Abstract
The pea lip1 (light-independent photomorphogenesis1) mutant shows many of the characteristics of light-grown development when grown in continuous darkness. To investigate the identity of LIP1, cDNAs encoding the pea homolog of COP1, a repressor of photomorphogenesis identified in Arabidopsis, were isolated from wild-type and lip1 pea seedlings. lip1 seedlings contained a wild-type COP1 transcript as well as a larger COP1′ transcript that contained an internal in-frame duplication of 894 bp. The COP1′ transcript segregated with the lip1 phenotype in F2 seedlings and could be translated in vitro to produce a protein of ∼100 kD. The COP1 gene in lip1 peas contained a 7.5-kb duplication, consisting of exons 1 to 7 of the wild-type sequence, located 2.5 kb upstream of a region of genomic DNA identical to the wild-type COP1 DNA sequence. Transcription and splicing of the mutant COP1 gene was predicted to produce the COP1′ transcript, whereas transcription from an internal promoter in the 2.5-kb region of DNA located between the duplicated regions of COP1 would produce the wild-type COP1 transcript. The presence of small quantities of wild-type COP1 transcripts may reduce the severity of the phenotype produced by the mutated COP1′ protein. The genomic DNA sequences of the COP1 gene from wild-type and lip1 peas and the cDNA sequences of COP1 and COP1′ transcripts have been submitted to the EMBL database under the EMBL accession numbers AJ276591, AJ276592, AJ289773, and AJ289774, respectively.
INTRODUCTION
Light has a dramatic effect on plant morphology during early development (McNellis and Deng, 1995). After growth in continuous darkness, seedlings show skotomorphogenic or etiolated development, having long hypocotyls, apical hooks, and low expression of nuclear photosynthesis genes (Holm and Deng, 1999). In contrast, growth in the light results in photomorphogenic development, characterized by a short hypocotyl, the absence of an apical hook, and high expression of nuclear photosynthesis genes (Holm and Deng, 1999).
Many mutants that show altered responses to light have been identified by the use of genetic screens (Chory, 1993). Photomorphogenic mutants can be broadly classified into two groups: those that show dark-grown development in the light, and those that show light-grown development in darkness (Staub and Deng, 1996). Mutants in positive regulators of light-grown development show an hy (hypocotyl-elongated) phenotype (Chory, 1993). This phenotype can be produced by mutations in genes encoding photoreceptors (Ahmad and Cashmore, 1993; Weller et al., 1996) or encoding downstream signaling components (Oyama et al., 1997; Chattopadhyay et al., 1998). Mutations in negative regulators of light-grown development lead to light-grown development in darkness (Kwok et al., 1996; Wei and Deng, 1996). In Arabidopsis, 11 essential CONSTITUTIVELY PHOTOMORPHOGENIC/DEETIOLATED/FUSCA (COP/DET/FUS) genes have been identified that are required for the repression of light-grown development in darkness, and many of the proteins encoded by these genes have been identified (Wei and Deng, 1996, 1999; Holm and Deng, 1999). COP1 encodes a protein showing homology to Gβ proteins and contains a zinc binding RING-finger domain, a coiled-coil domain, and at least five WD-40 repeat motifs (Deng et al., 1992). COP1 shows nuclear enrichment in darkness but not in light (von Arnim and Deng, 1994), an enrichment that requires COP9 and DET1 (Chamovitz et al., 1996). COP9 exists in a large nuclear-localized protein complex (>560 kD) with at least eight distinct subunits, called the COP9 signalosome (Kwok et al., 1998; Karniol et al., 1999; Serino et al., 1999). The COP9 signalosome shows some similarity to the non-ATPase regulatory subunits of the 26S proteasome complex, is highly conserved in multicellular organisms, and is likely to represent a conserved cellular and developmental regulator (Seeger et al., 1998; Wei et al., 1998; Holm and Deng, 1999). COP1 interacts with HY5, a bZIP transcription factor responsible for the activation of light-regulated genes (Oyama et al., 1997; Ang et al., 1998; Osterlund et al., 1999), and targets the protein for proteosome-mediated degradation in the nucleus (Osterlund et al., 2000). COP1 interaction with HY5 and other transcription factors in darkness prevents them from activating target gene expression, thereby inhibiting photomorphogenic development (Ang et al., 1998).
Photomorphogenic mutants have been isolated in several plants, including tomato and pea, in addition to Arabidopsis (von Arnim and Deng, 1996; Mustilli et al., 1999). The pea lip1 (light-independent photomorphogenesis1) mutant shows many of the characteristics associated with cop/det/fus mutants from Arabidopsis (Frances et al., 1992). Dark-grown lip1 seedlings have short stems and open and expanded shoots; the shoots contain partially developed chloroplasts and transcripts of the nuclear photosynthesis genes Lhcb1, Fed1, RbcS, PetE, and AtpC in quantities comparable with those found in light-grown seedlings (Frances et al., 1992; Sullivan and Gray, 1999). The dark-grown lip1 mutant contains 10-fold less spectrally detectable phytochrome than do wild-type seedlings, and this decrease correlates with a 10-fold reduction in phytochrome A (PHYA) apoprotein (Frances et al., 1992). However, the spectral properties of phytochrome found in dark-grown seedlings were indistinguishable from that found in wild-type seedlings (Sineschchekov et al., 1997). The recessive lip1 mutation maps to a single locus and causes pleiotropic effects throughout development, including dwarfism, which is associated with a decrease in the ratio of GA19 to GA20 (Sponsel et al., 1996), and the ability of 9-day-old plants to respond to darkness (Frances et al., 1992; Frances and Thompson, 1997). However, dark-grown lip1 seedlings still contain protochlorophyllide reductase at levels comparable with those found in wild-type seedlings (Seyyedi et al., 1999).
The aim of this study was to identify the nature of the pea lip1 mutant. To date, only one COP/DET/FUS gene, COP1, has been identified in peas (Zhao et al., 1998). Because the roots of light-grown lip1 seedlings were reported not to green, Frances et al. (1992) concluded that LIP1 was unlikely to be a homolog of either COP1 or DET1. However, the observation that the tomato hp2 mutant, which does not show deetiolated development when grown in darkness, is caused by a defect in the tomato homolog of DET1 suggests phenotypes of cop/det/fus mutations may be species-specific (Mustilli et al., 1999). Furthermore, given recent reports that light-grown lip1 roots contain increased amounts of several nuclear photosynthesis gene transcripts associated with the development of chloroplasts (Sullivan and Gray, 1999), the possibility that LIP1 is a pea homolog of COP1 cannot be ruled out.
To examine the possibility that LIP1 is a homolog of COP1, we isolated cDNAs encoding COP1 from both lip1 and wild-type peas. We discovered that the lip1 seedlings contained both a wild-type COP1 transcript and a larger transcript (designated COP1′) consisting of an in-frame internal duplication within the wild-type COP1 cDNA. Analysis suggested that the COP1′ transcript was caused by a single recessive mutation that segregated with the lip1 phenotype at both the RNA and DNA level. Furthermore, determination of the genomic DNA sequence of pea COP1 showed that lip1 peas also contained a large duplication within COP1 that, on transcription and splicing, would produce a COP1′ transcript. Transient assays and transgenic Arabidopsis studies demonstrated the presence of a promoter element within the duplicated COP1 that would be capable of producing the low amounts of wild-type COP1 transcript observed in lip1 seedlings. These results clearly demonstrate that the lip1 phenotype is caused by an unusual duplication within the pea homolog of COP1.
RESULTS
lip1 Seedlings Contain COP1 and COP1′ Transcripts
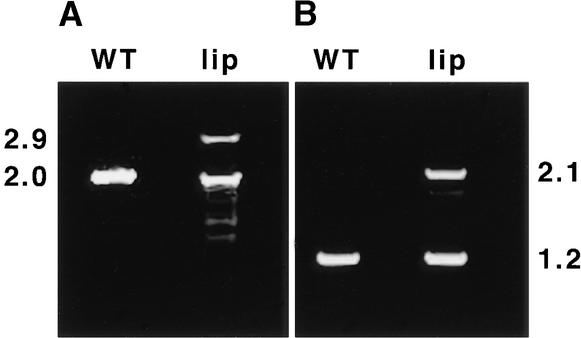
To investigate the possibility that lip1 results from a mutation in a pea COP1 homolog, cDNAs encoding pea COP1 were isolated from wild-type and lip1 peas by a reverse transcription–polymerase chain reaction (RT-PCR) procedure with primers designed from the published sequence of a pea COP1 cDNA (Zhao et al., 1998). Figure 1 shows the results of RT-PCR using two sets of primer pairs to amplify cDNA produced from total RNA extracted from wild-type and lip1 pea seedlings. After RT-PCR with both pairs of primers, bands corresponding to the expected pea COP1 cDNA fragments (2.0 and 1.2 kb) were observed in reactions with cDNA produced from wild-type and lip1 seedlings. However, a second major band, ∼900 bp larger than the expected COP1 cDNA fragments, was also observed in PCR reactions that used lip1 cDNA as a template. These larger cDNA fragments (2.9 and 2.1 kb) were observed with both pairs of primers (see Figure 1) and were produced consistently on cDNA derived from several RNA extractions from lip1 seedlings (data not shown). The larger cDNA was designated COP1′.
Figure 1.
RT-PCR of Wild-Type and lip1 Seedlings.
Wild-type (WT) and lip1 seedlings were grown for 7 days in continuous darkness. Shoot tissue was excised from the seedlings, and the total RNA was extracted and used to produce first-strand cDNA by using oligo(dT) primers and reverse transcriptase. The resulting cDNA was used as template in PCR reactions using two sets of primers.
(A) Primers PCCOPF and PCCOPR (see Methods), which are expected to amplify a 2016-bp fragment containing the entire pea COP1 coding region.
(B) Primers FS1 and RS1 (see Methods), which are expected to amplify a 1227-bp region within the pea COP1 open reading frame.
The PCR products produced were separated by electrophoresis in a 1% agarose gel. Numbers at left and right indicate the approximate sizes (in kilobases) of PCR products produced.
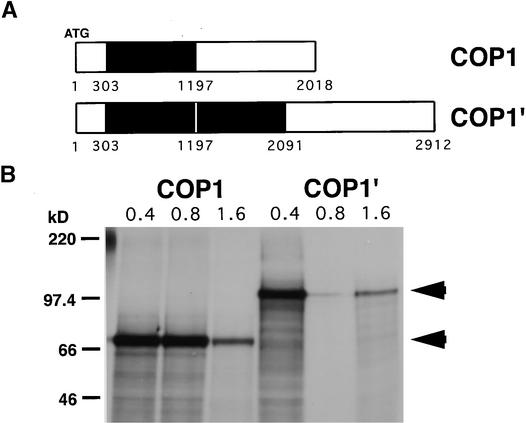
Sequence analysis showed that the PCR products produced from wild-type cDNA and the smaller of the two cDNAs produced from lip1 were identical, showing 99.4% sequence identity to the published sequence of pea COP1 (Zhao et al., 1998). These cDNAs contained an open reading frame of 672 codons producing a protein with a predicted molecular mass of 76 kD. Analysis of the larger DNA fragment produced from lip1 cDNA showed it to contain an internal duplication within the pea COP1 cDNA (see Figure 2A). This duplication consists of an 894-bp region (+303 to +1197, relative to the translation start) duplicated within the pea COP1′ cDNA, producing an open reading frame of 970 codons encoding a protein with a predicted molecular mass of 110 kD. Both COP1 and COP1′ cDNAs were transcribed and translated in vitro, producing single major translation products of 70 and 100 kD, respectively (see Figure 2B), similar to the molecular masses predicted from the DNA sequences.
Figure 2.
Schematic Representation and Translation in Vitro of COP1 and COP1′ Transcripts.
(A) Schematic representation of COP1 and COP1′ transcripts as determined from sequence analysis of the cDNAs amplified with PCCOPF and PCCOPR primers. The region in black indicates the region from +303 to +1197 (relative to start codon) duplicated in the COP1′ transcript.
(B) Cloned RT-PCR products were transcribed in vitro using T7 RNA polymerase, and increasing amounts (0.4 to 1.6 μg) of RNA were translated by using wheat germ extract and 35S-labeled methionine and cysteine. Translation products were separated by electrophoresis on an SDS–10% polyacrylamide gel and compared with markers of molecular mass, shown at left in kilodaltons. Arrowheads indicate major translation products of 100 and 70 kD.
The COP1′ Transcript Segregates with the lip1 Phenotype
To determine if the photomorphogenic phenotype of dark-grown lip1 seedlings was correlated with the presence of the COP1′ transcript, the segregation of the COP1′ transcript and the lip1 phenotype was investigated. F1 progeny from a cross between wild-type and lip1 seedlings showed a wild-type skotomorphogenic phenotype when grown in darkness, suggesting a recessive mutation, as showed previously by Frances et al. (1992). In 60 F2 progeny examined, the lip1 photomorphogenic phenotype segregated in dark-grown seedlings at a ratio of ∼3:1 (48:12 wild type:lip1), confirming that the lip1 phenotype is caused by a recessive mutation at a single locus.
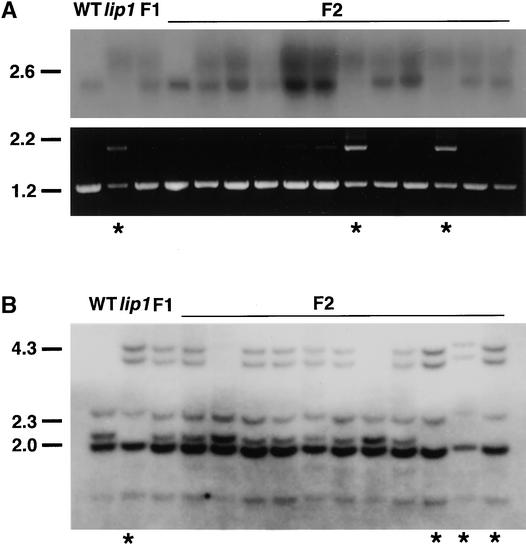
Total RNA was extracted from dark-grown wild-type, lip1, F1, and F2 seedlings and subjected to RNA gel blot analysis with a probe made from the pea COP1 cDNA (Figure 3A, top). In total RNA extracted from wild-type seedlings, only a single band of 2.1 kb that hybridized to a probe for pea COP1 mRNA was observed (Figure 3A, top). In contrast, in dark-grown lip1 seedlings a more diffuse region of hybridization, corresponding to transcripts ∼1 kb larger than that found in wild-type seedlings, was observed. In F1 seedlings (which showed a wild-type phenotype), both of the transcripts found in wild-type and lip1 seedlings were observed (Figure 3A, top). In F2 seedlings, the larger COP1′ transcript was the only transcript detected in seedlings showing the lip1 phenotype; the smaller COP1 transcript was present in all other seedlings, which showed a wild-type phenotype (Figure 3A, top).
Figure 3.
Segregation Analysis of Transcripts and Genomic DNA in F2 Progeny.
Wild-type (WT) and lip1 pea plants were crossed to produce F1 plants, which were allowed to self-fertilize to produce F2 seed. Parental wild-type, lip1, F1, and F2 seeds were germinated and grown for 7 days in darkness, at which time the lip1 phenotype was scored; asterisks indicate seedlings showing the lip1 phenotype.
(A) Transcript analysis. Total RNA was extracted from the shoots of parental wild-type, lip1, F1, and 12 F2 seedlings. Top, RNA gel blot analysis using 32P-labeled probe against pea COP1; bottom, RT-PCR products amplified by using primers FS1 and RS1 from first-strand cDNA produced from total RNA used for RNA gel blot analysis.
(B) DNA gel blot analysis. Genomic DNA was extracted from parental wild-type, lip1, F1, and 11 F2 seedlings (different from those shown in [A]). Genomic DNA was digested overnight with DraI, fractionated by electrophoresis on a 0.7% agarose gel, blotted to GeneScreen Plus membrane, and hybridized with a 32P-labeled COP1 probe.
Markers at left in (A) and (B) indicate the positions of size markers in kilobases.
RT-PCR analysis was also performed on the total RNA samples used for RNA gel blot analysis (Figure 3A, bottom). As previously demonstrated, parental wild-type seedlings gave only a single band of the expected size (1.2 kb) after PCR with primers designed from the pea COP1 cDNA sequence (Figure 3A, bottom). PCR with parental lip1 cDNA as a template, however, yielded two bands corresponding to the sizes of COP1 and COP1′ transcripts (1.2 and 2.1 kb, respectively) (Figure 3A, bottom). This indicates that small amounts of COP1 mRNA, which were not detectable by RNA gel blot analysis, were present in lip1 seedlings. PCR with cDNA from F1 seedlings produced only a single band of 1.2 kb, corresponding to the size of the COP1 transcript (Figure 3A, bottom). This observation suggests the preferential amplification of the smaller COP1 cDNA over the COP1′ cDNA during PCR. In F2 seedlings that showed a wild-type phenotype, PCR yielded only a single product of 1.2 kb, corresponding to the size of the predicted COP1 cDNA (Figure 3A, bottom). In contrast, PCR on cDNA produced from F2 seedlings that showed a lip1 phenotype gave two bands of 1.2 and 2.1 kb, corresponding to the predicted sizes of COP1 and COP1′ cDNAs, respectively (Figure 3A, bottom). Because only the COP1′ transcript was observed after RNA gel blot analysis of total RNA extracted from these seedlings, this again suggests the preferential amplification of the smaller COP1 cDNA over COP1′ during the PCR reaction.
The observation that the altered COP1′ transcript segregated with the lip1 phenotype is consistent with the hypothesis that the lip1 phenotype is caused by the presence of the COP1′ mRNA. To determine whether the altered COP1′ transcript resulted from a mutation in the pea COP1 gene, we subjected to DNA gel blot analysis the total genomic DNA from wild-type, lip1, F1, and F2 seedlings. Distinct differences in the pattern of bands that hybridized to a pea COP1 cDNA probe were observed between total genomic DNA from parental wild-type and lip1 seedlings (Figure 3B). lip1 DNA digested with DraI contained two extra bands, of 4.4 and 4.0 kb, and had lost a band of ∼2.2 kb. F1 seedlings showed a banding pattern consistent with a heterozygote containing both parental wild-type and lip1 genomic DNA (Figure 3B). In F2 seedlings, the parental lip1 genomic DNA banding pattern segregated with the lip1 phenotype, whereas a wild-type phenotype was observed in seedlings that showed either the parental wild-type or heterozygous banding pattern (Figure 3B). A ratio of wild-type:heterozygous:lip1 digestion patterns of 2:6:3 was observed in the F2 seedlings examined—not dissimilar to the 1:2:1 ratio expected from a mutation in a single recessive locus. These results demonstrate that differences exist between the COP1 genomic DNA of wild-type and lip1 seedlings. The segregation of these differences in COP1 genomic DNA in F2 seedlings is consistent with the hypothesis that the lip1 phenotype is caused by a recessive mutation in the pea COP1 gene.
Characterization of COP1 Genomic DNA from Wild-Type and lip1 Peas
Because analysis of the digestion patterns of genomic DNA containing COP1 indicated differences between wild-type and lip1 peas, the genomic DNA sequences of COP1 from wild-type and lip1 peas were compared. DNA fragments containing COP1 were amplified from genomic DNA from wild-type and lip1 peas in PCR reactions with primers designed from the COP1 cDNA sequence. DNA sequence was then obtained using a directed sequencing approach.
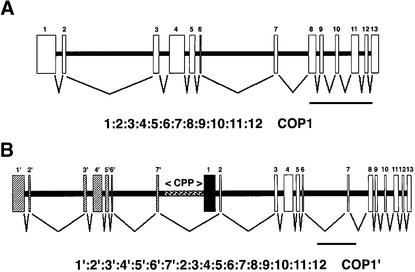
In wild-type peas, the coding region of COP1 was contained within a 9.5-kb region of pea genomic DNA, which was sufficient to generate the pattern of hybridizing bands observed in DNA gel blot analysis and was consistent with COP1 being present as a single copy in the pea genome (data not shown). Alignment of the COP1 cDNA sequence showed that the COP1 coding region was partitioned into 13 exons, separated by 12 introns (Figure 4A). All exons showed 100% identity to the corresponding regions of the COP1 cDNA, and all intron/exon boundaries (Table 1), except for the 3′ splice site of intron 1 and the 3′ splice site of intron 6, were predicted by NetPlantGene intron prediction software (Hebsgaard et al., 1996).
Figure 4.
Schematic Representation of COP1 Genes in Wild-Type and lip1 Peas and Predicted Pattern of Splicing.
(A) COP1 gene in wild-type peas. A 9.5-kb pea genomic DNA fragment produced using the primers PCCOPF and PCCOPR on wild-type genomic DNA was inserted into the vector pTOPO-XL, and the DNA sequence was obtained by using a directed sequencing approach. Boxes indicate regions of genomic DNA that contain the COP1 coding region; the pattern of splicing is shown below.
(B) COP1 gene in lip1 peas. A 20-kb region of overlapping DNA was obtained in PCR reactions on lip1 genomic DNA. A 9.5-kb fragment produced by use of the primers PCCOPF and PCCOR, a 3-kb fragment produced by the primers E7.F and E2.R, and an 8-kb fragment produced by the primers PCCOF and DREV2 were inserted into the vector pTOPOXL. The DNA sequence was obtained by using a directed sequencing approach, and a contig of overlapping DNA was produced. Boxes show regions of genomic DNA containing the COP1 coding region; hatched boxes indicate duplicated exon sequences. The 2.5-kb CPP region between exon 7′ and exon 1, used for subsequent promoter analysis, is also indicated by hatching. The pattern of splicing predicted from analysis of intron/exon boundaries is shown below. Exon 1 (black box) is skipped due to lack of recognized intron/exon boundary at the 5′ end.
 .
.
Table 1.
Intron/Exon Splice Site Junctions in the Pea COP1 Gene
| Introna | Nucleotide Positionb |
5′ Donor | 3′ Acceptorc |
|---|---|---|---|
| 1 | 307–648 | AAG GTTGGCTTCG | ACATTTGTAG CTA |
| 2 | 727–2938 | AAG GTAGGTGATA | CTTGAAACAG GGC |
| 3 | 3094–3374 | AAG GTGTGCTTGA | TCATCTGTAG GTG |
| 4 | 3732–4745 | CAG GTTATCTTAG | CTTGGTCCAG TTC |
| 5 | 4905–5010 | CAG GTACTACTAT | ATTGAAACAG CCG |
| 6 | 5076–6946 | AAG GTGAGTTATT | TGTATTGCAG CAT |
| 7 | 7026–7785 | GCG GTAAGATGGA | TGCTTTTCAG GTC |
| 8 | 7943–8057 | AAG GTAATTGTGA | CCTTTGGCAG AGT |
| 9 | 8166–8482 | AAG GTATTCTATA | CTTTGAGCAG GTC |
| 10 | 8587–8884 | GCA GTATGTTCCT | ATTTTTACAG GTT |
| 11 | 9074–9220 | CCA GTACGTAATC | ATGTGTACAG GTT |
| 12 | 9331–9411 | AAG GTTTGTGATC | TCCTTTGCAG GAA |
| 7′ | AAACCTCACC ATGd | ||
| Consensus | CorGAG GTAAGT | TTTTTTGCAG G |
Introns were identified by comparing the sequence of the COP1 cDNA identified using RT-PCR with COP1 genomic DNA.
Nucleotide position shown is relative to start codon.
Splice sites underlined were not identified as boundary elements by NetPlantGene intron prediction software.
Sequence of 3′ boundary of intron 7′ from the COP1 gene in lip1 peas.
In contrast, the coding region of the COP1 gene in lip1 peas was located within an ∼20-kb region of genomic DNA, which was also sufficient to explain the pattern of hybridization observed after DNA gel blot analysis and was consistent with a single-copy gene (data not shown). This region of genomic DNA contains a duplicated sequence of ∼7 kb, 99% identical to exons 1 to 7 and intron 1 to 6 of the wild-type COP1 genomic DNA and henceforth termed exons 1′ to 7′ and introns 1′ to 6′ (Figure 4B). All differences in DNA sequence between this duplicated sequence and wild-type COP1 genomic DNA, consisting of ∼70 bp of small deletions and point mutations, are located within introns (data not shown). Exons 1′ to 7′ are located upstream of a 9.5-kb region that shows 99% identity (the sequence differences being confined to introns) to wild-type COP1 (Figure 3B). Exons 1′ to 7′ are separated from exons 1 to 13 by ∼3 kb of DNA consisting of 720 bp of the wild-type intron 7 sequence (intron 7′) and by 2.5 kb of a DNA sequence (termed CPP) not found in wild-type COP1. This CPP region contains a 534-bp region with 76.2% identity to a portion of the upstream region from a gene encoding a disease response PR10 protein (GenBank accession number PS31669) and a 256-bp region with 85.9% identity to an RNaseH gene from a Tps8 retrotransposon (accession number psa243040) (data not shown). Analysis of exon boundaries with NetPlantGene intron prediction software predicted all intron/exon boundaries, except for the sequences identical to the 3′ splice site of intron 1 and the 3′ splice site of intron 6 of the wild-type COP1 sequence (Table 1). However, the 5′ boundary of exon 1 (Figure 4B and Table 1) was not predicted as an intron/exon boundary by the NetPlantGene software. The skipping of exon 1 during splicing of transcripts from the COP1 gene in lip1 peas would produce an mRNA identical to the COP1′ transcript found in lip1 seedlings (Figure 4B). These results demonstrate that lip1 peas contain a partial duplication within the COP1 genomic locus. The pattern of splicing based on predicted intron/exon boundaries produces a theoretical open reading frame identical to COP1′ cDNA isolated from lip1 seedlings.
The CPP Region Can Activate Transcription of a β-Glucuronidase Reporter Gene
Although lip1 peas contain only the mutated, partially duplicated COP1′ gene, RT-PCR analysis showed that a small amount of wild-type COP1 mRNA was present in addition to the larger COP1′ transcript. Sequence analysis of the COP1 genomic locus in lip1 peas predicted a pattern of splicing that would produce COP1′ but not COP1 transcripts. However, a COP1 transcript could be produced if transcription were initiated between exon 7′ and exon 1 (Figure 4B). After splicing, a transcript produced from such an internal initiation would contain an open reading frame indistinguishable from that contained in wild-type COP1 mRNA. To examine this possibility, we investigated the ability of CPP to activate expression of a uidA reporter gene in microprojectile bombardment transient expression assays in onion bulb epidermal cells and in transgenic Arabidopsis seedlings.
In onion bulb epidermal cells bombarded with a construct in which transcription of uidA is driven by CPP, multiple cells contained β-glucuronidase (GUS) activity, detected by histochemical staining (Figure 5B). The number of cells expressing GUS and the intensity of the histochemical reaction were similar to the amounts of expression observed when transcription of the uidA reporter gene was driven by the cauliflower mosaic virus (CaMV) 35S promoter (Figure 5A). In contrast, no GUS activity was observed after bombardment with a promoterless uidA construct (Figure 5C). GUS activity was high in transgenic Arabidopsis seedlings containing uidA driven by CPP, with GUS activity present in both root and cotyledon tissue (Figure 5D). These results demonstrate that the 2.5-kb CPP region within the partially duplicated COP1 genomic DNA sequence in lip1 peas can activate transcription of a GUS reporter gene in both onion bulb epidermal cells and in Arabidopsis seedlings. Thus, the small amount of wild-type COP1 mRNA found in lip1 peas probably results from internal initiation of transcription within the mutated COP1 genomic DNA.
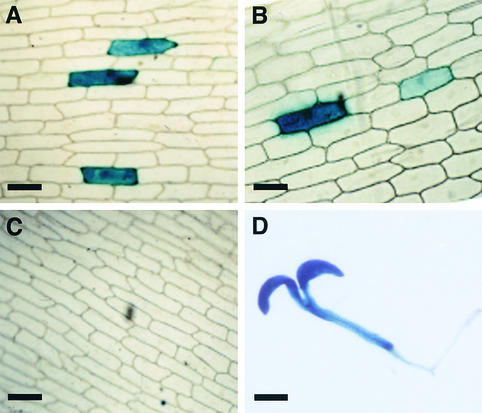
Figure 5.
Analysis of a Putative Promoter Region within COP1 in lip1 Peas.
The 2.5-kb CPP region located between exon 7′ and exon 1 (see Figure 4) was used to create a transcriptional fusion with the uidA reporter gene and a nos terminator sequence.
(A) GUS actively expressed from a CaMV 35S promoter in onion epidermal cells after microprojectile bombardment.
(B) GUS activity from the CPP region.
(C) GUS histochemical staining after bombardment with a promoterless construct.
(D) GUS activity in a representative 7-day-old transgenic Arabidopsis seedling containing uidA fused to CPP.
 ;
;  .
.
DISCUSSION
In this study, we have shown that lip1 pea seedlings contain an altered COP1 gene, consisting of a large duplication of ∼7 kb, containing exons 1 to 7 of the wild-type gene, located 2.5 kb upstream of a region showing 99% identity to wild-type COP1. After transcription and splicing, the altered COP1, which segregates with the lip1 phenotype, produces an altered COP1′ transcript containing an internal duplication of ∼900 bases. This COP1′ transcript, which can be translated in vitro to produce a protein of ∼100 kD, segregates with the lip1 phenotype in F2 seedlings. lip1 seedlings also contain small quantities of the wild-type COP1 transcript, produced from a promoter within the duplicated COP1 gene. Because the mutated COP1 gene segregates with the lip1 phenotype, and given the similarity between lip1 and cop1 mutants in Arabidopsis, we conclude that lip1 is caused by a duplication within the pea homolog of COP1.
lip1 Seedlings Share Many of the Characteristics Associated with Arabidopsis cop1 Seedlings
Dark-grown Arabidopsis cop1 mutants have short hypocotyls, open and expanded cotyledons, and chloroplast differentiation in cotyledons (Deng et al., 1991; Deng and Quail, 1992). Strong alleles of cop1 (such as cop1.1) accumulate anthocyanins in darkness, whereas weak alleles (such as cop1.4) do not (Deng and Quail, 1992; Torii and Deng, 1997). If grown in prolonged darkness, Arabidopsis cop1 mutants exhibit true leaf development similar to that observed in light-grown plants and, in contrast to other photomorphogenic mutants, are defective in the adaptive response of adult light-grown plants to darkness (Deng et al., 1991; Deng and Quail, 1992). Dark-grown lip1 seedlings also have short stems and open and expanded shoots, which contain chloroplast-like plastids (Frances et al., 1992). After prolonged growth in darkness, lip1 seedlings will develop true leaves similar to those found in light-grown plants (Frances et al., 1992). Furthermore, in common with Arabidopsis cop1 mutants, 9-day-old lip1 plants do not show an adaptive response to darkness (Frances et al., 1992; Frances and Thompson, 1997).
Interestingly, null alleles of cop1 in Arabidopsis are lethal (Torii and Deng, 1997), and the severity of the cop1 allele is correlated with the amount of residual COP1 activity (Deng and Quail, 1992). Because lip1 peas germinate and grow to produce viable seed, it seems likely that the lip1 mutation does not abolish all COP1 activity. Supporting this hypothesis is the observation that, as with weak Arabidopsis cop1 alleles, no visible increase in anthocyanin content was observed in dark-grown lip1 seedlings (data not shown). Perhaps, the apparent weak phenotype of lip1 seedlings is a result of small quantities of wild-type protein translated from the wild-type COP1 mRNA found in lip1 seedlings. Unfortunately, no cross-reaction was observed between antibodies raised against Arabidopsis COP1 protein and pea COP1 or COP1′ (data not shown), which makes it difficult to assess this possibility. However, given that the wild-type transcript was detectable in lip1 seedlings only after RT-PCR, but not by using RNA gel blots, probably only a very small proportion of COP1-like transcripts are wild type and would therefore produce only small quantities of wild-type protein capable of moderating any phenotype produced by the COP1′ protein.
Frances et al. (1992) concluded that the phenotype of the lip1 mutant was unlikely to be the result of mutations in the pea homologs of COP1 or DET1, because of differences in plastid development and expression of photosynthesis genes in roots of light-grown seedlings of lip1 in comparison with the Arabidopsis mutants. Recently, we showed the development of chloroplasts and the expression of several nuclear genes encoding photosynthesis proteins in roots of light-grown lip1 seedlings (Sullivan and Gray, 1999), indicating the similarity of the phenotypes of lip1 and cop1 mutants. This contrasts markedly with differences in the phenotypes of det1 mutants of tomato and Arabidopsis (Chory and Peto, 1990; Mustilli et al., 1999). The tomato high pigment-2 mutant is caused by a mutation in the tomato DET1 homolog, however, unlike Arabidopsis det1 mutants, hp2 plants do not show any visible phenotype in the dark and are dependent on active phytochrome for the high-pigment phenotype in the light (Mustilli et al., 1999).
The COP1′ Protein Contains Several Duplicated Motifs
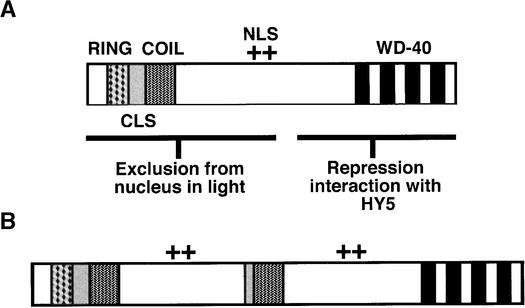
Arabidopsis COP1 contains several distinct structural motifs that have specific roles in the light-related control of seedling development (Torii et al., 1998; Osterlund et al., 1999; Stacey et al., 1999). A Zn binding RING domain and a coiled-coil domain at the N terminus of COP1 (see Figure 6A) are required for exclusion from the nucleus in the light (Torii et al., 1998). The RING finger domain may also recruit other COP1-interacting proteins bound at the C-terminal WD-40 motif region (Torii et al., 1998; Osterlund et al., 2000). A cytoplasmic localization signal that functions in a light-independent manner is also located between the coiled-coil and the WD-40 motifs (Stacey et al., 1999; see Figure 6A). The duplicated region in COP1′ contains the coiled-coil domain, part of the cytoplasmic localization signal, and the nuclear localization signal located between the coiled-coil and WD-40 motifs (Figure 6B). The N-terminal portion of Arabidopsis COP1 has recently been shown to maintain partial function and to act in a concentration-dependent manner (Stoop-Myer et al., 1999). Accordingly, perhaps the COP1′ protein still maintains residual activity associated with the intact protein domains at both the N and C termini. Such activity might explain the apparently weak nature of the lip1 phenotype as compared with Arabidopsis cop1 mutants.
Figure 6.
Duplicated Motifs within the COP1′ Protein.
(A) Motifs within Arabidopsis COP1 required for repression of photomorphogenic development in darkness (modified from that shown in Torii et al., 1998). COP1 contains a Zn binding RING domain (RING) and a coiled-coil domain (COIL) at the N terminus between which is found a cytoplasmic localization signal (CLS). A region containing at least five WD-40 repeats (WD-40) is found at the C terminus with a nuclear localization signal (NLS) located toward the middle of the protein.
(B) Motifs within pea COP1′. The duplicated region within COP1′ contains part of the cytoplasmic localization signal, the coiled-coil domain, and the region containing the nuclear localization signal. COP1′ contains intact N- and C-terminal motifs.
(+), position of NLS as shown in Figure 6A.
How Has the lip1 Mutation Arisen?
Exactly how the lip1 mutation of COP1 arose in wild-type peas is difficult to predict. Duplication of DNA sequences within a genome can occur through several mechanisms, including unequal crossover, translocation between nonhomologous chromosomes, and interstitial translocation (Pichersky, 1990). The duplication in pea COP1 could have been caused by an unequal crossover event during meiosis, producing one gamete with the lip1 mutation and a second with a severely, presumably lethal, truncated COP1. Although analysis of COP1 genomic DNA shows no obvious regions of sequence identity through which such an event could occur, the presence of a Tps8 retrotransposon RNaseH gene in the region between exon 7′ and exon 1 suggests that the lip1 mutation may be a result of recombination between retrotransposon DNA within the pea genome. Like the lip1 mutation, the L2 allele of the flax rust-resistance gene L is caused by a partial duplication, in this case within a leucine-rich region of the protein (Ellis et al., 1999). Partial duplications within the human dystrophin gene cause ∼6 to 7% of mutations leading to Duchenne muscular dystrophy (Hu et al., 1991). However, in all of these genes, tandem repeats, which are not present in COP1, act as targets for recombination. Isolation and analysis of the genomic DNA sequence that flanks the mutated COP1 gene in lip1 peas may help to indicate the cause of the duplication within pea COP1. However, attempts to isolate the 5′ upstream regions of the COP1 gene in wild-type or lip1 peas have so far been unsuccessful. This has also prevented further studies on the nature of the 5′ ends of the transcripts from the COP1 genes.
Benefits of Experiments with lip1
lip1 seedlings provide an extremely useful system for studying the regulation of photomorphogenic development. The large size of dark-grown lip1 and wild-type pea seedlings, in comparison with dark-grown Arabidopsis seedlings, allows the application of biochemical techniques that would not otherwise be practical in Arabidopsis. The discovery that the lip1 mutant is the result of a partial duplication of the pea COP1 homolog is likely to influence the design and interpretation of experiments with lip1 seedlings. Most previous publications on lip1 have described the phenotype of the plants (Frances et al., 1992; Sponsel et al., 1996; Frances and Thompson, 1997; Sineschchekov et al., 1997; Seyyedi et al., 1999), whereas in the future the known genetic defect in lip1 should allow more directed experiments on the regulation of photomorphogenesis.
METHODS
Plant Material and Growth Conditions
Seeds of wild-type pea plants (Pisum sativum cv Alaska) and the lip1 mutant were obtained from the John Innes Germ-Plasm collection (John Innes Centre, Norwich, UK) and from W.F. Thompson (North Carolina State University, Raleigh, NC). Seeds from these stocks were grown to maturity in a greenhouse and allowed to self-fertilize to produce seed used for subsequent experiments. Crosses between wild-type and lip1 plants were performed by emasculation of flowers in which the corolla tube had emerged 0.5 to 1.0 cm from the sepals and pollination with pollen from mature flowers. The resulting F1 plants were allowed to self-fertilize; the seeds obtained were surface-sterilized and grown in sterile magenta vessels for 7 days in continuous darkness at 22°C (Sullivan and Gray, 1999) before being scored for a wild-type or lip1 photomorphogenic phenotype.
RNA Methods
Total RNA was extracted from shoot tissue by using Tripure isolation reagent (Boehringer Mannheim) according to the manufacturer's protocol. First-strand cDNA synthesis reactions were performed in a total volume of 20 μL, consisting of 5 mM MgCl2; 5 mM each dATP, dCTP, dTTP, dCTP; 1 × MMLV buffer (Stratagene); 1 unit of RNasin (Promega); 5 μg of total RNA; 1 μg of oligo(dT)15 primer (Promega); 100 units of MMLV reverse transcriptase (Stratagene); and sterile distilled water to 20 μL. The mixture was prepared in the absence of MMLV reverse transcriptase and then heated to 65°C for 5 min and cooled to 42°C for 5 min before the MMLV reverse transcriptase was added. cDNA synthesis was performed at 42°C for 1 hr.
cDNAs for pea COP1 were amplified from first-strand cDNA by using the primers PCCOPF (5′-CCATGGAAGAGCACTCAGTAGGAC-3′) and PCCOPR (5′-GCAGCAAAGCACCAGCACTTTGATGG-3′), designed from the published sequence of pea COP1 (Zhao et al., 1998), to amplify a region of COP1 from position –2 to +2014 relative to the start codon. A second pair of primers FS1 (5′-GGTCATTACCTCACCAAC-3′) and RS1 (5′-TAGAGGGGTCCGTTCTTG-3′) were also designed to amply a region from +215 to +1442 (relative to start codon) within the COP1 cDNA. Polymerase chain reaction (PCR) products were inserted into the plasmid pBCSK (Stratagene). DNA sequences were obtained (Automated Sequencing Facility, Department of Biochemistry, University of Cambridge) for three independent clones for each PCR product and a consensus sequence was determined by using Seqed sequence analysis software (Perkin-Elmer).
For RNA gel blot analysis, ∼10 μg of total RNA was separated by electrophoresis on a 1.2% agarose gel and blotted to GeneScreen Plus membrane (New England Nuclear Research Products) as previously described (Sullivan and Gray, 1999). A radiolabeled probe was produced from a 2.0-kb COP1 cDNA (produced with PCCOPF and PCCOPR primers) by using random hexanucleotide primers and α-32P-dATP (Feinberg and Vogelstein, 1983). Autoradiographic images were obtained after exposure to X-ograph Blue x-ray autoradiography film (X-ograph Ltd, Tetbury, UK).
Transcription and Translation in Vitro
Full-length cDNAs encoding COP1 and COP1′ were inserted into the plasmid pBCSK (Stratagene), and transcription in vitro was performed by using ∼2 μg of unlinearized DNA template and T7 RNA polymerase (Boehringer Mannheim) as described by Melton et al. (1984). The resulting RNA was then used in a translation reaction in vitro with wheat germ extract (Promega) and 60 μCi of Promix (Amersham) as described previously (Knight and Gray, 1995). Translation products were separated by electrophoresis on an SDS–10% (w/v) polyacrylamide gel, fixed in a solution of 10% (v/v) propan-2-ol and 10% (v/v) acetic acid, and dried under vacuum with a Bio-Rad 583 gel dryer at 80°C. Autoradiographic images were obtained by exposing the dried gel to X-ograph Blue x-ray film for 16 hr at room temperature.
DNA Methods
Genomic DNA was extracted from ∼0.75 g of shoot tissue with a plant DNA miniprep protocol (Dellaporta et al., 1983). After digestion with DraI, genomic DNA was fractionated by electrophoresis on a 0.7% agarose gel, transferred to GeneScreen Plus membrane by using the manufacturer's alkaline salt transfer protocol, and hybridized (according to manufacturer's protocol) with the pea COP1 probe prepared as described above.
To isolate pea COP1, 500 ng of pea genomic DNA was used as template in PCR reactions with an Expand DNA polymerase kit (Boehringer Mannheim) and various combinations of the primers PCCOPF, PCCOPR, E7.F (5′-GCTACTGCTGGAGTTTCCCGACGTA-3′, +1149 to +1174 of pea COP1 cDNA relative to start codon), E2.R (5′-GCTCCACAGGAGAAGCCGTCTTTG-3′, +357 to +334, relative to pea COP1 cDNA), and DREV2 (5′-CGACAAGTGACCAAAGTGTTGTTGACTTTGAT-3′, +732 to +700 of the E7.F/E2.R PCR product). PCR reactions were performed on both wild-type and lip1 genomic DNA with the primer combinations PCCOPF and PCCOPR, E7.F and E2.R, and DREV2 and PCCOPF. PCR products were inserted into the vector pTOPO-XL by using a TOPO-XL cloning kit (Invitrogen, La Jolla, CA). The DNA sequence was obtained from three independent clones for each product by using directed sequencing; a consensus sequence was assembled and analyzed by using Seqed sequence analysis software and NetPlantGene intron prediction software (http://www.cbs.dtu.dk/netgene/cbsnetgene.announce.html).
Promoter Analysis
The primers PP5 (5′-GCTCCTACCAAGCTTTCAAAAGTC-3′) and PP3 (5′-GCTCTTGGATCCTGAGGTTTAGAG-3′), which contain additional HindIII and XbaI restriction sites, were used to amplify the region of DNA from lip1 genomic DNA located between exon 7′ and exon 1 (see Figure 4). The 2.5-kb fragment produced was inserted into the HindIII and XbaI sites of the plasmid pUCGUS, which contains the cauliflower mosaic virus (CaMV) 35S promoter, the β-glucuronidase (GUS) coding region, and a nopaline synthase (nos) terminator sequence derived from pBI121 (Jefferson et al., 1987) in pUC19, which replaces the CaMV 35S promoter and produces the plasmid pCPP-GUS-nos. A promoterless construct was also made from pUCGUS by removing the CaMV 35S promoter with HindIII and XbaI, incubating with the Klenow fragment of DNA polymerase I (New England Biolabs, Beverly, MA) and dNTPs, and religating the blunt ends.
To produce a vector suitable for Agrobacterium tumefaciens–mediated transformation of Arabidopsis thaliana (Columbia), a 3.5-kb HindIII-EcoRI fragment was removed from pCPP-GUS-nos and inserted into the plant binary transformation vector pBIN19 (Bevan, 1984). The resulting plasmid was subsequently used in a vacuum-infiltration transformation protocol as previously described by Bechtold et al. (1993). Transient expression assays were performed in onion epidermal cells by using a Bio-Rad PDS 1000 (He) gun in a protocol previously described by Varagona et al. (1992). Histochemical staining for GUS activity was performed on onion epidermis and ethanol-cleared 7-day-old transgenic Arabidopsis seedlings as described by Jefferson et al. (1987). Photographic images were obtained with a Nikon Optiphot2 microscope (Nikon Inc., Melville, NY) using transmitted white light.
Acknowledgments
We thank Bill Thompson and those at the John Innes Pea Germ-Plasm Centre for their gifts of wild-type Alaska and lip1 peas, and Xing-Wang Deng for antibodies against Arabidopsis COP1. We also thank Martyn Seekings for his invaluable help with the growth of the plants. J.A.S was supported by a research studentship from the Biotechnology and Biological Sciences Research Council.
References
- Ahmad, M., and Cashmore, A. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue light photoreceptor. Nature 360, 162–166. [DOI] [PubMed] [Google Scholar]
- Ang, L.H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., and Deng, X.-W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene-transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Ser. III 316, 1194–1199. [Google Scholar]
- Bevan, M. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12, 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamovitz, D.A., Wei, N., Osterlund, M.T., von Arnim, A., Staub, J.M., Matsui, M., and Deng, X.W. (1996). The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86, 115–121. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay, S., Ang, L.H., Puente, P., Deng, X.-W., and Wei, N. (1998). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory, J. (1993). Out of darkness: Mutants reveal pathways controlling light-regulated development in plants. Trends Genet. 9, 167–172. [DOI] [PubMed] [Google Scholar]
- Chory, J., and Peto, C.A. (1990). Mutations in the DET1 gene affect cell-type–specific expression of light-regulated genes and chloroplast development in Arabidopsis. Proc. Natl. Acad. Sci. USA 87, 8776–8780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA minipreparation. Version II. Plant Mol. Biol. Rep. 1, 19–21. [Google Scholar]
- Deng, X.-W., and Quail, P.H. (1992). Genetic and phenotypic characterization of cop1 mutants of Arabidopsis thaliana. Plant J. 2, 83–95. [Google Scholar]
- Deng, X.-W., Caspar, T., and Quail, P. (1991). cop1: A regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5, 1172–1182. [DOI] [PubMed] [Google Scholar]
- Deng, X.-W., Matsui, M., Wei, N., Wagner, D., Chu, A.M., Feldmann, K., and Quail, P. (1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a Gβ homologous domain. Cell 71, 791–801. [DOI] [PubMed] [Google Scholar]
- Ellis, J.G., Lawrence, G.J., Luck, J.E., and Dodds, P.N. (1999). Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg, A.P., and Vogelstein, B. (1983). A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132, 6–13. [DOI] [PubMed] [Google Scholar]
- Frances, S., and Thompson, W.F. (1997). The dark-adaptation response of the deetiolated pea mutant lip1 is modulated by external signals and endogenous programs. Plant Physiol. 115, 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frances, S., White, M.J., Edgerton, M.D., Jones, A.M., Elliott, R.C., and Thompson, W.F. (1992). Initial characterization of a pea mutant with light-independent photomorphogenesis. Plant Cell 4, 1519–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebsgaard, S.M., Korning, P.G., Tolstrup, N., Engelbrecht, J., Pouse, P., and Brunak, S. (1996). Splice site predication in Arabidopsis thaliana DNA by combining local and global sequence information. Nucleic Acids Res. 24, 3439–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, M., and Deng, X.-W. (1999). Structural organization and interactions of COP1, a light-regulated developmental switch. Plant Mol. Biol. 41, 151–158. [DOI] [PubMed] [Google Scholar]
- Hu, X., Ray, P.N., and Worton, R.G. (1991). Mechanisms of tandem duplication in the Duchenne muscular dystrophy gene include both homologous and nonhomologous intrachromosomal recombination. EMBO J. 10, 2471–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R., Kavanagh, T., and Bevan, M. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniol, B., Malec, P., and Chamovitz, D.A. (1999). Arabidopsis FUSCA5 encodes a novel phosphoprotein that is a component of the COP9 complex. Plant Cell 11, 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, J.S., and Gray, J.C. (1995). The N-terminal hydrophobic region of the mature phosphate translocator is sufficient for targeting to the chloroplast inner envelope. Plant Cell 7, 1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok, S., Piekos, B., Miséra, S., and Deng, X.-W. (1996). A complement of ten essential and pleiotropic Arabidopsis COP/DET/FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Physiol. 110, 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok, S.F., Solano, R., Tsuge, T., Chamovitz, D.A., Ecker, J.R., Matsui, M., and Deng, X.-W. (1998). Arabidopsis homologues of a c-Jun coactivator are present in both monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell 10, 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis, T.W., and Deng, X.-W. (1995). Light control of seedling morphogenetic pattern. Plant Cell 7, 1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton, D.A., Kreig, P.A., Rebagliati, M.R., Maniatis, T., Zinu, K., and Green, M.R. (1984). Efficient in vitro synthesis of biologically active RNA and RNA hybridisation probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 12, 7035–7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli, A.C., Fenzi, F., Ciliento, R., Alfano, F., and Bowler, C. (1999). Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell 11, 145–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund, M., Ang, L.H., and Deng, X.-W. (1999). The role of COP1 in repression of Arabidopsis photomorphogenic development. Trends Cell Biol. 9, 113–118. [DOI] [PubMed] [Google Scholar]
- Osterlund, M., Hardtke, C.S., Wei, N., and Deng, X.-W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. [DOI] [PubMed] [Google Scholar]
- Oyama, T., Shimura, Y., and Okada, K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11, 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky, E. (1990). Nomad DNA—A model for movement and duplication of DNA sequences in plant genomes. Plant Mol. Biol. 15, 437–448. [DOI] [PubMed] [Google Scholar]
- Seeger, M., Kraft, R., Ferrell, K., Bech-Otschir, D., Dumdey, R., Schade, R., Gordon, C., Naumann, M., and Dubiel, W. (1998). A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. FASEB J. 12, 469–478. [PubMed] [Google Scholar]
- Serino, G., Tsuge, T., Kwok, S.F., Matsui, M., Wei, W., and Deng, X.-W. (1999). Arabidopsis cop8 and fus4 mutations define the same locus that encodes subunit 4 of the COP9 signalosome. Plant Cell 11, 1967–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyyedi, M., Timko, M.P., and Sundqvist, C. (1999). Protochlorophyllide, POR and chlorophyll formation in the lip1 mutant of pea. Physiol. Plant. 106, 344–354. [Google Scholar]
- Sineschchekov, V.A., Frances, S., and White, M.J. (1997). Fluorescence and photochemical characterisation of phytochrome in deetiolated pea mutant lip1. J. Photochem. Photobiol. 28, 47–51. [Google Scholar]
- Sponsel, V.M., Ross, J.J., Reynolds, M.R., Symons, G.M., and Reid, J.B. (1996). The gibberellin status of lip1, a mutant of pea that exhibits light-independent photomorphogenesis. Plant Physiol. 112, 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey, M.C., Hicks, S.N., and von Arnim, A. (1999). Discrete domains mediate the light-responsive nuclear and cytoplasmic localization of Arabidopsis COP1. Plant Cell 11, 349–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub, J.M., and Deng, X.-W. (1996). Light signal transduction in plants. Photochem. Photobiol. 64, 897–905. [DOI] [PubMed] [Google Scholar]
- Stoop-Myer, C., Torii, K.U., McNellis, T.W., Coleman, J.E., and Deng, X.-W. (1999). The N-terminal fragment of Arabidopsis photomorphogenic mutant repressor COP1 maintains partial function and acts in a concentration-dependent manner. Plant J. 20, 713–717. [DOI] [PubMed] [Google Scholar]
- Sullivan, J.A., and Gray, J.C. (1999). Plastid translation is required for the expression of nuclear photosynthesis genes in the dark and in roots of the pea lip1 mutant. Plant Cell 11, 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii, K.U., and Deng, X.-W. (1997). The role of COP1 in light control of Arabidopsis seedling development. Plant Cell Environ. 20, 728–733. [Google Scholar]
- Torii, K.U., McNellis, T.W., and Deng, X.-W. (1998). Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. EMBO J. 17, 5577–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varagona, M.J., Schmidt, R.J., and Raikhel, N.V. (1992). Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell 4, 1213–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim, A., and Deng, X.-W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79, 1035–1045. [DOI] [PubMed] [Google Scholar]
- von Arnim, A., and Deng, X.-W. (1996). Light control of seedling development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 215–243. [DOI] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.-W. (1996). The role of COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol. 112, 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.-W. (1999). Making sense of the COP9 signalosome: A regulatory protein complex conserved from Arabidopsis to human. Trends Genet. 15, 98–103. [DOI] [PubMed] [Google Scholar]
- Wei, N., Tsuge, T., Serino, G., Dohmae, N., Takio, K., Matsui, M., and Deng, X.-W. (1998). The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. Curr. Biol. 8, 919–922. [DOI] [PubMed] [Google Scholar]
- Weller, J.C., Terry, M.J., and Rameau, C. (1996). The phytochrome-deficient pcd1 mutant of pea is unable to convert heme to biliverdin IXa. Plant Cell 8, 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, L., Wang, C., Zhu, Y., Zhao, J., and Wu, X. (1998). Molecular cloning and sequencing of the cDNA of COP1 gene from Pisum sativum. Biochim. Biophys. Acta 1395, 326–328. [DOI] [PubMed] [Google Scholar]