Abstract
Farnesylation of Ras proteins is necessary for transforming activity. Although farnesyl transferase inhibitors show promise as anticancer agents, prenylation of the most commonly mutated Ras isoform, K-Ras4B, is difficult to prevent because K-Ras4B can be alternatively modified with geranylgeranyl (C20). Little is known of the mechanisms that produce incomplete or inappropriate prenylation. Among non-Ras proteins with CaaX motifs, murine guanylate-binding protein (mGBP1) was conspicuous for its unusually low incorporation of [3H]mevalonate. Possible problems in cellular isoprenoid metabolism or prenyl transferase activity were investigated, but none that caused this defect was identified, implying that the poor labeling actually represented incomplete prenylation of mGBP1 itself. Mutagenesis indicated that the last 18 residues of mGBP1 severely limited C20 incorporation but, surprisingly, were compatible with farnesyl modification. Features leading to the expression of mutant GBPs with partial isoprenoid modification were identified. The results demonstrate that it is possible to alter a protein's prenylation state in a living cell so that graded effects of isoprenoid on function can be studied. The C20-selective impairment in prenylation also identifies mGBP1 as an important model for the study of substrate/geranylgeranyl transferase I interactions.
INTRODUCTION
Interest in prenylation has stemmed from the discovery that key proteins in multiple signal transduction cascades contain covalently attached isoprenoids (Casey, 1995). Perhaps the most notable examples are the Ras proteins. Mutated forms of Ras proteins are found in 30% of all human tumors (Lowy and Willumsen, 1993). However, these mutant Ras proteins are not oncogenic if they cannot be prenylated (Lowy and Willumsen, 1993). Prevention of Ras prenylation thus holds promise as a new tactic for cancer chemotherapy (Cox and Der, 1997). To this end, many prenylation inhibitors have been developed, several of which appear to be effective anticancer agents in animal studies and are undergoing clinical trials (Buss and Marsters, 1995; Kohl et al., 1995; Liu et al., 1998; Gelb et al., 1999).
It is currently presumed that, to be effective, these drugs will need to prevent isoprenoid modification of oncogenic Ras entirely. However, forms of Ras that are incompletely modified have received little study (Goalstone and Draznin, 1996; Gadbut et al., 1997), largely because of the assumption, based on direct physical studies, that prenyl proteins are fully and completely modified (Farnsworth et al., 1990; Page et al., 1990; Myung et al., 1999). It is still not known if all functions of oncogenic Ras require prenylation or if some effector pathways may remain active regardless of prenylation state.
The signals that permit isoprenoid attachment are known in some detail. Either a farnesyl (C15) or geranylgeranyl (C20) isoprenoid is attached through a thioether linkage to a cysteine residue at the C terminus of the protein (Zhang and Casey, 1996; Seabra, 1998; Gelb et al., 1999). Enzymes that catalyze isoprenoid modification of proteins are grouped into two major classes: geranylgeranyl transferase II, which recognizes C-terminal X-X-Cys-Cys, Cys-Cys-X-X, and Cys-X-Cys motifs of Rab proteins, and CaaX motif prenyl transferases. CaaX motifs consist of a cysteine followed by two amino acids that frequently are aliphatic, then the final amino acid of the protein, X. The X residue is currently believed to be the major factor determining which of the two CaaX protein prenyl transferases will modify the CaaX cysteine (Kinsella et al., 1991; Moores et al., 1991; Yokoyama et al., 1991). Farnesyl transferase (FTase) modifies proteins with X residues such as Met, Ser, Ala, Gln, or Asn. Geranylgeranyl transferase I (GGTase I) preferentially modifies CaaX proteins with X residues of Leu or Phe. However, in cells treated with an FTase inhibitor, the K-Ras4B protein, whose CaaX motif (CVIM) is usually modified by FTase, can become C20 modified (James et al., 1996; Rowell et al., 1997; Zhang et al., 1997; Sun et al., 1998). This ability of GGTase I to modify particular FTase substrates is a difficult problem for the design of drugs to prevent Ras protein farnesylation. Current inhibitors are designed to specifically inhibit FTase and can do little to prevent K-Ras4B cross-prenylation by GGTase I.
Despite the details identified regarding how isoprenoids are attached to proteins, little is known of the mechanisms that GGTase I or FTase use to exclude inappropriate proteins. FTase and GGTase I are heterodimers with a shared α subunit but distinct β subunits (Zhang and Casey, 1996). The β subunit binds the isoprenoid and is therefore responsible for the lipid specificity of each prenyl transferase. Recent crystallographic studies of FTase indicate that amino acids from both α and β subunits contribute to the site at which the CaaX motif of the protein substrate binds (Park et al., 1997; Strickland et al., 1998). This combination presents a further challenge for the design of CaaX-based inhibitors for FTase, which must bind FTase tightly yet avoid interactions with the GGTase I enzyme to preserve the function of critical proteins that are C20 modified.
Although Ras and other small GTPases are the most well-known prenyl proteins, several other classes of CaaX-containing proteins exist. One such group is the family of guanylate-binding proteins (GBPs). GBPs are 65-kDa proteins of still unknown function that are highly induced by interferons and that were originally characterized based on their ability to bind to guanine nucleotide affinity columns (Cheng et al., 1991; Wynn et al., 1991). Six of the eight GBPs identified thus far possess CaaX motifs (Cheng et al., 1991; Wynn et al., 1991; Asundi et al., 1994; Schwemmle et al., 1996; Han et al., 1998; Vestal et al., 1998). Only one of the GBPs (human hGBP1) has a C15-type CaaX (CTIS) and appears to be farnesylated, as predicted (Schwemmle and Staeheli, 1994; Nantais et al., 1996). Other GBP family members have C20-type CaaX boxes. The CaaX sequence (CTIL) of murine mGBP1 was predicted to be a good motif for isoprenoid attachment, based on the successful prenylation of five proteins with the same CTIL sequence (rat p67 GBP, murine GBP2, Gγ4, murine mRpgr protein, and a plant calmodulin [Kalman et al., 1995; Vestal et al., 1996, 1998; Yan et al., 1998; Rodriguez-Concepcion et al., 1999]). However, despite having the hallmarks thought to be necessary, even favorable, for prenylation, isoprenoid attachment to mGBP1 proved difficult to detect. Detailed examination of possible reasons for the meager incorporation of [3H]mevalonate ([3H]MVA) indicated that the defect was not the result of problems in cellular isoprenoid metabolism or prenyl transferase activity but arose from mGBP1 itself.
MATERIALS AND METHODS
Construction of DNAs
The 1.8-kilobase mGBP1 coding region in pRC/RSV (Wynn et al., 1991) was amplified by PCR and cloned directionally into the pcDNA3 vector (Invitrogen, Carlsbad, CA) with the use of NotI and XbaI sites included on the PCR primers. Additional mutants were generated from this mGBP1 gene with the use of reverse primers that harbored the desired mutations. The GBP:Ras chimera, Chim, was constructed with the use of a primer complementary to the coding strand of the vector, with 24 mGBP1 nucleotides plus 57 nucleotides corresponding to the final 18 codons of K-Ras4B, plus a stop codon and an XbaI site. Full-length hGBP1 was cloned into pcDNA3 after PCR amplification from the pHMG-SVpolyA vector (Cheng et al., 1991) with the use of 5′ and 3′ primers containing BamHI or NotI restriction sequences, respectively. Sequences of the mutated regions in all DNAs were verified before use in transfections.
Cell Culture, Transfection, and Interferon Treatment
COS-1 cells were maintained in DMEM (Life Technologies, Grand Island, NY) containing 10% calf serum (Hyclone, Logan, UT) supplemented with 2 mM l-glutamine, 1 mM sodium pyruvate, and penicillin/streptomycin. Purified plasmid DNA (1–2 μg) was introduced into COS-1 cells with the use of Lipofectamine (Life Technologies), and cells were harvested 48 h later. RAW264.7 cells were maintained in RPMI medium (Life Technologies) with similar additions but with 10% FBS (Hyclone). Endogenous mGBP1 was induced by treatment of RAW264.7 cells with interferon-γ (IFNγ; 300 U/ml) and lipopolysaccharide (LPS; 10 μg/ml) for 20 h.
Metabolic Labeling, Electrophoresis, and Fluorography
Transfected COS-1 cells were labeled overnight in medium containing 10% serum and 100 μCi/ml [3H]MVA (American Radiolabeled Chemicals, St. Louis, MO) in the presence of compactin (a generous gift from D. Graves, Iowa State University, Ames, IA) at 50 μM or as indicated in the figure legends. RAW264.7 cells were simultaneously exposed to IFNγ/LPS and labeled with 100 μCi/ml [3H]MVA or 50 μCi/ml [3H]geranylgeraniol (American Radiolabeled Chemicals) in the presence of 50 μM compactin. Cells were lysed directly in electrophoresis sample buffer, and samples were separated by SDS-PAGE. Proteins were transferred by electroblotting onto a polyvinylidene difluoride membrane (New England Nuclear, Boston, MA), the membrane was sprayed with En3Hance (DuPont/New England Nuclear), and the 3H-labeled proteins were detected by fluorographic exposure. After fluorography, blots were stripped of fluorographic enhancer by rinsing with methanol and Tris-buffered saline containing 0.5% Tween 20. GBPs were then detected directly by immunoblotting the same membrane used for fluorography.
Immunoblotting, Immunoprecipitation, and Subcellular Fractionation
GBPs were detected with the use of a rabbit polyclonal antibody to recombinant mGBP1 (provided by D. Paulnock, University of Wisconsin, Madison, WI) with a biotinylated secondary antibody and alkaline phosphatase (Vector Laboratories, Burlingame, CA). Endogenous H-Ras in COS-1 cells was detected similarly with the use of the H-Ras–specific mouse mAb 146-03E4 (Quality Biotech, Camden, NJ). Immunoprecipitates were isolated by solubilizing cells as described (Vestal et al., 1998) and incubating the clarified supernatant with anti-GBP–coated Pansorbin (Calbiochem, La Jolla, CA). Samples were washed and resuspended in electrophoresis sample buffer for analysis by SDS-PAGE. Membranes (P100) were separated from cytosol (S100) by centrifugation at 100,000 × g as described (Nantais et al., 1996). Proteins in both fractions were precipitated by the addition of 4 volumes of cold acetone, collected by centrifugation, and resuspended in electrophoresis sample buffer.
RESULTS
Endogenous mGBP1 Is Poorly Labeled by [3H]MVA in IFNγ-treated RAW264.7 Cells
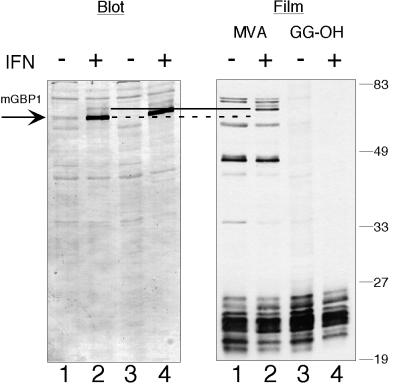
Our early studies on the effects of IFNγ/LPS on protein prenylation in murine bone marrow–derived macrophages had identified a 65-kDa prenyl protein (p65) that was strongly induced by this cytokine (Vestal et al., 1995). Incorporation of [3H]MVA into p65 was sensitive to the farnesyl transferase inhibitor BZA-5B, implying that p65 was farnesylated. Clear potential candidates for p65 were the 65-kDa IFN-inducible murine GBPs. However, both identified murine GBPs had C20-type CaaX motifs, and the mGBP2 protein appeared to be successfully C20 modified (Vestal et al., 1998). Therefore, mGBP1 was examined to determine if it would also be C20 modified or might instead be farnesylated. Unexpectedly, when the endogenous mGBP1 protein was induced by treatment of RAW264.7 cells with IFNγ (Figure 1), no prenyl protein was visible on the film at the correct molecular weight. Just above the position of mGBP1, two IFNγ-inducible proteins were clearly labeled with [3H]MVA. These minor proteins were also recognized by the polyclonal anti-GBP serum. This indicated that RAW264.7 cells were capable of attaching isoprenoids to IFNγ-induced proteins and suggested that some of these were potential GBP family members. Surprisingly, although the immunoblot showed that mGBP1 was abundantly expressed, the amount of isoprenoid it contained appeared to be below our level of detection.
Figure 1.
Incorporation of [3H]isoprenoid into mGBP1 cannot be detected in IFNγ-treated RAW264.7 cells. RAW264.7 cells were labeled for 18 h with either [3H]MVA (lanes 1 and 2) or [3H]GG-OH (lanes 3 and 4) in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of IFNγ and LPS. Total cell lysates were displayed by SDS-PAGE, and mGBP1 was detected by immunoblotting (Blot). “Film” indicates the fluorogram from the immunoblot shown, after 14 d of exposure. With either labeled precursor, no radiolabeled band of the correct size for the induced mGBP1 protein (dotted line) was detected after IFNγ/LPS. However, two new [3H]MVA-labeled proteins (solid line) of slightly slower mobility appeared after IFNγ/LPS treatment. Positions of molecular weight markers are shown on the right.
Because [3H]MVA is converted into both [3H]farnesyl pyrophosphate and [3H]geranylgeranyl pyrophosphate ([3H]GGPP), the possibility was tested that the poor labeling of mGBP1 resulted from difficulties in the transport of [3H]MVA or the production of [3H]GGPP in RAW264.7 cells. Examination of the other proteins present in the radiolabeled cell lysates did not support this theory, because many 21- to 28-kDa small GTPases of the Rho and Rab families, most of which are geranylgeranylated (Reese and Maltese, 1991; Zhang and Casey, 1996), were labeled successfully (Figure 1). To more directly examine geranylgeranyl utilization, cells were incubated with [3H]geranylgeraniol ([3H]GG-OH), an alcoholic isoprenoid that specifically labels C20-modified proteins (Crick et al., 1994). The several proteins between 30 and 80 kDa that had been labeled with [3H]MVA, including the two ∼66-kDa IFNγ-induced proteins, did not incorporate label from [3H]geranylgeraniol. This suggested that these larger proteins contained a C15 isoprenoid, as had been reported for other cell lines (James et al., 1994). The group of 21- to 28-kDa proteins was labeled effectively (Figure 1), demonstrating that IFN-treated RAW264.7 cells metabolize [3H]GG-OH to [3H]GGPP properly. However, once again, mGBP1 labeling was below detectable levels. These results indicated that IFN treatment and the presence of mGBP1 did not impair C20 isoprenoid incorporation into the appropriate proteins and thus could not explain mGBP1's poor labeling.
mGBP1 Also Displays Poor Isoprenoid Incorporation in COS-1 Cells
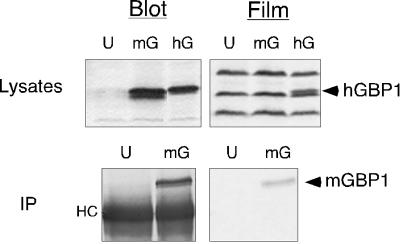
Two other possible explanations for the poor labeling of mGBP1 were that the poorly labeled IFNγ-induced protein was not actually mGBP1 or that the monocytic RAW264.7 cells specifically had difficulty with mGBP1 modification. These possibilities were tested by transiently expressing a molecular clone of authentic mGBP1 in COS-1 cells to produce an amount of mGBP1 similar to that attained naturally through IFNγ treatment. An endogenous prenyl protein that migrated just above mGBP1 (as well as the labeling of other prenyl proteins in the cell lysates) showed no differences in incorporation between mock-transfected cells and cells expressing mGBP1 (Figure 2, top). The positive control hGBP1 protein (Nantais et al., 1996) was clearly labeled (arrowhead). However, isoprenoid incorporation into mGBP1 was still below the level of detection. These results indicated that authentic mGBP1 also encountered difficulties in labeling in another cell type. Thus, the problem in the labeling of mGBP1 was not restricted to monocytic or IFN-treated cells.
Figure 2.
Poor incorporation of [3H]MVA occurs in cloned mGBP1 expressed in COS-1 cells. (Top) COS-1 cells were labeled for 18.5 h with [3H]MVA in the presence of 25 μM compactin, starting at 30 h after transfection with no DNA (U) or DNAs for mGBP1 (mG) or hGBP1 (hG). Cell lysates were resolved by SDS-PAGE, and radiolabeled proteins were detected by fluorographic exposure for 13 d. (Bottom) After transfection and labeling as described above, mGBP1 was isolated by immunoprecipitation (IP) and separated by SDS-PAGE. Incorporation of radiolabel into mGBP1 was detected after fluorographic exposure for 21 d. “HC” denotes the heavy chain of the anti-GBP antibody.
To allow us to examine mGBP1 more clearly, immunoprecipitation was used to isolate mGBP1 from the endogenous prenyl protein of COS-1 cells. With this technique, incorporation of small amounts of label into mGBP1 was detected (Figure 2, bottom). The use of immunoprecipitation to isolate larger amounts of mGBP1 from RAW264.7 cells also revealed [3H]MVA incorporation in IFNγ-induced mGBP1 after long film exposure times (our unpublished results). This ability to detect mGBP1 [3H]MVA incorporation in an immunoprecipitate did not derive from a more favored interaction of the GBP antiserum with prenylated forms of the protein, because the serum could capture uniform amounts of proteins from lysates even when the proteins showed up to eightfold differences in labeling (see below). Reciprocally, the poor labeling of mGBP1 seen directly in cell lysates (Figure 1) also indicated that immunoprecipitates had not selectively lost a prenylated form of mGBP1. Thus, the antiserum could recognize equally both prenylated and nonprenylated forms of mGBP1. Therefore, neither native, cytokine-induced, nor artificially expressed mGBP1 was totally devoid of isoprenoid, but each appeared to contain so little lipid that the small amounts present were difficult to detect unless the protein was concentrated and purified by immunoprecipitation.
Finally, to determine if the problem with mGBP1 labeling might be kinetic, and might arise from a slow equilibration of C20 pools or inefficient modification by prenyl transferase, the amount of compactin used to inhibit cellular hydroxymethylglutaryl-CoA reductase was decreased (see Figure 4, top) to avoid unintentional depletion of C20 isoprenoids (Rilling et al., 1993), and extended labeling periods (up to 52 h; our unpublished results) were used. Both approaches failed to improve mGBP1 [3H]MVA incorporation. These data indicated that mGBP1 underwent neither rapid but short-lived prenylation, nor slow but eventual prenylation, but was, at all times, poorly labeled.
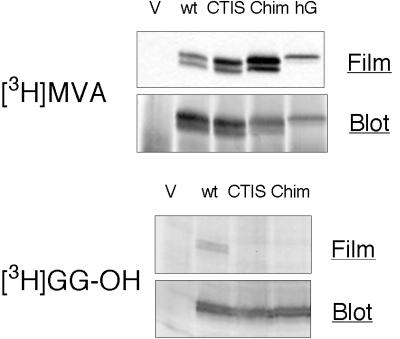
Figure 4.
[3H]MVA labeling improves in mGBP1 proteins with C15-type CaaX boxes. COS-1 cells were transfected with empty vector (V) or DNAs encoding wild-type mGBP1 (wt), hGBP1 (hG), or mGBP1 variants with C15-type CaaX motifs (CTIS and Chim). Cells were labeled with either 100 μCi/ml [3H]MVA plus 10 μM compactin or 50 μCi/ml [3H]GG-OH and no compactin, and immunoprecipitates (top panels) or cell lysates (bottom panels) were prepared and analyzed by SDS-PAGE. Incorporation of radiolabel was detected by fluorographic exposure for 22 d ([3H]MVA) or 28w ([3H]GG-OH), and then GBPs on the membranes were visualized by immunoblotting.
Poor Labeling of mGBP1 Is Not Due to Removal of a Prenylated C Terminus
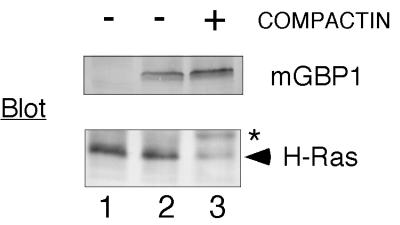
All of the experiments described above indicated that, despite the expectation that the mGBP1 CaaX motif should make the protein a good target, the difficulties underlying mGBP1's poor labeling did not involve isoprenoid metabolism or changes in prenyl transferase activity. Therefore, reasons for limited incorporation that might arise from special properties of the protein itself were considered. An initial hypothesis was that mGBP1 processing might resemble that of prelamin A. The prelamin A protein undergoes farnesylation but subsequently loses the isoprenoid when the C-terminal 18 amino acids are removed (Kilic et al., 1997). It was estimated that if a C-terminal domain of similar size were removed from mGBP1, the change in length should be detectable, because the 589-amino acid mGBP1 and the 592-amino acid hGBP1 proteins can be distinguished on our gel system (see Figure 2). When compactin was used to limit isoprenoid synthesis and force accumulation of precursors of prenylated proteins, the endogenous H-Ras protein in the COS-1 cells collected in its unprocessed form, but the mobility of the transfected mGBP1 protein did not change (Figure 3). The two forms of mGBP1 with slightly different mobilities also persisted after exposure to compactin and both incorporated [3H]isoprenoid (Figures 4 and 5) at the same low levels, indicating that neither protein was a nonprenylated precursor of the other. Presumably, some other modification or internal initiation of translation generates these two forms. Thus, compactin treatment failed to cause accumulation of any specific precursor form of mGBP1 and also failed to detect any potential isoprenoid modification that might have been difficult to observe with radiolabeling techniques.
Figure 3.
Compactin treatment does not alter mGBP1 mobility. COS-1 cells were transfected with DNA encoding mGBP1 with compactin (50 μM) added to the medium 5 h after transfection and remaining until samples were prepared 42 h after transfection. Cell lysates were separated by SDS-PAGE, and endogenous H-Ras or the transfected mGBP1 was detected by immunoblotting. Lane 1 is from cells transfected with empty vector, and lanes 2 and 3 are from mGBP1-transfected cells treated either in the absence or the presence of compactin. The asterisk (*) denotes unprocessed H-Ras; the arrowhead shows fully processed, farnesylated H-Ras.
Figure 5.
The C terminus of mGBP1 selectively hinders C20 modification. COS-1 cells were transfected with vector DNA or DNAs for various GBPs and labeled with [3H]MVA. Immunoprecipitates were formed and separated by SDS-PAGE. After fluorographic exposure for 22 d, GBPs present in the immunoprecipitates were visualized by immunoblotting.
mGBP1 with a C15-type CaaX Motif Is Modified Well
A second possibility was that the bulk of newly synthesized mGBP1 might be concealed in some location that was inaccessible to GGTase I and (because they share an α subunit) FTase. This possibility was tested by creating a chimeric mGBP1 (designated Chim) with a signal for farnesylation derived from K-Ras4B. [3H]MVA incorporation into this chimeric GBP:Ras protein showed a prominent increase above the native, wild-type mGBP1 (mGBP1wt) (Figure 4), showing that FTase had no difficulty gaining access to the Chim protein. Therefore, mGBP1 was not sequestered.
However, the loss of 18 residues and their replacement with a lysine-rich domain was a rather significant alteration of the mGBP1 C terminus. To address more precisely how well FTase could modify mGBP1, a less drastic mutant of mGBP1 was made with only the final X amino acid changed from leucine to serine. This mutant, designated CTIS, also incorporated more label than mGBP1wt (Figure 4). To determine if the increased labeling of the CTIS protein resulted simply from the change to FTase or also might result from an unintended improvement in presentation of the serine at the C terminus to a prenyl (farnesyl) transferase, an additional mutant was constructed. An alanine substitution in the X position of CaaX (CTIA) was chosen to more closely mimic the leucine present in the natural CaaX motif. The mGBP-CTIA protein also showed improved labeling, to an extent similar to that of the mGBP-CTIS protein (our unpublished results). Because this conservative change was unlikely to alter the C-terminal structure, it appeared that increased [3H]MVA incorporation resulted from the predicted change in the modifying prenyl transferase from GGTase I to FTase.
The size of isoprenoid attached to these proteins was verified with the use of [3H]GG-OH labeling. The mGBP1 protein incorporated small amounts of [3H]GG-OH (Figure 4, bottom), providing evidence that what little isoprenoid was incorporated into mGBP1 was of the expected, C20 type. The Chim and the CTIS proteins were not labeled by the [3H]C20 isoprenoid (Figure 4, bottom), indicating that these proteins were no longer substrates for GGTase I. Thus, the lipid detected on CTIS and Chim after [3H]MVA labeling was likely a C15 farnesyl moiety, as intended by their CaaX motifs. These results implied that mGBP1 was acceptable as a substrate for FTase as long as it had the appropriate CaaX motif. In addition, mGBP1 was not the still-elusive farnesylated p65 protein of bone marrow–derived macrophages.
The C Terminus of mGBP1 Interferes with C20 Modification
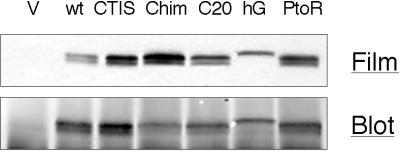
At this point, all of the data suggested the unexpected possibility that the problem with mGBP1 prenylation was C20-selective. To determine if it was the C terminus or a more distant part of mGBP1 that caused this difficulty, a new C20 version of Chim was constructed by remodeling the C15-type K-Ras4B CVIM motif of Chim back to CTIL. This C20-Chim would thus be modified by [3H]C20 isoprenoid of the same specific activity as mGBP1wt. The only differences between mGBP1wt and C20-Chim were in the C-terminal 14 amino acids that mimicked those of K-Ras4B, adjacent to the C20-CaaX motif. As shown in Figure 5, [3H]MVA labeling of C20-Chim was twofold to threefold higher than mGBP1wt. Therefore, mGBP1 modification by C20 isoprenoid could be improved by inserting 14 K-Ras4B amino acids directly upstream of the CaaX motif. This result identified this region of the C terminus as part of the mGBP1 structure that impaired C20 modification. Significantly, this replacement did not improve C20 prenylation to the same level as the C15-Chim, indicating that additional internal structures of mGBP1 contribute to and continue to impede C20 modification.
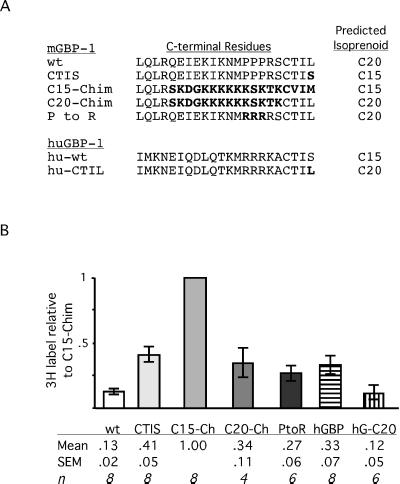
Among these C-terminal residues of mGBP1wt were three consecutive prolines (Figure 6). To determine if these prolines hindered C20 modification of mGBP1, a new mutant was constructed in which the CTIL motif was retained but the prolines were changed to arginines, the residues found in these positions in hGBP1. This P-to-R mutant was labeled approximately threefold better than mGBP1wt, to a level similar to that of the more extensively altered C20-Chim. Therefore, among the C-terminal residues of mGBP1wt, these prolines were responsible for at least a portion of the difficulty in C20 modification. It should be noted that these prolines may also impair FTase interaction, because the CTIS mutant, which retains the prolines, was also not prenylated as well as the C15-Chim protein. GGTase interaction appears to be hindered by both these prolines and more N-terminal regions and thus is more severely affected.
Figure 6.
C-terminal residues of GBPs and [3H]MVA incorporation relative to C15-Chim. (A) C-terminal amino acids of various GBP constructs are aligned. Changes from wild-type mGBP1 or hGBP1 are indicated in boldface type. (B) With the use of a fluorogram and immunoblot from the same membrane, [3H]MVA incorporation and protein amounts of each immunoprecipitated GBP were quantified by scanning, and values of [3H]MVA-derived label were corrected for variations in protein recovery. The amount of 3H for each GBP was then expressed relative to C15-Chim. Values shown are averages of these relative amounts ± SEM. The number of independent experiments is indicated by n. One-way analysis of variance indicated that all GBP proteins incorporated significantly greater amounts of 3H than mGBP1 (p < 0.05), with the exception of the C20-modified hGBP-CTIL (hG-C20), which was not significantly different from mGBP1.
Isoprenoid Modification of mGBP1 Is Incomplete
The consistent inequalities in labeling observed among these various GBPs suggested that these proteins might not be fully prenylated in mammalian cells (Figure 6A). Direct measurement of mGBP1's isoprenoid content by mass spectrometry was not practicable, because methods have not yet been developed for purification of sufficient amounts of mGBP1 from mammalian cells (where the impairment occurs), especially given the small portion of native protein that appeared to be prenylated. Therefore, the fraction of each protein that contained isoprenoid was determined by dividing the amounts of [3H]isoprenoid incorporated into the various proteins by the amount of each protein that was expressed. To increase the accuracy of this analysis, both 3H and protein measurements for each protein were drawn from a single immunoprecipitate blotted onto a single membrane. The membrane was first exposed to film for detection of 3H, then developed with GBP antiserum for quantification of protein. As an additional control, comparisons of immunoblots from total cell lysates and immunoprecipitates of cells expressing the various GBPs showed that all forms, including those well or poorly labeled, were immunoprecipitated with equal efficiency by the antibody. To determine the reliability of the calculations, the amounts of the C15-Chim proteins on the gel were varied to verify that the immunoblot measurements were in the linear range of detection. These experiments gave identical results.
These calculations indicated that the C15-Chim protein contained the most [3H]isoprenoid (Figure 6B). The 3H:protein ratio of C15-Chim, therefore, was set to 1 (because each protein molecule could contain no more than one isoprenoid, although it conceivably might contain less), and the ratios of the other GBP variants were expressed relative to this value. This analysis quantified the previous visual results and indicated that mGBP1wt contained only 13 ± 2% as much [3H]isoprenoid as C15-Chim (Figure 6B), implying that >85% of mGBP1 was not prenylated. Although the C20-Chim and P-to-R proteins were labeled more strongly than mGBP1wt, modification of these proteins also appeared to be incomplete (∼30% of C15-Chim). Thus, all of the mGBP variants with C20-type CaaX motifs appeared to be poorly prenylated (Figure 6B). Even the C15-type CTIS protein was modified only ∼40% as well as the C15-Chim. The fact that the CTIS and C15-Chim proteins had different levels of [3H]C15 labeling further confirmed that differences detected in the incorporation of 3H radioactivity reflected differences in the amount of isoprenoid attached. Thus, the level of isoprenoid modification of mGBP1 that occurred within an intact cell could be manipulated over an eightfold range by altering the last 18 residues. A more precise replacement of the three prolines with arginines could double the amount of C20-modified mGBP1, whereas simply switching to a C15-modified form could triple levels of prenylated mGBP1.
Interestingly, hGBP1 was also labeled less well than the C15-Chim. These results suggested that modification of hGBP1, although efficient enough to allow detection, was also incomplete. Finally, hGBP1 with the C20 motif CTIL was labeled as poorly as mGBP1 (12 ± 5% of C15-Chim). The hGBP-C20 protein provided a second example in which prenylation was far less than stoichiometric amounts. The threefold difference in labeling of native hGBP1 and hGBP-CTIL illustrated again that in a pair of proteins in which all non-CaaX structures were identical, C15 and C20 modification could be different.
Prenylation of GBPs Does Not Affect Membrane Association
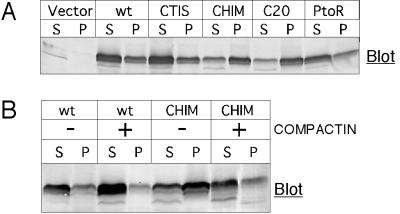
Because for some proteins isoprenoid modification enhances membrane binding, mutant GBPs with wide variations in prenylation were examined to determine if this might lead to variable extents of membrane binding. The mGBP1wt, CTIS, P-to-R, and hGBP1 proteins were largely soluble (∼70%; Figure 7A), even though their extents of isoprenoid modification differed threefold, from 15 to 45%. Earlier work had already found 75% of [3H]MVA-labeled hGBP1 in cytosolic fractions (Nantais et al., 1996), showing that the prenylated form of hGBP1 was not restricted to membranes. Prenylation thus had little impact on the membrane association of these GBPs.
Figure 7.
Membrane association of GBPs that differ in prenylation state. (A) COS-1 cells were transfected with DNAs for the indicated proteins and 48 h later separated into cytosolic (S) and membrane (P) fractions. Samples were separated by SDS-PAGE, and GBPs were detected by immunoblotting. (B) COS-1 cells were transfected with DNAs encoding mGBP1wt or C15-Chim. Treatment with 50 μM compactin was started 5 h later. After 48 h, lysates were separated into cytosolic (S) and membrane (P) fractions and separated by SDS-PAGE, and GBPs were detected by immunoblotting.
However, C15-Chim exhibited a distinct distribution, with significantly greater association with the membrane fraction (∼60%). When compactin was used to deplete the amount of farnesyl available for prenylation of C15-Chim (Figure 7B), the portion of C15-Chim present in the membrane fraction decreased. The compactin sensitivity of C15-Chim membrane interaction provided additional evidence, independent of radioactive labeling, that indicated the C15-Chim contained significant amounts of isoprenoid.
DISCUSSION
mGBP1 is the first example of a protein in which a seemingly adequate CaaX motif almost completely escapes modification. The RhoB GTPase has been shown to exist naturally as a mixture of C15- and C20-modified forms (Adamson et al., 1992), but the stoichiometry of isoprenoid attachment has been presumed to be nearly complete. The mGBP1 protein, rather than existing as a fully prenylated protein modified by one or the other isoprenoid, represents one of the few documented cases of incomplete prenylation. The ability of mGBP1 to evade prenylation is particularly unusual because it applies selectively to modification of the protein with C20 isoprenoid. Because the CTIL CaaX sequence can be C20 modified in other proteins, it is clear that this negative control of isoprenoid modification not only arises from regions of mGBP1 outside of its CaaX box but must be predominant over the otherwise acceptable CaaX motif's role.
Prenylation of mGBP1 Is Incomplete
Identification of mGBP1 as a protein with a severe deficit in [3H]MVA labeling but with an outwardly acceptable CaaX motif was unexpected. Careful examination of cellular isoprenoid utilization determined that difficulty with production of [3H]isoprenoid or IFN-induced alteration of prenyl transferase activity was not the cause of the poor incorporation.
Our results provide an important foundation for the notion that difficulty in detecting the incorporation of [3H]isoprenoid into a protein should not be ignored as an experimental or cellular flaw but may actually result from incomplete prenylation of the substrate protein. From comparison of labeling of mGBP1 and the C15-Chim protein, we calculate that only 15% of mGBP1wt molecules contain isoprenoid. This estimate is based on the assumption that the specific activity of the [3H]farnesyl pyrophosphate and [3H]geranylgeranyl pyrophosphate pools used to modify these proteins will be equal after >18 h of labeling in minimal compactin. Even if C20 and C15 isoprenoids were to continue to harbor differences in specific activities, the much better labeling of mGBP1's C20 counterpart, C20-Chim, indicates that, at best, only one-third of mGBP1wt is likely to be modified. Therefore, the bulk of mGBP1 within an intact cell genuinely appears to lack isoprenoid. Even this simple estimate gives mGBP1 the distinction of being the most poorly prenylated protein identified to date (Farnsworth et al., 1990; Page et al., 1990). There are reports that there may be some nonfarnesylated Ras proteins in cholesterol-deprived cardiac cells (Gadbut et al., 1997) and insulin-starved 3T3-L1 adipocytes (Goalstone and Draznin, 1996); additionally, the Rab24 protein also appears to be prenylated inefficiently by the Rab GGTase II (Erdman et al., 2000). Our work with mGBP1 and hGBP1 now suggests that serious deficits in prenylation can occur in other proteins and cell types and with all three classes of prenyl transferase.
Native mGBP1 thus appears to exist persistently in the cell as a mixture of C20-modified and (more predominantly) nonmodified forms. However, the amount of isoprenoid-modified mGBP1 could be increased up to eightfold by replacing mGBP1's 18 C-terminal residues. Furthermore, mutant proteins with varying degrees of C20 or C15 modification could be produced either by altering more specific residues of the C-terminal domain or by changing the prenyl transferase responsible for modification to FTase. Thus, the extent of mGBP1 prenylation can be manipulated through relatively modest changes in the C-terminal domain. Similarly, mutation of C-terminal residues of other proteins may allow mixed populations of prenyl and nonmodified proteins to be produced within a living cell and isoprenoid-sensitive functions studied. Experiments are under way to determine if replacing the C terminus of K-Ras4B with that of mGBP1 can produce a mixture of lipidated and nonlipidated forms of an oncogenic Ras. This would allow the study of responses that might be encountered if treatment with prenyl transferase inhibitors were only partially effective.
Mechanism Limiting Prenylation of mGBP1
The incomplete prenylation of mGBP1 does not appear to result from prelamin A–like proteolysis or from sequestration of the protein from cellular prenyl transferases. Simply changing the CaaX motif of mGBP1 to a form recognized by FTase significantly improved mGBP1 modification. This result also indicates that the CaaX motif of mGBP1 is not likely to be buried within the structure of the protein, because such masking would presumably impede interaction with either FTase or GGTase I.
Occupation of the mGBP1 CaaX cysteine by another modifying group remains a possibility, although we have not been able to detect incorporation of [3H]palmitate into mGBP1 (our unpublished results). ADP ribosylation of the mGBP1 CaaX is a theoretical possibility, because pertussis toxin modifies the α subunits of heterotrimeric Gi and Go proteins at a cysteine in the position analogous to the CaaX (West et al., 1985). However, in normal circumstances, these α subunit cysteines are unmodified (Jones and Spiegel, 1990) and available to interact with heptahelical receptors (Blahos et al., 1998; Yang et al., 1999). Additional information on the physiological function of mGBP1 will be needed to form a clearer picture of whether the CaaX cysteine of mGBP1 might undergo an alternative modification.
Notably, the mechanism that interferes with isoprenoid attachment to mGBP1 appears to differ from that of the G protein α subunits. For the pertussis toxin–sensitive Gi α proteins, the glycine in the second position of the “pseudo-CaaX“ sequence (CGLF) interferes with prenyl transferase interaction, making this sequence a very poor substrate (Jones and Spiegel, 1990). In contrast, the CTIL CaaX motif of mGBP1 has been shown to be a good substrate for prenylation in four other proteins (Kalman et al., 1995; Vestal et al., 1998; Yan et al., 1998; Rodriguez-Concepcion et al., 1999). Thus, the mechanism that negatively regulates mGBP1 prenylation is sufficiently strong to limit prenylation of an acceptable CaaX motif.
The Defect in mGBP1 Prenylation Is Selective for C20 Modification
The negative influence of C-terminal non-CaaX regions on mGBP1 prenylation is intriguing because it appears to selectively impair the ability of GGTase I to modify the protein. This is seen most clearly with the CTIS mutant, which retains all residues of the wild type except for the final residue that confers FTase recognition and that is modified three times as well as native mGBP1. One major structural impediment to GGTase I interaction appears to be in the C-terminal region of mGBP1, because replacement of 14 C-terminal residues (C20-Chim) or the somewhat unusual triplet of prolines (P-to-R) increases mGBP1's C20 modification approximately threefold. However, neither of these mutants of mGBP1 show the eightfold improvement in MVA incorporation seen with C15-Chim. Thus, N-terminal structures of mGBP1, in addition to its C terminus, appear to contribute to hindering GGTase I–mediated prenylation. The C20-selective impairment in prenylation identifies mGBP1 as an important model for the study of substrate/GGTase I interactions. With the use of mGBP1 as a platform, additional model structures that can produce this selective interference can be tested and help guide the design of compounds to better prevent C20 modification of K-Ras4B. Including non-CaaX elements modeled on mGBP1 may decrease a prenyl transferase inhibitor's affinity for GGTase I while retaining good FTase interaction.
It will be important to determine which distant residues outside of the C-terminal region also contribute to the difficulty in C20 modification. Work with chimeric Gγs of heterotrimeric G proteins has suggested that GGTase I recognizes protein sequences outside of the CaaX box (Kalman et al., 1995). In vitro prenylation assays and further testing of deletion mutants of mGBP1 in living cells will be necessary to clarify the location of these residues and whether they interfere directly with GGTase I interaction or bind another protein that impairs enzyme access.
An explanation of why mGBP1 might evade prenylation is not yet clear. Very little is known about the cellular function of any of the GBP proteins. One study indicates that hGBP1, like the more well-studied IFN-inducible MxA GTPase protein, may produce antiviral effects (Anderson et al., 1999). The recent solution of the three-dimensional structure of hGBP1 led to speculation that this protein might be structurally related to the dynamin family of GTPases (Prakash et al., 2000). Whether mGBP1 will show any natural variation in its prenylation or stay largely unmodified remains to be studied. Two somewhat divergent forms of murine GBPs (mag-2 and mGBP3) lack CaaX motifs and therefore will never be prenylated (Wynn et al., 1991; Han et al., 1998). The variety of mGBP1 proteins constructed here, especially the C15-Chim protein with its gain in prenylation and membrane association, may be useful for studies on GBP function.
The exceptionally poor isoprenoid modification of mGBP1 indicates that our understanding of protein prenylation in the intact cell is far from complete. Our results clearly show that full prenylation of a protein, even one with an excellent CaaX motif, is not automatic, suggesting that mechanisms that regulate isoprenoid attachment do exist but appear to function via the structure of the protein substrate rather than through changes in transferase activity. Such information is particularly needed for the newly described prenyl transferases of protozoa (Yokoyama et al., 1998) and fungi (Omer and Gibbs, 1994) and for isoprenoid-modified viral proteins such as hepatitis delta antigen (Glenn et al., 1998), in which it may be possible to exploit differences between pathogen and mammalian enzymes and their recognition of protein substrates in the treatment of infectious diseases.
ACKNOWLEDGMENTS
We thank Dave Lucas and Donna Paulnock for the original mGBP1 plasmid and polyclonal anti-GBP1 antiserum and also for helpful suggestions, Peter Staeheli for the hGBP1 plasmid, W. Robert Bishop and Deborah Vestal for comments on the manuscript, Molly Foster for technical assistance, and Michelle Booden for technical advice and assistance. These studies were funded by National Science Foundation Research Training Group award DBI960-2259 to J.T.S., Public Health Service award CA5189, and the Roy J. Carver Charitable Trust.
REFERENCES
- Adamson P, Marshall CJ, Hall A, Tilbrook PA. Post-translational modifications of p21rho proteins. J Biol Chem. 1992;267:20033–20038. [PubMed] [Google Scholar]
- Anderson SL, Carton JM, Lou J, Xing L, Rubin BY. Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology. 1999;256:8–14. doi: 10.1006/viro.1999.9614. [DOI] [PubMed] [Google Scholar]
- Asundi VK, Stahl RC, Showalter L, Conner KJ, Carey DJ. Molecular cloning and characterization of an isoprenylated 67 kDa protein. Biochim Biophys Acta. 1994;1217:257–265. doi: 10.1016/0167-4781(94)90284-4. [DOI] [PubMed] [Google Scholar]
- Blahos J, II, Mary S, Perroy J, de Colle C, Brabet I, Bockaert J, Pin J-P. Extreme C terminus of G protein α-subunit contains a site that discriminates between Gi-coupled metabotropic glutamate receptors. J Biol Chem. 1998;273:25765–25769. doi: 10.1074/jbc.273.40.25765. [DOI] [PubMed] [Google Scholar]
- Buss JE, Marsters JC., Jr Farnesyl transferase inhibitors: the successes and surprises of a new class of potential cancer chemotherapeutics. Chem Biol. 1995;2:787–791. doi: 10.1016/1074-5521(95)90083-7. [DOI] [PubMed] [Google Scholar]
- Casey PJ. Protein lipidation in cell signaling. Science. 1995;268:221–225. doi: 10.1126/science.7716512. [DOI] [PubMed] [Google Scholar]
- Cheng Y-SE, Patterson CE, Staeheli P. Interferon-induced guanylate-binding proteins lack an N(T)KXD consensus motif and bind GMP in addition to GDP and GTP. Mol Cell Biol. 1991;11:4717–4725. doi: 10.1128/mcb.11.9.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Der CJ. Farnesyltransferase inhibitors and cancer treatment: targeting simply Ras? Biochim Biophys Acta. 1997;1333:F51–F71. doi: 10.1016/s0304-419x(97)00011-5. [DOI] [PubMed] [Google Scholar]
- Crick DC, Waechter CJ, Andres DA. Utilization of geranylgeraniol for protein isoprenylation in C6 glial cells. Biochem Biophys Res Commun. 1994;205:955–961. doi: 10.1006/bbrc.1994.2758. [DOI] [PubMed] [Google Scholar]
- Erdman RA, Shellenberger KE, Overmeyer JH, Maltese WA. Rab24 is an atypical member of the Rab GTPase family: deficient GTPase activity, GDP dissociation inhibitor interaction, and prenylation of Rab24 expressed in cultured cells. J Biol Chem. 2000;275:3648–3656. doi: 10.1074/jbc.275.6.3848. [DOI] [PubMed] [Google Scholar]
- Farnsworth CC, Casey PJ, Howald WN, Glomset JA, Gelb MH. Structural characterization of prenyl groups attached to proteins. Methods. 1990;1:231–240. [Google Scholar]
- Gadbut AP, Wu L, Tang D, Papageorge A, Watson JA, Galper JB. Induction of the cholesterol metabolic pathway regulates the farnesylation of RAS in embryonic chick heart cells: a new role for Ras in regulating the expression of muscarinic receptors and G proteins. EMBO J. 1997;16:7250–7260. doi: 10.1093/emboj/16.24.7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb MH, Scholten JD, Sebolt-Leopold JS. Protein prenylation: from discovery to prospects for cancer treatment. Curr Opin Chem Biol. 1999;2:40–48. doi: 10.1016/s1367-5931(98)80034-3. [DOI] [PubMed] [Google Scholar]
- Glenn JS, Marsters JC, Jr, Greenberg HB. Use of a prenylation inhibitor as a novel antiviral agent. J Virol. 1998;72:9303–9306. doi: 10.1128/jvi.72.11.9303-9306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goalstone ML, Draznin B. Effect of insulin on farnesyltransferase activity in 3T3–L1 adipocytes. J Biol Chem. 1996;271:27585–27589. doi: 10.1074/jbc.271.44.27585. [DOI] [PubMed] [Google Scholar]
- Han BH, Park DJ, Lim RW, Im JH, Kim HD. Cloning, expression, and characterization of a novel guanylate-binding protein, GBP3, in murine erythroid progenitor cells. Biochim Biophys Acta. 1998;1384:373–386. doi: 10.1016/s0167-4838(98)00034-x. [DOI] [PubMed] [Google Scholar]
- James G, Goldstein JL, Brown MS. Resistance of K-RasBV12 proteins to farnesyltransferase inhibitors in Rat1 cells. Proc Natl Acad Sci USA. 1996;93:4454–4458. doi: 10.1073/pnas.93.9.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James GL, Goldstein JL, Pathak RK, Anderson RGW, Brown MS. PxF, a prenylated protein of peroxisomes. J Biol Chem. 1994;269:14182–14190. [PubMed] [Google Scholar]
- Jones TLZ, Spiegel AM. Isoprenylation of an inhibitory G protein alpha subunit occurs only upon mutagenesis of the carboxyl terminus. J Biol Chem. 1990;265:19389–19392. [PubMed] [Google Scholar]
- Kalman VK, Erdman RA, Maltese WA, Robishaw JD. Regions outside of the CAAX motif influence the specificity of prenylation of G protein gamma subunits. J Biol Chem. 1995;270:14835–14841. doi: 10.1074/jbc.270.24.14835. [DOI] [PubMed] [Google Scholar]
- Kilic F, Dalton MB, Burrell SK, Mayer JP, Patterson SD, Sinensky M. In vitro assay and characterization of the farnesylation-dependent prelamin A endoprotease. J Biol Chem. 1997;272:5298–5304. doi: 10.1074/jbc.272.8.5298. [DOI] [PubMed] [Google Scholar]
- Kinsella BT, Erdman RA, Maltese WA. Posttranslational modification of Ha-ras p21 by farnesyl versus geranylgeranyl isoprenoids is determined by the COOH-terminal amino acid. Proc Natl Acad Sci USA. 1991;88:8934–8938. doi: 10.1073/pnas.88.20.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl NE, et al. Inhibition of farnesyltransferase induces regression of mammary and salivary carcinomas in ras transgenic mice. Nat Med. 1995;1:792–797. doi: 10.1038/nm0895-792. [DOI] [PubMed] [Google Scholar]
- Liu M, et al. Antitumor activity of SCH 66336, an orally bioavailable tricyclic inhibitor of farnesyl protein transferase, in human tumor xenograft models and Wap-ras transgenic mice. Cancer Res. 1998;58:4947–4956. [PubMed] [Google Scholar]
- Lowy DR, Willumsen BM. Function and regulation of Ras. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- Moores SL, Schaber MD, Mosser SD, Rands E, O'Hara MB, Garsky VM, Marshall MS, Pompliano DL, Gibbs JB. Sequence dependence of protein isoprenylation. J Biol Chem. 1991;266:14603–14610. [PubMed] [Google Scholar]
- Myung C-S, Yasuda H, Liu WW, Harden TK, Garrison JC. Role of isoprenoid lipids on the heterotrimeric G protein γ subunit in determining effector activation. J Biol Chem. 1999;274:16595–16603. doi: 10.1074/jbc.274.23.16595. [DOI] [PubMed] [Google Scholar]
- Nantais DE, Schwemmle M, Stickney JT, Vestal DJ, Buss JE. Prenylation of an interferonγ-induced GTP-binding protein: the human guanylate binding protein, huGBP1. J Leukocyte Biol. 1996;60:423–431. doi: 10.1002/jlb.60.3.423. [DOI] [PubMed] [Google Scholar]
- Omer CA, Gibbs JB. Protein prenylation in eukaryotic microorganisms: genetics, biology and biochemistry. Mol Microbiol. 1994;11:219–225. doi: 10.1111/j.1365-2958.1994.tb00302.x. [DOI] [PubMed] [Google Scholar]
- Page MJ, Aitken A, Cooper DJ, Magee AI, Lowe PN. Use of the insect/baculovirus expression system to produce lipid-modified ras proteins for electrospray mass spectrometric studies. Methods. 1990;1:221–230. [Google Scholar]
- Park H-W, Boduluri SR, Moomaw JF, Casey PJ, Beese LS. Crystal structure of protein farnesyltransferase at 2.25 angstrom resolution. Science. 1997;275:1800–1804. doi: 10.1126/science.275.5307.1800. [DOI] [PubMed] [Google Scholar]
- Prakash B, Praefcke GJK, Renault L, Wittinghofer A, Herrmann C. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature. 2000;403:567–571. doi: 10.1038/35000617. [DOI] [PubMed] [Google Scholar]
- Reese JH, Maltese WA. Post-translational modification of proteins by 15-carbon and 20-carbon isoprenoids in three mammalian cell lines. Mol Cell Biochem. 1991;104:109–116. doi: 10.1007/BF00229810. [DOI] [PubMed] [Google Scholar]
- Rilling HC, Bruenger E, Leining LM, Buss JE, Epstein WW. Differential prenylation of proteins as a function of mevalonate concentration in CHO cells. Arch Biochem Biophys. 1993;301:210–215. doi: 10.1006/abbi.1993.1135. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Concepcion M, Yalovsky S, Zik M, Fromm H, Gruissem W. The prenylation status of a novel plant calmodulin directs plasma membrane or nuclear localization of the protein. EMBO J. 1999;18:1996–2007. doi: 10.1093/emboj/18.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowell CA, Kowalczyk JJ, Lewis MD, Garcia AM. Direct demonstration of geranylgeranylation and farnesylation of Ki-Ras in vivo. J Biol Chem. 1997;272:14093–14097. doi: 10.1074/jbc.272.22.14093. [DOI] [PubMed] [Google Scholar]
- Schwemmle M, Kaspers B, Irion A, Staeheli P, Schultz U. Chicken guanylate-binding protein. J Biol Chem. 1996;271:10304–10308. doi: 10.1074/jbc.271.17.10304. [DOI] [PubMed] [Google Scholar]
- Schwemmle M, Staeheli P. The interferon-induced 67-kDa guanylate-binding protein (HGBP1) is a GTPase that converts GTP to GMP. J Biol Chem. 1994;269:11299–11305. [PubMed] [Google Scholar]
- Seabra MC. Membrane association and targeting of prenylated Ras-like GTPases. Cell Signal. 1998;10:167–172. doi: 10.1016/s0898-6568(97)00120-4. [DOI] [PubMed] [Google Scholar]
- Strickland CL, Windsor WT, Syto R, Wang L, Bond R, Wu Z, Schwartz J, Le HV, Beese LS, Weber PC. Crystal structure of farnesyl protein transferase coupled with a CaaX peptide and farnesyl diphosphate analogue. Biochemistry. 1998;37:16601–16611. doi: 10.1021/bi981197z. [DOI] [PubMed] [Google Scholar]
- Sun J, Qian Y, Hamilton AD, Sebti SM. Both farnesyltransferase and geranylgeranyltransferase I inhibitors are required for inhibition of oncogenic K-Ras prenylation but each alone is sufficient to suppress human tumor growth in nude mouse xenografts. Oncogene. 1998;16:1467–1473. doi: 10.1038/sj.onc.1201656. [DOI] [PubMed] [Google Scholar]
- Vestal DJ, Buss JE, Kelner GS, Maciejewski D, Asundi VK, Maki RA. Rat p67 GBP is induced by interferon-gamma and isoprenoid-modified in macrophages. Biochem Biophys Res Commun. 1996;224:528–534. doi: 10.1006/bbrc.1996.1060. [DOI] [PubMed] [Google Scholar]
- Vestal DJ, Buss JE, McKercher SR, Jenkins NA, Copeland KG, Kelner GS, Asundi VK, Maki RA. Murine GBP-2: a new IFN-g-induced member of the GBP family of GTPases from macrophages. J Interferon Cytokine Res. 1998;18:977–985. doi: 10.1089/jir.1998.18.977. [DOI] [PubMed] [Google Scholar]
- Vestal DJ, Maki RA, Buss JE. Induction of a prenylated 65-kd protein in macrophages by interferon or lipopolysaccharide. J Leukocyte Biol. 1995;58:607–615. doi: 10.1002/jlb.58.5.607. [DOI] [PubMed] [Google Scholar]
- West RE, Jr, Moss J, Vaughan M, Liu T, Liu T-H. Pertussis toxin-catalyzed ADP-ribosylation of transducin. J Biol Chem. 1985;260:14428–14430. [PubMed] [Google Scholar]
- Wynn TA, Nicolet CM, Paulnock DM. Identification and characterization of a new gene family induced during macrophage activation. J Immunol. 1991;147:4384–4392. [PubMed] [Google Scholar]
- Yan D, Swain PK, Breuer D, Tucker RM, Wu W, Fujita R, Rehemtulla A, Burke D, Swaroop A. Biochemical characterization and subcellular localization of the mouse retinitis pigmentosa GTPase regulator (mRpgr) J Biol Chem. 1998;273:19656–19663. doi: 10.1074/jbc.273.31.19656. [DOI] [PubMed] [Google Scholar]
- Yang C-S, Skiba NP, Mazzoni MR, Hamm HE. Conformational changes at the carboxyl terminus of Ga occur during G protein activation. J Biol Chem. 1999;274:2379–2385. doi: 10.1074/jbc.274.4.2379. [DOI] [PubMed] [Google Scholar]
- Yokoyama K, Goodwin GW, Ghomashchi F, Glomset JA, Gelb MH. A protein geranylgeranyltransferase from bovine brain: implications for protein prenylation specificity. Proc Natl Acad Sci USA. 1991;88:5302–5306. doi: 10.1073/pnas.88.12.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K, Trobridge P, Buckner FS, Van Voorhis WC, Stuart KD, Gelb MH. Protein farnesyltransferase from Trypanosoma brucei. J Biol Chem. 1998;273:26497–26505. doi: 10.1074/jbc.273.41.26497. [DOI] [PubMed] [Google Scholar]
- Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- Zhang FL, et al. Characterization of Ha-Ras, N-Ras, Ki-Ras4A, and Ki-Ras4B as in vitro substrates for farnesyl protein transferase and geranylgeranyl protein transferase type I. J Biol Chem. 1997;272:10232–10239. doi: 10.1074/jbc.272.15.10232. [DOI] [PubMed] [Google Scholar]