Abstract
Multiple alleles controlling different gene-for-gene flax rust resistance specificities occur at the L locus of flax. At least three distinct regions can be recognized in the predicted protein products: the Toll/interleukin-1 receptor homology (TIR) region, a nucleotide binding site (NBS) region, and a leucine-rich repeat (LRR) region. Replacement of the TIR-encoding region of the L6 allele with the corresponding regions of L2 or LH by recombination changed the specificity of the allele from L6 to L7. Replacement of the TIR and most of the NBS-encoding region of L10 with the equivalent region of L2 or L9 generated recombinant alleles having a novel specificity. However, replacement of the L10 TIR-encoding region with the TIR-encoding region of L2 gave rise to an allele with no detectable specificity. These data indicate that non-LRR regions can determine specificity differences between allelic gene products and that functional specificity involves interactions between coadapted polymorphic regions in the protein products of the alleles. Evidence for the action of diversifying selection on the TIR region is observed.
INTRODUCTION
One of the key questions in the biology of plant disease resistance genes is the molecular basis of gene-for-gene resistance specificity. We have been addressing this problem through the analysis of 11 alleles at the L locus in flax that control different resistance specificities to flax rust. These alleles all encode Toll/interleukin-1 receptor homology–nucleotide binding site–leucine-rich repeat (TIR-NBS-LRR) proteins (Lawrence et al., 1995; Ellis et al., 1999). Several observations suggest that the LRR region of resistance proteins is involved in specific interactions with diverse ligands, probably of pathogen origin. First, the LRR-encoding region is the most variable part of resistance genes (Botella et al., 1998; McDowell et al., 1998; Meyers et al., 1998; Ellis et al., 1999). Second, after the initial molecular evolutionary analysis of the Cf-9 class of LRR resistance genes by Parniske et al. (1997), several groups (Botella et al., 1998; McDowell et al., 1998; Meyers et al., 1998) demonstrated that the regions encoding the predicted solvent-exposed residues of the LRR of NBS-LRR resistance genes, including flax L alleles (Dodds et al., 2000), are also subject to selection for diversification. In contrast, those authors found no evidence for diversifying selection acting on the non-LRR-encoding regions of these genes. These analyses, like similar analyses of the major histocompatability complex (MHC) genes in vertebrates (Hughes and Yeager, 1998), are consistent with the diversified regions being involved in specific ligand-recognition processes. Furthermore, we have shown experimentally that the LRR region is an important determinant of specificity differences between certain L alleles (Ellis et al., 1999). First, the L6 and L11 alleles differ only in this region. Second, when the L6 or L10 TIR and NBS domains were fused to the LRR region of L2, the chimeric genes encoded L2 specificity and not L6 or L10 specificity. However, we also provided (Ellis et al., 1999) initial evidence that the LRR region does not exclusively control differences in allelic specificity. For example, although L6 and L7 encode distinct resistance specificities, they have identical LRR regions and differ only in or near the TIR region.
Here, we investigate the specificity problem in further detail, particularly the involvement of non-LRR regions, by using a series of intragenic recombinants. In vivo recombinant alleles, involving six different L alleles (L2, L6, L9, L10, LH, and suppressed L10 [suL10]) (Table 1), were previously isolated among progeny of flax plants heterozygous for different L alleles (Shepherd and Mayo, 1972; Islam et al., 1991; Lawrence et al., 1995). In vitro exchanges involving L2, L6, L10, and suL10 alleles were tested in transgenic flax. These experiments demonstrate that in addition to the LRR region, variable regions in the resistance gene products outside the LRR region also influence expression and specificity of rust resistance.
Table 1.
Source of in Vivo Recombinant Alleles
| Recombinant Allele |
Parental Genotype |
Progeny Resistance Phenotype |
|---|---|---|
| LX | L2/L6 × LH/LHa | 8610 parental (L2 or L6) |
| 3 LX | ||
| 1 susceptible | ||
| LH/L6 selfedb | 15,714 parental (L6 or susceptible) | |
| 3 LX | ||
| LH/L6 × LH/LHc | 51 parental (L6 or susceptible) | |
| 1 LX | ||
| suL10 | L2/L10 × LH/LH | 3120 parental (L2 or L10) |
| 6 susceptible, including 2 suL10d | ||
| RL10e | suL10/L9 selfed | 27,817 parental (L9 or susceptible) |
| 27 RL10 |
Shepherd and Mayo (1972) and this study.
This study.
Recombinant D237; Lawrence et al. (1995).
Shepherd and Mayo (1972). Resistant progeny (RL10) were recovered from only two of the six susceptible plants, which were designated suL10.
Islam et al. (1991). All three RL10 alleles described in this article were recovered as independent revertants from the one suL10 allele also analyzed here.
RESULTS
The Role of Non-LRR Regions in Gene-for-Gene Specificity
We prepared a series of recombinant alleles (Figure 1) from the L rust resistance locus by using either in vivo crossover or in vitro recombination. These recombinant alleles were used to study the role of sequence polymorphisms in the TIR and NBS regions in the control of gene-for-gene resistance specificity. The in vivo exchanges (Table 1) had been isolated previously (Shepherd and Mayo, 1972; Islam et al., 1991; Lawrence et al., 1995) and were classified as putative recombinants based on rust resistance phenotypes that differed from those of the parental alleles. These alleles have now been confirmed as recombinants by analysis of flanking markers and by DNA sequence analysis (see details in Methods). Lines homozygous for each recombinant allele were tested with a set of flax rust strains to determine resistance specificity of each allele.
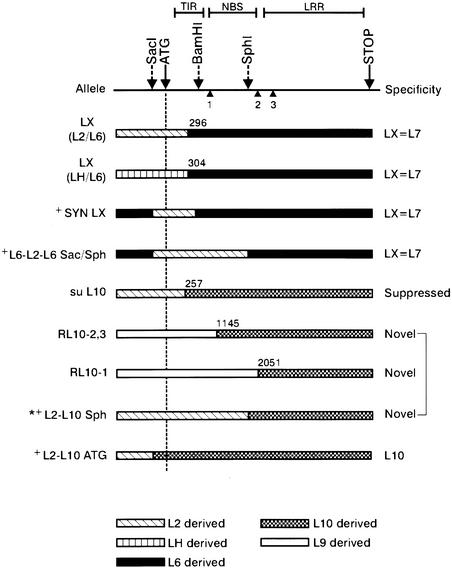
Figure 1.
Schematic Representation of in Vivo and in Vitro Recombinant Alleles.
Diagrams of the recombinant alleles show the regions of each recombinant that are derived from its respective parental L alleles. The positions of the start (ATG and indicated by the dashed vertical line) and end (STOP) of translation, the three introns (numbered triangles), and the regions encoding the TIR, NBS, and LRR domains are shown. Also shown are the positions of restriction sites SacI, BamHI, and SphI used for making in vitro recombinants (indicated by + next to the allele name). The sequence of the SacI-ATG interval (162 bp) is identical for all L alleles. The position of the last polymorphic nucleotide (numbered from the A in the ATG initiation codon and determined as described in Methods) derived from the 5′ sequence donor allele is indicated for recombinants that arose in vivo. Three independent LX alleles from L2/L6 were identical in sequence, and four independent LX alleles from LH/L6 were identical. The L2-L10Sph allele marked with a star is described in Ellis et al. (1999) and is included here for comparison and discussion. The rust resistance specificity expressed by each recombinant allele is indicated at the right. The three bracketed recombinant alleles have a novel and identical resistance specificity.
Recombinants Involving L6: The LX Specificity
Previously, three recombinant seedlings from test-cross progeny of L2/L6 heterozygotes had been identified as susceptible to rust strains that were avirulent to the L2 and L6 parents (Shepherd and Mayo, 1972). Follow-up tests with different rust strains indicated that these three lines expressed a resistance specificity, designated LX, that differed from those of L2 and L6 (Table 1 and Figure 2A; Methods). Lawrence et al. (1981) extensively characterized the resistance specificity of one of these LX lines by inoculating the LX line, and other lines carrying identified resistance specificities, with flax rust cultures derived from the sexual progeny of two rust strains. Eighty rust cultures derived from selfing strain CH5, 32 cultures from selfing strain I, and 27 cultures from intercrossing strain CH5 and strain I were used in these tests. Segregation for avirulence/virulence to LX and L7 occurred in all three rust families. Each rust culture avirulent to LX was avirulent to L7, and each culture virulent to LX was virulent to L7, which indicates that LX and L7 express the same specificity. In the CH5-selfed family, the data also indicate that the genotype at an avirulence locus segregating for A-L7/a-L7 and the genotype at an inhibitor locus segregating for I/i together determine pathogenicity to L7 plants. Segregants with the dominant avirulence allele (A-L7) are not avirulent to L7 if the dominant inhibitor allele (I) is also present, and only the genotype A-L7/−, i/i is avirulent to L7 (Lawrence et al., 1981).
Figure 2.
Rust Reactions of L2, L6, and an L2/L6 (LX) Recombinant.
(A) Rust reactions of leaves from plants homozygous for the L2, L6, and an in vivo recombinant LX allele to infection with rust strains 1 and 2 (strains CH5 F2-134 and CH5 F2-136, respectively) at 14 days after inoculation. L2 and L6 are completely resistant (R) to both strains and produce minute hypersensitive flecks at the point of infection. LX is susceptible (S) to rust strain 1, and orange uredospore pustules have erupted on the leaf surface. LX is completely resistant to rust strain 2 and shows a clear hypersensitive response at the sites of rust infection. The hypersensitive flecks on LX plants commonly are larger than those on L2 and L6.
(B) Reactions of leaves to rust infections (14 days postinoculation) to strain CH5 F2-136. The upper leaf, from the flax line Ward, shows the fully susceptible reaction; the lower leaf, from the flax line Ward homozygous for the L6-L2-L6Sac/Sph transgene, shows a partially resistant reaction.
Four additional recombinants, for which the resistance specificity with a limited set of rust strains was apparently LX, were recovered among the progeny of LH/L6 heterozygotes (Table 1; Methods). One of these four (recombinant D237; Lawrence et al., 1995) was tested with a subset of 25 CH5-selfed rust cultures, of which 12 were avirulent to L7 (genotype A-L7/−, i/i), five were fully virulent (genotype a-L7/a-L7, −/−), and eight showed slightly less than full virulence to L7 (genotype A-L7/−, I/−). The rust reactions of the LH/L6 recombinant allele and of L7 to these 25 strains were the same. In summary, rust testing indicates that in vivo recombinants from both L2/L6 and LH/L6 heterozygotes have the same specificity as the naturally occurring L7 allele.
The DNA sequences of long-range polymerase chain reaction products of the LX alleles were determined. The three independent LX recombinants from L2/L6 heterozygotes were identical, which indicated that in each case, crossover had occurred in the same region. The promoter and TIR regions were from L2, and the NBS and LRR regions were from L6 (Figure 1; Methods). Likewise, the four independent LX recombinants from LH/L6 parents were identical, with the promoter and TIR regions coming from LH and the NBS and LRR regions from L6 (Figure 1; Methods).
LX Specificity Results from Coding Region Changes to L6
The LX alleles derived from L2/L6 heterozygotes differ from L6 by polymorphic sites (derived from L2) in the promoter region and in the coding region. To distinguish the promoter and coding region effects, we replaced exon 1 (the TIR region) of L6 with the corresponding region of L2. The coding region of the recombinant gene (called synLX) is identical to that of LX alleles from L2/L6 heterozygotes, but the promoter is that of L6, not L2 (Figure 1). Fifteen independently transformed plants containing the synLX gene were inoculated with the rust strain CH5 F2-132, which is avirulent to L2, L6, and LX. Eight transgenic plants were fully resistant, and seven were fully susceptible. This result is consistent with our previous transformation experiments in which ∼20 to 50% of transgenic plants containing resistance transgenes did not express rust resistance—in some cases, because of cosuppression-like effects (P.N. Dodds, unpublished data). Cuttings from the resistant plants were also inoculated with rust strain CH5, which is avirulent to L2 and L6 and virulent to LX. The eight plants resistant to CH5 F2-132 were susceptible to the CH5 rust strain. Resistance to strain CH5 F2-132 and susceptibility to strain CH5 are consistent with the LX specificity but not L6 or L2 specificity. Therefore, we concluded that the change from L6 to LX specificity is the result of sequence alterations in the coding region and is not a promoter effect.
A similar construct, L6-L2-L6 Sac/Sph (Figure 1), in which the TIR and most of the NBS-encoding region of L6 was replaced by the equivalent L2 region, also expressed the LX-L7 specificity as demonstrated by testing transgenic plants homozygous for the transgene with a subset of 23 CH5-selfed rust cultures. However, the extent of resistance conferred by the L6-L2-L6 Sac/Sph allele was quantitatively less (light sporulation of avirulent rust strains on the youngest leaves) than the full resistance of LX and synLX alleles (Figure 2B). Three independent transgenic lines expressed the partial resistance phenotype. In summary, replacing either the TIR region or TIR and most of the NBS region of L6 with the equivalent region of L2 generated recombinant alleles with LX-L7 specificity. However, although alleles with the smaller TIR region exchange expressed full resistance, those with the larger exchange (TIR and most of the NBS) express less than full resistance.
Figure 3A shows the amino acid residues that differ between the predicted gene products of L6, the two classes of LX alleles (L2/L6 and LH/L6 recombinants), and the naturally occurring L7. These amino acid differences occur in the TIR domain. The LH-derived LX alleles encode six amino acid differences in comparison with L6, and five of these differences are also present in the L7 protein. The L2-derived LX alleles encode only three amino acid differences in comparison with L6, and all three differences also occur in both L7 and the LH-derived LX protein sequence. Thus, one or more of these three amino acid differences—S47P, S83F, and L99R (the L6 residue being listed first)—determine the specificity differences between LX-L7 and L6. The L6-L2-L6 Sac/Sph protein (Figure 3A) differs from L6 by the same three L2-derived TIR residues as LX, but also by seven differences in the NBS region. These additional seven differences affect the extent of resistance (partial versus full) but not the specificity. The positions of the polymorphic residues with respect to “landmark” sequence features of the L proteins are indicated in Figure 3B.
Figure 3.
Amino Acid Sequence Variation between L Protein Sequences.
(A) Shown are alignments of the amino terminal regions of naturally occurring L6 and L7 proteins together with recombinant proteins LX (parental alleles shown in parentheses) and L6-L2-L6 Sac/Sph. Only the polymorphic residues are shown, and sites are numbered from the first methionine residue (position 1) in the predicted L protein amino acid sequences. Residues that differ from the L6 sequence are shown in bold. The dashes at position 301 indicate where the Q residue in L2 is absent from the L6 and L7 sequence because of a codon deletion/insertion polymorphism in the corresponding DNA sequences. The position of intron 1 in the corresponding DNA sequence is indicated by an arrow.
(B) Consensus sequence of N-terminal regions of 11 L proteins (Ellis et al., 1999); residues polymorphic between the alleles in this study are shown in bold and underlined. The positions of conserved P-loop, kinase-2 (Kin2), and GLPL protein motifs are marked, as are the positions in the corresponding DNA sequence of intron 1 and the SphI restriction site used in the construction of some recombinants. The region having sequence similarity to the TIR region is overlined and is extended on each end with dashed lines to show where additional predicted protein structural similarity occurs (Parker et al., 1997; Rock et al., 1998).
(C) Alignments of polymorphic amino acid residues in the L10, suL10, RL10-2/3, and L2-L10Sph proteins. Polymorphic sites are numbered as in (A), with the residues that differ from L10 shown in bold. RL10-1 has the same nine polymorphisms as RL10-2/3 and an additional 18 polymorphisms extending to the end of the exon 2 product. The exact polymorphisms can be seen by comparing the amino acid sequences of the L9 and L10 gene products reported elsewhere (Ellis et al., 1999).
Recombinants Involving L2 and L10: suL10
Shepherd and Mayo (1972) described several possible recombinant alleles (called suL10 alleles) involving L2 and L10 which expressed neither L2 nor L10 resistance. We analyzed the sole surviving suL10 allele and found that this recombinant consists of the promoter and TIR-encoding region of L2 and the NBS- and LRR-encoding regions of L10 (Figure 1; see Methods for details). suL10 homozygotes were tested for reaction to six strains of flax rust, which represent all of the original, and unrelated, sources of rust strains available in our laboratory. Because all six strains were virulent to suL10, no resistance specificity associated with suL10 could be detected.
Reverse transcription–polymerase chain reaction analysis of mRNA (data not shown) demonstrated that the suL10 allele is expressed, suggesting that its lack of resistance is due to the recombinant coding region. To test this, we replaced the L2-derived coding region of suL10 with the L10 coding region (L2-L10ATG; Figure 1). L2-L10ATG is identical to L2 upstream of the ATG codon and identical to L10 downstream. Ten independent transgenic lines containing the chimeric gene were recovered, five of which were resistant to rust strain BS-1 (a strain recognized by L10 but not by suL10). One of these lines was also tested with several other rust strains that are recognized by L10. The reactions of the L2-L10ATG recombinant allele to these strains were identical to the reactions of the L10 allele, confirming the L10 specificity. Therefore, we concluded that the suppressed phenotype of suL10 is the result of sequence polymorphisms derived from exon 1 of L2 and is not related to the L2 promoter region.
Recombinants Involving L9 and suL10
We also characterized three recombinant alleles independently derived from L9/suL10 heterozygotes (Shepherd and Mayo, 1972; Islam et al., 1991; see also Methods). These alleles express resistance to rust strain BS-1 (Table 2). Initial testing with a limited set of rust strains (Shepherd and Mayo, 1972) indicated that the specificity of these recombinants was not L2 but could be L10 (suL10 is an L2/L10 recombinant). Consequently, these recombinant alleles were designated Revertant L10 (RL10). However, in more recent and extensive tests, we found that four rust strains avirulent to L10 and of a provenance different from that of strain BS-1 were virulent to all three of the RL10 lines (Table 2). Therefore, the specificity expressed by RL10 cannot be the same as that expressed by L10. The six rust strains in Table 2 that are virulent to RL10 alleles are collectively avirulent to all known L locus alleles. This suggests that the specificity expressed by RL10 is novel.
Table 2.
Reactions of Plants Containing L9, L10, or Recombinant L10 Genes to Rust Strains
| Rust Strainsa
|
|||||||
|---|---|---|---|---|---|---|---|
| Allele | C | H | I | CH5 × I(32) | J | BS-1 | BGSS-1 |
| L9 | + | + | + | + | + | + | − |
| L10 | + | − | − | − | − | − | + |
| RL10-1, -2, -3 | + | + | + | + | + | − | + |
| L2-L10Sphb | + | ± | + | + | ± | − | −b |
(+), fully susceptible; (−), fully resistant; (±), partially resistant.
The L2-L10Sph transgene (Ellis et al., 1999) was expressed in the flax line Ward, which contains the L9 resistance gene that accounts for the resistance of these plants to the rust strain BGSS-1. The partial resistance of these plants to rust strains H and J is also seen in nontransformed Ward plants, indicating that the resistance is due to factors in the Ward genetic background and not caused by the introduced transgene. Ward is fully susceptible to rust strains C, I and CH5 × I(32).
Sequence analysis of the RL10 alleles revealed that they are 5′-L9–3′-suL10 recombinants (Figure 1). Because the 3′-suL10 region in the interchange is entirely of L10 origin, the three RL10 alleles are effectively L9-L10 chimeras. Recombinants RL10-2 and RL10-3 are identical: each contains the promoter, the TIR-encoding region, and part of the NBS-encoding region from L9, with the remaining 3′ region coming from suL10 (Figure 1). The remaining recombinant, RL10-1, contains the promoter, the TIR-encoding region, and the complete NBS-encoding region from L9 and the LRR-encoding region from suL10 (Figure 1). A previously described in vitro recombinant L2-L10Sph (Ellis et al., 1999; Figure 1), which contains the TIR and most of the NBS region of L2 and the LRR of L10, also encodes a resistance specificity indistinguishable from that of the RL10 alleles (Table 2). Therefore, replacing the TIR and some or the entire NBS region of L10 with the equivalent region of either L2 or L9 generates recombinant alleles with apparently the same novel specificity. Because RL10 alleles are also susceptible to rust strain BGSS-1, which is avirulent to L9 (Table 2), we conclude that the L9-L10 chimeras, especially RL10-1, which contains the entire TIR- and NBS-encoding regions of L9, do not express L9 specificity.
Amino Acid Changes in RL10, L2-L10Sph, and suL10
The RL10-1 protein differs from L10 by 27 amino acid residues derived from L9, six in the TIR region and the rest in the NBS domain (data not shown). The RL10-2 and RL10-3 proteins, which are identical to each other (henceforth referred to as RL10-2/3), differ from L10 at nine amino acid positions, six in the TIR region and three in the NBS region (Figure 3C); consequently, one or more of these differences must contribute to the change in specificity. Further indications of which amino acids may be critical in modification of specificity come from examining the predicted sequence of the L2-L10Sph protein (Ellis et al., 1999), which expresses the same specificity as RL10-2/3 (Table 2). The L2-L10Sph protein differs from L10 at 10 amino acid positions, and five of these polymorphic residues (three in the TIR and two in the NBS regions) also occur in RL10-2/3 (Figure 3C). One or more of the five shared amino acid polymorphisms in RL10-2/3 and L2-L10Sph that distinguish them from the L10 protein potentially determine the difference in resistance specificity.
The suL10 protein differs from L10 by five amino acid residues, all in the TIR region: D62E, K74E, H82Q, C85R, and Y86R (the suL10 residue is listed second) (Figure 3C). One or more of these differences may alter the resistance specificity of the recombinant protein so that it does not recognize any of the tested rust strains, or they may interfere with the function of the recombinant protein or cause instability and degradation. The lack of suitable antibodies has kept us from testing the relative stability of suL10 and L10 proteins. The problem is compounded by the fact that proteins encoded from the M rust resistance locus are ∼80% identical to the L proteins (Anderson et al., 1997). Importantly, the TIR region of L2, which suppresses resistance of the suL10 protein, is functional in the context of the naturally occurring L2 protein and the experimentally derived L2-L10Sph and LX recombinant proteins.
Diversifying Selection Acts on Regions of the TIR Domain of L Alleles
Because amino acid changes in the TIR and NBS domains affect the resistance specificity encoded by L proteins, we examined whether diversifying selection may be acting on this region. Previous analysis had detected an excess of nonsynonymous over synonymous nucleotide substitutions within the xxLxLxx motif of the LRR domain of L alleles (Dodds et al., 2000), consistent with the action of diversifying selection on this region to generate new specificities. This pattern has also been observed for other R gene families. However, we did not detect any significant difference in the rates of synonymous (Ks) and nonsynonymous (Ka) substitution in the TIR or NBS domains as a whole (Table 3). We therefore looked at two smaller regions implicated here as being involved in the specificity changes we observed between recombinant L alleles. In fact, these two regions are relatively more polymorphic than the remainder of the TIR and NBS domains of L alleles (see Figure 2 in Ellis et al., 1999). The first region encodes amino acid residues 47 to 102, which includes those amino acid polymorphisms responsible for the L6/L7 specificity difference as well as the L10/suL10 difference. In this region we found that Ka was significantly greater than Ks (P < 0.05), suggesting that the amino acid changes here are the result of positive selection (Table 3); a similar result was obtained for the smaller region encoding amino acids 74 to 102 (Table 3). The second region encodes amino acids 182 to 222, which includes amino acids responsible for the RL10/suL10 differences and spans the site of the first intron, which separates the TIR and NBS domains. In this case, although Ka was greater that Ks, the difference was not statistically significant (Table 3); similarly, no significant difference was observed in a smaller region encoding amino acids 202 to 222 (Table 3).
Table 3.
Average Rates of Nucleotide Substitution between L Alleles per 100 Nonsynonymous (Ka) and Synonymous (Ks) Sites
| Region | Kaa | Ksb |
|---|---|---|
| Exon 1/TIR | 1.4 (0.3) | 1.2 (0.6) |
| Exon 2/NBS | 2.0 (0.3) | 3.2 (0.7) |
| Amino acids 47–102 | 3.5 (1.1) | 1.0 (0.7)b |
| Amino acids 74–102 | 5.4 (1.9) | 1.0 (1.0)b |
| Amino acids 182–222 | 4.4 (1.2) | 1.9 (1.9) |
| Amino acids 202–222 | 5.8 (2.0) | 3.9 (3.9) |
Standard errors of average Ka and Ks values are shown in parentheses.
Ka > Ks is significant at P < 5%.
DISCUSSION
The Non-LRR Region Plays a Role in Specificity Determination
This article details several examples in which the resistance specificity of alleles at the L locus in flax is altered to a different specificity by changes confined solely to the TIR- and NBS-encoding regions of the alleles; that is, alleles with the same LRR region but different TIR and NBS regions can encode different specificities. We have previously reported that the products of L6 and L7 have identical LRR regions but differ by 11 amino acids in the TIR region (Ellis et al., 1999). Here, we report sequence analyses of recombinant alleles derived from in vivo exchanges between L6 and either L2 or LH that, like two in vitro recombinants between L6 and L2, express a resistance specificity (originally designated LX) that is identical to L7. The product of the LH/L6 recombinant allele is identical to that of L6 except for six amino acid changes in the TIR region, whereas the products of the L2/L6 recombinants differ from L6 by just three amino acids in the TIR region. The three amino acid changes in the gene product of the L2/L6 recombinant are also found in the gene products of the LH/L6 recombinant and the L7 gene. Consequently, one or more of these three amino acids must be responsible for the alteration from L6 to L7 specificity (given the demonstration that the promoter region is not responsible for the differences).
Under the model that resistance proteins are receptors for molecules of pathogen origin, the identical LRR sequences in the L6 and L7 (LX) proteins suggest that they may interact with the same or very similar ligands. Data consistent with this supposition come from inheritance studies in flax rust, which show that the avirulence genes A-L6 and A-L7 map to the same locus (Flor, 1959, 1965; Lawrence et al., 1981), and from mutation studies, in which concomitant loss of both the A-L6 and A-L7 specificities has always been observed (Flor, 1956, 1958; J.G. Ellis, G.J. Lawrence, and K.W. Shepherd, unpublished data). If A-L6 and A-L7 are separate but closely linked genes, then these genes could be related if derived by duplication of an original gene. Alternatively, A-L6 and A-L7 could be the same avirulence gene, but with its product recognized differently by L6 and L7.
Given the probable similarity of the L6/A-L6 interaction and the L7/A-L7 interaction, it is interesting that a dominant inhibitor gene (I) present in some rust strains inhibits the L7/A-L7 interaction but not the L6/A-L6 interaction (Lawrence et al., 1981). The I gene effect also applies to the recombinant alleles with L7 specificity. The rust strains in our collection fall into three phenotypic classes—avirulent to both L6 and L7, virulent to both, or avirulent to L6 and virulent to L7. Rust cultures of the last-named class possess I (Lawrence et al., 1981). No strains have been described that are virulent to L6 and avirulent to L7. Importantly, the I gene does not affect the L2/A-L2 interaction, despite the presence in the L2 protein of the three critical amino acid polymorphisms in the TIR region that distinguish L6 and L7 specificity in the L2/L6 recombinants.
Further examples of changes in the TIR-NBS region that alter specificity come from in vivo and in vitro recombinant studies involving the L10 resistance allele. The product of the suL10 allele (a recombinant between L2 and L10) is identical to that of L10, except for five amino acid changes in the TIR region. These five changes, which are functional in the context of the L2 allele, result in the loss of the L10 specificity; in fact, suL10 apparently possesses no resistance specificity—it conferred no resistance to any of the rust strains in our collection. Subsequently, an in vivo recombination between suL10 and L9 yielded two identical recombinants (RL10-2 and RL10-3) that express a novel specificity and encode products that differ from the L10 protein by nine amino acid changes (derived from L9) in the TIR-NBS region. In another example, replacing the TIR-NBS-encoding region of L10 with the equivalent region of L2 generated a recombinant allele (the L2-L10Sph in vitro recombinant) with specificity identical to that of RL10-2 and RL10-3. The product of the L2-L10Sph recombinant differs from L10 by 10 amino acid changes in the TIR-NBS region, of which five are identical to changes in the L9-L10 (i.e., RL10) recombinant. Consequently, we think it likely that one or more of these five amino acid differences (four in the TIR and one at the beginning of the NBS) converts L10 to a novel specificity. Extending this line of reasoning, perhaps the three amino acid changes common to suL10 and RL10-2/3 (D62E, K74E, and Y86R) (Figure 3C) cause the apparent loss of function of suL10, whereas the additional two changes common to RL10-2/3 and L2-L10Sph (E209K and T214A) (Figure 3) result in the novel specificity. It is interesting that none of these five positions coincides with any of the three positions that determine the L6/L7 specificity difference.
Coadaptation of Regions within L Proteins
The findings outlined above, and those reported previously for L7 (Ellis et al., 1999), demonstrate that the TIR-NBS region has a role in specificity determination. This idea is supported by the observation of an excess of nonsynonymous nucleotide substitutions in the N-terminal region of the TIR domain (Table 3). This suggests that positive selection has acted on this region and that the allelic variation seen here has a role in the differences in function of the L proteins. Comparing the TIR domains of several resistance proteins indicates that the sites of amino acid variation between L alleles are in the nonconserved regions of the TIR (Figure 4). Homologs of RPP5 (Noël et al., 1999) and RPP1A (Botella et al., 1998) also contain polymorphic sites within the TIR, again falling within the nonconserved regions, although diversifying selection has not been reported in these cases.
Figure 4.
Alignment of TIR Domains of Resistance Proteins.
The TIR domains of L6 (Lawrence et al., 1995), M (Anderson et al., 1997), RPP1A (Botella et al., 1998), RPP5 (Parker et al., 1997), N (Whitham et al., 1994), and RPS4 (Gassmann et al., 1999) were aligned with the homologous region of the Drosophila Toll protein (Hashimoto et al., 1988) by using the Pileup program (see Methods). Residues with only conservative changes between all seven proteins are shaded in dark gray; those conserved between at least four sequences are shaded in light gray. The amino acid residues of L6 that correspond to the polymorphic sites shown in Figure 3 are given as white lettering on a black background and are marked by numbered asterisks above the sequences. Numbers at right indicate the amino acid position relative to the N-terminal methionine residue.
The findings outlined in the previous section also demonstrate that the appropriate matching of polymorphic TIR-NBS regions and polymorphic LRR regions is required for function specificity; that is, functioning requires coadapted TIR-NBS and LRR regions. The molecular basis of this coadaptation is unknown at present. In broad terms, the possibilities include (1) the presence of ligand contact points in both the LRR and non-LRR regions, the additive effects of which are required for functional specificity; (2) direct interaction between amino acid residues in the LRR and non-LRR regions (or TIR and NBS) that are essential for functional specificity or protein stability; and (3) the necessity of having different TIR-NBS regions for different LRR regions, to ensure that binding of the ligand to the resistance protein (even if this involves only the LRR region) results in the triggering of subsequent signaling processes and thus leads to the resistance reaction.
This study also provides evidence for coadaptation between regions within the TIR-NBS region. The TIR-NBS region of L2 is functional in combination with the LRR of L2 or the LRR region of L10 (as in the L2-L10Sph recombinant), although the specificity of L2-L10Sph is not L10 but a novel specificity. However, if only the TIR of L2 is transferred to L10, the resulting recombinant allele (suL10) is apparently nonfunctional. Similarly, the addition of six amino acid changes from the NBS domain of L2 into LX results in a decrease in resistance expressed by the L6-L2-L6 Sac/Sph allele but does not alter specificity. These observations suggest that coadaptation between polymorphic TIR and NBS regions is also necessary for functional specificity.
In summary, in this and a previous study (Ellis et al., 1999), we observed three classes of recombinant alleles: (1) resistant recombinants in which the specificity is identical to that of the LRR donor allele (e.g., L6-L2Sph and L10-L2Sph both expressing L2 specificity); (2) recombinants with specificities that differ from those of both parental alleles (e.g., the LX recombinants derived from L2 and L6 and the RL10 recombinants derived from L9 and suL10); and (3) recombinants that express no resistance specificity. This last class includes recombinant alleles derived from exchanges in the LRR region (e.g., L2-L6-L2 Xho/Bgl) (Ellis et al., 1999) and from exchanges in the non-LRR region (e.g., suL10). In general, the phenotypes of the recombinant alleles have proved to be unpredictable, a consequence of the coadaptation between different polymorphic regions of the gene product that evidently is required for functional specificity. We have now also provided molecular documentation of in vivo intragenic recombination events previously only inferred from genetic experiments involving plants heterozygous for different L alleles. These recombination events, which occur at frequencies of at least one per 1000 gametes (Shepherd and Mayo, 1972; Islam et al., 1991), are likely to be important in resistance gene evolution.
METHODS
Flax Rust Strains
The origins and descriptions of flax rust (Melampsora lini) strains used in this study are provided in Lawrence et al. (1981) and Ellis et al. (1999). BGSS-1 originates from a cross between strain WA and strain C. Rust infections were performed as described by Lawrence et al. (1981).
Origin of LX Recombinants
Shepherd and Mayo (1972) identified four putative recombinants within the L locus among 8614 test-cross progeny of L2/L6 heterozygotes (using Hoshangabad with the allele LH as the susceptible parent) that were susceptible to rust strain Fi, which is avirulent to both parental alleles L2 and L6 (Table 1). The susceptible reaction of these plants was confirmed by analysis of selfed progeny of the four recombinants. In subsequent analysis, several progeny plants from each of these recombinants were grown in isolation for seed multiplication to allow a search for rare resistant revertants, as had been found earlier for some of the susceptible putative recombinants detected in L2/L10 test-cross progeny (Shepherd and Mayo, 1972). In a pilot experiment, progeny families derived from one of the four recombinants (plant 10/3) were tested with another rust strain (BS-1) that, like strain Fi, was avirulent to both parental alleles L2 and L6. Unexpectedly, some of the progeny families contained plants that were fully resistant to BS-1. Three types of family were detected: one type contained only susceptible plants, another segregated 3:1 resistant:susceptible, and the third had only resistant plants. Clearly BS-1 was detecting a specificity not detected by Fi in these recombinant progeny, a specificity designated LX. Testing progeny of the other three recombinants with BS-1 revealed that two of them (plants Blr-2 and 52-2) also carried the LX specificity, whereas the other (plant Blr-1) was susceptible to both Fi and BS-1 rusts. The three LX alleles from plants 10/3, Blr-2, and 52-2 were recovered as homozygotes. The susceptibility allele from plant Blr-1 was not recovered.
Three progeny (75-948, 75-1508, and 75-1628) with a resistance specificity that was nonparental (subsequently shown to be LX) were also identified when 15,717 F2 progeny from selfing LH/L6 heterozygotes were successively inoculated with two rust strains (BS-1 and CH5), both of which were avirulent to L6 and virulent to LH. The LX progeny were resistant to strain BS-1 and susceptible to strain CH5. An additional plant with LX specificity (Table 1) was recovered in an independent experiment among 52 test-cross progeny of an LH/L6 heterozygote (Lawrence et al., 1995).
Origin of suL10 and RL10 Alleles
Shepherd and Mayo (1972) recovered six recombinants among 3126 test-cross progeny from L2/L10 heterozygotes that were susceptible to a rust strain avirulent to both L2 and L10 (Table 1). One of these alleles, designated suL10 (Shepherd and Mayo, 1972; Islam et al., 1991), has survived for molecular analysis. A line homozygous for suL10 was used in this study.
From 27,844 self-progeny of suL10/L9 heterozygotes, Islam et al. (1991) identified 27 revertants that were resistant to rust strain BS-1, which is virulent to suL10 and L9 but is avirulent to L2 and L10, the parent alleles of suL10 (Table 1). Three lines homozygous for revertant alleles, designated RL10-1, RL10-2, and RL10-3, were analyzed in the present study.
Resistance Gene Cloning and Manipulation and Plant Transformation
The methods for cloning rust resistance genes from flax (Linum usitatissimum), the manipulation and DNA sequence analysis of the genes, and their introduction into transgenic flax plants are described by Lawrence et al. (1995), Anderson et al. (1997), and Ellis et al. (1999). We used the flax line Ward, which is susceptible to the flax rust strains used in this study, for Agrobacterium-mediated transformation.
An EcoRI fragment containing the entire suL10 allele was cloned in the λ vector EMBL4, and the clone was detected by using probe Lu-1 (Lawrence et al., 1995). LX and RL10 alleles were cloned by long-range polymerase chain reaction as described by Ellis et al. (1999). To make the in vitro recombinant synLX, we replaced the 629-bp SacI-BamHI fragment from exon 1 of the L6 allele with the same fragment from the L2 allele. To make L6-L2-L6 Sac/Sph, we replaced the 1709-bp SacI-SphI fragment from exon 1 and exon 2 of L6 with the same fragment from L2. To make L2-L10ATG, we replaced the SacI-SphI fragment from exon 1 and exon 2 of suL10 with the same fragment from the L10 allele. The positions of these restriction enzyme cleavage sites are shown in Figure 1.
Analysis of Restriction Site Polymorphisms in L Alleles
Restriction site polymorphisms were detected by DNA gel blot analysis of DNA from recombinant and parent lines. Polymorphic EcoRV and KpnI sites upstream of the L alleles were detected using probe Lu-2 (Ellis et al., 1995); EcoRI, HindIII, and BglII polymorphisms in the leucine-rich repeat (LRR) region were detected using probe Lu-1 (Lawrence et al., 1995).
Molecular Analysis of LX Recombinants
DNA gel blot analysis demonstrated that LX alleles possessed 5′ restriction fragment length polymorphism (RFLP) markers from either L2 or LH, depending on their parents, and 3′ RFLP markers from L6. The sites of crossing over were determined by DNA sequence analysis of the recombinant alleles. The three independent LX alleles derived from L2/L6 recombination were identical, with 5′ L2 polymorphic sites up to position 296 (relative to the A residue in the ATG codon) in exon 1 and L6 polymorphic sites downstream from position 953 near the beginning of exon 2 (L2 and L6 are identical between positions 296 and 953). Four independent LX recombinants from LH/L6 heterozygotes were also identical, and crossing over had occurred between positions 304 and 953. The entire coding regions of two LX alleles, one from L2/L6 and one from LH/L6, were sequenced, and no other nonparental changes were detected. The data are summarized in Figure 1.
Molecular Analysis of suL10 and RL10 Alleles
The suL10 allele contains 5′ RFLP markers from L2 and 3′ RFLP markers from L10. The RL10 alleles derived from the suL10/L9 heterozygotes contain 5′ markers from L9 and 3′ markers from suL10. The DNA sequence of suL10 showed that apart from recombination, no other changes were found that could account for its suppressed resistance phenotype. The suL10 allele contains 5′ L2 polymorphic sites up to position 257 (relative to the A residue in the ATG codon) in exon 1 and L10 polymorphic sites downstream from position 407 (L2 and L10 are identical between positions 257 and 407).
Sequencing the regions containing the crossover sites in RL10 alleles showed that RL10-2 and RL10-3 have identical sequences, both containing 5′ L9 polymorphisms to position 1145 (relative to the ATG codon) and the L10 sequence after this position. This corresponds to the region just downstream of the P-loop codons in exon 2. Crossover in RL10-1 occurred ∼800 bp farther downstream. The last of the 5′ L9 polymorphisms occurs in intron 2, at 2051 bp downstream of the ATG, with the remaining 3′ sequence identical to L10. Thus, RL10-1 contains the entire toll/interleukin-1 receptor homology–nucleotide binding site (TIR-NBS) region from L9 (Figure 1).
Sequence Comparisons and DNA Evolutionary Analysis
Amino acid or DNA sequences in the TIR region were aligned using the Pileup program of the GCG (Genetics Computer Group, Madison, WI) software package. Rates of synonymous and nonsynonymous nucleotide substitutions between L allele coding sequences were determined using the Jukes–Cantor algorithm of the Molecular Evolutionary Genetics Analysis software version 1.02 (Kumar et al., 1993; http://evolgen.biol.metro-u.ac.jp/MEGA/). The significance of any difference in the average rates of synonymous and nonsynonymous substitutions was assessed by t test.
Acknowledgments
This research was supported by grants from the Grains Research and Development Corporation and the Australian Research Grants Scheme. Valerie Ryle and Patricia Atkinson provided excellent technical assistance.
References
- Anderson, P.A., Lawrence, G.J., Morrish, B.C., Ayliffe, M.A., Finnegan, E.J., and Ellis, J.G. (1997). Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell 9, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella, M.A., Parker, J.E., Frost, L.N., Bittner-Eddy, P.D., Beynon, J.L., Daniels, M.J., Holub, E.B., and Jones, J.D.G. (1998). Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10, 1847–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., Pryor, A.J., and Ellis, J.G. (2000). Genetic analysis and evolution of plant disease resistance genes. In Molecular Plant Pathology, M. Dickinson and J. Beynon, eds (Sheffield, UK: Sheffield Academic Press), in press.
- Ellis, J.G., Lawrence, G.J., Finnegan, E.J., and Anderson, P.A. (1995). Contrasting complexity of two rust resistance loci in flax. Proc. Natl. Acad. Sci. USA 92, 4185–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J.G., Lawrence, G.J., Luck, J.E., and Dodds, P.N. (1999). Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H.H. (1956). Mutations in flax rust induced by UV radiation. Science 124, 888–889. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1958). Mutation to wider virulence in Melampsora lini. Phytopathology 48, 297–301. [Google Scholar]
- Flor, H.H. (1959). Differential host range of the monokaryon and dikaryons of a eu-autoecious rust. Phytopathology 49, 794–795. [Google Scholar]
- Flor, H.H. (1965). Inheritance of smooth-spore-wall and pathogenicity in Melampsora lini. Phytopathology 55, 724–727. [Google Scholar]
- Gassmann, W., Hinsch, M.E., and Staskawicz, B.J. (1999). The Arabidopsis RPS4 bacterial resistance gene is a member of the TIR-NBS-LRR family of disease resistance genes. Plant J. 20, 265–277. [DOI] [PubMed] [Google Scholar]
- Hashimoto, C., Hudson, K.L., and Anderson, K.V. (1988). The toll gene of Drosophila, required for dorsal–ventral embryonic polarity, appears to encode a transmembrane protein. Cell 52, 269–279. [DOI] [PubMed] [Google Scholar]
- Hughes, A.L., and Yeager, M. (1998). Natural selection at major histocompatability complex loci of vertebrates. Annu. Rev. Genet. 32, 415–435. [DOI] [PubMed] [Google Scholar]
- Islam, M.R., Shepherd, K.W., and Mayo, G.M.E. (1991). An analysis of reversion among recombinants involving genes of the L group, conferring resistance to rust in flax. J. Genet. Breed. 45, 181–188. [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (1993). MEGA: Molecular Evolutionary Genetics Analysis, version 1.0. (University Park, PA: Pennsylvania State University).
- Lawrence, G.J., Mayo, G.M.E., and Shepherd, K.W. (1981). Interactions between genes controlling pathogenicity in the flax rust fungus. Phytopathology 71, 12–19. [Google Scholar]
- Lawrence, G.J., Finnegan, E.J., Ayliffe, M.A., and Ellis, J.G. (1995). The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell 7, 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, J.M., Dhandaydham, M., Long, T.A., Aarts, M.G., Goff, S., Holub, E.B., and Dangl, J.L. (1998). Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10, 1861–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Shen, K.A., Rohani, P., Gaut, B.S., and Michelmore, R.W. (1998). Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell 10, 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël, L., Moores, T.L., van der Biezen, E.A., Parniske, M., Daniels, M.J., Parker, J.E., and Jones, J.D.G. (1999). Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11, 2099–2111. [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E., Coleman, M.J., Szabo, V., Frost, L.N., Schmidt, R., van der Biezen, E.A., Moores, T., Dean, C., Daniels, M.J., and Jones, J.D.G. (1997). The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the Toll and Interleukin-1 receptors with N and L6. Plant Cell 9, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske, M., Hammond-Kosack, K.E., Goldstein, C., Thomas, C.W., Jones, D.A., Harrison, K., Wulff, B.B.H., and Jones, J.D.G. (1997). Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91, 821–832. [DOI] [PubMed] [Google Scholar]
- Rock, F.L., Hardiman, G., Timans, J.C., Kastelein, R.A., and Bazan, J.F. (1998). A family of human receptors structurally related to Drosophila Toll. Proc. Natl. Acad. Sci. USA 95, 588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd, K.W., and Mayo, G.M.E. (1972). Genes conferring specific plant disease resistance. Science 175, 375–380. [DOI] [PubMed] [Google Scholar]
- Whitham, S., Dinesh-Kumar, S.P., Choi, D., Hehl, R., Corr, C., and Baker, B. (1994). The product of the tobacco mosaic virus resistance gene N: Similarity to Toll and the interleukin-1 receptor. Cell 78, 1101–1115. [DOI] [PubMed] [Google Scholar]