Potato.
Bootstrap analysis (Felsenstein, 1985) was performed on aligned protein sequences of AtSUC1 (GenBank accession number X75365), AtSUC2 (X75382), AtSUT2 (AC004138), AtSUT4 (AF175321), AtSUT5 (AB016875), AtSUT6 (AC021665), AtSUT8 (AC005398), LeSUT1 (X82275), LeSUT2 (AF166498), LeSUT4 (AF176950), StSUT1 (X69165), StSUT2 (L. Barker and W.B. Frommer, unpublished data), StSUT4 (AF237780), and SpSUT1 (A. Reinders and J.M. Ward, unpublished data). The SpSUT1 sequence from Schizosaccharomyces pombe was included to root the tree.
SUT4 Expression and Function in Plants
The AtSUT4 promoter is active in minor veins in source leaves. This indicates a functional difference between the phloem in minor veins and in lower-order veins. High fluxes of sucrose are expected to occur in minor veins, the major site of phloem loading. Thus, LAHC transport activity may be specifically important for phloem loading, whereas high-affinity transporters may have an additional role in maintaining the sucrose gradient along the transport pathway. Phloem loading of sucrose is a regulated process, and LAHC transport may be important under conditions in which loading rates are increased. For example, in Arabidopsis, an increase in the rate of sucrose export from source leaves precedes and is required for the onset of flowering (Corbesier et al., 1998).
In Arabidopsis and tomato, SUT4 is also expressed in sink tissues (in sink leaves, flowers, and fruit). It is reasonable that high rates of sucrose uptake into sink cells would require a LAHC transport system. It is unlikely that in AtSUT4 promoter–GUS plants, the GUS accumulation in sink tissues represents GUS movement in the phloem. Imlau et al. (1999) showed that in AtSUC2 promoter–green fluorescent protein (GFP) plants, GFP accumulated in sink tissues. In AtSUC2 promoter–GUS plants, however, GUS protein was restricted to the phloem (Truernit and Sauer, 1995), possibly because GUS is larger (68 kD) than GFP (27 kD). In addition, the GUS expression in Arabidopsis driven by the AtSUT4 promoter is consistent with the LeSUT4 expression pattern in tomato shown by RNA expression analyses. In sink tissues, SUT4 could have the following functions: if SUT4 is expressed in sieve elements in sink tissue, then it could function in regulating extracellular sucrose by way of reuptake; or if SUT4 is expressed outside of the phloem, it could function directly in sucrose uptake into sink cells and have a role in determining sink strength.
In solanaceous species (e.g., potato, tomato, and tobacco), SUT1 protein has been localized to enucleate sieve elements of mature leaves and stems (Kühn et al., 1997). Immunolocalization of SUT4 in potato and tomato provides the second example of a transporter protein localized to the enucleate sieve elements. This raises the possibility that the two transporters, despite their difference in primary sequence, utilize a similar targeting mechanism to deliver the mRNA or protein to the sieve elements. Components of the mRNA transport machinery have been identified: the RNA binding protein CmPP16 has been shown to increase SUT1 mRNA mobility between mesophyll cells (Xoconostle-Cazares et al., 1999). However, structural determinants of mRNA that allow for cell-to-cell movement have not been demonstrated in plants.
Recently, Barker et al. (2000) identified a third sucrose transporter–like protein, SUT2 (Figure 8), with the features of a sugar sensor. This protein also has been localized to sieve elements, thus providing a third gene that may help to delineate cis-sequences involved in macromolecular trafficking. The following model is proposed for the function of the three SUT proteins in the plasma membrane of sieve elements: SUT2 functions as a receptor for extracellular sucrose and regulates the relative activities of the high-affinity SUT1 transporter and the low-affinity SUT4 transporter, either by controlling protein turnover or through signal transduction, resulting in transcriptional activation/repression in the companion cell.
In conclusion, differentially regulated expression of LAHC and HALC sucrose transporters in sieve elements could provide a mechanism for plants to regulate the rate of export of sucrose from source leaves both at the level of phloem loading (LAHC and SUT4) and in long-distance transport (HAL and SUT1). Transgenic plants and knock-out mutants affected in transport and sensing of sucrose will help to unravel this complex process.
METHODS
Isolation of SUT4 cDNAs
A tomato (Lycopersicon esculentum cv UC82b) cDNA library derived from flowers was screened with a 300-bp EcoRI-BglII fragment of the genomic clone of tobacco NtSUT3 (Lemoine et al., 1999) under reduced stringency. Three independent clones that were distinct from LeSUT1 were isolated and named LeSUT4 (GenBank accession number AF176950). The orthologous sequence isolated from potato by reverse transcription–polymerase chain reaction (PCR) with primers derived from the LeSUT4 sequence was named StSUT4 (GenBank accession number AF237780). StSUT4 was cloned as a PstI-NotI fragment into pBC SK− (Stratagene). It was then subcloned as an Xho-SacII fragment into the yeast expression vector pDR195 that contains a URA3 marker, a PMA1 promoter, and an ADH1 terminator (Rentsch et al., 1995). A closely related Arabidopsis thaliana genomic sequence was identified (GenBank accession number AC000132). A cDNA was amplified from an Arabidopsis seedling library (Minet et al., 1992) by PCR. Primers were designed on the basis of genomic sequence (5′-gactctgcagcgagaaatggctacttccg, 5′-taacctgcaggagaa-tctcatgggagagg). Each primer contains a PstI restriction site (underlined) and is designed to amplify the entire coding sequence of AtSUT4. The product of the expected size (1566 bp) was digested with PstI and ligated into the PstI site of vector pBC SK+ (Stratagene). AtSUT4 was subcloned into the PstI site of pDR196 (D. Rentsch, unpublished data). The AtSUT4 cDNA in pDR196 was sequenced in both directions (GenBank accession number AF175321). Ecotype differences between the SUT4 gene in Columbia (Col-0) and Landsberg erecta were confirmed by sequencing AtSUT4 from the latter strain (GenBank accession number AF175322).
Functional Analysis of SUT4
The yeast strain SUSY7/ura3 is a modified version of SUSY7 (Riesmeier et al., 1992); part of the URA3 gene is deleted in this strain, allowing selection for uracil auxotrophy. For assays of yeast growth on sucrose, media contained 1.7 g L−1 yeast nitrogen base without amino acids (Difco), 2% sucrose (Difco), 5 g L−1 ammonium sulfate, 20 mg L−1 tryptophan, and 1.5% agarose (SeaKem LE; FMC Bioproducts, Rockland, ME), adjusted to pH 5.0 with HCl.
For sucrose-uptake assays, yeast was grown to an OD623 of ∼0.8 in liquid minimal media containing glucose. Cells were collected by centrifugation, washed in 25 mM sodium phosphate buffer, pH 5.5, and suspended in the same buffer to an OD623 of 20. Uptake assays were initiated by adding glucose to a final concentration of 10 mM to yeast cells 1 min before adding 14C-sucrose. After incubation at 30°C with shaking, cells were collected (at 1, 2, 3, and 5 min) by vacuum filtration onto glass fiber filters (GF/C; Whatman) and washed twice with 4 mL of ice-cold 10 mM sucrose before radioactivity was determined by liquid scintillation counting. Data for kinetic analysis were analyzed by using Sigma Plot. Nonlinear regression was performed by using the Michaelis–Menten equation. Data for Km are presented as mean ±se.
RNA Isolation and RNase Protection Analysis
RNA was isolated from different organs of greenhouse-grown tomatoes (L. esculentum cv Moneymaker) according to the method of Schwacke et al. (1999). Reverse transcription was performed with the MAXIscript SP6/T7 in vitro Transcription Kit (Ambion Inc., Austin, TX), using α-32P-UTP. A 600-bp PCR product derived from pSport containing the 340-bp LeSUT4 fragment was used as template. The probe was not further purified, and the amount of product hybridized per sample contained 3.0 × 105 counts min−1. Samples were hybridized with 20 μg of RNA overnight at 45°C. After RNA digestion, the protected RNA was resolved on a 5% polyacrylamide gel (13 × 15 cm) at 150 mV. Gels were dried and exposed to x-ray film (Hyperfilm; Amersham).
AtSUT4 Promoter–β-Glucuronidase Constructs
AtSUT4 gene fragments upstream of the translational start codon were isolated by PCR using Arabidopsis Col-0 ecotype genomic DNA as a template and ExTaq (TaKaRa Shuzo Co., Ltd., Shiga, Japan) polymerase. The 2.2-kb fragment was obtained by using the primers forward: 5′-tcccccgggctcgcttcacagtcgtcgtggcgtag and reverse: 5′-acggtcgacagggtcgcatctcgatattatgg (restriction sites underlined). The 3.1-kb fragment was obtained by using the same reverse primer and a second primer (5′-gtttgttgtcgacgggcgaaatctcgcataacttc). Products were digested with SalI and SmaI and ligated into SalI-SmaI–digested pGPTV-HPT (Becker et al., 1992). Agrobacterium tumefaciens GV2260 was used to transform Arabidopsis plants (Col-0) by vacuum infiltration in 0.5 × Murashige and Skoog (1962) medium containing 5% sucrose, 0.05% Silwet, and 44 μM benzylaminopurine. Arabidopsis transformants were selected on sterile media containing 50 μg mL−1 hygromycin. β-Glucuronidase (GUS) staining was performed according to Martin et al. (1992). Tissues were cleared in 70% ethanol at 22°C with shaking. For the 2.2-kb promoter, eight of 12 showed staining; for the 3.1-kb promoter, 14 of 17 showed staining. The pattern was identical in all cases.
Generation of Anti-SUT4 Polyclonal Antisera
Rabbits were immunized with synthetic peptides that were coupled to keyhole limpet hemocyanin and corresponded to the central loop (GSSHTGEEIDESSHGQEEAFLW) of LeSUT4. Affinity purification of the antisera was performed as described previously (Lemoine et al., 1996) by using synthetic peptides coupled to CNBr-activated Sepharose 4B columns (Pharmacia).
Immunolocalization and Microscopy
Fluorescent immunodetection of SUT4 in potato and tomato was performed as previously described (Stadler et al., 1995) with the following modifications. Hand-cut sections (1 mm thick) from tomato or potato stems were fixed overnight under vacuum in Mops buffer (50 mM Mops-NaOH, pH 6.9, 5 mM EGTA, and 2 mM MgCl2) containing 0.1% glutaraldehyde and 6% formaldehyde. After three washes with Mops buffer on ice, the fragments were dehydrated by incubation in an ethanol series followed by two incubations in 96% ethanol. After incubation overnight in a 1:1 solution of ethanol:methacrylate mix (75% [v/v] butylmethacrylate, 25% [v/v] methylmethacrylate, 0.5% benzoine ethyl ether, and 10 mM DTT), the material was embedded in 100% methacrylate mix. Some sections were embedded in Unicryl (British BioCell International, Cardiff, UK), as indicated in the legend to Figure 7. Polymerization took place overnight under UV light (365 nm) at 4°C. Semithin sections (1 μm thick) were mounted on prewarmed Histobond slides (Camon Laborservice GMBH, Wiesbaden, Germany) and dried at 50°C.
For removal of methacrylate from the sections, slides were incubated for 30 sec in acetone, rehydrated by an ethanol series, and blocked for 1 hr with 2% BSA in PBS (100 mM sodium phosphate, pH 7.5, and 100 mM NaCl). Unicryl-embedded sections were incubated in 0.1 M HCl for 10 min and briefly washed with water. After overnight incubation with affinity-purified antibodies, the slides were washed twice in PBS containing 0.1% Tween (PBS-T) and once with PBS alone, followed by a 1-hr incubation with anti–rabbit IgG–fluorescein isothiocyanate conjugate. After three final washes with PBS-T, PBS, and distilled water, the slides were photographed under a fluorescence phase microscope (Axiophot; Zeiss, Jena, Germany) with an excitation light of 450 to 490 nm. For peptide competition experiments, affinity-purified antisera were incubated for 16 hr at 4°C with either the corresponding peptide used for immunization or an unrelated control peptide (CAFGDPIDSKLSR). The peptide concentration was 10-fold greater than was the antisera protein concentration.
Slides were stained with 4′6-diamidino-2-phenylindole (DAPI) after the immunostaining. Slides were incubated for 1 hr in 0.2 μg mL−1 DAPI. Fluorescence was detected with an excitation light of 365 nm. Sieve plate callose was stained with aniline blue. The slides were incubated for 10 min in 0.05% aniline blue in 50 mM sodium potassium phosphate buffer, pH 7.2. Aniline blue fluorescence was detected with an excitation light of 365 nm.
Acknowledgments
We thank Nicole Thiele for excellent technical assistance in the isolation of LeSUT4. This work was supported by grants from Deutsche Forschungsgemeinschaft (No. SFB446) and by KWS Kleinwanzlebener Saatzucht AG and Südzucker AG.
References
- Barker, L., Kühn, C., Weise, A., Schulz, A., Gebhardt, C., Hirner, B., Hellmann, H., Schulze, W., Ward, J.M., and Frommer, W.B. (2000). SUT2, a putative sucrose sensor in sieve elements. Plant Cell 12, 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20, 1195–1197. [DOI] [PubMed] [Google Scholar]
- Boorer, K.J., Loo, D.D.F., Frommer, W.B., and Wright, E.M. (1996). Transport mechanism of the cloned potato H+/sucrose transporter StSUT1. J. Biol. Chem. 271, 25139–25144. [DOI] [PubMed] [Google Scholar]
- Bürkle, L., Hibberd, J.M., Quick, W.P., Kühn, C., Hirner, B., and Frommer, W.B. (1998). The H+-sucrose cotransporter NtSUT1 is essential for sugar export from tobacco leaves. Plant Physiol. 118, 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier, L., Lejeune, P., and Bernier, G. (1998). The role of carbohydrates in the induction of flowering in Arabidopsis thaliana: Comparison between the wild type and a starchless mutant. Planta 206, 131–137. [DOI] [PubMed] [Google Scholar]
- Delrot, S. (1981). Proton fluxes associated with sugar uptake in Vicia faba leaf tissues. Plant Physiol. 68, 706–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delrot, S., and Bonnemain, J.-L. (1981). Involvement of protons as a substrate for the sucrose carrier during phloem loading in Vicia faba leaves. Plant Physiol. 67, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskandari, S., Wright, E.M., Kreman, M., Starace, D.M., and Zampighi, G.A. (1998). Structural analysis of cloned plasma membrane proteins by freeze-fracture electron microscopy. Proc. Natl. Acad. Sci. USA 95, 11235–11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791. [DOI] [PubMed] [Google Scholar]
- Fondy, B.R., and Geiger, D.R. (1977). Sugar selectivity and other characteristics of phloem loading in Beta vulgaris L. Plant Physiol. 59, 953–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahrtz, M., Stolz, J., and Sauer, N. (1994). A phloem-specific sucrose-H+ symporter from Plantago major L. supports the model of apoplastic phloem loading. Plant J. 6, 697–706. [DOI] [PubMed] [Google Scholar]
- Gahrtz, M., Schmelzer, E., Stolz, J., and Sauer, N. (1996). Expression of the PmSUC1 sucrose carrier gene from Plantago major L. is induced during seed development. Plant J. 9, 93–100. [DOI] [PubMed] [Google Scholar]
- Giaquinta, R. (1977). Phloem loading of sucrose: pH dependence and selectivity. Plant Physiol. 59, 750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms, K., Wöhner, R.V., Schulz, B., and Frommer, W.B. (1994). Isolation and characterization of P-type H+-ATPase genes from potato. Plant Mol. Biol. 26, 979–988. [DOI] [PubMed] [Google Scholar]
- Hirose, T., Imaizumi, N., Scofield, G.N., Furbank, R.T., and Ohsugi, R. (1997). cDNA cloning and tissue specific expression of a gene for sucrose transporter from rice (Oryza sativa L.). Plant Cell Physiol. 38, 1389–1396. [DOI] [PubMed] [Google Scholar]
- Hofmann, K., and Stoffel, W. (1993). TM-base—A database of membrane-spanning protein segments. Biol. Chem. Hoppe-Seyler 347, 166. [Google Scholar]
- Imlau, A., Truernit, E., and Sauer, N. (1999). Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11, 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn, C., Quick, W.P., Schulz, A., Sonnewald, U., and Frommer, W.B. (1996). Companion cell–specific inhibition of the potato sucrose transporter SUT1. Plant Cell Environ. 19, 1115–1123. [Google Scholar]
- Kühn, C., Franceschi, V., Schulz, A., Lemoine, R., and Frommer, W.B. (1997). Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science 275, 1298–1300. [DOI] [PubMed] [Google Scholar]
- Lalonde, S., Boles, E., Hellmann, H., Barker, L., Patrick, J.W., Frommer, W.B., and Ward, J.M. (1999). The dual function of sugar carriers: Transport and sugar sensing. Plant Cell 11, 707–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine, R., Kühn, C., Thiele, N., Delrot, S., and Frommer, W.B. (1996). Antisense inhibition of the sucrose transporter: Effects on amount of carrier and sucrose transport activity. Plant Cell Environ. 19, 1124–1131. [Google Scholar]
- Lemoine, R., Bürkle, L., Barker, L., Sakr, S., Kühn, C., Regnacq, M., Gaillard, C., Delrot, S., and Frommer, W.B. (1999). Identification of NtSUT3, a pollen-specific sucrose transporter in tobacco. FEBS Lett. 454, 325–330. [DOI] [PubMed] [Google Scholar]
- Lu, J.M.Y., and Bush, D.R. (1998). His-65 in the proton-sucrose symporter is an essential amino acid whose modification with site-directed mutagenesis increases transport activity. Proc. Natl. Acad. Sci. USA 95, 9025–9030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie, B., Loo, D.D.F., Panayotova-Heiermann, M., and Wright, E.M. (1996). Biophysical characteristics of the pig kidney Na+/glucose cotransporter SGLT2 reveal a common mechanism for SGLT1 and SGLT2. J. Biol. Chem. 271, 32678–32683. [DOI] [PubMed] [Google Scholar]
- Martin, T., Schmidt, R., Altmann, T., and Frommer, W.B. (1992). Non-destructive assay systems for detection of β-glucuronidase activity in higher plants. Plant Mol. Biol. Rep. 10, 37–46. [Google Scholar]
- Maynard, J.W., and Lucas, W.J. (1982). A reanalysis of the two-component phloem loading system in Beta vulgaris. Plant Physiol. 69, 734–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet, M., Dufour, M.-E., and Lacroute, F. (1992). Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 2, 417–422. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Overvoorde, P.J., Frommer, W.B., and Grimes, H.D. (1996). A soybean sucrose binding protein independently mediates nonsaturable sucrose uptake in yeast. Plant Cell 8, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch, D., Laloi, M., Rouhara, I., Schmelzer, E., Delrot, S., and Frommer, W.B. (1995). NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett. 370, 264–268. [DOI] [PubMed] [Google Scholar]
- Riesmeier, J.W., Willmitzer, L., and Frommer, W.B. (1992). Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J. 11, 4705–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier, J.W., Hirner, B., and Frommer, W.B. (1993). Potato sucrose transporter expression in minor veins indicates a role in phloem loading. Plant Cell 5, 1591–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier, J.W., Willmitzer, L., and Frommer, W.B. (1994). Antisense repression of the sucrose transporter affects assimilate partitioning in transgenic potato plants. EMBO J. 13, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, N., and Stolz, J. (1994). SUC1 and SUC2: Two sucrose transporters from Arabidopsis thaliana; expression and characterization in baker's yeast and identification of the histidine-tagged protein. Plant J. 6, 67–77. [DOI] [PubMed] [Google Scholar]
- Schwacke, R., Grallath, S., Breitkreuz, K.E., Stransky, E., Stransky, H., Frommer, W.B., and Rentsch, D. (1999). LeProT1, a transporter for proline, glycine betaine, and γ-butyric acid in tomato pollen. Plant Cell 11, 377–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya, R., and Sturm, A. (1998). Characterization of source- and sink-specific sucrose/H+ symporters from carrot. Plant Physiol. 118, 1473–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjölund, R.D. (1997). The phloem sieve element: A river runs through it. Plant Cell 9, 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler, R., and Sauer, N. (1996). The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Bot. Acta 109, 299–306. [Google Scholar]
- Stadler, R., Brandner, J., Schulz, A., Gahrtz, M., and Sauer, N. (1995). Phloem loading by the PmSUC2 sucrose carrier from Plantago major occurs into companion cells. Plant Cell 7, 1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler, R., Truernit, E., Gahrtz, M., and Sauer, N. (1999). The AtSUC1 sucrose carrier may represent the osmotic driving force for anther dehiscence and pollen tube growth in Arabidopsis. Plant J. 19, 269–278. [DOI] [PubMed] [Google Scholar]
- Tadege, M., Bucher, M., Staehli, W., Suter, M., Dupuis, I., and Kuhlemeier, C. (1998). Activation of plant defense responses and sugar efflux by expression of pyruvate decarboxylase in potato leaves. Plant J. 16, 661–671. [Google Scholar]
- Tegeder, M., Wang, X.D., Frommer, W.B., Offler, C.E., and Patrick, J.W. (1998). Sucrose transport into developing seeds of Pisum sativum. L. Plant J. 18, 151–161. [DOI] [PubMed] [Google Scholar]
- Truernit, E., and Sauer, N. (1995). The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of β-glucuronidase to the phloem: Evidence for phloem loading and unloading by SUC2. Planta 196, 564–570. [DOI] [PubMed] [Google Scholar]
- Ward, J.M., Kühn, C., Tegeder, M., and Frommer, W.B. (1998). Sucrose transport in higher plants. Int. Rev. Cytol. 178, 41–71. [DOI] [PubMed] [Google Scholar]
- Weber, H., Borisjuk, L., Heim, U., Sauer, N., and Wobus, U. (1997). A role for sugar transporters during seed development: Molecular characterization of a hexose and a sucrose carrier in fava bean seeds. Plant Cell 9, 895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weig, A., and Komor, E. (1996). An active sucrose carrier (Scr1) that is predominantly expressed in the seedling of Ricinus communis L. J. Plant Physiol. 147, 685–690. [Google Scholar]
- Xoconostle-Cazares, B., Xiang, Y., Ruiz-Medrano, R., Wang, H.L., Monzer, J., Yoo, B.C., McFarland, K.C., Franceschi, V.R., and Lucas, W.J. (1999). Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283, 94–98. [DOI] [PubMed] [Google Scholar]
- Zhou, J.-J., Theodoulou, F., Sauer, N., Sanders, D., and Miller, A.J. (1997). A kinetic model with ordered cytoplasmic dissociation for SUC1, an Arabidopsis H+-sucrose cotransporter expressed in Xenopus oocytes. J. Membr. Biol. 159, 113–125. [DOI] [PubMed] [Google Scholar]