Abstract
Microarray and RNA gel blot analyses were performed to identify Arabidopsis genes that responded to nitrate at both low (250 μM) and high (5 to 10 mM) nitrate concentrations. Genes involved directly or indirectly with nitrite reduction were the most highly induced by nitrate. Most of the known nitrate-regulated genes (including those encoding nitrate reductase, the nitrate transporter NRT1, and glutamate synthase) appeared in the 40 most strongly nitrate-induced genes/clones on at least one of the microarrays of the 5524 genes/clones investigated. Novel nitrate-induced genes were also found, including those encoding (1) possible regulatory proteins, including an MYB transcription factor, a calcium antiporter, and putative protein kinases; (2) metabolic enzymes, including transaldolase and transketolase of the nonoxidative pentose pathway, malate dehydrogenase, asparagine synthetase, and histidine decarboxylase; and (3) proteins with unknown functions, including nonsymbiotic hemoglobin, a senescence-associated protein, and two methyltransferases. The primary pattern of induction observed for many of these genes was a transient increase in mRNA at low nitrate concentrations and a sustained increase when treated with high nitrate concentrations. Other patterns of induction observed included transient inductions after both low and high nitrate treatments and sustained or increasing amounts of mRNA after either treatment. Two genes, AMT1;1 encoding an ammonium transporter and ANR1 encoding a MADS-box factor, were repressed by nitrate. These findings indicate that nitrate induces not just one but many diverse responses at the mRNA level in Arabidopsis.
INTRODUCTION
For more than three centuries, plants have been known to respond to nitrate. Saltpeter (KNO3) was first isolated in 1656 by Glauber and shown to enhance plant growth (cited in Glass, 1989). To induce this response, nitrate serves as a nutrient and as a signal (reviewed in Redinbaugh and Campbell, 1991; Hoff et al., 1994; Crawford, 1995; Koch, 1997; Forde and Clarkson, 1999; Stitt, 1999). As a nutrient, nitrate is reduced to ammonium and then is incorporated into amino acids. As a signal, nitrate reprograms metabolism of nitrogen and carbon, resource allocation, and root development. These responses involve rapid and marked changes in gene expression, but the underlying mechanisms are poorly understood in plants (reviewed in Crawford and Arst, 1993; Hoff et al., 1994; Huppe and Turpin, 1994; Koch, 1997; Stitt, 1999).
Nitrate assimilation begins with nitrate uptake, followed by the nitrate reductase (NR)–catalyzed reduction of nitrate to nitrite, the nitrite reductase (NiR)–catalyzed reduction of nitrite to ammonium, and then the incorporation of ammonium into amino acids, catalyzed primarily by glutamine synthetase (GS) and glutamate synthase (GOGAT) (reviewed in Crawford, 1995; Lam et al., 1996; Campbell, 1999; Stitt, 1999). Nitrate reduction occurs in the cytosol of cells in both shoots and roots and uses NAD(P)H as the source of reductant. Nitrite reduction occurs in chloroplasts of green tissues and in plastids of the roots and uses reduced ferredoxin (Fd) as reductant. When rates of nitrate reduction are high, this pathway becomes a major sink for reductant. In green tissues, reductant originates from photosynthetic electron transport; in nongreen tissues, reductant arises primarily from the oxidative pentose pathway. In addition to reductant, organic acids are needed for ammonium incorporation into amino acids and maintenance of cellular pH because nitrate reduction generates hydroxide ions.
As a signal, nitrate reprograms nitrogen and carbon metabolism and the expression of a select group of genes (reviewed in Crawford, 1995; Koch, 1997; Stitt, 1999). Metabolic processes stimulated by nitrate include nitrate uptake and reduction, ammonium assimilation, organic acid synthesis, Fd reduction, and glucose-phosphate oxidation (by way of the oxidative pentose phosphate pathway). Nitrate also suppresses starch synthesis to help mobilize carbon into organic acids.
These responses to nitrate involve direct induction of gene expression as shown by the rapid increase (within 0.5 to 1 hr) in mRNA concentrations of several nitrate and ammonium assimilatory genes in response to treatment with low concentrations of nitrate (10 to 250 μM) even in the presence of protein synthesis inhibitors. The most studied gene is that for NR, the first gene shown to be nitrate inducible (Tang and Wu, 1957; Cheng et al., 1986; Crawford et al., 1986). NR mRNA accumulates in plants within minutes after treatment with nitrate at concentrations from 10 μM to 50 mM (Melzer et al., 1989; Cheng et al., 1991; Gowri et al., 1992; Aslam et al., 1993; Tischner et al., 1993). At high nitrate concentrations (10 to 50 mM), NR induction is not inhibited by cycloheximide in maize (Gowri et al., 1992). Similarly, genes involved directly or indirectly in nitrate uptake or nitrite reduction are rapidly induced over a similar range of nitrate concentrations (reviewed in Koch, 1997; Crawford and Glass, 1998; Forde and Clarkson, 1999; Stitt, 1999; see also below). These genes encode nitrate transporters (NRT1 and NRT2), NiR, Fd, Fd NADP+ oxidoreductase (FNR), 6-phosphogluconate dehydrogenase (6PGDH), and S-adenosyl-l-methionine–dependent uroporphyrinogen III methyltransferase (UPM1). Genes involved in ammonium assimilation, encoding specific isoforms of GS and GOGAT, are also induced (reviewed in Lam et al., 1996; Koch, 1997; Stitt, 1999; see also below). For starch and organic acid metabolism, mRNA concentrations for phosphoenolpyruvate carboxylase (PEPC; involved in organic acid metabolism) increase and those for ADP-glucose pyrophosphorylase (AGPS2; involved in starch synthesis) decrease after 2 hr of treatment with 12 mM nitrate (Scheible et al., 1997a). Transcripts for other organic acid metabolic enzymes—cytosolic pyruvate kinase, citrate synthase, and NADP+–isocitrate dehydrogenase—were present in NR mutant plants in greater amounts than in wild-type plants grown in 12 mM nitrate, implying that these genes also respond to the nitrate signal (Scheible et al., 1997a).
Besides inducing metabolic genes, nitrate has other important effects on plants. Nitrate treatment increases nitrogen-to-carbon ratios, decreases root-to-shoot ratios, and delays flowering (reviewed in Marschner, 1995; Koch, 1997; Stitt, 1999). Discontinuities of nitrate in the soil affect root architecture, leading to preferential proliferation of lateral roots in zones of high nitrate concentrations (Drew, 1975; Granato and Raper, 1989; Zhang et al., 1999; Zhang and Forde, 2000). Complicating these analyses is the fact that other forms of nitrogen, including such downstream metabolites as ammonium and glutamine, can sometimes elicit the same response (such as in root branching [Drew, 1975]) or suppress nitrate inductions (Hoff et al., 1994; Quesada et al., 1997; Sivasankar et al., 1997; Dzuibany et al., 1998; Krapp et al., 1998; Zhuo et al., 1999). To help discern which responses involve specific sensing of nitrate, NR-deficient mutants have been used that make little of the downstream metabolites in the presence of nitrate. To show that nitrate affects resource allocation, tobacco NR mutants were treated with nitrate, resulting in a stimulation in shoot growth and an inhibition of overall root growth (Scheible et al., 1997b) and of lateral root formation (Stitt and Feil, 1999). In Arabidopsis wild-type plants, localized concentrations of nitrate but not ammonium or glutamine stimulate lateral root elongation but not initiation (Zhang et al., 1999; Zhang and Forde, 2000). In both wild-type Arabidopsis and NR-deficient mutants, high concentrations of nitrate systemically inhibit lateral root elongation (Zhang et al., 1999; Zhang and Forde, 2000).
Such a diversity of nitrate responses indicates that plants have intricate regulatory networks for integrating nitrate assimilation with the other metabolic and developmental pathways of the cell. Clues about nitrate regulatory mechanisms that mediate these processes are beginning to emerge. Ca2+ has been implicated in these mechanisms because treatments with the inhibitors La3+ or EGTA block the nitrate induction of the genes encoding (1) NR, NiR, UPM1, the plastidic form of GS (GS2), and NADH-GOGAT in detached maize leaves (Sakakibara et al., 1996, 1997) and (2) NR and NiR in excised barley leaves (Sueyoshi et al., 1999). Phosphatases and kinases may also play a role (Champigny and Foyer, 1992); treatments of maize leaves with okadaic acid reduced nitrate inductions of NR, NiR, and GS2 but not of Fd-GOGAT (Sakakibara et al., 1997), and treatments of excised barley leaves with okadaic acid, calyculin A, genisitein, quercetin, and curcumin inhibited NR and NiR induction (Sueyoshi et al., 1999). In addition to these physiological studies, two potential regulatory genes/mutants have been described. In the first, a chlorate-resistant Arabidopsis mutant, cr88, the nitrate and light induction of the NR gene NIA2 but not NIA1 is altered (Lin and Cheng, 1997). The second, ANR1, is a MADS-box gene of Arabidopsis; when it is repressed in antisense transgenic plants, systemic nitrate repression of lateral root growth and localized nitrate stimulation of lateral root growth are impaired (Zhang and Forde, 1998, 2000). This gene is induced by 2 mM nitrate in nitrogen-starved plants but not by potassium or phosphate deprivation (Zhang and Forde, 1998).
To elucidate nitrate regulatory mechanisms further and to establish a more complete inventory of genes and pathways that are responsive to nitrate, a microarray analysis of nitrate-induced gene expression was performed in Arabidopsis. These experiments were designed to reveal genes that (1) are rapid responders to low nitrate concentrations to identify possible primary response genes and (2) are induced with high nitrate concentrations over a longer time frame to generate a comprehensive list of responsive genes. Because approximately a dozen genes are already known to be nitrate induced, we could test the efficacy of this approach by recording which of the known genes were detected by the microarray analysis. Further analysis should then reveal previously unidentified nitrate-reduced genes as well as provide a comprehensive view of the patterns of nitrate regulation for the nitrate-responsive genes. The results of our analyses are presented below.
RESULTS
Strategy for Nitrate-Regulated Gene Analysis
Two treatments of wild-type Arabidopsis plants were used to identify nitrate-responsive genes. In the first treatment, plants were treated with 250 μM nitrate for 20 min to reveal genes that respond rapidly to low concentrations of nitrate. In the second treatment, plants were treated with 5 to 10 mM nitrate for 2 hr to provide a more comprehensive screen for nitrate-regulated genes. The latter conditions are most commonly reported for studies on nitrate-induced genes. Plants were grown in liquid culture containing a pH buffer and ample nitrogen in the form of ammonium ion (but no nitrate) for 10 days, after which either KNO3 (for nitrate induction) or KCl (for the control) was added to the cultures. After 20 min or 2 hr, whole plants were harvested and used to isolate mRNA for analysis. This approach should detect nitrate-induced genes expressed in roots or shoots, or both, if the extent of expression is great enough for detection.
mRNA was measured by using Arabidopsis GEM1 microarrays (Incyte Corp., Palo Alto, CA) containing 7942 cDNA clones corresponding to 5524 unique genes/clusters—or approximately one-quarter of the Arabidopsis genes (estimated to be ∼25,000 [Meyerowitz, 1999]). Two mRNA samples from two independent sets of cultures were prepared for each condition: 250 μM nitrate for the 20-min treatment (L-20min) and 10 mM nitrate for the 2-hr treatment (H-2hr). Three microarray hybridizations were performed for each condition (one for the first mRNA sample and two for the second mRNA sample), for a total of six hybridizations. For each nitrate-induced mRNA sample, a corresponding chloride-treated sample was prepared for the control. Labeled DNA copies were prepared from nitrate-treated and chloride-treated control plants and then were competitively hybridized to microarrays on glass slides, as described in Methods. An expression ratio for each DNA clone on the array was determined to indicate the relative amounts of the various mRNAs as a measure of how many multiples of induction or repression were induced. Lists of genes/clones were generated and ordered on the basis of the amplitude of these ratios. Specific genes from the microarray lists were then selected for further study by RNA gel blot analysis of mRNAs from plants treated with 250 μM nitrate for 20 min or 2 hr (L-20min and L-2hr) or with 5 mM nitrate for 20 min or 2 hr (H-20min and H-2hr).
Microarray Data
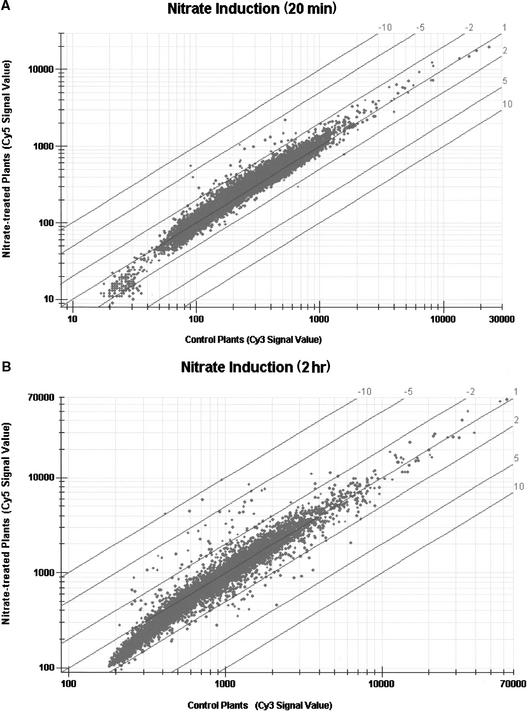
Plots showing signal intensities for each gene/clone on the microarray are shown in Figure 1 for two of the six hybridization experiments. Diagonal lines show the normalized signal ratios, with a value of 1 corresponding to no difference in mRNA expression between the two samples. Dots above the line (given negative ratios in this figure) indicate stronger expression in the nitrate-treated plants; dots below the line (given positive ratios) indicate the reverse. As shown in the graphs, most dots fall between the −2 and +2 lines. The −2 to +2 values are taken as approximate thresholds for significant differences because uniprobe controls, in which one RNA sample was used for making both probes so that ratios of 1.0 were expected for each DNA clone on the array, showed that the limit of detection for differential expression was 1.74 (see GEM Microarray Reproducibility Study; http://gem.incyte.com/gem/GEM-reproducibility.pdf). Some clones in Figure 1, however, fall outside these limits; it is these clones that we want to identify. One interesting feature of the two plots is that the scatter or deviation in Figure 1B is greater than that in Figure 1A, indicating that more genes/clones are differentially expressed in the H-2hr nitrate-treated samples than in the L-20min samples.
Figure 1.
Scatter Plots of Signal Values for Clones on Microarray.
Signal intensities for each clone on the microarray, plotted with the signals from the chloride-treated controls on the x axis and the signals from the nitrate-treated samples on the y axis. The diagonal lines represent normalized ratios, where 1 indicates no difference in signal between the two samples, −2 to −10 are ratios indicating greater signals for the nitrate-treated samples, and +2 to +10 are ratios indicating greater signals for the chloride controls.
(A) Plot for plants treated with 250 μM solute for 20 min.
(B) Plot for plants treated with 10 mM solute for 2 hr.
A complete list of clones on the microarray was prepared, including gene assignments made by tiered BLAST searches of GenBank databases. If no significant similarities were found for a given clone, either a genomic clone designation or an Incyte expressed sequence tag number was assigned. The list of clones is available at the following site: http://www.genomesystems.com/expression/clonelists/argem1.txt. Using the data from the microarray hybridization experiments, we then ordered this list on the basis of the amplitude of average signal ratios for each clone. The complete lists are available on request or at the following site: www.biology.ucsd.edu/publications/crawford.
Genes/clones at or near the top of both lists (with absolute ratios of ⩾2) were selected for further analysis. First, BLAST searches of the GenBank NR database were repeated, allowing additional gene assignments and chromosome locations to be determined. The refined lists, restricted to clones for which assignments could be made, are presented as Tables 1 and 2. The GenBank accession numbers for the Incyte DNA clone (Clone ID) and the best or most informative GenBank sequence alignment (GenBank Hit ID) are also given. Genes are ranked by their ratios: those having the highest values in the nitrate-treated plants in comparison with chloride controls (indicated here as positive ratios) are at the top of the list, and those showing the greatest decrease in mRNA content (indicated here as negative ratios) are at the bottom of the list. The signal ratios (column 1) were calculated as the arithmetic average (mean) by using values from the three microarray hybridizations performed for each condition, and the error analysis is given as the standard deviation (column 2) for each signal ratio.
Table 1.
Arabidopsis Genes That Respond to Low Nitrate Concentrations (20 Min)
| Ratio | sd | Gene | Chromosome | Clonea | GenBank Hitb |
|---|---|---|---|---|---|
| 11.4 | 3.8 | Uroporphyrin III methyltransferase (UPM1) | V | AW004380 | L47479 |
| 10.5 | 5.4 | Nitrite reductase (NiR) | II | AI998242 | D14824 |
| 6.8 | 2.4 | 6-Phosphogluconate dehydrogenase (6PGDH) | V | AI997770 | BAA22812 |
| 6.4 | 3.1 | Glucose-6-phosphate dehydrogenase (G6PDH) | I | AI998472 | X84229 |
| 6.2 | 1.2 | Ferredoxin NADP oxidoreductase (FNR) | IV | AI994434 | CAA67796 |
| 5.5 | 1.0 | Nitrate reductase (NIA1) | I | AW004371 | Z19050 |
| 5.5 | 1.6 | Ferredoxin NADP oxidoreductase (FNR) | I | AI995147 | AAF19753 |
| 5.1 | 1.7 | Glucose-6-phosphate dehydrogenase (G6PDH-E5) | ? | AW004529 | X84229 |
| 3.6 | 1.0 | Ferredoxin (Fd) | II | AI998909 | S62722 |
| 3.5 | 0.2 | Nitrate reductase (NIA2) | I | AI996208 | J03240 |
| 3.1 | 0.1 | Senescence-associated protein (SAG21) | IV | AI995253 | AF053065 |
| 2.7 | 1.0 | Nitrate transporter (NRT1/CHL1) | I | AI994210 | L10357 |
| 2.6 | 0.2 | Putative anthranilate N-benzoyltransferase | IV | AI998980 | BAA87043 |
| 2.4 | 0.3 | Nitrate transporter (NRT1/CHL1) | I | AW004538 | L10357 |
| 2.4 | 0.1 | Putative sugar transporter | IV | AI997793 | AAC36683 |
| 2.4 | 0.9 | Putative auxin-induced protein | II | AI997322 | T17020 |
| 2.3 | 0.3 | Histidine decarboxylase | I | AI994252 | BAA78331 |
| 2.3 | 0.3 | Transaldolase (TAL1) | V | AW004093 | U95923 |
| 2.3 | 0.3 | High-capacity calcium antiporter (CAX1) | II | AI999789 | U57411 |
| 2.3 | 0.9 | Putative NPK1-related protein kinase | II | AI998827 | T04812 |
| 2.2 | 0.1 | Transaldolase (TAL1) | V | AI994809 | U95923 |
| 2.2 | 0.1 | Transaldolase (TAL1) | V | AI992945 | U95923 |
| 2.2 | —c | NAD-dependent malate dehydrogenase, chloroplast | III | AW004089 | Y13987 |
| 2.2 | 0.2 | High-capacity calcium antiporter (CAX1) | II | AW004488 | U57411 |
| 2.1 | 0.2 | Serine acetyltransferase (Sat5) | I | AI994235 | L34076 |
| 1.9 | 0.1 | NADH-dependent glutamate synthase (GOGAT) | II | AI993750 | Q03460 |
| 1.8 | 0.1 | MYB transcription factor | V | AI992511 | AJ006404 |
| 1.8 | 0.1 | Class 1 nonsymbiotic hemoglobin (AHB1) | II | AI998560 | U94998 |
GenBank accession numbers for the Incyte DNA clone.
Best or most informative GenBank sequence alignments.
This ratio is based on a single measurement.
Table 2.
Arabidopsis Genes That Respond to High Nitrate Concentrations (2 Hr)
| Ratio | sd | Gene | Chromosome | Clonea | GenBank Hitb |
|---|---|---|---|---|---|
| 16.0 | 4.3 | Nitrite reductase (NiR) | II | AI998242 | D14824 |
| 13.3 | 2.4 | Uroporphyrin III methyltransferase (UPM1) | V | AW004380 | L47479 |
| 9.0 | 1.1 | Ferredoxin NADP oxidoreductase (FNR) | I | AI995147 | AAF19753 |
| 8.3 | 0.1 | Ferredoxin NADP oxidoreductase (FNR) | IV | AI994434 | CAA67796 |
| 7.1 | 0.8 | 6-Phosphogluconate dehydrogenase (6PGDH) | V | AI997770 | BAA22812 |
| 6.3 | 1.6 | Nitrate reductase (NIA1) | I | AW004371 | Z19050 |
| 6.3 | 1.1 | Glucose-6-phosphate dehydrogenase (G6PDH) | I | AI998472 | X84229 |
| 5.8 | 1.5 | Class 1 nonsymbiotic hemoglobin (AHB1) | II | AI998560 | U94998 |
| 4.9 | 0.1 | Transaldolase (TAL1) | V | AW004093 | U95923 |
| 4.9 | 0.1 | Transaldolase (TAL1) | V | AI992945 | U95923 |
| 4.9 | 0.5 | Nitrate reductase (NIA2) | I | AI996208 | J03240 |
| 4.8 | 1.5 | Asparagine synthetase (ASN2) | V | AI997088 | AF095453 |
| 4.6 | 0.3 | Transaldolase (TAL1) | V | AI994809 | U95923 |
| 4.5 | 0.2 | Senescence-associated protein (SAG21) | IV | AI995253 | AF053065 |
| 4.5 | 0.5 | Ferredoxin (Fd) | II | AI998909 | S62722 |
| 4.4 | 0.8 | Glucose-6-phosphate dehydrogenase (G6PDH-E5) | ? | AW004529 | X84229 |
| 4.2 | 0.1 | Putative sterol methyltransferase (1) | I | AI995048 | AAF24830 |
| 4.2 | 0.6 | Putative Ser/Thr protein kinase | II | AI995848 | CAA73067 |
| 4.1 | 1.1 | Putative sterol methyltransferase (2) | I | AI992732 | AAF24830 |
| 4.0 | 1.3 | Glutathione S-transferase | ? | AI998434 | AC005309 |
| 3.7 | 1.2 | Putative auxin-induced protein | II | AI997322 | T17020 |
| 3.2 | 0.5 | NADH-dependent glutamate synthase (GOGAT) | II | AI993750 | Q03460 |
| 3.2 | 0.2 | Nicotianamine synthase | V | AI993200 | AB021934 |
| 3.2 | 0.3 | Putative CoA-ligase | III | AI996670 | CAB62011 |
| 2.9 | 0.5 | GAST1-like protein | I | AI993388 | AAF15937 |
| 2.9 | 0.9 | Response regulator ARR6 | V | AI993209 | AB008489 |
| 2.8 | 0.2 | Transketolase | III | AI998240 | L76554 |
| 2.8 | 0.0 | Ketol-acid reductoisomerase subunit | III | AI993038 | X68150 |
| 2.7 | 0.3 | Histidine decarboxylase | I | AI994252 | BAA78331 |
| 2.7 | 0.4 | Putative nodulin-like protein | IV | AI998079 | AAD24599 |
| 2.7 | 2.2 | Putative carbonic anhydrase | I | AI994046 | L19255 |
| 2.6 | 0.5 | ATP phosphoribosyl transferase (PRT1) | ? | AI992914 | AB025249 |
| 2.5 | 0.1 | Glutamine synthetase | V | AI999231 | S69727 |
| 2.4 | 0.3 | MYB transcription factor | V | AI992511 | AJ006404 |
| 2.4 | 0.2 | High-capacity calcium antiporter (CAX1) | II | AI999789 | U57411 |
| 2.4 | 0.2 | GF14 protein χ-chain | IV | AI992781 | L09112 |
| 2.3 | 0.4 | ATP phosphoribosyl transferase (PRT1) | ? | AI995495 | AB025249 |
| 2.3 | 0.7 | NADH-dependent glutamate synthase (GOGAT) | II | AI997600 | Q03460 |
| 2.2 | 0.2 | High-capacity calcium antiporter (CAX1) | II | AW004488 | U57411 |
| 2.1 | 0.3 | Sulfite reductase | V | AI993054 | Z49217 |
| 2.1 | 0.3 | GF14 protein χ-chain | IV | AW004585 | L09112 |
| −2.5 | 0.3 | Osmotin | IV | AI998685 | X89008 |
| −2.7 | 0.3 | Homeobox-leucine zipper gene (ATHB-12) | III | AI994027 | AF001949 |
| −2.7 | 0.5 | Phosphoglycerate dehydrogenase | ? | AW004434 | AB010407 |
| −2.8 | 0.2 | Putative auxin/aluminum-regulated gene | ? | AI996702 | AB012110 |
| −2.9 | 0.2 | Vacuolar processing enzyme (γ-VPE) | IV | AI994531 | D61395 |
| −3.2 | 0.2 | Putative copper amine oxidase | IV | AI995612 | AJ009825 |
| −3.2 | 0.2 | Ammonium transporter AMT1;1 | IV | AI996147 | X75879 |
| −4.1 | 0.6 | Homolog of No Apical Meristem gene | ? | AI992865 | AJ222713 |
GenBank accession numbers for the Incyte DNA clone.
Best or most informative GenBank sequence alignments.
Known Nitrate-Induced Genes
Our first analysis of the microarray data focused on genes known to be nitrate-induced in plants. The GEM1 microarray included almost all known nitrate-regulated genes. Table 3 summarizes the data from the six microarray hybridizations. RNA gel blot analyses were performed on selected genes, and the results—signal ratios for each nitrate-treated sample divided by the average signal from the 20-min– and 2-hr– treated chloride controls—are also included in Table 3. Below, the results for each gene are discussed in more detail. Unless stated otherwise, there was only one clone on the array for each gene under discussion.
Table 3.
Known Nitrate-Responsive Genes: Microarray and RNA Gel Blot Data
| Microarrayb
|
RNA Gel Blotb
|
|||||
|---|---|---|---|---|---|---|
| Genea | L-20min | H-2hr | L-20min | L-2hr | H-20min | H-2hr |
| Nitrate uptake and assimilation | ||||||
| NR (NIA1) | 5.5 (1.0) | 6.3 (1.6) | 5.6 | 2.3 | 12 | 9.6 |
| NR (NIA2) | 3.5 (0.2) | 4.9 (0.5) | 8.9 | 2.4 | 16 | 31 |
| NiR | 11 (5.4) | 16 (4.3) | 140 | 40 | 130 | 130 |
| UPM1 | 11 (3.8) | 13 (2.4) | 13 | 2.2 | 17 | 17 |
| NRT1;1 (CHL1)c | 2.6 (0.5) | 1.6 (0.0) | 4.4 | 3.4 | 2.8 | 2.7 |
| NRT2 | NA | NA | 2.0 | 2.1 | 2.3 | 18 |
| Fd reduction | ||||||
| Fd | 3.6 (1.0) | 4.5 (0.5) | 5.2 | 3.2 | 3.9 | 3.9 |
| FNR chromosome IV | 6.2 (1.2) | 8.3 (0.1) | 6.4 | 7.6 | 17 | 17 |
| FNR chromosome I | 5.5 (1.6) | 9.0 (1.1) | 56 | 36 | 43 | 69 |
| Oxidative pentose phosphate pathway | ||||||
| G6PDH (clone E5) | 5.1 (1.7) | 4.4 (0.8) | 20 | 4.0 | 35 | 36 |
| G6PDH (chromosome I) | 6.4 (3.1) | 6.3 (1.6) | 18 | 3.6 | 12 | 9.5 |
| 6PGDH | 6.8 (2.4) | 7.1 (0.8) | ND | ND | ND | ND |
| Ammonium assimilation | ||||||
| Glutamine synthetase | 1.2 (0.1) | 2.5 (0.1) | ND | ND | ND | ND |
| Fd-GOGAT | NS | NS | ND | ND | ND | ND |
| NADH-GOGATc | 1.7 (0.1) | 2.8 (0.5) | ND | ND | ND | ND |
| Organic acid metabolism | ||||||
| PEP carboxylasec | 1.3 (0.1) | 1.1 (0.1) | ND | ND | ND | ND |
Gene symbols: NRT1;1 (CHL1), dual-affinity nitrate transporter; NRT2, high-affinity nitrate transporter; G6PDH, glucose-6-P-dehydrogenase; 6PGDH, 6-phosphogluconate dehydrogenase. Chr., chromosome.
L refers to inductions with 250 μM. H refers to inductions with 10 mM nitrate for the arrays and 5 mM for the RNA gel blots. Values for the arrays are averages for all clones and hybridizations for each gene. Numbers in parentheses are standard deviations. NA, not on microarray; ND, not determined; NS, on array but no signal.
Indicates more than one clone was present on the array so that several values were averaged to give the number in the table.
Nitrate Reductase Genes
The most extensively studied nitrate-induced genes encode NR. Arabidopsis has two NR genes, both of which are nitrate inducible (Cheng et al., 1988; Crawford et al., 1988; Wilkinson and Crawford, 1991, 1993). Both genes show strong inductions and rank in the top 11 identifiable genes at L-20min and H-2hr. The blot data corroborate the microarray results in that both genes show strong inductions with nitrate (the first and fourth columns of the blot data in Table 3 correspond to the first and second columns of the microarray data, respectively). The absolute values in these two data sets, however, did not exactly match. Care should be taken in interpreting the very high values (>10) for the blot data because these are the result of very low background values in the chloride-treated samples where accuracy is low. As reported previously, both genes respond quickly and markedly to low concentrations of nitrate. In addition, the induction in conditions of low nitrate concentrations is transient, the mRNA ratios at 2 hr being lower than the ratios at 20 min. This behavior was observed for many genes and is discussed below in more detail.
Nitrite Reductase
The next gene in the nitrate assimilation pathway is NiR, which is also rapidly induced by nitrate (Lahners et al., 1988). This gene displays strong induction on both microarrays and has the first or second highest value of all listed genes. On RNA gel blots, NiR showed the strongest induction of any gene we tested. These high values reflect the absence of any background signal in the chloride-treated controls and are a low estimate. Only one NiR gene is known in Arabidopsis (Tanaka et al., 1994).
Uroporphyrin III Methyltransferase
Next to NiR on both microarray lists is the UPM1 gene, which encodes UPM1. This protein catalyzes the branch point step in the biosynthesis of siroheme, an essential cofactor for NiR (Sakakibara et al., 1996; Leustek et al., 1997). UPM1 is strongly induced in maize within 2 hr of 16 mM nitrate treatment (Sakakibara et al., 1996). The RNA gel blot data also show very strong inductions.
Nitrate Transporters
Two clones corresponding to NRT1 (also known as CHL1) were included in the array but none corresponding to NRT2. NRT1 showed good induction at L-20min but little increase at H-2hr. The blot data indicate modest but significant inductions at all time points. It was surprising to find such low ratios at H-2hr on the array, but both clones gave very similar results and were in the top 500 genes/clones. Nitrate inductions of NRT1 are complicated by the fact that NRT1 mRNA also responds to changes in the pH of the medium (Tsay et al., 1993).
Although no clone corresponding to NRT2 was on the array, RNA gel blots were performed; they showed weak inductions at L-20min, L-2hr, and H-20min but a strong induction at H-2hr. This gene is induced rapidly and strongly with nitrate, but it is also repressed by ammonium (Trueman et al., 1996; Quesada et al., 1997; Amarasinghe et al., 1998; Krapp et al., 1998; Wang et al., 1998; Zhuo et al., 1999). Because our induction conditions included high concentrations of ammonium in the growth medium, the weak response at low nitrate concentrations and at early times in our experiments most likely results from the antagonistic effect of ammonium.
Ferredoxin and Ferredoxin NADP+ Oxidoreductase
Nitrite reduction requires Fd as reductant. In photosynthetic cells, Fd is reduced in photosystem I. In roots and nongreen tissues, Fd is reduced in plastids by FNR, which uses NADPH as reductant. Both Fd and FNR genes are induced by nitrate in maize roots. FNR mRNA accumulates within 1 hr of 10 mM nitrate treatment, even in the presence of cycloheximide (Ritchie et al., 1994). As little as 10 μM nitrate is effective in inducing FNR after 2 hr. The amounts of Fd mRNA encoding the maize Fd IV isoform increase markedly in response to 16 mM nitrate after 2 hr (Matsumura et al., 1997). These findings indicate that nitrate increases redox potential in part by inducing the genes that encode Fd and FNR in roots.
The microarray contained five clones corresponding to four Fd genes. Only one (on chromosome II) had a high induction ratio. This gene showed good inductions on both the array and the blots and displayed the typical transient expression at low nitrate. Thus, within this multigene family, only one gene showed detectable induction in whole plants—similar to the results reported for maize (Matsumura et al., 1997).
For FNR, two clones were present, corresponding to two distinct genes on different chromosomes. The microarray data showed very strong induction for both genes, which were in the top seven at L-20min and in the top four at H-2hr. The genes also displayed very strong induction on the RNA gel blots. One of them showed a sustained increase instead of a transient one at low nitrate concentrations.
Oxidative Pentose Phosphate Pathway
To support generation of reduced Fd in roots, NADPH is produced by the oxidative pentose phosphate pathway, which converts glucose 6-phosphate into ribose 5-phosphate and reduces two NADP+ molecules (Bowsher et al., 1992). Activities of two key enzymes in the pathway—glucose-6-phosphate dehydrogenase (G6PDH) and 6PGDH—increase 12- to 27-fold in maize root plastids after 24 hr of treatment with 10 mM nitrate (Redinbaugh and Campbell, 1998). One of three 6PGDH maize genes displays a primary response, being strongly induced by 0.1 to 10 mM nitrate within 1 hr in maize roots with or without cycloheximide (Redinbaugh and Campbell, 1998). mRNA studies of G6PDH have not been reported.
The microarray has two G6PDH clones that correspond to two different genes. Both show strong inductions on the array and very strong inductions on the blots. Although no nitrate induction of plant G6PDH mRNA has been previously reported, the increase in mRNA we observed corresponds well with the increase in enzyme activity reported for maize. Both genes show a very significant drop in mRNA concentrations at L-2hr.
For 6PGDH, two clones were present, corresponding to two distinct genes. Only one (on chromosome V) showed induction. The induction of this gene was very strong; its signal ratio was in the top five genes for both L-20min and H-2hr. RNA gel blot analysis was not performed for this gene.
ANR1, Encoding MADS Box Protein
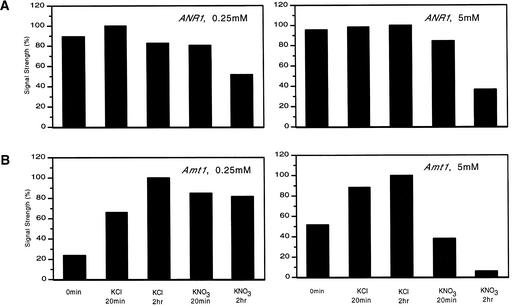
An Arabidopsis gene expressed specifically in roots and encoding a MADS-box protein was shown to be induced within 0.5 hr after treatment of nitrogen-starved plants with 2 mM nitrate (Zhang and Forde, 1998). We tested whether this gene would be induced under our conditions, in which nitrate is added to nitrogen-replete plants. No clone corresponding to this gene was included on the array. RNA gel blot analysis (Figure 2A) showed no increase in ANR1 mRNA quantities but in fact showed a decrease, which was most pronounced at 5 mM nitrate. Our results indicate that nitrate induction of ANR1 either is suppressed by ammonium or requires nitrogen starvation.
Figure 2.
Nitrate Responses for ANR1 and AMT1 Genes.
Histograms of mRNA signals from RNA gel blots at 0 and 20 min and 2 hr after plants were treated with the indicated amounts of KNO3 and KCl. ANR1 encodes a MADS-box factor, and AMT1 encodes an ammonium transporter.
Ammonia Assimilatory Genes
Other genes shown to be nitrate-induced include those that encode GS and GOGAT, which are involved in ammonia assimilation. Both GS and GOGAT have multiple forms in different compartments of the cell, and these forms show different regulation (reviewed in Lam et al., 1996). In maize roots, Fd-GOGAT and GS2 are induced within 30 min by 10 mM nitrate (with or without cycloheximide) and within 2 hr by 10 μM nitrate (Redinbaugh and Campbell, 1993). In contrast, no induction was observed in leaves of maize seedlings (Redinbaugh and Campbell, 1993). In another study using detached maize leaves, GS2, Fd-GOGAT, and NADH-GOGAT were all induced by 16 mM nitrate after 2 hr (Sakakibara et al., 1997). However, the gene encoding the major isoform of cytosolic GS, GS1a, was not induced under these conditions (Sakakibara et al., 1997). In tobacco NR mutants, GS1, GS2, and Fd-GOGAT transcripts were found to be more plentiful in plants grown with 12 mM nitrate than in plants grown with 0.2 mM nitrate (Scheible et al., 1997a).
The Arabidopsis microarray contained two clones corresponding to the same NADH-dependent GOGAT on chromosome II and one clone for Fd-dependent GOGAT. The latter clone did not produce a detectable signal. The two NADH-GOGAT clones gave very similar results, showing nitrate induction at both L-20min and H-2hr. Of the five clones for GS, only one showed marked induction, and that was at H-2hr only.
One gene involved in ammonia assimilation that is not known to be nitrate-regulated encodes the high-affinity ammonium transporter AMT1;1, which is induced by nitrogen starvation (von Wiren et al., 1997; Gazzarrini et al., 1999). We find this gene to be less abundant in the high nitrate-treated plants than in the controls (Figure 2B), which accounts for the strong negative ratio seen in the microarray list for the H-2hr plants (Table 2). In fact, AMT1 is the second most suppressed identifiable gene in the H-2hr list. These results reflect both an increase in AMT1 mRNA in the chloride controls over time and the decrease in mRNA in the nitrate-treated plants.
Carbon Metabolic Genes
The last group of known nitrate-regulated genes includes those genes involved in organic acid and starch metabolism, including PEPC and AGPS (Stitt, 1999). Previous work showed that PEPC mRNA increases, whereas AGPS2 mRNA decreases after a 2-hr treatment with 12 mM nitrate (Scheible et al., 1997a). Two clones for PEPC and no clones for AGPS were present on the microarray. None of the PEPC clones showed a marked nitrate response. The values shown in Table 3 are the averages from both clones. Two explanations could account for this result: either PEPC is not induced under these conditions or the induced isoforms are not present on the microarray.
Novel Nitrate-Responsive Genes
As shown in Tables 1 and 2, many of the genes we detected are novel in this setting in that they are not known to be responsive to nitrate. We selected specific genes for further study, focusing on those that were rapidly induced with low nitrate (Table 1) or are potentially regulatory proteins. Table 4 summarizes the data, from both microarray analyses and RNA gel blot, for these genes. The genes were grouped according to potential function and are described below in more detail.
Table 4.
New Nitrate-Responsive Genes: Microarray and RNA Gel Blot Data
| Microarrayb
|
RNA Gel Blotb
|
|||||
|---|---|---|---|---|---|---|
| Genea | L- 20min | H-2hr | L-20min | L-2hr | H-20min | H-2hr |
| Nonoxidative pentose pathway | ||||||
| Transaldolasec | 2.2 (0.1) | 4.8 (0.1) | 5.1 | 6.3 | 2.2 | 14 |
| Transketolase | 1.6 (0.1) | 2.8 (0.2) | 2.6 | 4.3 | 2.6 | 4.3 |
| Calcium transport | ||||||
| Ca-antiporter/CAX1c | 2.4 (0.1) | 2.3 (0.1) | 5.4 | 2.1 | 5.3 | 12 |
| Transcription factors | ||||||
| Myb protein | 1.8 (0.1) | 2.4 (0.3) | 2.0 | 1.7 | 4.3 | 4.0 |
| LHY-myb protein | −1.1 | −1.1 | 1.8 | 2.8 | −2.0 | 1.6 |
| Protein phosphorylation | ||||||
| Putative S/T PKase | 1.6 (0.0) | 4.2 (0.6) | 3.6 | 1.9 | 2.7 | 5.0 |
| NPK1-like PKase | 2.3 (0.9) | 1.3 (0.1) | 4.9 | −1.7 | 20 | 4.2 |
| Additional genes | ||||||
| SAG21 | 3.1 (0.1) | 4.5 (0.2) | 12 | 4.0 | 4.1 | 5.2 |
| Auxin-induced gene | 2.4 (0.9) | 3.7 (1.2) | 7.2 | 2.5 | 13 | 8.1 |
| Sugar-transporter | 2.4 (0.1) | 1.7 (0.1) | 5.1 | 1.7 | 3.6 | −1.3 |
| His decarboxylase | 2.3 (0.3) | 2.7 (0.3) | 4.2 | 1.5 | 3.3 | 3.6 |
| Malate dehydrogenase | 2.2d | 1.8 (0.1) | 3.9 | 2.2 | 2.8 | 2.4 |
| Hemoglobin/AHB1 | 1.8 (0.1) | 5.8 (1.5) | 3.2 | 3.2 | 2.6 | 4.7 |
| Asn synthetase 2 | 1.3 (0.2) | 4.8 (1.5) | 1.9 | 1.7 | 17 | 33 |
| Methyltransferase 1 | 1.2 (0.1) | 4.2 (0.1) | 1.9 | 5.4 | 1.4 | 5.6 |
Gene symbols: CAX1, high-capacity calcium antiporter; LHY, late elongated hypocotyl; SAG21, senescence-associated protein; S/T Pkase, serine/threonine protein kinase; AHB1, class 1 nonsymbiotic hemoglobin.
L refers to inductions at 250 μM nitrate. H refers to inductions at 10 mM nitrate for the arrays and 5 mM for the RNA gel blots. Numbers in parentheses are standard deviations.
Indicates more than one clone was present on the array so that several values were averaged to give the number in the table.
This ratio was based on a single measurement.
Nonoxidative Pentose Pathway
As described above, two key genes in the oxidative pentose pathway, G6PDH and 6PGDH, are strongly induced by nitrate (Table 3). This pathway, which oxidizes glucose 6-phosphate to ribose 5-phosphate, produces NADPH for Fd reduction. There is also a nonoxidative branch to the pentose pathway, which recycles ribose 5-phosphate back into glucose 6-phosphate or into other sugars that can be used in biosynthesis of aromatic amino acids. Clones for two key genes in this branch of the pathway, transaldolase and transketolase, are present on the microarray. Three clones are included for transaldolase, and they most likely correspond to the same gene (Table 4). All three transaldolase clones show strong increases in response to nitrate and are in the top 22 identifiable genes for L-20min and in the top 13 for H-2hr. RNA gel blots corroborate these results. For transketolase, two clones are present, corresponding to two distinct genes (Table 4); only one of the two showed induction, however (weakly at L-20min and moderately at H-2hr). Both transaldolase and transketolase show a sustained increase in mRNA content at the low nitrate concentration. This differs from what was observed for G6PDH and most of the other known nitrate-responsive genes.
Calcium Transport
As described above, calcium has been implicated in nitrate signaling. The microarray had two clones for the same calcium transporter (CAX1) gene. Both clones showed moderate inductions on the microarray at both nitrate conditions. The blots showed good to strong inductions, the increase being transient at low concentrations of nitrate and sustained at high concentrations. CAX1 encodes a tonoplast, high-capacity calcium antiporter (Hirschi et al., 1996). In Arabidopsis, CAX1 expression is strongly induced with 80 mM Ca2+, weakly induced with 80 mM NaCl or 10% PEG, and not induced by abscisic acid (Hirschi, 1999). Overexpression of CAX1 in tobacco leads to plant sensitivity to salt and cold (Hirschi, 1999).
Transcription Factor
The only transcription factor known to respond to nitrate is ANR1, the MADS-box factor that is induced by nitrate after nitrogen starvation. As described above, we found no induction of ANR1 under our conditions. Thus, identification of a transcription factor that is nitrate-induced in the presence of ammonium would be important. The microarray data revealed a factor inducible by nitrate. This gene encodes a protein that is >70% identical over most of its sequence to two Arabidopsis MYB DNA binding proteins, LHY and CCA1. The genes encoding these two proteins are involved in light regulation; they show circadian regulation and control hypocotyl elongation and flowering time (Schaffer et al., 1998; Wang and Tobin, 1998). The induction ratios from the microarray for the new MYB clone are modest. By RNA gel blot analysis, this clone shows modest induction at low nitrate concentration but good induction at high nitrate content. Because of the close similarity between this new MYB clone and LHY and CCA1, we tested the nitrate-responsive behavior of LHY and CCA1. Both LHY and CCA1 are represented on the microarray, and neither shows any significant induction ratios. On blots, CCA1 shows no evidence of induction (data not shown), but LHY shows modest inductions at low concentrations of nitrate but not at high concentrations; at H-20min, a decrease in LHY RNA is observed.
Protein Kinases
One clone encoding a putative serine/threonine protein kinase ranked high on the H-2hr list (rank 18, ratio 4.2) and further down on the L-20min list (rank 70, ratio 1.6). This clone is most similar to a family of sorghum kinase genes similar to the yeast SNF1 protein kinase (Annen and Stockhaus, 1998). The similarity does not include the kinase domain itself but a C-terminal region. RNA gel blot analysis showed substantial inductions of the Arabidopsis gene that increased at high concentrations of nitrate. More than 130 clones correspond to putative protein kinases on the microarray; this particular clone, however, is notable in that it is only one of two that showed any marked nitrate response. Its behavior is similar to that of the other known nitrate-responsive genes that show transient induction at low nitrate concentrations (Figure 3).
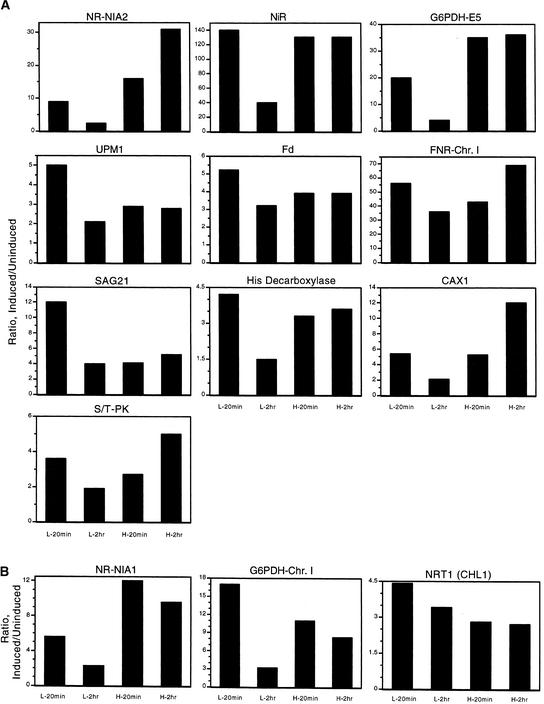
Figure 3.
Nitrate Responses for Genes Showing Major Nitrate Response.
Histograms of the nitrate/control ratios of mRNA expression in plants treated with 250 μM nitrate for 20 min or 2 hr (L-20min or L-2hr, respectively) and 5 mM nitrate for 20 min or 2 hr (H-20min or H-2hr, respectively). Ratios were determined from RNA gel blots by dividing the signal for the nitrate-treated sample by the averaged signals from the chloride-treated controls for that concentration of nitrate/chloride.
(A) Genes with induction patterns that are transient at low concentrations of nitrate and sustained or increasing at high concentrations.
(B) Genes similar to those in (A) but for which the induction pattern is borderline with other patterns. Chr., chromosome; S/T-PK, serine/threonine protein kinase.
Another putative protein kinase was identified that is similar in sequence to NPK1, a tobacco kinase similar to mitogen-activated protein kinase kinase kinases (Banno et al., 1993; Kishihama et al., 1997). The similarity includes the most C-terminal 20 amino acids of the kinase domain. This clone shows modest induction at L-20min and no induction at H-2hr—the opposite of the outcome seen for the first putative kinase. On RNA gel blots, this gene shows sharp increases after treatment for 20 min with either low or high nitrate and a diminution of signal after 2 hr of treatment.
Response Regulator
One of the moderately induced genes at H-2hr was the response regulator ARR6 (signal ratio 2.9; Table 2). This gene encodes a two-domain protein containing a receiver domain and an output domain; it is induced by nutrient deprivation (Coello and Polacco, 1999). In Arabidopsis plants deprived of nitrogen for 11 days, ARR6 mRNA decreases, but it recovers when 7 mM nitrate is added to the medium (Taniguchi et al., 1998). This gene shows no evidence of induction at L-20min on the microarray (data not shown).
Additional Genes
Additional genes that did not fit into the above categories are included on the lists. We examined specific clones in more detail, selecting those that were either very high on one of the lists or prominent on both lists. Table 4 includes the microarray and blot data for these genes.
Senescence-Associated Protein
Senescence-associated protein (SAG21) showed strong inductions on both the microarrays and the RNA gel blots. A member of a gene family that is induced during leaf senescence in Arabidopsis (Weaver et al., 1998), SAG21 is notable as the first gene of this group that is turned on in aging leaves, suggesting that it might be involved in the regulation of the other SAG genes. SAG21 expression also responds to dark and ethylene treatments but not to abscisic acid. SAG21 has substantial similarity to proteins that are abundant in late embryogenesis. The response of SAG21 to nitrate follows that of most other nitrate-responsive genes in that its mRNA decreases after 2 hr at low nitrate concentrations but is maintained in a high nitrate environment.
Putative Auxin-Induced Gene
One of the nitrate-induced genes is highly similar to many auxin-induced genes from several species, although this particular gene is not known to be auxin-induced in Arabidopsis. The GenBank Hit ID is given for the most similar known auxin-induced gene, which is from apple tree. This gene shows good to strong nitrate inductions with transient responses at both low and high nitrate concentrations.
Putative Sugar Transporter
This gene is similar to a large group of sugar transporter genes, encoding mostly glucose and hexose carriers. The gene is induced at 20 min but not at 2 hr for both low and high concentrations of nitrate. This transient induction at both concentrations is also observed for the putative NPK1-related protein kinase and the putative auxin-induced gene.
Histidine Decarboxylase
Although showing modest ratios on the microarray, the gene encoding histidine carboxylase shows good inductions on the RNA gel blots. The response is typical of nitrate-induced genes—transient at low nitrate and sustained at high nitrate.
Malate Dehydrogenase
A chloroplast-targeted, NAD+-dependent malate dehydrogenase gene is moderately induced at all time points and nitrate conditions. For the L-20min array, no data were obtained for the second two hybridizations, so only one value and no standard deviation are given in Tables 1 and 3. Four additional clones on the microarray encode malate dehydrogenases: two for the microbody, one for the mitochondrial form, and one unspecified. None of these shows any indication of induction. Malate dehydrogenase catalyzes the interconversion of malate and oxaloacetate. Nitrate has a large effect on malate pools in plants. Tobacco plants grown on 12 mM nitrate have 10 times the amount of malate in leaves and roots than do plants grown on 0.2 mM nitrate (Scheible et al., 1997a). Perhaps the increase in malate dehydrogenase in response to nitrate is contributing to the increases in organic acid metabolism and metabolites.
AHB1 and AHB2
AHB1, an interesting gene that is moderately induced by nitrate, is one of two nonsymbiotic hemoglobin genes of Arabidopsis. AHB1 encodes a class I nonsymbiotic hemoglobin that binds oxygen with very high affinity; it is induced by low oxygen content in roots and leaves (Trevaskis et al., 1997). AHB2, a member of the class II genes, encodes a protein that is related to the symbiotic hemoglobins and binds oxygen with lower affinity than does AHB1; it is induced by low temperatures but not by low oxygen content (Trevaskis et al., 1997). AHB1 is the gene in this pair that is induced by nitrate. On the microarray, the highest ratio is at H-2hr. On RNA gel blots, good inductions are seen at all times of treatment. Unlike the majority of nitrate-induced genes, the AHB1 mRNA did not decrease at 2 hr when treated with a low concentration of nitrate. AHB2 was also included on the microarray, but its ratios showed no evidence of induction. We can only speculate why AHB1 is induced by nitrate. Perhaps it is binding something besides oxygen, a binding that is needed during nitrate reduction. Alternatively, oxygen concentrations may have to be decreased when a lot of NR is synthesized, given that NR and its molybdenum cofactor can be sensitive to oxygen.
Methyltransferases
Two similar methyltransferase clones (89% identical at the nucleotide level) show inductions at 2 hr but not at 20 min of treatment. Both gene products are highly related to a spinach phosphoethanolamine N-methyltransferase (the first being 80% identical over 140 amino acids and the second 72% identical over 171 amino acids). From the microarray data, both clones show no induction at L-20min but good induction at H-2hr (Tables 2 and 4). On the RNA gel blots, only the first clone gave enough signal to quantify. Inductions of this clone were low at 20 min regardless of the nitrate concentration, but at 2 hr, inductions were strong for both nitrate concentrations (Table 4). This pattern is similar to that of only one other gene: transketolase (Figure 4).
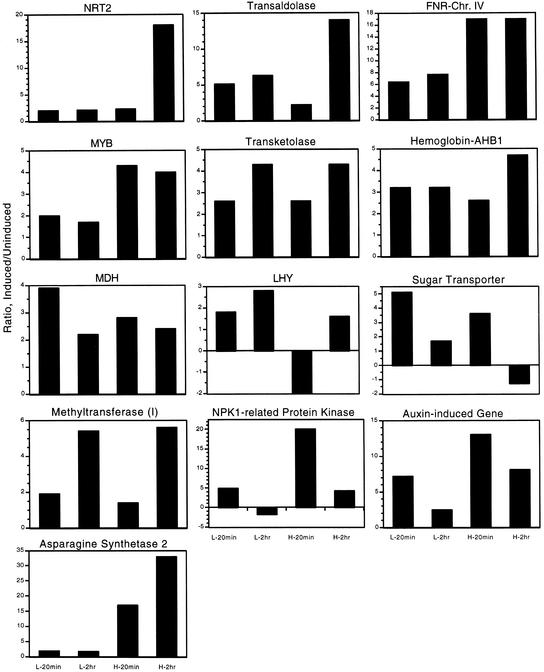
Figure 4.
Nitrate Responses for Genes Showing Diverse Nitrate Responses.
Histograms of the nitrate/control ratios of mRNA expression for plants receiving L-20min, L-2hr, H-20min, and H-2hr nitrate/chloride treatments (see legend to Figure 3). Ratios were determined from RNA gel blots by dividing the signal for the nitrate-treated sample by the averaged signals from the chloride-treated controls for that concentration of nitrate/chloride. Chr., chromosome; MDH, malate dehydrogenase.
Asparagine Synthetase 2
The ASN2 gene shows little to no induction at 20 min but strong inductions at 2 hr for both nitrate concentrations. One of three Arabidopsis ASN genes included on the array, ANS2, is induced by light and sucrose but repressed by amino acids, just the opposite of the regulation for ASN1 (Lam et al., 1998). The nitrate regulation of this gene was not known previously.
DISCUSSION
Our analysis has identified >15 new nitrate-induced genes in Arabidopsis, more than doubling the number known to this point. The nitrate regulation of these genes was verified by several methods. The results from multiple microarray hybridizations (three hybridizations using two independent mRNA preparations per time point) were compared with previously published reports of nitrate-induced genes and found to be consistent. Most of the known nitrate-regulated genes were among the top genes on at least one of the microarray lists (PEPC being the only exception). Select genes from the microarray lists were then further investigated by RNA gel blot analysis, the data from which corroborated the array results. Every gene we tested that showed nitrate induction on the microarrays also showed induction on the RNA gel blot assays. Although the absolute values often did not match, the overall pattern was maintained such that the most induced genes on the microarray lists usually showed the greatest induction on the blots.
One of the interesting aspects of this genomic analysis is that the relative amounts of induction of a large number of genes could be compared simultaneously. The overall finding is that the most highly nitrate-induced genes are those devoted to nitrite reduction, with those encoding NiR and UPM1 being the most affected. Genes encoding Fd, FNR, transaldolase, G6PDH, and 6PGDH were also very highly induced—all genes that provide reductant for nitrite reduction. These results are not surprising in hindsight, given that nitrite is toxic to plants and is not allowed to accumulate. Plants may reduce their risk of nitrite exposure by imposing a sensitive regulatory system on the nitrite reduction system, which results in substantial induction in response to low concentrations of nitrate.
Interestingly, some of the nitrate-responsive genes are also known to be induced by some form of stress: SAG21 by senescence, ethylene, and dark (Weaver et al., 1998); AHB1 by low oxygen (Trevaskis et al., 1997); and CAX1 by 80 mM NaCl (Hirschi, 1999). Neither SAG21 nor CAX1 is induced by abscisic acid (Weaver et al., 1998; Hirschi, 1999). It is possible that SAG21, AHB1, and CAX1 induction during the nitrate treatment is due to a stress response, but we favor the hypothesis that nitrate is directly inducing these genes because they respond to 250 μM KNO3 and not to 250 μM KCl after only 20 min. Perhaps they are being induced to help protect the plant from the toxic effects of nitrate or peroxynitrite, which are produced by NR (Yamasaki and Sakihama, 2000).
Examination of the plots of the nitrate-induced gene expression (Figures 3 and 4) indicates that many genes show a transient induction at a low concentration of nitrate and a sustained or increasing induction at a high concentration (Figure 3A). This list includes many of the known nitrate-induced genes, including those encoding NR, NiR, UPM1, Fd, FNR, and G6PDH. This “classic” or characteristic nitrate response for these nitrate conditions is also the response of such genes as those encoding SAG21, His decarboxylase, CAX1, and the putative Ser/Thr protein kinase, all of which can be grouped with this class of responding genes. The decline in relative mRNA quantities at low nitrate concentrations could result from the loss of nitrate from the media, the production of metabolites such as glutamine that suppress mRNA, or both. Our data suggest that both effects are in operation. Nitrate concentrations measured in the low-concentration nitrate cultures at 20 min and at 2 hr showed depletion to 22 μM at 2 hr (Figure 5). This concentration of nitrate is low but still high enough to induce some nitrate-responsive genes; thus, the decline in mRNA content is not simply attributable to the loss of nitrate from the media, and an increase in some repressing metabolite may also be responsible. On the other hand, no decline in mRNA was observed after 2 hr of treatment with high nitrate concentrations, so the mere accumulation of a repressing metabolite similarly is not sufficient.
Figure 5.
Nitrate Concentration in Media for Low Nitrate Cultures.
Samples taken from the culture media at the indicated time points for plants treated with 250 μM nitrate were analyzed by HPLC as described in Methods. Error bars indicate sd.
Inspection of these plots also reveals that almost half of the nitrate-induced genes we examined show different patterns of induction (Figure 4). Several known nitrate-induced genes (encoding NR-NIA1, one form of G6PDH, and NRT1) show a transient response at low nitrate but show some decrease when exposed to high nitrate concentrations (Figure 3B). Some genes show no suppression of mRNA at low concentrations of nitrate, including those that encode one of the FNR proteins (FNR on chromosome IV), transaldolase, transketolase, LHY, AHB1, and methyltransferase1. Another common response pattern is a transient increase in mRNA at both low and high concentrations of nitrate. These genes encode malate dehydrogenase, the putative sugar transporter, the putative NPK1-related protein kinase, and the putative auxin-induced gene. For these genes, the suppression of mRNA is independent of the nitrate concentration. The genes shown in Figure 3B are borderline in the types of response they show, but they too may fall into this class. Three genes had unique responses: NRT2, which is known to be suppressed by ammonium and showed induction only at H-2hr; LHY, which showed a twofold decrease at H-20min; and ASN2, which showed almost no induction at low nitrate concentration and very strong induction at high nitrate concentration (Figure 4). Two genes were repressed by nitrate, ANR1 and AMT1. ANR1 showed a decrease in mRNA in response to nitrate relative to the chloride controls (Figure 2). This gene has been reported to be nitrate-induced (Zhang and Forde, 1998), but the reported induction was performed with nitrogen-starved plants instead of with the ammonium-replete plants used in our study. AMT1;1, a high-affinity, starvation-induced ammonium transporter (von Wiren et al., 1997; Gazzarrini et al., 1999), is also repressed by nitrate treatment at high concentrations but not at low ones (Figure 2). All of these patterns of expression indicate that specific subsets of genes have different regulatory responses to the changing pools of metabolites in the cell. Therefore, plants have multiple responses to nitrate that provide a regulatory diversity capable of tailoring the expression of individual genes.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia was grown in liquid culture for 10 days under constant illumination with 10 mM ammonium succinate as the sole nitrogen source, as previously described (Wang et al., 1998).
Nitrate Treatment of Arabidopsis Seedlings
Ten-day-old seedlings grown in 10-mL flasks containing 4 mL of medium and ∼100 seedlings were supplemented with a KNO3 solution to give a final concentration of 0.25 mM (for the low nitrate treatment) and 5 or 10 mM nitrate (for the high nitrate treatments) or the equivalent amount of KCl for the controls. We found that responses were essentially the same for 5 and 10 mM nitrate, so these conditions were used interchangeably (data not shown). Flasks were incubated for 0, 20, or 120 min (2 hr). Whole seedlings from three to five flasks for each condition were combined and frozen in liquid nitrogen. Nitrate concentrations in the media were determined as described previously (Wang et al., 1998).
RNA Preparation and RNA Gel Blot Analysis
Total RNA was extracted from whole seedlings, and poly(A)+ RNA was prepared as previously described (Tsay et al., 1993). For RNA gel blot analysis, 1 μg of poly(A)+ RNA was loaded onto 1.5% agarose gels containing formaldehyde and processed as described by Liu and Crawford (1998). Radiolabeled DNA (probe) for each gene was prepared as described by Feinberg and Vogelstein (1983). DNA was isolated from individual DNA clones or from polymerase chain reaction (PCR) amplification of genomic DNA from Arabidopsis Columbia whole-cell DNA as template, with use of primers generated from sequence data deposited with GenBank by Incyte Pharmaceuticals (Palo Alto, CA). A list of accession numbers for all clones on the Arabidopsis GEM1 array is available at http://www.genomesystems.com/expression/clonelists/argem1.txt. RNA gel blots were autoradiographed, the film was scanned, and hybridization signals were quantitated using Adobe Photoshop 4.0 (Adobe Systems, Mountain View, CA).
Microarray Preparation
The Arabidopsis GEM1 microarrays were prepared on modified glass slides by Incyte Pharmaceuticals. The sequences used for the fabrication were generated by PCR. The PCR products were purified by gel filtration with Sephacryl-400 (Amersham Pharmacia Biotech, Piscataway, NJ) equilibrated in 0.2 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate). The filtrates were dried and rehydrated in water for arraying. The DNA solutions were arrayed by robotics on the modified glass slides as described previously (Schena et al., 1995, 1996). Briefly, glass slides were coated with 3-aminopropyltrimethoxysilane under polymerizing conditions and then allowed to cure overnight. Ten thousand DNA elements were then arrayed onto the glass slides by high-speed robotics over a 1.8 × 1.8-cm area. The glass slides were irradiated in a Stratalinker model 2400 (Stratagene, San Diego, CA) at 254 nm to fix the DNA. The slides were then washed for 2 min in 0.2% SDS and rinsed three times in water for 1 min each at room temperature. Slides were treated with 0.2% I-Block (Tropix, Bedford, MA) dissolved in Dulbecco's phosphate-buffered saline (Life Technologies, Gaithersburg, MD) at 60°C for 30 min and then rinsed in 0.2% SDS for 2 min, followed by three 1-min washes in water. The microarrays were dried by a brief centrifugation.
Fluorescent Labeling of Probe
Isolated mRNA was reverse-transcribed with 5′ Cy3- or 5′ Cy5-labeled random 9-mers (Operon Technologies, Alameda, CA). Reactions were incubated for 2 hr at 37°C with 200 ng of poly(A)+ RNA, 200 units of M-MLV reverse transcriptase (Life Technologies), 4 mM DTT, 1 unit of RNase Inhibitor (Ambion, Austin, TX), 0.5 mM dNTPs, and 2 μg of labeled 9-mers in 25 μL of enzyme buffer supplied by Operon Technologies. The reactions were terminated by incubation at 85°C for 5 min. The paired reactions were combined and purified with a TE-30 column (Clontech, Palo Alto, CA), diluted to 90 μL with water, and precipitated with 2 μL of 1 mg/mL glycogen, 60 μL of 5 M ammonium acetate, and 300 μL of ethanol. After centrifugation, the pellet was resuspended in 24 μL of hybridization buffer (5 × SSC containing 0.2% SDS and 1 mM DTT).
Hybridization on Microarrays
Probe solutions were thoroughly resuspended by incubating at 65°C for 5 min with mixing. The probe was applied to the array, which was then covered with a 22 × 22-mm glass cover slip and placed in a sealed chamber to prevent evaporation. After hybridization at 60°C for 6.5 hr, the slides were washed three times in washes of consecutively decreasing ionic strength.
Scanning Microarrays
Microarrays were scanned in both Cy3 and Cy5 channels with Axon (Foster City, CA) GenePix scanners at a 10-μm resolution. The signal was converted into 16-bits-per-pixel resolution, yielding a 65,536 count dynamic range.
Normalization and Ratio Determination
Incyte GEMtools software was used for image analysis. A gridding and region detection algorithm was used to determine the elements. The local background was calculated based on the area surrounding each element image. Background-subtracted element signals were used to calculate Cy3/Cy5 ratios. The average of the resulting total Cy3 and Cy5 signals yielded the ratio that was used to balance or normalize the signals.
Acknowledgments
We thank Mingsheng Chen for help in analysis of the gene assignments. This work was supported by Grant No. GM40672 from the National Institutes of Health.
References
- Amarasinghe, B.H., DeBruxelles, G.L., Braddon, M., Onyeocha, I., Forde, B.G., and Udvardi, M.K. (1998). Regulation of GmNRT2 expression and nitrate transport in roots of soybean (Glycine max). Planta 206, 44–52. [DOI] [PubMed] [Google Scholar]
- Annen, F., and Stockhaus, J. (1998). Characterization of a Sorghum bicolor gene family encoding putative protein kinases with a high similarity to the yeast SNF1 protein kinase. Plant Mol. Biol. 36, 529–539. [DOI] [PubMed] [Google Scholar]
- Aslam, M., Travis, R., and Huffaker, R.C. (1993). Comparative induction of nitrate and nitrite uptake and reduction systems by ambient nitrate and nitrite in intact roots of barley seedlings. Plant Physiol. 102, 811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banno, H., Hirano, K., Nakamura, T., Irie, K., Nomoto, S., Matsumoto, K., and Machida, Y. (1993). NPK1, a tobacco gene that encodes a protein with a domain homologous to yeast BCK1, STE11, and Byr2 protein kinases. Mol. Cell. Biol. 13, 4745–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowsher, C.G., Boulton, E.L., Rose, J., Nayagam, S., and Emes, M.J. (1992). Reductant for glutamate synthase is generated by the oxidative pentose phosphate pathway in non-photosynthetic root plastids. Plant J. 2, 893–898. [Google Scholar]
- Campbell, W.H. (1999). Nitrate reductase structure, function and regulation: Bridging the gap between biochemistry and physiology. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 277–303. [DOI] [PubMed] [Google Scholar]
- Champigny, M.-L., and Foyer, C. (1992). Nitrate activation of cytosolic protein kinases diverts photosynthetic carbon from sucrose to amino acid biosynthesis. Plant Physiol. 100, 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, C.-L., Dewdney, J., Kleinhofs, A., and Goodman, H.M. (1986). Cloning and nitrate induction of nitrate reductase mRNA. Proc. Natl. Acad. Sci. USA 83, 6825–6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, C.-L., Dewdney, J., Nam, H.-G., Den Boer, B.G.W., and Goodman, H.M. (1988). A new locus (NIA1) in Arabidopsis thaliana encoding nitrate reductase. EMBO J. 7, 3309–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, C.-L., Acedo, G.N., Dewdney, J., Goodman, H.M., and Cankling, M.A. (1991). Differential expression of the two Arabidopsis nitrate reductase genes. Plant Physiol. 96, 275–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coello, P., and Polacco, J.C. (1999). ARR6, a response regulator from Arabidopsis, is differentially regulated by plant nutritional status. Plant Sci. 143, 211–220. [Google Scholar]
- Crawford, N.M. (1995). Nitrate: Nutrient and signal for plant growth. Plant Cell 7, 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, N.M., and Arst, H.N.J. (1993). The molecular genetics of nitrate assimilation in fungi and plants. Annu. Rev. Genet. 27, 115–146. [DOI] [PubMed] [Google Scholar]
- Crawford, N.M., and Glass, A.D.M. (1998). Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3, 389–395. [Google Scholar]
- Crawford, N.M., Campbell, W.H., and Davis, R.W. (1986). Nitrate reductase from squash: cDNA cloning and nitrate regulation. Proc. Natl. Acad. Sci. USA 83, 8073–8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, N.M., Smith, M., Bellissimo, D., and Davis, R.W. (1988). Sequence and nitrate regulation of the Arabidopsis thaliana mRNA encoding nitrate reductase, a metalloflavoprotein with three functional domains. Proc. Natl. Acad. Sci. USA 85, 5006–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew, M.C. (1975). Comparison of the effects of a localized supply of phosphate, nitrate, ammonium and potassium on the growth of the seminal root system, and the shoot, in barley. New Phytol. 75, 479–490. [Google Scholar]
- Dzuibany, C., Haupt, S., Fock, H., Biehler, K., Migge, A., and Becker, T.W. (1998). Regulation of nitrate reductase transcript levels by glutamine accumulating in the leaves of a ferredoxin-dependent glutamate synthase–deficient gluS mutant of Arabidopsis thaliana, and by glutamine provided via the roots. Planta 206, 515–522. [DOI] [PubMed] [Google Scholar]
- Feinberg, A.P., and Vogelstein, B. (1983). A technique for radiolabeling restriction endonuclease fragments to high specific activity. Anal. Biochem. 132, 6–13. [DOI] [PubMed] [Google Scholar]
- Forde, B.G., and Clarkson, D.T. (1999). Nitrate and ammonium nutrition of plants: Physiological and molecular perspectives. Adv. Bot. Res. 30, 1–90. [Google Scholar]
- Gazzarrini, S., Lejay, T., Gojon, A., Ninnemann, O., Frommer, W.B., and von Wiren, N. (1999). Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 11, 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass, A.D.M. (1989). Plant Nutrition: An Introduction to Current Concepts. (Boston: Jones and Bartlett).
- Gowri, G., Kenis, J.D., Ingemarsson, B., Redinbaugh, M.G., and Campbell, W.H. (1992). Nitrate reductase transcript is expressed in the primary response of maize to environmental nitrate. Plant Mol. Biol. 18, 55–64. [DOI] [PubMed] [Google Scholar]
- Granato, T.C., and Raper, C.D. (1989). Proliferation of maize (Zea mays L.) roots in response to localized supply of nitrate. J. Exp. Bot. 40, 263–275. [DOI] [PubMed] [Google Scholar]
- Hirschi, K.D. (1999). Expression of Arabidopsis CAX1 in tobacco: Altered calcium homeostasis and increased stress sensitivity. Plant Cell 11, 2113–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi, K.D., Zhen, R.G., Cunningham, K.W., Rea, P.A., and Fink, G.R. (1996). Cax1, an H+/Ca2+ antiporter from Arabidopsis. Proc. Natl. Acad. Sci. USA 93, 8782–8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff, T., Truon, H.-M., and Caboche, M. (1994). The use of mutants and transgenic plants to study nitrate assimilation. Plant Cell Environ. 17, 489–506. [Google Scholar]
- Huppe, H.C., and Turpin, D.H. (1994). Integration of carbon and nitrogen metabolism in plant and algal cells. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45, 577–607. [Google Scholar]
- Kishihama, R., Banno, H., Kawahara, E., Irie, K., and Machida, Y. (1997). Possible involvement of differential splicing in regulation of the activity of Arabidopsis ANP1 that is related to mitogen-activated protein kinase kinase kinases (MAPKKKs). Plant J. 12, 39–48. [DOI] [PubMed] [Google Scholar]
- Koch, K.E. (1997). Molecular crosstalk and the regulation of C- and N-responsive genes. In A Molecular Approach to Primary Metabolism in Higher Plants, C.H. Foyer and W.P. Quick, eds (London: Taylor and Francis), pp. 105–124.
- Krapp, A., Fraisier, V., Scheible, W.R., Quesada, A., Gojon, A., Stitt, M., Caboche, M., and Daniel-Vedele, F. (1998). Expression studies of Nrt2:1Np, a putative high-affinity nitrate transporter: Evidence for its role in nitrate uptake. Plant J. 14, 723–731. [Google Scholar]
- Lahners, K., Kramer, V., Back, E., Privalle, L., and Rothstein, S. (1988). Molecular cloning of complementary DNA encoding maize nitrite reductase. Plant Physiol. 88, 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, H.M., Coschigano, K.T., Oliveira, I.C., Melooliveira, R., and Coruzzi, G.M. (1996). The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 569–593. [DOI] [PubMed] [Google Scholar]
- Lam, H.-M., Hsieh, M.-H., and Coruzzi, G. (1998). Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis thaliana. Plant J. 16, 345–353. [DOI] [PubMed] [Google Scholar]
- Leustek, T., Smith, M., Murillo, M., Singh, D.P., Smith, A.G., Woodcock, S.C., Awan, S.J., and Warren, M.J. (1997). Siroheme biosynthesis in higher plants: Analysis of an S-adenosyl-l-methionine–dependent uroporphyrinogen III methyltransferase from Arabidopsis thaliana. J. Biol. Chem. 272, 2744–2752. [DOI] [PubMed] [Google Scholar]
- Lin, Y., and Cheng, C.L. (1997). A chlorate-resistant mutant defective in the regulation of nitrate reductase gene expression in Arabidopsis defines a new HY locus. Plant Cell 9, 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D., and Crawford, N.M. (1998). Characterization of the putative transposase mRNA of Tag1, which is ubiquitously expressed in Arabidopsis and can be induced by Agrobacterium-mediated transformation with dTag1 DNA. Genetics 149, 693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner, H. (1995). Mineral Nutrition of Higher Plants. (San Diego, CA: Academic Press).
- Matsumura, T., Sakakibara, H., Nakano, R., Kimata, Y., Sugiyama, T., and Hase, T. (1997). A nitrate-inducible ferredoxin in maize roots—Genomic organization and differential expression of two nonphotosynthetic ferredoxin isoproteins. Plant Physiol. 114, 653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer, J.M., Kleinhofs, A., and Warner, R.L. (1989). Nitrate reductase regulation: Effects of nitrate and light on nitrate reductase mRNA accumulation. Mol. Gen. Genet. 217, 341–346. [Google Scholar]
- Meyerowitz, E.M. (1999). Today we have naming of parts. Nature 402, 731–732. [DOI] [PubMed] [Google Scholar]
- Quesada, A., Krapp, A., Trueman, L.J., Daniel-Vedele, F., Fernandez, E., Forde, B.G., and Caboche, M. (1997). PCR-identification of a Nicotiana plumbaginifolia cDNA homologous to the high affinity nitrate transporters of the crnA family. Plant Mol. Biol. 34, 265–274. [DOI] [PubMed] [Google Scholar]
- Redinbaugh, M.G., and Campbell, W.H. (1991). Higher plant responses to environmental nitrate. Physiol. Plant. 82, 640–650. [Google Scholar]
- Redinbaugh, M.G., and Campbell, W.H. (1993). Glutamine synthetase and ferredoxin-dependent glutamate synthase expression in the maize (Zea mays) root primary response to nitrate. Plant Physiol. 101, 1249–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redinbaugh, M.G., and Campbell, W.H. (1998). Nitrate regulation of the oxidative pentose phosphate pathway in maize (Zea mays L.) root plastids: Induction of 6-phosphogluconate dehydrogenase activity, protein and transcript levels. Plant Sci. 134, 129–140. [Google Scholar]
- Ritchie, S.W., Redinbaugh, M.G., Shiraishi, N., Vrba, J.M., and Campbell, W.H. (1994). Identification of a maize root transcript expressed in the primary response to nitrate—Characterization of a cDNA with homology to ferredoxin-NADP(+) oxidoreductase. Plant Mol. Biol. 26, 679–690. [DOI] [PubMed] [Google Scholar]
- Sakakibara, H., Takei, K., and Sugiyama, T. (1996). Isolation and characterization of a cDNA that encodes maize uroporphyrinogen III methyltransferase, an enzyme involved in the synthesis of siroheme, which is a prosthetic group of nitrite reductase. Plant J. 10, 883–892. [DOI] [PubMed] [Google Scholar]
- Sakakibara, H., Kobayashi, K., Deji, A., and Sugiyama, T. (1997). Partial characterization of the signaling pathway for the nitrate-dependent expression of genes for nitrogen-assimilatory enzymes using detached maize leaves. Plant Cell Physiol. 38, 837–843. [Google Scholar]
- Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carre, I.A., and Coupland, G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229. [DOI] [PubMed] [Google Scholar]
- Scheible, W.R., GonzalezFontes, A., Lauerer, M., MullerRober, B., Caboche, M., and Stitt, M. (1997. a). Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9, 783–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible, W.R., Lauerer, M., Schulze, E.D., Caboche, M., and Stitt, M. (1997. b). Accumulation of nitrate in the shoot acts as a signal to regulate shoot–root allocation in tobacco. Plant J. 11, 671–691. [Google Scholar]
- Schena, M., Shalon, D., Davis, R.W., and Brown, P.O. (1995). Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470. [DOI] [PubMed] [Google Scholar]
- Schena, M., Shalon, D., Heller, R., Chai, A., Brown, P.O., and Davis, R.W. (1996). Parallel human genome analysis: Microarray-based expression monitoring of 1000 genes. Proc. Natl. Acad. Sci. USA 93, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasankar, S., Rothstein, S., and Oaks, A. (1997). Regulation of the accumulation and reduction of nitrate by nitrogen and carbon metabolites in maize seedlings. Plant Physiol. 114, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt, M. (1999). Nitrate regulation of metabolism and growth. Curr. Opin. Plant Biol. 2, 178–186. [DOI] [PubMed] [Google Scholar]
- Stitt, M., and Feil, R. (1999). Lateral root frequency decreases when nitrate accumulates in tobacco transformants with low nitrate reductase activity: Consequences for the regulation of biomass partitioning between shoots and root. Plant Soil 215, 143–153. [Google Scholar]
- Sueyoshi, K., Mitsuyama, T., Sugimoto, T., Kleinhofs, A., Warner, R.L., and Oji, Y. (1999). Effects of inhibitors for signaling components on the expression of the genes for nitrate reductase and nitrite reductase in excised barley leaves. Soil Sci. Plant Nutr. 45, 1015–1019. [Google Scholar]
- Tanaka, T., Ida, S., Irifune, K., Oeda, K., and Morikawa, H. (1994). Nucleotide sequence of a gene for nitrite reductase from Arabidopsis thaliana. DNA Sequence 5, 57–61. [DOI] [PubMed] [Google Scholar]
- Tang, P.S., and Wu, H.Y. (1957). Adaptive formation of nitrate reductase in rice seedlings. Nature 179, 1355–1356. [Google Scholar]
- Taniguchi, M., Kiba, T., Sakakibara, H., Ueguchi, C., Mizuno, T., and Sugiyama, T. (1998). Expression of Arabidopsis response regulator homologs is induced by cytokinins and nitrate. FEBS Lett. 429, 259–262. [DOI] [PubMed] [Google Scholar]
- Tischner, R., Waldeck, B., Goyal, S., and Rains, W.D. (1993). Effect of nitrate pulses on the nitrate-uptake rate, synthesis of mRNA coding for nitrate reductase, and nitrate reductase activity in the roots of barley seedlings. Planta 189, 533–537. [Google Scholar]
- Trevaskis, B., Watts, R.A., Andersson, C.R., Llewellyn, D.J., Hargrove, M.S., Olson, J.S., Dennis, E.S., and Peacock, W.J. (1997). Two hemoglobin genes in Arabidopsis thaliana: The evolutionary origins of leghemoglobins. Proc. Natl. Acad. Sci. USA 94, 12230–12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueman, L.J., Richardson, A., and Forde, B.G. (1996). Molecular cloning of higher plant homologues of the high-affinity nitrate transporters of Chlamydomonas reinhardtii and Aspergillus nidulans. Gene 175, 223–231. [DOI] [PubMed] [Google Scholar]
- Tsay, Y.-F., Schroeder, J.I., Feldmann, K.A., and Crawford, N.M. (1993). A herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72, 705–713. [DOI] [PubMed] [Google Scholar]
- von Wiren, N., Gazzarrini, S., and Frommer, W.B. (1997). Regulation of mineral nitrogen uptake in plants. Plant Soil 196, 191–199. [Google Scholar]
- Wang, R., Liu, D., and Crawford, N.M. (1998). The Arabidopsis CHL1 protein plays a major role in high affinity nitrate uptake. Proc. Natl. Acad. Sci. USA 95, 15134–15139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217. [DOI] [PubMed] [Google Scholar]
- Weaver, L.M., Gan, S.S., Quirino, B., and Amasino, R.M. (1998). A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol. Biol. 37, 455–469. [DOI] [PubMed] [Google Scholar]
- Wilkinson, J.Q., and Crawford, N.M. (1991). Identification of the Arabidopsis CHL3 gene as the nitrate reductase structural gene NIA2. Plant Cell 3, 461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, J.Q., and Crawford, N.M. (1993). Identification and characterization of a chlorate resistant mutant of Arabidopsis with mutations in both NIA1 and NIA2 nitrate reductase structural genes. Mol. Gen. Genet. 239, 289–297. [DOI] [PubMed] [Google Scholar]
- Yamasaki, H., and Sakihama, Y. (2000). Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: In vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Lett. 468, 89–92. [DOI] [PubMed] [Google Scholar]
- Zhang, H.M., and Forde, B.G. (1998). An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279, 407–409. [DOI] [PubMed] [Google Scholar]
- Zhang, H., and Forde, B.G. (2000). Regulation of Arabidopsis root development by nitrate availability. J. Exp. Bot. 51, 51–59. [PubMed] [Google Scholar]
- Zhang, H.M., Jennings, A., Barlow, P.W., and Forde, B.G. (1999). Dual pathways for regulation of root branching by nitrate. Proc. Natl. Acad. Sci. USA 96, 6529–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo, D., Okamoto, J., Vidmar, J.J., and Glass, A.D.M. (1999). Regulation of a putative high-affinity nitrate transporter (Nrt2;1At) in roots of Arabidopsis thaliana. Plant J. 17, 563–568. [DOI] [PubMed] [Google Scholar]