Abstract
Despite the differences in flower form, the underlying mechanism in determining the identity of floral organs is largely conserved among different angiosperms, but the details of how the functions of A, B, and C are specified varies greatly among plant species. Here, we report functional analysis of a Gerbera MADS box gene, GRCD1, which is orthologous to AGL2-like MADS box genes. Members of this group of genes are being reported in various species in growing numbers, but their functions remained largely unsettled. GRCD1 expression is detected in all four whorls, but the strongest signal is seen in the developing stamen and carpel. Downregulating GRCD1 expression by antisense transformation revealed that lack of GRCD1 caused homeotic changes in one whorl only: sterile staminodes, which normally develop in whorl 3 of marginal female florets, were changed into petals. This indicates that the GRCD1 gene product is active in determining stamen identity. Transgenic downregulation of GRCD1 causes a homeotic change similar to that in the downregulation of the Gerbera C function genes GAGA1 and GAGA2, but one that is limited to whorl 3. Downregulation of GRCD1 expression does not reduce expression of GAGA1 or GAGA2, or vice versa; and in yeast two-hybrid analysis, GRCD1 is able to interact with GAGA1 and GAGA2. We propose that a heterodimer between the GRCD1 and GAGA1/2 gene products is needed to fulfill the C function in whorl 3 in Gerbera.
INTRODUCTION
The study of floral homeotic mutants in Arabidopsis and Antirrhinum has shown that despite their differences in flower form, the underlying mechanism of flower organ identity determination is largely conserved. The outcome of such studies is a simplified ABC model. Sepal development is determined by expression of the A function alone; the combination of the A and B functions or the B and C functions specifies petal or stamen development, respectively; and expression of the C function alone determines carpel formation (Coen and Meyerowitz, 1991). Most of the ABC function genes are shown to encode MADS-box transcription factors, which form homo- or heterodimers to perform their functions (see, e.g., Yanofsky et al., 1990; Jack et al., 1992; Goto and Meyerowitz, 1994). Plant MADS box genes (>200 known) have been divided into subgroups (clades) mainly by phylogenetic analyses of conserved parts of their predicted amino acid sequences (Doyle, 1994; Purugganan et al., 1995; Tandre et al., 1995; Theissen et al., 1996). Most clade members, although originating from different plant species, share similar expression patterns and have related developmental functions; the MADS box genes providing A, B, and C functions also fall into separate clades. However, the AGAMOUS (AG) clade, for example, comprises not only typical C function genes that show similar mutant or loss-of-function phenotypes—such as Arabidopsis AGAMOUS, Antirrhinum PLENA and FARINELLI, and Gerbera GAGA1 and GAGA2—but also genes that determine ovule identity—such as Petunia FBP7 and FBP11 (Bowman et al., 1989; Schwarz-Sommer et al., 1990; Angenent et al., 1995; Davies et al., 1999; Yu et al., 1999).
Two models, which may act in combination, have been presented for Arabidopsis regarding how the different C function activities (stamen specification, carpel specification, and flower meristem determinacy) can be separated (Sieburth et al., 1995). For the single C function gene of Arabidopsis, AGAMOUS, the quantitative model predicts that the amount of gene product needed varies for each of those activities. On the basis of the phenotypes of the ag-4 and AG-MET205 mutations, the highest concentration of AG protein has been suggested to be required for determinacy, whereas mid-range amounts are needed for carpel specification, and the lowest concentration is sufficient for stamen specification. According to the protein–protein interaction model, the AGAMOUS gene product would make heterodimers with different partners in different whorls. This model is based on the finding that ag-4 and AG-MET205 mutations occur in the coiled-coil motif called the K box, which is thought to be important in protein–protein interactions (Sieburth et al., 1995; Fan et al., 1997). Yeast two-hybrid analysis and in vitro immunoprecipitation experiments in Arabidopsis suggest that in addition to the well-characterized ability to make homodimers, AG is also able to interact with four other MADS domain proteins—AGL2, AGL4, AGL6, and AGL9—all encoded by genes belonging to the AGL2 clade. Fan et al. (1997) proposed (but did not demonstrate) that the interaction between AG and these proteins could contribute to the functional specificity of the AG protein. This is in line with the protein–protein interaction model for separation of the C function in Arabidopsis.
Here, we report direct mechanistic evidence from Gerbera hybrida bearing on the quantitative versus protein–protein interaction models. Gerbera, a readily transformable model system in the sunflower family (Asteraceae), has proven useful for shedding light on novel as well as incompletely understood plant gene functions (see, e.g., Eckermann et al., 1998; Kotilainen et al., 1999). Previous work on Gerbera indicated functions for several MADS box gene components of the ABC system (Yu et al., 1999). High sequence similarity, identical expression patterns, and similar transgene phenotypes for GAGA1 and GAGA2 (AG/PLE orthologs) imply that these C-class genes participate equally in C functions. For example, in transgenic lines with reduced GAGA2 expression, petal-like organs formed in whorl 3. In whorl 4 of these transgenic lines, green organs with carpelloid- and sepaloid-like traits developed, and inside these altered whorl 4 organs were extra sepal and petal whorls (Yu et al., 1999).
Here, we report the isolation and characterization of GRCD1 (Gerbera Regulator of Capitulum Development 1), a cDNA molecule representing a Gerbera member of the AGL2 clade of MADS box genes. The AGL2 lineage includes several genes that are expressed during flower development but whose precise functions in plant development remain obscure. Thus far, two functional analyses of AGL2-like genes have been published. Downregulation of petunia FBP2 and tomato TM5 genes disturbed normal development of flower organs in whorls 2 to 4 in petunia and tomato, respectively (Angenent et al., 1994; Pnueli et al., 1994a). During flower organ differentiation, GRCD1 expression is detected in all whorls but is strongest in stamen and carpel primordia, suggesting that GRCD1 (gene product) could especially play a role in stamen and carpel development. Downregulation of GRCD1 expression revealed that lack of GRCD1 caused a homeotic transformation in one whorl only: sterile staminodes, which normally develop in whorl 3 of outermost female ray flowers, changed into petals. In sexually perfect central disc florets, downregulation of GRCD1 expression had only minor effects on stamen development, and fertile pollen was produced (although not released). Downregulation of GRCD1 did not affect normal development of carpels in whorl 4. The transgenic phenotypes suggest that the GRCD1 gene product is active in determining stamen identity during Gerbera flower development. Downregulation of the expression of the C function genes, GAGA1 or GAGA2, did not markedly reduce GRCD1 expression, and lack of GRCD1 did not reduce the expression of GAGA1 or GAGA2. Furthermore, yeast two-hybrid analysis results showed that the gene product of GRCD1 was able to make heterodimers with both the GAGA1 and GAGA2 gene products. These results indicate that GRCD1 has a role in the C function of whorl 3.
RESULTS
Isolation of a GRCD1 cDNA by Differential Screening
During studies of region-specific control of gene expression along the longitudinal axis of Gerbera petals, differential screening and sequence comparisons showed that one of the cDNAs, the expression of which is strongest at early stages of petal development, belongs to the MADS box gene family. Given this expression pattern and the results of transgenic functional analysis (see below), we named this cDNA GRCD1.
DNA gel blot analysis with a highly specific probe (a 200-bp-long 3′ fragment of the GRCD1 cDNA; 80% noncoding) recognized two bands at the stringency used in the RNA gel blot (data not shown). This indicates that the RNA gel blot analyses presented below correspond most probably to a transcript of a single locus, and the two bands found are the result of restriction site polymorphism in the heterozygous cultivar. However, the existence of a second very similar gene in the Gerbera genome cannot be ruled out.
GRCD1 Groups inside the AGL2 Clade
The GRCD1 cDNA is most similar to members of the AGL2 group, for example, AGL2, AGL4, and AGL9 of Arabidopsis; DEFH49, DEFH72, and DEFH200 of Antirrhinum; and FBP2 of petunia. To address the evolutionary relationships between GRCD1 and other MADS box genes, a phylogenetic analysis of nucleotide sequences of the MADS and K box regions was performed (see Yu et al., 1999). The analysis revealed that GRCD1 groups inside the AGL2 clade and that it pairs with another Gerbera gene, GRCD2 (GRCD1 and GRCD2 cDNAs have 64% identity at nucleotide level). Gene pairs or groups are typical of the AGL2 clade, for example, in Antirrhinum (DEFH200 and DEFH72) and in Arabidopsis (AGL2 and AGL4) (Figure 1).
Figure 1.
Phylogenetic Analysis of Selected MADS Box Genes.
Previously published plant MADS box genes were retrieved from the EMBL and GenBank databases.
Spatiotemporal Expression Pattern of GRCD1 during Flower Development
In situ hybridization analysis revealed that during flower development, GRCD1 expression is first detected before any flower organ primordia emerge, thus coinciding with the onset of expression of Gerbera B- and C-class MADS box genes. As with Gerbera B-class genes (Yu et al., 1999), induction of GRCD1 expression was asymmetrical within the plug-like flower primordia (see Hill et al., 1998, regarding Arabidopsis) and proceeded centripetally in the capitulum (Figure 2A).
Figure 2.
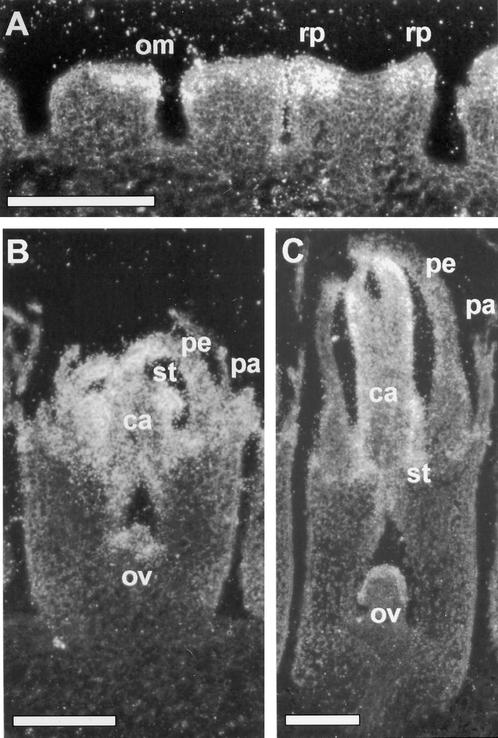
Analysis of GRCD1 Expression during Early Stages of Flower Development Using in Situ Hybridization.
(A) Three flower primordia at progressive developmental stages. Onset of GRCD1 expression takes place before flower organ determination, coinciding with the onset of expression of Gerbera B- and C-class MADS box genes (see Yu et al., 1999). GRCD1 signal, like that of Gerbera B-class MADS box genes, was first detected in the outer margins of floret primordia, where ring primordia (origin of perianth) later develop.
(B) A single flower primordium at early differentiation stages of flower organ primordia development. GRCD1 is expressed evenly in all four whorls and in the developing ovule.
(C) As the differentiation of flower organs progresses, GRCD1 expression is focused on stamen and carpel primordia, particularly in the outer integuments of ovules.
ca, carpel; om, outer margin; ov, ovule; pa, pappus bristles; pe, petal; rp, ring primordium; st, stamen.  .
.
Later in flower development, when all flower organ primordia had been determined, GRCD1 expression could be detected in all four whorls. As shown in Figure 2B, GRCD1 was also expressed in all whorls at early phases of flower organ differentiation; but at later phases, GRCD1 expression was strongest in stamen and carpel primordia, especially within carpel in the outer integument of the ovule. A weaker signal was also detected in differentiating petals (Figure 2C). This description of GRCD1 expression applies to all floret types. In the sexually perfect disc florets, GRCD1 signal in stamens was concentrated in anther connectives later in stamen development (data not shown).
The expression pattern of GRCD1 during the late developmental stages of different plant organs was studied using RNA gel blot analysis. The expression of GRCD1 was determined to be flower-specific; it was observed in all four flower organs—pappus bristles (simplified whorl 1 organs that lack vascular bundles), petals, stamens, and carpels (data not shown).
Downregulation of GRCD1 Expression Changes Whorl 3 Identity from Stamens to Petals
Detailed expression analyses showed that GRCD1 expression was concentrated in whorls 3 and 4 during the midstages of floral organ differentiation (Figure 2C), which might indicate that the GRCD1 protein plays a functional role in stamen and carpel development. To test this hypothesis, we generated transgenic plants in which GRCD1 expression is downregulated by introducing a nearly full-length GRCD1 cDNA (20 nucleotides missing in the 5′ end) in antisense orientation under the control of the cauliflower mosaic virus 35S promoter.
In the wild type, at early stages, the stamen primordia of different floret types are morphologically indistinguishable; the feminization of marginal florets occurs later in development, when stamen development in marginal ray and trans florets arrests. Anther tissues do not differentiate, and eventually they dry and wither, resulting in the formation of nonfunctional staminal rudiments or staminodes.
We obtained three independent antisense lines, t3, t7, and t8, which expressed much less GRCD1 during capitula development (Figure 3A). Downregulation of GRCD1 expression did not have an effect on normal carpel development in whorl 4 but did affect different floret types in whorl 3 differently. All three transgenic lines with reduced GRCD1 expression had petals in place of stamen rudiments in whorl 3 of the marginal ray and trans florets (Figures 4 and 5). Typical petal characteristics, such as anthocyanin pigmentation, shape, and cuticular striation of epidermal cells, were seen in whorl 3 organs of these transgenic lines. As reported by Yu et al. (1999), arrest of whorl 3 organs depends on organ identity, not on whorl position. Similarly, the GRCD1-downregulated lines did not show any signs of developmental arrest in whorl 3 petal development. In accordance with the ABC model, the whorl 3 identity change from stamens to petals was also detected in marginal florets of transformant lines in which either of the two functionally similar Gerbera C-class MADS box genes, GAGA1 or GAGA2, was downregulated (Figures 3 and 4C; Yu et al., 1999). The parallelism in downregulation phenotypes in whorl 3 among GAGA1/2 and GRCD1 suggests that GRCD1 participates in the normal C function for whorl 3.
Figure 3.
RNA Gel Blot Hybridization Analysis of Expression of GRCD1 and Gerbera C-Class MADS Genes (GAGA1 and GAGA2) in GRCD1 Antisense Lines, in GAGA2 Antisense Lines, in a GAGA1 Antisense Line, and in a Nontransformed Control Line.
Capitula covering several early developmental stages (both at determination and differentiation stages of flower organ development) were examined. The probe for each blot is indicated at left.
(A) The expression of GRCD1 was downregulated in all three antisense GRCD1 lines (t3, t7, and t8), but downregulation of both GAGA2 and GAGA1 in the antisense GAGA2 lines did not affect GRCD1 expression.
(B) The expression of GAGA2 was downregulated in three anti-GAGA2 lines, but downregulation of GRCD1 did not affect GAGA2 expression.
(C) Because of high sequence similarity between GAGA1 and GAGA2, the expression of GAGA1 was also downregulated in anti-GAGA2 lines (t17, t20, and t21), with the homeotic changes being most severe in whorls 3 and 4 (Yu et al., 1999). However, downregulation of GRCD1 did not affect GAGA1 expression.
(D) GRCD1 was expressed in GAGA1 antisense line t5.
(E) GAGA1 expression was downregulated in GAGA1 antisense line t5.
(F) GAGA2 expression was markedly reduced in a GAGA1 antisense line t5.
a-s, antisense; WT, wild type.
Figure 4.
Downregulation of GRCD1 Expression Caused Homeotic Transformation of Anther Rudiments into Petals in Whorl 3 in Ray Florets of the Anti-GRCD1 Lines.
(A) Inflorescence of the anti-GRCD1 line t3.
(B) An individual marginal ray floret of t3. Petals in whorl 3 were seen as light-colored, strip-like organs (arrow).
(C) The whorl 3 organs of marginal ray floret in a nontransformed control line, in GRCD1 downregulation line t3, and in GAGA1 downregulation line t5. The two petal rudiments were removed to uncover the whorl 3 organs (arrows). a-s, antisense.
Figure 5.
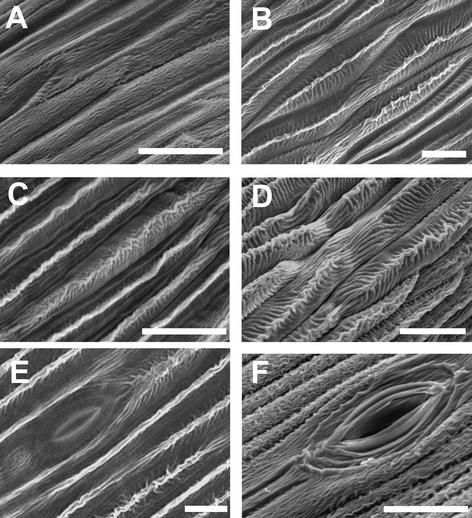
Scanning Electron Microscopy of the Whorl 2 and 3 Organs of Anti-GRCD1, Anti-GAGA1, and Nontransformed Lines.
(A) Surface of whorl 3 (stamen rudiment) in a nontransformed control line.
(B) Adaxial surface of whorl 2 (petal) in a nontransformed control line.
(C) Adaxial surface of whorl 3 organ in the anti-GRCD1 line t3.
(D) Adaxial surface of whorl 3 organ in the anti-GAGA1 line t5.
(E) Abaxial surface of whorl 2 organ (petal) in a nontransformed control line.
(F) Abaxial surface of whorl 3 organ in the anti-GRCD1 line t5 showing stomatum of petal-like development. During wild-type flower development, stomata develop only on the abaxial surface of petal.
 .
.
In the central disc florets, downregulation of GRCD1 expression had only slight effects on the development of fertile stamens in whorl 3. During normal flower development in Gerbera, stomata develop only on the abaxial surfaces of petals (Figure 5E). In the transformants, however, stomata also developed on the abaxial surface of anthers (Figure 5F). Pollen was not released from pollen sacs as it is in nontransformed control lines, indicating that normal anther development was disturbed. However, differentiation of tissues took place normally, and the pollen produced was fertile (data not shown). Thus, in the perfect disc florets, downregulation of GRCD1 expression causes minor disturbances in whorl 3 development, which is keeping with the homeotic transformation of whorl 3 organs to petals seen in marginal florets.
RNA gel blot analysis of capitula of anti-GRCD1 transformants showed that downregulation of GRCD1 had no effect on the extent of expression or spatial distribution of GAGA1 and GAGA2. These results, verified by in situ hybridization (data not shown), indicate that the gene product of GRCD1 is not needed for the expression of the C-class genes and that GRCD1 is directly involved in stamen development. Likewise, in anti-GAGA1 or anti-GAGA2 plants, which phenocopy the homeotic change in whorl 3 of ray florets (petals instead of staminodes) in anti-GRCD1 plants, GRCD1 expression was at the same amount as in nontransformed controls (Figure 3, and data not shown). This demonstrates that the gene products of GAGA1 or GAGA2 (Gerbera C-class genes) are not necessary for GRCD1 expression.
Yeast Two-Hybrid Analysis Showed That GRCD1 Protein Can Pair with C Function Proteins GAGA1 and GAGA2
Earlier, GAGA1 and GAGA2 were shown to code in Gerbera for C function, which is needed for proper development of stamen and carpel (Yu et al., 1999). Transgenic downregulation of GRCD1 phenocopies the downregulation of GAGA1/2 in whorl 3, and because GRCD1 does not seem to be upstream or downstream of GAGA1 or GAGA2 and because MADS proteins are known to form homo- and heterodimers to specify their functions, we hypothesized that perhaps GRCD1 participates in the C function by directly interacting with GAGA1 or GAGA2 in Gerbera. To study the putative protein–protein interactions among GRCD1, GAGA1, and GAGA2, we performed yeast two-hybrid analysis, finding that, indeed, GRCD1 made heterodimers with both GAGA1 and GAGA2 proteins. Interactions among GAGA1 and GAGA2 proteins were substantially weaker, and the GRCD1 protein did not homodimerize in this assay (Figure 6).
Figure 6.
Interaction between GRCD1, GAGA1, and GAGA2 Proteins in the Yeast Two-Hybrid Assay.
LacZ reporter activity in yeast cells for the pairwise combinations of GRCD1 (R1), GAGA1 (G1), and GAGA2 (G2) is shown. The activity in the negative control (neg. contr.; containing vectors pLEXA and pB42AD) represents the background value in yeast cells. Error bars indicate sd.
DISCUSSION
GRCD1 Participates in the C Function
GRCD1, a Gerbera AGL2-like gene, has flower-specific expression that was first detected in all four whorls and later found to have its strongest expression in stamen and carpel primordia. To study the role of GRCD1 in Gerbera flower development, we generated transgenic plants in which GRCD1 expression was downregulated by introducing an antisense GRCD1 cDNA into the Gerbera genome. Lack of the GRCD1 gene product especially affected stamen development in whorl 3. In marginal florets, a complete homeotic transformation of the sterile staminodes to petals took place, suggesting that GRCD1 is needed for determining stamen identity in Gerbera. In central disc florets, the stamens of anti-GRCD1 lines remained fertile but had petal-like characteristics (see Results). Downregulation of GRCD1 did not affect normal development of carpels in whorl 4.
As reported earlier (Yu et al., 1999), both expression patterns and analysis of transgenic plants reveal that Gerbera C-class MADS box genes GAGA1 and GAGA2 have similar functions. In Gerbera, downregulation of these C function genes caused whorl 4 organs to develop sepal-like characteristics. Inside these altered whorl 4 organs, extra whorl 1 and 2 organs were observed. Furthermore, and in accordance with the ABC model of flower development, downregulation of C function genes also caused a homeotic transformation of whorl 3 organs into petals in all flower types of Gerbera (Yu et al., 1999). As presented above, downregulation of GRCD1 expression phenocopies the homeotic transformation of whorl 3 marginal florets that is obtained by downregulation of GAGA1 or GAGA2. The similarity of GRCD1 function and GAGA1 and GAGA2 function in whorl 3 suggests that GRCD1 participates in the C function in whorl 3.
AGL2-like MADS Box Genes Are Abundant among Different Plant Species
Phylogenetic analysis revealed that GRCD1 is grouped into the AGL2 clade, members of which are increasingly being discovered in seed plants. Gerbera GRCD1 expression, like the expression of its characterized orthologs, varies spatially as flowers develop. The onset of GRCD1 expression takes place simultaneously with that of B- and C-class MADS box genes. This is different, for example, from the expression of the Arabidopsis orthologs AGL2, AGL4, and AGL9 and the Antirrhinum orthologs DEFH72 and DEFH200, which precedes the accumulation of transcripts of B- and C-class MADS box genes, although the expression and accumulation overlap temporally to some extent (Flanagan and Ma, 1994; Savidge et al., 1995; Davies et al., 1996; Mandel and Yanofsky, 1998).
As with GRCD1, the expression of many angiosperm AGL2 clade members is inflorescence specific, and all AGL2-like eudicot genes share similarities in their spatial expression patterns in flower development. For example, petunia FBP2, tomato TM5, pea MTF1, and Arabidopsis AGL2, AGL4, and AGL9 are expressed mainly in petal, stamen, and carpel primordia, which permits these genes to participate in controlling the development of those organs (Pnueli et al., 1991; Angenent et al., 1992; Flanagan and Ma, 1994; Savidge et al., 1995; Buchner and Boutin, 1998; Mandel and Yanofsky, 1998).
Two functional analyses of AGL2-like MADS box genes have been reported. Cosuppression of FBP2 in petunia resulted in developmental changes in whorls 2 to 4, causing the development of greenish and smaller than usual petals in whorl 2; stamens were replaced by green petaloid structures; and the size of the inner whorl was reduced, with no ovules or placenta forming, but new inflorescences developed in the axis of the carpels (Angenent et al., 1994). Similarly, organ-specific changes in antisense TM5 tomato plants included evergreen, cauline petals; greenish sterile stamens; and defective carpels with sepaloid or petaloid characteristics (Pnueli et al., 1994a). Thus, downregulation of the expression of petunia FBP2 and tomato TM5 disturbed the normal development of petal, stamen, and carpel but did not cause complete homeotic transformation of any particular organ. The minor effect of GRCD1 downregulation on stamen development in the perfect disc floret is in keeping with the partial downregulation phenotypes of petunia FBP2 and tomato TM5 floral organs.
Alternative Models for GRCD1 Function
GRCD1 and GAGA1/2 are both needed for C function in whorl 3, given that a similar homeotic transformation of whorl 3 organs in the marginal ray florets from rudimentary anthers into true petals in all three antisense lines occurs (Figures 4 and 5; Yu et al., 1999). Three separate results suggest that the involvement of GRCD1 in the C function is direct and might occur through heterodimerization with GAGA1 and GAGA2. First, GRCD1 expression correlates temporally and spatially with the expression patterns of C function genes, GAGA1 and GAGA2, during stamen development (Figure 2 and Yu et al., 1999). Second, the expression analyses of antisense GRCD1, antisense GAGA1, and antisense GAGA2 plants show that the gene product of GRCD1 is not needed for upregulating the expression of GAGA1 or GAGA2 or vice versa (Figure 3). Third, GRCD1 can pair both with GAGA1 and GAGA2 proteins in yeast two-hybrid analyses. Interestingly, interactions among GAGA1 and GAGA2 proteins are much weaker than is their interaction with GRCD1 (Figure 6). This suggests that the GAGA1 and GAGA2 proteins could pair with different and alternative proteins in planta to perform the C function during flower development. Thus, we hypothesize that GRCD1 makes a heterodimer with GAGA1 and GAGA2 and participates in the C function specifically during stamen development.
Three different hypotheses of GRCD1 function during flower development can be drawn from the results presented. First, GRCD1 could participate in separation of the C function between whorls 3 and 4, thus supporting the protein–protein interaction model for separation of the C function in Gerbera (see above, Introduction). This would require that the GRCD1/GAGA heterodimers are active in whorl 3 only because of post-transcriptional regulation of GRCD1 or because of other regulatory factors involved. Second, GRCD1 function could be redundant with that of another gene, including a complete redundancy outside the stamen whorl. Good candidates would be the close ortholog GRCD2 or a hypothetical duplicate locus of GRCD1 (see Results). Because GRCD2 is expressed in the antisense GRCD1 lines (data not shown), it is possible that the ortholog might substitute for the GRCD1 function partially in whorl 3 of disc florets and fully in whorl 4. Third, the GRCD1 antisense plants may not be complete loss-of-function lines; that is, some residual GRCD1 activity might suffice for normal GRCD1 function outside whorl 3. Bearing in mind the partial downregulation phenotypes of petunia FBP2 and tomato TM5 (see above), if either of the two latter hypotheses of GRCD1 function is true, GRCD1 would also have a function in whorls other than stamen whorls.
In tomato, ectopic expression of the tomato C function gene TAG1 changes the identity of petals in whorl 2 to sterile stamens (Pnueli et al., 1994b). Interestingly, Lifschitz (1996) mentions that in a double transformant in which TAG1 was ectopically expressed and TM5 (a tomato AGL2-like gene) was downregulated, the identity of whorl 2 reverted into petals. Because TM5 is normally expressed in whorl 2, TM5 protein is required for the homeotic conversion of petals to stamens by the TAG1 gene in lines expressing TAG1 ectopically. Like Gerbera GRCD1 and GAGA1/2, TM5 and TAG1 are known to pair in yeast two-hybrid analysis (Lifschitz, 1996). These results suggest that as in Gerbera, a heterodimer between AGL2-like and C-class MADS box gene products could be needed to fulfill the C function in whorl 3 in tomato. Generalization of these findings in Gerbera and tomato to other angiosperms (e.g., Arabidopsis, Antirrhinum, rice, and maize) would lead to an advanced understanding of the generality of the ABC model, enhanced by including interactions with other regulatory functions within the large family of MADS box factors.
METHODS
Plant Material
Gerbera hybrida var Terra Regina used in this research was obtained from Terra Nigra BV, De Kwakel, Holland. Plants were grown under standard greenhouse conditions.
Isolation of Plant DNA and RNA
Plant DNA was isolated by the method of Dellaporta et al. (1983). Total RNA was isolated as described in Jones et al. (1985) or by using the RNeasy Plant total RNA kit (Qiagen, Chatsworth, CA). Poly(A)+ RNA was isolated by using oligo(dT) cellulose affinity chromatography (Sambrook et al., 1989).
Construction and Differential Screening of a Petal cDNA Library
Polyadenylated RNA (5 μg), extracted from the proximal part of ray floret petals at developmental stages 5 to 9 (Helariutta et al., 1993), was used to construct a cDNA library in the λ ZAPII vector (ZAP-cDNA synthesis kit; Stratagene, La Jolla, CA). From the nonamplified cDNA library, ∼50,000 plaques were plated and transferred onto replica nylon membranes and then screened differentially with radiolabeled first-strand cDNA pools from the proximal part of the ray floret tube region and the distal part of the ligule (First-Strand cDNA synthesis kit; Amersham Corp.).
GRCD1 cDNA was isolated as a clone expressed at early stages of petal development. A nearly full-length (20 nucleotides missing from 5′ end) cDNA clone was isolated, subcloned into a pUC18 derivative, and sequenced using the AutoRead kit (Pharmacia, Uppsala, Sweden). The missing 5′ nucleotides were amplified by single-sided polymerase chain reaction (PCR) from genomic DNA, and the full-length cDNA was amplified once more by PCR from first-strand cDNA corresponding to RNA isolated from young capitula and sequenced. The GRCD1 gene has been submitted to the EMBL database (accession number AJ400623).
RNA and DNA Gel Blot Analyses and in Situ Hybridization
To each lane was loaded 15 μg of total RNA or 10 μg of digested total genomic DNA. The electrophoresis and hybridizations were as described in Sambrook et al. (1989). The 200-bp-long 3′ fragment (of which 149 bp is from a noncoding region) served as the probe. Washing conditions of 0.2 × SSC (1 × SSC is 0.15 M NaCl, 0.015 M sodium citrate) and 0.1% SDS at 58°C were applied in all RNA and DNA gel blot analyses. In situ hybridization was performed as described previously (Kotilainen et al., 1994) using 35S-CTP–labeled antisense and sense (control) RNA probes. The probes were transcribed from the same fragment used in blot studies under the T7 promoter in vector pSP73 (SP6/T7 transcription kit; Roche Diagnostics, Mannheim, Germany).
Plant Transformation
Gerbera transformation was performed with Agrobacterium-mediated gene transfer basically as described previously (Elomaa et al., 1993, 1998). Three transformants with lowered GRCD1 expression and changed phenotype as described in Results were obtained. Transformation was verified by DNA gel blot analysis, and the antisense effect was demonstrated by RNA gel blot and in situ analyses showing downregulation of GRCD1 expression in young, developing capitula of the transformants. The analyses were performed on clones of the original transgenic plants (T0).
Scanning Electron Microscopic Analysis
After the samples were fixed overnight in a buffer of 50% ethanol, 5% acetic acid, and 2% formaldehyde, they were transferred through ethanol series to 10% ethanol, critical-point–dried (Balzers CPD 020 critical point dryer; Bal-Tec, Balzers, Liechtenstein), and coated with platinum/palladium (agar sputter coater; Agar Scientific, Stansel, UK). Specimens were mounted on aluminum stubs by using graphite adhesive or tape and examined with a scanning electron microscope (digital scanning microscope model DSM 962; Karl Zeiss, Oberkochen, Germany) at the Institute of Biotechnology, Electron Microscopy Laboratory, University of Helsinki.
Phylogenetic Analyses
The parsimony jackknife approach to tree construction was used (Farris et al., 1996). The computer application Xac, written by J.S. Farris, was used to compute the parsimony jackknife support tree. The 1000 jackknife replicates were tested with branch-swapping and five random restarts per replicate. Previously published plant MADS box gene sequences were retrieved from the GenBank database: DEF (accession number X52023), DEFH49 (X95467), DEFH72 (X95468), DEFH200 (X95469), GLO (X68831), PLE (S53900), and SQUA (X63701) from Antirrhinum majus; AG (X53579), AGL2 (M55551), AGL4 (M55552), AGL6 (M55554), AGL8 (U33473), AGL9 (AF015552), AGL15 (U22528), AP1 (Z16421), AP3 (D21125), and PI (D30807) from Arabidopsis thaliana; OM1 (X69107) from Aranda × Deborah; AGL15-1 (U22665) from Brassica napus; EGM1 (AF029975) and EGM3 (AF029977) from Eucalyptus grandis; GAGA1 (AJ009722), GAGA2 (AJ009723), GDEF1 (AJ009724), GDEF2 (AJ009725), GGLO1 (AJ009726), and GSQUA1 (AJ009727) from Gerbera hybrida; MDMADS1 (U78947), MDMADS3 (AF068722), MDMADS4 (U78950), and MDMADS11 (AJ000763) from Malus domestica; TM3 (X60756), TM4 (X60757), TM5 (X60480), TM6 (X60759), and TM8 (X60760) from Lycopersicon esculentum; NSMADS3 (AF068722) from Nicotiana sylvestris; TOBMADS (X76188) from N. tabacum; OSMADS8 (U78892) and OSMADS45 (U31994) from Oryza sativa; FBP2 (M91666) and FBP7 (X81651) from Petunia hybrida; DAL1 (X80902) and DAL3 (X79281) from Picea abies; PRMADS2 (U42400) from Pinus radiata; PEAMTF1 (AJ223318) from Pisum sativum; RBPI-1 (AF052859) from Rumex acetosa; SLM2 (X80489) and SLM5 (X80492) from Silene latifolia; SAMADSD (Y08626) from Sinapsis alba; ZAG3 (L46397), ZAG5 (L46398), ZMM8 (Y09303), and ZMM14 (AJ005338) from Zea mays.
Yeast Two-Hybrid Analysis
Yeast two-hybrid analysis was performed by using the MATCHMAKER LexA two-hybrid system (Clontech, Palo Alto, CA). The fusion protein plasmids were constructed as follows: the full coding sequences of genes GRCD1, GAGA1, and GAGA2 were amplified by PCR with cDNA clones as templates. PCR fragments were purified (High Pure PCR product purification kit; Roche Diagnostics) and digested at their ends by restriction enzymes for subcloning into plasmids. Yeast transformation and selection of transformants were done according to the manufacturer's instructions. The transformants were grown on Gal/Raff induction plates (MATCHMAKER LexA two-hybrid system; Clontech, Palo Alto, CA), suspended into Z buffer (100 mM Na-PO4, 10 mM KCl, 1 mM Mg SO4, 50 mM B-mercaptoethanol, pH 7.0), and assayed for β-galactosidase activity with o-nitrophenyl β-d-galactopyranoside as substrate according to the manufacturer's protocol. β-Galactosidase was assayed in 7 to 12 individual transformants for each combination. β-Galactosidase activity was calculated according to Miller (1992).
NOTE ADDED IN PROOF
While this manuscript was in press, Pelaz et al. (Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E., and Yanovsky, M.F. [2000]. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203) published that the MADS-box genes SEP1/2/3 (formerly known as AGL2/4/9) are required for the B and C floral organ identity functions in Arabidopsis flower development.
Acknowledgments
This work was supported by the Academy of Finland (Grant No. 44315). The Xac parsimony jackknifing application was kindly provided by its author, James S. Farris. Yrjö Helariutta and two anonymous reviewers provided useful comments on the manuscript. We thank Eija Takala, Marja Huovila, and Anu Rokkanen for excellent technical assistance. We also thank Jaap Molenaar (Terra Nigra B.V. Holland) for providing a supply of plant material; Sanna Peltola, Eija Saarikko, and Anne Aaltonen for greenhouse care of plant material; Jyrki Juhanoja for his help in electron microscopy; and Deyin Guo for his comments on β-galactosidase assays.
References
- Angenent, G.C., Bussher, M., Franken, J., Mol, J.N.M., and van Tunen, A. (1992). Differential expression of two MADS box genes in wild-type and mutant petunia flowers. Plant Cell 4, 983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent, G.C., Franken, J., Busscher, M., Weiss, D., and van Tunen, A.J. (1994). Cosuppression of the petunia homeotic gene FBP2 affects the identity of the generative meristem. Plant J. 5, 33–44. [DOI] [PubMed] [Google Scholar]
- Angenent, G.C., Franken, J., Busscher, M., van Dijken, A., van Went, J.L., Dons, H.J.M., and van Tunen, A.J. (1995). A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell 7, 1569–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner, P., and Boutin, J.-P. (1998). A MADS box transcription factor of the AP1/AGL9 subfamily is also expressed in the seed coat of pea (Pisum sativum) during development. Plant Mol. Biol. 38, 1253–1255. [DOI] [PubMed] [Google Scholar]
- Coen, E.S., and Meyerowitz, E.M. (1991). The war of the whorls: Genetic interaction controlling flower development. Nature 353, 31–37. [DOI] [PubMed] [Google Scholar]
- Davies, B., Egea-Cortines, M., de Andrade Silva, E., Saedler, H., and Sommer, H. (1996). Multiple interactions amongst floral homeotic MADS box proteins. EMBO J. 15, 4330–4343. [PMC free article] [PubMed] [Google Scholar]
- Davies, B., Motte, P., Keck, E., Saedler, H., Sommer, H., and Schwarz-Sommer, Z. (1999). PLENA and FARINELLI: Redundancy and regulatory interactions between two Antirrhinum MADS box factors controlling flower development. EMBO J. 18, 4023–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA minipreparation. Version II. Plant Mol. Biol. Rep. 1, 19–21. [Google Scholar]
- Doyle, J.J. (1994). Evolution of a plant homeotic multigene family: Toward connecting molecular systematics and molecular developmental genetics. Syst. Biol. 43, 307–328. [Google Scholar]
- Eckermann, S., Schröder, G., Schmidt, J., Strack, D., Edrada, R.A., Helariutta, Y., Elomaa, P., Kotilainen, M., Kilpeläinen, I., Proksch, P., Teeri, T.H., and Schröder, J. (1998). New pathway to polyketides in plants. Nature 396, 387–390. [Google Scholar]
- Elomaa, P., Honkanen, J., Puska, R., Seppänen, P., Helariutta, Y., Mehto, M., Kotilainen, M., Nevalainen, L., and Teeri, T.H. (1993). Agrobacterium-mediated transfer of antisense chalcone synthase cDNA to Gerbera hybrida inhibits flower pigmentation. Bio/Technology 11, 508–511. [Google Scholar]
- Elomaa, P., Mehto, M., Kotilainen, M., Helariutta, Y., Nevalainen, L., and Teeri, T.H. (1998). A bHLH transcription factor mediates organ, region and flower type specific signals on dihydroflavonol-4-reductase (dfr) gene expression in the inflorescence of Gerbera hybrida (Asteraceae). Plant J. 16, 93–100. [DOI] [PubMed] [Google Scholar]
- Fan, H.-Y., Hu, Y., Tudor, M., and Ma, H. (1997). Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant J. 12, 999–1010. [DOI] [PubMed] [Google Scholar]
- Farris, J.S., Albert, V.A., Källersjö, M., Lipscomb, D., and Kluge, A.G. (1996). Parsimony jackknifing outperforms neighbor-joining. Cladistics 12, 99–124. [DOI] [PubMed] [Google Scholar]
- Flanagan, C.A., and Ma, H. (1994). Spatially and temporally regulated expression of the MADS box gene AGL2 in wild-type and mutant Arabidopsis flowers. Plant Mol. Biol. 26, 581–595. [DOI] [PubMed] [Google Scholar]
- Goto, K., and Meyerowitz, E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene pistillata. Genes Dev. 8, 1548–1560. [DOI] [PubMed] [Google Scholar]
- Helariutta, Y., Elomaa, P., Kotilainen, M., Seppänen, P., and Teeri, T.H. (1993). Cloning of cDNA coding for dihydroflavonol-4-reductase (DFR) and characterization of dfr expression in the corollas of Gerbera hybrida var. Regina (Compositae). Plant Mol. Biol. 22, 183–193. [DOI] [PubMed] [Google Scholar]
- Hill, T.A., Day, C.D., Zondlo, S.C., Thackeray, A.G., and Irish, V.F. (1998). Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development 125, 1711–1721. [DOI] [PubMed] [Google Scholar]
- Jack, T., Brockman, L.L., and Meyerowitz, E.M. (1992). The homeotic gene apetala3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G., Duismuir, P., and Bedbrook, J. (1985). High level expression of introduced chimeric genes in regenerated transformed plants. EMBO J. 4, 2411–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotilainen, M., Helariutta, Y., Elomaa, P., Paulin, L., and Teeri, T.H. (1994). A corolla- and carpel-abundant, non-specific lipid transfer protein gene is expressed in the epidermis and parenchyma of Gerbera hybrida var. Regina (Compositae). Plant Mol. Biol. 26, 971–978. [DOI] [PubMed] [Google Scholar]
- Kotilainen, M., Helariutta, Y., Mehto, M., Pöllänen, E., Albert, V.A., Elomaa, P., and Teeri, T.H. (1999). GEG participates in the regulation of cell and organ shape during corolla and carpel development in Gerbera hybrida. Plant Cell 11, 1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz, E. (1996). Flowers, leaves and inflorescences: An integrated approach. Flowering Newsl. 21, 28–33. [Google Scholar]
- Mandel, M.A., and Yanofsky, M.F. (1998). The Arabidopsis AGL9 MADS box gene is expressed in young flower primordia. Sex. Plant Reprod. 11, 22–28. [Google Scholar]
- Miller, J.H. (1992). A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Pnueli, L., Abu-Abeid, M., Zamir, D., Nacken, W., Schwarz-Sommer, Z., and Lifschitz, E. (1991). The MADS box gene family in tomato: Temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant Cell 1, 255–266. [PubMed] [Google Scholar]
- Pnueli, L., Hareven, D., Broday, L., Hurwitz, C., and Lifschitz, E. (1994. a). The TM5 MADS box gene mediates organ differentiation in the three inner whorls of tomato flowers. Plant Cell 6, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli, L., Hareven, D., Rounsley, S.D., Yanofsky, M.F., and Lifschitz, E. (1994. b). Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell 6, 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan, M.D., Rounsley, S.D., Schmidt, R.J., and Yanofsky, M.F. (1995). Molecular evolution of flower development: Diversification of the plant MADS-box regulatory gene family. Genetics 140, 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Savidge, B., Rounsley, S.D., and Yanofsky, M.F. (1995). Temporal relationship between the transcription of two Arabidopsis MADS box genes and the floral organ identity genes. Plant Cell 7, 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer, Z., Huijser, P., Nacken, W., Saedler, H., and Sommer, H. (1990). Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 250, 931–936. [DOI] [PubMed] [Google Scholar]
- Sieburth, L.E., Running, M.P., and Meyerowitz, E.M. (1995). Genetic separation of third and fourth whorl functions of AGAMOUS. Plant Cell 7, 1249–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandre, K., Albert, V.A., Sundås, A., and Engström, P. (1995). Conifer homologues to genes that control floral development in angiosperms. Plant Mol. Biol. 27, 69–78. [DOI] [PubMed] [Google Scholar]
- Theissen, G., Kim, J.T., and Saedler, H. (1996). Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 43, 484–516. [DOI] [PubMed] [Google Scholar]
- Yanofsky, M.F., Ma, H., Bowman, J.L., Drews, G.N., Feldman, K.A., and Meyerowitz, E.M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. [DOI] [PubMed] [Google Scholar]
- Yu, D., Kotilainen, M., Pöllänen, E., Mehto, M., Elomaa, P., Helariutta, Y., Albert, V.A., and Teeri, H. (1999). Organ identity genes and modified patterns of flower development in Gerbera hybrida (Asteraceae). Plant J. 17, 51–62. [DOI] [PubMed] [Google Scholar]