Abstract
We have established an Arabidopsis protoplast model system to study plant cell death signaling. The fungal toxin fumonisin B1 (FB1) induces apoptosis-like programmed cell death (PCD) in wild-type protoplasts. FB1, however, only marginally affects the viability of protoplasts isolated from transgenic NahG plants, in which salicylic acid (SA) is metabolically degraded; from pad4-1 mutant plants, in which an SA amplification mechanism is thought to be impaired; or from jar1-1 or etr1-1 mutant plants, which are insensitive to jasmonate (JA) or ethylene (ET), respectively. FB1 susceptibility of wild-type protoplasts decreases in the dark, as does the cellular content of phenylalanine ammonia-lyase, a light-inducible enzyme involved in SA biosynthesis. Interestingly, however, FB1-induced PCD does not require the SA signal transmitter NPR1, given that npr1-1 protoplasts display wild-type FB1 susceptibility. Arabidopsis cpr1-1, cpr6-1, and acd2-2 protoplasts, in which the SA signaling pathway is constitutively activated, exhibit increased susceptibility to FB1. The cpr6-1 and acd2-2 mutants also constitutively express the JA and ET signaling pathways, but only the acd2-2 protoplasts undergo PCD in the absence of FB1. These results demonstrate that FB1 killing of Arabidopsis is light dependent and requires SA-, JA-, and ET-mediated signaling pathways as well as one or more unidentified factors activated by FB1 and the acd2-2 mutation.
INTRODUCTION
Plant cell death is often the consequence of plant–pathogen interactions in both compatible and incompatible relationships (Greenberg, 1997). A notable example is localized cell collapse, called the hypersensitive response (HR), which is induced rapidly in a resistant plant at the infection site of an avirulent pathogen (Staskawicz et al., 1995; Bent, 1996; Dangl et al., 1996; Hammond-Kosack and Jones, 1996). Hypersensitive cell death, which is distinct from necrosis caused by metabolic toxins or severe trauma, is genetically programmed (programmed cell death [PCD]) and requires active host cell metabolism (Mittler and Lam, 1996; Morel and Dangl, 1997; Pennell and Lamb, 1997; Gilchrist, 1998; Gray and Johal, 1998; Heath, 1998; Richberg et al., 1998). In fact, plant cells undergoing the HR display several molecular and morphological markers characteristic of animal apoptosis (a specialized form of PCD), including systematic DNA degradation and formation of apoptotic-like bodies, which suggests that the terminal steps in PCD are well conserved in animals and plants (Levine et al., 1996; Mittler et al., 1996; Ryerson and Heath, 1996; Wang et al., 1996). It remains to be determined, however, whether the signal transduction mechanisms leading to the onset of PCD are also equally conserved between the two kingdoms.
Salicylic acid (SA) is the best-characterized signaling molecule in plant defense responses (Ryals et al., 1996; Delaney, 1997; Durner et al., 1997). Application of exogenous SA or SA analogs activates the expression of a variety of pathogenesis-related (PR) genes and enhances resistance to a variety of pathogens (White, 1979; Ward et al., 1991; Uknes et al., 1992). Conversely, transgenic plants expressing the bacterial nahG gene, which encodes an enzyme that degrades SA to catechol, display reduced or no expression of PR genes and fail to establish systemic acquired resistance (SAR; Gaffney et al., 1993; Delaney et al., 1994). Phenylalanine ammonia-lyase (PAL) is a key enzyme in the synthesis of SA (Bate et al., 1994; Mauch-Mani and Slusarenko, 1996; Coquoz et al., 1998; Smith-Becker et al., 1998), and genes encoding PAL are induced by a variety of pathogens and pathogen-derived elicitors (Lois et al., 1989; Wanner et al., 1995, and the references therein).
An important advance in our understanding of SA-mediating signaling response pathways was the isolation of Arabidopsis npr1 (nim1, sai1) mutants, which fail to express PR genes or fail to induce SAR in response to exogenously supplied SA or its analogs (Cao et al., 1994; Delaney et al., 1995; Shah et al., 1997). The recessive nature of these mutations suggests that NPR1 is a positive regulator that transmits SA signals to downstream components in a signaling pathway. In addition to NPR1, at least one other defense-related gene, PAD4, is directly involved in SA signaling. pad4 (phytoalexin deficient) mutants, which were isolated by screening for enhanced susceptibility to virulent strains of Pseudomonas syringae or Erysiphe orontii (Glazebrook et al., 1996; Jirage et al., 1999), exhibit decreases in PR-1 expression and SA accumulation after pathogen infection (Reuber et al., 1998; Zhou et al., 1998). Because the pad4 mutant shows no defects in defense responses when infected with avirulent pathogens that elicit a strong HR, PAD4 is thought to amplify weak SA signals, such as those resulting from infection by a virulent pathogen, to higher amounts sufficient for activation of SA signaling (Zhou et al., 1998; Jirage et al., 1999). In contrast to the npr1 and pad4 mutants, a different group of defense-related mutants, including cpr1 (for constitutive expresser of PR genes; Bowling et al., 1994), cpr6 (Clarke et al., 1998), and acd2 (for accelerated cell death; Greenberg et al., 1994), accumulates greater amounts of SA constitutively and expresses SAR and PR genes continuously. As a consequence, these mutants exhibit enhanced resistance to various pathogens.
Recent evidence shows that in addition to SA, jasmonate (JA) and ethylene (ET) also play important roles as signal molecules mediating disease resistance, especially in response to necrotrophic fungal pathogens (Penninckx et al., 1996; Dong, 1998; Reymond and Farmer, 1998; Thomma et al., 1998; Glazebrook, 1999; Lam et al., 1999; Pieterse and van Loon, 1999). Interestingly, most of the JA/ET-dependent defense responses analyzed to date are SA independent. For instance, the PDF1.2 gene encoding an antifungal plant defensin is induced concomitantly by the JA and ET signaling pathways (Penninckx et al., 1998), and the induction is enhanced in transgenic NahG plants (Penninckx et al., 1996). Induced systemic resistance (ISR) in Arabidopsis, triggered by the biocontrol bacterium P. fluorescens in association with Arabidopsis roots, also depends on both JA and ET but is independent of SA (Pieterse and van Loon, 1999). Interestingly, although ISR is SA independent, it is dependent on NPR1, indicating that the JA/ET- and SA-dependent resistance pathways share at least one key regulatory component (Pieterse et al., 1998; Pieterse and van Loon, 1999). Additional evidence for interaction of the SA and JA/ET pathways is that the cpr6 and acd2 mutations cause constitutive expression of the JA/ET-responsive PDF1.2 gene as well as the SA-responsive PR genes (Penninckx et al., 1996; Clarke et al., 1998).
Useful model systems for the study of plant cell death in pathogen response pathways use PCD-eliciting mycotoxins such as AAL toxin or fumonisin B1 (FB1; Gilchrist et al., 1995; Gilchrist, 1997, 1998). AAL toxin is secreted by the tomato pathogen Alternaria alternata f sp lycopersici, and the structurally related toxin FB1 is produced by the maize pathogen Fusarium moniliforme. Gilchrist and co-workers have shown that these toxins induce apoptosislike cell death in tomato leaflets and protoplasts and that the cell death in the leaflets induced by AAL toxin is suppressed by inhibitors of ET biosynthesis or its action (Moussatos et al., 1994; Wang et al., 1996; Moore et al., 1999). They have further demonstrated that leaflets of the tomato never ripe mutant, which is deficient in ET perception, display fewer lesions in response to these toxins (Moore et al., 1999). These results indicate that an ET-dependent pathway is involved in the tomato cell death signaling triggered by AAL toxin or FB1. Using FB1, we have developed a pathogen-free model system to study disease response pathways in Arabidopsis plants (Stone et al., 2000). Leaves infiltrated with FB1 or seedlings transferred to FB1-containing media develop HR-like lesions and express PR genes.
In this article, we describe an Arabidopsis protoplast model for the analysis of defense-related cell death in Arabidopsis. We demonstrate that FB1 induces light-dependent PCD in Arabidopsis protoplasts and that this PCD requires SA, JA, and ET as well as one or more unknown factors activated by FB1 and the acd2 mutation.
RESULTS
Induction of Apoptosis-like Cell Death in Arabidopsis Protoplasts
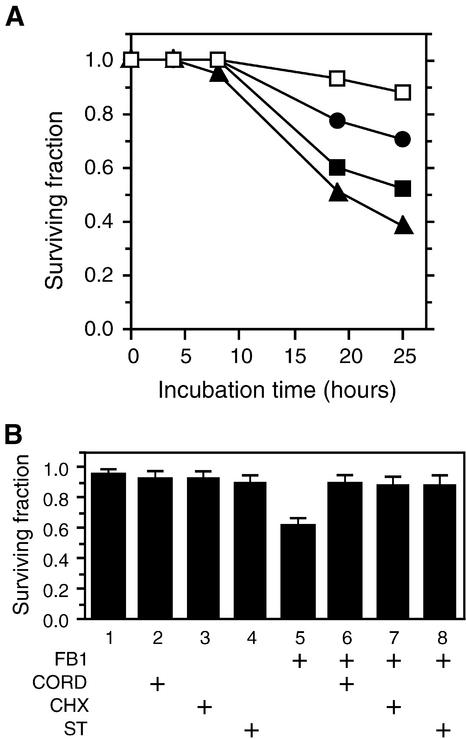
Protoplasts isolated from Arabidopsis leaves were treated with different concentrations of FB1 and analyzed for viability after various incubation times by using an Evans blue staining assay. Evans blue dye is excluded from viable cells and early stage apoptotic cells that retain intact plasma membranes (Gaff and Okong'O-Ogola, 1971; Turner and Novacky, 1974; O'Brien et al., 1997). As shown in Figure 1A, although a little cell death was observed in the absence of FB1, the toxin reduced protoplast viability in a dose-dependent manner. In the following experiments, protoplasts were treated with 70 nM FB1 for 16 hr before Evans blue staining. These conditions were chosen because they consistently reduced protoplast viability by 30 to 40% and because this proportion of cell death permitted detection of both increases and decreases in death caused by various treatments and mutations analyzed in this work.
Figure 1.
FB1-Induced PCD in Arabidopsis Protoplasts.
(A) Time and dose dependency of FB1-induced cell death. Protoplasts were prepared from wild-type plants and incubated in the light with various concentrations of FB1 (open squares, 0 nM; filled circles, 0.7 nM; filled squares, 70 nM; and filled triangles, 7 μM). Samples were withdrawn at 4, 8, 19, and 25 hr after the addition of FB1, and surviving fractions were determined as described in Methods. For control protoplasts (0 nM FB1), methanol was added instead of the FB1 solution at the final concentration of 0.1%. The methanol did not affect protoplast viability (not shown). The largest standard error in this experiment was 0.05.
(B) Requirement of plant cell activities for FB1-induced cell death. Protoplasts isolated from wild-type plants were incubated in the light for 16 hr in the presence of 70 nM FB1 (+FB1) or various inhibitors (100 μM cordycepin [+CORD], 10 μM cycloheximide [+CHX], and 1 μM staurosporine [+ST]), alone or in combination. In experiments 1 to 5, methanol was added instead of FB1 or inhibitor solutions. The final concentrations of methanol were as follows: experiment 1, 0.101%; experiments 2 to 4, 0.001%; and experiment 5, 0.1%. Surviving fractions were determined as described above. Error bars indicate sd.
FB1 killing was efficiently suppressed by cordycepin and cycloheximide, inhibitors of transcription and translation, respectively, and by staurosporine, a broad-spectrum inhibitor of protein kinases (Figure 1B). These results suggest that FB1 does not induce necrosis in Arabidopsis protoplasts. Rather, FB1 most likely elicits cell death by activating a controlled program that requires active plant metabolism. As with penicillin, however, FB1 may require active host functions to express its killing effect (discussed in Greenberg et al., 1994). In the following sections, we show that protoplasts isolated from certain defense-related mutants are as insensitive to FB1 as protoplasts treated with the above inhibitors. Unlike the inhibitor-treated protoplasts, the mutant protoplasts would not be expected to exhibit the global shutoff of cellular metabolism. Thus, we conclude that FB1 triggers PCD in Arabidopsis protoplasts.
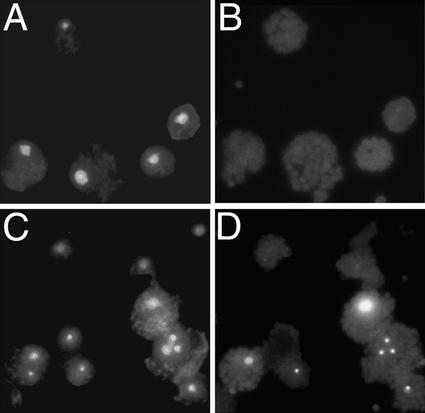
One hallmark of apoptosis is the fragmentation of nuclear DNA with disintegration of the nucleus and the subsequent formation of distinct apoptotic bodies containing the fragmented DNA (Martin et al., 1994). Typically, the TUNEL method, which labels free 3′-OH groups of DNA, is used to detect fragmented DNA and visualize apoptotic bodies in situ (Gorczyca et al., 1993). We applied this method to Arabidopsis protoplasts treated with FB1. As shown in Figure 2A, control protoplasts incubated for 6 hr without FB1 contained a single nucleus in each cell, as visualized by nonspecific DNA staining with Hoechst 33342 (Wang et al., 1996). Specific labeling of fragmented DNA, however, was undetectable in the control protoplasts (Figure 2B). In contrast, protoplasts treated with 70 nM FB1 contained nuclei of various sizes and shapes, frequently accompanied by multiple distinct bodies containing DNA (Figure 2C). Furthermore, application of the TUNEL method to the same FB1-treated protoplasts clearly detected free 3′-OH groups of DNA within the nuclei and the distinct bodies (Figure 2D). The TUNEL assays described here were conducted 6 hr after FB1 treatment. Because FB1-elicited Evans blue staining did not occur until later (Figure 1A), nuclear DNA fragmentation apparently precedes the loss of membrane integrity, suggesting that the observed DNA fragmentation was caused by apoptotic rather than necrotic cell death (Martin et al., 1994; O'Brien et al., 1997). FB1-treated Arabidopsis leaves displaying systemic FB1-induced lesions (Stone et al., 2000) also showed TUNEL-positive cells (data not shown). Similar results were reported by Wang et al. (1996) after treating tomato leaves and protoplasts with an FB1 analog, AAL toxin. Although the presence of 3′-OH groups should not be used as a sole criterion for the nonrandom DNA fragmentation characteristic of apoptosis (Collins et al., 1992), these results are consistent with the conclusion that FB1 induces PCD in Arabidopsis protoplasts.
Figure 2.
Detection of Fragmented DNA in FB1-Treated Protoplasts.
Protoplasts were prepared from wild-type plants and incubated for 6 hr in the light in the presence of 0.001% methanol or 70 nM FB1. DNA molecules in the protoplasts then were stained with Hoechst 33342, and free 3′-OH groups in the molecules were labeled by the TUNEL technique as described in Methods.
(A) Hoechst 33342 staining of methanol-treated protoplasts.
(B) TUNEL staining of methanol-treated protoplasts.
(C) Hoechst 33342 staining of FB1-treated protoplasts.
(D) TUNEL staining of FB1-treated protoplasts.
As shown in Figure 1A, ∼40% of the protoplasts maintained membrane integrity, even after a 25-hr incubation with 7 μM FB1; the addition of greater concentrations of FB1 did not markedly reduce the survival rate (data not shown). Although FB1-elicited killing could not be correlated directly with nuclear fragmentation (because the TUNEL assay was performed several hours before the protoplasts became Evans blue positive), we think it likely that ∼40% of the cells may be unable to respond to FB1 or FB1-induced signals. In tomato, fully expanded leaves produce fewer lesions in response to AAL toxin than do rapidly expanding leaves (Moussatos et al., 1993a, 1993b). Thus, Arabidopsis protoplasts derived from older leaves may be less sensitive to FB1 than are protoplasts from younger leaves.
Mutational Analysis of Cell Death Signaling
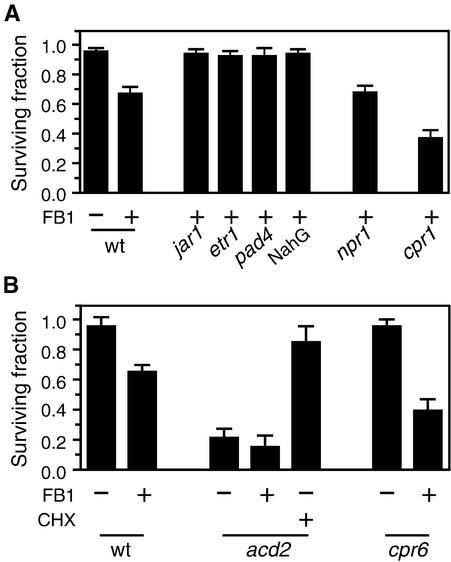
To determine whether JA-, ET-, or SA-dependent signaling pathways are involved in FB1-induced cell death, we used the JA-insensitive mutant jar1-1 (JA-resistant; Staswick et al., 1992), the ET-insensitive mutant etr1-1 (ET-resistant; Bleecker et al., 1988), the SA-depleted mutant pad4-1, and the SA-depleted transgenic plant NahG. As shown in Figure 3A, protoplasts isolated from all four of these plants exhibited much less susceptibility to FB1, suggesting that all three signaling pathways are required for cell death. In contrast, npr1-1 protoplasts were just as susceptible as wild-type protoplasts to FB1 (Figure 3A), even though NPR1 is required for both SAR and ISR, which are SA-dependent and JA/ET-dependent defense responses, respectively. These results suggest that FB1-elicited cell death requires SA and JA/ET signaling components upstream of NPR1 and are consistent with the observation that the npr1-1 mutant develops the HR normally when infected with avirulent bacterial pathogens (Cao et al., 1994). Similar to FB1-elicited cell death, the constitutive expression of PR genes conferred by the cpr6-1 and ssi1 (for suppressor of SA insensitivity) mutations and the induction of PDF1.2 by Alternaria brassicicola are also NPR1 independent, although they require the SA and JA/ET signaling pathways, respectively (Penninckx et al., 1996; Clarke et al., 1998; Shah et al., 1999).
Figure 3.
Mutational Analysis of PCD.
(A) Effects of various defense-related mutations on FB1-induced cell death. Protoplasts isolated from the indicated plants were incubated for 16 hr in the light, and surviving fractions were determined as described in Methods. FB1 was added at the final concentration of 70 nM (+FB1). To the control experiment (wild type [wt], −FB1), methanol was added instead of FB1 at the final concentration of 0.001%. In the presence of methanol alone, surviving fractions of these mutant protoplasts were similar to that of wild-type protoplasts (not shown).
(B) Effects of the acd2-2 and cpr6-1 mutations on protoplast viability. Protoplasts were isolated from the indicated plants and incubated for 16 hr in the light in the presence of 70 nM FB1 (+FB1) or 0.001% methanol (−FB1). Cycloheximide was added at the final concentration of 10 μM (+CHX). Surviving fractions were determined as described in Methods.
Error bars indicate sd.
The cpr1-1 mutant accumulates high amounts of SA constitutively (Bowling et al., 1994). As shown in Figure 3A, FB1-induced cell death of cpr1-1 protoplasts was substantially greater than with wild-type protoplasts. When treated with 70 nM FB1, only 40% of the cpr1-1 protoplasts survived, whereas 65% of the wild type survived. As described above, ∼40% of the total protoplasts are not killed even with higher concentrations of FB1 and may be FB1 insensitive. Thus, one interpretation of the result obtained with cpr1-1 protoplasts is that although SA, JA, and ET are all required for FB1-induced PCD, SA may be the limiting factor. This idea is compatible with the observation that in soybean cell suspensions, addition of exogenous SA greatly potentiates hypersensitive cell death triggered by a pathogen (Shirasu et al., 1997; Tenhaken and Rübel, 1997). In our protoplast system, however, exogenous SA did not enhance FB1-induced cell death (T. Asai and F.M. Ausubel, unpublished data), suggesting that exogenous SA may not always increase cellular SA concentrations.
Several lines of evidence suggest that the JA-, ET-, and SA-dependent signaling pathways necessary for FB1-induced cell death are constitutively activated in the cpr6-1 and acd2-2 mutants (Penninckx et al., 1996; Clarke et al., 1998). In addition, similar to cpr1-1, SA accumulates constitutively to high concentrations in the cpr6-1 and acd2-2 mutants (Greenberg et al., 1994; Clarke et al., 1998). Therefore, in light of the results described above indicating that JA, ET, and SA are all required for FB1-induced PCD and that SA was most likely the limiting factor, we tested whether FB1 is dispensable for the death of protoplasts isolated from cpr6-1 and acd2-2 leaves. As predicted and as shown in Figure 3B, acd2-2 protoplasts showed only ∼20% of the viability of wild-type protoplasts, even in the absence of FB1. The toxin-independent death of acd2-2 protoplasts is a programmed event because it was effectively suppressed by cycloheximide (Figure 3B). In contrast to acd2-2, however, the viability of cpr6-1 protoplasts was similar to that of wild-type protoplasts in the absence of FB1. These results are consistent with the observation that the acd2-2 mutant, but not the cpr6-1 mutant, spontaneously develops HR-like lesions (Greenberg et al., 1994; Clarke et al., 1998). Enhanced FB1 susceptibility of cpr6-1 protoplasts most probably results from high concentrations of SA in this mutant (see above). We conclude that induction of PCD in Arabidopsis protoplasts requires, in addition to SA, JA, and ET, a cell death factor or factors that are activated by FB1 or the acd2-2 mutation but are missing in cpr6-1 protoplasts (see Discussion).
Because of the toxin-independent cell death of acd2-2 protoplasts, it is not clear whether high amounts of SA in acd2-2 plants also potentiate FB1-induced cell death (Figure 3B). We therefore infiltrated FB1 into acd2-2 plants and monitored the development of lesions. In these experiments, we infiltrated healthy leaves of acd2-2 plants that had developed spontaneous lesions on the lower leaves (Greenberg et al. [1994] has demonstrated that SA contents are increased in such healthy leaves). For a control, we also infiltrated FB1 into wild-type plants. As shown in Figure 4, acd2-2 leaves collapsed after 3 days, whereas wild-type leaves took 6 days to collapse. This accelerated lesion formation was not simply the result of the spontaneous cell death phenotype of the acd2-2 mutant or of wounding (Greenberg et al., 1994). When infiltrated with 0.14% methanol (carrier for the FB1 infiltrate), acd2-2 plants showed little lesion development after 3 days (Figure 4A). Moreover, in contrast to the acd2-2 mutant, SA-depleted NahG plants infiltrated with FB1 produced much less severe lesions than the corresponding wild-type plants (Figure 4B). These observations are consistent with the results obtained with Arabidopsis protoplasts and suggest that SA is an important regulator that determines the extent of cell death in Arabidopsis leaves.
Figure 4.
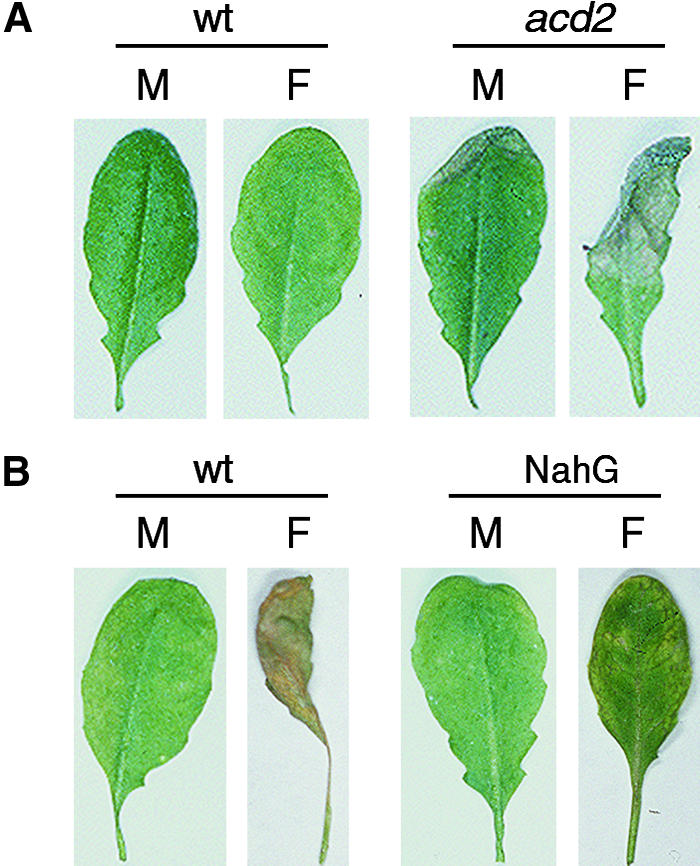
Lesion Formation by FB1 in Arabidopsis Leaves.
Five-week-old plants (wild type [wt], acd2-2, and NahG) were infiltrated with 10 μM FB1 (F) or 0.14% methanol (M) as described by Greenberg et al. (1994) and incubated under a 12-hr light/dark cycle. Leaves were excised 3 or 6 days after infiltration for photographing.
(A) After 3 days.
(B) After 6 days.
Induction of JA-, ET-, and SA-Responsive Genes in FB1-Treated Protoplasts
The results described above suggested that FB1 increases cellular contents of JA, ET, and SA. Direct measurement of cellular concentrations of these molecules, however, is not practical, given the difficulty of obtaining a sufficient number of protoplasts from Arabidopsis leaves. Therefore, as an alternative to direct measurement of JA, ET, and SA concentrations in protoplasts, we monitored the expression of JA-, ET-, and SA-regulated genes instead. The rationale of this approach is that if FB1 increases the cellular concentrations of JA, ET, and SA, genes that are responsive to these molecules should be induced in FB1-treated protoplasts. To obtain a profile of FB1-induced gene expression in protoplasts, we created a subtracted cDNA library to represent mRNA species that are present at relatively high amounts in FB1-treated protoplasts but in relatively low amounts in untreated protoplasts (see Methods). As listed in Table 1, among the ∼200 clones from this library that were sequenced, eight corresponded to genes (or homologs of genes from other species) described in the literature as being induced by SA, JA, or ET. Although it is still necessary to confirm that the genes identified by subtractive hybridization actually are induced by FB1, the data in Table 1 are consistent with the conclusion that FB1 increases cellular concentrations of JA, ET, and SA. Interestingly, several of the FB1-induced genes encode proteins that are expressed during senescence, in which ET plays an important role (Pennell and Lamb, 1997; Gray and Johal, 1998; Johnson and Ecker, 1998). Some of these senescence-related genes induced by FB1 also are listed in Table 1. A description of the genes in the subtracted library that are not known to be SA, JA, or ET inducible will be published separately.
Table 1.
Examples of FB1-Induced Genes and SA, JA, ET, and Senescence Inducibility
| Inducibilitya
|
||||||
|---|---|---|---|---|---|---|
| Gene | Identity/Similarity | SA | JA | ET | Senescence | Reference |
| 2B11 | Glucosyltransferase | + | + | ND | ND | Horvath and Chua (1996) |
| 2E8 | Extensin | + | + | ND | ND | Merkouropoulos et al. (1999) |
| 2E11 | α-Vacuolar processing enzyme | + | − | + | + | Kinoshita et al. (1999) |
| 1C11 | Proteinase inhibitor I | ND | + | − | ND | Heitz et al. (1999) |
| 1G9 | Polyphenol oxidase | ND | + | ND | ND | Constabel et al. (1995) |
| 2H5 | ACC oxidase | ND | ND | + | + | Johnson and Ecker (1998) |
| 5G5 | Senescence-associated gene 21 | ND | ND | + | + | Weaver et al. (1998) |
| 1A5 | Blue copper binding protein | ND | ND | + | + | Weaver et al. (1998) |
| 1A6 | Metallothionein | ND | ND | − | + | Butt et al. (1998) |
| 1H12 | ATP sulfurylase | ND | ND | ND | + | Buchanan-Wollaston (1997) |
| 3E1 | B12D protein | ND | ND | ND | + | To et al. (1999) |
(+), induced; (−), not induced; ND, not determined.
One of the genes that was present in the subtracted library was the Arabidopsis glucosyltransferase gene (Table 1), the homologs of which in tobacco are known to be induced by SA treatment with immediate-early kinetics (Horvath and Chua, 1996). To substantiate the conclusion that the glucosyltransferase gene is induced in protoplasts by FB1, we used a protoplast transient expression system (Kovtun et al., 2000) in which a luciferase (LUC) reporter gene was fused to the glucosyltransferase promoter to monitor the amounts of FB1-induced expression. To normalize the LUC activity for variation in transfection efficiency and cell viability, we cotransfected the protoplasts with a control plasmid carrying the constitutive ubiquitin1 (UBQ1) promoter (Holtorf et al., 1995) fused to the β-glucuronidase (GUS) reporter gene (Kovtun et al., 2000). The relative LUC/GUS activities shown in Figure 5 therefore represent relative promoter activities per viable protoplast. When wild-type protoplasts were incubated with 70 nM FB1 for 16 hr in the light, the promoter activity increased approximately fivefold. In NahG protoplasts, on the other hand, FB1 did not affect the promoter activity. Moreover, the basal promoter activity was reduced in NahG protoplasts. Although various factors could be affecting the expression of the glucosyltransferase promoter in this system, the results are consistent with the idea that induction of the glucosyltransferase promoter is SA dependent and that FB1 increases SA concentrations in wild-type protoplasts.
Figure 5.
Inducibility of an SA-Responsive Promoter by FB1.
Activity of the Arabidopsis glucosyltransferase promoter was determined in wild-type (wt) and NahG protoplasts by using a transient expression system as described in Methods. The protoplasts were incubated for 16 hr in the light or in the dark in the presence of 70 nM FB1 (+FB1) or 0.001% methanol (−FB1). Promoter activities were normalized to the value obtained with wild-type protoplasts incubated in the light without FB1. Error bars indicate sd.
The FB1-mediated accumulation of SA in Arabidopsis protoplasts does not appear to require de novo synthesis of PAL1 or PAL2 mRNAs because the PAL1 and PAL2 promoters fused to the same reporter gene were not induced substantially by FB1 (data not shown). As a control, we showed that the PAL1 and PAL2 promoters were strongly induced in the same assay by a different elicitor, Flg22 (Felix et al., 1999), derived from bacterial flagellin (T. Asai and F.M. Ausubel, unpublished data). At this time, how FB1 increases the intracellular SA concentrations is unclear. FB1 may activate PAL enzymes post-translationally. Or perhaps FB1 indirectly stimulates the hydrolysis of the glucose-conjugated form of SA (SA β-glucoside), increasing the cellular SA concentrations independently of PAL.
Effects of Light on Cell Death
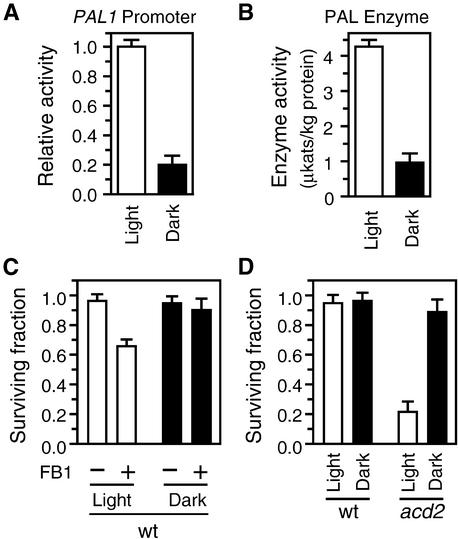
Light has been shown to activate expression of the Arabidopsis PAL genes (Ohl et al., 1990). In fact, as shown in Figure 6A, PAL1 promoter activity decreased approximately fivefold when wild-type protoplasts were incubated in the dark. The decrease in the promoter activity was reflected in a decrease in PAL enzyme activity (Figure 6B). SA concentrations in protoplasts as monitored by the activity of the glucosyltransferase promoter also were notably reduced in the dark, and the FB1 inducibility of the promoter was abolished (Figure 5). Indeed, the activity of the glucosyltransferase promoter in the dark was similar to that obtained in NahG protoplasts. This suggested that incubated in the dark wild-type protoplasts should be less susceptible to FB1, and indeed, this was demonstrated by the experiment shown in Figure 6C. Interestingly, we also found that the toxin-independent death of acd2-2 protoplasts is light dependent (Figure 6D).
Figure 6.
Effects of Light on PAL Expression, PAL Activity, and Cell Death.
(A) PAL1 promoter activity in wild-type protoplasts analyzed with a transient expression system. Protoplasts transformed with a PAL1 promoter–LUC construct were divided into two identical portions; one was incubated in the light and the other in the dark. LUC activities were assayed 16 hr after transformation, and the activity obtained in the dark was normalized to the activity in the light. See Methods for details of this assay system.
(B) PAL enzyme activities in wild-type protoplasts. Protoplasts isolated from wild-type plants were divided into two identical portions; one was incubated in the light and the other in the dark. After 16 hr, samples were taken and the PAL enzyme activity in each sample was determined.
(C) Effects of light on FB1-induced cell death. Protoplasts isolated from wild-type (wt) plants were incubated for 16 hr in the light or dark in the presence of 70 nM FB1 (+FB1) or 0.001% methanol (−FB1). Surviving fractions were determined as described in Methods.
(D) Effects of light on toxin-independent cell death. Protoplasts were isolated from wild-type (wt) and acd2-2 plants and incubated for 16 hr in the light or dark before determination of the surviving fractions.
Error bars indicate sd.
DISCUSSION
In this article, we have shown that the Arabidopsis protoplast system is a useful tool for characterizing plant cell death signaling related to defense responses. By using the pathogen-derived cell death elicitor FB1, we can control the extent of cell death and create a situation in which cell death is semisynchronized. The number of protoplasts undergoing cell death is sufficient for pharmacological and molecular biological analyses. Unlike whole-plant systems, cell death can be quantified in the protoplast system similarly to its quantification in plant suspension cell systems. The advantage of the protoplast system over the suspension cell system is that the role of a variety of Arabidopsis defense-related genes in PCD can be readily assessed by preparing protoplasts from the corresponding mutants.
We propose the model shown in Figure 7 for the signal transduction pathways leading to cell death in Arabidopsis protoplasts. The conclusion that SA mediates FB1-induced PCD is based on the result that NahG protoplasts exhibit markedly less susceptibility to FB1, which is consistent with the observation that the HR against avirulent pathogens observed in wild-type Arabidopsis is impaired in transgenic NahG plants (Delaney et al., 1994). The nahG gene also suppresses spontaneous lesion formation in certain defense-related mutants such as lsd6 and lsd7 (for lesion-simulating disease; Weymann et al., 1995), ssi1 (Shah et al., 1999), and acd6 (Rate et al., 1999). FB1 also appears to elicit an increase in SA concentrations in protoplasts, because it induces the SA-dependent glucosyltransferase promoter. However, because FB1 susceptibility is decreased in pad4 protoplasts, in which a regulatory mechanism that amplifies SA signals is thought to be compromised (Zhou et al., 1998; Jirage et al., 1999), our results are also consistent with the conclusion that FB1 can induce only relatively small amounts of SA accumulation. This latter conclusion is supported by the observation that FB1 susceptibility increases in protoplasts isolated from the cpr1, cpr6, and acd2 mutants, in which SA is overproduced (Bowling et al., 1994; Greenberg et al., 1994; Clarke et al., 1998).
Figure 7.
A Model for Cell Death Signaling in Arabidopsis Protoplasts.
Weak and strong SA signals are denoted by smaller and larger letters, respectively. As alternatives to the light-dependent PAL-mediated generation of SA shown in the model, SA might be produced from SA β-glucoside through a pathway independent of PAL and light, and the primary role of light in PCD may be to generate ROS rather than to activate the transcription of PAL genes. See text for details.
Induction of cell death by FB1 and the acd2 mutation requires light, suggesting that FB1- and acd2-inducible signals are insufficient to activate cell death signaling. The gener-ation of reactive oxygen species (ROS) has been implicated in the HR as well as in lesion formation in the Arabidopsis lsd1 mutant (Jabs et al., 1996; Lam et al., 1999; McDowell and Dangl, 2000). Therefore, one explanation for the observed light requirement for FB1-induced PCD is that ROS generated during photosynthesis are involved, either directly or indirectly, in the cell death process. For example, increased amounts of SA elicited by FB1 and the acd2 mutation may inhibit H2O2-scavenging enzymes such as ascorbate peroxidase and catalase, leading to the enhanced sensitivity of protoplasts to the ROS produced during photosynthesis (Durner et al., 1997). A different, but not mutually exclusive, model for the light requirement of FB1-induced PCD is presented in Figure 7. As shown in Figure 5, FB1 appears to increase SA contents in wild-type protoplasts incubated in the light. However, under the same conditions, FB1 does not substantially induce the PAL promoter (data not shown). Thus, SA-mediated cell death triggered by FB1 may depend on light-induced PAL enzyme activity. Consistent with this hypothesis, wild-type protoplasts incubated in the dark display a concomitant reduction in PAL activity and FB1 susceptibility. The importance of PAL in SA-mediated cell death has been demonstrated previously (Mauch-Mani and Slusarenko, 1996; Shirasu et al., 1997).
The model shown in Figure 7 predicts that FB1 should induce SA-responsive PR genes in addition to the SA-responsive genes such as those that encode glucosyltransferase that were identified in the subtracted cDNA library (Table 1). This was confirmed in FB1-treated seedlings, in which PR-1, PR-2, and PR-5 mRNAs accumulated to high amounts 4 days after treatment (Stone et al., 2000). Preliminary experiments in protoplasts, however, showed that induction of a PR1-LUC fusion was undetectable within a 20-hr incubation with FB1—the longest treatment time feasible before a substantial number of the FB1-treated protoplasts died (T. Asai and F.M. Ausubel, unpublished data). In addition, PR-1, PR-2, and PR-5 were not among the putative FB1-induced genes identified in the subtracted cDNA library (Table 1). In an accompanying paper (Stone et al., 2000), we analyzed the pattern of expression of the PR-1 gene with a transgenic plant harboring a PR-1 promoter–GUS fusion construct and found that PR-1 expression was restricted to the cells surrounding the FB1-induced lesions. This suggests that PR gene expression may occur in response to a signal (or signals) emerging from dead or dying cells, rather than from within the cells undergoing cell death. Unless the signal is diffusible, the PR gene induction would not be expected in protoplast suspensions, in which individual cells are not directly attached to each other.
In addition to the SA signaling, Figure 3 shows that FB1-induced cell death requires the JA- and ET-dependent signal transduction pathways. To our knowledge, this is the only defense-related response for which induction clearly requires the concomitant activation of the SA-, JA-, and ET-dependent pathways. As is the case with SA, our data and the model shown in Figure 7 predict that FB1 should cause the cellular contents of JA and ET to increase. Indeed, in a separate study, we have demonstrated that the JA/ET-responsive PDF1.2 gene is induced in FB1-treated seedlings 4 days after treatment (Stone et al., 2000). Moreover, analysis of cDNA molecules derived from FB1-induced mRNAs suggests that FB1 induces a variety of JA- and ET-responsive genes within 20 hr (Table 1; see Methods), although we have not identified the PDF1.2 gene among the ∼200 cDNA molecules sequenced to date. Involvement of ET in FB1-induced cell death is further supported by the observation that the ET action inhibitor Ag+ suppresses FB1-mediated cell death in protoplasts (T. Asai and F. Ausubel, unpublished data) and that FB1 induces the ET-forming enzyme 1-aminocyclopropane-1-carboxylic acid (ACC) oxidase (Table 1). Finally, induction of ET and its biosynthetic genes by AAL toxin and FB1 has been reported in tomato leaflets (Moussatos et al., 1994; Moore et al., 1999). ET has been well documented to be involved in developmentally controlled PCD such as senescence (Pennell and Lamb, 1997; Gray and Johal, 1998; Johnson and Ecker, 1998). Interestingly, a fraction of the FB1-induced genes in protoplasts encode senescence-related proteins (Table 1), suggesting that senescence and pathogen-elicited PCD may share a common molecular basis (Butt et al., 1998; Navarre and Wolpert, 1999; Pontier et al., 1999; Quirino et al., 1999).
The signaling pathways described above that are necessary for FB1-induced cell death are activated in the cpr6 and acd2 mutants. However, unlike the acd2 mutant, the cpr6 mutant does not exhibit spontaneous lesions, and protoplasts isolated from the mutant still require FB1 to undergo cell death. Perhaps in the cpr6 mutant, activation of the pathways may not be strong enough to induce cell death. Another possibility is that the acd2 mutation, but not the cpr6 mutation, can activate an additional cell death factor or factors in the absence of FB1, bypassing the requirement of this elicitor for triggering cell death. This putative factor is denoted by the letter X in Figure 7, which cannot be constitutively activated in acd2-2 plants because the mutant is viable. In acd2 plants, spontaneous HR-like lesions develop primarily on the cotyledons and the older leaves (Greenberg et al., 1994). Therefore, in the absence of pathogens or pathogen-derived elicitors such as FB1, the unknown cell death factor may be regulated developmentally in a sporadic fashion. The factor could be a component of a signaling pathway normally involved in senescence. In the presence of the acd2 mutation, the factor may be overproduced in the older tissue, accelerating natural senescence. This idea is consistent with the observation that FB1 induces many senescence-related proteins (see above). Greenberg et al. (1994) also have demonstrated that HR-like lesions can be initiated in acd2 plants by mechanical stress such as wounding. Because plant tissue suffers mechanical stress during the course of protoplast isolation, the cell death factor is probably overproduced in acd2 protoplasts, explaining why they are not viable.
The HR is closely correlated with disease resistance. However, a determinative role of the HR in restricting pathogen growth and spread has not been conclusively demonstrated (Richael and Gilchrist, 1999). An Arabidopsis mutant has been isolated in which effective pathogen resistance is expressed in the absence of HR cell death (Yu et al., 1998). Likewise, it is unclear whether the cell death signaling described here is involved in actual pathogen resistance. However, protoplasts isolated from the plants with enhanced pathogen resistance exhibit enhanced FB1 susceptibility, and protoplasts from the plants that show enhanced pathogen susceptibility often exhibit reduced FB1 susceptibility. These results may indicate that the cell death signaling identified in our protoplast system plays a critical role in developing defense responses in the entire plant. Protoplast systems are ideal for characterizing plant signaling components such as mitogen-activated protein kinases (Kovtun et al., 2000). Identification of the signaling pathways located upstream and downstream of SA, JA, and ET should help elucidate the role of PCD in pathogen resistance.
METHODS
Plants and Growth Conditions
All plants used in this work were Arabidopsis thaliana ecotype Columbia (Col-0) and its derivatives. Transgenic NahG plants were constructed in our laboratory (Reuber et al., 1998). The pad4-1 (Glazebrook et al., 1996) and acd2-2 (Greenberg et al., 1994) mutants were isolated in our laboratory; the npr1-1 (Cao et al., 1994), cpr1-1 (Bowling et al., 1994), and cpr6-1 (Clarke et al., 1998) mutants were obtained from X. Dong (Duke University, Durham, NC); and the jar1-1 (Staswick et al., 1992) and etr1-1 (Bleecker et al., 1988) mutants were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus). Plants were grown in a greenhouse as described by Reuber et al. (1998).
Chemicals
Stock solutions of fumonisin B1 (FB1) and other chemicals were prepared as follows: FB1, 7 mM in methanol; cordycepin, 50 mM in 50% ethanol; cycloheximide, 0.8 M in methanol (all from Sigma); and staurosporine (Calbiochem, San Diego, CA), 1 mM in methanol. Stock solutions were diluted with water (FB1 and cycloheximide) or 50% ethanol (cordycepin).
Protoplast Preparation and Determination of Surviving Fractions
Protoplasts were isolated, under normal light conditions, from a mixture of rapidly expanding and fully expanded young leaves of ∼4-week-old plants. We used the protocol described by Kovtun et al. (2000) except that leaf strips were digested for 3 hr in 1.5% cellulase and 0.4% Macerozyme (both from Yakult Pharmaceutical Co., Tokyo, Japan). Isolated protoplasts were suspended in Murashige and Skoog complete medium (Gibco BRL, Gaithersburg, MD) supplemented with 0.4 M glucose. After taking three samples for the initial cell count, we treated the protoplast suspension with FB1 or other chemicals, singly or in combination, and incubated them at 22°C either in the light (40 μmol m−2 sec−1) or in the dark. After incubation, three samples were withdrawn, and Evans blue dye (Sigma) was added to the samples to a final concentration of 0.04%. The suspension was incubated for 10 min, and the number of unstained cells was determined under the light microscope with a hemacytometer. The fraction of surviving cells was calculated by dividing the number of viable cells after incubation by the initial cell count.
Detection of Nuclear DNA Fragmentation and Apoptotic Bodies
Total cellular DNA was stained with Hoechst 33342 (Sigma; 5 μg mL−1 in PBS), and the free 3′-OH groups in the DNA were labeled by the TUNEL method, using the In Situ Cell Death Detection Kit (Boehringer Mannheim) as instructed by the manufacturer (see also Wang et al., 1996). DNA was visualized with a fluorescent microscope.
Construction of a Subtracted cDNA Library
Protoplasts isolated from wild-type plants were divided into two identical portions; one was treated with 70 nM FB1 and the other with 0.001% methanol. The protoplasts were incubated in the light as described above, and samples were withdrawn at 6, 12, and 20 hr after treatment. A mixture of these three samples was used to prepare total cellular RNA with the RNeasy kit (Qiagen, Valencia, CA), using the protocol for animal cells. The RNA then was treated with DNase I (amplification grade; Gibco BRL), and 1 μg of this DNase-treated RNA was used for cDNA synthesis with the SMART polymerase chain reaction (PCR) cDNA Synthesis kit (Clontech, Palo Alto, CA). To construct a cDNA library representing FB1-induced genes, we mixed cDNA molecules derived from FB1-treated protoplasts with excess cDNA from methanol-treated protoplasts, and subtraction was performed by using a PCR-based technique with the PCR-Select cDNA Subtraction kit (Clontech) as instructed by the manufacturer. Unsubtracted cDNA molecules then were ligated to a vector plasmid (pT-Adv; Clontech) and introduced into Escherichia coli cells by electroporation. The transformants were picked individually and used to amplify inserted cDNA molecules by PCR. (The PCR primers were provided in the above-described subtraction kit.) The amplified cDNA molecules were arrayed identically onto two nylon membranes with a slot blot manifold (PR648; Hoefer Scientific Instruments, San Francisco, CA); differential expression of these cDNA clones in FB1-treated protoplasts was confirmed by using the PCR-Select Differential Screening kit (Clontech). DNA fragments containing a part of the UBQ10 coding sequence also were arrayed and used for an internal control. The fragments were amplified by PCR with the following primers: 5′-GGAGGTGGAGAGTTCTGACACCATT-3′ and 5′-CCTTGACGTTGTCAATGGTGT-CGGA-3′. Identities of FB1-induced cDNA clones then were obtained by DNA sequence analysis and the advanced BLAST program (Altschul et al., 1997).
Protoplast Transient Expression Assays
The DNA regions (GenBank accession number AC006248) immediately upstream from the translation start sites of the glucosyltransferase (GenBank accession number AAD17393) and PAL1 genes were amplified by PCR from Arabidopsis (Col-0) genomic DNA with the following oligonucleotide primers: glucosyltransferase, 5′-GGA-AAAGCTTAATGGTAGAAAGTAGAGG-3′ and 5′-GGCCATGGTTAAGAGTGGAATCATG-3′; and PAL1, 5′-CGGGATCCGGAGGCTTTTGCC-TGCTAGT-3′ and 5′-CCGTTAATGTCGACTTAGACTTTTGATC-3′. The sizes of the amplified fragments were 1400 and 1832 bp, respectively. The fragments were cloned in a LUC plasmid vector, pJD301 (Luehrsen et al., 1992), between the BamHI and the SalI sites. Protoplasts (2 × 105 cells) were transfected with the resulting plasmids (20 μg) by a polyethylene glycol procedure (Kovtun et al., 2000) and incubated in Murashige and Skoog complete medium containing 0.4 M glucose. LUC activity was determined as described by Luehrsen et al. (1992). To normalize the LUC activity for variation in transformation efficiency and cell viability, we cotransfected the protoplasts with a control plasmid (30 μg) carrying the constitutive UBQ1 promoter (Holtorf et al., 1995) fused to the GUS reporter gene (Kovtun et al., 2000). The UBQ1 promoter fragment (2312 bp) was amplified by PCR with the following primers: 5′-GTACCTCTCTCTGTTCCAAACCATGG-3′ and 5′-CTTTTGGGATCCGTCTTCTCTCAC-GTAG-3′. GUS activities were determined as described previously (Jefferson, 1987).
Phenylalanine Ammonia-Lyase Enzyme Activity
Phenylalanine ammonia-lyase (PAL) enzyme activity was determined as described previously (Edwards and Kessmann, 1992).
Acknowledgments
We thank Xinnian Dong for the nahG construct, and Xinnian Dong and the Arabidopsis Biological Resource Center for mutant seeds. We also thank Julia Dewdney and Mary Wildermuth for critical reading of the manuscript. This work was supported by National Institutes of Health Grant No. R01-GM48707 awarded to F.M.A. and by United States Department of Agriculture Grant No. NRICGP-97-35306-4581 and National Science Foundation (NSF) Grant No. IBN-9723610 awarded to J.S. T.A. was supported in part by fellowships from the Toyobo Biotechnology Foundation and the Uehara Memorial Foundation; J.M.S. was supported by NSF fellowship No. DBI-9750297; and J.E.H. was a Department of Energy/Energy Biosciences Research Fellow of the Life Sciences Research Foundation.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate, N.J., Orr, J., Ni, W., Meromi, A., Nadler-Hassar, T., Doerner, P.W., Dixon, R.A., Lamb, C.J., and Elkind, Y. (1994). Quantitative relationship between phenylalanine ammonia-lyase levels and phenylpropanoid accumulation in transgenic tobacco identifies a rate-determining step in natural product synthesis. Proc. Natl. Acad. Sci. USA 91, 7608–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A. (1996). Plant disease resistance genes: Function meets structure. Plant Cell 8, 1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker, A.B., Estelle, M.A., Somerville, C., and Kende, H. (1988). Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241, 1086–1089. [DOI] [PubMed] [Google Scholar]
- Bowling, S.A., Guo, A., Cao, H., Gordon, A.S., Klessig, D.F., and Dong, X. (1994). A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6, 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston, V. (1997). The molecular biology of leaf senescence. J. Exp. Bot. 48, 181–199. [Google Scholar]
- Butt, A., Mousley, C., Morris, K., Beynon, J., Can, C., Holub, E., Greenberg, J.T., and Buchanan-Wollaston, V. (1998). Differential expression of a senescence-enhanced metallothionein gene in Arabidopsis in response to isolates of Peronospora parasitica and Pseudomonas syringae. Plant J. 16, 209–221. [DOI] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1998). Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominant Arabidopsis cpr6–1 mutant. Plant Cell 10, 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, R.J., Harmon, B.V., Gobe, G.C., and Kerr, J.F.R. (1992). Internucleosomal DNA cleavage should not be the sole criterion for identifying apoptosis. Int. J. Radiat. Biol. 61, 451–453. [DOI] [PubMed] [Google Scholar]
- Constabel, C.P., Bergey, D.R., and Ryan, C.A. (1995). Systemin activates synthesis of wound-inducible tomato leaf polyphenol oxidase via the octadecanoid defense signaling pathway. Proc. Natl. Acad. Sci. USA 92, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquoz, J.-L., Buchala, A., and Métraux, J.-P. (1998). The biosynthesis of salicylic acid in potato plants. Plant Physiol. 117, 1095–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L., Dietrich, R.A., and Richberg, M.A. (1996). Death don't have no mercy: Cell death programs in plant–microbe interactions. Plant Cell 8, 1793–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P. (1997). Genetic dissection of acquired resistance to disease. Plant Physiol. 113, 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., Ward, E., and Ryals, J. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X. (1998). SA, JA, ethylene, and disease resistance in plants. Curr. Opin. Plant Biol. 1, 316–323. [DOI] [PubMed] [Google Scholar]
- Durner, J., Shah, J., and Klessig, D.F. (1997). Salicylic acid and disease resistance in plants. Trends Plant Sci. 2, 266–274. [Google Scholar]
- Edwards, R., and Kessmann, H. (1992). Isoflavonoid phytoalexins and their biosynthetic enzymes. In Molecular Plant Pathology: A Practical Approach, S. Gurr, M. McPherson, and D. Bowles, eds (Oxford, UK: Oxford University Press), pp. 45–62.
- Felix, G., Duran, J.D., Volko, S., and Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. [DOI] [PubMed] [Google Scholar]
- Gaff, D.F., and Okong'O-Ogola, O. (1971). The use of non-permeating pigments for testing the survival of cells. J. Exp. Bot. 22, 756–758. [Google Scholar]
- Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., Ward, E., Kessmann, H., and Ryals, J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Gilchrist, D.G. (1997). Mycotoxins reveal connections between plants and animals in apoptosis and ceramide signaling. Cell Death Differ. 4, 689–698. [DOI] [PubMed] [Google Scholar]
- Gilchrist, D.G. (1998). Programmed cell death in plant disease: The purpose and promise of cellular suicide. Annu. Rev. Phytopathol. 36, 393–414. [DOI] [PubMed] [Google Scholar]
- Gilchrist, D.G., Wang, H., and Bostock, R.M. (1995). Sphingosine-related mycotoxins in plant and animal diseases. Can. J. Bot. 73, S459–S467. [Google Scholar]
- Glazebrook, J. (1999). Genes controlling expression of defense responses in Arabidopsis. Curr. Opin. Plant Biol. 2, 280–286. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., Rogers, E.E., and Ausubel, F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorczyca, W., Gong, J., and Darzynkiewicz, Z. (1993). Detection of DNA strand breaks in individual apoptotic cells by the in situ terminal deoxynucleotidyl transferase and nick translation assays. Cancer Res. 53, 1945–1951. [PubMed] [Google Scholar]
- Gray, J., and Johal, G.S. (1998). Programmed cell death in plants. In Arabidopsis, M. Anderson and J.A. Roberts, eds (Sheffield, UK: Sheffield Academic Press), pp. 360–394.
- Greenberg, J.T. (1997). Programmed cell death in plant–pathogen interactions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 525–545. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., Guo, A., Klessig, D.F., and Ausubel, F.M. (1994). Programmed cell death in plants: A pathogen-triggered response activated coordinately with multiple defense functions. Cell 77, 551–563. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K., and Jones, J.D.G. (1996). Resistance gene-dependent plant defense responses. Plant Cell 8, 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath, M.C. (1998). Apoptosis, programmed cell death and hypersensitive response. Eur. J. Plant Pathol. 104, 117–124. [Google Scholar]
- Heitz, T., Geoffroy, P., Fritig, B., and Legrand, M. (1999). The PR-6 family: Proteinase inhibitors in plant–microbe and plant–insect interactions. In Pathogenesis-Related Proteins in Plants, S.K. Datta and S. Muthukrishnan, eds (Boca Raton, FL: CRC Press), pp. 131–155.
- Holtorf, S., Apel, K., and Bohlmann, H. (1995). Comparison of different constitutive and inducible promoters for the overexpression of transgenes in Arabidopsis thaliana. Plant Mol. Biol. 29, 637–646. [DOI] [PubMed] [Google Scholar]
- Horvath, D.M., and Chua, N.-H. (1996). Identification of an immediate-early salicylic acid–inducible tobacco gene and characterization of induction by other compounds. Plant Mol. Biol. 31, 1061–1072. [DOI] [PubMed] [Google Scholar]
- Jabs, T., Dietrich, R.A., and Dangl, J.L. (1996). Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273, 1853–1856. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. [Google Scholar]
- Jirage, D., Tootle, T.L., Reuber, T.L., Frost, L.N., Feys, B.J., Parker, J.E., Ausubel, F.M., and Glazebrook, J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96, 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, P.R., and Ecker, J.R. (1998). The ethylene gas signal transduction pathway: A molecular perspective. Annu. Rev. Genet. 32, 227–254. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Yamada, K., Hiraiwa, N., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (1999). Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetative tissues during senescence and under various stress conditions. Plant J. 19, 43–53. [DOI] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.-L., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, E., Pontier, D., and del Pozo, O. (1999). Die and let live—Programmed cell death in plants. Curr. Opin. Plant Biol. 2, 502–507. [DOI] [PubMed] [Google Scholar]
- Levine, A., Pennell, R.I., Alvarez, M.E., Palmer, R., and Lamb, C. (1996). Calcium-mediated apoptosis in a plant hypersensitive disease resistance response. Curr. Biol. 6, 427–437. [DOI] [PubMed] [Google Scholar]
- Lois, R., Dietrich, A., Hahlbrock, K., and Schulz, W. (1989). A phenylalanine ammonia-lyase gene from parsley: Structure, regulation and identification of elicitor and light responsive cis-acting elements. EMBO J. 8, 1641–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen, K.R., de Wet, J.R., and Walbot, V. (1992). Transient expression analysis in plants using firefly luciferase reporter gene. Methods Enzymol. 216, 397–414. [DOI] [PubMed] [Google Scholar]
- Martin, S.J., Green, D.R., and Cotter, T.G. (1994). Dicing with death: Dissecting the components of the apoptosis machinery. Trends Biochem. Sci. 19, 26–30. [DOI] [PubMed] [Google Scholar]
- Mauch-Mani, B., and Slusarenko, A.J. (1996). Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of Arabidopsis to Peronospora parasitica. Plant Cell 8, 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, J.M., and Dangl, J.L. (2000). Signal transduction in the plant immune response. Trends Biol. Sci. 25, 79–82. [DOI] [PubMed] [Google Scholar]
- Merkouropoulos, G., Barnett, D.C., and Shirsat, A.H. (1999). The Arabidopsis extensin gene is developmentally regulated, is induced by wounding, methyl jasmonate, abscisic and salicylic acid, and codes for a protein with unusual motifs. Planta 208, 212–219. [DOI] [PubMed] [Google Scholar]
- Mittler, R., and Lam, E. (1996). Sacrifice in the face of foes: Pathogen-induced programmed cell death in plants. Trends Microbiol. 4, 10–15. [DOI] [PubMed] [Google Scholar]
- Mittler, R., Shulaev, V., Seskar, M., and Lam, E. (1996). Inhibition of programmed cell death in tobacco plants during a pathogen-induced hypersensitive response at low oxygen pressure. Plant Cell 8, 1991–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, T., Martineau, B., Bostock, R.M., Lincoln, J.E., and Gilchrist, D.G. (1999). Molecular and genetic characterization of ethylene involvement in mycotoxin-induced plant cell death. Physiol. Mol. Plant Pathol. 54, 73–85. [Google Scholar]
- Morel, J.-B., and Dangl, J.L. (1997). The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 4, 671–683. [DOI] [PubMed] [Google Scholar]
- Moussatos, V.V., Lucas, W.J., and Gilchrist, D.G. (1993. a). AAL toxin-induced physiological changes in Lycopersicon esculentum Mill: Differential sucrose transport in tomato lines isogenic for the Asc locus. Physiol. Mol. Plant Pathol. 42, 359–371. [Google Scholar]
- Moussatos, V.V., Witsenboer, H., Hille, J., and Gilchrist, D.G. (1993. b). Behaviour of the disease resistance gene Asc in protoplasts of Lycopersicon esculentum mill. Physiol. Mol. Plant Pathol. 43, 255–263. [Google Scholar]
- Moussatos, V.V., Yang, S.F., Ward, B., and Gilchrist, D.G. (1994). AAL-toxin induced physiological changes in Lycopersicon esculentum Mill: Roles for ethylene and pyrimidine intermediates in necrosis. Physiol. Mol. Plant Pathol. 44, 455–468. [Google Scholar]
- Navarre, D.A., and Wolpert, T.J. (1999). Victorin induction of an apoptotic/senescence-like responses in oats. Plant Cell 11, 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, I.E.W., Reutelingsperger, C.P.M., and Holdaway, K.M. (1997). Annexin-V and TUNEL use in monitoring the progression of apoptosis in plants. Cytometry 29, 28–33. [PubMed] [Google Scholar]
- Ohl, S., Hedrick, S., Chory, J., and Lamb, C.J. (1990). Functional properties of a phenylalanine ammonia-lyase promoter from Arabidopsis. Plant Cell 2, 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell, R.I., and Lamb, C. (1997). Programmed cell death in plants. Plant Cell 9, 1157–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Eggermont, K., Terras, F.R.G., Thomma, B.P.H.J., De Samblanx, G.W., Buchala, A., Métraux, J.-P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid–independent pathway. Plant Cell 8, 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Thomma, B.P.H.J., Buchala, A., Metraux, J.-P., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J., and van Loon, L.C. (1999). Salicylic acid–independent plant defense pathways. Trends Plant Sci. 4, 52–58. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J., van Wees, S., van Pelt, J., Knoester, M., Laan, R., Gerrits, H., Weisbeek, P., and van Loon, L. (1998). A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10, 1571–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier, D., Gan, S., Amasino, R.M., Roby, D., and Lam, E. (1999). Markers for hypersensitive response and senescence show distinct patterns of expression. Plant Mol. Biol. 39, 1243–1255. [DOI] [PubMed] [Google Scholar]
- Quirino, B.F., Normanly, J., and Amasino, R.M. (1999). Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol. Biol. 40, 267–278. [DOI] [PubMed] [Google Scholar]
- Rate, D.N., Cuenca, J.V., Bowman, G.R., Guttman, D.S., and Greenberg, J.T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11, 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber, T.L., Plotnikova, J.M., Dewdney, J., Rogers, E.E., Wood, W., and Ausubel, F.M. (1998). Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 16, 473–485. [DOI] [PubMed] [Google Scholar]
- Reymond, P., and Farmer, E.E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1, 404–411. [DOI] [PubMed] [Google Scholar]
- Richael, C., and Gilchrist, D. (1999). The hypersensitive response: A case of hold or fold? Physiol. Mol. Plant Pathol. 55, 5–12. [Google Scholar]
- Richberg, M.H., Aviv, D.H., and Dangl, J.L. (1998). Dead cells do tell tales. Curr. Opin. Plant Biol. 1, 480–485. [DOI] [PubMed] [Google Scholar]
- Ryals, J.A., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.-Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryerson, D.E., and Heath, M.C. (1996). Cleavage of nuclear DNA into oligonucleosomal fragments during cell death induced by fungal infection or abiotic treatments. Plant Cell 8, 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., Tsui, F., and Klessig, D.F. (1997). Characterization of a salicylic acid insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant-Microbe Interact. 10, 69–78. [DOI] [PubMed] [Google Scholar]
- Shah, J., Kachroo, P., and Klessig, D.F. (1999). The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expression salicylic acid dependent. Plant Cell 11, 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu, N., Nakajima, H., Rajasekhar, V.K., Dixon, R.A., and Lamb, C.J. (1997). Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 9, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Becker, J., Marois, E., Huguet, E.J., Midland, S.L., Sims, J.J., and Keen, N.T. (1998). Accumulation of salicylic acid and 4-hydroxybenzoic acid in phloem fluids of cucumber during systemic acquired resistance is preceded by a transient increase in phenylalanine ammonia-lyase activity in petioles and stems. Plant Physiol. 116, 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz, B.J., Ausubel, F.M., Baker, B.J., Ellis, J.G., and Jones, J.D.G. (1995). Molecular genetics of plant disease resistance. Science 268, 661–667. [DOI] [PubMed] [Google Scholar]
- Staswick, P.E., Su, W., and Howell, S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89, 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, J.M., Heard, J.E., Asai, T., and Ausubel, F.M. (2000). Simulation of fungal-mediated cell death by fumonisin B1 and selection of fumonisin B1-resistant (fbr) Arabidopsis mutants. Plant Cell 12, 1811–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenhaken, R., and Rübel, C. (1997). Salicylic acid is needed in hypersensitive cell death in soybean but does not act as a catalase inhibitor. Plant Physiol. 115, 291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P.H.J., Eggermont, K., Penninckx, I.A.M.A., Mauch-Mani, B., Vogelsang, R., Cammue, B.P.A., and Broekaert, W.F. (1998). Separate jasmonate-dependent and salicylate-dependent defense–response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95, 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To, K.-Y., Suen, D.-F., and Chen, S.-C.G. (1999). A sweet potato leaf cDNA encoding barley and rice B12D homolog. Plant Physiol. 120, 933.10454902 [Google Scholar]
- Turner, J.G., and Novacky, A. (1974). The quantitative relation between plant and bacterial cells involved in the hypersensitive reaction. Phytopathology 64, 885–890. [Google Scholar]
- Uknes, S., Mauch-Mani, B., Moyer, M., Potter, S., Williams, S., Dincher, S., Chandler, D., Slusarenko, A., Ward, E., and Ryals, J. (1992). Acquired resistance in Arabidopsis. Plant Cell 4, 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Li, J., Bostock, R.M., and Gilchrist, D.G. (1996). Apoptosis: A functional paradigm for programmed plant cell death induced by a host-selective phytotoxin and invoked during development. Plant Cell 8, 375–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner, L.A., Li, G., Ware, D., Somssich, I.E., and Davis, K.R. (1995). The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol. Biol. 27, 327–338. [DOI] [PubMed] [Google Scholar]
- Ward, E.R., Uknes, S.J., Williams, S.C., Dincher, S.S., Wiederhold, D.L., Alexander, D.C., Ahl-Goy, P., Métraux, J.-P., and Ryals, J.A. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, L.M., Gan, S., Quirino, B., and Amasino, R.M. (1998). A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol. Biol. 37, 455–469. [DOI] [PubMed] [Google Scholar]
- Weymann, K., Hunt, M., Uknes, S., Neuenschwander, U., Lawton, K., Steiner, H.Y., and Ryals, J. (1995). Suppression and restoration of lesion formation in Arabidopsis lsd mutants. Plant Cell 7, 2013–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, R.F. (1979). Acetylsalicylic acid (aspirin) induces resistance to tobacco mosaic virus in tobacco. Virology 99, 410–412. [DOI] [PubMed] [Google Scholar]
- Yu, I.-C., Parker, J., and Bent, A.F. (1998). Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl. Acad. Sci. USA 95, 7819–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, N., Tootle, T.L., Tsui, F., Klessig, D.F., and Glazebrook, J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10, 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]