Abstract
Squash genes (SLW1 and SLW3) induced systemically after silverleaf whitefly feeding were identified. Differences in the local and systemic expression of SLW1 and SLW3 after feeding by the closely related silverleaf and sweetpotato whiteflies were observed. Temporal and spatial studies showed that SLW1 and SLW3 were induced when second, third, and fourth nymphal instars were feeding. Although only barely detected after wounding and bacterial infection, SLW1 and SLW3 RNAs were abundant during water-deficit stress. Treatments with wound/defense signal molecules showed that SLW1 RNAs accumulated in response to methyl jasmonate and ethylene, whereas SLW3 was not regulated by known wound/defense signals, suggesting utilization of a novel mechanism for defense signal transduction. SLW1 RNAs accumulated during floral and fruit development, whereas SLW3 RNAs were not detected during vegetative or reproductive development. The potential roles of SLW1, an M20b peptidase–like protein, and SLW3, a β-glucosidase–like protein, in defense and the leaf-silvering disorder are discussed.
INTRODUCTION
Although the signal transduction pathways that are activated by mechanical wounding and pathogen invasion have been studied intensively (Ryals et al., 1996; Ryan and Pearce, 1998; Dempsey et al., 1999; Pieterse and van Loon, 1999), there is limited understanding of changes in plant gene expression in response to insect feeding. Given the similarities of mechanical wounding and the damage incurred by insects that masticate foliage, it is not surprising that wound-response transcripts and proteins (i.e., proteinase inhibitors, polyphenol oxidases, and leucine aminopeptidase) accumulate locally and systemically in response to caterpillar feeding (Green and Ryan, 1982; Pautot et al., 1993; Stout et al., 1996; Karban and Baldwin, 1997). However, insect feeding and wounding are not equivalent. Feeding by Manduca sexta larvae, for example, enhances expression of wound-response genes relative to wounding but also induces expression of a novel set of plant genes that are not induced by wounding alone (Korth and Dixon, 1997). Although these herbivory-induced genes have not been identified, some of their gene products may be involved in biosynthesis or release of volatiles that are important in mediating plant–herbivore–predator interactions (Páre and Tumlinson, 1999).
Far less is known about the changes in gene expression in response to insects that use other modes of feeding (Stout et al., 1994). Only a few recent studies have investigated plant responses to phloem-feeding whiteflies. Unlike insects that consume foliage, whiteflies do not induce the wound-response genes that are modulated by the octadecanoid pathway. Whitefly feeding on tomatoes primarily induces the salicylic acid (SA)–independent, jasmonic acid (JA)/ethylene–dependent cascade of defense signal transduction (Chao et al., 1999; Pieterse and van Loon, 1999; D.P. Puthoff and L.L. Walling, submittedmanuscript ; D.P. Puthoff, C.S. LeVesque, T.M. Perring, and L.L. Walling, manuscript in preparation). This causes increases in some pathogenesis-related (PR) protein RNAs and PR protein activities (Mayer et al., 1996; D.P. Puthoff, C.S. LeVesque, T.M. Perring, and L.L. Walling, manuscript in preparation). In addition, genes important for lignin and SA biosynthesis (i.e., phenylalanine ammonia lyase) and the oxidative burst (i.e., Wfi1, a gp91-phox homolog) are also induced (D.P. Puthoff and L.L. Walling, manuscript submitted; D.P. Puthoff, C.S. LeVesque, T.M. Perring, and L.L. Walling, manuscript in preparation).
To determine if novel genes are expressed in response to phloem-feeding insects, squash responses to the sweetpotato whitefly (Bemisia tabaci Type A) and silverleaf whitefly (B. argentifolii) were studied. These closely related whitefly species are known to cause distinct phenotypic changes in plants (Perring et al., 1993; Brown et al., 1995). Unlike the sweetpotato whitefly, the silverleaf whitefly provokes distinct developmental disorders in several plants, including irregular ripening of tomato and green pepper fruit; stalk blanching of broccoli, kai choy, and lettuce; and leaf silvering in squash (Maynard and Cantliffe, 1989; Costa et al., 1993a; Summers and Estrada, 1996). Squash leaf silvering is a systemic response induced by as few as three silverleaf whitefly nymphs; symptoms are proportional to the number of feeding nymphs, whereas high-density feeding by silverleaf or sweetpotato whitefly adults or by sweetpotato whitefly nymphs does not induce leaf silvering (Yokomi et al., 1989; Hoelmer et al., 1991; Schuster et al., 1991; Costa et al., 1993a).
Transmission and grafting studies have indicated that squash leaf silvering is not caused by a pathogenic agent (Yokomi et al., 1990), but the chemical nature of the factor that induces silvering has remained elusive. The translocated silvering factor may be a component of the silverleaf whitefly saliva egested during whitefly feeding, or it may be produced by the squash plant as a response to silverleaf whitefly feeding. A search for chemicals that mimic the silvering factor showed that the gibberellic acid (GA3)–biosynthesis inhibitor (chlormequat chloride [CCC]) induced leaf silvering in squash that was partially ameliorated after GA3 application (Yokomi et al., 1995). Studies of the fungus Stereum purpureum, which induces leaf silvering in apples and plums (Miyairi et al., 1977), identified the apple-silvering factor as a fungal endo-polygalacturonase (Miyairi, 1988), suggesting that modification of cell wall polysaccharide polymers might be important for the development of silvering symptoms.
Consistent with a role of the plant extracellular matrix in apple silvering, silvered squash leaves have large airspaces that are correlated with altered mesophyll cell self-adherence and adherence to the adaxial epidermis (Jiménez et al., 1995; Ramos et al., 1995). The silvering factor may directly antagonize cell adhesion or influence squash gene expression to alter the timing or abundance of gene products that control cell adhesion. Finally, silvered squash leaves have normal concentrations of chlorophyll a and b and normal chloroplast development (Jiménez et al., 1995)—unlike the Arabidopsis silvering mutant pale cress (Reiter et al., 1994).
The unique and species-specific nature of the leaf-silvering disorder in squash and the dearth of information about local and systemic defense responses to phloem-feeding insects stimulated a search for genes expressed in apical, silvered leaves. SLW1 and SLW3 were expressed in apical leaves after silverleaf whitefly feeding but not after feeding by the closely related sweetpotato whitefly. SLW1 and SLW3 genes were expressed locally in response to both whitefly species. Here, the expression programs of SLW1 and SLW3 during development and in response to hormones and abiotic stress treatments are reported. Exogenous treatments with wound and defense signals, such as methyl jasmonate (MeJA), SA, abscisic acid (ABA), ethylene, H2O2, and nitric oxide (NO), indicate that distinct signal transduction pathways are used to activate SLW1 and SLW3 gene expression. Finally, SLW1 and SLW3 encode gene products that have not previously been implicated in plant–insect interactions; the possible roles of these products in defense and the silverleaf disorder are discussed.
RESULTS
Silverleaf Symptom Development and Expression of SLW1 and SLW3
The interaction of insects and plants is inherently complex. In the studies presented here, gene expression in infested leaves and in noninfested leaves at various distances from infested leaves was examined. The experimental system was set up as follows. In all experiments, three sets of plants were monitored. Plants with two true leaves were encased in a nylon mesh bag and were infested with either silverleaf whiteflies or sweetpotato whiteflies or were left as noninfested controls. During the first 3 to 5 days, the adult whiteflies fed, mated, and oviposited; after this time, the adults and/or the nylon mesh bags were removed from infested and control plants. Leaves 1 and 2 were infested with whiteflies. All leaves older than leaves 1 and 2 were not infested (apical, noninfested leaves). The leaves closest to the infested leaves (leaves 3 to 7), and another group farther from the infested leaves (8 to 12), were referred to as proximal and distal noninfested leaves, respectively. Between days 3 and 5, the eggs hatched and first instar nymphs searched for a feeding site. Once a site was established, the second to early fourth instar nymphs fed almost continuously from a minor vein. In most experiments, whitefly nymph development was not synchronous (Figures 1 and 2). However, in a few experiments (such as those described in Figures 3 and 4), a degree of synchrony in insect development was observed, allowing changes in gene expression to be correlated with insect instar.
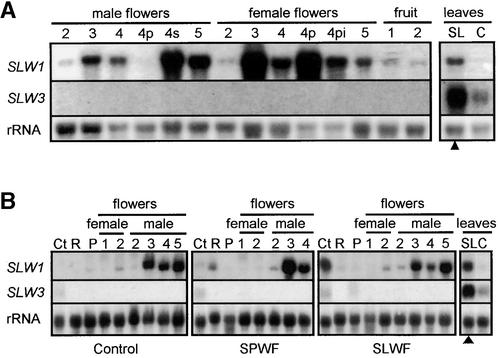
Figure 1.
Silvering Symptoms and Expression of SLW1 and SLW3 Genes in Squash Leaves.
(A) Silvering of leaves from silverleaf whitefly–infested plants. RNA was isolated from distal silvered leaves (young, leaves 13 and 14; intermediate, leaves 11 and 12; and mature, leaves 9 and 10) and from mature infested leaves (leaves 1 and 2). The four leaf stages were also examined from noninfested plants and from plants infested with sweetpotato whiteflies. Shown are the intermediate-silvered, mature-silvered, and mature-infested leaves from silverleaf whitefly–infested plants. The young leaves are not visible because they are shielded by the leaf canopy.
(B) SLW1 and SLW3 RNA accumulation in infested and distal, noninfested leaves. Total RNA (15 μg per lane) was isolated from three different distal leaf stages (young, intermediate, and mature) and from mature infested leaves. Samples were taken 4 weeks after infestation with 120 adult silverleaf whiteflies (SL) or with 120 adult sweetpotato whiteflies (SP) or from noninfested control (C) plants. Silverleaf whitefly– and sweetpotato whitefly–infested plants had 60 and 40 first- to fourth-stage instars per infested leaf at the time of harvest, respectively. RNA gel blots were hybridized with 32P-labeled SLW1 and SLW3 cDNA probes. Hybridization of blots with an 18 S rRNA probe was used as control for RNA loading and transfer. Leaves infested with whiteflies (open triangles) and those that were completely silvered (filled triangles) are indicated. Sizes of SLW1 and SLW3 RNAs are indicated in kb.
Figure 2.
Expression of SLW1 and SLW3 during Squash Development.
Blots were hybridized with 32P-labeled SLW1 and SLW3 cDNA probes. Hybridization with 18S rRNA was used as control to assess RNA loading and transfer. Mature silvered leaves from silverleaf whitefly–infested plants (SL; filled triangles) and mature green leaves from noninfested control plants (C) were included as controls.
(A) SLW1 and SLW3 RNA levels in flowers and fruit. Total RNA (15 μg per lane) was isolated from male and female flowers and fruit from noninfested control plants.
(B) SLW1 and SLW3 RNA levels in vegetative and reproductive organs after whitefly infestations. Total RNA (15 μg per lane) was isolated from cotyledons (Ct), roots (R), petioles (P), and female and male flowers of squash 4 weeks after infestation with silverleaf whiteflies (SLWF) or sweetpotato whiteflies (SPWF) or of noninfested control plants. Infested plants had first- to fourth-stage instars feeding at the time of harvest.
The floral stages shown are as follows: stage 1, bud size <1.5 cm; 2, buds 1.5 to 3 cm; 3, buds 3 to 4.5 cm; 4, open flowers; and 5, senescing flower. Petals (4p), stamens (4s), or pistils (4pi) were dissected from stage 4 flowers. Fruit stages 1 and 2 correspond to fruits 3 to 4 cm in diameter and 12 to 15 cm in diameter, respectively.
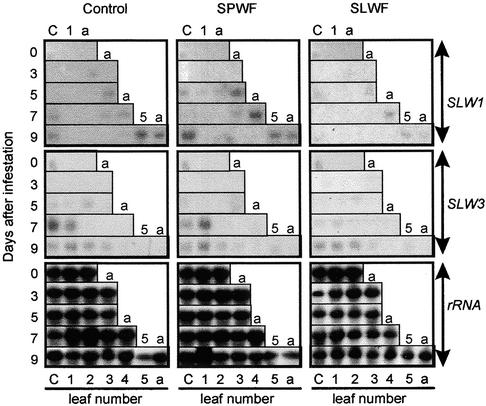
Figure 3.
Expression of SLW1 and SLW3 during Adult and First Instar Nymph Feeding.
Total RNA (15 μg per lane) was isolated from cotyledons (C), leaves (1 to 5), and the shoot apex (a) at days 0, 1, 5, 7, and 9 after infestation with 120 adult sweetpotato whiteflies (SPWF) or 120 adult silverleaf whiteflies (SLWF) or from noninfested control plants. At the time of infestation, the squash plants had one true leaf. After 9 days, five leaves had developed, leaf 5 being ∼2 cm long. Adult whiteflies were allowed to feed and oviposit during days 0 to 5, after which they were removed. From days 5 to 9, eggs hatched and first instar nymphs were observed. Blots were hybridized with 32P-labeled SLW1 and SLW3 cDNA probes. Hybridization with 18 S rRNA was used as a control to assess RNA loading and transfer.
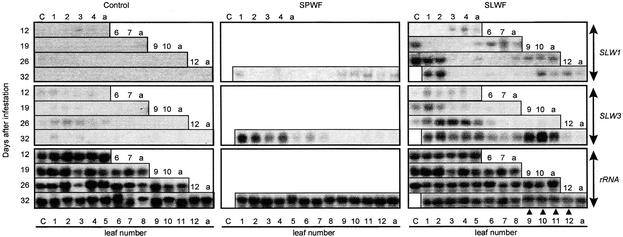
Figure 4.
Expression of SLW1 and SLW3 during Second to Fourth Instar Nymph Feeding.
Total RNA (15 μg per lane) was isolated from cotyledons (C), leaves (1 to 12), and the shoot apex (a) at days 0, 1, 5, 7, and 9 of infestation with 120 adult sweetpotato whiteflies (SPWF) or 120 adult silverleaf whiteflies (SLWF) or from noninfested control plants. At the time of infestation, the squash plants had one true leaf. After 32 days, 12 leaves had developed, leaf 12 being ∼2 cm long. Adult whiteflies were allowed to feed and oviposit for 5 days, after which they were removed. At days 12 and 19, the majority of nymphs (∼400) were in their second instar; at day 26, the majority (∼400) were in third instars; and by day 32, most (∼400 silverleaf whitefly nymphs and ∼300 sweetpotato whitefly nymphs) were in their third and fourth instars. Blots were hybridized with 32P-labeled SLW1 and SLW3 cDNA probes. Hybridization with 18 S rRNA was used as a control for RNA loading and transfer. Completely silvered leaves are indicated (filled triangles).
Squash plants infested with silverleaf whiteflies exhibited the leaf-silvering disorder (Figure 1A). During these studies, the exquisite sensitivity of squash plants to the silverleaf whitefly was noted. As previous investigators had observed (Yokomi et al., 1989; Costa et al., 1993b), only a few feeding silverleaf whiteflies were sufficient for development of silvering (data not shown). The silvering disorder was reproducibly detected in leaves that initiated development after nymph feeding had begun. In the experiments shown in Figure 1, seedlings with two leaves were infested with 120 silverleaf whiteflies. Leaves 3 to 5 were initiated during the period of adult feeding, mating, oviposition, and egg maturation. These leaves were green and were not infested with whiteflies. Leaves 6 and 7, which initiated after nymphs began to feed, were partially silvered with blanching of the veins at the basal end of the lamina. Leaf 8 showed vein clearing over the entire lamina. In all subsequent leaves, interveinal silvering was progressively more severe until the entire lamina was silvered. Leaf petioles were also blanched. Squash plants infested with sweetpotato whiteflies or noninfested controls remained free of symptoms. The timing, specificity, and nature of the silverleaf symptoms were similar to those previously described (Hoelmer et al., 1991; Schuster et al., 1991).
To identify genes correlated with leaf silvering or systemic defense responses, genes that were expressed in apical, noninfested leaves from silverleaf whitefly–infested, sweetpotato whitefly–infested, and noninfested plants were evaluated by using differential RNA display (Liang and Pardee, 1995). After screening ∼18% of the squash leaf RNA population (28 primer-pair combinations), SLW1 and SLW3 were identified. These genes were preferentially expressed in apical leaves after silverleaf whitefly feeding but not after sweetpotato whitefly feeding (Figure 1B). SLW1 RNAs were most abundant in silverleaf whitefly–infested leaves (leaves 1 and 2) (Figure 1B). SLW1 RNAs were also detected in the distal, noninfested but completely silvered leaves (leaves 9 to 14). SLW1 transcripts were not detected in leaves (Figure 1B) or petioles (Figure 2B) of noninfested controls. A low level of SLW1 RNA was detected in the sweetpotato whitefly–infested leaves, but SLW1 RNA did not accumulate systemically in these plants (Figure 1B).
Unlike SLW1, which showed distinct responses to the two whitefly species both locally and systemically, species-specific differences in SLW3 expression were detected only in distal leaves. SLW3 transcripts were most abundant in the distal, silvered leaves (leaves 9 to 14) from silverleaf whitefly–infested plants, and systemic induction was not observed after sweetpotato whitefly feeding. In contrast, RNAs accumulated to similar quantities in both silverleaf whitefly– and sweetpotato whitefly–infested leaves (leaves 1 and 2). Low levels of SLW3 RNAs were detected in the noninfested controls.
Expression of SLW1 and SLW3 during Squash Development
The accumulation of SLW1 and SLW3 RNAs in reproductive and vegetative organs from noninfested squash plants was determined (Figures 2A and 2B). SLW1 transcripts were detected in both male and female flowers. SLW1 RNAs were undetectable or present in low levels in floral buds <3 cm long (stages 1 and 2) but were abundant in more mature buds (stage 3) and in open flowers (stages 4 and 5). SLW1 RNAs were also detected in small quantities in squash fruit and occasionally in roots (Figure 2B). SLW1 RNAs were low or undetectable in control plants but accumulated in cotyledons and leaves after silverleaf whitefly infestation (Figure 2B). It was not clear whether the expression in cotyledons was a local or a systemic response. Adult whiteflies fed on cotyledons, but cotyledons were not optimal for oviposition because few feeding nymphs were detected on this organ.
In contrast to SLW1, SLW3 transcripts were not detected in any organ from healthy plants other than leaves and cotyledons, in which low levels of SLW3 RNAs were detected. Except in leaves (and in cotyledons for SLW1), the expression of SLW1 and SLW3 during development was not enhanced or suppressed by silverleaf whitefly or sweetpotato whitefly feeding (Figure 2B).
Impact of Adult and Nymph Feeding on the Expression of SLW1 and SLW3
Squash responds differently to feeding by silverleaf whitefly nymphs and adults: as few as three silverleaf whitefly nymphs induce silvering (Yokomi et al., 1989), and large numbers of feeding adults do not induce this disorder (Costa et al., 1993b). To determine whether SLW1 and SLW3 gene expression was correlated with nymph or adult whitefly feeding, the abundance of SLW1 and SLW3 RNAs was assessed in leaves, cotyledons, and the shoot apex at different times after silverleaf whitefly and sweetpotato whitefly infestation and in noninfested control plants (Figures 3 and 4). Adult whiteflies were allowed to feed and oviposit for 5 days (Figure 3). After 5 days, adults were removed, and from 5 to 9 days after infestation, their eggs hatched and the first and second instars were observed.
The levels of SLW1 and SLW3 RNAs in leaves from healthy, noninfested plants were either low or undetectable (Figure 3). A low level of SLW1 RNA was observed in cotyledons and in the shoot apex at day 7 and in cotyledons, apex, and very young leaves (leaf 5; 2 cm long) at day 9. Low levels of SLW3 RNA accumulated in cotyledons and first leaf during days 7 to 9. During the first 5 days of infestation, when 120 whitefly adults were feeding, and during days 6 to 9, when ∼400 second instars were feeding, there were no increases in SLW1 and SLW3 RNAs in leaves, cotyledons, or shoot apex that could be attributed to whitefly nymph feeding.
To determine whether SLW1 and SLW3 RNA accumulated in the apical nonsilvered leaves of silverleaf whitefly–infested plants or whether SLW1 and SLW3 gene expression was strictly correlated with the silvering disorder, the abundance of SLW1 and SLW3 RNAs in individual leaves at four different times over a 32-day period was assayed (Figure 4). In control plants, SLW1 RNAs were not detected in cotyledons, leaves, or in the shoot apex throughout the 32 days (Figure 4). In contrast, SLW3 RNAs were detected at low levels in cotyledons and leaves 1 to 5 during this period. Both SLW1 and SLW3 RNAs were detected in cotyledons and leaves of silverleaf whitefly–infested plants. On days 12 and 19, when the majority of nymphs were in the second instar, and on day 26, when the majority of insects were in their third instar, SLW1 and SLW3 RNAs accumulated in cotyledons. Cotyledons were not assayed at day 32 because they had senesced and the RNA yields were insufficient for RNA gel blot analysis.
After 26 and 32 days of infestation, when most of the insects were in their third and fourth instars, SLW1 transcripts were abundant in the infested leaves (leaves 1 and 2). SLW1 had a distinct program of expression in apical tissues. SLW1 RNAs were detected in the two or three most distal leaves and in the shoot apex but were not detected in lower, noninfested leaves. For example, at 12 days after infestation, SLW1 RNAs accumulated in leaves 3 and 4 and the apex; likewise, after 26 days, SLW1 RNAs were detected in leaves 8 to 10 and the apex. At day 32, leaves 6 to 8 were partially silvered and leaves 9 to 12 were already, or soon would become, completely silvered. By this time, leaves 10 and younger were partially expanded. Finally, 32 days after sweetpotato whitefly infestation, very low levels of SLW1 RNAs were detected in infested leaves. In addition, low levels of SLW1 RNAs were noted in the apex and in the three to four most distal leaves (leaves 9 to 12).
SLW3 regulation differed from that of SLW1. SLW3 RNAs accumulated primarily in infested leaves (leaves 1 and 2) of silverleaf whitefly–infested plants during second instar feeding (days 12 and 19). By days 26 and 32, when third and fourth instars were feeding, SLW3 RNAs were present in high levels in infested leaves and in adjacent noninfested leaves (leaves 3 to 5). SLW3 RNAs were most abundant in leaves 9 to 11, which were completely silvered, but did not accumulate in the shoot apex. Analysis of RNAs from sweetpotato whitefly–infested plants at day 32 indicated that sweetpotato whitefly also caused SLW3 RNAs to accumulate in infested leaves and in the proximal leaves (leaves 3 to 7). However, unlike silverleaf whitefly infestations, SLW3 RNAs were not detected in the more distal leaves (leaves 8 to 12) after sweetpotato whitefly infestation.
Expression of SLW1 and SLW3 in Response to Wound, Defense, and Stress Signals
To determine whether SLW1 and SLW3 genes responded to known modulators of wound and defense signal transduction pathways, the accumulation of SLW1 and SLW3 transcripts in response to wounding, Pseudomonas syringae pv syringae infection, and defense signaling molecules was determined. Because the age of leaves can impact the wound/defense response (Alarcon and Malone, 1995; Stout et al., 1996), changes in SLW1 and SLW3 gene expression were monitored in young, expanding leaves as well as in mature leaves from 3-week-old squash plants.
Very low levels of SLW1 RNAs were detected in wounded leaves 24 hr after the leaf lamina was crushed and in mature leaves 19 hr after infiltration with P. s. syringae (Figure 5A), but none accumulated in apical nonwounded or noninfected leaves. In comparison, Rasmussen et al. (1995) found a local induction of acidic peroxidases between 18 and 24 hr after P. s. syringae infiltration. SLW3 transcripts did not accumulate locally after wounding and either were not detected or were present only in low levels in P. s. syringae–infected leaves. SLW3 transcripts did not accumulate systemically after wounding or P.s. tomato infection.
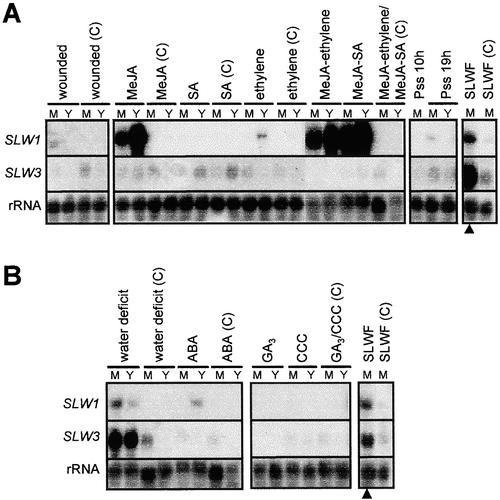
Figure 5.
Expression of SLW1 and SLW3 in Response to Wound, Defense, and Stress Signals.
Blots were hybridized with 32P-labeled SLW1 and SLW3 cDNA probes. Hybridization with 18S rRNA (rRNA) was used as a control for RNA loading and transfer. Mature silvered leaves from silverleaf whitefly–infested plants (SLWF; filled triangles) and mature green leaves from noninfested control plants (SLWF(C)) were included as controls.
(A) SLW1 and SLW3 RNA quantities in response to defense and wound signals. Total RNA (15 μg per lane) was isolated from mature (M) and young (Y) leaves of squash plants after different treatments and from their controls (C). Mature leaves of 3- to 4-week-old plants were wounded. After 24 hr, mature wounded and young, apical, nonwounded leaves were excised. Excised shoots of 3- to 4-week-old plants were treated for 24 hr with MeJA (10 μM), SA (500 μM), ethylene (10 ppm), MeJA and ethylene, or MeJA and SA. Mature leaves of 3-week-old squash plants were infiltrated with 106 colony-forming units/mL P. s. pv syringae (Pss) and harvested at 10 or 19 hr (10h and 19h) postinfection. Young noninfected leaves were harvested at 19 hr postinfection.
(B) SLW1 and SLW3 RNA quantities in response to water deficit or treatment with ABA, CCC, or GA3. Total RNA (15 μg per lane) was isolated from mature (M) and young (Y) leaves of squash plants after different treatments or from controls (C). Three-week-old squash plants were subjected to 3 days of water-deficit stress. The relative water content of the leaves from stressed plants was 75 to 79% at the time of harvest. Excised shoots of 3- to 4-week-old plants were treated with ABA (100 μM) for 24 hr. One-week-old plants were sprayed twice at 7-day intervals with CCC (920 μL/L) or were sprayed with GA3 (50 μL/L). Plants were harvested 4 weeks after the initial treatment.
A wide variety of molecules have been implicated as defense signals, including systemin, SA, ethylene, MeJA, and ABA (Peña-Cortés et al., 1991; Ryan and Pearce, 1998; Chao et al., 1999; Dempsey et al., 1999; Pieterse and van Loon, 1999). These signals can act alone or synergistically (Xu et al., 1994; O'Donnell et al., 1996; Chao et al., 1999). An excised shoot assay was used to determine whether SLW1 and SLW3 gene transcripts accumulated in squash plants after exposure to wound/defense signals (Figure 5). SLW1 RNAs were abundant in mature and young leaves from MeJA-treated plants (Figure 5A). In contrast, SLW1 RNAs did not accumulate in response to exogenous SA, and only young leaves accumulated low levels of SLW1 RNAs after ethylene treatments. There was no evidence for a synergistic effect of MeJA and ethylene, or of MeJA and SA, in young or mature leaves, because SLW1 RNA quantities after these treatments were similar to those after treatment with MeJA alone. Although the data presented here suggest that SLW1 RNAs were greater in MeJA/SA treatments than in treatments with MeJA alone, replicate experiments showed that MeJA/SA and MeJA alone caused similar increases in SLW1 transcripts. Unlike SLW1, SLW3 RNAs did not increase in response to any of the known wound- or defense-signal molecules or combinations of signals (Figure 5A).
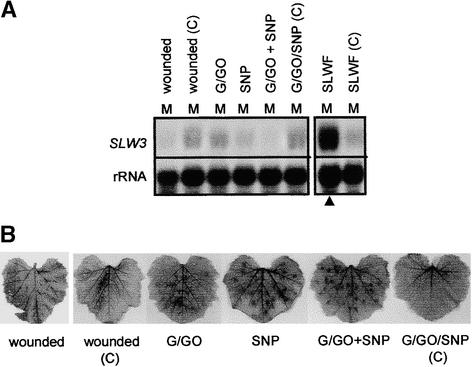
The reactive oxygen species (ROS), superoxide anion and hydrogen peroxide, and NO have been shown to activate defense gene expression (Levine et al., 1994; Delledonne et al., 1998; Desikan et al., 1998; Durner et al., 1998). To test whether ROS or NO induced SLW3 expression, excised leaves (Figure 6) or leaves from excised shoots (data not shown) were infiltrated with glucose and glucose oxidase (G/GO) to generate H2O2, with sodium nitroprusside (SNP) to generate NO, or with a mixture of G/GO and SNP. Although these treatments were shown to generate ROS, as evidenced by visualization of the brown diaminobenzidine (DAB) polymers (Figure 6B), SLW3 transcripts did not accumulate (Figure 6A). These data support the idea that a novel signal was being generated and perceived in the silverleaf whitefly–squash interaction to induce the local and systemic accumulation of SLW3 transcripts.
Figure 6.
SLW3 Expression in Response to ROS and NO.
(A) SLW3 RNA quantities after treatment with H2O2 or NO or both. Mature leaves were excised and incubated in 1 mg/mL DAB or H2O for 10 hr. Leaves were wounded or served as nonwounded controls (wound (C)). Leaves were infiltrated with G/GO, SNP, G/GO and SNP, or phosphate buffer as a control (G/GO/SNP (C)). After 4 hr of treatment or wounding, leaves were harvested for RNA isolation or decolorized to visualize DAB polymers. Blots (15 μg of total RNA per lane) were hybridized with a 32P-labeled SLW3 cDNA probe. Hybridization with 18S rRNA (rRNA) was used as a control for RNA loading and transfer. Mature silvered leaves from silverleaf whitefly–infested plants (SLWF; filled triangle) and mature green leaves from noninfested control plants (SLWF (C)) were included as controls.
(B) H2O2-generated DAB polymer visualized in leaves after treatments described in (A).
Because leaf silvering is exacerbated by drought (Paris et al., 1993), it was of interest to determine whether SLW1 or SLW3 genes were induced by water deficit. The relative water content of the mature and young leaves from well-watered or water-deficit-stressed plants was used as a measure of water-deficit stress. The mature and young leaves from control plants had relative water contents of 89 and 82%, respectively, values typical of well-watered plants, whereas the mature and young leaves from water-deficit-stressed plants had relative water contents of 75 and 79%, respectively. Water deficits of this magnitude induce water-deficit response genes (Bray, 1997). SLW1 RNAs accumulated to low levels in both young and mature leaves enduring a water deficit, and these values were well correlated with the small increases in SLW1 RNAs after treatment with ABA (Figure 5B), a known modulator of water-deficit response genes (Bray, 1997). Although SLW3 was not induced by wounding, pathogen infection, or defense signals (Figure 5A), SLW3 RNAs accumulated to high levels in mature and young leaves during water-deficit stress (Figure 5B). Because ABA treatments did not cause an increase in SLW3 transcripts, SLW3 must use an ABA-independent signaling mechanism (Shinozaki et al., 1998).
Yokomi et al. (1995) found that CCC, an inhibitor of gibberellic acid (GA3) biosynthesis, caused squash leaves to silver, whereas GA3 ameliorated CCC-induced silvering. CCC-induced silvering was less severe than silverleaf whitefly–induced silvering, and other pleiotropic effects were also noted. To determine whether CCC or GA3 regulated SLW1 or SLW3 RNAs accumulation, we treated young squash plants with GA3 or CCC. Neither SLW1 nor SLW3 transcripts were induced by these treatments (Figure 5B). In contrast to Yokomi et al. (1995), CCC-induced silvering was not observed.
SLW1 Is an M20B Metallopeptidase-like Protein
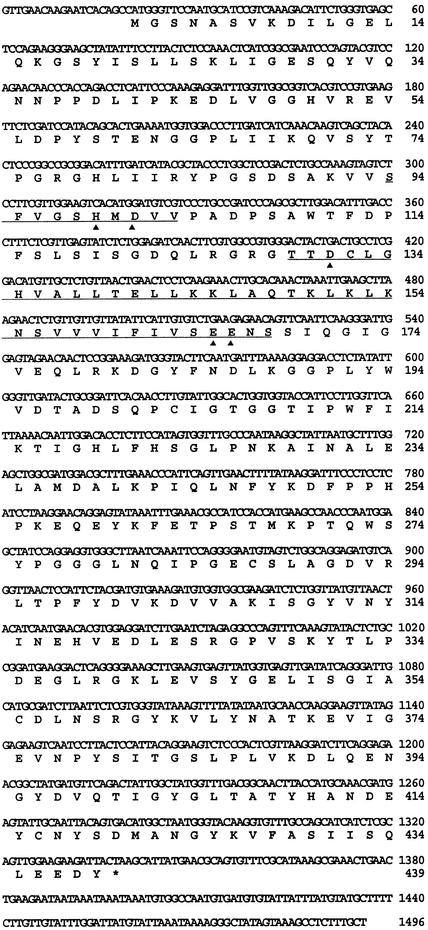
Near full-length clones of SLW1 and SLW3 cDNA were isolated and characterized by DNA sequence analysis. Figure 7 shows the nucleotide sequence and the deduced amino acid sequence of SLW1. The open reading frame of SLW1 was 1318 bp and encoded a 45-kD protein. SLW1 was predicted to be a cytosolic, soluble protein because it lacked transit or signal peptide sequences and transmembrane domains. Database searches and sequence comparisons showed that SLW1 shares strong sequence similarity (67%) with a Dictyostelium discoideum protein (DdArgE) that belongs to the M20B metallopeptidase family. Database searches also identified a SLW1 homolog in Arabidopsis (AtSLW1). The AtSLW1 cDNA clone was incomplete, and two bacterial artificial chromosome clones containing nonoverlapping portions of the AtSLW1 gene were also identified. Using SLW1-specific primers, the complete Arabidopsis gene and cDNA sequence for AtSLW1 was obtained. The Arabidopsis SLW1 protein has 74% similarity with the squash SLW1.
Figure 7.
Nucleotide and Deduced Amino Acid Sequence of SLW1.
The nucleotide sequence of the near full-length SLW1 cDNA is displayed. The deduced amino acid sequence (single-letter designation) is shown below each codon, and the stop codon is indicated by an asterisk. The cDNA sequence nucleotide and amino acid residue coordinates are indicated at right. The two M20B consensus sequences (underlined) are more thoroughly described in Figure 8. The invariant histidine, aspartic acids, and glutamic acids are indicated by arrowheads below the peptide sequence. The SLW1 sequence has GenBank accession number AF170086.
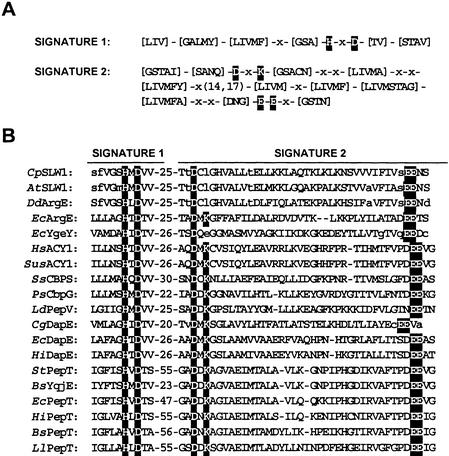
The M20B peptidases are dimeric metallopeptidases that are grouped together on the basis of two regions of strong sequence conservation (Figure 8A). The plant and Dictyostelium proteins were most closely related to each other (Figure 8B). Although they did not match the M20B signatures completely, they did share four of five conserved charged residues, and the conserved histidine may be involved in the binding of metal ions (Boyen et al., 1992). Outside these regions of conservation, the sequences of the M20B peptidases diverged.
Figure 8.
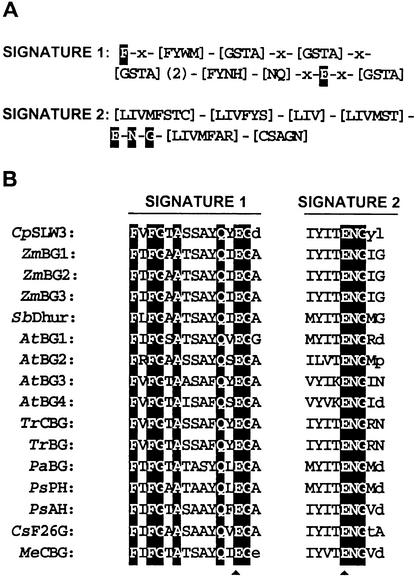
Alignment of the Consensus Sequence of Two Conserved Regions in the M20B (Gly-X Carboxypeptidase) Metallopeptidase Family.
(A) Conserved motifs in M20B. The 10–amino acid M20B signature 1 contains a conserved histidine (H), which could be involved in binding of metal ions, and a conserved aspartic acid (D). The second signature contains 12- and 13-residue regions, which are separated by 14 to 17 residues. Four conserved charged residues are found within signature 2—two aspartic acids (D), a lysine (K), and a glutamic acid (E) residues. The conserved H, D, K, and E residues are shaded. Several positions in the signature motifs allow for more than one type of residue at a position. The alternative residue choices are indicated in brackets.
(B) Signature 1 and 2 motifs for 18 M20B peptide sequences. These sequences include: Cucurbita pepo SLW1 (CpSLW1); the Arabidopsis SLW1 homolog (AtSLW1); ArgE proteins from D. discoideum (DdArgE; SwissProt accession number P54638) and Escherichia coli (EcArgE; P23908); an M20B-like hydrolase from E. coli (EcYgeY; Q46805); ACY1 from humans (HsACY1; Q03154) and swine (SusACY1; P37111); yeast carboxypeptidase-S (SsCBPS; P27614) and Pseudomonas sp strain RS-16 carboxypeptidase G2 (PsCbpG; P06621); Lactobacillus delbrueckii aminoacyl-histidine dipeptidase (LdPepV; P45494); DapE enzymes from Corynebacterium glutamicum (CgDapE; Q59284), E. coli (EcDapE; P24176), and Haemophilus influenzae (HiDapE; P44514); aminotripeptidase (PepT) proteins from Salmonella typhimurium (StPepT; P26311), E. coli (EcPepT; P29745), H. influenzae (HiPepT; P45172), Bacillus subtilis (BsPepT; P55179), and Lactococcus lactis (LlPepT;P42020); and a B. subtilis PepT–like protein (BsYqjE; P54542). The conserved H, D, K, and E residues are shaded. The residues that do not match the consensus sequences are in lowercase letters. The numbers between the signature motifs 1 and 2 indicate the number of residues separating these conserved regions. The M20B signature motifs 1 and 2 were found by using ProSite searches; signature 1 was identified by ProSite, but signature 2 motifs for CgDapE and EcYgeY were identified by visual inspection.
SLW3 Is a β-Glucosidase–like Protein
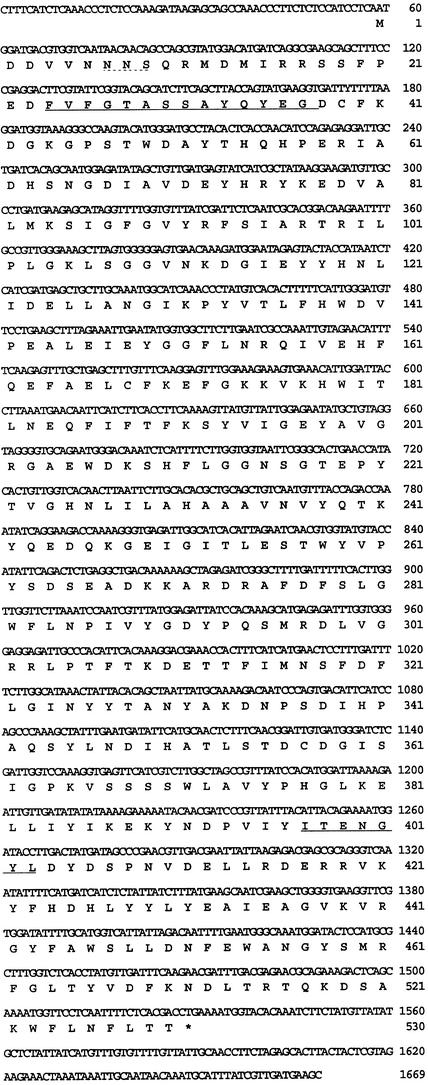
Figure 9 shows the near full-length cDNA sequence and the deduced amino acid sequence of SLW3. The open reading frame of SLW3 was 1470 bp and encoded a 52-kD protein with an acidic pI (5.25). The PSORT program did not identify any organellar targeting signals in SLW3. SLW3 had one N-glycosylation site (N-X-S/T) and shared 56% identity with the Trifolium repens cyanogenic β-glucosidase (Oxtoby et al., 1991). Similar extents of identity were found with plant β-glucosidases that hydrolyze other substrates and belong to the glycosyl hydrolase family 1 (Rojas and Romeu, 1996). The members of this family have been grouped together on the basis of sequence similarities and share two conserved regions (Figure 10A). Figure 10B compares these regions of SLW3 and of the 15 glycosyl hydrolases that were most similar to SLW3 (57 to 66% identity). Although SLW3 does not match the two signatures perfectly, it does contain the two glutamic acid residues that act as a proton donor and a nucleophile. These two residues are required for the enzymatic hydrolysis of the glycosidic bond via general acid catalysis (Davies and Henrissat, 1995).
Figure 9.
Nucleotide and Deduced Amino Acid Sequence of SLW3.
The nucleotide sequence of the near full-length SLW3 cDNA is displayed, with the deduced amino acid sequence shown (single-letter designations) below each codon. The stop codon is indicated with an asterisk. The cDNA sequence nucleotide and amino acid residue coordinates are indicated at right. The two glycosyl hydrolase family 1 consensus sequences 1 and 2 are underlined, and one putative N-glycosylation site is underlined with dots. The SLW3 sequence has the GenBank accession number AF170087.
Figure 10.
Alignment of the Consensus Sequences of Two Conserved Regions in Glycosyl Hydrolases Family 1 (Clan GH-A).
(A) Conserved motifs of the clan GH-A. The locations of signatures 1 and 2 in SLW3 are indicated in Figure 9. Signature 1 is 15 amino acid residues long and contains a conserved glutamic acid (E), which is thought to be a proton donor, and an invariant phenylalanine residue (F). Signature motif 2 is nine amino acid residues long with an invariant ENG motif. The conserved glutamic acid (E) is thought to be directly involved in glycosidic bond cleavage by acting as a nucleophile. Residues that are invariant are shaded. Several positions in the signature motifs allow for more than one type of residue at a position, and the alternative residue choices are indicated in brackets.
(B) Conservation of the signatures 1 and 2 in 15-plant β-glucosidases. The peptide sequences correlating with the glycosyl hydrolase family 1 conserved motifs are shown in alignment with 15 glucosyl hydrolase peptide sequences most closely related to C. pepo SLW3 (CpSLW3). The β-glucosidases included are: the maize chloroplast β-glucosidase (ZmBG1; SwissProt accession number P49235), cytokinin-glucosyl hydrolase in root meristems (ZmBG2; PIR accession number A48860), and an etiolated shoot β-glucosidase (ZmBG3; GenBank accession number U44087); dhurrinase from Sorghum bicolor (SbDhur; GenBank U33817); four putative β-glu-cosidases from Arabidopsis thaliana (AtBG1, GenPept accession number AAC16091; AtBG2, GenPept AAC16094; AtBG3, GenPept AAC16092; AtBG4, GenPept AAC16093); the T. repens (white clover) cyanogenic (TrCBG; PDB accession number 1311386) and noncyanogenic (TrBG; SwissProt P26205) β-glucosidases; a Prunis avium (sweet cherry) β-glucosidase (PaBG; GenBank U39228); P. seratina (black cherry) prunasin hydrolase (PsPH; GenBank U50201) and amygdalin hydrolase (PsAH; GenBank U26025); Costus speciosis furostanol-glycoside 26-O-β-glucosidase (CsF26B; GenBank D83177); and a Manihot esculenta (cassava) cyanogenic β-glucosidase (MeCBG; SwissProt S23940). Invariant residues are shaded. The residues that do not match the consensus sequence are in lowercase letters. The glutamic acid residues important for catalysis are indicated with arrowheads.
DISCUSSION
Study of whitefly–squash interactions provides a unique opportunity to dissect the changes in plant gene expression that accompany infestation by phloem-feeding insects. The fact that two closely related whitefly species, B. argentifolii and B. tabaci Type A, cause distinct phenotypic changes in squash leaves has allowed several questions to be addressed: Do different whitefly species cause similar or different changes in plant gene expression? Do whiteflies induce changes in gene expression in both infested and noninfested leaves? Does nymph and adult feeding affect plant gene expression in a similar manner? What types of signals are important in the activation of genes induced by whiteflies? And finally, can the changes in squash gene expression be correlated with a defense response or with developmental disorder induction or maintenance?
To initiate these studies, genes preferentially expressed in apical leaves from silverleaf whitefly–infested plants were identified using differential RNA display, because silvering occurred only in apical leaves and because defense-response genes that have important roles in induced resistance to pathogens and pests are expressed systemically (Karban and Baldwin, 1997; Ryan and Pearce, 1998; Dempsey et al., 1999; Pieterse and van Loon, 1999). The differential expression of SLW1 and SLW3 in response to feeding by these closely related insect species suggests that the silverleaf and sweetpotato whitefly have diverged with respect to the molecules that determine the nature of their plant–insect interactions. This idea is also supported by the diversity of developmental disorders that are induced only by the silverleaf whitefly (Maynard and Cantliffe, 1989; Brown and Costa, 1992; Costa et al., 1993a; Summers and Estrada, 1996).
Similar to the silverleaf disorder that is induced by a small number of silverleaf whitefly nymphs (Yokomi et al., 1989), SLW1 and SLW3 genes were expressed when nymph feeding began. Three to five days of feeding by whitefly adults was insufficient to activate SLW1 or SLW3. Apparently, like the leaf-silvering disorder itself, the elicitors for SLW1 and SLW3 expression were produced by nymphs and not adults. The influence of the stage of insect development on plant responses to herbivores has also been noted for volatile production in the maize–Pseudaletia separata interaction (Takabayashi et al., 1995).
The fact that plant genes were preferentially induced by a specific whitefly species indicates that we are just beginning to uncover the complexity of plant–insect interactions. As other plant interactions with insects that use alternate modes of feeding are investigated, an increasing diversity in plant gene expression responses will most likely be revealed. This idea is supported by several studies that show the differential accumulation of peroxidase, polyphenol oxidase, proteinase inhibitor, and lipoxygenase activities after damage incurred by caterpillars (Helicoverpa zea and M. sexta), beetles (Leptinotarsa decemlineata), leaf miners (Liriomyza trifolii), and mites (Aculops lycopersici) (Green and Ryan, 1982; Stout et al., 1994). More recently, differences in tomato responses to aphids and whiteflies were noted. Although transcripts for the tomato Wfi1 gene that encodes a JA/ethylene–regulated subunit of the NADPH oxidase (gp91-phox) were detected after whitefly feeding, Wfi1 transcripts were not detected in plants infested with pink potato aphid (D.P. Puthoff and L.L. Walling, manuscript submitted).
The differences in the local and systemic signals being generated or perceived by the squash plant during silverleaf and sweetpotato whitefly feeding are notable. For example, in response to silverleaf whitefly, SLW1 RNAs accumulated to high levels in infested leaves, in the most distal leaves, and in the shoot apices. SLW1 transcripts were detected only in low levels in the sweetpotato whitefly–infested leaves, and systemic expression was not seen. In contrast, SLW3 RNAs accumulated to high levels in both silverleaf whitefly– and sweetpotato whitefly–infested leaves, but long-range systemic expression was observed only in silverleaf whitefly–infested plants.
SLW1 and SLW3 were expressed in low levels, or not at all, after P. s. syringae infection and severe mechanical wounding. However, treatments with defense signals showed that SLW1 RNA concentrations increased in response to MeJA and ethylene. These data are consistent with the fact that the JA/ethylene–responsive PR genes and Wfi1 are expressed in tomatoes after whitefly feeding (Chao et al., 1999; D.P. Puthoff and L.L. Walling, submittedmanuscript ; D.P. Puthoff, C.S. LeVesque, T.M. Perring, and L.L. Walling, manuscript in preparation). Unlike SLW1, none of the known wound and defense signals, including ROS and NO, alone or in combination, induced SLW3 RNA accumulation. These data indicated that SLW3 is regulated by an unknown defense signal transduction mechanism. It is not clear if this novel signaling pathway is also used by other insects. However, the preferential, long-range systemic expression of SLW3 by the silverleaf whitefly suggests that the silverleaf whitefly may generate novel signals, larger amounts of a systemic elicitor, or a more potent systemic signal than is produced by the sweetpotato whitefly.
The initial signal or signals perceived by the plant to induce SLW1 and SLW3 gene expression are not known. Although only a small amount of physical damage and mechanical stress occurs during whitefly feeding, these stresses could produce signals that activate gene expression. Such stresses include the rare destruction of an epidermal cell or mesophyll cell, the disruption of the plant extracellular matrix as whitefly stylets move between cells of the leaf lamina, and the puncturing of the minor vein when establishing a feeding site (Cohen et al., 1998). Although these physical and mechanical stresses have the potential to generate signals that contribute to the recognition of insects that use a piercing mode of feeding, these mechanisms cannot account for the differences in temporal and spatial expression of SLW1 and SLW3 gene expression induced by the silverleaf and sweetpotato whitefly. Because stylet probing and feeding site selection occur only with adults and first instar nymphs (Cohen et al., 1998) and because SLW1 and SLW3 gene expression correlates with the feeding of second to fourth instar nymphs, it seems unlikely that these physical stresses are generating the critical signal. Furthermore, the method of probing and feeding is thought to be similar in silverleaf and sweetpotato whiteflies; therefore, the species-specific differences in SLW1 and SLW3 expression cannot be accounted for by these mechanical signaling mechanisms.
More probably, a component of the nymph's digestive or sheath saliva may be the elicitor for SLW gene expression. The importance of insect regurgitant in the induction of wound- and herbivory-response genes and volatile production has been documented (Korth and Dixon, 1997; Páre and Tumlinson, 1999). The whitefly elicitor or elicitors may be synthesized by the insect or may be a product of the different endosymbiotic bacterial species harbored in the silverleaf and sweetpotato whitefly mycetomes (Costa et al., 1995). Perhaps this elicitor is generated by biochemical activities of the insect and the plant in concert, similar to what has been observed in the synthesis of volicitin, an inducer of the production of terpenoid volatiles (Páre et al., 1998). If components in the saliva are elicitors of SLW1 and SLW3 gene expression, the composition of the salivas of the silverleaf whitefly adults must be different from the salivas of second to fourth instars. To account for the limited induction of SLW1 and SLW3 by the sweetpotato whitefly, it must be proposed that the sweetpotato whitefly produced small quantities of this elicitor, a less potent elicitor, or a less mobile elicitor.
The whitefly elicitor probably stimulates the plant to produce JA/MeJA or ethylene (or all of these) to activate SLW1 expression. Alternatively, silverleaf whitefly saliva might contain JA, MeJA, or a MeJA mimic. JA mimics, such as coronatine, are produced by the pathogens (Palmer and Bender, 1995). Coronatine activates expression of wound-response genes but does not affect JA-responsive PR genes (Wasternack et al., 1998; Chao et al., 1999; V. Pautot, F.M. Holzer, J. Chaufaux, and L.L. Walling, submittedmanuscript ). Whether insects produce JA or JA mimics is at present unknown.
Water deficit also generated signals that activated SLW1 and SLW3 gene expression, suggesting that water deficit and responses to phloem feeding may share some signaling elements. At least four signal transduction pathways are important in mediating water-deficit responses, including the ABA-dependent and -independent responses (Bray, 1997; Shinozaki et al., 1998). Because SLW1 transcripts increased in response to ABA, SLW1 is probably regulated by an ABA-dependent signaling pathway. In contrast, SLW3 is regulated by an ABA-independent pathway. At present, it is not clear whether the signals generated during water-deficit stress were similar to or distinct from those generated by the feeding of silverleaf whiteflies. An interesting correlation is that low soil moisture is also known to accentuate squash leaf silvering (Paris et al., 1993). Water-deficit stress in squash may, if its response is similar to that of cotton, increase numbers of eggs and nymphs (Flint et al., 1994). Because the degree of silvering is proportional to the number of feeding insects, this could account for the enhanced silvering during water deficit. Alternately, both water-deficit stress and whitefly feeding may induce the expression of genes that potentiate or cause leaf silvering.
The systemic induction of SLW1 RNA by the silverleaf whitefly was probably not a response to silvering, because SLW1 was expressed before the development of silvering. Given that SLW1 transcripts accumulated only in leaves that were still expanding, perhaps the SLW1 gene product potentiated silvering. The SLW1 polypeptide had two signature peptide motifs found in proteins belonging to the M20B peptidase family. M20B proteins have diverse roles in cellular metabolism, including deacetylating peptides and amino acids or acting as carboxypeptidases (Boyen et al., 1992). These enzymes influence arginine metabolism (ArgE), lysine metabolism and prokaryotic cell wall biosynthesis (DapE), catabolism of acetylated amino acid residues and deacetylation of bioactive peptides that control cell regulation (ACY1), purine and pyrimidine biosynthesis by catabolizing folic acid (CPG2), processing of C-terminal residues from peptides and proteins (yscS; CPG2), and processing of N-terminal amino acids from dipeptide or tripeptide substrates by aminopeptidases (PepV and PepT, respectively). At present, it is not clear whether SLW1 is a new member of the M20B family or encodes one of the established activities. Current efforts are focused on determining the activity associated with the SLW1 protein by using the well-defined biochemical and functional complementation assays for the M20B enzymes. These efforts have been hampered by difficulties in overexpressing the full-length SLW1 in E. coli; other expression systems are being pursued.
SLW3 expression was not as strictly correlated with leaf silvering, because SLW3 RNAs accumulated to nearly equivalent levels in leaves infested with silverleaf and sweetpotato whiteflies and in the apical leaves in close proximity to the infested leaves. However, because SLW3 transcripts were abundant in completely silvered leaves but not in similar leaves from control or sweetpotato whitefly–infested plants, it is not clear whether SLW3 has a role in defense, development of silvering disorder, or both. SLW3 showed a high degree of similarity with plant β-glucosidases. β-Glucosidases impact plant defense responses and development in several ways, depending on the substrates on which they act. β-Glucosidases influence plant development by catabolizing glycosylated forms of phytohormones (Brzobohaty et al., 1993) and fruit maturation by hydrolyzing cell wall polysaccharides (Wallner and Walker, 1975). In addition, many molecules with roles in plant defense (i.e., phenols, isoflavanoids, SA, and cyanogenic compounds) are released from a glucosylated storage form by β-glucosidases (Miller, 1973). Given the diversity of potential molecules that can be β-glucosidase substrates, assigning a role to the SLW3 β-glucosidase–like protein would be only speculative. Current efforts are focused on expressing the full-length SLW3 protein in E. coli and yeast. Finally, several experiments suggest that an insect β-glucosidase may be an elicitor for the release of volatiles that attract herbivore predators and parasitoids (Hopke et al., 1994; Mattiacci et al., 1995). Given the identification of a plant β-glucosidase–like gene that is expressed locally and systemically by insect feeding, the potential role of SLW3 in the release of volatiles is intriguing. At present, little is known about volatiles generated by homopteran insects such as aphids and whiteflies (Quiroz et al., 1997; Bernasconi et al., 1998; Du et al., 1998). Transgenic plant strategies are being pursued to test the possible role of SLW3 in this process.
METHODS
Plant and Insect Materials
Cucurbita pepo cv Chefini were grown in soil supplemented with osmocote in a growth chamber with a 14-hr-light (24.5°C)/10-hr-dark (21.5°C) cycle. Plants were watered daily and fed with a solution of Miracle Gro (concentrated 18-18-21; Stern's) once a week. In all insect infestation studies and plant treatments, leaves were numbered from the base of the plant stem, with leaf 1 being the oldest leaf. Plants were monitored weekly to ensure that they were not contaminated with additional insects. Bemisia argentifolii Bellows and Perring (Bemisia tabaci Type B, the silverleaf whitefly) and B. tabaci Gennadius (B. tabaci Type A, the sweetpotato whitefly) were reared in separate greenhouses in insect-proof cages on Phaseolus vulgaris. The Bemisia cultures were assayed periodically for isoenzyme variants to ensure culture purity. Phosphoglucomutase and phosphoglucoisomersase isoenzyme patterns readily distinguish B. argentifolii and B. tabaci Type A (Perring et al., 1992).
Insect Infestations
One-week-old squash plants were transferred to insect-proof cages within a greenhouse whose temperatures ranged from 40°C (day) to 10°C (night), and the plants were monitored continuously. One day after transfer, three plants per treatment were infested with 100 female and 20 male silverleaf whiteflies or sweetpotato whiteflies per plant. The whiteflies were confined to each plant by placing an insect-proof mesh bag over the entire plant. Control (noninfested) plants were also covered with a mesh bag. The adult whiteflies were allowed to oviposit for 3 to 5 days, after which all adult whiteflies and the mesh bags were removed. The eggs were allowed to hatch and develop from the first to fourth nymphal instars. At 4 to 5 weeks after infestation, just before the new generation of adult whiteflies would have emerged, the experiment was terminated. At the end of the experiment, sweetpotato whitefly–infested leaves had an average of 40 nymphs feeding per leaf, whereas silverleaf whitefly–infested leaves averaged 60 nymphs per leaf; all stages of development were represented for both species. Mature infested leaves (leaves 1 and 2) and distal silvered leaves were harvested. Three stages of silvered leaf development were studied: young (leaves 13 and 14), intermediate (leaves 11 and 12), and mature, fully expanded leaves (leaves 9 and 10). Cotyledons, petioles, roots, flowers, and fruit were also excised. All tissues were placed directly in liquid nitrogen and stored at −80°C. Plant materials at the same developmental stage (three plants per treatment) were pooled. Each experiment was repeated three times. Older female flowers and fruit were harvested from noninfested plants that were allowed to continue growing for an additional 2 to 3 weeks.
For the time-course experiments, cotyledons, leaves, and the shoot apex were excised from infested or control plants and placed in liquid nitrogen at 0, 3, 5, 7, 9, 12, 19, 26, and 32 days after infestation. Individual leaves of the same developmental stage from three plants per treatment were pooled. The time-course experiments were repeated twice. Because of the slowed development of whiteflies in cooler temperatures, a partial synchrony of whitefly development was achieved. From days 12 to 19, most nymphs (∼400 silverleaf whiteflies per plant) were in their second instar stage. By days 26 and 32, there were ∼400 silverleaf whitefly and 300 sweetpotato whitefly nymphs per plant, most in their third and fourth instars, respectively. The sweetpotato whitefly colony was not large enough to allow sampling at four times after infestation in the experiment presented. Therefore, leaves 1 to 12 were assayed only once, at 32 days after sweetpotato whitefly infestation. These data obtained were consistent with previous time-course experiments.
Treatments to Induce Wound, Defense, and Stress Signals
Mature leaves (leaves 3 to 5) from 3-week-old squash plants were wounded as described by Pautot et al. (1991). Leaf lamina were crushed ∼30 times with needle-nosed pliers. Leaves from control plants were not wounded. The wounded mature leaves (leaves 3 to 5) and apical nonwounded young leaves (leaves 6 to 8) were harvested and placed in liquid nitrogen 24 hr after wounding.
Pseudomonas syringae pv syringae treatments were performed as described by Smith et al. (1991). Mature infected leaves and apical, noninfected young leaves were excised and placed in liquid nitrogen 10 and 19 hr after P. s. syringae infiltration; by 19 hr, defense genes were induced (Rasmussen et al., 1995).
Shoots from 3-week-old squash plants were excised just above the second true leaf for treatment with salicylic acid (SA), methyl jasmonate (MeJA), abscisic acid (ABA), or ethylene. The 24-hr MeJA (10 μM) and control treatments were performed as described by Gu et al. (1996). The SA (0.5 mM), ABA (100 μM), ethylene (1 μL/L), and control treatments were performed as described by Chao et al. (1999). Mature leaves (leaves 3 to 5) from two plants per treatment were pooled; the young leaves (leaves 6 to 8) were pooled separately. The chlormequat chloride (CCC), gibberellic acid (GA3), and control treatments were performed as described by Yokomi et al. (1995).
Three-week-old squash plants were subjected to water-deficit stress by withholding water for 3 days, after which mature and young leaves were excised and placed in liquid nitrogen. Control plants were watered daily. Some of the leaves were used for measuring relative water content. Leaves were excised and weighed (FW), allowed to fully hydrate in water for 16 hr, and weighed again (HW). The hydrated leaves were fully desiccated at 45°C for 16 hr and weighed (DW). Relative water content was calculated as ([FW − DW] / [HW − DW]) × 100%.
For treatments that generated reactive oxygen species (ROS), mature leaves from 2- to 3-week-old squash plants submerged in water were excised at the base of the petiole with a razor blade. Leaves were immediately placed in a solution of 1 mg/mL 3,3-diaminobenzidine (DAB), pH 3.8, for 10 hr under continuous light at 25°C, according to Orozco-Cardenas and Ryan (1999). Leaves were wounded as described above or served as nonwounded controls. Glucose and glucose oxidase (G/GO) were used to generate H2O2 as described by Alvarez et al. (1998). Leaves were infiltrated with 25 U/mL Aspergillus niger GO (Calbiochem) in a solution of 25 mM d-glucose in 20 mM sodium phosphate buffer, pH 6.5. Nitric oxide (NO) was generated after sodium nitroprusside (SNP) treatments. Leaves were infiltrated with 25 mM SNP (Sigma) in 20 mM sodium phosphate buffer. Leaves were also infiltrated with a combination of G/GO and SNP or with 20 mM sodium phosphate buffer alone (control). Leaves were kept in the DAB solution for an additional 4 hr, after which they were harvested into liquid nitrogen for RNA isolations or decolorized in boiling ethanol (95%) for 10 min to visualize the brown DAB polymers formed in response to ROS (Orozco-Cardenas and Ryan, 1999). These experiments were repeated with seedlings, excising the shoots just above cotyledons and treating them similarly to the excised leaves. All experiments were repeated twice. Leaves from three plants per treatment were pooled for RNA isolations.
RNA Isolation and Gel Blot Analysis
Total RNA was isolated from all plant materials by the procedure described in Martienssen et al. (1989). RNA gel blot hybridizations were performed with 15 μg of total RNA according to Pautot et al. (1991). Probes were labeled with α-32P-dCTP (NEN, Boston, MA) by using the Prime-a-Gene labeling system (Promega).
Differential RNA Display and cDNA Characterization
Differential RNA display profiles from noninfested apical leaves from whitefly–infested or noninfested plants were evaluated. DNAs from mature, silvered leaves from silverleaf whitefly–infested plants and mature, green leaves of sweetpotato whitefly–infested and noninfested plants were compared using the Hieroglyph RNA profile kit and Genomyx LR sequencer (Genomyx, Foster City, CA). Many changes in gene expression accompanied squash leaf development; therefore, all leaf samples were examined at the same developmental stage to ensure that the cDNAs identified reflected changes in gene expression correlated with whitefly feeding and not with developmental cues. Twenty-eight primer–pair combinations identified 11 differentially expressed cDNA bands. These cDNAs were eluted according to the vendor's (Genomyx) instructions. cDNAs were used as templates to generate 32P-labeled probes for RNA gel blot analysis to confirm the differential expression of each gene (Sambrook et al., 1989). Of the 11 bands excised, eight failed to detect a RNA in total RNA gel blots or were false positives, but the remaining three SLW genes were differentially expressed. These SLW cDNAs were cloned into pGEM-T Easy (Promega) and sequenced by using the T7-Sequenase system (version 2.0; Amersham) or fmol DNA sequencing system (Promega) with T7 promoter and M13 reverse primers. Sequence analysis showed that SLW2 and SLW3 encoded the same gene product.
Near full-length SLW1 and SLW3 cDNA sequences were obtained using the Universal GenomeWalker kit (Clontech, Palo Alto, CA). Poly(A+) mRNA was isolated from total RNA from mature, silvered leaves by using Dynabeads (Dynal, Great Neck, NY) according to the manufacturer's instructions. Double-stranded cDNA was synthesized by using reverse transcriptase and the RiboClone cDNA synthesis system (Promega). The GenomeWalker kit adapter was ligated to the cDNA according to the instructions for generating a cDNA/adapter library. Oligonucleotides corresponding to the 5′ sequences of the differential display cDNAs for SLW1 primer 1 (5′-ATCTTCGCC-ACCACATCTTTCACATCGTAG-3′) and SLW3 primer 1 (5′-AGTCAAGGTATCCATTTTCTGTAA-3′) were synthesized (Genosys, The Woodlands, TX). The GenomeWalker adapter primers (as 5′ primers) were used with SLW1 primer 1 or SLW3 primer 1 (as 3′ primers) to amplify the 5′ end of the SLW1 and SLW3 cDNAs from the cDNA/adapter library. The amplified products were sequenced. Near full-length cDNA clones from SLW1 and SLW3 were obtained through a similar strategy, using the GenomeWalker adapter primers and the 3′-terminal primers for SLW1 (SLW1 primer 2, 5′-ACATAATCCAAATACAACAAGAAAAGCATA-3′) and SLW3 (SLW3 primer 2, 5′-TCAGGTCGTGAGAAAATTGAGGAAC-3′). The amplified products were cloned into pGEM-T Easy and sequenced. The near full-length SLW1 and SLW3 cDNAs were used as probes in RNA gel blot analyses to confirm differential expression.
Sequence Analyses
Database searches were performed using NCBI BLAST and deduced amino acid sequences from the nucleotide sequences by using the TRANSLATE program (version 6; Genetics Computer Group, Madison, WI). Amino acid sequence alignments were performed using PILE UP from GCG. Consensus sequences were extracted from the PROSITE data bank (PROSITE: A dictionary of protein sites and patterns; http://expasy.hcuge.ch/). Deduced polypeptide sequences were submitted to PSORT to predict the presence of signals for targeting proteins to organelles.
Acknowledgments
We thank Dr. Jennifer Becker for providing the P. s. pv syringea strain and for help with infiltrations; Arthur Cooper for help with whitefly infestations and immature whitefly counts; Nhung Nuygen, Paula Huang, and Wanida Ruangsiriluk for laboratory assistance; and members of the Walling laboratory, especially David P. Puthoff, for helpful discussions. This research was partially supported by a University of California Biotechnology Grant, U.S. Department of Agriculture (USDA) Grant No. 95-37301-2081 to T.M.P. and L.L.W., and USDA Grant No. 99-35301-8077 to L.L.W.
References
- Alarcon, J.J., and Malone, M. (1995). The influence of plant age on wound induction of proteinase inhibitors in tomato. Physiol. Plant. 95, 423–427. [Google Scholar]
- Alvarez, M.E., Pennell, R.I., Meijer, P.J., Ishikawa, A., Dixon, R.A., and Lamb, C. (1998). Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92, 773–784. [DOI] [PubMed] [Google Scholar]
- Bernasconi, M.L., Turlings, T.C.J., Ambrosetti, L., Bassetti, P., and Dorn, S. (1998). Herbivore-induced emissions of maize volatiles repel the corn leaf aphid, Rhopalosiphum maidis. Entomol. Exp. Appl. 87, 133–142. [Google Scholar]
- Boyen, A., Charlier, D., Charlier, J., Sakanyan, V., Mett, I., and Glansdorff, N. (1992). Acetylornithine deacetylase, succinyldiaminopimelate desuccinylase and carboxypeptidase-G2 are evolutionarily related. Gene 116, 1–6. [DOI] [PubMed] [Google Scholar]
- Bray, E.A. (1997). Plant responses to water deficit. Trends Plant Sci. 2, 48–54. [Google Scholar]
- Brown, J.K., and Costa, H.S. (1992). First report of whitefly-associated squash silverleaf disorder of Cucurbita in Arizona and of white streaking disorder of Brassica species in Arizona and California. Plant Dis. 76, 426–426. [Google Scholar]
- Brown, J.K., Frohlich, D.R., and Rosell, R.C. (1995). The sweetpotato or silverleaf whiteflies: Biotypes of Bemisia tabaci or a species complex? Annu. Rev. Entomol. 40, 511–534. [Google Scholar]
- Brzobohaty, B., Moore, I., Kristoffersen, P., Bako, L., Campos, N., Schell, J., and Palme, K. (1993). Release of active cytokinin by a β-glucosidase localized to the maize root meristem. Science 262, 1051–1054. [DOI] [PubMed] [Google Scholar]
- Chao, W.S., Gu, Y.-Q., Pautot, V., Bray, E.A., and Walling, L.L. (1999). Leucine aminopeptidase mRNAs, proteins and activities increase in response to drought, salinity and the wound signals—Systemin, methyl jasmonate, and abscisic acid. Plant Physiol. 120, 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, A.C., Chu, C.C., and Henneberry, T.J. (1998). Feeding biology of the silverleaf whitefly (Homoptera: Aleyrodidae). Chin. J. Epidemiol. 18, 65–82. [Google Scholar]
- Costa, H.S., Ullman, D.E., Johnson, M.W., and Tabashnik, B.E. (1993. a). Association between Bemisia tabaci density and reduced growth, yellowing, and stem blanching of lettuce, and kai choy. Plant Dis. 77, 969–972. [Google Scholar]
- Costa, H.S., Ulmanh, D.E., Johnson, M.W., and Tabashnik, B.E. (1993. b). Squash silverleaf symptoms induced by immature but not adult Bemisia tabaci. Phytopathology 83, 763–766. [Google Scholar]
- Costa, H.S., Westcot, D.M., Ullman, D.E., Rosell, R., Brown, J.K., and Johnson, M.W. (1995). Morphological variation in Bemisia endosymbionts. Protoplasma 189, 194–202. [Google Scholar]
- Davies, G., and Henrissat, B. (1995). Structures and mechanisms of glycosyl hydrolases. Structure 3, 853–859. [DOI] [PubMed] [Google Scholar]
- Delledonne, M., Xia, Y., Dixon, R.A., and Lamb, C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394, 585–588. [DOI] [PubMed] [Google Scholar]
- Dempsey, D.M.A., Shah, J., and Klessig, D.F. (1999). Salicylic acid and disease resistance in plants. Crit. Rev. Plant Sci. 18, 547–575. [Google Scholar]
- Desikan, R., Reynolds, A., Hancock, J.T., and Neill, S.J. (1998). Harpin and hydrogen peroxide both initiate programmed cell death but have differential effects on defence gene expression in Arabidopsis suspension cultures. Biochem. J. 330, 115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Y., Poppy, G.M., Powell, W., Pickett, J.A., Wadhams, L.J., and Woodcock, C.M. (1998). Identification of semiochemicals released during aphid feeding that attract parasitoid Aphidius ervi. J. Chem. Ecol. 24, 1355–1368. [Google Scholar]
- Durner, J., Wendehenne, D., and Klessig, D.F. (1998). Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 95, 10328–10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint, H.M., Wilson, F.D., Hendrix, D., Leggett, J., Naranjo, S., Henneberry, T.J., and Radin, J.W. (1994). The effect of plant water stress on beneficial and pest insects including the pink bollworm and the sweetpotato whitefly in two short-season cultivars of cotton. South. Entomol. 19, 11–22. [Google Scholar]
- Green, T.R., and Ryan, C.A. (1982). Wound-induced proteinase inhibitors in plant leaves: A possible defense mechanism against insects. Science 175, 776–777. [DOI] [PubMed] [Google Scholar]
- Gu, Y.Q., Chao, W.S., and Walling, L.L. (1996). Localization and post-translational processing of the wound-induced leucine aminopeptidase proteins of tomato. J. Biol. Chem. 271, 25880–25887. [DOI] [PubMed] [Google Scholar]
- Hoelmer, K.A., Osborne, L.S., and Yokomi, R.K. (1991). Foliage disorders in Florida associated with feeding by sweetpotato whitefly, Bemisia tabaci. Fla. Entomol 74, 162–166. [Google Scholar]
- Hopke, J., Donath, J., Blechert, S., and Boland, W. (1994). Herbivore-induced volatiles: The emission of acyclic homoterpenes from leaves of Phaseolus lunatus and Zea mays can be triggered by a β-glucosidase and jasmonic acid. FEBS Lett. 352, 146–150. [DOI] [PubMed] [Google Scholar]
- Jiménez, D.R., Yokomi, R.K., Mayer, R.T., and Shapiro, J.P. (1995). Cytology and physiology of silverleaf whitefly–induced squash silverleaf. Physiol. Mol. Plant Pathol. 46, 227–242. [Google Scholar]
- Karban, R., and Baldwin, I.T. (1997). Induced Responses to Herbivory. (Chicago: University of Chicago Press).
- Korth, K.L., and Dixon, R.A. (1997). Evidence for chewing insect-specific molecular events distinct from a general wound response in leaves. Plant Physiol. 115, 1299–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, A., Tenhaken, R., Dixon, R., and Lamb, C. (1994). H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79, 583–593. [DOI] [PubMed] [Google Scholar]
- Liang, P., and Pardee, A.B. (1995). Recent advances in differential display. Curr. Opin. Immunol. 7, 274–280. [DOI] [PubMed] [Google Scholar]
- Martienssen, R.A., Barkan, A., Freeling, M., and Taylor, W.C. (1989). Molecular cloning of a maize gene involved in photosynthetic membrane organization that is regulated by Robertson's Mutator. EMBO J. 8, 1633–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiacci, L., Dicke, M., and Posthumus, M.A. (1995). β-Glucosidase: An elicitor of herbivore-induced plant odor that attracts host-searching parasitic wasps. Proc. Natl. Acad. Sci. USA 92, 2036–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, R.T., McCollum, T.G., McDonald, R.E., Polston, J.E., and Doostdar, H. (1996). Bemisia feeding induces pathogenesis-related proteins in tomato. In Bemisia 1995: Taxonomy, Biology, Damage, Control and Management, D. Gerling and R.T. Mayer, eds (Andover, UK: Intercept Ltd.), pp. 179–188.
- Maynard, D.N., and Cantliffe, D.J. (1989). Squash Silverleaf and Irregular Ripening: New Vegetable Disorders in Florida. Veg Crops Facts Sheet Fla Coop Ext Serv Inst. Food Agric. Sci. (Gainesville, FL: University of Florida).
- Miller, L.P. (1973). Glycosides. In Phytochemistry. The Process and Products of Photosynthesis, L.P. Miller, ed (New York: Van Nostrand Reinhold), pp. 297–375.
- Miyairi, K. (1988). Biochemical studies on the silver-leaf inducing substance of apple silver-leaf disease. Bull. Fac. Agric. Hirosaki Univ. 49, 61–148. [Google Scholar]
- Miyairi, K., Fujita, K., Okuno, T., and Sawai, K. (1977). A toxic protein causative of silver-leaf disease symptoms on apple trees. Agric. Biol. Chem. 41, 1897–1902. [Google Scholar]
- O'Donnell, P.J., Calvert, C., Atzorn, R., Wasternack, C., Leyser, H.M.O., and Bowles, D.J. (1996). Ethylene as a signal mediating the wound response of tomato plants. Science 274, 1914–1917. [DOI] [PubMed] [Google Scholar]
- Orozco-Cardenas, M., and Ryan, C.A. (1999). Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc. Natl. Acad. Sci. USA 96, 6553–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxtoby, E., Dunn, M.A., Pancoro, A., and Hughes, M.A. (1991). Nucleotide and derived amino acid sequence of the cyanogenic β-glucosidase (linamarase) from white clover (Trifolium repens L.). Plant Mol. Biol. 17, 209–219. [DOI] [PubMed] [Google Scholar]
- Palmer, D.A., and Bender, C.L. (1995). Ultrastructure of tomato leaf tissue treated with the pseudomonad phytotoxin coronatine and comparison with methyl jasmonate. Mol. Plant-Microbe Interact. 8, 683–692. [Google Scholar]
- Páre, P.W., and Tumlinson, J.H. (1999). Plant volatiles as a defense against insect herbivores. Plant Physiol. 121, 325–331. [PMC free article] [PubMed] [Google Scholar]
- Páre, P.W., Alborn, H.T., and Tumlinson, J.H. (1998). Concerted biosynthesis of an insect elicitor of plant volatiles. Proc. Natl. Acad. Sci. USA 95, 13971–13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris, H.S., Stoffella, P.J., and Powell, C.A. (1993). Sweetpotato whitefly drought stress and leaf silvering of squash. Hortic. Sci. 28, 157–158. [Google Scholar]
- Pautot, V., Holzer, F.M., and Walling, L.L. (1991). Differential expression of tomato proteinase inhibitor-I and inhibitor-II genes during bacterial pathogen invasion and wounding. Mol. Plant-Microbe Interact. 4, 284–292. [DOI] [PubMed] [Google Scholar]
- Pautot, V., Holzer, F.M., Reisch, B., and Walling, L.L. (1993). Leucine aminopeptidase: An inducible component of the defense response in Lycopersicon esculentum (tomato). Proc. Natl. Acad. Sci. USA 90, 9906–9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Cortés, H., Willmitzer, L., and Sanchez-Serrano, J.J. (1991). Abscisic acid mediates wound induction but not developmental-specific expression of the proteinase inhibitor-II gene family. Plant Cell 3, 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perring, T.M., Cooper, A., and Kazmer, D.J. (1992). Identification of the poinsettia strain of Bemisia tabaci (Homoptera, Aleyrodidae) on broccoli by electrophoresis. J. Econ. Entomol. 85, 1278–1284. [Google Scholar]
- Perring, T.M., Cooper, A.D., Rodriguez, R.J., Farrar, C.A., and Bellows, T.S., Jr. (1993). Identification of a whitefly species by genomic and behavioral studies. Science 259, 74–77. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J., and van Loon, L.C. (1999). Salicylic acid–independent plant defense pathways. Trends Plant Sci. 4, 52–58. [DOI] [PubMed] [Google Scholar]
- Quiroz, A., Pettersson, J., Pickett, J.A., Wadhams, L.J., and Niemeyer, H.M. (1997). Semiochemicals mediating spacing behavior of bird cherry-oat aphid, Rhopalosiphum padi feeding on cereals. J. Chem. Ecol. 23, 2599–2607. [Google Scholar]
- Ramos, L.J., Bharanthan, N., McMillan, R.T., and Narayanan, K.R. (1995). Histopathological changes associated with silverleaf syndrome in squash. Plant Pathol. 44, 316–324. [Google Scholar]
- Rasmussen, J.B., Smith, J.A., Williams, S., Burkhart, W., Ward, E., Somerville, S.C., Ryals, J., and Hammerschmidt, R. (1995). cDNA cloning and systemic expression of acidic peroxidases associated with systemic acquired resistance to disease in cucumber. Physiol. Mol. Plant Pathol. 46, 389–400. [Google Scholar]
- Reiter, R.S., Coomber, S.A., Bourett, T.M., Bartley, G.E., and Scolnik, P.A. (1994). Control of leaf and chloroplast development by the Arabidopsis gene pale cress. Plant Cell 6, 1253–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas, A., and Romeu, A. (1996). A sequence analysis of the β-glucosidase subfamily B. FEBS Lett. 378, 93–97. [DOI] [PubMed] [Google Scholar]
- Ryals, J.A., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, C.A., and Pearce, G. (1998). Systemin: A polypeptide signal for plant defensive genes. Annu. Rev. Cell Dev. Biol. 14, 1–17. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schuster, D.J., Kring, J.B., and Price, J.F. (1991). Association of the sweetpotato whitefly with a silverleaf disorder of squash. HortScience 26, 155–156. [Google Scholar]
- Shinozaki, K., et al. (1998). Molecular responses to water stress in Arabidopsis thaliana. J. Plant Res. 111, 345–351. [Google Scholar]
- Smith, J.A., Hammerschmidt, R., and Fulbricht, D.W. (1991). Rapid induction of systemic resistance in cucumber by Pseudomonas syringae pv. syringae. Mol. Plant Pathol. 38, 223–235. [Google Scholar]
- Stout, M.J., Workman, J., and Duffey, S.S. (1994). Differential induction of tomato foliar proteins by arthropod herbivores. J. Chem. Ecol. 20, 2575–2594. [DOI] [PubMed] [Google Scholar]
- Stout, M.J., Workman, K.V., Workman, J.S., and Duffey, S.S. (1996). Temporal and ontogenetic aspects of protein induction in foliage of the tomato, Lycopersicon esculentum. Biochem. Syst. Ecol. 24, 611–625. [Google Scholar]
- Summers, C.G., and Estrada, D. (1996). Chlorotic streak of bell pepper: A new toxicogenic disorder induced by feeding of the silverleaf whitefly, Bemisia argentifolii. Plant Dis. 80, 822. [Google Scholar]
- Takabayashi, J., Takahashi, S., Dicke, M., and Posthumus, M.A. (1995). Developmental stage of herbivore Pseudaletia separata affects production of herbivore-induced synomone by corn plants. J. Chem. Ecol. 21, 273–287. [DOI] [PubMed] [Google Scholar]
- Wallner, S.J., and Walker, J.E. (1975). Glycosidases in cell wall–degrading extracts of ripening tomato fruits. Plant Physiol. 55, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C., Ortel, B., Miersch, O., Kramell, R., Beale, M., Greulich, F., Feussner, I., Hause, B., Krumm, T., Boland, W., and Parthier, B. (1998). Diversity in octadecanoid-induced gene expression of tomato. J. Plant Physiol. 152, 345–352. [Google Scholar]
- Xu, Y., Chang, P.F.L., Liu, D., Narasimhan, M.L., Raghothama, K.G., Hasegawa, P.M., and Bressan, R.A. (1994). Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell 6, 1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomi, R.K., Osborne, L.S., and Hoelmer, K.A. (1989). Squash silverleaf and its association with the sweetpotato whitefly. Phytopathology 79, 1161–1162. [Google Scholar]
- Yokomi, R.K., Hoelmer, K.A., and Osborne, L.S. (1990). Relationships between the sweetpotato whitefly and the squash silverleaf disorder. Phytopathology 80, 895–900. [Google Scholar]
- Yokomi, R.K., Jimenez, D.R., Osborne, L.S., and Shapiro, J.P. (1995). Comparison of silverleaf whitefly–induced and chlormequat chloride–induced leaf silvering in Cucurbita pepo. Plant Dis. 79, 950–955. [Google Scholar]