Abstract
Interactions between the nuclear matrix and special regions of chromosomal DNA called matrix attachment regions (MARs) have been implicated in various nuclear functions. We have identified a novel protein from wheat, AT hook–containing MAR binding protein1 (AHM1), that binds preferentially to MARs. A multidomain protein, AHM1 has the special combination of a J domain–homologous region and a Zn finger–like motif (a J-Z array) and an AT hook. For MAR binding, the AT hook at the C terminus was essential, and an internal portion containing the Zn finger–like motif was additionally required in vivo. AHM1 was found in the nuclear matrix fraction and was localized in the nucleoplasm. AHM1 fused to green fluorescent protein had a speckled distribution pattern inside the nucleus. AHM1 is most likely a nuclear matrix component that functions between intranuclear framework and MARs. J-Z arrays can be found in a group of (hypothetical) proteins in plants, which may share some functions, presumably to recruit specific Hsp70 partners as co-chaperones.
INTRODUCTION
The nuclear matrix, operationally defined, is the dynamic fibrogranular structure forming the skeletal framework that surrounds and penetrates the interphase nucleus; it has been implicated in most nuclear functions, including replication, repair, transcription, RNA processing, and RNA transport (Berezney and Jeon, 1995). The chromosomal DNAs are known to be associated with the nuclear matrix at specific regions called matrix attachment regions (MARs) and are thereby thought to be organized into topologically constrained loops, each of which represents a sort of functional or structural domain (or both) (Laemmli et al., 1992; Bode et al., 1996). In animals, interactions between the nuclear matrix and MARs have also been shown to be involved in DNA replication and repair and in various aspects of gene regulation, thus playing a key role in the essential functions of the nucleus (Boulikas, 1995; Bode et al., 1996).
MARs consist of AT-rich sequences extending over at least a few hundred base pairs and containing various AT-rich motifs as well as structural motifs such as base-unpairing regions and intrinsically curved portions (Boulikas, 1995). In animals, MAR binding activity has been found in a wide range of structurally and functionally diverse proteins (listed in Boulikas, 1995). These include topoisomerase II (Adachi et al., 1989); filament proteins such as lamins and NuMA (Ludérus et al., 1992, 1994); ARBP, identical to a methylated CpG binding protein (von Kries et al., 1991; Weitzel et al., 1997); hnRNP-U/SAF1, an RNA binding protein involved in RNA processing (Fackelmayer et al., 1994; von Kries et al., 1994); SATB1, a tissue-specific transcription factor with high affinity for base-unpairing regions (Dickinson et al., 1992; de Belle et al., 1998); and architectural chromatin proteins such as histone H1 and HMG-I/Y (Zhao et al., 1993).
In plants, MARs have been found in intergenic regions, often in or close to the regulatory regions of genes, and close to the integration sites of transgenes (Holmes-Davis and Comai, 1998). Mapping MARs along a long stretch of chromosomal DNA in cereals and Arabidopsis showed that each of the potential loops defined by two MARs contained a single gene or a subset of genes—findings that support the loop model (Avramova et al., 1995, 1998; van Drunen et al., 1997; Tikhonov et al., 2000). Transgenic studies have indicated that MARs improve the amount of expression, and sometimes the stability, of flanking transgenes, although the results have depended on the systems used and the MARs examined (reviewed in Spiker and Thompson, 1996; Holmes- Davis and Comai, 1998). Growing evidence indicates that nuclear matrix–MAR interactions are also important in plants at higher levels of gene regulation as well as chromosomal organization.
Plant nuclear matrices are morphologically similar to vertebrate matrices (reviewed in Moreno Díaz de la Espina, 1995) and contain proteins antigenically related to animal nuclear matrix components, such as topoisomerase II, NuMA, and lamins (e.g., Mínguez and Moreno Díaz de la Espina, 1993; Yu and Moreno Díaz de la Espina, 1999). However, plant nuclear matrices are thought to be organized differently from vertebrate matrices, at least in part, based on the complex protein composition and greater stability of plant matrices and the distribution of some intermediate filament-related proteins (reviewed in Moreno Díaz de la Espina, 1995). Two kinds of filament-like proteins, tomato MFP1 (Meier et al., 1996; Gindullis and Meier, 1999) and carrot NMCP1 (Masuda et al., 1997), and the MFP1-interacting protein MAF1 (Gindullis et al., 1999) have been identified as nuclear matrix proteins localized to the peripheral region of the nucleus. MFP1 is also a MAR binding protein and is thought to be involved in the organization of chromosomal DNAs (Meier et al., 1996; Gindullis and Meier, 1999). Recently, two MAR binding proteins with homology to nucleolar proteins have been found in the pea nuclear matrix (Hatton and Gray, 1999). Thus, our knowledge of the molecular basis of the nuclear matrix–MAR interactions is still fragmentary. To further understand the nuclear matrix–MAR interactions in plants, we screened for nuclear matrix–localized MAR binding proteins. Here, we report the isolation and characterization of a novel type of nuclear protein, AT hook–containing MAR binding protein1 (AHM1), which could potentially function to connect the nuclear framework and MARs.
RESULTS
Isolation of cDNA and Genomic Clones of AHM1
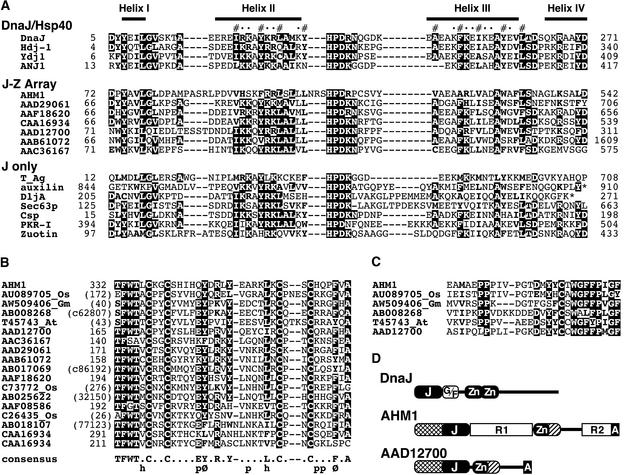
The WCI-3b MAR found in the upstream region of a winged bean chymotrypsin inhibitor gene has characteristics typical of MARs (Minami et al., 1999). This MAR was radiolabeled and used as a probe to isolate wheat cDNAs encoding potential MAR binding proteins by screening a λgt11 expression library. Thirteen positive phages were isolated from ∼106 recombinants. The cDNA-encoded proteins were expressed in lysogens as β-galactosidase fusions, separated by SDS-PAGE, and subjected to a MAR binding assay. Seven clones exhibited MAR binding activity on a filter, from which two that expressed larger β-galactosidase fusions were finally selected (λAH8 and λAH10). Sequencing revealed that both encoded novel proteins contained an AT hook, so we named them AHM1 and AHM2 (Figure 1). As a result of the structural novelty and the MAR binding activity displayed in the yeast one-hybrid assay (detailed below), we focused on AHM1.
Figure 1.
Amino Acid Sequences of AHM1 and AHM2.
(A) Amino acid sequence of AHM1 deduced from the sequences of the genomic and cDNA clones. A J domain–like region (positions 72 to 143), a Zn finger–like motif (positions 332 to 366), and a C-terminal basic region (positions 517 to 539) containing an AT hook are boxed. Key amino acids in their respective motifs are highlighted in black. Direct repeats (R1, positions 151 to 315; R2, positions 439 to 516) are indicated with arrows. Internal extra amino acids in the first and second repeats of R2 are indicated with broken lines. A conserved region (positions 367 to 393; see Figure 2C) following the Zn finger–like motif is double underlined.
(B) Partial amino acid sequence of AHM2 deduced from the sequence of a cDNA clone. A direct repeat (positions 138 to 221) and an AT hook (positions 220 to 231) are marked as in (A).
Using the cDNA insert of λAH8 as a probe, we isolated and sequenced additional cDNA clones and a corresponding genomic clone. The transcription initiation site was deduced from the sequence of the longest clones, which were repeatedly isolated in independent experiments of rapid amplification of cDNA 5′ ends (5′ RACE). Comparisons between the genomic and cDNA sequences revealed that the Ahm1 gene contains no introns and encodes a 542–amino acid protein, assuming the first ATG in the sequence is the initiation codon. This estimation was in good agreement with the size of the immunologically detected AHM1 protein. In DNA gel blot analysis, two to four bands hybridizing to the 3′ half of the Ahm1 gene (corresponding to amino acid positions 332 to 527) were detected when the genomic DNA was first digested with BamHI, EcoRI, or HindIII (data not shown). Because wheat is hexaploid, Ahm1 appears to be a single-copy gene per haploid. No typical TATA sequence was found at an appropriate distance from the deduced transcription initiation site. The DDBJ/EMBL/GenBank accession numbers for the genomic sequence of AHM1 and for the cDNA sequence of AHM2 are AB030708 and AB030709, respectively.
AHM1 I a Member of the Plant-Specific J Domain Protein Subfamily
The amino acid sequence of the AHM1 protein deduced from the nucleotide sequence has several distinct regions, as shown in Figure 1. A considerable part (∼44%) of the molecule consists of two kinds of imperfect tandem repeats: R1 (positions 151 to 315), containing 11 copies of a 15–amino acid unit [consensus sequence PPPQSSPL(Q/R)AATQPP], and R2 (positions 439 to 516), containing five copies of a 14–amino acid unit (consensus sequence EVPAAPEPVMPTRV). The first and second repeats of R2 have an extra four amino acids internally. No related repeats could be found in the databases. The second repeat of R2 is followed immediately by a short basic region containing an AT hook, which is known to interact with the minor groove of AT-rich stretches of B-form DNA (Reeves and Nissen, 1990; Huth et al., 1997). Such a combination—a tandem repeat plus an AT hook—is also found in AHM2, although the sequence of its repeat unit (consensus sequence GKEPISSETTKQ) differs from that of R2 (Figure 1B).
The N-terminal (positions 14 to 143) and middle (positions 332 to 393) portions of AHM1, just flanking R1, are similar to the respective portions of a group of (hypothetical) proteins in Arabidopsis, rice, soybean, and others (Figure 2). The former includes a region of ∼70 amino acids (positions 72 to 143; Figure 2A) with similarity to so-called J domains. The J domains, known to interact with Hsp70/Hsc70 chaperones, are found in the DnaJ/Hsp40 family members and also in other structurally and functionally different proteins (J only, in Figure 2A) (Kelley, 1998; Mayer and Bukau, 1998). As evidenced from the alignment shown in Figure 2A, the J domain–like region of AHM1 has considerably diverged from the canonical J domains of Escherichia coli DnaJ and its close relatives. However, the divergence is similar to the difference seen with the J domains of simian virus 40 (SV40) and other polyomaviral T antigens, which can functionally substitute for the DnaJ J domain in E. coli (Kelley and Georgopoulos, 1997). Furthermore, secondary structure prediction for AHM1 did not deny the folding pattern resolved with J domain polypeptides derived from DnaJ and human Hdj-1 (Pellecchia et al., 1996; Qian et al., 1996). In particular, hydrophobic residues participating in the formation of a short coiled-coil between helices II and III and the three–amino acid sequence HPD in the resulting loop are well conserved (Figure 2A). The helix II and HPD-containing loop have been shown to interact directly with the ATPase domain of the Hsp70 partner (Greene et al., 1998; Suh et al., 1998). Taken together, these findings support AHM1 as a member of the J domain protein family.
Figure 2.
Comparisons of AHM1-Related Proteins.
(A) An alignment of J domains. J domain proteins can be divided into three groups: DnaJ/Hsp40, which includes the canonical DnaJ and closely related homologs; J-Z array, which includes AHM1 and selected Arabidopsis proteins (shown with their GenBank accession numbers) having the Zn finger–like motif shown in (B); and J only, a diverse set of proteins that have no important similarity outside of the J domain. Above the alignment, four α-helical regions resolved in DnaJ (Pellecchia et al., 1996) are shown. Hydrophobic residues participating in the short coiled-coil structure between helices II and III and solvent-accessible residues are indicated with (#) and (•), respectively. Conserved residues (>50% in the alignment) are highlighted, and related residues (hydrophobic or basic) are in boldface. Asterisks denote the ends of proteins. Amino acid positions of the N-terminal residue in the alignment are shown at left. The total length of each protein is shown at right. GenBank accession numbers are as follows: DnaJ, P08622; human Hdj-1, P25685; yeast Ydj1, P25491; Atriplex nummularia ANJ1, P43644; SV40 T antigen (T_Ag), P03070; bovine auxilin, Q27974; E. coli DljA, P31680; yeast Sec63p, P14906; mouse cysteine-string protein (Csp), P54101; bovine double-stranded RNA-induced protein kinase inhibitor (PKR-I), AAA17795; and yeast zuotin, P32527.
(B) An alignment of the Zn finger–like motif. Names of hypothetical proteins are denoted by GenBank accession numbers. Expressed sequence tag clones from rice, soybean, and Arabidopsis are indicated with the respective suffixes Os, Gm, and At after their accession numbers. The others were predicted from the Arabidopsis genomic sequence. Conserved residues are highlighted. The consensus sequence deduced is shown at the bottom; h, p, and Ø denote hydrophobic, polar, and aromatic residues, respectively. Sequences of hypothetical proteins that do not have GenBank accession numbers are indicated with nucleotide positions shown in parentheses. c denotes translation of the complementary strand.
(C) An alignment of the conserved region immediately adjoining the Zn finger–like motif. The proteins aligned are the same as the first six proteins shown in (B). No amino acids are present between the sequences shown in (B) and (C) (see Figure 1A for AHM1). Conserved and related residues are highlighted as in (A).
(D) Topological comparison of DnaJ, AHM1, and an Arabidopsis protein (AAD12700). Homologous regions of AHM1 and AAD12700, adjacent to the N terminus of the J domainlike region and adjacent to the C terminus of the Zn fingerlike motif, are indicated with the same hatch patterns. A, AT hook; Gly/Phe-rich linker region; J, J domain–like region; G/F, Zn, Zn finger–like motif.
The middle portion includes a region (positions 332 to 366) with four Cys residues arranged in the pattern CX2CX16CX2C, reminiscent of a Zn finger motif, although the primary sequence around this motif did not fulfill any of the known Zn finger motif signatures. Each member of a group of proteins predicted from the genomic and expressed sequence tag sequences of Arabidopsis, soybean, and rice has a region homologous to the Zn finger–like motif found in AHM1, in which several residues are highly conserved in addition to the four Cys residues (Figure 2B). Thus, the Zn finger–like motif is expected to act as a structurally distinct module. Most of these predicted proteins have a J domain–like region toward the N terminus of the Zn finger–like motif with or without a linker-like region in between, which suggests that these two regions may function in a cooperative manner. Such a combination, hereafter referred to as a J-Z array, represents a subset of plant-specific J domain proteins. The proteins containing a J-Z array probably share a common function, possibly serving as co-chaperones.
Some of the putative proteins with a Zn finger–like motif share additional similarity to AHM1 in the region immediately following the motif (Figure 2C). Interestingly, an AT hook was found in one of these proteins (GenBank accession number AAD12700) at the C terminus (Figure 2D; see Discussion).
MAR Binding Activity of AHM1
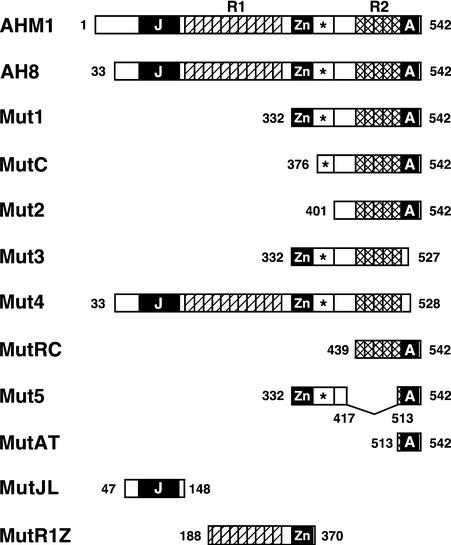
Based on the structural features described above, various portions of AHM1 (Figure 3) were expressed in E. coli as a His-tagged protein to determine the region or regions relevant to MAR binding. Expressed proteins were purified and subjected to an in vitro MAR binding assay with WCI-3b MAR as a probe. As shown in Figure 4A, all derivatives with the same C terminus as AHM1 were capable of binding to the MAR (see His-AH8, -Mut1, -MutC, -Mut2, -MutRC, and -Mut5), whereas a deletion of only 14 or 15 amino acids from the C terminus (His-Mut3 and -Mut4), which disrupts the AT hook, abolished MAR binding activity. Thus, the C-terminal basic region was the only portion absolutely required for MAR binding activity of AHM1 in vitro, and the rest—including the J domain–like region, R1, the Zn finger–like motif, and R2—were dispensable. However, a very faint signal was detected for His-Mut3 in some experiments.
Figure 3.
Schematic Representation of AHM1 and Its Derivatives.
The molecular organization of AHM1 is shown schematically at the top. Shown below are AHM1 derivatives expressed in E. coli as a His-tagged protein and/or in yeast as a fusion with the Gal4p activation domain. Amino acid positions at the junctions and ends are indicated. The J domain–like region (J), the Zn finger–like motif (Zn), and the C-terminal basic region containing an AT hook (A) are indicated by black boxes; each unit of direct repeats (R1 and R2) is indicated by a hatched or cross-hatched box. The conserved region immediately following the Zn finger–like motif is indicated by an asterisk.
Figure 4.
MAR Binding Activity of AHM1 in Vitro.
(A) Dissection of AHM1. His-tagged AHM1 derivatives were purified and separated on an SDS–12% polyacrylamide gel and then visualized with Coomassie blue staining (CBB) or subjected to a filter binding assay with WCI-3b MAR as a probe (MAR binding). Positions of molecular mass markers are shown at left in kilodaltons. The positions of His-Mut3 and His-Mut4 are marked with arrowheads.
(B) Importance of the AT hook in MAR binding. Total extracts of E. coli expressing His-Mut1 and His-Mut1(R528K) were treated as in (A). IPTG denotes the extract derived from uninduced E. coli cells harboring pET-Mut1. The position of His-Mut1 is indicated by the arrowhead at right.
To clarify the involvement of the AT hook in MAR binding, we introduced a point mutation to substitute Lys for Arg528, located at the center of the absolutely conserved three–amino acid sequence, GRP, in AT hooks (Aravind and Landsman, 1998). The corresponding Arg residue in HMG- I/Y has been shown to participate directly in the interaction with AT base pairs (Huth et al., 1997). The resulting mutant protein, His-Mut1(R528K), showed much less MAR binding activity (Figure 4B); therefore, the AT hook, not simple basicity, is responsible for the ability of AHM1 to bind to MAR.
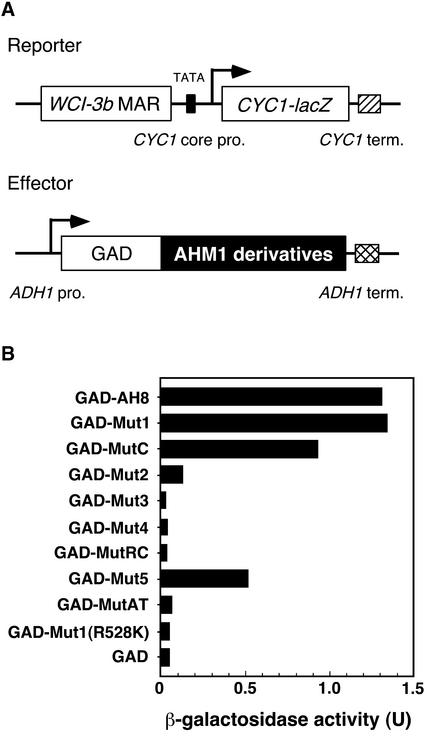
To examine MAR binding activity in vivo, we expressed AHM1 derivatives fused to the activation domain of Gal4p (GAD) in a yeast strain carrying the CYC1–lacZ reporter with the WCI-3b MAR upstream of the promoter (Figure 5A). If a GAD–AHM1 fusion protein binds to the MAR and activates the promoter, then the amount of β-galactosidase activity detected is increased. As shown in Figure 5B, the presence of the AT hook was essential (see GAD-Mut3, -Mut4, and -Mut1[R528K]), but the J domainlike region and R1 were dispensable (cf. GAD-AH8 and -Mut1), consistent with the results of the in vitro binding assay. However, as the deletion was extended from position 332 toward the C terminus, the amount of reporter gene expression decreased (cf. GAD- Mut1, -MutC, and -Mut2), being reduced to almost background values (cf. GAD-MutRC and -MutAT with GAD). When the internal portion was fused directly to the AT hook (GAD-Mut5), considerable reporter gene expression was detected. Thus, in yeast, the AT hook alone was insufficient, and the internal portion between the two repeat regions R1 and R2 was also required for MAR binding. Consistent with these results, the MAR binding activity of AHM2, which has a single AT hook immediately following a tandem repeat, was detected in vitro but not in yeast (data not shown).
Figure 5.
MAR Binding Activity of AHM1 in Yeast.
(A) Schematic representation of the effector and reporter genes. The CYC1-lacZ reporter gene carries the WCI-3b MAR upstream of the CYC1 minimum promoter (CYC1 core pro.). The effectors used are the activation domain of Gal4p (GAD) alone or GAD fused with AHM1 derivatives shown in Figure 3. The effector genes are under the control of the ADH1 promoter (ADH1 pro.). Terminators (term.) are shown by a hatched or cross-hatched box.
(B) β-Galactosidase activity in yeast cells carrying the reporter construct and an indicated effector construct. U, specific activity of β-galactosidase expressed as nanomoles catalyzed per minute per microgram of protein.
Mode of the AHM1–MAR Interaction
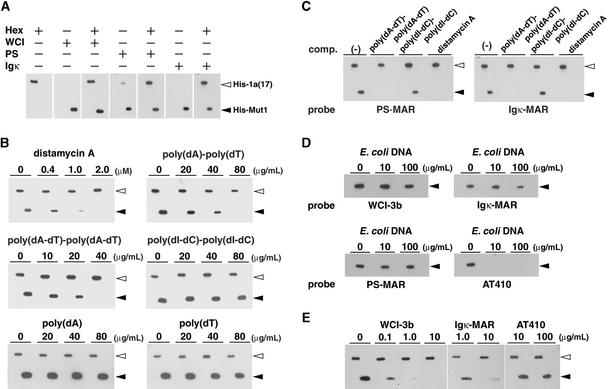
To determine the binding specificity of AHM1, we examined the effects of AT-rich polynucleotide competitors and distamycin A, an antibiotic known to interact specifically with the minor groove of AT-rich DNA with little conformational change (Kopka et al., 1985). To compare the results from different filters, we used the interaction between the wheat basic leucine zipper (bZIP) protein HBP1a(17) and its high-affinity binding site Hex (CCACGTCA) (Meshi et al., 1998) as a reference. As shown in Figure 6A, WCI-3b MAR and Hex probes specifically bound to His-Mut1 and His-tagged HBP1a(17) (His-1a[17]), respectively.
Figure 6.
DNA Binding Specificity of AHM1.
(A) MAR binding activity of AHM1. His-tagged HBP1a(17) (His-1a[17]) and His-Mut1, 0.1 μg each, were electrophoresed side by side and subjected to the filter binding assay with the 32P-labeled, 31-bp-long oligonucleotide Hex31, which contained Hex (CCACGTCA), a high-affinity binding site for HBP-1a(17), with or without WCI-3b MAR (WCI), the MAR-1 of Arabidopsis PS locus (PS), or Igκ MAR (Igκ) as a probe. The probes included in the reaction are indicated with (+). The length of MARs used and their AT content (in parentheses) are as follows: WCI-3b MAR, 0.88 kb (81%); PS MAR, 1.3 kb (70%); and Igκ MAR, 0.44 kb (72%).
(B) Effects of competitors. The MAR binding assay was performed as in (A) in the presence of an increasing amount of distamycin A, poly(dA)·poly(dT), poly(dA-dT)·poly(dA-dT), poly(dI-dC)·poly(dI-dC), poly(dA), or poly(dT). The probes used were a mixture of labeled Hex31 and WCI-3b MAR.
(C) Binding specificity of AHM1 for PS MAR and Igκ MAR. The MAR binding assay was performed as in (A). The probes used were a mixture of Hex31 and either PS MAR or Igκ MAR. Competitors included were 40 μg/mL poly(dA-dT)·poly(dA-dT), 40 μg/mL poly(dI-dC)·poly(dI-dC), or 1 μM distamycin A.
(D) Specific binding of AHM1 to MARs. The binding assay was performed with WCI-3b MAR, Igκ MAR, PS MAR, or AT410 as a probe in the presence of an increasing amount of E. coli DNA. AT410 is a 0.41-kb-long, AT-rich non-MAR fragment (63% AT richness). Only His-Mut1 (0.3 μg) was included in this experiment because the binding of His-1a(17) to Hex31 was inhibited by excess E. coli DNA.
(E) Competition of MAR binding of AHM1 by MARs. The probes used were a mixture of labeled Hex31 and WCI-3b MAR. The MAR binding assay was performed with 0.05 μg each of His-1a(17) and His-Mut1 in the presence of 20 ng/mL E. coli DNA plus an increasing amount of WCI-3b MAR, Igκ MAR, or AT410.
The positions of His-1a(17) and His-Mut1 are shown with open and closed arrowheads, respectively.
When distamycin A was included in the binding reaction, the MAR binding of His-Mut1 was strongly inhibited at ∼1 μM (Figures 6B and 6C). On the other hand, the binding of His-1a(17) was little affected at 2 μM (Figure 6B) or 10 μM (data not shown), consistent with the fact that the bZIP DNA binding domain interacts with the major groove of its recognition site (e.g., Ellenberger et al., 1992). Therefore, AHM1 interacts with the minor groove of DNA. When synthetic polynucleotides with similar minor groove surfaces were included in the binding reaction (Figure 6B), poly(dA-dT)·poly(dA-dT) and poly(dA)·poly(dT) inhibited the binding of His-Mut1 at ∼40 μg/mL, whereas little, if any, competition was observed with poly(dI-dC)·poly(dI-dC) at 80 μg/mL. No competition was observed with single-stranded polynucleotide poly(dA) or poly(dT) (Figure 6B).
His-Mut1 also interacted with the MARs derived from the Arabidopsis plastocyanin gene (PS MAR) and the mouse immunoglobulin κ gene (Igκ MAR) with similar specificity (Figures 6A and 6C). The binding of His-Mut1 to MARs was barely inhibited by excess amounts of E. coli DNA, whereas the binding to an AT-rich, non-MAR fragment (AT410) was effectively inhibited (Figure 6D). The MAR binding efficiently competed with the cognate WCI-3b MAR and Igκ MAR but not with AT410 (Figure 6E). Taken together, AHM1 is a MAR binding protein and is also an AT-rich sequence-directed double-stranded DNA binding protein with some sequence specificity, conformational preference, or both.
Expression and Subcellular Localization of AHM1
Ahm1 mRNA was detected in the poly(A)+ RNA-enriched fraction extracted from wheat seedlings but could not be detected in the total RNA fraction (data not shown). Therefore, the mRNA is likely to be expressed in low amounts under normal growth conditions. The size of the mRNA on an RNA gel blot was ∼2.1 kb, consistent with the sum of the cloned cDNAs. The Ahm1 transcript was detected in both roots and leaves of young seedlings by reverse transcription–polymerase chain reaction analysis (data not shown).
Expression of the AHM1 protein was examined by immunoblot analysis. As shown in Figure 7A (lanes 5 and 8), anti-AHM1 antibodies that had been affinity-purified by means of His-MutJL or His-MutR1Z (Figure 3) detected two closely migrating proteins in the nuclear fraction with an apparent molecular mass of 67 kD. Their mobility was almost the same as that of His-AH8 (Figure 7A, lanes 4 and 7), which is only two amino acids longer than AHM1 predicted from the nucleotide sequence. Therefore, the two bands must represent AHM1. The presence of two bands may result from modification of the protein or may represent a protein derived from a different gene (genome) in wheat. The faster migrating minor bands may be derived from cross-reacting proteins with a similar epitope or from degradation products of AHM1. No signal could be detected in total protein extracts of wheat roots (data not shown).
Figure 7.
Immunoblot Analysis of AHM1.
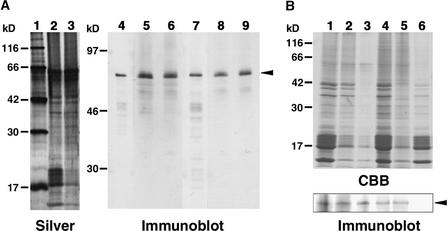
(A) Immunodetection of AHM1. Wheat nuclear proteins and nuclear matrix proteins were extracted 40 hr after imbibition. Proteins were separated on an SDS–12% polyacrylamide gel for silver staining (lanes 1 to 3) or on an SDS–10% polyacrylamide gel for immunoblotting (lanes 4 to 9). Lane 1, molecular mass markers given at left in kilodaltons; lanes 2, 5, and 8, total nuclear proteins; lanes 3, 6, and 9, lithium diiodosalicylate (LIS)–extracted nuclear matrix proteins; and lanes 4 and 7, affinity-purified His-AH8. The amounts of proteins loaded were derived from 1 and 3 A260 units of nuclei for silver staining and immunoblot, respectively. Anti-AHM1 antibodies were affinity-purified with His-MutJL (lanes 4 to 6) or His-MutR1Z (lanes 7 to 9) and used to detect AHM1. An arrowhead indicates the position of AHM1.
(B) AHM1 in the nuclear matrix fraction. Nuclei were prepared 5 days (lanes 1 to 3) or 40 hr (lanes 4 to 6) after imbibition. Proteins were separated on an SDS–12% polyacrylamide gel for Coomassie blue staining (top, CBB) or on an SDS–10% polyacrylamide gel for immunoblotting with the IgG fraction of the anti-AHM1 antibody (bottom, Immunoblot). Lane 1, total nuclear proteins; lanes 2 and 3, LIS-extracted nuclear matrix proteins prepared with (lane 2) or without (lane 3) heat stabilization; lane 4, nuclear proteins after DNase I treatment; lane 5, nuclear matrix proteins prepared from DNase I–treated nuclei by the high-salt extraction method; and lane 6, nuclear proteins solubilized during the high-salt treatment. The amounts of proteins loaded for immunoblot were equivalent to 6 A260 units (lanes 1 to 3) or 4 A260 units (lanes 4 to 6) of nuclei; for CBB staining, they were equivalent to 2 A260 units (lanes 1 to 3) or 1.3 A260 units (lanes 4 to 6) of nuclei. An arrowhead indicates the position of AHM1. Positions of molecular mass markers are indicated at left in kilodaltons.
To determine whether AHM1 is localized in nuclear matrix, we prepared wheat nuclear matrix from the apical portion of young roots by two different methods. As shown in Figures 7A (lanes 6 and 9) and 7B (lane 2), AHM1 was recovered in the lithium diiodosalicylate–extracted nuclear matrix. Most of AHM1 remained in the nuclear matrix fraction that had been prepared without heat stabilization, from which most proteins had been removed (Figure 7B, lanes 1 to 3). The high-salt extraction method yielded a different protein composition for the nuclear matrix (Figure 7B, top, lanes 4 to 6). AHM1 was also recovered in this nuclear matrix fraction but was undetectable in the soluble protein fraction (Figure 7B, bottom, lanes 4 to 6). It is therefore likely that AHM1 is a component of the wheat nuclear matrix.
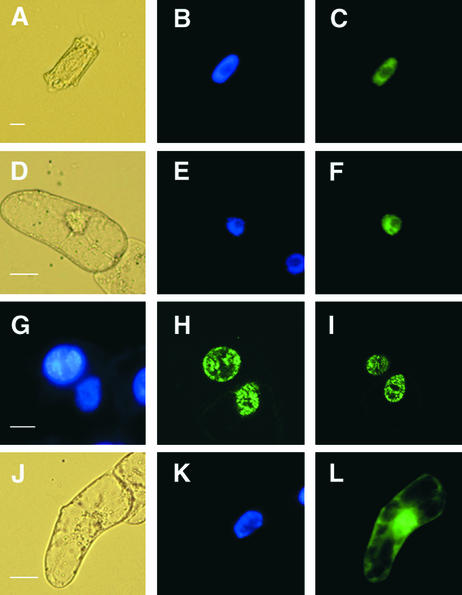
The nuclear localization of AHM1 was confirmed by in situ immunofluorescence microscopy. As shown in Figures 8A to 8C, when permeabilized root cells of wheat seedlings were incubated sequentially with the affinity-purified antibody and a fluorescein isothiocyanate (FITC)–conjugated secondary antibody, the green signal was seen mainly in the nucleoplasm and outside the nucleoli, in good agreement with the pattern obtained by 4′,6-diamidine-2-phenylindole (DAPI) staining. Control staining with the secondary antibody alone showed a low amount of diffuse staining throughout the cell (data not shown).
Figure 8.
Subcellular Localization of AHM1.
(A) to (C) Nuclear localization of AHM1 in a root apical cell of wheat. AHM1 was detected by using an anti-AHM1 antibody (affinity-purified with His-MutJL) and an FITC-conjugated secondary antibody. (A) Phase-contrast image; (B) DAPI staining; (C) FITC-derived fluorescent emission.
(D) to (F) Localization of the G3GFP–AH8 fusion protein in a tobacco BY2 cell. (D) Phase-contrast image; (E) DAPI staining; (F) GFP-derived fluorescent emission.
(G) to (I) Subnuclear distribution of GFP–AH8 in two adjacent BY2 cells. (G) DAPI staining; (H) and (I) two different optical sections of the same field.
(J) to (L) Localization of G3GFP in a BY2 cell. (G) Phase-contrast image; (H) DAPI staining; (I) GFP-derived fluorescent emission.
 ;
;  ;
; 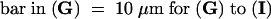 ;
; 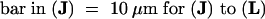 .
.
When a green fluorescent protein (GFP)–AH8 fusion gene driven by the 35S promoter was introduced into tobacco BY2 cells by particle bombardment, the GFP-derived green signal was almost exclusively detected in the nucleoplasm (Figures 8D to 8F), consistent with the results of the immunolocalization. A speckled distribution of GFP–AH8 was observed by confocal microscopy (Figures 8G to 8I). These observations suggest that AHM1 is associated with the intranuclear framework. In contrast, GFP alone was detected in both the cytoplasm and the nucleus (Figures 8J to 8L).
DISCUSSION
AHM1 Is a Single AT Hook Protein
In this work, we have identified a novel type of nuclear matrix protein, AHM1, with high affinity for MARs. We found that a single AT hook at the C terminus is required for binding to MARs both in vitro and in vivo. In addition, an internal portion of the molecule, including the Zn finger–like motif and its C-terminal flanking region, was required for AHM1 to have full binding activity in vivo. Likewise, the MAR binding activity of AHM2, which also has a single AT hook, was detected in vitro but not in yeast. AHM1 was found to interact with the minor groove of MARs, and the central Arg residue in the three–amino acid sequence, GRP, which is conserved among AT hooks, was critical for MAR binding. This provides additional evidence that the Arg side chain is involved in the recognition of AT base pairs in the minor groove (e.g., Reardon et al., 1993; Huth et al., 1997). MAR binding proteins sometimes have an affinity for single-stranded nucleic acids, as exemplified by hnRNP-U and lamins (Fackelmayer et al., 1994; Ludérus et al., 1994); however, the MAR binding of AHM1 was not influenced by excess poly(dA) or poly(dT) in the reaction.
Numerous chromatin proteins have been found to contain single or multiple AT hooks (listed in Aravind and Landsman, 1998). In general, an AT hook is thought to function as an auxiliary protein motif that cooperates with other DNA binding regions (either in the same or in a different molecule) or facilitates changes in the structure of the DNA (Aravind and Landsman, 1998). In the case of HMG-I/Y, which contains three AT hooks, multivalent interactions between the AT hooks and appropriately spaced AT-rich tracts are required for specific high-affinity binding; a single AT hook in a polypeptide or a single AT tract on the target DNA results in the loss of interaction or a greatly reduced interaction (Maher and Nathans, 1996; Yie et al., 1997). Furthermore, the C-terminal acidic region and the spacer regions between AT hooks have also been shown to facilitate the binding (Yie et al., 1997; Frank et al., 1998). A requirement for more than one AT hook in DNA binding has also been reported for rice PF1, an HMG-I–like protein (Nieto-Sotelo et al., 1994). These observations are consistent with our finding that the cooperation of the internal portion and the C-terminal AT hook is necessary for MAR binding in vivo. In the in vitro assay, a single MAR probe may be able to bind to multiple AT hooks, each derived from an AHM1 derivative fixed on the filter, resulting in the ability to bind to MAR without the internal portion.
AHM1 Is a Novel Nuclear Matrix Protein
We located AHM1 in the operationally defined nuclear matrix fraction as a 67-kD protein. It was localized in situ in the nucleoplasm, and transiently expressed GFP–AH8 had a speckled distribution pattern inside the nucleus. Similar distribution patterns have been reported for nuclear matrix–localized snRNPs, including a protein capable of binding to MARs (Mattern et al., 1999). The mRNA encoding AHM1 was expressed at a low level. Furthermore, AHM1 does not have the structural features of filament proteins such as MFP1 (Meier et al., 1996). Therefore, AHM1 does not appear to be an architectural component of the nuclear framework; rather, it seems to connect the intranuclear framework and MARs. It may also be involved in specific nuclear activities that depend on nuclear matrix–MAR interactions. In this connection, possible functions of the J domain–like region and the Zn finger–like motif are discussed below.
Possible Functions of AHM1 as a J-Z Array–Containing MAR Binding Protein
An interesting feature of the AHM1 protein is its J-Z array, the special combination of a J domain–homologous region and a novel type of Zn finger–like motif, conserved among a subset of plant-specific J domain proteins. The J domain was first recognized as an ∼70–amino acid region conserved among the members of the DnaJ/Hsp40 family (Silver and Way, 1993; Kelley, 1998). Homologous regions have also been found in a diverse set of proteins (see Figure 2A) that are not necessarily related to each other in terms of structure and function, for example, auxilin, functioning in the uncoating of clathrin-coated vesicles (Ungewickell et al., 1995); SV40 T antigen, involved in viral replication and functional modulation of cell cycle regulators (Campbell et al., 1997; Zalvide et al., 1998); Sec63p, a part of the machinery involved in protein translocation through the endoplasmic membrane (Lyman and Schekman, 1997); and cysteine string proteins that are involved in exocytosis of synaptic vesicles (Zinsmaier et al., 1994). These J domain–only proteins as well as the DnaJ/Hsp40 family members commonly utilize their J domains to recruit a specific Hsp70/Hsc70 chaperone partner or partners and to facilitate the processes involving conformational changes of proteins or protein–protein interactions (Kelley, 1998; Mayer and Bukau, 1998). Considering these facts, AHM1 probably also functions by interacting through its J domain–like region with an Hsp70 member in the nucleus.
Interestingly, a parallel molecular organization can be found between AHM1 and DnaJ, as illustrated in Figure 2D. DnaJ and closely related homologs have a J domain at or near the N terminus, followed by a Gly/Phe-rich linker region, C4-type Zn fingers consisting of CXXCXGXG repeats, and a C-terminal unique region (Silver and Way, 1993; Kelley, 1998). AHM1 is composed of a J domain–like region near the N terminus, a linker-like tandem repeat region (R1), a Zn finger–like motif with four conserved Cys residues, and a C-terminal region containing the combination of a tandem repeat (R2) plus an AT hook. In DnaJ and Ydj1p, the Zn finger and C-terminal regions have been shown to be involved in substrate binding (Szabo et al., 1996; Lu and Cyr, 1998); likewise, we determined that the corresponding regions of AHM1 were needed for binding to the substrates, that is, the MARs. The AHM1-related proteins with a J-Z array have various C-terminal regions. Among them, an Arabidopsis protein (AAD12700) is of particular interest because an AT hook is found at the C terminus and because the region immediately adjacent to the C-terminal end of the Zn finger–like motif is also homologous to the corresponding region of AHM1 (Figures 2C and 2D); these regions of AHM1 were found to be required for MAR binding.
The nucleus is known as the most heat-labile organelle, and the nuclear matrix is thought to be a primary target for heat shock effects (reviewed in Roti Roti et al., 1997). In animal cells, Hsp70 is known to be relocated into the nucleus in heat shock, and this change is thought to be involved in the disaggregation of proteins that precipitate on the nucleoskeleton at high temperatures (e.g., Neri et al., 1995). Although little is known about the heat stress response of the plant nucleus, an interesting possibility is that AHM1 may be involved in the recovery from lesions caused at high temperatures; at the same time, nuclear matrix–MAR interactions may be involved in such a process.
Interactions between the nuclear matrix and MAR binding proteins as well as nuclear heat shock proteins, including Hsp70 and Hsp40, have been implicated in gene regulation in animals, as exemplified by hormone receptor–mediated transcriptional regulation (e.g., Lauber et al., 1995; Dittmar et al., 1998). Thus, it is also possible that AHM1 is involved in MAR-mediated gene regulation. Nuclear matrix–MAR interactions are thought to be a sort of mass binding phenomenon (Zuckerkandl and Villet, 1988); in other words, the specific function of a MAR may be achieved through a special multiprotein–DNA complex, resulting from the sum of the low-affinity binding of individual components (Bode et al., 1996). AHM1 might function as a co-chaperone during the formation of such a multiprotein–DNA complex by recruiting the Hsp70 chaperone machinery around the MARs. Further analyses of AHM1, including its interacting partners, may provide a clue for understanding the molecular basis of elusive MAR functions from a novel aspect.
METHODS
Isolation of cDNA and Genomic Clones
All recombinant DNA manipulations were performed according to standard techniques (Sambrook et al., 1989), unless otherwise specified. The screening of a wheat (Triticum aestivum cv HY1) expression cDNA library in λgt11 with a DNA probe was performed as described by Tabata et al. (1989), except that the binding buffer contained 25 mM Tris-HCl, pH 7.5, 136.9 mM NaCl, 2.68 mM KCl, 0.25% nonfat milk, 5 ng/mL 32P-labeled WCI-3b matrix attachment region (MAR; 1.9 kb long; Minami et al., 1999), and 50 μg/mL sonicated Escherichia coli carrier DNA. The cDNA inserts of positive phages (λAH series) were recloned into the EcoRI site of pBluescript SK+ to yield pAH series plasmids and then sequenced. A wheat genomic library (Sakamoto et al., 1993) and cDNA library (5 × 106 plaque-forming units) were screened by using the insert of pAH8 as a probe to yield a genomic clone (λg50) and some partial cDNA clones. The 5′ end of the transcript was determined by the rapid amplification of cDNA 5′ ends (5′ RACE) method, essentially as described by Frohman et al. (1988), with modification (Chen, 1996). RNAs were isolated from wheat embryos 40 hr after imbibition or from tissues of 5-day-old seedlings using the TRIzol LS reagent (Life Technologies, Grand Island, NY). Gene-specific primers used were 5′-GAGGGCGGACTTAAGT-3′ for cDNA synthesis and 5′-GAGGGCGGACTT-AAGT-3′ and 5′-AGAGCCGGCGGAACTTGGAAT-3′ for the first and second polymerase chain reaction (PCR) amplification.
Plasmid Constructions
The cDNA insert (EcoRI fragment) of pAH8 was recloned into pGAD424 (Clontech, Palo Alto, CA) and pET28a (Novagen, Madison, WI) to yield pGAD-AH8 and pET-AH8, which were used for expression of GAD-AH8 in yeast and His-AH8 in E. coli, respectively. pGAD-Mut1 and -Mut3 were constructed by inserting the 0.72-kb (partial digestion) and 0.59-kb HincII fragments of pAH8 into the SmaI site of pGAD424. The 0.81-kb BstYI-EcoRI fragment of pAH8 was blunt-ended and cloned into the filled-in BamHI site of pGAD424 to yield pGAD-Mut2. pGAD-Mut4 was created by deleting a SalI fragment from pGAD-AH8. pGAD-Mut5 was constructed by replacing an NcoI-SalI fragment of pGAD-Mut1 with a synthetic double-stranded oligonucleotide, resulting in an in-frame deletion. pGAD-MutRC and -MutC were created with PCR-amplified fragments. Site-directed mutagenesis was performed with a PCR-based kit method (Takara Shuzo, Otsu, Japan) to create pGAD-Mut1(R528K). pET-Mut series plasmids were generated by replacing the vector of the pGAD-Mut series with pET15b. As for pET-MutJL and -MutR1Z, PCR-amplified fragments were directly cloned into pET15b. The lacZ reporter plasmid pLGM was constructed by placing the 0.88-kb fragment of WCI-3b MAR (fragment M; Minami et al., 1999) between the SmaI and XhoI sites of pLGΔ-312 (Guarente and Mason, 1983). The G3GFP gene (Kawakami and Watanabe, 1997) was slightly modified (A. Tamai and T. Meshi, unpublished data) and cloned into pBI221 (Clontech) in place of the β-glucuronidase gene to yield pBIG3GFP. In pAH8G3GFP, a derivative of pBIG3GFP, the termination codon of the original G3GFP gene (Kawakami and Watanabe, 1997) was replaced with a synthetic linker (encoding GGGGVDLEGGP) plus the EcoRI fragment of pAH8. All of the PCR-amplified fragments and all ligation junctions were sequenced before use.
Expression of Recombinant Proteins in E. coli
His-tagged recombinant proteins were expressed in E. coli BL21(DE3)pLysS and purified with a HiTrap chelating column (Pharmacia), essentially as described previously (Meshi et al., 1998), in the presence of 6 M urea. His-AH8 carried vector (pET28a)-derived 34 amino acids at the N terminus and lacked the N-terminal 32 amino acids of AT hook–containing MAR binding protein1 (AHM1). The other derivatives had the same N-terminal His-tag sequence derived from pET15b, MGSSHHHHHHSSGLVPRGSH, plus one to six extra amino acids: KFP for His-Mut1, -Mut3, and -Mut5; EIPG for His-Mut2; KFPGIE for His-Mut4; and M for His-MutRC and -MutC. His-Mut3 and -Mut4 carried vector-derived C-terminal sequences GSVDLQRSMLEDPAANKARKEAELAAATAEQ and AEIYARGSGC, respectively.
In Vitro DNA Binding Assay
Proteins (usually 0.1 μg each) were separated on SDS–10% polyacrylamide gels and electrophoretically transferred onto a nitrocellulose filter. Binding was performed in a sealed bag in a DNA binding buffer containing 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 4 μg/mL sheared E. coli DNA, and 2 ng/mL end-labeled MAR fragment in the presence or absence of competitors, unless otherwise specified. Synthetic oligonucleotides and distamycin A were obtained from Pharmacia and Sigma, respectively. After incubation for 15 hr at room temperature, the filters were washed three times with the DNA binding buffer minus probe and autoradiographed. The probes used were a 0.88-kb fragment of WCI-3b MAR (fragment M; Minami et al., 1999), a 0.44-kb fragment containing the mouse Igκ locus MAR (Cockerill and Garrard, 1986), and a 1.3-kb fragment corresponding to MAR-1 in the Arabidopsis plastocyanin locus (van Drunen et al., 1997). The latter two were PCR-amplified and subcloned. A 205-bp fragment of pUC19 (nucleotides 1421 to 1625) was dimerized and used as an AT-rich, non-MAR fragment. Preparation of His-tagged HBP-1a(17) and the synthetic oligo Hex31 was as described earlier (Meshi et al., 1998).
One-Hybrid Assay in Yeast
Yeast techniques used were essentially those described by Kaiser et al. (1994). The yeast strain YPH499 (Sikorski and Hieter, 1989) was transformed with pLGM and a pGAD series plasmid, and the β-galactosidase activity expressed in the transformants was determined according to method I described by Kaiser et al. (1994).
Isolation of Nuclei and Nuclear Matrix
Wheat seedlings were harvested ∼1.5 or 5 days after imbibition, and the roots were frozen in liquid N2. After the specimens had been pulverized to a fine powder, the cells were lysed at 4°C for 5 min in a buffer containing 25 mM Tris-HCl, pH 8.5, 5 mM MgCl2, 2.5% Ficoll, 5% Dextran T-40, 0.1% Triton X-100, 0.44 M sucrose, 2.5 mM DTT, 0.05 mM spermine, and 0.125 mM spermidine, supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 μM trans-epoxysuccinyl-l-leucylamido-(4-guanidino)butane (E-64). The lysate was filtered sequentially through two layers of Miracloth (Calbiochem) and 60-μm (pore size) nylon mesh. The crude nuclei were collected by centrifugation and washed twice in the lysis buffer without Triton X-100. Further purification of nuclei by Percoll gradient centrifugation and preparation of the nuclear matrix with lithium diiodosalicylate (either with or without heat stabilization) were performed as described previously (Minami et al., 1999). High-salt extraction of nuclear matrix was performed essentially as described by Masuda et al. (1993). Briefly, nuclei were suspended in buffer A (50 mM Mes, 5 mM MgCl2, 0.25 M sucrose, and 10% glycerol, pH 6.0) plus 1 mM PMSF and incubated at 12°C for 1 hr with DNase I (Takara Shuzo) at a final concentration of 50 μg/mL. After the nuclei were washed three times, they were suspended in buffer A. The salt concentration of the suspension was gradually increased to 2 M by adding an equal amount of buffer A containing 4 M NaCl. The suspension was then incubated for 30 min on ice. Nuclei were washed twice, and the final precipitable fraction was used as nuclear matrix. The salt-extracted proteins were recovered from soluble fractions with Protein Concentrate (Geno Technology, St. Louis, MO). One A260 unit of nuclei was defined as the amount that corresponded to 50 μg of nuclear DNA.
Immunological Analysis
Affinity-purified His-AH8 was used to immunize a rabbit. The IgG fraction was prepared by ammonium sulfate precipitation and HiTrap Q (Pharmacia) column chromatography. Polyclonal antibodies were affinity-purified with His-MutJL or His-MutR1Z. Immunoreactive proteins on protein gel blots were detected with the ProtoBlot II AP System (Promega). Wheat roots were fixed for 45 min at room temperature by immersion in 7.4% formaldehyde in a buffer of 60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 2 mM MgCl2, pH 6.8. The specimens were digested in a solution containing 2% Cellulase Onozuka RS (Yakult Pharmaceutical, Tokyo, Japan), 0.1% Pectolyase Y-23 (Seishin Pharmaceutical Co., Tokyo, Japan), 5 mM EGTA, and 0.4 M mannitol for 20 min and placed onto poly-l-Lys–coated cover slips. The samples were incubated with affinity-purified anti-AHM1 antibody for 60 min at room temperature. The cover slips were washed three times (5 min each) and incubated with goat anti–rabbit IgG coupled to fluorescein isothiocyanate (Molecular Probes, Eugene, OR) for 1 hr at room temperature. The samples were washed three times (5 min each), incubated briefly with 4′,6-diamidine-2-phenylindole, and examined under a microscope (Axioskop; Carl Zeiss, Jena, Germany) equipped with epifluorescence illumination. Images were obtained by a color-chilled charge-coupled device camera (model C5810; Hamamatsu, Photonics, Hamamatsu, Japan).
Expression and Detection of GFP and Its Fusion Product
Suspension-cultured cells of tobacco, Nicotiana tabacum cv Bright Yellow 2, were evenly distributed over the surface of a paper filter (No. 6; Whatman, Maidstone, UK) with the aid of a Buchner funnel. The paper filter was placed on a Petri dish containing solid Murashige and Skoog (1962) medium. Plasmids (pBIG3GFP and pAH8G3GFP) were adsorbed to 1-μm-diameter gold particles and then bombarded into the cells on the paper filter with PDS1000/He (Rio-Rad Laboratories, Hercules, CA), essentially according to the manufacturer's instructions. Cells were cultured at 26°C in the dark. Ten to 24 hr after bombardment, cells were examined under a fluorescence microscope or under a confocal laser scanning microscope (model LSM410; Carl Zeiss) equipped with a filter set for fluorescence.
Acknowledgments
We thank Dr. Leonard Guarente for pLGΔ-312, Dr. Masahiro Okanami for Igκ MAR, and Dr. Yuichiro Watanabe for the G3GFP gene. This work was supported in part by grants-in-aid from the Ministry of Education, Science, Sports, and Culture and from the Ministry of Agriculture, Forestry, and Fisheries of Japan.
References
- Adachi, Y., Käs, E., and Laemmli, U.K. (1989). Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J. 8, 3997–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L., and Landsman, D. (1998). AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 26, 4413–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramova, Z., SanMiguel, P., Georgieva, E., and Bennetzen, J.L. (1995). Matrix attachment regions and transcribed sequences within a long chromosomal continuum containing maize Adh1. Plant Cell 7, 1667–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramova, Z., Tikhonov, A., Chen, M., and Bennetzen, J.L. (1998). Matrix attachment regions and structural colinearity in the genomes of two grass species. Nucleic Acids Res. 26, 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney, R., and Jeon, K.W. (1995). Nuclear Matrix. Structural and Functional Organization. (San Diego, CA: Academic Press).
- Bode, J., Stengert-Iber, M., Kay, V., Schlake, T., and Dietz-Pfeilstetter, A. (1996). Scaffold/matrix-attached regions: Topological switches with multiple regulatory functions. Crit. Rev. Eukaryot. Gene Expr. 6, 115–138. [DOI] [PubMed] [Google Scholar]
- Boulikas, T. (1995). Chromatin domains and prediction of MAR sequences. Int. Rev. Cytol. 162A, 279–388. [DOI] [PubMed] [Google Scholar]
- Campbell, K.S., Mullane, K.P., Aksoy, I.A., Stubdal, H., Zalvide, J., Pipas, J.M., Silver, P.A., Roberts, T.M., Schaffhausen, B.S., and DeCaprio, J.A. (1997). DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 11, 1098–1110. [DOI] [PubMed] [Google Scholar]
- Chen, Z. (1996). Simple modifications to increase specificity of the 5′ RACE procedure. Trends Genet. 12, 87–88. [DOI] [PubMed] [Google Scholar]
- Cockerill, P.N., and Garrard, W.T. (1986). Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell 44, 273–282. [DOI] [PubMed] [Google Scholar]
- de Belle, I., Cai, S., and Kohwi-Shigematsu, T. (1998). The genomic sequences bound to special AT-rich sequence-binding protein 1 (SATB1) in vivo in Jurkat T cells are tightly associated with the nuclear matrix at the bases of the chromatin loops. J. Cell Biol. 141, 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, L.A., Joh, T., Kohwi, Y., and Kohwi-Shigematsu, T. (1992). A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell 70, 631–645. [DOI] [PubMed] [Google Scholar]
- Dittmar, K.D., Banach, M., Galigniana, M.D., and Pratt, W.B. (1998). The role of DnaJ-like proteins in glucocorticoid receptor·hsp90 heterocomplex assembly by the reconstituted hsp90·p60·hsp70 foldsome complex. J. Biol. Chem. 273, 7358–7366. [DOI] [PubMed] [Google Scholar]
- Ellenberger, T.E., Brandl, C.J., Strul, K., and Harrison, S.C. (1992). The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: Crystal structure of the protein–DNA complex. Cell 71, 1223–1237. [DOI] [PubMed] [Google Scholar]
- Fackelmayer, F.O., Dahm, K., Renz, A., Ramsperger, U., and Richter, A. (1994). Nucleic-acid–binding properties of hnRNP-U/SAF-A, a nuclear-matrix protein which binds DNA and RNA in vivo and in vitro. Eur. J. Biochem. 221, 749–757. [DOI] [PubMed] [Google Scholar]
- Frank, O., Schwanbeck, R., and Wisniewski, J.R. (1998). Protein footprinting reveals specific binding modes of a high mobility group protein I to DNAs of different conformation. J. Biol. Chem. 273, 20015–20020. [DOI] [PubMed] [Google Scholar]
- Frohman, M.A., Dush, M.K., and Martin, G.R. (1988). Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindullis, F., and Meier, I. (1999). Matrix attachment region binding protein MFP1 is localized in discrete domains at the nuclear envelope. Plant Cell 11, 1117–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindullis, F., Peffer, N.J., and Meier, I. (1999). MAF1, a novel plant protein interacting with nuclear matrix attachment region protein MFP1, is located at the nuclear envelope. Plant Cell 11, 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, M.K., Maskos, K., and Landry, S.J. (1998). Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc. Natl. Acad. Sci. USA 95, 6108–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente, L., and Mason, T. (1983). Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell 32, 1279–1286. [DOI] [PubMed] [Google Scholar]
- Hatton, D., and Gray, J.C. (1999). Two MAR DNA-binding proteins of the pea nuclear matrix identify a new class of DNA-binding proteins. Plant J. 18, 417–429. [DOI] [PubMed] [Google Scholar]
- Holmes-Davis, R., and Comai, L. (1998). Nuclear matrix attachment regions and plant gene expression. Trends Plant Sci. 3, 91–97. [Google Scholar]
- Huth, J.R., Bewley, C.A., Nissen, M.S., Evans, J.N.S., Reeves, R., Gronenborn, A.M., and Clore, G.M. (1997). The solution structure of an HMG-I(Y)–DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 4, 657–665. [DOI] [PubMed] [Google Scholar]
- Kaiser, C., Michaelis, S., and Mitchell, A. (1994). Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Kawakami, S., and Watanabe, Y. (1997). Use of green fluorescent protein as a molecular marker tag of protein movement in vitro. Plant Biotechnol. 14, 127–130. [Google Scholar]
- Kelley, W.L. (1998). The J-domain family and the recruitment of chaperone power. Trends Biochem. Sci. 23, 222–227. [DOI] [PubMed] [Google Scholar]
- Kelley, W.L., and Georgopoulos, C. (1997). The T/t common exon of simian virus 40, JC, and BK polyomavirus T antigens can functionally replace the J-domain of the Escherichia coli DnaJ molecular chaperone. Proc. Natl. Acad. Sci. USA 94, 3679–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka, M.L., Yoon, C., Goodsell, D., Pjura, P., and Dickerson, R.E. (1985). The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc. Natl. Acad. Sci. USA 82, 1376–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K., Käs, E., Poljak, L., and Adachi, Y. (1992). Scaffold-associated regions: Cis-acting determinants of chromatin structural loops and functional domains. Curr. Opin. Genet. Dev. 2, 275–285. [DOI] [PubMed] [Google Scholar]
- Lauber, A., Sandhu, N.P., Schuchard, M., Subramaniam, M., and Spelsberg, T.C. (1995). Nuclear matrix acceptor binding sites for steroid hormone receptors: A candidate nuclear matrix acceptor protein. Int. Rev. Cytol. 162B, 337–376. [DOI] [PubMed] [Google Scholar]
- Lu, Z., and Cyr, D.M. (1998). The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J. Biol. Chem. 273, 5970–5978. [DOI] [PubMed] [Google Scholar]
- Ludérus, M.E.E., de Graaf, A., Mattia, E., den Blaauwen, J.L., Grande, M.A., de Jong, L., and van Driel, R. (1992). Binding of matrix attachment regions to lamin B1. Cell 70, 949–959. [DOI] [PubMed] [Google Scholar]
- Ludérus, M.E.E., den Blaauwen, J.L., de Smit, O.J.B., Compton, D.A., and van Driel, R. (1994). Binding of matrix attachment regions to lamin polymers involves single-stranded regions and the minor groove. Mol. Cell. Biol. 14, 6297–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman, S.K., and Schekman, R. (1997). Binding of secretory precursor polypeptides to a translocon subcomplex is regulated by BiP. Cell 88, 85–96. [DOI] [PubMed] [Google Scholar]
- Maher, J.F., and Nathans, D. (1996). Multivalent DNA-binding properties of the HMG-I proteins. Proc. Natl. Acad. Sci. USA 93, 6716–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda, K., Takahashi, S., Nomura, K., Arimoto, M., and Inoue, M. (1993). Residual structure and constituent proteins of the peripheral framework of the cell nucleus in somatic embryos from Daucus carota L. Planta 191, 532–540. [Google Scholar]
- Masuda, K., Xu, Z.-J., Takahashi, S., Ito, A., Ono, M., Nomura, K., and Inoue, M. (1997). Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long α-helical domain. Exp. Cell Res. 232, 173–181. [DOI] [PubMed] [Google Scholar]
- Mattern, K.A., van der Kraan, I., Schul, W., de Jong, L., and van Driel, R. (1999). Spatial organization of four hnRNP proteins in relation to sites of transcription, to nuclear speckles, and to each other in interphase nuclei and nuclear matrices of HeLa cells. Exp. Cell. Res. 246, 461–470. [DOI] [PubMed] [Google Scholar]
- Mayer, M.P., and Bukau, B. (1998). Hsp70 chaperone systems: Diversity of cellular functions and mechanism of action. Biol. Chem. 379, 261–268. [PubMed] [Google Scholar]
- Meier, I., Phelan, T., Gruissem, W., Spiker, S., and Schneider, D. (1996). MFP1, a novel plant filament-like protein with affinity for matrix attachment region DNA. Plant Cell 8, 2105–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshi, T., Moda, I., Minami, M., Okanami, M., and Iwabuchi, M. (1998). Conserved Ser residues in the basic region of the bZIP-type transcription factor HBP-1a(17): Importance in DNA binding and possible targets for phosphorylation. Plant Mol. Biol. 36, 125–136. [DOI] [PubMed] [Google Scholar]
- Minami, M., Morisawa, G., Hayashi, M., Sakata, Y., Habu, Y., Iwabuchi, M., and Meshi, T. (1999). High affinity of the 5′-upstream region of a winged bean chymotrypsin inhibitor gene for nuclear matrix. Plant Cell Physiol. 40, 109–113. [Google Scholar]
- Mínguez, A., and Moreno Díaz de la Espina, S. (1993). Immunological characterization of lamins in the nuclear matrix of onion cells. J. Cell Sci. 106, 431–439. [DOI] [PubMed] [Google Scholar]
- Moreno Díaz de la Espina, S. (1995). Nuclear matrix isolated from plant cells. Int. Rev. Cytol. 162B, 75–139. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Neri, L.M., Riederer, B.M., Marugg, R.A., Capitani, S., and Martelli, A.M. (1995). Analysis by confocal microscopy of the behavior of heat shock protein 70 within the nucleus and of a nuclear matrix polypeptide during prolonged heat shock response in HeLa cells. Exp. Cell Res. 221, 301–310. [DOI] [PubMed] [Google Scholar]
- Nieto-Sotelo, J., Ichida, A., and Quail, P.H. (1994). PF1: An A-T hook–containing DNA binding protein from rice that interacts with a functionally defined d(AT)-rich element in the oat phytochrome A3 gene promoter. Plant Cell 6, 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellecchia, M., Szyperski, T., Wall, D., Georgopoulos, C., and Wüthrich, K. (1996). NMR structure of the J-domain and the Gly/Phe-rich region of the Escherichia coli DnaJ chaperone. J. Mol. Biol. 260, 236–250. [DOI] [PubMed] [Google Scholar]
- Qian, Y.Q., Patel, D., Hartl, F.U., and McColl, D.J. (1996). Nuclear magnetic resonance solution structure of the human Hsp40 (HDJ-1) J-domain. J. Mol. Biol. 260, 224–235. [DOI] [PubMed] [Google Scholar]
- Reardon, B.J., Winters, R.S., Gordon, D., and Winter, E. (1993). A peptide motif that recognizes A·T tracts in DNA. Proc. Natl. Acad. Sci. USA 90, 11327–11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves, R., and Nissen, M.S. (1990). The A·T-DNA–binding domain of mammalian high mobility group I chromosomal proteins. J. Biol. Chem. 265, 8573–8582. [PubMed] [Google Scholar]
- Roti Roti, J.L., Wright, W.D., and VanderWaal, R. (1997). The nuclear matrix: A target for heat shock effects and a determinant for stress response. Crit. Rev. Eukaryot. Gene Expr. 7, 343–360. [DOI] [PubMed] [Google Scholar]
- Sakamoto, A., Minami, M., Huh, G.H., and Iwabuchi, M. (1993). The putative zinc-finger protein WZF-1 interacts with a cis-acting element of histone genes. Eur. J. Biochem. 217, 1049–1056. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver, P.A., and Way, J.C. (1993). Eukaryotic DnaJ homologs and the specificity of Hsp70 activity. Cell 74, 5–6. [DOI] [PubMed] [Google Scholar]
- Spiker, S., and Thompson, W.F. (1996). Nuclear matrix attachment regions and transgene expression in plants. Plant Physiol. 110, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh, W.-C., Burkholder, W.F., Lu, C.Z., Zhao, X., Gottesman, M.E., and Gross, C.A. (1998). Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc. Natl. Acad. Sci. USA 95, 15223–15228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo, A., Korszun, R., Hartl, F.U., and Flanagan, J. (1996). A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J. 15, 408–417. [PMC free article] [PubMed] [Google Scholar]
- Tabata, T., Takase, H., Takayama, S., Mikami, K., Nakatsuka, A., Kawata, T., Nakayama, T., and Iwabuchi, M. (1989). A protein that binds to a cis-acting element of wheat histone genes has a leucine zipper motif. Science 245, 965–967. [DOI] [PubMed] [Google Scholar]
- Tikhonov, A.P., Bennetzen, J.L., and Avramova, Z.V. (2000). Structural domains and matrix attachment regions along colinear chromosomal segments of maize and sorghum. Plant Cell 12, 249–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewickell, E., Ungewickell, H., Holstein, S.E.H., Lindner, R., Prasad, K., Barouch, W., Martin, B., Greene, L.E., and Eisenberg, E. (1995). Role of auxilin in uncoating clathrin-coated vesicles. Nature 378, 632–635. [DOI] [PubMed] [Google Scholar]
- van Drunen, C.M., Oosterling, R.W., Keultjes, G.M., Weisbeek, P.J., van Driel, R., and Smeekens, S.C.M. (1997). Analysis of the chromatin domain organisation around the plastocyanin gene reveals an MAR-specific sequence element in Arabidopsis thaliana. Nucleic Acids Res. 25, 3904–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kries, J.P., Buhrmester, H., and Strätling, W.H. (1991). A matrix/scaffold attachment region binding protein: Identification, purification, and mode of binding. Cell 64, 123–135. [DOI] [PubMed] [Google Scholar]
- von Kries, J.P., Buck, F., and Strätling, W.H. (1994). Chicken MAR binding protein p120 is identical to human heterogeneous nuclear ribonucleoprotein (hnRNP)U. Nucleic Acids Res. 22, 1215–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzel, J.M., Buhrmester, H., and Strätling, W.H. (1997). Chicken MAR-binding protein is homologous to rat methyl-CpG-binding protein MeCP2. Mol. Cell. Biol. 17, 5656–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yie, J., Liang, S., Merika, M., and Thanos, D. (1997). Intra- and intermolecular cooperative binding of high-mobility-group protein I(Y) to the beta-interferon promoter. Mol. Cell. Biol. 17, 3649–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, W., and Moreno Díaz de la Espina, S. (1999). The plant nucleoskeleton: Ultrastructural organization and identification of NuMA homologues in the nuclear matrix and mitotic spindle of plant cells. Exp. Cell Res. 246, 516–526. [DOI] [PubMed] [Google Scholar]
- Zalvide, J., Stubdal, H., and DeCaprio, J.A. (1998). The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol. Cell. Biol. 18, 1408–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, K., Käs, E., Gonzales, E., and Laemmli, U.K. (1993). SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG- I/Y is enriched in H1-depleted chromatin. EMBO J. 12, 3237–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinsmaier, K.E., Eberle, K.K., Buchner, E., Walter, N., and Benzer, S. (1994). Paralysis and early death in cysteine string protein mutants of Drosophila. Science 263, 977–980. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl, E., and Villet, R. (1988). Generation of high specificity of effect through low-specificity binding of proteins to DNA. FEBS Lett. 231, 291–298. [DOI] [PubMed] [Google Scholar]