Abstract
Parasitic strategies are widely distributed across the angiosperms and are estimated to have evolved at least eight different times. Within the obligate hemiparasitic and holoparasitic members, elaborate strategies for host selection have emerged. Here, we demonstrate that in the parasitic Scrophulariceae Striga asiatica, for which signal-mediated host detection is critical, expansin mRNA provides a reliable and accurate downstream molecular marker for the transition to the parasitic mode. Three different expansin genes, saExp1, saExp2, and saExp3, are regulated by xenognostic quinones. saExp3 appears to function as a seedling expansin, and its mRNA is depleted within minutes after induction of the host attachment organ. saExp1 and saExp2 share less homology with the known expansins, and their transcripts accumulate linearly over a critical induction period. The regulation of these genes suggests that the resources for developmental commitment must accumulate to a defined threshold before commitment to organogenesis is terminal. When the induction signal is removed prematurely, the accumulated message decays with a time constant that correlates with the time required for additional signal exposures to reinduce parasitic development. These results suggest that sophisticated controls exist for the accumulation of the necessary components for terminal commitment to the parasitic mode. Furthermore, building on the redox dependence of the inducing signal, they suggest a model akin to a “molecular capacitor” for clocking organogenesis in S. asiatica.
INTRODUCTION
Organizing the complex cellular array of eukaryotic organs requires precise spatial and temporal commitments during organogenesis. The parasitic plants constitute a special situation in which organogenesis can be coupled to signals originating from the epidermal cells of the host plant (Kim et al., 1998). In the obligate and holoparasitic plants, viability is intimately linked with signal perception and ultimate host attachment (Kuijt, 1969; Press and Graves, 1995). Development of the host attachment organ, the haustorium, in the parasitic Scrophulariceae Striga asiatica, involves a recommitment of the cells of the root meristem (Riopel and Baird, 1987). Rapid arrest of root elongation, a redirection of cellular expansion from longitudinal to radial dimensions in the cells just distal to the root tip, and finally the development and growth of haustorial hairs centrifugal to the swelling root tip constitute the stages of haustorial organogenesis. Premature commitment of the root meristem to haustorial development in this obligate parasite restricts further seedling growth, which limits host attachment and viability.
Molecular signals are both necessary and sufficient to induce this transition from vegetative growth to haustorial development in Striga. Structural evaluation of synthetic analogs of the xenognostic quinones has established that signal perception correlates with electromotive potential (Smith et al., 1996). The active quinones appear to depend on both reductive and oxidative events, defining a redox window between −250 and 0 mV. These data, together with inhibitors specifically designed to exploit the reactivity of a semiquinone intermediate, have led to a model in which the quinones serve as one-electron carriers in a redox reaction that induces haustorial development (Smith et al., 1996).
Signal perception also has proven to be time dependent, with quinone exposures of several hours generally being required (Chang and Lynn, 1986; Smith et al., 1990, 1996). Although prior histological studies demonstrated that a halt in cell division and elongation occurred immediately after xenognosin exposure (Riopel and Baird, 1987), the signal exposure time for terminal commitment to haustorial development was quinone specific (Smith et al., 1996). At times that were less than half the maximal exposure time (t1/2) needed for each xenognostic quinone, meristematic growth stalled in the presence of the inducing signal but resumed immediately after the signal was removed. Although no mature attachment organs formed in response to these short exposures, radial enlargement could be detected at the point along the growing root axis that had been exposed to signal (Smith et al., 1990). Signal removal/reexposure experiments showed that multiple exposure times could be summed, as long as the time between exposures was short. With longer delays after an initial exposure, this “clock” was gradually reset so that ≥6 hr after signal removal, a full exposure time was required for haustorial induction (Smith et al., 1990). In an obligate parasite, in which premature commitment of the root meristem to haustorial development can seriously compromise host attachment, the timing of signal exposure before commitment could serve to increase the precision in host commitment. Therefore, the mechanism that underlies this apparent ability to clock signal exposure is of interest.
Because haustorial development is critically dependent on cellular expansion, we evaluate here the regulation of the cell wall–loosening protein, expansin, during signal exposure. Elaborate transcriptional control is exerted on three expansin genes. Analysis of the regulation of these marker genes has led us to propose a mechanistic model for the control of developmental commitment in the vegetative/parasitic transition of S. asiatica.
RESULTS
Haustorial Development
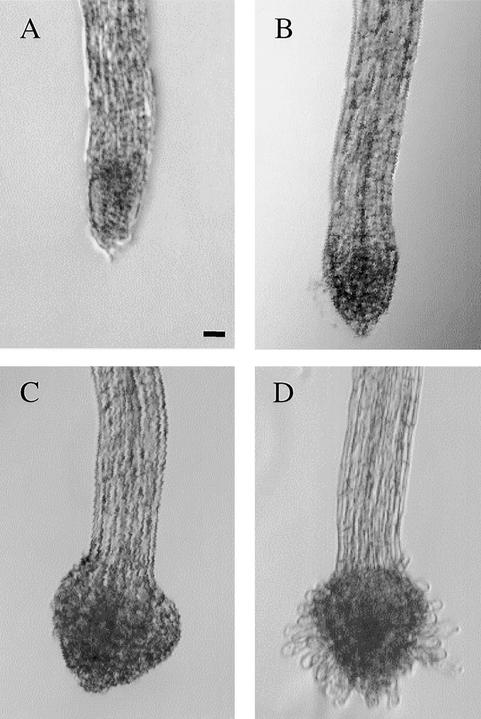
The stages of haustorial induction in S. asiatica with 2,6-dimethoxy-p-benzoquinone (DMBQ) are marked initially by the arrest of normal root elongation (cf. Figures 1A and 1B). As early as 6 hr after exposure, radial swelling of the meri-stem can be detected; by 8 hr, cells distal to the root meristem are visibly enlarged (Figure 1B). This swelling continues, producing the large bulbous root tip and the haustorial hairs that emerge around the periphery of the swollen tip by 16 hr (Figure 1C). By 22 hr, the haustorial hairs have extended, and development of the mature attachment-competent organ is complete (Figure 1D). In Agalinis purpurea and Triphysaria spp, haustorial hairs appear before the radial swelling (Baird and Riopel, 1984; Yoder, 1997), reinforcing our current beliefs that these morphological changes are common among the parasitic Scrophulariceae and that the order of their appearance may vary.
Figure 1.
Haustorial Development.
Magnification (×20) of 2-day-old S. asiatica seedlings exposed to 10 μM DMBQ for 0 hr (A), 8 hr (B), 16 hr (C), and 22 hr (D). Similar morphologies are observed after 24 hr when seedlings are exposed for 2 hr to 10 μM or for 3 hr to 2 μM DMBQ (B) or for 6 hr to 10 μM or for 10 hr to 2 μM DMBQ (C).  .
.
Expansins of S. asiatica
The apparent centrality of cell enlargement in the morphological changes of haustorial development was tested qualitatively by measuring the incorporation of thymidine into DNA (Hoy et al., 1990). The observations that incorporation halted within minutes of DMBQ exposure and did not recover during the swelling process further supported the critical position of cell expansion in haustorial development (data not shown). A novel class of proteins called expansins has been implicated in such cell enlargement events (McQueen-Mason, 1995; Shieh and Cosgrove, 1998; Cosgrove, 1999). However, attempts to monitor expansins directly with protein or RNA gel blots were complicated by the small size of the S. asiatica seedling (one-tenth the size of the Arabidopsis seedling), and degenerate primers based on regions of homology between 10 known expansin sequences were used in a reverse transcription–polymerase chain reaction (RT-PCR) procedure. The RT-PCR products from RNA isolated from DMBQ-treated (10 μM for 4 hr) and untreated seedlings were cloned, and 40 of the clones were sequenced, revealing the presence of three seedling-expressed expansins, SaExp1, SaExp2, and SaExp3. Using the same degenerate primers, we isolated 10 additional expansin sequences from cloned PCR products amplified from genomic DNA, eight of which encoded unique Striga expansin sequences. These eight genomic expansins, which differed from the three saExp genes, demonstrate that the degenerate expansin primers are not specific to a subset of expansin genes and, importantly, that, of the large expansin gene family present in S. asiatica, only a small subset are expressed in Striga seedlings.
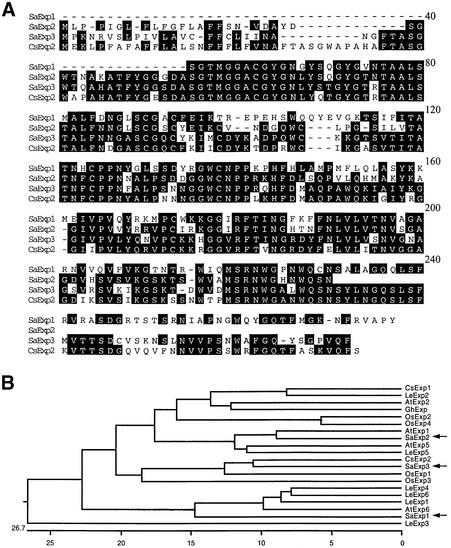
Rapid amplification of cDNA ends (RACE; Frohman, 1995) was used to obtain the untranslated 3′ region of SaExp1 and SaExp3 and the 5′ region of SaExp2 and SaExp3. A comparison of the translated regions of the complete sequences, including the SaExp2 and SaExp3 signal peptide sequences, is shown in Figure 2A. The three expansins show high sequence identity (>60%) to each other and to expansins found in other species. Figure 2B, in a dendrogram generated by DNASTAR, compares expansins in S. asiatica seedlings with those from other species. SaExp3 is most similar to a cucumber hypocotyl expansin (CsExp2)—with 90% identity excluding the variable 20 C-terminal residues—and to two rice root–localized expansins (OsExp1 and OsExp3). SaExp2 is most related to known Arabidopsis (AtExp1 and AtExp5) and tomato (LeExp5) expansins.
Figure 2.
Deduced Striga Expansin Amino Acid Sequences.
(A) SaExp1, SaExp2, and SaExp3 sequences are compared with each other and with the cucumber expansin CsExp2 by using the DNASTAR (Madison, WI) Megalign Clustal program. Conserved residues are shaded.
(B) Dendrogram of the coding regions of 20 different expansins without signal peptides. The positions of the SaExp sequences are indicated in the dendrogram by arrows. The GenBank accession numbers are as follows: AtExp1 (U30476), AtExp2 (U030481), AtExp5 (U30478), and AtExp6 (U30480) from Arabidopsis thaliana; CsExp1 (U30382) and CsExp2 (U30476) from Cucumis sativus; LeExp1 (U82123), LeExp2 (AJ239068), LeExp3 (AF059487), LeExp4 (AF059488), LeExp5 (AF059489), and LeExp6 (AAD13635) from Lycopersicon esculentum; OsExp1 (Y07782), OsExp2 (U30477), OsExp3 (U30479), and OsExp4 (U85246) from Oryza sativa; and GhExp1 (AF043284) from Gossypium hirsutum.
SaExp1 is an outlier, in part because of its amino acid sequence from positions 97 to 113 amino acids, which has little homology to other known expansins in the consensus sequence shown in Figure 2A. Normally, this region contains two of the eight cysteines conserved in all other α-expansins. However, this pair of cysteines, the fourth and fifth from the N terminus, are missing from SaExp1, suggesting that a conserved intramolecular disulfide linkage may exist between these cysteines in other expansins. Interestingly, β-expansins also share only six of the eight cysteines conserved in α-expansin, but they lack the next pair, at positions six and seven from the N terminus (Cosgrove et al., 1997).
Quantification of SaExp Transcripts
A competitive RT-PCR procedure, similar to that described by Wang et al. (1989), was developed to circumvent the limited sensitivity afforded by the small Striga seedlings. Gene-specific SaExp1, SaExp2, and SaExp3 primers were designed from the divergent untranslated regions. At positions between these primer pairs, a 30-bp insertion in SaExp1 and SaExp2 and a 50-bp deletion in SaExp3 were constructed by complementary PCR (Ho et al., 1989). In a PCR amplification with the gene-specific primers, wild-type saExp1, saExp2, and saExp3 give products of 150, 200, and 300 bp, respectively, and the synthetic saExp constructs give products of 180, 230, and 250 bp, respectively. The synthetic saExp constructs were cloned into a vector containing the T7 RNA polymerase promoter and transcribed into sense RNA that could be quantified using UV irradiation of the PAGE gel following ethidium bromide staining.
Because the wild-type and synthetic SaExp constructs share identical primer sites and only a small size difference, the efficiency of RT-PCR amplification should be comparable for the two species. Indeed, when these synthetic SaExp RNAs were added in a dilution series to three aliquots of 0.5 μg of total Striga RNA, reverse-transcribed into cDNA with the antisense gene-specific primers, and PCR-amplified with 32P-labeled gene-specific primers, the standard curves had regression analyses correlation coefficients of 0.98. As shown in the Figure 3 insert, three band intensities from the wild-type product can be used as controls for tube-to-tube variability under these conditions. Using the ratio of the wild-type and synthetic band intensities to generate the standard curve eliminated tube-to-tube variations in amplification (Pfaffl et al., 1998). With this quantitative expression assay, it was possible to evaluate accurately the transcriptional regulation of expansin message during haustorial development.
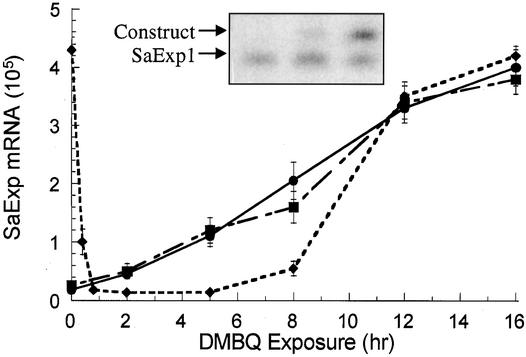
Figure 3.
Quantification of SaExp mRNA.
Quantities of SaExp1 (circles), SaExp2 (squares), and SaExp3 (diamonds) transcripts in the seedlings exposed to 2 μM DMBQ as determined by competitive RT-PCR. The inset gel was subjected to imaging with a PhosphoImager and contains the RT-PCR product for SaExp1 at 12-hr induction. An aliquot of 0.5 μg of seedling RNA was combined with each of three separate samples containing a dilution series, 20-fold each, of a synthetic SaExp1 RNA with a 30-bp insertion (construct). Each RNA sample was reverse-transcribed, PCR-amplified with 32P-labeled SaExp1 primers, and used to determine the wild-type concentration from a standard curve by using the ratiometric method described by Pfaffl et al. (1998). Tube-to-tube variation showed R > 0.98; errors based on variation between triplicate runs are expressed as ±sd.
Figure 3 shows the changes in the three xenognosin-regulated expansins in seedlings exposed to DMBQ (2 μM) over a 16-hr period. SaExp1 and SaExp2 have similar basal values, and both were upregulated by DMBQ. After 12 hr of DMBQ exposure, the level of each mRNA plateaued ∼15-fold higher than the corresponding basal values. In contrast, SaExp3 decreased 30-fold within 15 min of DMBQ exposure, remained this low for at least 6 hr, and then returned to its preinduction value by 12 hr. This second upregulation occurred in the apparent absence of any new external stimuli.
SaExp Regulation in Xenognosis
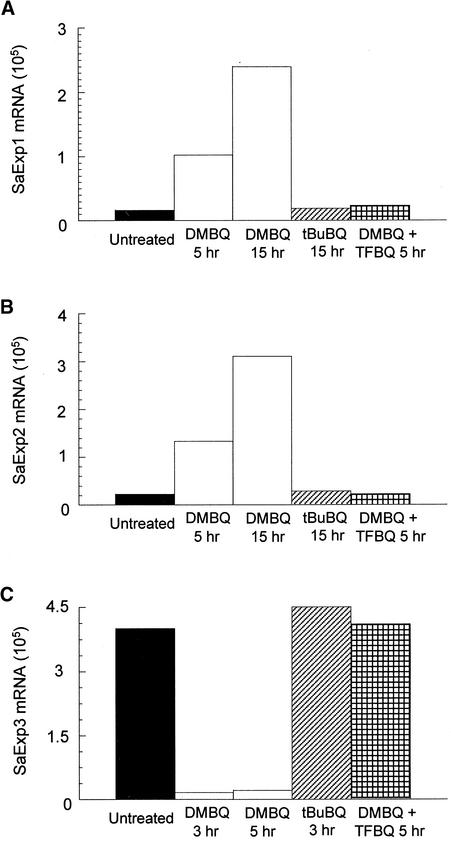
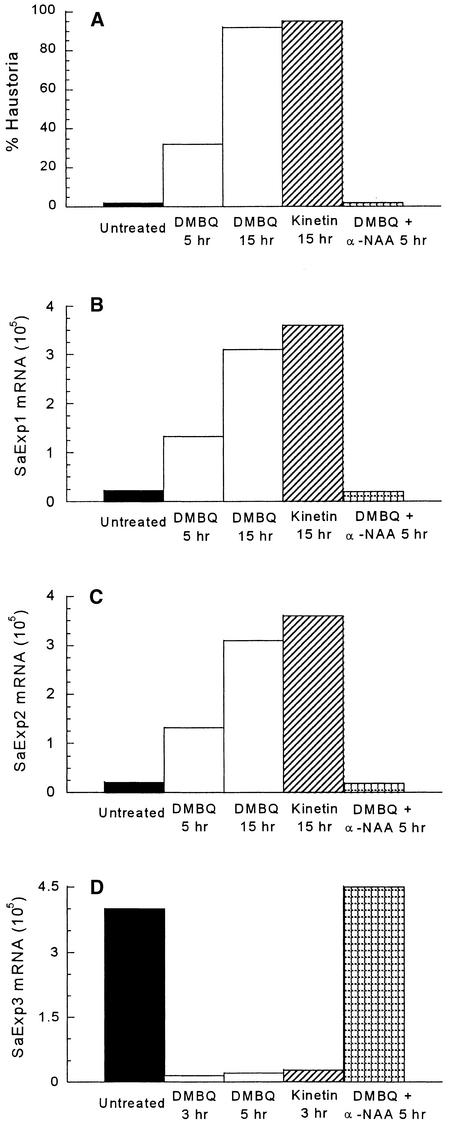
The chemistry of xenognosin-regulated haustorial development is well developed in S. asiatica (Smith et al., 1996; Zeng et al., 1996) and was exploited in evaluating the specificity of SaExp accumulation. Binding site requirements appear to restrict the xenognostic benzoquinones to one or two small substituents, and tert-butyl benzoquinone (tBuBQ) is inactive (Smith et al., 1996). As shown in Figure 4, the inactive tBuBQ does not stimulate any accumulation of SaExp1 and SaExp2 within 15 hr or any depletion of SaExp3 in 3 hr, in both cases giving results identical with those for the water control. This absence of an effect with tBuBQ is striking because its Em is approximately −70 mV, well within the active redox window.
Figure 4.
Effect of Various Quinones on SaExp mRNA Accumulation.
SaExp quantities were measured in a 0.5-μg RNA sample from 2-day-old seedlings treated with water alone (Untreated), DMBQ (10 μM), tBuBQ (10 μM), or DMBQ (10 μM) plus tetrafluorobenzoquinone (TFBQ; 1 μM).
This redox window was further exploited with tetrafluorobenzoquinone (TFBQ) (Smith et al., 1996). TFBQ is sterically similar to benzoquinone, but its Em exceeds +500 mV. Therefore, TFBQ should be accommodated sterically by a benzoquinone site and readily reduced as a one-electron carrier but not reoxidized within the oxidative limit of the defined window. Consistent with this analysis, TFBQ functions as a specific, competitive, and reversible inhibitor of haustorial induction (Smith et al., 1996), and, as shown in Figure 4, the DMBQ (10 μM)-induced depletion of SaExp3 and accumulation of SaExp1 and SaExp2 transcripts are completely blocked by coincubating the seedlings with TFBQ (1 μM).
In addition to quinone inducers, Striga seedlings are responsive to certain plant hormones. Kinetin induces haustorial development in parasitic Scrophulariceae (Riopel and Baird, 1987), and a 10-μM concentration is sufficient for quantitative induction in S. asiatica (Figure 5A). In contrast, auxins proved to be very potent inhibitors of haustorial induction: the 0.1 μM α-naphthalene acetic acid (α-NAA) was sufficient to completely inhibit induction by DMBQ (10 μM; Figure 5A). The effects of kinetin (10 μM) on SaExp1 and SaExp2 accumulation as well as the characteristic decrease in SaExp3 after short exposures are the same as DMBQ, in terms of rates and total amounts (Figures 5B to 5D). The α-NAA effect mimics that of the quinone inhibitor TFBQ and prevents DMBQ-induced changes in transcript amounts (Figures 5B to 5D). Taken together, SaExp regulation correlates tightly with the induction of haustorial organogenesis.
Figure 5.
Effect of Plant Hormones on Haustoria and SaExp mRNA Accumulation.
(A) Two-day-old seedlings (20 to 30 per well) were incubated at 30°C with water (Untreated), DMBQ (10 μM), kinetin (10 μM), or DMBQ (10 μM) plus α-naphthalene acetic acid (α-NAA; 0.1 μM); washed; and transferred to water at the times shown. After 24 hr of incubation, the seedlings were scored for haustorial development.
(B) and (C) SaExp1 and SaExp2 transcripts, respectively, were measured in 2-day-old seedlings treated for the times shown with water (Untreated), DMBQ (10 μM), kinetin (10 μM), or DMBQ (10 μM) plus α-NAA (0.1 μM).
(D) SaExp3 measured 3 or 5 hr after the same treatments described in (B) and (C).
Time Dependence of SaExp Regulation
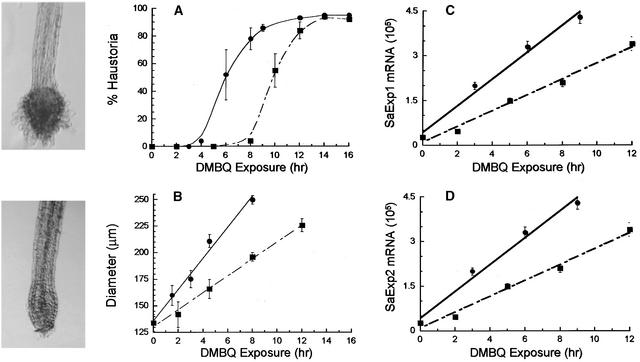
As might be expected for an obligate parasite, the overall time required for haustorial development is short. The required signal exposure time for induction, however, is long and, even more remarkably, varies with xenognosin concentration. Figure 6 quantifies the time dependence for signal exposure by using exposure/removal sequences with two different DMBQ concentrations, 2 and 10 μM. From the data presented in Figure 6A, t1/2 was determined as 10.5 hr with 2 μM and 6.3 hr with 10 μM DMBQ. Therefore, within the active concentration range, the greater the concentration of the quinone, the shorter the required exposure time.
Figure 6.
Concentration Dependence of Haustorial Induction and SaExp Accumulation.
Two-day-old S. asiatica seedlings (20 to 30 per well) were incubated with 10 μM (circles) or 2 μM (squares) DMBQ in 24-well culture plates; approximately every 2 hr, the DMBQ solution was replaced with double-distilled water in triplicate sets of wells.
(A) and (B) At 24 hr, seedlings were scored for either (A) haustorial development, with an enlarged tip and hairs, as in the photograph at top left (after 8-hr exposure to 10 μM DMBQ), or (B) root tip diameter, as in the photograph at bottom left (after 3-hr exposure to 10 μM DMBQ).
(C) and (D) SaExp1 and SaExp2, respectively, were measured in 2-day-old seedlings incubated with 10 μM (circles) or 2 μM (squares) DMBQ for the indicated times. Errors (based on variation between triplicate runs) are expressed as ±sd.
Short DMBQ exposure, that is, less than t1/2, did not lead to development of a viable attachment organ (see Figure 6A, photograph); however, visible radial enlargement orthogonal to normal vegetative elongation was apparent along the root axis (see Figure 6B, photograph). In Figure 6B, the degree of this radial expansion, expressed as the average increase in root meristem diameter, is shown to be a linear function of quinone exposure time. The fivefold greater DMBQ concentration results in a 1.7-fold increase in the rate of this radial swelling, the same fold increase as t1/2 for commitment to haustorial development shown in Figure 6A. Therefore, the spherical swelling of the root tip appears to build over time to a maximal enlargement, a function of both xenognosin concentration and signal exposure time, beyond which the seedling commits irreversibly to haustorial development.
Measurements of the rates of message accumulation with either 10 or 2 μM DMBQ for both SaExp1 and SaExp2 are shown in Figures 6C and 6D, respectively. Both expansins accumulate linearly with time, but the rate of increase is the same: 1.7 times faster at the higher DMBQ concentration. Thus, expansin accumulation and cellular enlargement show the same relative dependence on inducer concentration. At t1/2 for 10 and 2 μM DMBQ, the absolute number of transcripts per 0.5 μg of total RNA is essentially identical at both concentrations for SaExp1 (2.5 × 105) and for SaExp2 (3.0 × 105 and 2.8 × 105, respectively).
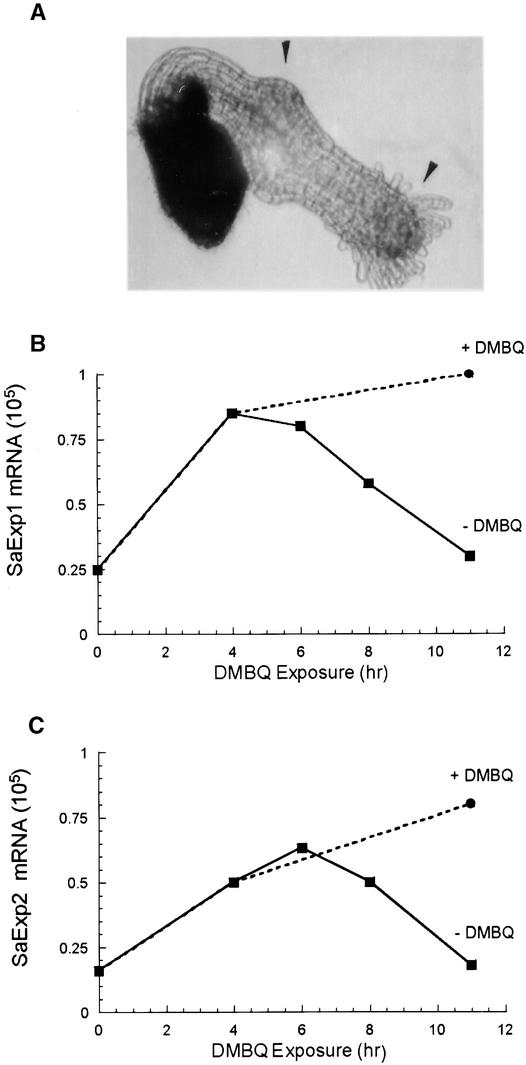
Previous signal exposure/removal experiments had established that multiple exposures were additive for haustorial induction as long as the delay between reexposures was <2 hr (Smith et al., 1990). Longer delays of ⩽6 hr showed a gradual increase in the exposure time required for the second exposure. The extent to which a connection exists between the absolute number of expansin mRNA transcripts and haustorial commitment could therefore be further tested by measuring SaExp after signal removal. Message was isolated from seedlings that were untreated, DMBQ treated for 4 or 10 hr, or DMBQ treated for 4 hr and then washed and incubated for 2, 4, or 6 hr in water (Figures 7A and 7B). The 4- and 10-hr–induced samples show the expected linear accumulation of SaExp1 and SaExp2. In the samples in which DMBQ was replaced with water, SaExp1 and SaExp2 decreased to their preinduction values by 6 hr after inducer removal. With a much longer (10 hr) exposure, inducer removal results in a similar 6-hr return to preinduction values (data not shown), suggesting that the increased quantities of SaExp1 and SaExp2 mRNA have a 6-hr lifetime in the absence of inducer, independent of the absolute increase in message.
Figure 7.
Effect of Delay Times on SaExp1 and SaExp2 Accumulation.
(A) Two-day-old S. asiatica seedlings were exposed to DMBQ (10 μM) for 2 hr, washed, incubated for 10.5 hr in water, and reexposed to DMBQ. The photograph was taken 48 hr after the start of the experiment. Arrowheads indicate areas of induced swelling.
(B) and (C) SaExp1 and SaExp2, respectively, in seedlings treated with 10 μM DMBQ for 0, 4, and 11 hr (circles). A subset of the seedlings exposed for 4 hr was washed and placed in water for 2, 4, or 6 hr (squares) before being measured.
The apparent 2-hr overshoot of SaExp1 and SaExp2 accumulation seen in Figures 7A and 7B correlates with the previous DMBQ removal/readdition experiments; after a 2-hr removal time, the normal 6-hr time frame for haustorial commitment is required. Likewise, after 6 hr without DMBQ, the amounts of SaExp1 and SaExp2 are back to seedling control values, consistent with the requirement of an additional 6 hr of exposure to accumulate sufficient message for commitment to haustorial development. Therefore, not only does accumulation of SaExp1 and SaExp2 messages correlate with a threshold for commitment to haustorial development, but message stability suggests a mechanism to regulate commitment in the face of multiple signal exposures of variable frequency.
DISCUSSION
A plant cell's size is static when the outward pressure of turgor is less than the resistance of the cell wall. During cell growth, both forces are altered, but current models suggest that a decrease in cell wall resistance initiates changes in both cell size and cell shape (Cosgrove, 1987, 1997; Carpita and Gibeaut, 1993). A novel class of proteins, the expansins, induce cell enlargement by changing wall rheology in the network of intertwined saccharide polymers (McQueen-Mason et al., 1992; reviewed in McQueen-Mason, 1995; Shieh and Cosgrove, 1998; Cosgrove, 1999). At the low pH values associated with plant cell growth, expansins do not appear to have hydrolytic activity; instead, they are proposed to disrupt hydrogen bonding between cellulose microfibrils and the polysaccharide matrix in which they are embedded. This alteration allows slippage of load-bearing cellulose microfibrils and results in cell wall expansion (McQueen-Mason and Cosgrove, 1994).
Expansins are both widely distributed across divergent plant species and highly conserved (Shcherban et al., 1995). Their expression is upregulated by the same environmental and hormonal signals that increase the growth rate of rice internodes and the ripening of tomato fruit (Cho and Kende, 1997; Rose et al., 1997), and in situ hybridization experiments have localized their expression to growing cells (Cho and Kende, 1998; Reinhardt et al., 1998). In tomato, expansin upregulation precedes the initiation of new leaf primordia (Reinhardt et al., 1998), and applying expansin to the apical meristem during leaf generation is sufficient to trigger out-of-sequence development of leaf primordia (Fleming et al., 1997). These data argue that expansin is a necessary component for cell enlargement and suggest that it may even be a sufficient early stimulus for initiation of developmental commitments.
The xenognosin-regulated expansin genes identified in S. asiatica—saExp1, saExp2, and saExp3—are all members of a larger gene family and are expressed in the seedling during vegetative growth; nonetheless, the messages of the different expansins accumulate in very different ways after xenognosin exposure. Several lines of evidence suggest that saExp3 plays a role in normal vegetative seedling growth of Striga. First, saExp3 is the predominant expansin expressed in the seedling; accumulation of this message is very rapidly reduced by haustorial induction, and this depletion corresponds in time with the halt in vegetative elongation. Second, the SaExp3 sequence is ∼75% identical with OsExp1 and OsExp3, which are expressed in rice root meristem (Cho and Kende, 1997); in contrast, SaExp1 and SaExp2 show less similarity to expansins expressed in the root apex, and in fact, both fall into outlying groups, with SaExp1 being one of the most divergent of the α-expansins. Finally, the return of SaExp1 message to preinduction quantities appears to be synchronized with the exposure time necessary for terminal differentiation. For a 2-μM DMBQ exposure, SaExp1 recovery is ∼12 hr, whereas after exposure to a 10-μM inducer concentration, recovery occurs in half that time (data not shown), corresponding to t1/2 for haustorial induction at that concentration. Although high DMBQ concentrations can arrest seedling growth after haustorial induction (Smith et al., 1990), a new meristem can emerge from the terminal haustorium. It has even been argued that the new meristem serves as the infection thread for host penetration (Smith et al., 1990). Therefore, SaExp1 accumulation may be critical for the reestablishment of longitudinal growth in this next stage of parasite development.
The xenognosin-induced accumulation of SaExp1 and SaExp2 correlates specifically with the radial enlargement at the root apex. During vegetative growth, expression of SaExp1 and SaExp2 is low relative to SaExp3, but when induced, SaExp1 and SaExp2 accumulate at a constant rate. It is important to remember that radial enlargement does not occur immediately in this system (the exposure times are quantified only 24 hr later), but initial swelling can be detected as early as 6 hr. This delay between signal exposure and observed swelling is longer than the 3.5-hr lag between OsExp4 accumulation and the increase in growth rate of submerged rice internodes (Cho and Kende, 1997), possibly the result of a change in the direction of growth.
A 1-hr exposure to DMBQ is sufficient to remove the accumulated SaExp3 and stall longitudinal growth of the seedling, but meristematic growth is then reinitiated, and only a small radial bulge remains behind the elongating meristem. Correspondingly, this short exposure results in only a doubling of the SaExp1 and SaExp2 transcripts. For longer exposures, both message accumulation and commitment to radial enlargement show a linear increase with exposure time and likewise show the same relative rate dependence on the DMBQ concentration. Beyond a certain threshold in the accumulation of SaExp1 and SaExp2, which is a function of both time and DMBQ concentration, terminal commitment to haustorial development cannot be arrested by either the designed quinone inhibitors or DMBQ removal (Smith et al., 1996).
One intriguing aspect of these observations is the coupling of the redox reaction of the xenognostic quinones with the accumulation of SaExp transcripts. The processes of oxidative phosphorylation and photosynthesis both require quinones as electron carriers in the reduction of O2 and the oxidation of H2O, respectively. At least in the case of bacterial photosynthesis, quinone substitution has little effect on binding affinity (Warncke and Dutton, 1993), much as is seen for the mono- and disubstituted xenognostic quinones. In addition, changes in the redox potential of the quinones or site occupancy in photosynthesis can have a similar effect on the reaction rate (Graige et al., 1996), in which increased rates are observed with an increase in driving force or an increase in site saturation. At this point, neither the receptors responsible for perceiving the quinones nor the nature of the coupling between the redox reaction and the regulation of SaExp1 and SaExp2 accumulation and SaExp3 depletion in Striga has been determined; nor is it clear whether the observed changes in message accumulation are the result of increased transcription or reduced turnover, particularly considering that cytokinin has been seen to regulate soybean β-expansin by message stabilization (Downes and Crowell, 1998). Nevertheless, in combining these observations, it appears that SaExp1 and SaExp2 transcript accumulation is somehow gated to the molecular electron transfer reaction of the xenognostic quinone.
More generally with respect to SaExp1 and SaExp2 accumulation, the translated proteins should serve as functional markers for the degree of radial swelling in the meristem, and their accumulation to a critical threshold is necessary for the ultimate commitment to haustorial development. Unlike other mRNA detection methods, quantitative RT-PCR determines the absolute numbers of transcripts in an RNA sample, and because each seedling contributes 3.3 ng to an RNA extraction, the number of SaExp3 transcripts per seedlings can be estimated at 4000 before induction and decreasing to 100 after a 45-min exposure. For SaExp1 and SaExp2, ∼140 transcripts are present in a seedling before induction, and at t1/2, the critical threshold for haustorial commitment, ∼1800 transcripts are present per seedling. From a coarse estimate of the in vivo translation rate of five proteins per transcript per minute (Pavlov and Ehrenberg, 1996), a t1/2 exposure would produce 2 × 107 SaExp1 and SaExp2 proteins, or ∼10 pg. In one seedling with a dry weight of ∼2.5 μg, of which only ∼1 to 5% is growing cell wall, the SaExp1 and SaExp2 proteins would be between 1:2500 and 1:12,500 (w/w) of the cell wall. Previous measurements of expansin bound to isolated cell walls (1:5000) and of the amount of expansin necessary to double the rate of wall extension (1:12,500; McQueen-Mason, 1995) are within the range of these approximations. Therefore, 1.5 × 108 SaExp1 and SaExp2 proteins would contribute to an increase in volume of 1.5 × 106 μm3, a volume increase of 107 nm3 per expansin molecule, corresponding to an uptake of 3 × 108 molecules of water per molecule of expansin. This stoichiometry of almost 109:1 for the swelling process is certainly an impressive estimate for the role of the expansin proteins.
In summary, Striga seedlings irreversibly commit to the vegetative/parasitic transition only after exposure to the host- derived signal at a sufficient concentration over a sufficient period of time. From the point of the parasite, this ability to clock signal exposure could certainly improve precision of commitment to a viable host and ensure the success of the obligate parasitic strategy. In a better understood molecular clock, that of circadian rhythms, periodicity is generated by accumulating gene products that repress their own transcription (Dunlap, 1999; Hardin and Sehgal, 1999). Similarly, in haustorial development, there exists a linear accumulation of factors, as indicated by SaExp1 and SaExp2 accumulation, until a chemical or physical threshold is attained. Short gaps in signal exposure do not perturb the timing, and SaExp1 and SaExp2 transcripts continue to accumulate normally. With longer intervals between signal exposures, the clock is gradually reset over a 6-hr period (Smith et al., 1990), and correspondingly, the SaExp1 and SaExp2 expressions drop to the preinduced values. This clock, however, displays no periodicity or rhythm but instead charges like a capacitor, and at least part of that charge includes the expansins. Before this threshold is attained, the molecular charge is not stable and dissipates once the charging current has been switched off by removal of the quinones. The charging current can be switched on repeatedly by addition of the circuit-completing xenognostic quinones. Such a model is important specifically to the precision exhibited during host selection by the parasitic plants but also has profound implications for understanding the mechanisms that control plant developmental commitments generally.
METHODS
Reagents and Plant Material
Striga asiatica seeds, obtained from R.E. Eplee (U.S. Department of Agriculture Witchweed Methods Development Laboratory; Oxford, NC), were pretreated and germinated as previously described (Smith et al., 1990), except that 10−9 M strigol was used instead of sorghum root exudate as germination stimulant. Quinones were prepared as previously outlined (Smith et al., 1996). Twice-distilled water and appropriate glassware were sterilized in a Sterilmatic autoclave (Market Forge; Everett, MA) at 104 kPa at 121°C for 25 min.
Serial Exposure
As previously described (Smith et al., 1990), germinated 2-day-old S. asiatica seedlings (20 to 30) were placed in each of 24 wells of a microtiter plate with 900 μL of sterile, twice-distilled water. At time zero, 100 μL of either 100 or 20 μM 2,6-dimethoxy-p-benzoquinone (DMBQ) solution was added to each well. Every 2 hr, the seedlings in three of the wells at each DMBQ concentration were washed with water three times and covered with 1 mL of water. Haustoria were counted with an inverted microscope (Carl Zeiss; Oberkochen, Germany) 24 hr after the experiment began and scored only as those seedlings with clearly developed haustorial hair formation. All assays were performed in triplicate wells, and values are expressed as ±sd.
Photography
Seedlings were photographed in culture wells at a ×20 magnification with a black-and-white Sony CCD camera. The root tip diameter of the serial exposed seedlings was measured at ×5 magnification with an accuracy of ±5 μm. The average diameter was calculated for individual wells, and the difference between three wells was expressed as ±sd.
Isolation of SaExp cDNA from S. asiatica Seedlings
Total RNA was extracted from 50 to 100 mg of untreated and 4-hr DMBQ-treated S. asiatica seedlings using the RNeasy kit (Qiagen; Chatsworth, CA). The total quantity of RNA was determined by UV light. Denaturing PAGE further confirmed the concentration and provided a qualitative check on the RNA integrity. Of the total Striga RNA from treated and untreated seedlings, 2.5 μg was digested with 2.5 U (1 U/μg) of DNase (Gibco BRL) and used to prepare cDNA by reverse transcription (Superscript II; Gibco BRL) with 500 nM oligo(dT)12–18 (Pharmacia) in a 20-μL reaction volume. One microliter of cDNA products was amplified in 20-μL polymerase chain reactions (PCRs) (an initial 10 min at 94°C was followed by 35 cycles of 45 sec at 94°C, 1 min at 54°C, and 1 min at 72°C, concluding with a final 7 min at 72°C for extension) with a degenerate primer pair based on regions of high conservation in nine expansins (5′-TCIGGA/GACIATGGGNGGIGCNTGT/CGGITA-3′ and 5′-GAGTTIGATTGCCAA/GTTIT/GG/CICCCCAA/GTT-3′). The initial 10-min 94°C step is to inactivate an inhibitor of the Taq polymerase used (Amplitaq Gold; Perkin-Elmer), thereby allowing for a “hot start” reaction. The reaction reagent composition is 20 mM Tris-HCl, pH 8.0, 50 mM KCl, 2.5 mM MgCl2, 250 nM each primer, 200 μM each dNTP, and 0.05 U of Taq polymerase. These same reaction and thermocycling conditions were used in subsequent PCR amplifications unless otherwise specified.
As determined by 6% agarose gel electrophoresis, reverse transcription (RT)–PCR on RNA from both DMBQ-treated and untreated seedlings gave multiple products of ∼500 bp, the predicted size of the products of the expansin primers. These products were cloned into the pCR vector with T4 ligase (TA cloning kit; Invitrogen; Carlsbad, CA), and 20 colonies were selected from each of the DMBQ-treated and untreated samples. M13 forward and reverse sites in the plasmid were used to prime a sequencing reaction with ABI Prism fluorescent dye terminators (Perkin-Elmer). Reactions were separated by electrophoresis and detected with an Applied Biosystems sequencer (model 377; Perkin-Elmer).
Genomic DNA was isolated from 100 mg of S. asiatica seedlings with the Qiagen DNeasy plant mini kit. One microgram of the isolate was used as a PCR template in a reaction sequence identical to that described above, and the products were cloned and sequenced as described above.
Rapid Amplification of cDNA Ends
The 3′ rapid amplification of cDNA ends (RACE) system (Gibco BRL) was used to determine 3′ untranslated sequences (Frohman, 1995). In a 20-μL reaction volume, total RNA (5 μg) was reverse-transcribed with the adapter primer specific for the poly(A) tail of the mRNA. One microliter of this cDNA was used as template in a 20-μL PCR, including a primer complementary to the adapter primer and a gene-specific sense primer for either saExp1 (5′-GCCCGAACATTCTTGGCAGCAGTACGAGGTT-3′) or saExp2 (5′-GAAATCAAGTGTGAC-TATGACTCC-3′). The reaction was assessed by electrophoresis, and promising samples were cloned and sequenced as previously described. SaExp1 and SaExp3 sequences, up to and including the poly(A) tail, were determined.
The 5′ RACE was used to determine the 5′ sequences of the cDNAs. saExp2 (5′-TATGAAGATGGAGGTCTTCCCACC-3′) and saExp3 (5′-GGTTATAGTGACGGAAGTACCCTTCTT-3′) antisense primers were used to prepare cDNA from 5 μg of total RNA from untreated seedlings. Following the manufacturer's instructions, we added a poly(C) tail to the cDNA with TdT and purified the product. One microliter of purified product was amplified by PCR with a poly(G) primer and a nested gene-specific primer for saExp2 (5′-TGCTGCCAAGAATGTTCGGGCTCTCTTGT-3′) or saExp3 (5′-TGCTGCCAAGAATGTTCGGGCTCTCTTGT-3′). Products were cloned and sequenced as previously described, providing untranslated sequences up to and including the signal peptide.
Quantitative RT-PCR
Sufficient sequence data were obtained from 5′ and 3′ RACE to design gene-specific primers for quantitative analyses. Gene-specific primers for saExp1, saExp2, and saExp3 were designed from the untranslated regions of each saExp gene to give products of 150, 130, and 300 bp, respectively. The sense and antisense sequences were as follows: saExp1, 5′-CTTTAGGTCCGGGCCTTGGTT-3′ and 5′- TAATCCCTACACAAACCCCATT-3′; saExp2, 5′-CTCTTCTTCTCGAGGCCTA-3′ and 5′-GCTATAAGCATCGACATTTGA-3′; and saExp3, 5′-TTGAAGCTGCTAATTGGGCTT-3′ and 5′-TTCATGCTCCTTATT-TTCAATGT-3′. By complementary PCR (Ho et al., 1989), synthetic saExp genes were engineered such that in the region between each pair of untranslated primers, 30-bp insertions in saExp1 and saExp2 and a 50-bp deletion in saExp3 were constructed. All three synthetic cDNAs were ligated into a pCR vector containing a T7 polymerase promoter. Two micrograms of each plasmid was linearized by Sca1 digestion (Promega) and used in the transcription of single-stranded sense RNA with T7 polymerase (Gibco BRL). The plasmid was digested with Dnase, and the synthetic RNA was recovered by ethanol precipitation. The concentrations of the synthetic SaExp RNAs were determined by both UV irradiation and gel chromotography (with ethidium bromide staining).
Each synthetic RNA was diluted into three aliquots of 106, 5 × 104, and 2.5 × 103 molecules per microliter, and 1 μL of each aliquot was added to tubes containing 0.5 μg total RNA from the tissue samples. The wild-type and the synthetic RNA were reverse-transcribed with the antisense primer by using the Gibco first strand reaction kit.
One-tenth of the cDNA mixture was PCR-amplified for 19 to 23 cycles of 45 sec at 94°C, 1 min at 54°C, and 1 min at 72°C, followed by a final 7 min at 72°C for extension. The gene-specific primers used in the PCR were labeled with T4 kinase (Promega) according to the manufacturer's directions; unreacted nucleotide was removed with a Qiagen QiQuick Column. The products were run on a 10% agarose gel (Invitrogen) at 100 V for 2 hr and dried in a gel drier. Products were detected with a PhosphorImager (Molecular Dynamics; Sunnyvale, CA) and quantified with ImageQuant software (Molecular Dynamics). Analysis of the data was performed as described by Pfaffl et al. (1998).
Acknowledgments
We are indebted to Rebecca Norris at the U.S. Department of Agriculture laboratory (Oxford, NC) for providing seeds of S. asiatica, and we are grateful to the U.S. Department of Energy (Grant No. ER20024) and the Rockefeller Foundation for financial support.
References
- Baird, W., and Riopel, J. (1984). Experimental studies of haustorium initiation and early development in Agalinis purpurea (L.) Raf (Scrophulariaceae). Am. J. Bot. 71, 803–814. [Google Scholar]
- Carpita, N.C.C., and Gibeaut, D.M. (1993). Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3, 1–30. [DOI] [PubMed] [Google Scholar]
- Chang, M., and Lynn, D.G. (1986). The haustorium and the chemistry of host recognition in parasitic angiosperms. J. Chem. Ecol. 12, 561–579. [DOI] [PubMed] [Google Scholar]
- Cho, H.T., and Kende, H. (1997). Expression of expansin is correlated with growth in deepwater rice. Plant Cell 9, 1661–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H.T., and Kende, H. (1998). Tissue localization of expansins in deepwater rice. Plant J. 15, 805–812. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (1987). Wall relaxation and the driving forces for cell expansive growth. Plant Physiol. 84, 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove, D.J. (1997). Relaxation in a high stress environment: The molecular bases of extensible cell walls and cell enlargement. Plant Cell 9, 1031–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove, D.J. (1999). Enzymes and other agents that enhance cell wall extensibility. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 391–417. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J., Bedinger, P.A., and Durachko, D.M. (1997). Group I allergens of grass pollen as cell wall loosening agents. Proc. Natl. Acad. Sci. USA 94, 6559–6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes, B.P., and Crowell, D.N. (1998). Cytokinin regulates the expression of a soybean β-expansin gene by a post-transcriptional mechanism. Plant Mol. Biol. 37, 437–444. [DOI] [PubMed] [Google Scholar]
- Dunlap, J.C. (1999). Molecular bases for circadian clocks. Cell 96, 271–290. [DOI] [PubMed] [Google Scholar]
- Fleming, A.J., McQueen-Mason, S., Mandel, T., and Kuhlemeier, C. (1997). Induction of leaf primordia by the cell wall protein expansin. Science 276, 1415–1418. [Google Scholar]
- Frohman, M.A. (1995). Rapid amplification of cDNA ends. In PCR Primer: A Laboratory Manual, C.W. Dieffenbach and G.S. Dveksler, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 381–409.
- Graige, M.S., Paddock, M.L., Bruce, J.M., Feher, G., and Okamura, M.Y. (1996). Mechanism of proton-coupled electron transfer for quinone (QB) reduction in reaction centers of Rb. sphaeroides. J. Am. Chem. Soc. 118, 9005–9016. [Google Scholar]
- Hardin, P.E., and Sehgal, A. (1999). Molecular components of a model circadian clock: Lessons from Drosophila. In Handbook of Behavioral State Control: Cellular and Molecular Mechanisms, R. Lydic and H.A. Baghdoyan, eds (Boca Raton, FL: CRC Press), pp. 61–74.
- Ho, S.N., Hunt, H.D., Horton, R.M., Pullen, J.K., and Pease, L.R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Hoy, C.A., Lewis, E.D., and Schimke, R.T. (1990). Perturbation of DNA replication and cell cycle progression by commonly used [3H]thymidine labeling protocols. Mol. Cell Biol. 10, 1584–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.J., Kocz, R., Boone, L., Keyes, W.J., and Lynn, D.G. (1998). On becoming a parasite: Evaluating the role of wall oxidases in parasitic plant development. Chem. Biol. 5, 103–117. [DOI] [PubMed] [Google Scholar]
- Kuijt, J. (1969). The Biology of Parasitic Flowering Plants. (Berkeley, CA: University of California Press).
- McQueen-Mason, S.J. (1995). Expansins and cell wall expansion. J. Exp. Bot. 46, 1639–1650. [Google Scholar]
- McQueen-Mason, S.J., and Cosgrove, D.J. (1994). Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc. Natl. Acad. Sci. USA 91, 6574–6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason, S.J., Durachko, D.M., and Cosgrove, D.J. (1992). Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov, M.Y., and Ehrenberg, M. (1996). Rate of translation of natural mRNAs in an optimized in vitro system. Arch. Biochem. Biophys. 328, 9–16. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M., Meyer, H.H.D., and Saurwein, H. (1998). Quantification of insulin-like growth factor-1 (IGF-1) mRNA: Development and validation of an internally standardized competitive reverse transcription–polymerase chain reaction. Endocrinol. Diabetes 106, 506–513. [DOI] [PubMed] [Google Scholar]
- Press, M.C., and Graves, J.D. (1995). Parasitic Plants. (London: Chapman and Hall).
- Reinhardt, D., Wittwer, F., Mandel, T., and Kuhlemeier, C. (1998). Localized upregulation of a new expansin gene predicts the site of leaf formation in the tomato meristem. Plant Cell 9, 1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riopel, L.J., and Baird, W.V. (1987). Morphogenesis of the early development of primary haustoria in Striga asiatica. In Parasitic Weeds in Agriculture, L.J. Musselman, ed (New York: CRC Press), pp. 107–125.
- Rose, J.K.C., Lee, H.H., and Bennett, A.B. (1997). Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc. Natl. Acad. Sci. USA 94, 5955–5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherban, T.Y., Shi, J., Durachko, D.M., Guiltinan, M.J., McQueen-Mason, S.J., Shieh, M., and Cosgrove, D.J. (1995). Molecular cloning and sequence analysis of expansins—A highly conserved, multigene family of proteins that mediate cell wall extension in plants. Proc. Natl. Acad. Sci. USA 92, 9245–9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh, M.W., and Cosgrove, D.J. (1998). Expansins. J. Plant Res. 111, 149–157. [DOI] [PubMed] [Google Scholar]
- Smith, C.E., Dudley, M., and Lynn, D.G. (1990). Vegetative/parasitic transition: Control and plasticity in Striga development. Plant Physiol. 93, 208–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.E., Ruttledge, T., Zeng, Z., O'Malley, R.C., and Lynn, D.G. (1996). A mechanism for inducing plant development: The genesis of a specific inhibitor. Proc. Natl. Acad. Sci. USA 93, 6986–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, A.M., Doyle, M.V., and Mark, D.F. (1989). Quantitation of mRNA by polymerase chain reaction. Proc. Natl. Acad. Sci. USA 86, 9717–9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warncke, K., and Dutton, P.L. (1993). Influence of Q(A)-site redox cofactor structure on equilibrium binding, in situ electrochemistry, and electron-transfer performance in the photosynthetic reaction center protein. Biochemistry 32, 4769–4779. [DOI] [PubMed] [Google Scholar]
- Yoder, J.I. (1997). A species-specific recognition system directs haustorium development in the parasitic plant Triphysaria (Scrophulariaceae). Planta 202, 407–413. [DOI] [PubMed] [Google Scholar]
- Zeng, Z., Cartwright, C.H., and Lynn, D.G. (1996). Chemistry of cyclopropyl-p-benzoquinone: A specific organogenesis inhibitor in plants. J. Am. Chem. Soc. 118, 1233–1234. [Google Scholar]