Abstract
We analyzed the 4-kb intragenic control region of the AGAMOUS (AG) gene to gain insight into the mechanisms controlling its expression during early flower development. We identified three major expression patterns conferred by 19 AG::reporter gene constructs: the normal AG pattern, a stamen-specific pattern, and a predominantly carpel pattern. To determine whether these three expression patterns were under negative control by APETALA2 (AP2) or LEUNIG (LUG), we analyzed β-glucuronidase staining patterns in Arabidopsis plants homozygous for strong ap2 and lug mutations. Our results indicated that the stamen-specific pattern was independent of AP2 but dependent on LUG; conversely, the carpel-specific pattern was independent of LUG but dependent on AP2. These results lead to a model of control of AG expression such that expression in each of the two inner whorls is under independent positive and negative control.
INTRODUCTION
Spatial and temporal control of expression of genes responsible for floral organ identity is critical for normal flower development. Three classes of organ identity genes (classes A, B, and C) have been characterized both genetically and molecularly (reviewed in Irish, 1999; Ng and Yanofsky, 2000). Phenotypic analyses of plants that are mutant for an organ identity gene have shown that genes from each of these classes contribute to the specification of organ identity in two adjacent whorls. Furthermore, except for APETALA2 (AP2), RNA coding for the organ identity genes also is predominantly expressed in a two-whorl domain. The importance of restricting expression to the normal domains is illustrated by the altered identities of organs observed in flowers of transgenic plants engineered to misexpress organ identity genes (Mandel et al., 1992; Mizukami and Ma, 1992; Krizek and Meyerowitz, 1996; Jack et al., 1997).
Expression patterns of the class C organ identity gene AGAMOUS (AG) have been carefully documented (Bowman et al., 1991b; Drews et al., 1991). Early in flower development, AG RNA is first detected in the central apical region of late stage 3 floral meristems. As the organs emerge, AG RNA is found throughout the third- and fourth-whorl organ primordia (which give rise to stamens and carpels, respectively) but not in the two outer whorls (the sources of sepals and petals). As the floral organs are maturing, AG continues to be expressed in the third- and fourth-whorl organs. However, rather than the uniform pattern observed during early stages, by the time the flower opens, AG RNA is found only in specific types of stamen and carpel cells.
How is the early flower expression pattern of AG RNA achieved? Genes that control AG expression were first identified on the basis of mutant phenotypes that suggested AG misexpression (Bowman et al., 1991a; Weigel et al., 1992; Weigel and Meyerowitz, 1993; Liu and Meyerowitz, 1995; Goodrich et al., 1997). Subsequent in situ hybridization analyses confirmed altered expression of AG and led investigators to distinguish between genes that function as either positive or negative regulators of AG expression. LEAFY (LFY) was first implicated as a positive regulator of AG in the floral meristem because AG RNA is decreased in lfy mutants (Weigel and Meyerowitz, 1993). Recently, this role in AG activation was shown to be direct: LFY interacts with binding sites within the AG control region, which in turn leads to activation of AG expression (Busch et al., 1999).
Two negative regulators of AG early flower expression have also been identified. AP2 is a negative regulator of AG; in strong ap2 mutants, AG RNA accumulates not only in the inner two whorls but also in the outer two whorls (Drews et al., 1991). Similarly, in leunig (lug) mutants, AG RNA accumulates ectopically in outer-whorl organs (Liu and Meyerowitz, 1995). Mechanisms by which these genes effect negative regulation are unknown.
The simplest model for negative regulation is to have AP2 and LUG function as transcriptional repressors. Transcriptional repressors have been characterized from several systems (reviewed in Cai et al., 1996; Hanna-Rose and Hansen, 1996). Repressor binding sites can overlap with those of the activator, repressing transcription by direct competition for binding, or they can be located at a distance from the activator cis sequence. Repressors that bind at a distance bring about negative regulation by interacting with other cellular components, such as the basal transcriptional machinery, specific histones (to bring about a repressive chromatin structure), specific transcriptional activators, or corepressors. The presence of putative nuclear localization signals, protein–protein interaction domains, and DNA binding domains are consistent with the possibility that AP2 is a transcriptional repressor (Jofuku et al., 1994; Weigel, 1995); however, these data do not rule out other possible mechanisms. Whether LUG also has motifs suggestive of a transcriptional repressor awaits its molecular characterization.
Our goal was to gain insight into possible mechanisms controlling AG expression during early flower development by analyzing the AG control region. Previously, we determined that sequences within the AG gene itself, largely second intron sequences, were required for normal AG activity and contained the cis sequences used by both AP2 and LUG to restrict AG expression to the inner two whorls. In this study, we have extended the reporter gene approach to analyze the AG intragenic sequences, and we have compared reporter gene expression patterns in wild-type and ap2 and lug mutants. We found that sequences derived from intron 2 can function as enhancer elements and confer separable carpel- and stamen-specific expression patterns. In addition, AP2 and LUG each provide negative regulation to only one of these expression patterns.
RESULTS
We characterized the patterns of floral staining by β-glucuronidase (GUS) conferred by the 19 different AG::GUS reporter gene constructs shown in Figure 1; these constructs differed in the amount, position, and orientation of their AG DNA content. Five of the constructs—pMD200, pMD221, pMD222, pMD227, and pMD228—are derivatives of the previously described pAG-I::GUS construct (Sieburth and Meyerowitz, 1997). The remaining reporter gene constructs were generated by placing AG DNA fragments upstream of a deletion derivative (−60) of the cauliflower mosaic virus (CaMV) 35S promoter (Benfey and Chua, 1990). This minimal promoter does not itself confer a GUS staining pattern and thus has been a useful tool in enhancer-trap experiments (Campisi et al., 1999).
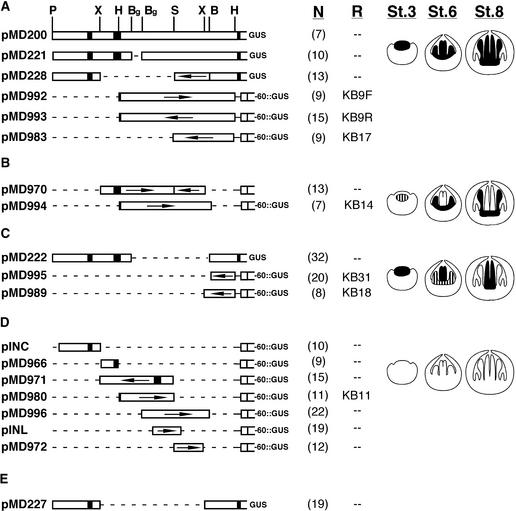
Figure 1.
Structure of AG::GUS Reporter Gene Constructs.
(A) Reporter gene constructs that conferred a normal AG pattern to the reporter gene during early flower development (stages 1 to 9).
(B) Reporter gene constructs that conferred a stamen expression pattern to the reporter gene during early flower development (stages 1 to 9).
(C) Reporter gene constructs that conferred a carpel expression pattern to the reporter gene during early flower development (stages 1 to 9).
(D) Reporter gene constructs that failed to confer reporter gene expression during early flower development (stages 1 to 9).
(E) Reporter gene construct that gave variable GUS staining patterns.
White bars indicate noncoding sequences; black boxes indicate the first two AG exons. Arrows indicate orientation of fragments; where arrows are omitted, the fragments are in their normal 5′ to 3′ orientation. Letters above pMD200 in (A) indicate restriction sites used for construction of the reporter gene constructs. P indicates PstI, and for convenience we refer to this as nucleotide position 1. X, XbaI, with sites at positions 1272 and 3936; H, HindIII, with sites at positions 1753 and 4762; Bg, BglII, with sites at 2127 and 2455; S, SpeI, with a site at position 3109; B, BamHI, with a site at position 4018. Numbers beneath N (within parentheses) indicate the number of independently transformed lines examined for each construct. Designations beneath R, when present, indicate the construct number containing the same AG DNA used in studies by Busch et al. (1999) and Bomblies et al. (1999). Diagrams at right represent the GUS staining patterns of stage (St.) 3, stage 6, and stage 8 flowers; black areas indicate strong GUS staining, and striped areas indicate weak GUS staining.
We transformed each of these constructs into Arabidopsis (ecotype Landsberg erecta) and characterized the GUS staining pattern of the transgenic plants (Figure 1). We observed some variability in stain intensity between independently transformed plants. For each line except pMD995, we describe the major GUS staining pattern seen in more than half of the independently transformed lines. The staining pattern described for plants containing pMD995 was observed in only eight of 20 lines; the remaining 12 lines produced no floral GUS staining. pMD995 contains the same AG DNA as the KB31 GUS reporter gene described by Busch et al. (1999), who also obtained a similar ratio of expressing and nonexpressing lines.
Six of the constructs—pMD200, pMD221, pMD228, pMD992, pMD993, and pMD983—contained sequences that conferred floral GUS staining in a normal AG pattern for floral stages 3 to 9 (stages according to Smyth et al., 1990). Seven constructs—pINC, pMD966, pMD971, pMD980, pMD996, pINL, and pMD972—did not confer floral GUS staining, and six constructs—pMD970, pMD994, pMD222, pMD995, pMD989, and pMD227—conferred a floral GUS staining pattern that did not match the normal AG expression pattern. Many of these constructs also produced GUS staining during seedling stages; because these vegetative patterns were variable, they are not characterized in detail.
Second Intron Sequences Are Sufficient To Confer a Normal AG Expression Pattern
We observed similar GUS staining patterns in developing flowers (up to stage 9) of transgenic plants carrying any one of six AG::GUS constructs: pMD200, pMD221, pMD228, pMD992, pMD993, and pMD983 (Figure 1A). Representative GUS staining patterns are shown in Figures 2A to 2I. In these flowers, we first observed strong GUS staining in the central floral meristem region of late stage 3 flowers (Figures 2A, 2C, 2D, 2F, 2H, and data not shown). As the flowers developed, we observed strong and uniform GUS staining in third-whorl stamen primordia and in fourth-whorl carpel primordia, but no GUS staining in the first-whorl sepal or second-whorl petal primordia (Figures 2A to 2C, and 2E to 2I). These GUS staining patterns matched the pattern of AG RNA observed by in situ hybridization in wild-type flowers (Drews et al., 1991) and that seen in wild-type plants carrying the previously described pAG-I::GUS gene fusion construct (Sieburth and Meyerowitz, 1997). These observations suggest that the assembly of these constructs removed no sequences that were essential for the normal early flower AG expression pattern.
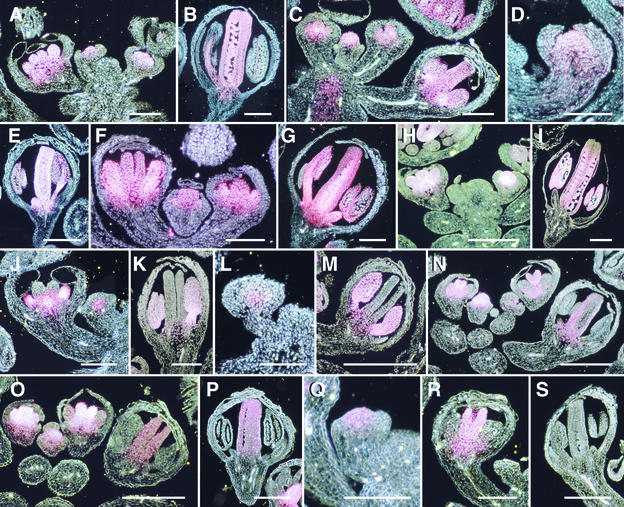
Figure 2.
GUS Staining Patterns in Wild-Type Plants carrying AG::GUS Reporter Gene Constructs.
(A) and (B) GUS staining in transgenic plants carrying pMD200. (A) In a longitudinal section through the inflorescence meristem, stage 3, 4, and 8 flowers show GUS staining in the central region of the floral meristem and in the developing stamen and carpel primordia. (B) This stage 12 flower shows GUS staining in the stamens and carpels but not in the sepals or petals.
(C) GUS staining in a transgenic plant carrying pMD221. GUS staining in the stage 4, 6, and 10 flowers is restricted to the central region of the floral meristem and the developing stamens and carpels.
(D) and (E) GUS staining in transgenic plants carrying pMD992. (D) This stage 4 flower shows GUS staining in the central region of the floral meristem. (E) This stage 10 flower shows GUS staining in the stamens and carpels but not in the sepals and petals.
(F) and (G) GUS staining in transgenic plants carrying pMD993. (F) Stage 5, 7, and 8 flowers show GUS staining in the developing stamens and carpels. (G) This stage 9 flower shows GUS staining in the stamens and carpels but not in the sepals and petals.
(H) and (I) GUS staining in transgenic plants carrying pMD983. (H) These stage 4 and 7 flowers show GUS staining in the floral meristem and in the stamen and carpel primordia, respectively. No GUS staining was detected in the inflorescence meristem (IM) or the developing sepals. (I) GUS staining in this stage 12 flower is in the stamens and carpels but not in the sepals and petals.
(J) and (K) GUS staining in transgenic plants carrying pMD970. (J) The stage 3 flower shows GUS staining in a small domain in the core of the floral meristem; the stage 7 flower has GUS staining in stamen primordia and the receptacle but not in the carpel or sepal primordia. (K) This stage 9 flower shows GUS staining in the developing stamens but not in the sepals or carpels.
(L) and (M) GUS staining in transgenic plants carrying pMD994. (L) GUS staining in this stage 4 flower is restricted to a small domain in the core of the floral meristem. (M) This stage 9 flower shows GUS staining in the stamens but not in the carpels, sepals, or petals.
(N) GUS staining in a transgenic plant carrying pMD222. GUS staining in the stage 4 flower is strong throughout the central apical region of the floral meristem. In the two stage 7 flowers, GUS staining is strong only in the carpel primordia; weak GUS staining can be detected in the abaxial proximal regions of the developing stamens. In the stage 9 flower, GUS staining is present in the carpels and receptacle only.
(O) and (P) GUS staining in transgenic plants carrying the pMD989 transgene. (O) GUS staining in the stage 4 flower is restricted to the central region of the floral meristem. In the stage 6 flower, GUS staining is present in both stamen and carpel primordia. In the stage 7 flower, GUS staining is strong only in the carpels. In the stage 9 flower, GUS staining is detected only in the carpels. (P) GUS staining in this stage 10 flower is present in the carpels but absent in the sepals and stamens.
(Q) to (S) GUS staining in transgenic plants carrying pMD995. (Q) GUS staining is present in the central apical region of the floral meristem of this stage 3 flower. (R) GUS staining is present in the carpel primordia but not in the sepal or stamen primordia of this stage 7 flower. (S) GUS staining is lost from all floral organs by stage 10.
Numbers at the base of the flowers indicate their floral stage; IM, the inflorescence meristem. 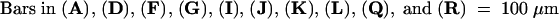 ;
; 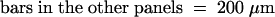 .
.
Comparing the AG sequences included in each of these constructs (Figure 1A), we found that pMD983 contained the smallest AG DNA fragment conferring a normal AG expression pattern. This fragment consisted of 1653 bp of DNA from the 3′ end of the large second intron, and its sequence was common to all of the constructs that gave a normal AG expression pattern. Apparently, this 1653-bp DNA fragment contains cis elements that are sufficient for normal early-flower expression of AG.
Stamen-Specific Expression
When transformed into plants, six of the AG::GUS constructs conferred GUS staining in the flower but not in the typical AG pattern (Figures 1B and 1C). One of these constructs, pMD227 (Figure 1E), gave variable GUS staining results and will not be considered further. The remaining five constructs conferred one of two different GUS staining patterns.
Both pMD970 and pMD994 conferred a floral GUS staining in late stage 3 flowers that was weak and appeared in a different position than in the normal stage 3 to 4 floral meristem domain (Figures 2J to 2M). The normal domain for AG RNA in a stage 3 or 4 flower is a large dome of cells internal to the sepal primordia (Figures 2A, 2C, 2D, and 2H). In contrast, pMD970 and pMD994 conferred GUS staining to a small spherical domain embedded in the center of the floral meristem (Figures 2J and 2L). The AG DNA contained in pMD994 is identical to that contained in another reporter gene construct (KB14) recently described (Busch et al., 1999). However, Busch et al. (1999) observed a normal early expression pattern for AG in transgenic plants carrying KB14. Possible reasons for the different staining patterns are considered in the Discussion.
At later flower development stages, transgenic plants carrying either pMD970 or pMD994 showed strong GUS staining in stamen primordia and in the receptacle (including beneath the carpel primordia) but not in carpel primordia (Figure 2J and data not shown). Stamens continued to show strong GUS staining through stage 9, but no GUS staining was detected in carpels, sepals, or petals (Figures 2K and 2M). This pattern resembled the normal AG expression pattern, in that GUS staining was present in the late stage 3 floral meristem, developing stamens, and receptacle and was not present in sepals and petals. However, the staining pattern differed from the normal AG expression pattern in that GUS staining at early stages (3 to 4) was weak and was in a smaller domain, and no GUS staining occurred in developing carpels. Such observations suggest that during the assembly of these two constructs, we removed sequences that contribute to normal early induction and that are required for carpel-specific expression.
We compared the AG sequences contained in constructs that conferred the stamen-specific staining pattern with those that conferred the typical AG pattern (cf. pMD983 in Figure 1A with Figure 1B). Both pMD994 and pMD970 lacked sequences from the 3′ end of intron 2 (they were missing 744 and 826 bp, respectively), whereas constructs that conferred the full AG GUS staining pattern all contained those sequences (cf. Figure 1B with Figure 1A). These observations indicate that carpel-specific expression requires sequences from the 3′ end of intron 2, whereas sequences within the 5′ part of intron 2 are sufficient to activate expression in the meristem (albeit weakly) and to confer stamen expression.
Because pMD970 and pMD994 each contained a large fragment of AG DNA (Figure 1), we had expected that further delineation of the cis element(s) in this region would be simple. We generated five AG::GUS fusion constructs containing sequences derived from this region: pMD971, pMD980, pMD996, pINL, and pMD972 (Figure 1D). However, none of the transgenic plants carrying any of these five constructs produced GUS staining in stage 1 to stage 9 flowers (data not shown), suggesting that the patterns of expression observed in pMD970 and pMD994 require multiple dispersed cis elements. Similarly, pMD983 conferred a normal AG expression pattern, but removal of 827 bp of DNA from its 5′ end (to generate pMD989; expression pattern described below) resulted in the loss of most stamen expression. This observation indicates that the 827-bp fragment removed contains cis elements that confer stamen expression; however, the 827-bp fragment alone (pMD972) and the 827-bp fragment with additional 5′ and 3′ sequences (pMD996) both failed to confer stamen expression (Figure 1D). Taken together, these observations indicate that stamen expression requires two or more dispersed elements. In addition, we have previously shown that stamen-specifying enhancer elements also reside upstream of AG (Sieburth and Meyerowitz, 1997).
Carpel-Predominant Expression
Three of the AG::GUS reporter gene constructs—pMD222, pMD995 (equivalent to KB31; Busch et al., 1999), and pMD989 (equivalent to KB18; Busch et al., 1999)—conferred similar floral GUS-staining patterns (Figures 2N to 2S, and data not shown). Transgenic plants bearing these constructs produced strong GUS staining in the central region of the floral meristem during late stage 3 (Figures 2N, 2O, and 2Q, and data not shown). As primordia for the floral organs became distinct, we observed strong GUS staining in the developing carpels; this carpel staining remained strong through stage 9 for plants bearing pMD989 and pMD222; however, carpel GUS staining was rapidly lost from pMD995 plants starting at approximately stage 8 (Figures 2N, 2P, and 2S). For all three constructs, GUS staining in stamen primordia of floral stage 5 flowers was weak. By stage 7, stamen GUS staining was reduced to a small region on the abaxial side of the stamens (Figure 2N). This GUS staining pattern was absent by stage 8. No GUS staining was observed in developing sepals and petals (Figures 2N, 2O, and 2R, and data not shown). These GUS staining patterns resembled the normal AG expression pattern with regard to the timing and position of GUS stain in the floral meristem, the GUS staining in the developing carpels, and the absence of GUS staining in sepals and petals. However, the rapid loss of GUS staining as stamens developed differed from the normal AG pattern. These observations suggest that in assembling pMD995, pMD989, and pMD222, we removed sequences that contribute to normal stamen expression.
We compared the AG sequences contained in constructs that conferred the carpel-specific staining pattern with those that conferred a normal AG staining pattern (cf. pMD983 in Figure 1A with Figure 1C). pMD222, pMD989, and pMD995 lacked between 827 and 909 bp, corresponding to the center of the second intron. This result suggests that sequences required for high stamen-specific expression fall within the removed fragments. It also indicates that sequences sufficient for floral meristem activation and expression in developing carpels are present in the 744-bp fragment contained by pMD995. Activation of expression in the floral meristem by this AG DNA fragment (construct KB31) was also noted in a recent study by Busch et al. (1999).
Characterization of AG::GUS Expression Patterns in ap2 and lug Mutants
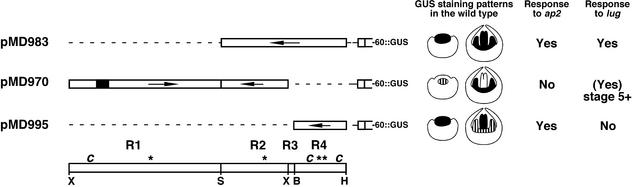
Previously, we showed that a 4-kb DNA fragment from within the AG gene contains the cis elements used by AP2 and LUG to restrict AG gene expression to whorls three and four (Sieburth and Meyerowitz, 1997). We were curious whether the subfragments derived from this region that conferred expression specifically to either third-whorl stamens or fourth-whorl carpels were also under negative regulation by AP2 and LUG. To address this question, we crossed three AG::GUS reporter gene constructs to plants containing strong ap2 or lug mutations and analyzed the GUS staining in the developing flowers of the homozygous transgenic mutants. The three constructs used in this experiment are depicted in Figure 3. To simplify the discussion of GUS staining patterns in relationship to the corresponding AG DNA, we refer to four regions (R1 to R4) contained within this 3490-bp region of AG. R1 contains 1837 bp (XbaI to SpeI), begins at the 5′ end of this region, and is present only in construct pMD970. R2 contains 827 bp (SpeI to XbaI) and is present in both pMD970 and pMD983. R3 contains 82 bp (XbaI to BamHI) and is present only in pMD983. R4 contains 744 bp (BamHI to HindIII) and is present in pMD970 and pMD995.
Figure 3.
Summary of Negative Regulation Experiments and Results.
Depicted are the three constructs (pMD983, pMD970, and pMD995) used in experiments examining GUS staining in ap2 and lug mutants; beneath them, the regions R1 to R4 are indicated. R1 begins at 1272 (relative to the upstream PstI site as 1) and extends to 3109 (SpeI site). R2 begins at 3110 and extends to 3936 (XbaI site). R3 begins at 3937 and extends to 4018 (BamHI). R4 extends from 4019 to 4762 (HindIII). The c and the asterisk indicate approximate positions of CArG boxes and LFY binding sequences, respectively. pMD983 contains R2, R3, and R4; pMD970 contains R1 and R2; and pMD995 contains only R4. Cartoons depicting the staining patterns of stage 3 and 6 flowers are presented with each construct. At right are indicated the constructs that showed an expansion of the GUS staining domain to include outer-whorl organs when present in either an ap2 or a lug mutant background. Other symbols and abbreviations are as given in Figure 1.
For analysis of AP2 interactions, we used the previously described ap2-2 and ap2-9 alleles (Bowman et al., 1989, 1991a). In plants homozygous for either allele, medial first-whorl organs typically differentiate as carpels or staminoid carpels, lateral first-whorl organs are typically absent or differentiate as leaves, and second-whorl organs are absent (Bowman et al., 1991a). Many of these outer-whorl defects are the result of ectopic AG expression, because in ap2-2 ag-1 double mutants, outer-whorl organ numbers are mostly restored and many carpelloid attributes are lost. Ectopic expression of AG RNA in flowers of ap2-2 mutants was confirmed by in situ hybridization analyses (Drews et al., 1991). In ap2-2 flowers, AG RNA is detected first in stage 2 flowers and as the flowers develop is maintained in medial first-whorl organs and all internal organs.
To analyze LUG regulation, we used the lug-1 allele. Plants homozygous for this mutation have flowers in which the outer-whorl organs are sepals that often have staminoid sectors, the second-whorl organs are fewer than usual and typically differentiate as petals, the third-whorl organs are fewer than usual and differentiate as stamens, and the fourth-whorl organs are two carpels with an unfused apex and hornlike extensions (Liu and Meyerowitz, 1995). In situ hybridization studies with this mutant allele have shown that AG RNA is expressed precociously in stage 2 flowers and in a patchy pattern in older outer-whorl organs.
We examined the GUS staining patterns of transgenic mutant flowers between stages 2 and 7. If the stamen- and carpel-specific GUS staining patterns lacked outer-whorl GUS staining because of negative regulation by AP2 or LUG, then we expected to observe GUS staining in medial first-whorl organs and organ primordia of transgenic mutant flowers. Alternatively, if the restricted pattern were independent of AP2 or LUG regulation, then we expected the GUS staining pattern in transgenic mutants to match the restricted expression domain observed in the wild type.
Negative Regulation by AP2
ap2 mutants carrying the pMD983 GUS reporter gene produced intense GUS staining in all floral whorls, as shown in Figure 4C. This pattern matched that of ap2-2 pAG-I::GUS flowers and the AG in situ hybridization pattern for ap2-2 mutants (Drews et al., 1991; Sieburth and Meyerowitz, 1997). This result indicates that the pMD983 GUS staining pattern in wild type is under negative regulation by AP2.
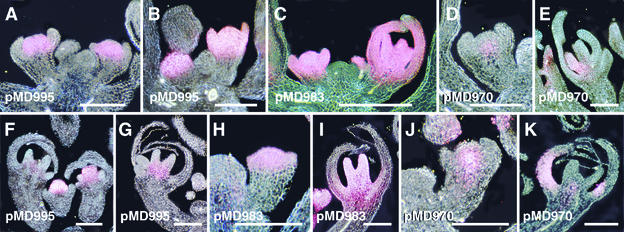
Figure 4.
GUS Staining Patterns in Flowers of Transgenic ap2 and lug Plants.
(A) to (E) GUS staining in strong ap2 mutants. (A) and (B) GUS staining in ap2-2 plants carrying the pMD995 transgene. (A) Pattern seen in approximately half the plants examined, in which intense GUS staining is restricted to the central region of the floral meristem in stage 4 flowers and is not present in the sepal primordia of these pMD995 ap2-2 plants. (B) Pattern seen in the remaining plants (also ∼50%), in which intense GUS staining is seen throughout the central floral meristem and the sepal primordia of the late stage 4 flower, as well as throughout the apical portion of the early stage 3 floral meristem. (C) GUS staining in ap2-2 plants carrying the pMD983 transgene, in which the stage 4 and 7 flowers have intense GUS staining through all floral organ primordia. (D) and (E) GUS staining in ap2-9 plants carrying the pMD970 transgene. (D) GUS staining in a stage 4 flower is restricted to a small domain in the center of the floral meristem. (E) GUS staining in this stage 7 flower is observed only in a developing stamen and not in any outer-whorl organs.
(F) to (K) GUS staining patterns in lug-1 mutants. (F) and (G) GUS staining in lug-1 plants carrying the pMD995 transgene. GUS staining is present in the central floral meristem of the stage 4 flower, in the carpel primordia of the stage 6 flower. In stage 8 flowers, GUS staining was highly reduced (F), and when present, was restricted to carpels (G). (H) and (I) GUS staining in lug-1 plants carrying the pMD983 transgene. GUS staining in an early stage 3 flower is present in sepal primordia as well as the floral meristem (H) but in this stage 8 flower is present in developing carpels and in proximal regions of the sepals (I). (J) and (K) GUS staining in lug-1 plants carrying the pMD970 transgene. GUS staining in this stage 3 floral meristem is restricted to the central core (J), whereas in this stage 8 flower it is present in outer-whorl sepals (K).
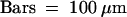 .
.
We observed variable patterns of floral GUS staining in ap2 mutants carrying the pMD995 GUS reporter gene. In approximately half the plants (eight of 19), GUS staining was restricted to the normal AG pattern in the floral meristem and was not observed in the outer-whorl organs (Figure 4A). In the remaining plants (11 of 19), we observed GUS staining in all whorls of developing flowers (Figure 4B). This variability was in contrast to our observations of pMD983 ap2 flowers, in which the expanded floral expression pattern was present in all flowers (12 of 12 plants examined in detail). Using a reporter gene with identical AG DNA (KB31), Bomblies et al. (1999) also observed ectopic expression when the transgene was present in strong ap2 mutants.
ap2 mutants carrying the pMD970 GUS reporter gene showed a low amount of GUS staining that did not extend to outer-whorl organs (Figures 4D and 4E). In stage 3 flowers, the GUS stain was restricted to the floral meristem core (Figure 4D); at later stages, the GUS stain was present only in stamens. At no stage did the GUS staining pattern extend into first-whorl organs. This analysis indicates that the pMD970 GUS staining pattern in the wild type is not under negative regulation by AP2.
We compared the AG sequences contained by these three constructs (Figure 3). Because we observed the expanded pattern of GUS staining in pMD983- and pMD995-containing ap2 plants, R4 must contain regulatory sequences that are used by AP2.
Negative Regulation by LUG
lug mutants carrying the pMD983 reporter gene showed GUS staining that extended from the center of the floral meristem to the sepal primordia of stage 3 flowers (Figure 4H) and at later stages of development was present in an irregular pattern in sepals (Figure 4I). This pattern matched that of lug-1 pAG-I::GUS flowers (Sieburth and Meyerowitz, 1997) and the AG in situ hybridization pattern in lug-1 mutants (Liu and Meyerowitz, 1995). These results indicate that the pMD983 GUS staining pattern in wild type is under negative regulation by LUG.
GUS staining in lug mutants carrying the pMD995 reporter gene is shown in Figures 4F and 4G. The stage 4 flower in Figure 4F shows strong GUS staining in the central dome of the floral meristem, a pattern that matched the normal AG expression pattern and that matched pMD995 in the wild type. As the internal floral organs differentiated, GUS staining was confined to the innermost whorl (Figures 4F and 4G). These patterns matched the pMD995 GUS staining pattern in wild type but not the pattern of AG RNA in lug mutants (Liu and Meyerowitz, 1995). These results suggest that the pMD995 GUS staining pattern in wild type is not under negative regulation by LUG.
The GUS staining pattern of pMD970 in lug mutants is shown in Figures 4J and 4K. At early stages of development, GUS staining was restricted to the core of the floral meristem (Figure 4J), which matched the pMD970 GUS staining pattern in wild type (Figure 2J) but not the AG RNA pattern in lug mutants. However, at slightly later stages, when pMD970 in the wild type would be conferring GUS staining to developing stamens, we observed GUS staining in outer whorls of lug flowers (Figure 4K). This outer-whorl pattern matched that of AG RNA in lug mutants (Liu and Meyerowitz, 1995). Taken together, these results suggest that expression from pMD970 is under negative regulation by LUG during floral stages 5 and beyond but that the floral meristem expression pattern at stages 3 to 4 is independent of LUG regulation.
We compared the AG sequences contained in these three constructs (Figure 3). Because we observed the expanded pattern of GUS staining in pMD983 (R2 to R3 to R4) and pMD970 (R1 to R2) lug flowers, negative regulation by LUG probably requires sequences within R2. However, we are unable to confirm this because R2 alone does not confer floral GUS staining (pMD972). Alternatively, LUG could use multiple dispersed elements, such that sufficient elements are contained only in specific combinations of intron fragments.
CArG Boxes, CCG Boxes, and LFY Binding Sequences
To link the AG enhancer–containing regions we had identified with known flower-related transcription factors, we analyzed AG genomic sequences between the upstream PstI site and the start of exon 3. We searched for three different sequence motifs. First, we analyzed this region for sequences that serve as DNA binding sites for the products of MADS box genes. MADS box gene products dimerize and bind to a conserved motif called the CArG box (Schwarz-Sommer et al., 1992; H. Huang et al., 1993; Shiraishi et al., 1993; J. Huang et al., 1996). Looking for at least a nine in 10 match (to the consensus sequence CC[A/T]6GG), we identified four potential CArG boxes (Table 1). Second, to identify possible AP2 binding sites, we searched for the GCC box motif (AGCCGCC)—the consensus binding sequence for the Ethylene Response Factor family (Fujimoto et al., 2000), of which AP2 is a member (Weigel, 1995). We found no sequences in which at least six of the nucleotides matched this seven-member sequence. Third, we searched for the consensus LFY binding sequence (CCANTG) (Parcy et al., 1998). Previously, two LFY binding sites had been identified within R4 (Busch et al., 1999). In addition to those, we found two more sequence matches (Table 1 and Figure 4). The possible relevance of these CArG box and LFY binding sequences to our observations of AG DNA fragments that function to regulate AG expression are considered below.
Table 1.
CArG Boxes and LFY Binding Sequences
| Positiona | Sequence Motif | Sequence | |
|---|---|---|---|
| 383 | U | CArG box | CCTTAATAAG |
| 1907 | R1 | CArG box | CCGATTTAGG |
| 2594 | R1 | LFY binding site | CCATTG |
| 3750 | R2 | LFY binding site | CCAATG |
| 4231 | R4 | CArG box | CCAAATAAGG |
| 4304b | R4 | LFY binding site | CCAATG |
| 4351c | R4 | LFY binding site | CCAATG |
| 4673 | R4 | CArG box | GCTTTTAAGG |
Positions relative to upstream PstI site is 1; U indicates upstream; and intron regions R1 to R4 are shown in Figure 3.
Same as AG I wild type described by Busch et al. (1999).
Same as AG II wild type described by Busch et al. (1999).
DISCUSSION
AG Intron Functions as an Enhancer Element
The location of the AG control region within a large intron raised the possibility that it functioned either as a transcriptional enhancer or as a post-transcriptional determinant of RNA abundance. We found that intron-derived fragments could confer the normal AG expression pattern to a reporter gene when placed upstream of a heterologous promoter (minimal −60 35S) regardless of orientation (pMD992 and pMD993; Figure 1). Because positioning these sequences upstream of the transcriptional start site removed the possibility of post-transcriptional control, these results indicate that the cis elements residing in the intron function transcriptionally, and because they function independently of orientation and position, their function matches the formal definition of an enhancer element. These results are in agreement with observations from another study of AG activation published recently on KB9F and KB9R (Busch et al., 1999).
Functions of the 3′ Activation Domain (R4)
Our analyses allowed us to define two regions of the AG second intron capable of activating expression in a stage 3 floral meristem (5′ and 3′ activation domains); again, our observations generally agree with those published recently by Busch et al. (1999). The Busch study identified two LFY binding sites within the 3′ activation domain (contained within R4) and showed that deletions of those binding sites led to a strong reduction in activation. We found that, in addition, two CArG boxes are present within this 3′ activation domain (R4) (Table 1 and Figure 3). Although we do not know whether these putative CArG boxes are required for AG expression, dimerized MADS box gene products could bind here, which could contribute to activation of AG expression. If so, that might account for the residual GUS activity observed in floral meristems of KB46 and MX68, in which the LFY binding sites of the inserted R4::GUS reporter gene had been either deleted or mutated (Busch et al., 1999). Candidate MADS box genes for interaction with these CArG boxes include AGL2, AGL4, AGL9, and AGL8 (Mandel and Yanofsky, 1995, 1997; Savidge et al., 1995).
As flower development progressed, this same region (R4) conferred expression that was largely confined to developing carpels but was also weakly and transiently present in developing stamens. One possible explanation for the stamen expression patterns is that residual GUS enzyme activity remains in cells recruited from the floral meristem (1.5 days between when stamen primordia arise and the beginning of stage 7; Smyth et al., 1990). Although a long half-life for GUS is known anecdotally, in this context it may not be so stable, given that GUS enhancer trap experiments have identified lines with floral GUS staining that disappears after stage 5 (Campisi et al., 1999). More likely, the dynamic stamen staining pattern indicates that constructs containing R4 confer activation of expression in stamen primordia and that maintenance requires sequences residing elsewhere. Separate regulatory mechanisms for activation and maintenance of the floral organ identity genes PISTILLATA (PI) and APETALA3 (AP3) have been observed (Schwarz-Sommer et al., 1992; Goto and Meyerowitz, 1994; Jack et al., 1994; Zachgo et al., 1995; Krizek and Meyerowitz, 1996).
Functions of the 5′ Activation Domain (R1 + R2)
The 5′ activation domain was contained by two constructs in our study (pMD970 and pMD994) and by three constructs in the Busch et al. (1999) study (KB11, KB13, and KB14). We identified two sequences matching the LFY binding domain within this region (Table 1 and Figure 4). However, these sequences are unlikely to be sufficient for activation, because the AG DNA in construct pMD996 contained both LFY binding sequences but did not activate expression within the floral meristem (Figure 1). We also identified one CArG box in R1. In addition to LFY, perhaps molecules interacting with the CArG domain are also required for activation.
Although both this study and that of Busch et al. (1999) observed activation with this 5′ region (R1 + R2), we observed different early GUS staining patterns. For both pMD970 and pMD994, we detected GUS staining only in a small internal zone of the floral meristem of stage 3 flowers, whereas Busch et al. (1999) reported a typical AG stage 3 pattern for KB14. KB14 and pMD994 contained identical AG DNA but differed in their minimal promoters. (Both studies used minimal promoters derived from the CaMV 35S promoter, but Busch et al. used the −46 derivative, whereas we used the −60 derivative.) Perhaps, the 14 bp contained in the −60 minimal promoter, and absent from the −46 promoter, supplied a repressor function. To assess possible repressor binding, we used the 14-bp sequence to search for similarities to other transcription factor binding sequences. Two proteins have been reported to bind to sequences contained by these 14 bp: the potato MybSt1 gene product and the Activating Sequence Factor 2 (ASF-2) of tobacco (Lam and Chua, 1989; Baranowskij et al., 1994). Although we do not know the activity of putative Arabidopsis homologs, we consider it unlikely that in this instance they would reduce expression. Another possible explanation for the different expression patterns in floral primordia is that the genomic insertion sites for pMD970 and pMD994 transgenes were in low-expressing regions of the genome; had the construct been inserted into a high-expressing region, the pattern presumably would have more closely resembled the normal AG pattern. However, we observed the same expression pattern in all 20 independently isolated lines, and that all would have been inserted into low-expression regions of the genome seems unlikely. Moreover, Busch et al. (1999) reported that most lines containing the 3′ enhancer also showed low expression, yet those authors observed the normal AG pattern. Thus, the reason for the difference in early expression patterns remains unexplained.
Later in development, plants containing the 5′ enhancer (R1 + R2) conferred strong GUS staining in stamens but no GUS staining in developing carpels. Because AP3, PI, and AG are all required for stamen development, possibly the stamen expression observed in experiments using the pMD970 and pMD994 constructs is derived from AP3/PI heterodimer binding to this region of the AG intron. Even if true, however, this interaction would not be sufficient for activation, because constructs that contained the CArG box alone (pMD971) did not confer floral expression.
Negative Regulation
Our first clue that negative regulation might not be a straightforward story was our failure to obtain ectopic staining of outer whorls from any deletion derivative expressed in wild-type plants. Instead, deletion derivatives that conferred floral staining either matched the normal AG pattern or conferred the abbreviated patterns described above: predominant staining in either only the third or only the fourth whorl. The second indication that negative regulation is complicated came from our analysis of GUS staining in pMD970 and pMD995 flowers in plants homozygous for either ap2 or lug mutations. We found that AP2 negatively regulated expression from the 3′ activating region (R4, contained by pMD995), whereas LUG negatively regulated expression from the 5′ activating region (R1 + R2, contained by pMD970). The third clue was that we were unable to identify any GCC boxes, with which ethylene responsive–element binding factors (such as AP2) interact. This result indicates either that AP2 has a different DNA binding specificity or, more likely, that negative regulation by AP2 is indirect.
Our conclusion that AP2 does not negatively regulate expression of constructs containing the 5′ activating region is in contrast with the findings of another recent study (Bomblies et al., 1999), in which constructs containing the 5′ enhancer (KB14) were identified as responding to the loss of AP2 as visualized by ectopic GUS staining. There are several possible explanations for these differences. As argued for the reduced GUS staining domain for stage 3 flowers of R1 + R2 constructs, if the additional 14 bp contained by the −60 CaMV minimal promoter (cf. with the −46 used in the Bomblies et al. [1999] and the Busch et al. [1999] studies) conferred negative regulation, it could have countered weak ectopic expression. However, other constructs containing the −60 CaMV promoter were able to respond strongly to an ap2 mutant background as shown by GUS staining in the outermost whorl.
Another potentially significant difference is that our two studies used overlapping, but not identical, AG sequences to test the 5′ enhancer (R1 + R2) for negative regulation by AP2. KB14 and pMD970 overlapped by 2183 bp, but our pMD970 contained an additional 481 bp at its 5′ end and lacked 82 bp of 3′ sequences that were contained in KB14. Thus, the cis elements used by AP2 may be contained by KB14 and be missing from pMD970. Because no sequences expected for AP2 binding sites were detected within the AG control region, suggesting indirect negative regulation, we are currently unable to test this possibility. Finally, we note that the GUS staining pattern shown for KB14 in an ap2 mutant appeared to be restricted to the central floral meristem for the stage 3 flower shown (Bomblies et al., 1999). Although this essentially matched their AG in situ hybridization for an ap2-9 flower at a similar stage, it did not match the GUS staining patterns for ap2-9 flowers containing KB9 (like our pMD992), KB18 (like our pMD989), or KB31 (like our pMD995). For these latter three constructs, GUS staining was clearly present in outer-whorl organs of ap2-9 flowers in early stages of development. We think it is possible that AP2 negative regulation of KB14 occurs later than stage 3.
Models for AG Regulation
Our analysis of AG enhancer elements that are sufficient to confer expression in developing flowers indicates that expression is activated independently in the third and fourth whorls. Separable cis elements that can activate expression independently in whorls two and three have been shown for the promoter of the class B gene for floral organ identity, AP3 (Hill et al., 1998; Tilly et al., 1998), but not for the other class B gene, PI (Honma and Goto, 2000). In addition, our study suggests that whorls three and four have separable negative regulation. Although we could identify CArG box and LFY binding sequences within AG DNA fragments conferring floral expression, we still do not know what mechanisms are involved for negative regulation. Does loss of negative regulation allow cryptic positive regulatory sequences to be used, or do negative regulators interact, directly or indirectly, with the positive regulators? Resolution of these questions awaits molecular characterization of LUG and increased biochemical understanding of AP2.
METHODS
Plasmid Construction
All reporter genes were constructed by standard molecular biology techniques. Five of the AG::GUS constructs—pMD200, pMD221, pMD222, pMD227, and pMD228—were derived from the previously described pAG-I::GUS (Sieburth and Meyerowitz, 1997). The remaining constructs were made by using a minimal cauliflower mosaic virus (CaMV) promoter (−60) driving expression of the reporter gene encoding β-glucuronidase (GUS), a generous gift from T. Jack (Tilly et al., 1998). All constructs used the pCGN1547 plant transformation vector (McBride and Summerfelt, 1990). The orientation of fragments inserted upstream of the minimal promoter was determined by using restriction mapping and the polymerase chain reaction.
Plant Transformation and Growth Conditions
Arabidopsis thaliana plants of the ecotype Landsberg erecta were transformed by using the vacuum infiltration method (Bechtold et al., 1993) after placement of the plasmid into the Agrobacterium tumefaciens strain ASE. We recovered transgenic plants by selecting for kanamycin resistance and assessed copy number by monitoring the segregation of resistance to kanamycin. All plants were grown under continuous illumination (∼130 μE) at 20°C.
GUS Staining and Microscopy
Inflorescences and seedlings were stained for GUS activity as described previously (Sieburth and Meyerowitz, 1997). The number of transgenic lines examined for each construct is given in Figure 1. For cellular resolution of GUS staining patterns, we obtained 10-μm-thick sections of Paraplast-embedded tissue, dewaxed them, mounted them in Permount (Fisher), and viewed them by dark-field illumination on an Olympus BX50 (Olympus, Melville, NY). Photomicrographs were digitized, and color balance and contrast were adjusted by using Adobe Photoshop version 4.0 (Adobe Systems, San Jose, CA).
DNA Database Search
AG sequences between 47,000 and 53,000 on Genbank accession AL021711 were compared with CArG and ethylene response factor sequences by using GCG version 10.0 (Genetics Computer Group, Madison, WI). The PstI site that we reference as position 0 corresponds to 47,074 of this bacterial artificial chromosome. Searches for CArG box–related sequences allowed a nine of 10 or 10 of 10 match to the CC(A/T)6GG consensus sequence. Searches for ethylene response factor sequences sought a match of six of seven or seven of seven to the consensus sequence of AGCCGCC. No sequences matching this criterion were identified. Searches for LFY binding sites required a match to the consensus CCANTG. Searches for possible repressor binding sites in the 5′-most 14 bp of the −60 CaMV promoter used the PLACE database.
Acknowledgments
We thank Tom Jack for the generous gift of the plasmid pD991 and Candace Waddell, Gary Drews, and Jack Okamuro for thoughtful discussions during the course of this work. In addition, we thank Sheryl Bisgrove, Whitney Hable, and Corey Christensen for critical reading of the manuscript and Betles Yowcel for assistance at the end of this project. Funds supporting this work came from National Sciences and Engineering Research Council of Canada (NSERC) (to L.E.S.), an NSERC PGSB fellowship awarded to M.K.D., and a National Science Foundation grant (No. 998276) to L.E.S.
References
- Baranowskij, N., Frohberg, C., Prat, S., and Willmitzer, L. (1994). A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J. 13, 5383–5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis plants. C. R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Benfey, P.N., and Chua, N.-H. (1990). The cauliflower mosaic virus 35S promoter: Combinatorial regulation of transcription in plants. Science 250, 959–966. [DOI] [PubMed] [Google Scholar]
- Bomblies, K., Dagenais, N., and Weigel, D. (1999). Redundant enhancers mediate transcriptional repression of AGAMOUS by APETALA2. Dev. Biol. 216, 260–264. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1991. a). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Drews, G.N., and Meyerowitz, E.M. (1991. b). Expression of the Arabidopsis floral homeotic gene agamous is restricted to specific cell types late in flower development. Plant Cell 3, 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch, M.A., Bomblies, K., and Weigel, D. (1999). Activation of a floral homeotic gene in Arabidopsis. Science 285, 585–587. [DOI] [PubMed] [Google Scholar]
- Cai, H.N., Arnosti, D.N., and Levine, M. (1996). Long-range repression in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 93, 9309–9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi, L., Yang, Y., Yi, Y., Heilig, E., Herman, B., Cassista, A.J., Allen, D.W., Xiang, H., and Jack, T. (1999). Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 17, 699–707. [DOI] [PubMed] [Google Scholar]
- Drews, G.N., Bowman, J.L., and Meyerowitz, E.M. (1991). Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65, 991–1002. [DOI] [PubMed] [Google Scholar]
- Fujimoto, S.Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box–mediated gene expression. Plant Cell 12, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, J., Puangsomlee, P., Martin, M., Long, D., Meyerowitz, E.M., and Coupland, G. (1997). A polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386, 44–51. [DOI] [PubMed] [Google Scholar]
- Goto, K., and Meyerowitz, E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548–1560. [DOI] [PubMed] [Google Scholar]
- Hanna-Rose, W., and Hansen, U. (1996). Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 12, 229–234. [DOI] [PubMed] [Google Scholar]
- Hill, T.A., Day, C.D., Zondlo, S.C., Thackeray, A.G., and Irish, V.F. (1998). Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development 125, 1711–1721. [DOI] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2000). The Arabidopsis floral homeotic gene PISTILLATA is regulated by discrete cis-elements responsive to induction and maintenance signals. Development 127, 2021–2030. [DOI] [PubMed] [Google Scholar]
- Huang, H., Mizukami, Y., and Ma, H. (1993). Isolation and characterization of the binding sequence for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 21, 4769–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J., Tudor, M., Su, T., Zhang, Y., Hu, Y., and Ma, H. (1996). DNA binding properties of two Arabidopsis MADS domain proteins: Binding consensus and dimer formation. Plant Cell 8, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish, V. (1999). Patterning the flower. Dev. Biol. 209, 211–220. [DOI] [PubMed] [Google Scholar]
- Jack, T., Fox, G.L., and Meyerowitz, E.M. (1994). Arabidopsis homeotic gene APETALA3 ectopic expression: Transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76, 703–716. [DOI] [PubMed] [Google Scholar]
- Jack, T., Sieburth, L.E., and Meyerowitz, E.M. (1997). Targeted misexpression of AGAMOUS in whorl 2 of Arabidopsis flowers. Plant J. 11, 825–839. [DOI] [PubMed] [Google Scholar]
- Jofuku, K.D., den Boer, B.G.W., Van Montagu, M., and Okamuro, J.T. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6, 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek, B.A., and Meyerowitz, E.M. (1996). The Arabidopsis homeotic genes apetala3 and pistillata are sufficient to provide the B class organ identity function. Development 122, 11–22. [DOI] [PubMed] [Google Scholar]
- Lam, E., and Chua, N.H. (1989). ASF-2: A factor that binds to the cauliflower mosaic virus 35S promoter and a conserved GATA motif in cab promoters. Plant Cell 1, 1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., and Meyerowitz, E.M. (1995). LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121, 975–991. [DOI] [PubMed] [Google Scholar]
- Mandel, A.M., and Yanofsky, M.F. (1995). The Arabidopsis AGL8 MADS-box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1. Plant Cell 7, 1763–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel, A.M., and Yanofsky, M.F. (1997). The Arabidopsis AGL9 MADS-box gene is expressed in young flower primordia. Sex. Plant Reprod. 11, 22–28. [Google Scholar]
- Mandel, M.A., Bowman, J.L., Kempin, S.A., Ma, H., Meyerowitz, E.M., and Yanofsky, M.F. (1992). Manipulation of flower structure in transgenic tobacco. Cell 71, 133–134. [DOI] [PubMed] [Google Scholar]
- McBride, K.E., and Summerfelt, K.R. (1990). Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 14, 269–276. [DOI] [PubMed] [Google Scholar]
- Mizukami, Y., and Ma, H. (1992). Ectopic expression of the floral homeotic gene agamous in transgenic Arabidopsis plants alters floral organ identity. Cell 71, 119–131. [DOI] [PubMed] [Google Scholar]
- Ng, M., and Yanofsky, M. (2000). Three ways to learn the ABCs. Curr. Opin. Plant Biol. 3, 47–52. [DOI] [PubMed] [Google Scholar]
- Parcy, F., Nilsson, O., Busch, M.A., Lee, I., and Weigel, D. (1998). A genetic framework for floral patterning. Nature 395, 561–566. [DOI] [PubMed] [Google Scholar]
- Savidge, B., Rounsley, S.D., and Yanofsky, M.F. (1995). Temporal relationship between the transcription of two Arabidopsis MADS-box genes and the organ identity genes. Plant Cell 7, 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer, Z., Hue, I., Huijser, P., Flor, P.J., Hansen, R., Tetens, F., Lonnig, W.-E., Saedler, H., and Sommer, H. (1992). Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: Evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 11, 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi, H., Okada, K., and Shimura, Y. (1993). Nucleotide sequences recognized by the AGAMOUS MADS domain of Arabidopsis thaliana in vitro. Plant J. 4, 385–398. [DOI] [PubMed] [Google Scholar]
- Sieburth, L.E., and Meyerowitz, E.M. (1997). Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9, 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly, J.J., Allen, D.W., and Jack, T. (1998). The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 125, 1647–1657. [DOI] [PubMed] [Google Scholar]
- Weigel, D. (1995). The APETALA2 domain is related to a novel type of DNA binding domain. Plant Cell 7, 388–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., and Meyerowitz, E. (1993). Activation of floral homeotic genes in Arabidopsis. Science 261, 1723–1726. [DOI] [PubMed] [Google Scholar]
- Weigel, D., Alvarez, J., Smyth, D.R., Yanofsky, M.F., and Meyerowitz, E.M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. [DOI] [PubMed] [Google Scholar]
- Zachgo, S., de Andrade Silva, E., Motte, P., Tröbner, W., Saedler, H., and Schwarz-Sommer, Z. (1995). Functional analysis of the Antirrhinum floral homeotic DEFICIENS gene in vivo and in vitro by using a temperature-sensitive mutant. Development 121, 2861–2875. [DOI] [PubMed] [Google Scholar]