Abstract
Root hairs are a major site for the uptake of water and nutrients into plants and form an increasingly important model system for studies of development of higher plants and cell biology. We have identified loss-of-function mutations in eight new genes required for hair growth in Arabidopsis: SHAVEN1 (SHV1), SHV2, and SHV3; CENTIPEDE1 (CEN1), CEN2, and CEN3; BRISTLED1 (BST1); and SUPERCENTIPEDE1 (SCN1). We combined mutations in 79 pairs of genes to determine the stages at which these and six previously known genes contribute to root hair formation. Double mutant phenotypes revealed roles for several genes that could not have been predicted from the single mutant phenotypes. For example, we show that TIP1 and RHD3 are required much earlier in hair formation than previous studies have suggested. We present a genetic model for root hair morphogenesis that defines the roles of each gene, and we suggest hypotheses about functional relationships between genes.
INTRODUCTION
Root hairs contribute as much as 77% of the root surface area of cultivated crops, forming the major point of contact between the plant and the rhizosphere. Each hair is a long, narrow tube produced from a single cell by tip growth (the deposition of new membrane and cell wall material at a growing tip). Large collections of viable and fertile Arabidopsis root hair mutants are available (Schiefelbein and Somerville, 1990; Masucci and Schiefelbein, 1994; Grierson et al., 1997; Pitts et al., 1998) with all of the advantages of the Arabidopsis genome project. In Arabidopsis, root hairs form in a predictable pattern, so developing hair cells can be observed from the beginning of their life in the root meristem throughout the developmental process. Mechanisms that control this patterning have been identified (Dolan et al., 1994; Galway et al., 1994; Tanimoto et al., 1995; DiCristina et al., 1996; Masucci et al., 1996; Masucci and Schiefelbein, 1996; Schneider et al., 1997, 1998; Wada et al., 1997; Dolan and Scheres, 1998; Lee and Schiefelbein, 1999).
The selection of an initiation site within the hair cell depends on the gene RHD6 and is influenced by the plant growth regulators auxin and ethylene (Masucci and Schiefelbein, 1994, 1996). Once an initiation site has been selected, a small swelling forms (sometimes called a bulge). This requires local acidification of the cell wall (Bibikova et al., 1998) and is thought to be due at least in part to local wall loosening. Mutations in the RHD1 and TIP1 genes result in hairs with larger swellings at their bases than those of wild-type hairs. This is thought to result from increased loosening of the wall around the base of the hairs and suggests that the degree of cell wall loosening in wild-type plants is restricted by the RHD1 (Schiefelbein and Somerville, 1990) and TIP1 (Ryan et al., 1998) gene products.
Once a swelling has formed, the hair begins to elongate by tip growth. This transition to tip growth takes place when the hair is between 20 and 40 μm long (Dolan et al., 1994). Elongation is accompanied by the generation of a tip-high calcium gradient that can be observed throughout the remainder of root hair growth (Schiefelbein et al., 1992; Wymer et al., 1997). Mutations at the RHD2 locus result in the formation of swellings that do not elongate successfully (Schiefelbein and Somerville, 1990). Calcium gradients are not observed in swellings on most hair cells on rhd2 mutant plants (Wymer et al., 1997), supporting the idea that the calcium gradient is associated with the ability to elongate.
Plants homozygous for mutations in COW1 and TIP1 produce short, wide root hairs, and sometimes multiple hairs form at a single site of hair formation (Schiefelbein et al., 1993; Grierson et al., 1997; Ryan et al., 1998). Plants homozygous for mutations in RHD3 and RHD4 have short, wavy root hairs (Schiefelbein and Somerville, 1990; Galway et al., 1997, 1999; Wang et al., 1997). The RHD3 gene has been characterized at the molecular level. RHD3 encodes a protein with GTP binding motifs that may be required during vacuole enlargement (Wang et al., 1997). The phenotypes of double mutants have previously suggested that COW1 acts after RHD2 and that TIP1 and RHD3 act in parallel with COW1 (Grierson et al., 1997).
To identify more genes involved in root hair formation and to isolate new alleles of previously known genes, we performed a new screen for mutants that affect root hair shape. This identified eight new loci and additional alleles of two previously characterized genes. We combined pairs of mutations at 14 genes to determine the stages at which each is required during hair cell morphogenesis and to identify the genetic interactions that take place as root hairs develop. The double mutant phenotypes revealed new roles for genes that could not have been predicted from their single mutant phenotypes and suggested hypotheses about functional relationships between genes. We combined our results with previously published data, in a model that defines roles for 15 genes during root hair morphogenesis.
RESULTS
Eight New Loci Involved in Root Hair Morphogenesis
We isolated 29 new mutants affecting root hair morphogenesis, 20 of which defined eight new loci: SHAVEN1 (SHV1), SHV2, and SHV3; CENTIPEDE1 (CEN1), CEN2, and CEN3; BRISTLED1 (BST1); and SUPERCENTIPEDE1 (SCN1). All of our new mutants formed root hairs on a similar proportion and arrangement of epidermal cell files and in a similar location on each hair-forming cell as in wild-type plants (data not shown), suggesting that processes governing the patterning of hair cells and the location of hairs on cells were not affected by the new alleles. We chose one allele at each locus to study in more detail or chose multiple alleles if they had obviously different phenotypes.
Figure 1A shows mature root hairs on homozygous mutant plants, and Table 1 lists phenotypes of mutants at each locus included in this study. The new mutants fell into two phenotypic groups.
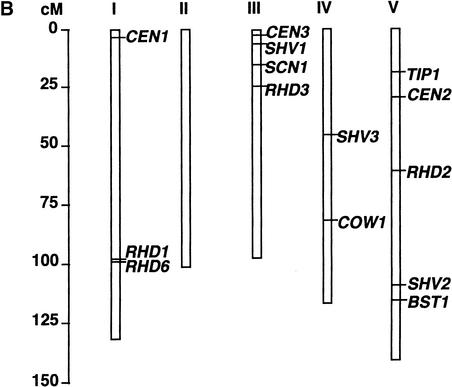
Figure 1.
Mutant Phenotypes and Map Positions of Genes Involved in Root Hair Morphogenesis.
(A) Root hairs of wild-type and single mutant seedlings. Mature root hairs of mutants at each new locus are shown alongside those of plants mutated at previously identified loci (rhd6, rhd2-1, rhd3-1, tip1-2, and cow1-1).
(B) Locations on the Arabidopsis genetic map of genes involved in root hair formation. Shown for comparison are previously published map positions of other loci included in this study: RHD1, RHD2, and RHD3 (Schiefelbein and Somerville, 1990); RHD6 (Masucci and Schiefelbein, 1994); TIP1 (Ryan et al., 1998); and COW1 (Grierson et al., 1997).
Table 1.
Root Hair Phenotypes of Arabidopsis Lines
| Phenotypeb
|
|
|
|
|
|---|---|---|---|---|
| Genotypea | % Hairs ⩾40 μm Longc |
Length of Hairs ⩾40 μm Long (% Wild-Type Length)d |
% Multiple Hairsc,e | Other Featuresf |
| Wild type | 96 sd ± 4 (5) | 100 (5) | 0 (5) | Straight; about half of epidermal cells form hairs |
| bst1 | 100 (6) | 19.2 sd ± 3.8 (6) | 4 sd ± 2 (6) | Straight |
| cen1-1 | 100 (5) | 25.9 sd ± 0.1 (5) | 0 (5) | Sometimes curled |
| cen1-2 | 95 sd ± 7 (5) | 12.6 sd ± 1.0 (5) | 0 (5) | Sometimes curled |
| cen2-1 | 100 (8) | 29.1 sd ± 6.1 (6) | 7 sd ± 4 (8) | Sometimes curled |
| cen3-1 | 100 (5) | 21.5 sd ± 6.1 (5) | 14 sd ± 10 (5) | Sometimes bulbous bases |
| cen3-2 | 100 (8) | 23.4 sd ± 4.2 (8) | 8 sd ± 5 (8) | Some bulbous bases; sometimes curled |
| cow1-1 | 100 (5) | 25.7 sd ± 2.3 (5) | 7 sd ± 3 (5) | Sometimes slightly crooked; wide |
| cow1-2 | 100 (6) | 20.4 sd ± 3.2 (6) | 20 sd ± 2 (6) | Sometimes slightly crooked; wide |
| rhd1 | ND | ND | ND | Large swelling at base |
| rhd2-1 | 0 (8) | None | 0 (5) | |
| rhd2-2 | 100 (5) | 8.2 sd ± 1.2 (5) | 0 (5) | Straight; wide |
| rhd2-3 | 22 sd ± 3 (5) | 13.0 sd ± 1.3 (5) | 0 (5) | Straight |
| rhd3-1 | 100 (6) | 15.6 sd ± 1.3 (6) | 0 (6) | Wavy; crooked |
| rhd3-7 | 100 (8) | 21.7 sd ± 0.8 (8) | 0 (8) | Wavy; crooked |
| rhd6 | 100 (5) | 89 sd ± 18 (5) | ND | Very few epidermal cells form hairs |
| scn1-1 | 100 (5) | 26.8 sd ± 2.8 (5) | 61 sd ± 7 (5) | Sometimes curled; wide |
| shv1-1 | 15 sd ± 5 (5) | 10.3 sd ± 2.3 (5) | 0 (5) | Straight |
| shv1-4 | 4 sd ± 2 (8) | 29.8 sd ± 8.3 (5) | 0 (8) | Straight |
| shv2-1 | 18 sd ± 9 (6) | 15.8 sd ± 1.8 (6) | 0 (6) | Straight |
| shv3 | 30 sd ± 12 (6) | 68.2 sd ± 11.6 (5) | 0 (6) | Straight |
| tip1-1 | 97 sd ± 6 (5) | 19.1 sd ± 5.4 (5) | 26 sd ± 4 (5) | Straight; wide |
| tip1-2 | 91 sd ± 13 (5) | 14.6 sd ± 3.7 (5) | 53 sd ± 6 (5) | Straight; wide |
Mutants are listed in alphabetical order to facilitate comparison with other tables in this article.
The numbers within parentheses are number of roots. ND, not determined.
Fifty mature hairs were assessed on each root, except rhd6, where three hairs were assessed on each root.
Ten mature hairs were measured on each root, except on rhd6, where two hairs were measured on each root. Wild-type length was ∼900 μm.
Multiple hairs that appeared at a single site of hair formation.
At least 1000 roots of each genotype were examined.
The first group included mutants in the RHD2, SHV1, SHV2, and SHV3 genes. Root hairs on all mutants at these loci except rhd2-2 seldom were ⩾40 μm long, indicating that they failed to develop beyond the transition to tip growth, which takes place when hairs are 20 to 40 μm long (Dolan et al., 1994). This suggests that the RHD2, SHV1, SHV2, and SHV3 genes are required for tip growth to be successfully established. When hairs ⩾40 μm long formed on mutants at the RHD2, SHV1, and SHV2 loci, they were much shorter than wild-type hairs (Figure 1A and Table 1), supporting the conclusion that this group of genes is required for tip growth.
The second group included mutants in the BST1, CEN1, CEN2, CEN3, RHD3, and SCN1 genes. Root hairs on these plants were ⩾40 μm, suggesting that they could develop beyond the transition to tip growth. These hairs were short and often had different shapes from those of the wild type (Figure 1A and Table 1), suggesting that these genes are required to control the shape of the root hair during tip growth. Several mutants in this group sometimes produced multiple hairs (Table 1), implicating the SCN1, BST1, CEN2, and CEN3 genes in mechanisms that restrict the number of root hairs produced by tip growth from each hair-forming site.
Where we have quantitative data for more than one allele (Table 1), we can draw conclusions about allele strength. Root hair length is more strongly affected by the cen1-2 mutation than by cen1-1, suggesting that cen1-2 is the stronger allele. The proportion of root hairs ⩾40 μm indicated that the rhd2-2 and rhd2-3 alleles are weaker than rhd2-1. Root hairs were longer on rhd3-7 plants than on rhd3-1, suggesting that rhd3-7 is the weaker allele. A higher proportion of multiple hairs formed on plants homozygous for tip1-2 than tip1-1, suggesting that the former was the stronger allele.
In a statistical test for 3:1 segregation in F2 populations, 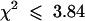 (and hence
(and hence 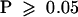 ) for shv1-1, shv1-4, shv2-1, cen1-1, cen1-2, cen1-3, cen2-1, cen3-1, cen3-2, scn1-1, bst1-1, rhd2-3, and rhd3-7; similar results were produced whether the mutant parent was male or female. This suggests that all of these alleles are recessive, single locus, nuclear mutations. shv3 was difficult to score because the phenotypic range was close to that of wild-type plants; the segregation of F2 populations (e.g., 175 wild type:94 Shv3−) probably included some wild-type plants misscored as Shv3−, but we cannot rule out the possibility that a Shv3− phenotype might be exhibited by a minority of heterozygous plants.
) for shv1-1, shv1-4, shv2-1, cen1-1, cen1-2, cen1-3, cen2-1, cen3-1, cen3-2, scn1-1, bst1-1, rhd2-3, and rhd3-7; similar results were produced whether the mutant parent was male or female. This suggests that all of these alleles are recessive, single locus, nuclear mutations. shv3 was difficult to score because the phenotypic range was close to that of wild-type plants; the segregation of F2 populations (e.g., 175 wild type:94 Shv3−) probably included some wild-type plants misscored as Shv3−, but we cannot rule out the possibility that a Shv3− phenotype might be exhibited by a minority of heterozygous plants.
Mutations in the RHD3 and TIP1 loci affect the growth of many cell types (Wang et al., 1997; Ryan et al., 1998), whereas the COW1 locus appears to be specific for root hair cells (Grierson et al., 1997). TIP1 affects pollen tube growth, and both TIP1 and RHD3 affect the stature and architecture of the plant (Galway et al., 1997; Wang et al., 1997; Ryan et al., 1998). Our segregation data show that none of the mutations in the eight new genes exhibited reduced transmission, which suggests that they do not affect pollen tube function, although we cannot exclude the possibility that these mutations impair pollen tube growth in a way that does not affect pollen competition during fertilization. The results of an extensive survey of plant architecture showed that mutations at the new loci did not have strong pleiotropic phenotypes (data not shown). Under our growth conditions, which provided high levels of nutrients and water, the stature and vigor of plants homozygous for mutations in each of the eight new genes were similar to those of the wild type.
Figure 1B shows the map locations of the new loci along with those of other loci included in this study. Representatives of each new complementation group map to a different position on the Arabidopsis genome, confirming that each represents a different genetic locus.
Double Mutant Analysis
We combined pairs of mutations at the eight new loci with each other and with those at six previously characterized loci affecting root hair morphogenesis (RHD1, RHD2, RHD3, RHD6, TIP1, and COW1) to make 79 double mutant combinations. We grouped the double mutants according to type of genetic interaction.
Epistatic Interactions at the Beginning of Root Hair Formation and during the Transition to Tip Growth
Epistatic interactions, where the phenotype of a double mutant was essentially the same as that of a single mutant parent, are listed in Table 2. Photomicrographs of a subset of the results are shown in Figure 2. Because the mutations used in our study are recessive, they are probably loss-of-function alleles. Loss-of-function mutations in a gene that is required early in the process might be expected to be epistatic to those in genes that act later.
Table 2.
Combinations of Mutations Producing Epistatic Phenotypes
| Gene | Phenotypea | Double Mutants with the Phenotype |
|---|---|---|
| RHD6 | Very few epidermal cells form hairs | rhd6 rhd2-2, rhd6 shv1-4, rhd6 shv3, rhd6 tip1-2, rhd6 cen2-1, rhd6 rhd3-7, rhd6 rhd3-9, rhd6 cen3-2, rhd6 bst1, rhd6 cen1-2, rhd6 scn1-1 |
| RHD2 | Very few hairs exceed 40 μm long; those that do are short |
shv3 rhd2-1, rhd2-3 cow1-1, rhd2-1 cen3-2, rhd2-1 cen2-1, rhd2-3 cen1-1, rhd2-1 cen1-1, rhd2-1 bst1 |
| SHV1 | Very few hairs exceed 40 μm long; those that do are short |
shv1-1 shv3, shv2-1 shv1-1, cen3-1 shv1-4, shv1-1 cow1-2, rhd3 shv1-1, bst1 shv1-4, cen2-1 shv1-4 |
| SHV2 | Very few hairs exceed 40 μm long; those that do are short |
shv2-1 cen1-2, shv2-1 cen3-2 |
| SHV3 | Few hairs exceed 40 μm long | rhd3 shv3, bst1 shv3, cen1-2 shv3, cen3-1 shv3 |
See Table 1.
Figure 2.
Mature Root Hairs of Double Mutant Plants with Epistatic Phenotypes.
Root hairs of wild-type and single mutant plants are shown for comparison. The rhd6 rhd2-2 double mutant is mostly bald, as is the rhd6 single mutant. This is an example of epistasis, where the rhd6 mutation is epistatic to rhd2-2. Other results shown here demonstrate that shv1-1 is epistatic to both shv2-1 and shv3 and that shv2-1 is epistatic to cen3-2.
The rhd6 mutation was epistatic to mutations in all 10 genes we tested (Table 2), suggesting that RHD6 acts before any of these genes to permit hair formation.
Mutations in RHD2, SHV1, SHV2, and SHV3 were epistatic to mutations in several other genes, resulting in double mutant roots with very few hairs ⩾40 μm long (Table 2 and Figure 2; Schiefelbein and Somerville, 1990; Grierson et al., 1997). These results support our conclusion from the single mutant phenotypes that these genes are required for the transition from swelling formation to tip growth. That rhd2-1 is epistatic to shv3 suggests RHD2 may be required earlier in this transition than SHV3. Similarly, epistasis of shv1-1 to both shv2-1 and shv3-1 suggests that SHV1 may be required earlier than SHV2 or SHV3.
Partially Suppressed Phenotypes Implicate TIP1, RHD3, CEN1, and SCN1 in the Transition from Swelling Formation to Tip Growth
Results in Figure 3 and Table 3 show that a second set of genes, TIP1, RHD3, CEN1, and SCN1, are also involved in the transition to tip growth. When mutations of some members of this group are present, a higher proportion of hairs on Rhd2−, Shv1−, and Shv2− plants grow ⩾40 μm long, partially suppressing the phenotypes of Rhd2−, Shv1−, and Shv2− single mutants.
Figure 3.
Mature Root Hairs of Double Mutant Plants with Partly Suppressed Phenotypes.
Root hairs on tip1-1 rhd2-3 double mutant roots are frequently longer than those on rhd2-3 single mutant roots. This phenotype suggests that the wild-type function of TIP1 contributes to the short root hair phenotype of the rhd2-3 mutant. Results shown here demonstrate similar roles for RHD3 in the shv2-1 phenotype, and for SCN1 in the shv1-1 and shv2-1 phenotypes. The phenotypes of tip1-2 and shv2-1 tip1-2 are almost indistinguishable, suggesting that tip1-2 is almost completely epistatic to shv2-1. The shv1-1 and shv2-1 single mutant phenotypes are shown in Figure 2.
Table 3.
Partly Suppressed Root Hair Phenotypes of Double Mutants
| Phenotypea
|
||||
|---|---|---|---|---|
| Genotype | % Hairs ⩾40 μm Longb |
Length of Hairs ⩾40 μm Long (% Wild-Type Length)c |
% Multiple Hairsb,d | Other Features |
| tip1-2 rhd2-1 | 29 sd ± 8 (5) | 11.3 sd ± 2.0 (5) | 36 sd ± 6 (5) | Straight; wide |
| tip1-1 rhd2-3 | 36 sd ± 4 (5) | 12.6 sd ± 4.7 (5) | 0 (5) | Straight; wide |
| rhd3-1 shv2-1 | 94 sd ± 9 (5) | 9.4 sd ± 0.1 (5) | 2 sd ± 3 (5) | Wavy; crooked |
| cen1-1 shv1-4 | 25 sd ± 2 (5) | 9.6 sd ± 3.2 (5) | 0 (5) | Sometimes curled |
| tip1-2 shv2-1 | 75 sd ± 5 (5) | 13.6 sd ± 3.8 (5) | 54 sd ± 6 (5) | Straight; wide |
| tip1-1 shv2-1 | 58 sd ± 1 (5) | 10.2 sd ± 2.8 (5) | ND (5) | Straight; wide; bulbous tips |
| shv1-1 scn1-1 | 96 sd ± 6 (5) | 13.7 sd ± 2.0 (5) | 43 sd ± 25 (5) | Sometimes curled; wide |
| shv2-1 scn1-1 | 96 sd ± 6 (5) | 28.9 sd ± 5.4 (5) | 25 sd± 1 (5) | Sometimes curled; wide; crinkled |
Features in boldface are closer to the wild type than they were in one of the parental lines. Numbers within parentheses are number of roots.
Fifty mature hairs were assessed on each root.
Ten mature hairs were measured on each root. Wild-type length was ∼900 μm.
Multiple hairs that appeared at a single site of hair formation. ND, not determined.
In three cases (rhd3-1 shv2-1, shv1-1 scn1-1, and shv2-1 scn1-1), the proportion of hairs ⩾40 μm long was similar to that of the wild type. This was not considered to be epistasis because some effects of the shv1-1 and shv2-1 mutations were evident in the shapes of hairs (Figure 3, and Tables 1 and 3).
The tip1-2 and tip1-2 shv2-1 phenotypes were indistinguishable except that the proportion of hairs ⩾40 μm long was slightly less in tip1-2 shv2-1 (Tables 1 and 3, and Figure 3). This almost complete epistasis suggests that the growth of root hairs on tip1-2 plants does not require SHV2.
Three Classes of Additive Phenotypes Revealed Genetic Relationships Affecting the Transition to Tip Growth, the Number of Hairs Produced at Each Hair-Forming Site, Hair Length, and Hair Shape
Genes that function independently of each other can be identified because the effects of mutant alleles combine in double mutant individuals to give an additive phenotype. By comparing data about double mutants with the single mutant phenotypes in Table 1, we identified three categories of additive double mutants. In class 1 additive double mutants, all of the phenotypes of both single mutant parents were present and were combined to some extent. Class 2 phenotypes had some additive and some synergistic features. Double mutants in class 3 lacked at least one phenotypic feature of a parental line, although other aspects of both parental phenotypes were present. Double mutants in each class are listed in Table 4, and example phenotypes are shown in Figure 4.
Table 4.
Additive Root Hair Phenotypes of Double Mutants
| Phenotypeb
|
|||||||
|---|---|---|---|---|---|---|---|
| Genotypea | % Hairs ⩾40 μm Longc |
Length of Hairs ⩾40 μm Long (% Wild-Type Length)d |
% Multiple Hairsc,e | Other Featuresf | |||
| Class 1g | |||||||
| cow1-2 bst1 | 96 sd ± 6 (5) | 10 sd ± 0.6 (5) | 23 sd ± 2 (5) | Sometimes crooked or curled | |||
| cow1-2 cen2-1 | 90 sd ± 9 (6) | 11.2 sd ± 2.1 (5) | 34 sd ± 5 (6) | Often crooked or curled | |||
| cen3-2 cow1-1 | 100 (5) | 15.8 sd ± 2.9 (5) | 8 sd ± 3 (5) | Some bulbous bases; sometimes crooked or curled; wide | |||
| rhd3-7 cow1-1 | 100 (5) | 13.6 sd ± 2.2 (5) | 8 sd ± 6 (5) | Wavy; crooked; wide | |||
| tip1-1 cen3-2 | 100 (5) | 13.4 sd ± 1.8 (5) | 27 sd ± 9 (5) | Some bulbous bases; sometimes curled | |||
| tip1-2 shv1-4 | 9 sd ± 3 (5) | 14.4 sd ± 8.7 (5) | ND | Straight; wide; length variable | |||
| Class 2 | |||||||
| cen1-1 bst1 | 100 (5) | 9.2 sd ± 1.5 (5) | 10 sd ± 3 (5) | Sometimes curled; some bulbous bases | |||
| cen3-1 cen1-1 | 51 sd ± 11 (5) | 9 sd ± 0.8 (5) | 18 sd ± 9 (5) | Sometimes curled | |||
| cen2-1 cen3-2 | 50 sd ± 2 (5) | 8.0 sd ± 2.9 (5) | 36 sd ± 4 (5) | Sometimes curled; occasional bulbous bases | |||
| tip1-1 bst1 | 71 sd ± 11 (5) | 8.3 sd ± 2.6 (5) | 44 sd ± 2 (5) | Bulbous bases; hair diameter varies | |||
| scn1-1 bst1 | 19 sd ± 5 (5) | 6.7 sd ± 0.9 (5) | 0 (5) | Most hairs bloated, formless balloons | |||
| scn1-1 cen1-1 | 13 sd ± 4 (5) | 7.0 sd ± 0.5 (5) | 100 (5) | Multiple hairs almost merged into one large balloon | |||
| cen2-1 scn1-1 | 12 sd ± 4 (5) | 5.2 sd ± 0.2 (5) | 100 (5) | Multiple hairs almost merged into one large balloon | |||
| scn1-1 cen3-2 | 100 (5) | 7.3 sd ± 1.8 (5) | 100 |
Some hairs have side branches or branch at tip; wide; sometimes curled |
|||
| scn1-1 cow1-1 | 100 (5) | 10.3 sd ± 0.6 (5) | 100 (5) | Wide; crooked; curled | |||
| scn1-1 shv3 | 1 sd ± 1 (5) | ND | ND | Single, large swelling | |||
| shv2-1 bst1 | 5 sd ± 1 (5) | 8.2 sd ± 2.5 (5) | ND | Often crooked | |||
| shv2-1 cow1-2 | 19 sd ± 4 (5) | 9.5 sd ± 4.2 (5) | 29 sd ± 6 (5) | Crooked or curled; sometimes wide toward the tip | |||
| tip1-1 scn1-1 | 37 sd ± 5 (5) | 13.4 sd ± 1.9 (5) | 98 sd ± 3 (5) | Some hairs bloated, others spiky | |||
| Class 3 | |||||||
| bst1 cen2-1 | 99 sd ± 1 (5) | 20.6 sd± +>± (5) | 5 sd ± 3 (6) | Wide; often curled; sometimes crooked | |||
| cen2-1 cen1-2 | 100 (5) | 15.0 sd± −>′ (5) | 0 (5) | Often curled | |||
| cen2-1 rhd3-7 | 100 (6) | 22.2 sd± + (6) | 5 sd ± 4 (6) | Wavy; crooked; not curled | |||
| rhd3-1 bst1 | 100 (6) | 13.8 sd± +>° (5) | 3 sd ± 5 (6) | Wavy; crooked; hair length variable | |||
| rhd3-1 cen1-2 | 100 (5) | 10.4 sd± +>° (5) | 0 (5) | Wavy; sometimes curled | |||
| rhd3-1 cen3-2 | 100 (5) | 16.1 sd± ″>× (5) | 3 sd ± 1 (5) | Some bulbous bases, sometimes curled; not wavy | |||
| rhd3-1 tip1-2 | 100 (5) | 14.9 sd± −>+ (5) | 44 sd ± 10 (5) | Wide; occasionally crooked; not wavy | |||
Single mutant phenotypes for comparison are listed in Table 1.
Features in boldface are synergistic. Features in italic are less severe than one would predict from combining the parental phenotypes (although some influence of both parental mutations can still be seen). Features underscored are much closer to those of one parent than one would predict from combining the parental phenotypes. Numbers within parentheses are number of roots. ND, not determined.
Fifty mature hairs were assessed on each root.
Ten mature hairs were measured on each root. Wild-type length was ∼900 μm.
Multiple hairs at a single site of hair formation.
Between 50 to 500 roots of each genotype were examined.
rhd1 scn1-1, rhd1 cen1-2, rhd1 rhd2-3, rhd1 cen2-1, rhd1 shv3, rhd1 cen3-2, rhd1 tip1-2, rhd1 shv2-1, rhd1 bst1, rhd1 rhd3-7, and rhd1 shv1-4 all had additive phenotypes with large swellings characteristic of rhd1 at the base of hairs that were similar to the hairs on the other parental line.
Figure 4.
Mature Root Hairs of Double Mutant Plants with Additive Phenotypes.
Examples of three categories of additive double mutants are shown. cow1-2 bst1 and cow1-2 cen2-1 have class 1 additive phenotypes, in which the defects of the parental lines affecting hair length, hair shape, and the production of multiple hairs from a single site of hair formation are combined in the double mutants. scn1-1 shv3 has a class 2 phenotype; synergistic effects greatly reduce the proportion of hairs >40 μm long, and a very high proportion of cells produce only a large swelling. rhd3-1 bst1 and rhd3-1 cen1-2 have class 3 additive phenotypes; both mutations affect hair length, but root hair lengths on double mutants are similar to that of one of the single mutant parents. Comparable pictures of the wild type and shv3 are shown in Figure 2, and of scn1-1 and rhd3-1 in Figure 3.
Class 2 additives had synergistic features affecting three phenotypic traits. The first was a decrease in the proportion of hairs ⩾40 μm long, so that fewer proceeded to successful tip growth. For example, only half of the root hairs on cen3-1 cen1-1 and cen2-1 cen3-2 double mutants exceeded 40 μm (Table 4), whereas in the corresponding single mutants, the proportion of hairs ⩾40 μm long was similar to that of the wild type (Table 1). Double mutants with this phenotype (Table 4 and Figure 4) included mutations in the CEN1, TIP1, and SCN1 genes, supporting our conclusions from the partly suppressed phenotypes (Table 3) that these genes are involved in the transition from swelling formation to tip growth. The cen1-1 bst1, tip1-1 bst1, scn1-1 bst1, cen2-1 scn1-1, and shv2-1 bst1 double mutants also had this phenotype, which suggests that BST1, CEN2, and CEN3 can also affect this step.
The second trait affected by synergy was the production of multiple hairs from hair-forming sites. For example, 61% of hair cells on scn1-1 single mutant roots had multiple hairs, and cen1-1 single mutants had no multiple hairs (Table 1), but when these mutations were combined, every hair cell on scn1-1 cen1-1 double mutant roots produced multiple hairs. Other double mutants with synergistic phenotypes affecting this trait had mutations in the CEN2, CEN3, COW1, and SCN1 genes, confirming our conclusions from single mutant phenotypes that these genes are required to restrict the number of hairs formed at each hair-forming site.
The third feature affected by synergy was hair shape. In the most dramatic cases, plants carrying combinations of the scn1-1 mutation with the bst1, cen1-1, or cen2-1 mutations produced hairs that were almost shapeless balloons (Table 4). Double mutants with synergistic phenotypes affecting hair shape were homozygous for mutations in the CEN1, CEN2, CEN3, SCN1, COW1, TIP1, BST1, SHV2, and SHV3 genes, confirming conclusions from the single mutant phenotypes that CEN1, CEN2, CEN3, SCN1, TIP1, and COW1 are involved in this process (Table 1; Schiefelbein et al., 1993; Grierson et al., 1997; Ryan et al., 1998) and also implicating BST1, SHV2, and SHV3.
In class 3 additive double mutants, both parental lines had short hairs, but these effects did not combine in the double mutants. Instead, double mutant hairs were similar in length to the hairs of one parental line (cf. results in Tables 1 and 4). For example, root hairs on cen2-1 cen1-2 plants were of similar length (Table 4) to those on cen1-2 single mutants (Table 1), suggesting that CEN2 may not be able to contribute to root hair elongation in the absence of the CEN1 gene. The results in Table 4 suggest a similar dependency of CEN2 on BST1 and RHD3, of CEN3 on RHD3, and of RHD3 on TIP1. In three cases, hair shape was also closer to one parent than the other, such that the curled (cen2-1 rhd3-7) or wavy (rhd3-1 cen3-2 and rhd3-1 tip1-2) phenotype of a parent was not seen in the double mutant.
Role of TIP1 during Swelling Formation
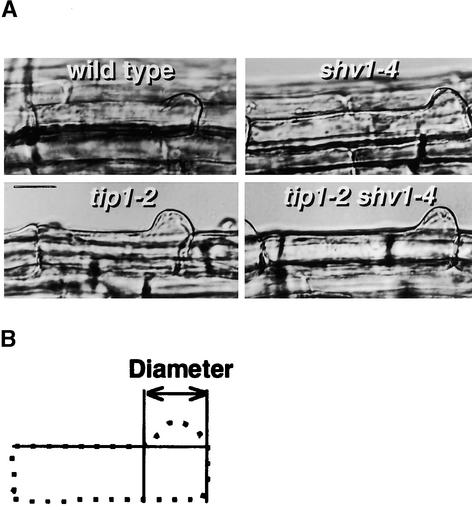
Ryan et al. (1998) previously suggested that mutations in the TIP1 gene could increase the size of the swelling formed during root hair initiation. Figure 5 shows swellings formed on wild-type and on tip1-2, shv1-4, and tip1-2 shv1-4 mutant plants. Statistical analysis (Figure 5) confirmed that the swellings formed on tip1-2 and tip1-2 shv1-4 mutant plants were markedly larger than those on wild-type or shv1-4 plants. The root hairs that we measured had not undergone elongation growth, demonstrating that increased swelling caused by the tip1-2 mutation had been produced early in root hair development and probably in the absence of tip growth.
Figure 5.
tip1-2 Affects the Diameter of the Swelling Formed during Root Hair Initiation.
(A) Young root hairs of wild-type, shv1-4, tip1-2, and tip1-2 shv1-4 plants. Most root hairs on shv1-4 (4%, sd ± 2%) and tip1-2 shv1-4 (9%, sd ± 3) plants did not progress beyond the stage shown (see Tables 1 and 4). For this character, shv1-4 tip1-2 plants had the shv1-4 phenotype (Table 1). 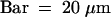 .
.
(B) Measurement of the diameters of young root hairs similar to those shown in (A). The shape of a root hair cell is shown with a dotted line. A horizontal line indicates the shape of the cell that had no hair formed, identifying the limits of the swelling that had formed at the beginning of hair formation. Student's t tests showed that the diameters of wild-type (21.55 μm, 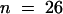 ) and shv1-4 (21.82 μm,
) and shv1-4 (21.82 μm, 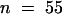 ) hairs were not significantly different from each other but were significantly smaller than those of tip1-2 (27.29 μm,
) hairs were not significantly different from each other but were significantly smaller than those of tip1-2 (27.29 μm, 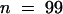 ) and shv1-4 tip1-2 (24.73 μm,
) and shv1-4 tip1-2 (24.73 μm, 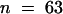 ). These results show that before growth ceases (a characteristic of shv1-4), root hairs on shv1-4 tip1-2 plants have an increased diameter, which is a characteristic of tip1-2.
). These results show that before growth ceases (a characteristic of shv1-4), root hairs on shv1-4 tip1-2 plants have an increased diameter, which is a characteristic of tip1-2.
To investigate the relationship between TIP1 and RHD1 during swelling formation, we measured the length (along the root axis) and height (perpendicular to the root) of five swellings on each of five roots of rhd1 (length 92.5 μm, sd ± 16.3; height 82.0 μm, sd ± 13.2) and tip1-2 rhd1 (length 83.7 μm, sd ± 13.7; height 62.1 μm, sd ± 9.7) plants. The results suggest that the increase in swelling size conferred by the tip1-2 mutation (Figure 5) is not seen in the presence of the rhd1 mutation, indicating that TIP1 may restrict swelling size by a mechanism that requires RHD1.
Synergistic Phenotypes Affect the Beginning of Root Hair Formation
The most dramatic results of our study are the three synergistic double mutant phenotypes shown in Figure 6. In all three cases, the double mutant roots are almost hairless, even though the corresponding single mutants have normal numbers and arrangements of hair cells. These results indicate that the TIP1, CEN2, RHD3, SCN1, and SHV3 genes have critical roles affecting the very beginning of root hair formation.
Figure 6.
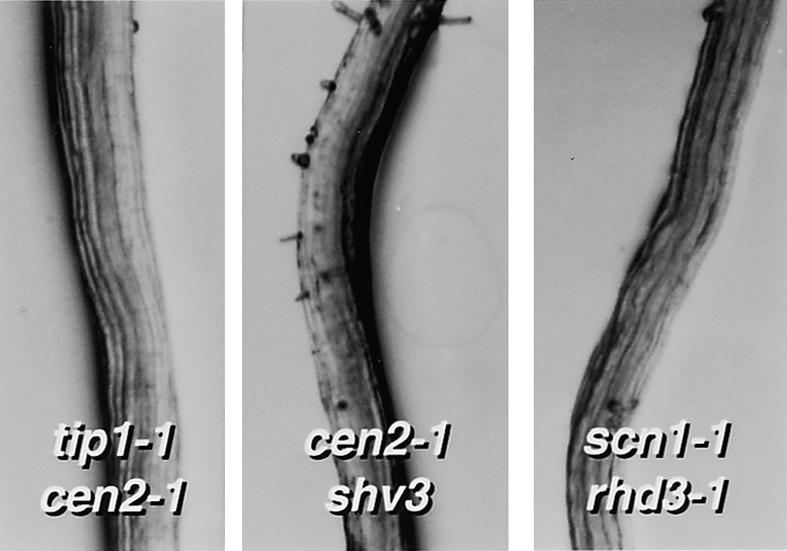
Synergistic Phenotypes Affecting the Beginning of Hair Formation.
The double mutants shown have a severe hairless phenotype that is not seen in the parental single mutant lines. Comparable pictures of the wild type and shv3 are shown in Figure 2, tip1-1 and scn1-1 in Figure 3, and cen2-1 in Figure 4.
DISCUSSION
Eight New Genes Affecting Root Hair Morphogenesis
In a large, visual screen, we identified 29 new mutants and eight new loci involved in root hair morphogenesis. The new mutants identify recessive, nuclear, single-gene mutations with strong root hair phenotypes. Single mutant phenotypes suggest roles for RHD2, SHV1, SHV2, and SHV3 in the transition from swelling formation to tip growth; for BST1, CEN1, CEN2, CEN3, RHD3, and SCN1 in controlling hair shape and length; and for BST1, CEN2, CEN3, and SCN1 in preventing the formation of multiple hairs at each hair-forming site (Table 1). Visual screens have not reached saturation (no more than nine alleles from all screens so far for RHD3; Wang et al., 1997; this work), which suggests that more genes remain to be found.
Double Mutant Analysis Reveals a Complex Genetic Network Controlling Root Hair Morphogenesis
The phenotypes of double mutant plants confirmed roles for genes suggested by the single mutant phenotypes, revealed additional roles that were not apparent from single mutant phenotypes, and suggested hypotheses about the functional relationships between genes. In some cases, the mutations cause incomplete loss of function, and our interpretations are models that can be tested as new molecular and genetic tools become available.
The Beginning of Root Hair Formation
Previous studies have shown that RHD6 acts downstream of TTG and GL2, and it is believed to be one of the first genes to act in root hair cells to promote hair formation (Masucci and Schiefelbein, 1996). The rhd6 mutation was epistatic to mutations in all 10 genes we tested (Table 2), which suggests that our new genes act downstream of RHD6, and hence probably downstream of TTG, GL2, and CPC (Masucci and Schiefelbein, 1996).
We observed synergistic effects between mutations in the TIP1 and CEN2, CEN2 and SHV3, and SCN1 and RHD3 pairs of genes that prevented root hair formation (Figure 6). The results demonstrate that those genes can influence root hair development much earlier in the process than has previously been suspected. For example, RHD3 had previously been placed after RHD2 in a genetic pathway of root hair formation (Schiefelbein and Somerville, 1990). However, the roots of scn1-1 rhd3-1 double mutant plants are almost completely bald (Figure 6), indicating that in the absence of SCN1 function, RHD3 is required at the very beginning of hair formation. tip1-1 cen2-1 and cen2-1 shv3 double mutant plants have phenotypes similar to scn1-1 rhd3-1 (Figure 6), showing that TIP1, CEN2, and SHV2 can also affect root hair formation much earlier than suggested by their single mutant phenotypes. Further anatomical and genetic (triple mutant) studies are required to clarify the relationships between these genes and other genes affecting the beginning of hair formation.
We have two alternate hypotheses concerning this group of double mutants. In the first, two genes could have overlapping functions and partly compensate for each other's absence; but in double mutants, where the functions of both genes are impaired, they might fail to compensate for each other, and a severe root hair phenotype would result. In the second, the mutant alleles used to make each double mutant are not complete loss of function, but because they affect the same linear pathway, the function of the pathway is critically impaired in each double mutant, resulting in a severe root hair phenotype. Further genetic and molecular information is needed to enable us to distinguish between these hypotheses.
Swelling Formation
Root hairs first appear as swellings on the surface of hair-forming cells. Previous research has suggested that these swellings are formed by cell wall loosening and that the TIP1 and RHD1 genes restrict the amount of loosening in wild-type plants (Schiefelbein and Somerville, 1990; Ryan et al., 1998). We showed that the tip1-2 shv1-4 double mutant has an additive phenotype (Figure 5), confirming that TIP1 acts during swelling formation to limit the size of the swelling. We also made a series of double mutants that included the rhd1 mutation. These had additive phenotypes (Table 4), suggesting that RHD1 acts independently of the other genes studied here. The increase in swelling size conferred by the tip1-2 mutation (Figure 5) is not seen in the presence of the rhd1 mutation, indicating that TIP1 may restrict swelling formation by a mechanism that involves RHD1. A more careful analysis of swelling volume and shape is required to verify these observations.
Transition from Swelling Formation to Tip Growth
Previous work has suggested that swelling formation does not involve the same mechanisms as root hair elongation and might require different genes (Schiefelbein and Somerville, 1990; Grierson et al., 1997). This is supported by physiological evidence that a calcium gradient is not established until after the swelling forms and that the gradient does not form in Rhd2− cells (Wymer et al., 1997). Our results uphold and expand on this view. Eleven genes (RHD2, SHV1, SHV2, SHV3, TIP1, BST1, RHD3, CEN1, CEN2, CEN3, and SCN1) are involved at the transition to tip growth, and six of these (RHD2, SHV1, SHV2, BST1, CEN1, and CEN3) are involved for the first time at this stage.
The RHD2 gene was previously shown to be required for the transition from swelling formation to tip growth (Schiefelbein and Somerville, 1990; Wymer et al., 1997). We have identified a similar requirement for SHV1, SHV2, and SHV3. Epistasis (Figure 2 and Table 2) suggests that SHV2 contributes later than SHV1 and that SHV3 contributes later than RHD2. Because the phenotypes were so similar, it was not possible to differentiate double mutants carrying mutations in the RHD2 gene and in the SHV1 or SHV2 genes, so the relationship between RHD2 and SHV1 and SHV2 could not be elucidated.
The requirement for RHD2, SHV1, and SHV2 is partly overcome by mutations in a second set of genes—TIP1, RHD3, CEN1, and SCN1 (Table 3 and Figure 3). In the case of TIP1 and SHV2, the tip1-2 mutation is almost completely epistatic to shv2-1 (Table 2 and Figure 3), suggesting that a function of SHV2 may be to negatively regulate TIP1. If this is the case, the phenotype of Shv2− mutants may be attributed to unregulated TIP1 activity. This model predicts that a double mutant carrying shv2-1 and a weak loss-of-function allele of TIP1 would have a phenotype closer to the shv2-1 single mutant phenotype because of the greater levels of residual TIP1 activity. The tip1-1 shv2-1 double mutant carries the weaker of the two Tip1− alleles; its partly suppressed phenotype is closer to that of shv2-1 than is the tip1-2 shv2-1 phenotype (Tables 1 and 3). This is consistent with our hypothesis. In other cases of partial suppression, we did not observe epistasis, suggesting that the corresponding genes are not involved in the same biochemical pathway. In those cases, we hypothesize that each partly suppressed phenotype identifies two genes with opposing effects on a process required for tip growth.
In some double mutants with class 2 additive phenotypes, we observed synergistic effects that reduced the proportion of hairs proceeding to successful tip growth to ⩾40 μm long (Table 4). This provided further evidence that CEN1, TIP1, and SCN1 are involved during the transition to tip growth and showed that the BST1, CEN2, and CEN3 genes can also affect this step.
Number of Hairs Produced from Each Site of Hair Formation
Tip1− and Cow1− mutants have previously been shown to produce multiple hairs from a proportion of hair-forming sites (Schiefelbein et al., 1993; Grierson et al., 1997; Ryan et al., 1998). This suggests that TIP1 and COW1 prevent multiple hairs from forming on wild-type plants. Single and double mutant phenotypes show that BST1, CEN2, CEN3, and SCN1 are also required to repress the production of multiple hairs (Tables 1 and 4). Synergistic phenotypes (Table 4, class 2 additives) also implicate CEN1. The proportions of multiple hairs on single and double mutants carrying the scn1-1 mutation were especially high (Tables 1 and 4), suggesting that SCN1 plays a particularly important role in preventing the production of multiple hairs.
The cen2-1 cen1-2 double mutant produced no multiple hairs (Table 4), in contrast to the cen2-1 single mutant phenotype in which ∼7% of the hair-forming sites produced multiple hairs (Table 1). These results raise the intriguing possibility that either CEN2 or CEN1 must be present for multiple hairs to form.
Root Hair Elongation by Tip Growth
Previous studies identified roles for RHD3, RHD4, TIP1, and COW1 during root hair elongation. The cow1-1 mutation was shown to be epistatic to rhd4, suggesting that RHD4 cannot contribute to tip growth in the absence of the COW1 gene (Grierson et al., 1997). We did not find any other examples of epistasis affecting tip growth. This indicates that most of the genes in our study that are involved in tip growth contribute at least partly independently during root hair elongation. That said, class 3 additive phenotypes identified several pairs of genes that do not contribute independently to hair length (Table 4). Many double mutants with mutations in the RHD3 gene had phenotypes in this class (Table 4 and Figure 4), suggesting that RHD3 contributes to tip growth by a process that involves several other genes, including BST1, CEN1, CEN2, CEN3, and TIP1. Similar results suggest that the contribution of CEN2 to hair length is dependent on CEN1 and BST1 (Table 4).
When tip growth took place in double mutants homozygous for mutations in the COW1 gene, the phenotypes were often class 1 additive. These results suggest that COW1 acts independently of most other genes. We found no evidence that COW1 contributes to hair formation at any stage other than to control the shape and number of hairs formed during tip growth.
Broader Features of the Network That Underpins Root Hair Morphogenesis
Mutations in the TIP1, CEN1, CEN2, CEN3, and BST1 genes formed double mutants in many categories, with phenotypes affecting a range of stages of hair formation (Figures 3 to 6 and Tables 1, 3, and 4). This suggests that these genes directly or indirectly affect multiple processes throughout root hair development.
The synergistic phenotypes of class 2 additive double mutants identify either functional redundancy between the corresponding genes or incomplete loss-of-function mutations in components of a linear pathway. We are not able to distinguish between these interpretations without more genetic or molecular information about the mutations involved. However, combinations of mutant alleles of CEN3 with CEN1, CEN2 with CEN3, and TIP1 with BST1 all have strong phenotypes (Table 4) that suggest functional relationships and that merit further analysis.
Mutations in the SCN1 gene had particularly strong effects. rhd6 was epistatic to scn1-1, preventing root hair formation (Table 2), but all of the other double mutants that included scn1-1 had partly suppressed, class 2 additive, or synergistic phenotypes (Tables 3 and 4, and Figure 6). Apparently SCN1 either takes part in direct interactions or has overlapping functions with many other genes, including RHD3, BST1, CEN1, CEN2, CEN3, COW1, SHV3, and TIP1.
Conclusions
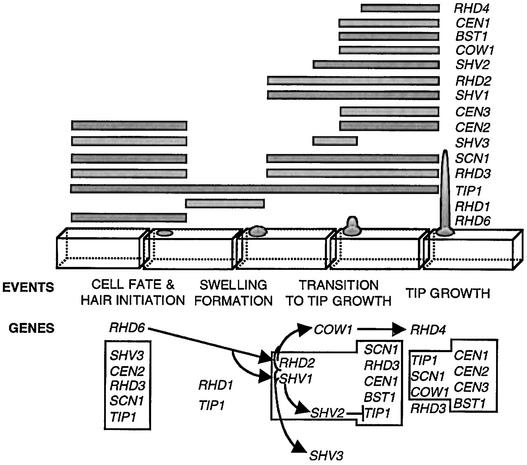
Figure 7 summarizes contributions of 15 genes to root hair growth. Hair formation requires RHD6. TIP1, CEN2, RHD3, SCN1, and SHV3 are active at the beginning of hair formation. In the absence of tip growth, TIP1 acts to restrict the size of the swelling that forms at the beginning of hair growth, possibly by a mechanism that requires RHD1. The transition to tip growth requires RHD2, SHV1, SHV2, and SHV3. The effects of RHD2, SHV1, and SHV2 oppose those of genes from the group TIP1, RHD3, CEN1, and SCN1 on the same processes of tip growth. We hypothesize that SHV2 negatively regulates TIP1 during tip growth. BST1, CEN2, and CEN3 are also required for tip growth. Mechanisms that restrict the number of root hairs produced from each site of hair formation require TIP1, COW1, BST1, CEN2, CEN3, and SCN1. CEN1, CEN2, CEN3, RHD3, SCN1, TIP1, COW1, BST1, SHV2, and SHV3 are all required to control hair shape. RHD3 contributes to tip growth by a process that involves other genes, including BST1, CEN1, CEN2, CEN3, and TIP1. The contribution of CEN2 to hair length is dependent on CEN1 and BST1. COW1 acts largely independently during elongation and does not contribute to any other stage of root hair development. In contrast, SCN1 and TIP1 interact with many other genes and affect many stages of morphogenesis.
Figure 7.
Genetic Contributions to Root Hair Development.
This diagram combines the information obtained from this study with that from previous publications (Schiefelbein and Somerville, 1990; Schiefelbein et al., 1993; Masucci and Schiefelbein, 1994; Grierson et al., 1997; Ryan et al., 1998). Wild-type development is summarized by a line drawing. Horizontal bars represent stages at which each gene is involved. The sequence of gene action is indicated below. A mutation in a gene immediately before an arrow is epistatic to a mutation in the gene immediately after an arrow (complete epistasis results are listed in Table 2). A blunted line indicates hypothetical negative regulation of TIP1 by SHV2 during the transition to tip growth. Boxed areas indicate double mutant results that identify functional relationships between genes during a particular stage of morphogenesis.
We will test our specific hypotheses about the functions of genes as soon as molecular tools become available. The genetic framework that we have provided sets the scene and provides genetic resources for future studies of root hairs, a cell type that is very well suited to research into cell biology and cell physiology and that plays critical roles affecting the human food chain.
METHODS
Arabidopsis Lines
The isolation of tip1-2 (Ryan et al., 1998) and cow1-1 and cow1-2 (Grierson et al., 1997) has been described previously. bst1 was isolated from a fast-neutron-mutagenized population obtained from Lehle Seeds (Round Rock, TX). To isolate other mutants, we used the chemical mutagen ethyl methanesulfonate to induce heritable changes in the genome of 6000 Arabidopsis thaliana ecotype Columbia seeds (treated in a 0.5% solution for 12 hr). Approximately 75,000 M2 seedlings were screened for root hair abnormalities. Heritable mutants were crossed to the wild type and self-fertilized to demonstrate that we had isolated 29 new recessive, single-locus, nuclear mutations. Lines with heritable root hair phenotypes were grouped according to phenotype and intercrossed with other members of the same group to test for complementation. Representatives of each complementation group were then intercrossed with each other to confirm the final number of complementation groups. Additional alleles of two monoallelic loci (scn1-2 and shv2-2) were identified by crossing to unpublished lines kindly provided by Katharina Schneider and John Schiefelbein. The 29 lines isolated in this study were shv3; cen3-1, cen3-2, cen3-3; shv1-1, shv1-2, shv1-3, shv1-4, shv1-5; shv2-1, shv2-2; bst1; cen2-1, cen2-2; scn1-1, scn1-2; cen1-1, cen1-2, cen1-3, cen1-4; cow1-3, cow1-4, cow1-5; rhd2-2, rhd2-3, rhd2-4; and rhd3-7, rhd3-8, rhd3-9. cen1 alleles have a trichome phenotype and map near to WAVY/SINGED, but no test for allelism has been conducted. In a side-by-side comparison, the phenotypes of our mutants were noticeably different from those of axr1-12, axr3, axr4-2, etr1, and aux1-7.
Mutant lines generated after two rounds of backcrossing to the wild type were used for phenotypic characterization. Root hair phenotypes of >1000 5-day-old seedlings of each line were assessed and photographed by using bright-field and dark-field microscopy with a Leica MZ6 stereozoom microscope. Root hairs on photographs were counted and measured to generate quantitative data. The proportion of hairs ⩾40 μm long was counted to indicate whether hairs could develop beyond the transition to tip growth, which takes place when hairs are 20 to 40 μm long (Dolan et al., 1994). Hairs ⩾40 μm long were presumed to be tip growing, and their length at maturity was measured to assess their ability to elongate. The lengths of five mature hairs on each of five wild-type roots grown under the same conditions as the mutants were also measured, and the results were used to calculate the mutants' percentage of the wild-type length. Several of the new mutants produced multiple hairs at a proportion of hair-forming sites. We counted the proportion of hair-forming sites producing multiple hairs to allow us to compare the strengths of these phenotypes. The Landsberg erecta ecotype was obtained from the Nottingham Arabidopsis Stock Centre. Seeds of rhd1-1, rhd2-1, rhd3-1, tip1-1, and rhd6 were the kind gift of John Schiefelbein. Mutants represented 10 complementation groups, two of which (RHD2 and RHD3) have been described before (Schiefelbein and Somerville, 1990). A mutant that is the only known representative of its complementation group is referred to without an allele extension (e.g., rhd6, not rhd6-1).
We recorded the number of days to bolting, the number of rosette leaves at bolting, trichome shape and density, the shape and area of the largest leaf, the number and length of main and cauline shoots and branches, the number and length of fruiting and nonfruiting stems, the distance between siliques, and silique length. Replicate observations were made of six homozygous mutant plants from each line after two rounds of backcrossing to the wild type. Because mutations at one locus, CEN1, strongly affected the shape of trichomes, the possibility that this locus is allelic to the WAVY locus (Marks and Esch, 1992) should be investigated. Mutations at SHV1 and CEN2 reduced the apparent hairiness of leaves by approximately half, although the shape of the trichomes that were present appeared to be normal.
Growth Conditions
Seedlings shown in Figure 1A were grown on Murashige and Skoog medium (Flow Laboratories, Irvine, Scotland, UK) containing 3% sucrose and 1% agarose (unless otherwise stated); those shown in Figures 2 to 4 and 6 were grown on another medium (Wymer et al., 1997) containing 1% sucrose and 1% agarose. Seeds were germinated with the plates in a vertical orientation at 22°C with constant illumination. For self-fertilization and crossing experiments, plants were transferred at the eight-leaf stage to compost supplemented with slow-release fertilizer (Osmocote; Sierra UK, Nottingham, UK) and kept well watered; the seedpods were bagged before seeds were shed.
Genetic Mapping
Mutants of the Columbia cultivar of Arabidopsis were crossed with wild-type plants of the Landsberg cultivar. DNA of mutant seedlings from the second (F2) generation was analyzed using cleaved amplified polymorphic sequences (CAPS) and microsatellite markers to determine the location of each mutated gene. CAPS primers (Research Genetics, Huntsville, AL) were used as described (Konieczny and Ausubel, 1993). The results were analyzed using Mapmaker 3.0 (Lander et al., 1987; Lincoln et al., 1992) and the Kosambi function. Loci were linked to markers as follows: shv1-1, 1.9 centimorgans (cM) from GAPC; shv2-1, between LFY3 (13.3 cM) and nga139 (35.2 cM); shv3, between AG (15.6 cM) and DHS1 (21.0 cM); scn1-1, 6.8 cM from GAPC; cen1-1, no recombinants with NCC1 in 26 plants; cen2-1, between nga151 (12.8 cM) and nga139 (10.9 cM); cen3-2, between GAPC (5.5 cM) and GL1 (31.2 cM); and bst1, 24 cM south of DFR.
Isolation and Characterization of Double Mutants
Mutant plants were intercrossed, and F2 progeny that displayed one of the mutant phenotypes were self-fertilized to produce multiple F3 families that segregated for the double mutant phenotype. Confirmation of the identities of cen3-2 cow1-1, scn1-1 cen1-1, cen2 scn1-1, rhd3 scn1-1, scn1-1 bst1, bst1 cen2, cen1-1 bst1, rhd3 cen3-2, scn1-1 shv1-1, shv2-1 tip1-2, and tip1-1 cen3-2 was obtained by backcrossing to single mutants and by crossing to the wild type and examining the F2 progeny for both parental single mutant phenotypes. The latter test was used to confirm the identities of cen3-2 scn1-1 and tip1-1 scn1-1. Null alleles are ideal for determining epistasis. We used the strongest loss-of-function alleles available to us for most of the double mutants described here. While this work was ongoing, Wang et al. (1997) published the sequence of rhd3-1, revealing that although it is the strongest loss-of-function allele of RHD3, it is not null.
Root hair phenotypes of 50 to 500 5-day-old seedlings of each double mutant were assessed and photographed by using bright-field and dark-field microscopy with a Leica MZ6 stereozoom microscope. Root hairs on photographs were counted and measured to generate quantitative data.
The diameters of initiating root hairs of wild-type, tip1-2, shv1-4, and tip1-2 shv1-4 double mutant plants were measured using differential interference photomicroscopy of roots grown on glass slides, as previously described (Grierson et al., 1997).
Acknowledgments
C.S.G. thanks all who have supported and encouraged her during this project, including the other authors, Malcolm Bennett, John Gray, David Gubb, Patrick Hussey, Jane Langdale, and Ottoline Leyser. She also thanks the reviewers and coeditor for helpful suggestions. The work was funded by a Royal Society Dorothy Hodgkin Research Fellowship to C.S.G. and by Biotechnology and Biological Sciences Research Council grants (Nos. AT 208/583 and 7/G06940). IACR–Long Ashton and John Innes Centre receive grant-aided support from the Biotechnology and Biological Sciences Research Council of the United Kingdom.
References
- Bibikova, T.N., Jacob, T., Dahse, I., and Gilroy, S. (1998). Localized changes in apoplastic and cytoplasmic pH are associated with root hair development in Arabidopsis thaliana. Development 125, 2925–2934. [DOI] [PubMed] [Google Scholar]
- DiCristina, M., Sessa, G., Dolan, L., Linstead, P., Baima, S., Ruberti, I., and Morelli, G. (1996). The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 10, 393–402. [DOI] [PubMed] [Google Scholar]
- Dolan, L., and Scheres, B. (1998). Root pattern: Shooting in the dark? Semin. Cell Dev. Biol. 9, 201–206. [DOI] [PubMed] [Google Scholar]
- Dolan, L., Duckett, C.M., Grierson, C., Linstead, P., Schneider, K., Lawson, E., Dean, C., Poethig, S., and Roberts, K. (1994). Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120, 2465–2474. [Google Scholar]
- Galway, M.E., Masucci, J.D., Lloyd, A.M., Walbot, V., Davis, R.W., and Schiefelbein, J.W. (1994). The TTG gene is required to specify epidermal-cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166, 740–754. [DOI] [PubMed] [Google Scholar]
- Galway, M.E., Heckman, J.W., and Schiefelbein, J.W. (1997). Growth and ultrastructure of Arabidopsis root hairs: The rhd3 mutation alters vacuole enlargement and tip growth. Planta 201, 209–218. [DOI] [PubMed] [Google Scholar]
- Galway, M.E., Lane, D.C., and Schiefelbein, J.W. (1999). Defective control of growth rate and cell diameter in tip-growing root hairs of the rhd4 mutant of Arabidopsis thaliana. Can. J. Bot. 77, 494–507. [Google Scholar]
- Grierson, C.S., Roberts, K., Feldmann, K.A., and Dolan, L. (1997). The COW1 locus of Arabidopsis acts after RHD2, and in parallel with RHD3 and TIP1, to determine the shape, rate of elongation, and number of root hairs produced from each site of hair formation. Plant Physiol. 115, 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using codominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Green, P., Abrahamson, J., Barlow, A., Daly, M.J., Lincoln, S.E., and Newburg, L. (1987). MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181. [DOI] [PubMed] [Google Scholar]
- Lee, M.L., and Schiefelbein, J.W. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473–483. [DOI] [PubMed] [Google Scholar]
- Lincoln, S., Daly, M., and Lander, E. (1992). Constructing Genetic Maps with Mapmaker/Exp 3.0, 3rd ed. (Cambridge, MA: Whitehead Institute).
- Marks, M.D., and Esch, J.J. (1992). Trichome formation in Arabidopsis as a genetic model system for studying cell expansion. Curr. Top. Plant Biochem. Physiol. 11, 131–142. [Google Scholar]
- Masucci, J.D., and Schiefelbein, J.W. (1994). The rhd6 mutation of Arabidopsis thaliana alters root hair initiation through an auxin-associated and ethylene-associated process. Plant Physiol. 106, 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci, J.D., and Schiefelbein, J.W. (1996). Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8, 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci, J.D., Rerie, W.G., Foreman, D.R., Zhang, M., Galway, M.E., Marks, M.D., and Schiefelbein, J.W. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122, 1253–1260. [DOI] [PubMed] [Google Scholar]
- Pitts, R.J., Cernac, A., and Estelle, M. (1998). Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 16, 553–560. [DOI] [PubMed] [Google Scholar]
- Ryan, E., Grierson, C.S., Cavell, A., Steer, M., and Dolan, L. (1998). TIP1 is required for both tip growth and non-tip growth in Arabidopsis. New Phytol. 138, 49–58. [Google Scholar]
- Schiefelbein, J.W., and Somerville, C. (1990). Genetic-control of root hair development in Arabidopsis thaliana. Plant Cell 2, 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein, J.W., Shipley, A., and Rowse, P. (1992). Calcium influx at the tip of growing root hair cells of Arabidopsis thaliana. Planta 187, 455–459. [DOI] [PubMed] [Google Scholar]
- Schiefelbein, J., Galway, M., Masucci, J., and Ford, S. (1993). Pollen-tube and root hair tip growth is disrupted in a mutant of Arabidopsis thaliana. Plant Physiol. 103, 979–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, K., Wells, B., Dolan, L., and Roberts, K. (1997). Structural and genetic analysis of epidermal cell differentiation in Arabidopsis primary roots. Development 124, 1789–1798. [DOI] [PubMed] [Google Scholar]
- Schneider, K., Mathur, J., Boudonck, K., Wells, B., Dolan, L., and Roberts, K. (1998). The ROOT HAIRLESS 1 gene encodes a nuclear protein required for root hair initiation in Arabidopsis. Genes Dev. 12, 2013–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto, M., Roberts, K., and Dolan, L. (1995). Ethylene is a positive regulator of root hair development in Arabidopsis thaliana. Plant J. 8, 943–948. [DOI] [PubMed] [Google Scholar]
- Wada, T., Tachibana, T., Shimura, Y., and Okada, K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277, 1113–1116. [DOI] [PubMed] [Google Scholar]
- Wang, H.Y., Lockwood, S.K., Hoeltzel, M.F., and Schiefelbein, J.W. (1997). The ROOT HAIR DEFECTIVE3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes Dev. 11, 799–811. [DOI] [PubMed] [Google Scholar]
- Wymer, C.L., Bibikova, T.N., and Gilroy, S. (1997). Cytoplasmic free calcium distributions during the development of root hairs of Arabidopsis thaliana. Plant J. 12, 427–439. [DOI] [PubMed] [Google Scholar]