Abstract
An Arabidopsis protein was found to interact specifically with the capsid protein (CP) of turnip crinkle virus (TCV) through yeast two-hybrid screening. This protein, designated TIP (for TCV-interacting protein), was found to be a member of the recently recognized NAC family of proteins. NAC proteins have been implicated in the regulation of development of plant embryos and flowers. TIP alone was able to activate expression of reporter genes in yeast if fused to a DNA binding domain, suggesting that it may be a transcriptional activator. The TIP binding region in the TCV CP has been mapped to the N-terminal 25 amino acids. Site-directed mutagenesis within this region revealed that loss of the TIP–CP interaction in the yeast two-hybrid assay correlated with loss of the ability of TCV to induce the hypersensitive response and resistance in the TCV-resistant Arabidopsis ecotype Dijon (Di-0 and its inbred line Di-17). These data suggest that TIP is an essential component in the TCV resistance response pathway.
INTRODUCTION
The susceptibility and resistance response of Arabidopsis to turnip crinkle virus (TCV) have been the subject of several studies because TCV is one of the few well-characterized plant viruses known to infect this model plant (Simon et al., 1992; Dempsey et al., 1993, 1997; Oh et al., 1995). TCV is a small icosahedral virus with a 4-kb RNA genome encoding five viral proteins, including two that are required for replication (p28 and p88) and two that function to facilitate the cell-to-cell movement (p8 and p9). The fifth gene encodes a 38-kD capsid protein (CP) that packages the viral RNA into a 30-nm icosahedral particle of determined structure (Hogle et al., 1986; Sorger et al., 1986; Carrington et al., 1987). Previous work (Dempsey et al., 1993, 1997) has established that an inbred line of Arabidopsis derived from ecotype Dijon-0 (Di-0), designated Di-17, exhibits the hypersensitive response (HR) in the inoculated leaves and systemic resistance in the whole plant when inoculated with TCV. A single dominant locus responsible for the HR, termed HRT, has been mapped to chromosome 5 of the Arabidopsis genome (Dempsey et al., 1997). Additional studies implicated the TCV CP as the elicitor of this resistance response (Oh et al., 1995; Wobbe and Zhao, 1998). Whether the HRT product actually interacts with TCV CP, however, has yet to be determined. In this report, we have identified an Arabidopsis protein that specifically interacts with TCV CP, and we have mapped the interacting domain to the N-terminal 25 amino acids. Single amino acid replacements within this TCV CP domain that resulted in loss of the specific protein–protein interaction also led to loss of resistance. These results demonstrate that this N-terminal domain is the elicitor and implicate the identified Arabidopsis protein in the resistance response of Di-17 plants to TCV. This protein, which we named TIP (for TCV-interacting protein), belongs to the developmentally important NAC family of proteins and acts as a transcriptional activator in yeast cells. To our knowledge, the TIP–TCV CP interaction is only the second example in which the direct interaction between the avirulence (Avr) gene product and a host protein has been demonstrated.
RESULTS
Isolation of the TCV CP–Interacting Arabidopsis Protein
To isolate Arabidopsis proteins capable of interacting with TCV CP, we screened an Arabidopsis cDNA library, using the yeast two-hybrid system with TCV CP as the bait. Only two positive colonies were identified in >2 × 107 transformants screened. The cDNA-containing pGAD10 derivatives were recovered from these two colonies, and the cDNA inserts were sequenced. The sequences of the two ∼1.5-kb cDNA fragments are identical, indicating that one single cDNA was isolated. Its deduced amino acid sequence revealed that three stop codons were present between the activation domain and the encoded protein, precluding the production of a fusion protein. However, the specific positive interaction between the TCV CP and this protein was confirmed by performing the yeast two-hybrid assay with a construct in which the intervening sequence between the activation domain and the protein had been removed. This result suggested that this Arabidopsis protein might encode its own transcriptional activation domain, which was confirmed in later experiments.
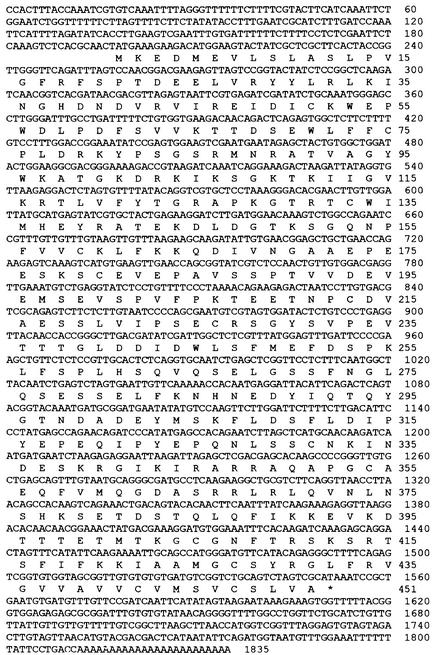
Sequence analysis also showed that the 3′ end of the cDNA was incomplete because the derived open reading frame contained no stop codon. Reverse transcription–polymerase chain reaction (RT-PCR) was performed to obtain the sequence of the 3′ end. The polyadenylated RNAs purified from Arabidopsis plants of both the TCV-susceptible Columbia (Col-0) and TCV-resistant Di-17 lines were used as templates. The deoxyoligonucleotide primers used were an oligo(dT) primer and a primer with the sequence identical to nucleotides 1436 to 1454 of the cDNA. The complete sequence of TIP cDNA, deduced from the combined sequence of the RT-PCR products and the 1.5-kb sequence (Figure 1), encodes an open reading frame of 451 amino acids. The amino acid sequence of TIP was subjected to a database search (GenBank, release 12/98) with the BLAST program (GCG package, version 9.1; Genetics Computation Group, Madison, WI). The N-terminal half of the TIP was found to share substantial homology with all members of the recently identified NAC family of proteins (Souer et al., 1996; Aida et al., 1997; Sablowski and Meyerowitz, 1998) and thus should be considered a new member of the family. The definitive members of this protein family include NAM (for NO APICAL MERISTEM) from petunia; ATAF1 and ATAF2, CUC2 (for CUP-SHAPED COTYLEDONS2), and NAP (for NAC-LIKE, ACTIVATED BY AP3/PI) from Arabidopsis; and SENU5 (for SENESCENCE UPREGULATED5) from tomato. Both NAM and CUC2 play important roles in establishing the shoot apical meristem and separating cotyledons and floral organs. NAP, being activated by the homeotic protein heterodimer AP3/PI, presumably functions in the transition between active cell division and cell expansion during flower development. Thus, all three NAC proteins with known functions have a critical role in the plant developmental process. TIP is most similar to NAP, with sequence similarity extending from amino acid position 5 to 288. Within the NAC domain defined by amino acid positions 17 to 169 of CUC2, TIP shares 55.0, 53.3, and 51.7% identity and 72.8, 70.4, and 68.2% similarity with NAM, CUC2, and NAP, respectively (Figure 2).
Figure 1.
The mRNA and Derived Amino Acid Sequences of TIP.
The cDNA sequence of the TIP mRNA and the aligned amino acid sequence of TIP (GenBank accession number AF281062).
Figure 2.
TIP Sequence Compared with That of Other Members of the NAC Protein Family.
The N-terminal half of TIP is aligned against the regions of similarity in the CUC2, NAP, NAM, and ATAF1 and ATAF2 sequences. The identical amino acids are highlighted. The dashes denote amino acid residues that are not present in the corresponding proteins.
TIP Activates Transcription of the Reporter Gene in Yeast Cells
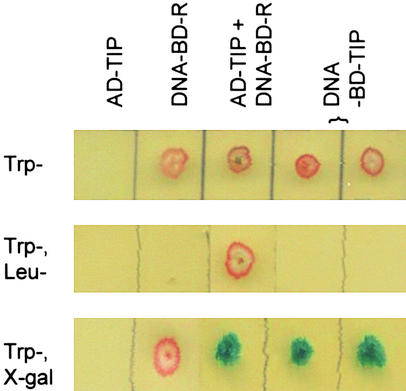
Two other Arabidopsis members of the NAC family of proteins, ATAF1 and ATAF2, were found to activate the 35S promoter of cauliflower mosaic virus in yeast cells (cited by Souer et al. [1996] as a personal communication). That suggests these two proteins may be transcription factors, although their functions in planta are unknown. As stated above, we suspected that TIP was also capable of transcriptional activation in yeast cells. To test this, the TIP coding sequence was cloned into the DNA binding domain vector pAS2-1 so that a DNA binding domain–TIP fusion protein would be produced when the yeast cells were transformed. The results in Figure 3 show that the DNA binding domain–TIP fusion protein is capable of activating the expression of the reporter gene in yeast in the absence of a complementing activation domain vector. This result provides direct evidence for the presence of an activation domain within TIP itself and suggests that TIP is a transcriptional activator.
Figure 3.
Interaction between TIP and the R Domain of TCV CP and Demonstration That TIP Is a Transcriptional Activator.
The result of one set of yeast two-hybrid assays is shown. The selection markers incorporated into the yeast growth media are shown at left: media at top lacked tryptophan (Trp–), media at center lacked both tryptophan and leucine (Trp–, Leu–), and media at bottom were Trp– and contained X-gal. The yeast strain (Y190) was deficient in producing both tryptophan and leucine (Trp–, Leu–). The activation domain vector (pGAD10) was Leu+ and Trp–, and the DNA binding domain vector (pAS2-1) was Leu– and Trp+. The first column shows that Y190 transformed with the activation domain vector containing TIP fused in frame with activation domain (AD-TIP) did not grow on Trp– medium. Y190 transformed with the DNA binding domain vector containing the R domain of TCV CP fused in frame with DNA binding domain (DNA-BD-R) did grow (second column). The third column shows that only AD-TIP in combination with DNA-BD-R was able to grow on the Trp–, Leu– medium. The third column also shows that activation of the reporter gene leading to the blue color in the X-gal–containing medium occurred only when both AD-TIP and DNA-BD-R were present. Columns four and five demonstrate the ability of TIP alone to activate the reporter gene. In these assays, TIP was fused in frame with DNA binding domain (DNA-BD-TIP) and transformed into Y190. Two separate colonies were chosen.
Characterization of the TIP Gene
The cDNA sequence of TIP was also subjected to a database search (GenBank, release 12/98) with the BLAST program. The TIP cDNA sequence could be identified as several discontinuous regions (from 5′ to 3′: 375, 281, 258, 459, and 439 bp) in the sequence of a genomic DNA TAC clone (K18P6) derived from chromosome 5 of Arabidopsis. The four interrupting regions between the coding regions (from 5′ to 3′: 192, 92, 85, and 79 bp) are apparently the introns. We concluded that this portion of clone K18P6 is the TIP gene.
Mapping the TIP-Interacting Region in the TCV CP
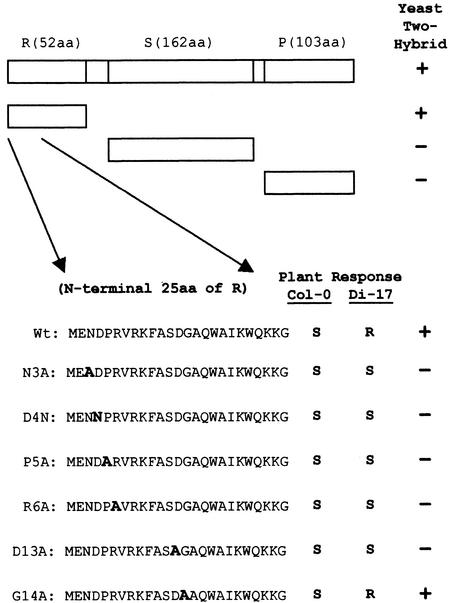
The yeast two-hybrid screen with which we identified the TIP–CP interaction used the entire CP coding sequence. This complex viral structural protein consists of three well-defined structural domains (Hogle et al., 1986; Sorger et al., 1986; Carrington et al., 1987): a basic N-terminal R domain that extends into the interior of the virus particle where it interacts with the viral RNA, an S domain that constitutes the virion shell, and a P domain that projects outward from the virion surface. To map the region of the CP responsible for specific interaction with TIP, we tested R, S, and P domains separately for their interaction with TIP, using the yeast two-hybrid system (Figure 4). The results established that the N-terminal 52–amino acid region that makes up the R domain of the CP was the only region required for specific interaction with TIP. Additional deletion analysis further mapped the interacting site to the N-terminal 25 amino acids of the R domain. We then introduced six single amino acid changes shown in Figure 4 into this region and tested the resulting mutants for interaction with TIP. Five out of the six mutants resulted in loss of the specific interaction with TIP. The only exception was at amino acid position 14 of TCV CP, where changing a glycine residue to an alanine did not affect the interaction (Figure 4).
Figure 4.
Correlation between TIP–CP Interaction and TCV Resistance.
The diagram shows the full-length TCV CP with different structural domains (R domain, S domain, and P domain) and identifies their lengths. The two small unlabeled intervals are the arm (29 amino acids) and the hinge (five amino acids) (see text for details). The regions of the CP tested are shown as the open boxes. The first 25 amino acids are shown for each of the CP mutants in which single amino acid changes were made, with the exact substitution in the respective mutant highlighted by bold letter A or N, representing alanine or asparagine residues, respectively. The interaction of the mutant CP with TIP in yeast two-hybrid assays is shown in the right column. The response of susceptible (Col-0) and resistant (Di-17) Arabidopsis plants to each of the tested mutants is shown in the left and middle columns, respectively. aa, amino acid; (+), positive interaction; (−), no interaction detected; R, typical resistance response; S, typical susceptible infection; Wt, wild type.
Effect of Single Amino Acid Changes on the Infectivity of TCV in Both Susceptible and Resistant Arabidopsis Plants
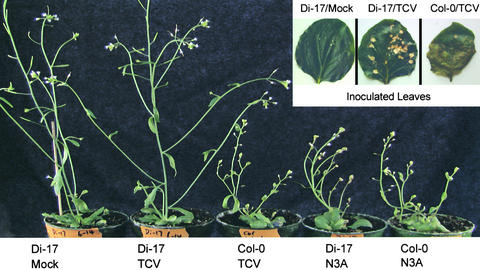
The TCV CP has previously been identified as the elicitor of the TCV resistance in the ecotype Di-0 and its inbred line Di-17 (Oh et al., 1995; Wobbe and Zhao, 1998). The resistance was found to function at the level of long-distance movement (Simon et al., 1992; Oh et al., 1995), which is thought to require assembled virions. The fact that TIP interacts primarily with the R domain of the CP, a domain not usually accessible in the assembled virion, raised the prospect that TIP might participate in the resistant response by preventing virus assembly. We therefore evaluated the effect of these R domain mutations on viral infection in protoplasts as well as Arabidopsis plants. The same single amino acid mutations were introduced into the full-length TCV infectious clone, and infectious transcripts of each clone were produced. Each mutant was first tested for its ability to replicate and assemble into virus particles in protoplasts (data not shown). None was found to be defective in either replication or virion assembly. We then inoculated both susceptible and resistant Arabidopsis plants with these mutants. As shown in Figure 5, typical symptoms of TCV infection were evident in Col-0 within 2 weeks of inoculation, whereas under the same conditions, the TCV-resistant Di-17 plants mounted an HR on the inoculated leaves and showed no visible signs of virus infection in the rest of the plant. As summarized in Figure 4 and illustrated in Figure 5, the mutants that resulted in loss of TIP interaction infected both the susceptible and resistant lines systemically. Only the mutant G14A, which retained TIP interaction, showed the same infection pattern as wild-type TCV. These results show that loss of resistance in Di-17 was absolutely correlated with loss of TIP–CP interaction in the yeast two-hybrid system (Figure 4). The positive correlation between the ability of TCV CP to interact with TIP and the ability of TCV to induce resistance in Di-17 implicates TIP as an important component of the resistance process and strongly suggests that the TIP–CP interaction may be the initiating event in the resistance response.
Figure 5.
TCV-Infected Arabidopsis Plants.
Shown is the typical response of the resistant (Di-17) and susceptible (Col-0) plants to infection with wild-type TCV at 3 weeks after inoculation. The insert shows the typical HR on the inoculated leaves of the resistant plants (Di-17/TCV) at 7 days after inoculation; this is absent in the inoculated leaves of the susceptible Col-0 plants (Col-0/TCV). Di-17/Mock refers to a leaf of a Di-17 plant that was inoculated with the inoculation buffer only. The two plants at right were inoculated with the representative mutant, N3A. Both the Di-17 and Col-0 plants show identical symptoms typical of wild-type TCV infection. Also, as with wild-type plants, the HR was not observed on the inoculated leaves (data not shown).
TIP Gene Comparison and Expression in the TCV-Susceptible and TCV-Resistant Plants
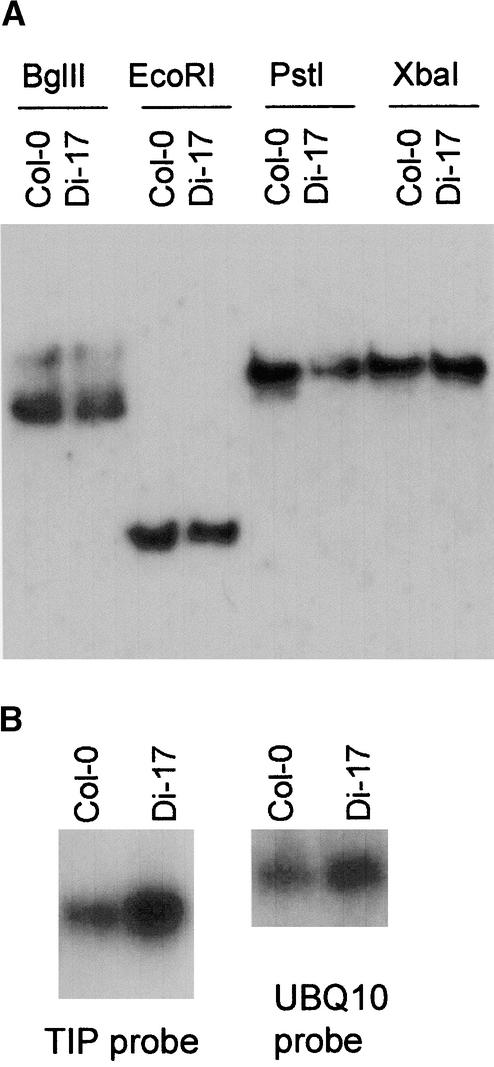
Having established TIP as an important participant in the resistance process, we sought to determine if there were any differences in the TIP gene in the resistant and susceptible lines. Equivalent amounts of genomic DNA were isolated from Col-0 and Di-17 plants; digested with EcoRI, BglII, PstI, and XbaI; and subjected to DNA gel blot hybridization with a TIP-specific probe. As Figure 6A shows, the TIP gene is most likely a single-copy gene in both ecotypes. We examined for sequence variation between the TIP genes of both ecotypes by sequencing the PCR fragments amplified from Col-0 and Di-17 genomic DNA. Sequence comparison of the PCR products showed that the genes from both plants were identical except for a single A→G change in the middle of the first intron, which should not affect the coding sequence. The sequences of the TIP mRNAs in these two Arabidopsis lines were also found to be identical by analyzing their RT-PCR products. Examination by RNA gel blot analysis of the TIP mRNA contents of the polyadenylated RNAs isolated from both uninfected and infected Col-0 and Di-17 plants indicated that TIP mRNA did not appear to be induced by TCV infection (data not shown). The Di-17 plants appears to have more TIP mRNA than do the Col-0 plants (Figure 6B). Given the likelihood that TIP expression may be developmentally regulated, more careful evaluation by using plant tissues of different ages and from different plant organs or sections is needed.
Figure 6.
Analysis of the TIP Gene and Its Expression.
(A) DNA gel blot analysis of TIP genomic DNA isolated from Col-0 and Di-17 plants. (B) RNA gel blot analysis of the TIP mRNA isolated from 4-week-old uninoculated Col-0 and Di-17 plants. Equal amount of poly(A)+ RNAs were also probed with a ubiquitin 10 (UBQ10) probe to serve as control.
DISCUSSION
We have identified an Arabidopsis protein (TIP) that specifically interacts with the coat protein of TCV. This interaction is positively correlated with the resistance phenotype in the TCV-resistant Di-17 plants, thus implicating TIP as an important participant in the resistance response. We also confirmed TCV CP as the elicitor of TCV resistance and further delineated the functional region to the N-terminal 25–amino acid residues. Together, these results strongly suggest that TIP is the intracellular receptor of the TCV elicitor, as defined in the elicitor–receptor model (Gabriel and Rolfe, 1990). This model, generally used to explain the molecular basis of the gene-for-gene hypothesis of resistance (Flor, 1971), postulates that to initiate the resistance response, the Avr gene product of the pathogen must be recognized by the corresponding R gene product of the plant through a specific ligand–ligand interaction, most commonly a protein–protein interaction. However, several lines of evidence suggest that a more complex model, similar to the Pto/Prf–mediated resistance response in tomato to bacterial speck disease, may be more relevant for the TCV–Arabidopsis system.
First, most of the characterized R gene products share structural motifs. Among them, nucleotide binding sites (NBS) and leucine-rich repeats (LRR) have been most commonly described. TIP lacks both NBS and LRR motifs. However, the product of HRT, a newly characterized gene involved in the TCV resistance response, contains both NBS and LRR motifs (Cooley et al., 2000). We think it is highly likely that both TIP and HRT are needed for the TCV resistance. We suggest that the TIP functions in a manner analogous to the product of the tomato R gene Pto, which confers resistance to the bacteria pathogen Pseudomonas syringae pv tomato. Pto protein is analogous to TIP in several ways. It does not possess NBS and LRR motifs but rather functions in concert with an NBS/LRR–type resistance gene, Prf (Salmeron et al., 1996). In addition, as does TIP, Pto specifically interacts with the pathogen elicitor, and that specific interaction is required for resistance (Scofield et al., 1996; Tang et al., 1996). These features prompted Van der Biezen and Jones (1998b) to propose a “guard” hypothesis in which they suggest that Pto functions to promote a basal level of resistance by activating generic defense responses in the host. In this model, the pathogen elicitor AvrPto (Avr gene product) functions as a virulence factor by binding to Pto to prevent the induction of basal resistance. The role of the resistance gene Prf, in this model, is to “guard” Pto by recognizing the AvrPto–Pto complex and initiating a defense response.
LRR motifs have been shown to play a more general role in protein–protein interactions (Kobe and Deisenhofer, 1995). Originally it was proposed that this motif possibly served as the receptor of the elicitor produced by various pathogens (Bent, 1996; Baker et al., 1997). However, direct interaction between the LRR-containing R gene product and the corresponding Avr gene product has yet to be reported. Recent evidence suggests that the LRR motif in some R gene products might interact instead with other signal transduction components (Van der Biezen and Jones, 1998a; Warren et al., 1998). If so, a mediating component should be present in plants to bridge the Avr gene product and the LRR-containing R gene product. We speculate, therefore, that both TIP and Pto may have similar roles, the primary difference being that TIP may directly activate the transcription of genes involved in the basal resistance, whereas Pto does so through a more elaborate cascade (Zhou et al., 1995, 1997).
The observation that the TIP gene is present in both TCV-susceptible and TCV-resistant plants is not unprecedented. A similar situation was observed in the Avr9-Cf9 system, where a high-affinity binding site for Avr9, the elicitor of the fungus Cladosporium fulvum, is present on the plasma membrane of both resistant and susceptible tomato genotypes (Kooman et al., 1996, 1998). As in the TIP–TCV CP interaction, the binding affinity of this membrane-anchored binding site with Avr9 is positively correlated with the induction of necrosis (HR) in the resistant plants. Although the molecular nature of this Avr9 binding site remains to be elucidated, it is unlikely to be the product of the corresponding resistance gene Cf9, which possesses an LRR motif.
TIP is also unusual in its capability for transcriptional activation in yeast cells. Interestingly, two other NAC proteins, ATAF1 and ATAF2, activate transcription in yeast cells as well (cited by Souer et al. [1996] as a personal communication). Recently, putative nuclear localization signals have been found in NAC proteins (Kikuchi et al., 2000). These observations suggest that the NAC proteins are transcription factors. If it can be proven that TIP is a transcriptional activator, then it would be the first example of a transcription factor serving as a pathogen receptor. This raises the possibility that TIP may directly activate pathogenesis-related (PR) genes or other genes involved in the resistance pathway (Zhou et al., 1997; Zhang et al., 1999) to establish a basal level of resistance, consistent with the guard hypothesis (Van der Biezen and Jones, 1998b).
In summary, the identification of TIP as a component of Arabidopsis resistance response to TCV is consistent with the guard hypothesis proposed by Van der Biezen and Jones (1998b) if we modify the model as follows: TIP functions through transcriptional activation to promote a basal level of resistance in the plant. The viral CP, produced in infected cells in very high concentrations during infection, functions as a virulence factor by binding to TIP to reduce basal resistance and to promote rapid systemic infection. Resistant plants expressing the HRT protein may well guard the TIP protein by detecting a change in TIP caused by the TIP–CP interaction, which then results in a stronger, HR-mediated resistance response.
TIP is a member of the NAC family of proteins. The fact that both of the well-characterized NAC proteins from Arabidopsis (CUC2 and NAP) are involved in the development of the plant suggests that TIP may play an important developmental role as well. The resistance response is developmentally regulated (Dixon and Lamb, 1990; Crowell et al., 1992; Hammond-Kosack et al., 1994; Honee et al., 1998), and the resistance pathway is also well known to share similarity with developmental signal transduction pathways. For example, a subset of R gene products represented by the tobacco N gene product (Whitman et al., 1994) possess N-terminal domains with homology to the cytoplasmic domain of the Toll protein of Drosophila. Another example is the structural similarity between CLAVATA1 gene product of Arabidopsis (Clark et al., 1997) and Xa21 protein in rice, which confers resistance to the bacterial pathogen Xanthomonas oryzae pv oryzae (Song et al., 1995). TIP is a novel example, however, of a presumed developmental protein being implicated directly in pathogen recognition.
METHODS
Yeast Two-Hybrid Screen
The Arabidopsis cDNA library was purchased from Clontech (Palo Alto, CA). The library was constructed in the yeast activation domain vector pGAD10, resulting in the expression of fusion proteins between the activation domain of the GAL4 transcriptional activator and the polypeptides encoded by the cDNAs upon transformation of the yeast cells. The library was made from 3-week-old Arabidopsis thaliana plants, ecotype Columbia (Col-0). The original library contained 3 × 106 independent clones, which should represent >99% of all Arabidopsis mRNA, taking into account that half of the cDNA might fuse to the activation domain in the reverse direction and that only one-third would be expected to be in frame with activation domain. The gene encoding the turnip crinkle virus capsid protein (TCV CP) was cloned into the yeast DNA binding domain vector pAS2-1 to produce pAS-TCV-CP, which encodes the fusion protein of the DNA binding domain of the Gal4 transcriptional activator and TCV CP when the yeast cells are transformed. The library was amplified and then transformed to yeast strain Y190 containing pAS-TCV-CP. The transfomants were cultured for as long as 10 days at 30°C.
Polymerase Chain Reaction and Reverse Transcription–Polymerase Chain Reaction
To obtain the 3′ end of the TIP cDNA, we used the following oligodeoxyribonucleotide primers: oligo(dT), GGTCTAGAT22(A/C/G); and full 5′, CAGGACTAGTTTCATATTC. RT-PCR was performed using the Titan One Tube RT-PCR kit from Roche Molecular Biochemicals (Mannheim, Germany). To amplify the full-length TIP gene by PCR, the following two oligodeoxyribonucleotide primers were used: TIP 5′, GCGGATCCACTTTACCAAATCGTGTC (identical to nucleotides 1 to 20 of TIP cDNA); and TIP full 3′, GGTCTAGACGGATTTATGCGACTAG (complementary to nucleotides 1558 to 1542). The genomic DNA templates were prepared from both Col-0 and Di-17 plants. The same primer set was also used to amplify the cDNAs from polyadenylated RNAs by RT-PCR.
Plant Material
The susceptible line Col-0 and the resistant line Di-17 (kindly provided by Drs. D. Klessig and K. Wobbe) were grown in a growth chamber at 20°C with a 14-hr daylength. They were inoculated at a plant age of 18 to 20 days.
DNA and RNA Gel Blot Analyses
Standard procedures were followed for preparation of the Arabidopsis genomic DNA and DNA gel blot analysis by using a 32P-random-labeled TIP probe (Ausubel et al., 1993). mRNAs were prepared from Arabidopsis plants by using a commercial kit (Promega). A 32P-labeled (−) strand RNA probe of TIP was used to probe the RNA blots. For probing the ubiquitin 10 mRNA, the following 32P-end-labeled oligodeoxyribonucleotide probe was used: 5′-TTACATGAAACGAAACATTGAACTTCTTAAGCATAACAGAGACGAGATTTAG-3′.
Acknowledgments
We thank Drs. K. Wobbe and D. Klessig for providing us with the Di-17 seeds; Dr. Klessig is also thanked for sharing his manuscripts before publication. We thank Drs. Roy French and Audry Atkin for helpful discussions. We also thank Drs. Martin Dickman and Jim Carrington for critical reading of the manuscript. Part of this research was supported by a U.S. Department of Energy grant (No. DE-FG03-98ER20315).
References
- Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1993). Current Protocols in Molecular Biology. (New York: Greene Publishing Association/Wiley Interscience).
- Baker, B., Zambryski, P., Staskawicz, B., and Dinesh-Kumar, S.P. (1997). Signaling in plant–microbe interactions. Science 276, 726–733. [DOI] [PubMed] [Google Scholar]
- Bent, A.F. (1996). Plant disease resistance genes: Function meets structure. Plant Cell 8, 1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C., Morris, T.J., Stockley, P.G., and Harrison, S.C. (1987). Structure and assembly of turnip crinkle virus. IV. Analysis of the coat protein gene and implications of the subunit primary structure. J. Mol. Biol. 194, 265–276. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Cooley, M.B., Pathirana, S., Wu, H.-J., Kachroo, P., and Klessig, D.F. (2000). Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 12, 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell, D.N., John, M.E., Russell, D., and Amasino, R.M. (1992). Characterization of a stress-induced, developmentally regulated gene family from soybean. Plant Mol. Biol. 18, 459–466. [DOI] [PubMed] [Google Scholar]
- Dempsey, D.A., Wobbe, K.K., and Klessig, D.F. (1993). Resistance and susceptible responses of Arabidopsis thaliana to turnip crinkle virus. Phytopathology 83, 1021–1029. [Google Scholar]
- Dempsey, D.A., Pathirana, M.S., Wobbe, K.K., and Klessig, D.F. (1997). Identification of an Arabidopsis locus required for resistance to turnip crinkle virus. Plant J. 11, 301–311. [DOI] [PubMed] [Google Scholar]
- Dixon, R.A., and Lamb, C.J. (1990). Molecular communication in interactions between plants and microbial pathogens. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41, 339–367. [Google Scholar]
- Flor, H.H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Gabriel, D.W., and Rolfe, B.G. (1990). Working models of specific recognition in plant–microbe interactions. Annu. Rev. Phytopathol. 28, 365–391. [Google Scholar]
- Hammond-Kosack, K.E., Harrison, K., and Jones, J.D.G. (1994). Developmentally regulated cell death on expression of the fungal avirulence gene Avr9 in tomato seedlings carrying the disease-resistance gene Cf-9. Proc. Natl. Acad. Sci. USA 91, 10445–10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogle, J.M., Maeda, A., and Harrison, S.C. (1986). Structure and assembly of turnip crinkle virus. I. X-ray crystallographic structure analysis at 3.2Å resolution. J. Mol. Biol. 191, 625–638. [DOI] [PubMed] [Google Scholar]
- Honee, G., Buitink, J., Jabs, T., De Kloe, J., Sijbolts, F., Apotheker, M., Weide, R., Sijen, T., Stuiver, M., and De Wit, P.J.G.M. (1998). Induction of defense-related responses in Cf9 tomato cells by the Avr9 elicitor peptide of Cladosporium fulvum is developmentally regulated. Plant Physiol. 117, 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, K., Ueguchi-Tanaka, M., Yoshida, K.T., Nagato, Y., Matsusoka, M., and Hirano, H.-Y. (2000). Molecular analysis of the NAC gene family in rice. Mol. Gen. Genet. 262, 1047–1051. [DOI] [PubMed] [Google Scholar]
- Kobe, B., and Deisenhofer, J. (1995). A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature 374, 183–186. [DOI] [PubMed] [Google Scholar]
- Kooman, G.M., Honee, G., Bonnema, G., and De Wit, P.J.G.M. (1996). A high-affinity binding site for the Avr9 peptide elicitor of Cladosporium fulvum is present on plasma membranes of tomato and other solanaceous plants. Plant Cell 8, 929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooman, G.M., Vogelsang, R., Van Den Vossen, P.H.H.W., Mahe, E., Honee, G., and De Wit, P.J.G.M. (1998). Correlation between binding affinity and necrosis-inducing activity of mutant Avr9 peptide elicitors. Plant Physiol. 117, 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, J.-W., Kong, Q., Song, C., Carpenter, C.D., and Simon, A.E. (1995). Open reading frames of turnip crinkle virus involved in satellite symptom expression and incompatibility with Arabidopsis thaliana ecotype Dijon. Mol. Plant-Microbe Interact. 8, 979–987. [DOI] [PubMed] [Google Scholar]
- Sablowski, R.W.M., and Meyerowitz, E.M. (1998). A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92, 93–103. [DOI] [PubMed] [Google Scholar]
- Salmeron, J.M., Oldroyd, G.E.D., Rommens, C.M.T., Scofield, S.R., Kim, H.-S., Lavelle, D.T., Dahlbeck, D., and Staskawicz, B.J. (1996). Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 86, 123–134. [DOI] [PubMed] [Google Scholar]
- Scofield, S.R., Tobias, C.M., Rathjen, J.P., Chang, J.H., Lavelle, D.T., Michelmore, R.W., and Staskawicz, B.J. (1996). Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274, 2063–2065. [DOI] [PubMed] [Google Scholar]
- Simon, A.E., Li, X.H., Lew, J.E., Stange, R., Zhang, C., Polacco, M., and Carpenter, C.D. (1992). Susceptibility and resistance of Arabidopsis thaliana to turnip crinkle virus. Mol. Plant-Microbe Interact. 5, 496–503. [Google Scholar]
- Song, W.-Y., Wang, G.-L., Chen, L.-L., Kim, H.-S., Pi, L.-Y., Holsten, T., Gardner, J., Wang, B., Zhai, W.-X., Zhu, L.-H., Fauquet, C., and Ronald, P. (1995). A receptor-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. [DOI] [PubMed] [Google Scholar]
- Sorger, P.K., Stockley, P.G., and Harrison, S.C. (1986). Structure and assembly of turnip crinkle virus. II. Mechanism of in vitro assembly. J. Mol. Biol. 191, 375–383. [DOI] [PubMed] [Google Scholar]
- Souer, E., van Houwelingen, A., Kloos, D., Mol, J., and Koes, R. (1996). The No Apical Meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed as meristem and primordia boundaries. Cell 85, 159–170. [DOI] [PubMed] [Google Scholar]
- Tang, X., Frederick, R.D., Zhou, J., Halterman, D.A., Jia, Y., and Martin, G.B. (1996). Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274, 2060–2063. [DOI] [PubMed] [Google Scholar]
- Van der Biezen, E.A., and Jones, J.D.G. (1998. a). The NB-ARC domain: A novel signaling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 8, 226–227. [DOI] [PubMed] [Google Scholar]
- Van der Biezen, E.A., and Jones, J.D.G. (1998. b). Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23, 454–456. [DOI] [PubMed] [Google Scholar]
- Warren, R.F., Henk, A., Mowery, P., Holub, E., and Innes, R.W. (1998). A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell 10, 1439–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman, S., Dinesh-Kumar, S.P., Choi, D., Hehl, R., Corr, C., and Baker, B. (1994). The product of the tobacco mosaic virus resistance gene N: Similarity to Toll and the interleukin-1 receptor. Cell 78, 1101–1115. [DOI] [PubMed] [Google Scholar]
- Wobbe, K.K., and Zhao, Y. (1998). Avirulence determinant of turnip crinkle virus localized to the N-terminus of the coat protein. In Abstracts of the 17th Annual Meeting of American Society for Virology (Vancouver: American Society for Virology), p. 169, P6–20.
- Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96, 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Loh, Y.-T., Bressan, R.A., and Martin, G.B. (1995). The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell 83, 925–935. [DOI] [PubMed] [Google Scholar]
- Zhou, J., Tang, X., and Martin, G.B. (1997). The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 16, 3207–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]