Abstract
Ribonucleotide reductase (RNR) is a key enzyme involved in the DNA synthesis pathway. The RNR-encoded genes are cell cycle regulated and specifically expressed in S phase. The promoter of the RNR2 gene encoding for the small subunit was isolated from tobacco. Both in vivo and in vitro studies of the DNA–protein interactions in synchronized BY2 tobacco cells showed that two E2F-like motifs were involved in multiple specific complexes, some of which displayed cell cycle–regulated binding activities. Moreover, these two elements could specifically interact with a purified tobacco E2F protein. Involvement of the E2F elements in regulating the RNR2 promoter was checked by functional analyses in synchronized transgenic BY2 cells transformed with various RNR2 promoter constructs fused to the luciferase reporter gene. The two E2F elements were involved in upregulation of the promoter at the G1/S transition and mutation of both elements prevented any significant induction of the RNR promoter. In addition, one of the E2F elements sharing homology with the animal E2F/cell cycle–dependent element motif behaved like a repressor when outside of the S phase. These data provide evidence that E2F elements play a crucial role in cell cycle regulation of gene transcription in plants.
INTRODUCTION
The G1/S transition of the cell cycle is a crucial step before entry into the S phase, in which DNA replication takes place (Johnson, 1992). One of the key processes in this phase is the biosynthesis of deoxyribonucleotides. Ribonucleotide reductase (RNR) is an essential enzyme for de novo synthesis of deoxyribonucleotides, catalyzing the reduction of the four ribonucleotide diphosphates to their corresponding deoxyribonucleotides (Reichard, 1988). The active enzyme consists of two different homodimeric subunits: the R1 large subunit, involved in the allosteric regulation of the enzyme, and the R2 small subunit, involved in the catalytic activity (Thelander et al., 1980).
In yeast and mammals, both RNR activity and RNR gene expression are tightly regulated throughout the cell cycle, with maximal values in the S phase (Elledge et al., 1992; Greenberg and Hilfinger, 1996). In yeast, regulation of RNR gene expression has been studied mainly at the transcriptional level. Periodic RNR1 gene expression was suggested to be controlled by GC-rich Mlu I boxes (Elledge et al., 1992; Lowndes et al., 1992), which apparently also mediate transcription of several other S phase–specific yeast genes (Verma et al., 1991). One near-match Mlu I sequence was found on the RNR2 promoter (Elledge and Davis, 1987), but its role remains unclear. In mammals, for example, two broad regions that interact with nuclear proteins were necessary for upregulation of the mouse RNR1 promoter at the G1/S transition (Johansson et al., 1995). Surprisingly, the mouse RNR2 promoter was activated at an earlier stage, when quiescent cells started to proliferate, and three regions were involved in this activation (Filatov and Thelander, 1995). However, S phase–specific expression was achieved by an S phase–specific release of a block in transcription elongation which occurs in the first intron during G0 and G1 phases (Björklund et al., 1992). Additionally, a proximal CCAAT element was required for the RNR2 promoter activity to persist through the S phase (Filatov and Thelander, 1995).
In animals, specific G1/S induction of other genes involved in the DNA synthesis pathway, such as thymidine kinase (Dou and Pardee, 1996), dihydrofolate reductase (Slansky et al., 1993), and DNA polymerase α (Pearson et al., 1991), is mediated primarily by the E2F transcription factor (Slansky and Farnham, 1996a). Molecular mechanisms of gene induction mediated by the E2F factor involve both phosphorylation and protein–protein interactions, which are tightly regulated in the cell cycle. E2F is active when associated to its partner protein DP, whereas this heterodimer is transcriptionally inactive when complexed to pocket proteins, such as Rb, p107, and P130 (Dyson, 1998). As the cell cycle progresses, hyperphosphorylation of the pocket proteins by specific cyclin–CDK (cyclin-dependent kinase) complexes triggers the E2F activation of target genes. In this regulatory pathway, cooperation sometimes was observed between E2F and additional transcription factors, such as SP1, that bind to adjacent or overlapping sites (Slansky and Farnham, 1996b; Rotheneder et al., 1999). Moreover, the role of E2F in the regulation of several promoters, such as cdc2, cyclin A, and cdc25C, was influenced by a bipartite cell cycle–regulated repressor element consisting of a variant E2F site called CDE (cell cycle–dependent element) and a CHR element (cell cycle genes homology region) located a few nucleotides downstream (Zwicker and Müller, 1997).
In plants, only a few studies have investigated the cis elements involved in S phase–specific gene expression. Two cis elements resembling the GC-rich conserved motifs found in the promoter regions of auxin-regulated genes were shown to be essential for the meristematic tissue–specific activity of the proliferating cell nuclear antigen promoter (Kosugi et al., 1995). Various elements, such as a GC-rich octamer specific to plant histone promoters, a nonamer, and an ATF-related hexameric motif, were shown to be involved in driving specific induction of histone genes in proliferating cells (Terada et al., 1995; Chaubet et al., 1996) and in S phase of synchronized cells (Taoka et al., 1999).
In the synchronized BY2 tobacco cell suspension, RNR2 gene expression was shown to increase sharply at the G1/S transition (Philipps et al., 1995; Chabouté et al., 1998). However, the mechanism and the cis elements involved in this induction have remained largely unknown. In this article, we present structural and functional analyses of the BY2 tobacco RNR2 promoter. In vivo footprinting experiments and bandshift assays demonstrate that two E2F-like motifs are protected differently during the cell cycle progression and that they interact with specific protein complexes, some of which have maximal binding activities in S phase. These E2F-like motifs are able to interact with a purified tobacco E2F protein. Functional experiments in transgenic cells show that both E2F elements act synergistically to direct maximal promoter induction in S phase and that one of the E2F elements behaves like the animal E2F/CDE repressor outside S phase. Therefore, these results provide evidence that E2F-like transcription factors mediate cell cycle–regulated gene expression in plants.
RESULTS
Sequence Analysis of the Tobacco RNR2 Gene Promoter
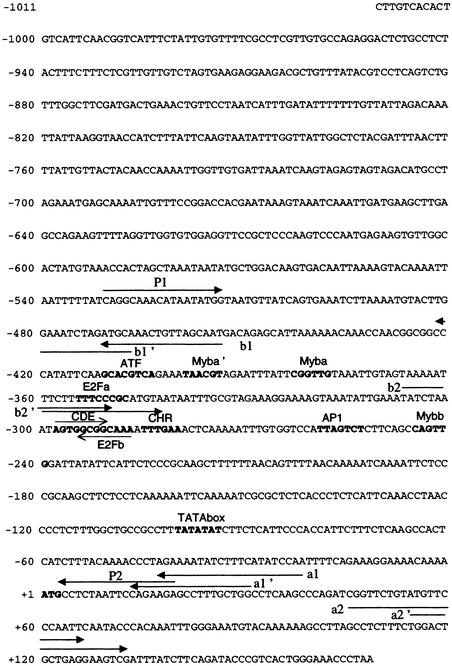
The RNR2 gene promoter was isolated by inverse polymerase chain reaction (PCR) from genomic DNA of BY2 tobacco cells, as described in Methods. The promoter sequence (EMBL accession number AJ 276,622) is shown in Figure 1. In the 1031-bp promoter region, a potential TATA box was found at −100 bp from the ATG codon. Two sequences with homology with the binding site of the mammalian E2F transcription factor (TTTG/CG/CCGC; Slansky and Farnham, 1996a) were found: E2Fa (TTTCCCGC) at −355 bp from the ATG on the direct strand, and E2Fb/CDE, composed of two overlapping E2F motifs located on the opposite strands, E2Fb (TTTGCCGC) at −294 bp on the reverse strand and a variant E2F site (AGTGGCGG-5 nucleotides [nt]-TTTGAA) on the direct strand, a sequence arrangement very similar to the CDE-CHR repressor elements described in animals (core: C/GGCGG-4 nt-C/TTGAA; Zwicker and Müller, 1997). Other motifs were found in the vicinity of the E2F-like sequences. Three sequences differing from the Myb binding consensus motif (C/TAACT/GG; Faisst and Meyer, 1992; Jin and Martin, 1999) by no more than one mismatch were found at −399, −383, and −245 bp from the ATG, in direct orientation (TAACGT: Myba) or in reverse orientation (CA/GGTTG: Myba' and Mybb). Additionally, two binding sites for AP1- and ATF-like leucine zipper factors were found at −259 and −404 bp from the ATG, on the upper or lower strands, respectively. The AP1-like site (TTAGTCT) differed by one base from the AP1 consensus (TG/TAGTCA), and the ATF-like site (TGACGTGC) differed by two mismatches from the animal ATF consensus (TGACGC/TC/AG/A; Faisst and Meyer, 1992) and by one mismatch from the plant composite ATF-like, G-box element (TGACGTGG; Schindler et al., 1992).
Figure 1.
Nucleotide Sequence of the Tobacco RNR2 Promoter.
The arrows indicate the position of the primers used in inverse PCR to amplify the sequence of the RNR2 promoter (a1/a2, a1'/a2' and b1/b2, b1'/b2') and primers used to subclone the RNR2 promoter (P1 and P2). Putative cis elements are in boldface characters, and the overlapping E2Fb/CDE motifs are indicated by thin lines.
Cell Cycle in Vivo Occupancy of Cis Elements in the RNR2 Promoter
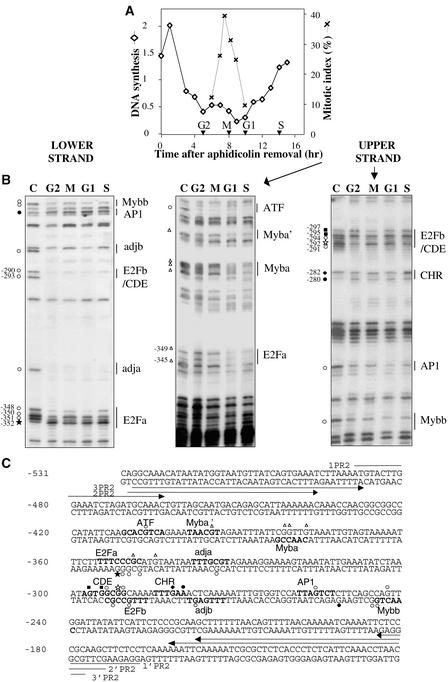
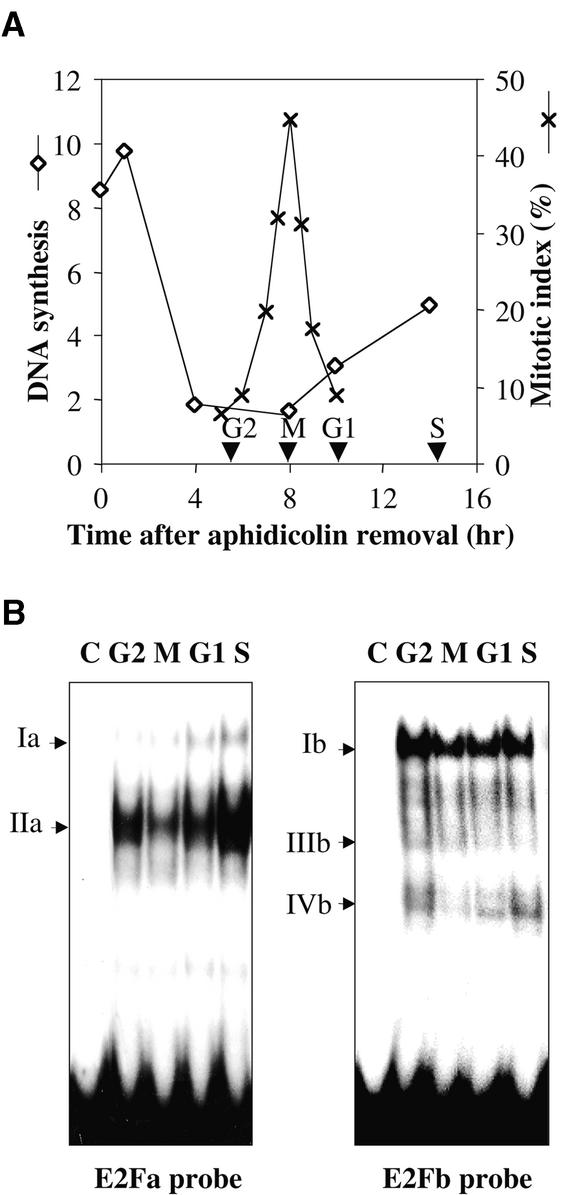
To identify the cis elements involved in the cell cycle regulation of the RNR2 gene, we first investigated the protein–DNA interactions on the RNR2 promoter at different phases of the cell cycle in synchronized BY2 cells. Synchronization was performed by a 24-hr treatment with aphidicolin, which results in a cell cycle arrest in early S phase (Nagata et al., 1992). After removal of the drug, the cells progressed synchronously through the cell cycle, reaching a maximal mitotic index at 8 hr (Figure 2A). The DMS (dimethyl sulfate)/ ligation-mediated polymerase chain reaction (LMPCR) in vivo footprinting method was used with primers that allowed analysis of the region between −489 and −107, which contains all the remarkable motifs mentioned above. Most of the consensus motifs were protected, thereby suggesting they have a functional significance (Figure 2B). The protected or hypersensitive sites were reported on the promoter sequence (Figure 2C). On the upper strand, both the E2Fa motif and the Myba and Myba' elements were protected primarily during G1 and S phases. Other motifs, such as ATF-like, AP1-like, and Mybb, displayed protections throughout the cell cycle. In contrast, some G residues were differentially reactive in the E2Fb/CDE motif: in addition to two constitutively protected G residues (−294 and −291), the G residue at −292 was protected in G2 and S phases, and the G residues at −297 and −295 were hyperreactive in G2 phase. The CHR element displayed two hypersensitive sites: one constitutive (A residue at −280) in the minor groove, and one S phase–specific (G residue at −282) in the major groove.
Figure 2.
In Vivo Footprinting Analysis of the Tobacco RNR2 Promoter during the Cell Cycle.
(A) Tobacco BY2 cells were synchronized by treatment with aphidicolin, and cell cycle progression was monitored by measuring DNA synthesis (lozenges: cpm × 102/μg protein) and mitotic index (crosses). Cells were harvested in G2, M, G1, or S phases (arrowheads) of the cell cycle.
(B) Cells harvested in G2, M, G1, or S phases of the cell cycle were treated in vivo with DMS, and LMPCRs were performed as described in Methods. Control LMPCRs were performed on in vitro DMS-treated genomic DNA (lane C). Protected and hyperreactive residues are indicated by open or filled symbols, respectively. Signals present in all steps of the cell cycle are represented by circles, in G2 by squares, in G2 and S by stars, in G1 and S by triangles, and in S by diamonds. Putative cis elements are indicated.
(C) Protected and hyperreactive G residues were reported on the RNR2 promoter sequence. Putative cis elements are in boldface characters. The primers used in LMPCR are indicated by arrows. Symbols are the same as those used in (B).
On the lower strand, the two E2Fa and E2Fb motifs displayed protection during all the steps of the cell cycle, except for the −352 G residue in the E2Fa motif, which presented hypersensitivity in G2 and S phases. Additionally, two homologous sequences, adja and adjb (TTTGA/CGT), located 9 and 7 bp from E2Fa and E2Fb sites, respectively, also were protected throughout the cell cycle. A constitutive protection also was observed for the Mybb-like motif, whereas the AP1-like element displayed hypersensitivity throughout the cell cycle. Together, these data show that E2F-like and other motifs were involved in binding proteins during cell cycle progression, thereby suggesting that they function as regulators of RNR2 gene expression. Furthermore, at least two types of nuclear complexes appear to interact with each E2F element, one constitutively and the other one more periodically during the cell cycle.
Nuclear Proteins from BY2 Cells Can Bind Specifically the E2F Elements
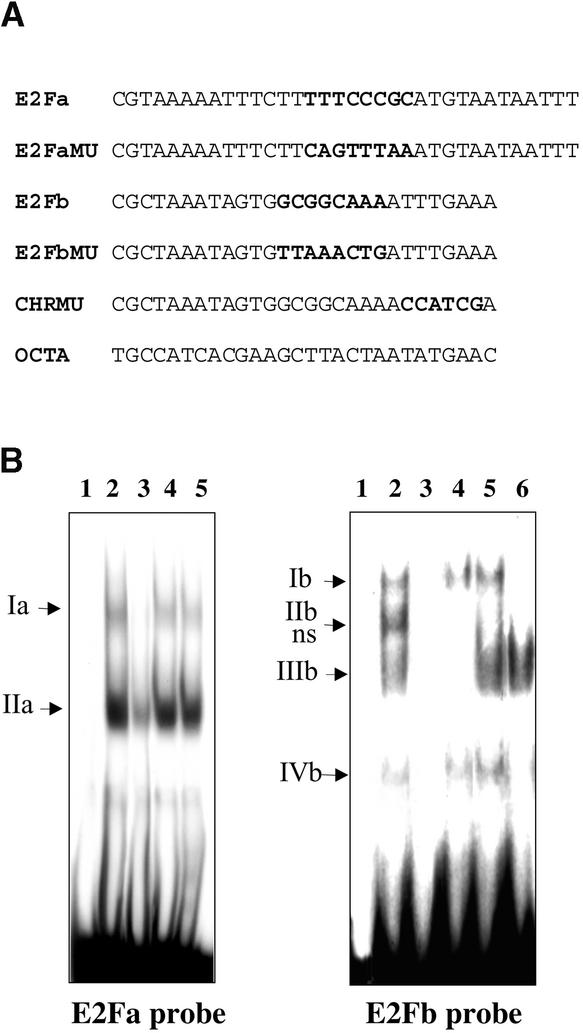
Considering the crucial role of E2F cis elements in the specific G1/S induction of cell cycle–regulated genes in animals and the fact that E2F motifs are absent from the animal RNR promoters, we focused our study on these elements in the tobacco RNR2 promoter. To analyze the capacity of E2F sites to bind nuclear complexes, electrophoretic mobility-shift assays (EMSAs) were performed with nuclear extracts from mid-log-phase BY2 cells and with 32P-labeled double-stranded oligonucleotides carrying the E2Fa or E2Fb/CDE-CHR sites (Figure 3A).
Figure 3.
Specific Binding of Nuclear Proteins to E2F-like Elements of the Tobacco RNR2 Promoter.
(A) DNA fragments used in EMSAs (only the upper strand sequences are presented).
(B) EMSA of nuclear extracts from mid-log-phase BY2 cells with the 32P-labeled E2Fa and E2Fb DNA probes: free probes (lanes 1); complexes with 6 μg of nuclear extract (lanes 2); competitions with a 100-fold molar excess of competitions with a 100-fold molar excess of the unlabelled probes (lanes 3), of the E2F site-mutated oligonucleotides (E2FMU, lanes 4), of an unrelated oligonucleotide (OCTA; lanes 5), and of the CHR site-mutated oligonucleotide (CHRMU, lane 6). Nuclear complexes are indicated by arrows. A nonspecific complex is indicated (ns).
Different patterns of protein binding were observed with the two probes (Figure 3B). Two retarded bands of different intensity were detected with the E2Fa probe (lane 2) and corresponded to complexes we called Ia and IIa. Specificity of the binding was demonstrated by competition experiments with the oligonucleotides presented in Figure 3A. Excess unlabeled wild-type E2Fa oligonucleotide competed markedly for the protein binding (lane 3), but neither an unrelated oligonucleotide (OCTA, lane 5) nor the oligonucleotide mutated in the whole E2Fa-like motif (E2FaMU, lane 4) did.
Multiple DNA–protein complexes were obtained with the E2Fb/CDE-CHR probe (Ib to IVb, lane 2). These complexes were titrated out by an excess of unlabeled E2Fb oligonucleotide (lane 3) but not by the unrelated oligonucleotide OCTA (lane 5), except for complex IIb, which therefore was designated as nonspecific. Complexes Ib and IVb were not displaced by the E2FbMU oligonucleotide mutated in the whole E2Fb/CDE-like motif (lane 4), thus indicating that these two complexes were specific to the E2Fb/CDE site. Complex IIIb was titrated out by the E2FbMU but not by the CHRMU oligonucleotide carrying the mutated version of the CHR motif (lane 6), demonstrating that this complex is related to the CHR motif.
Binding of E2F Proteins to the E2F Elements of the RNR2 Promoter
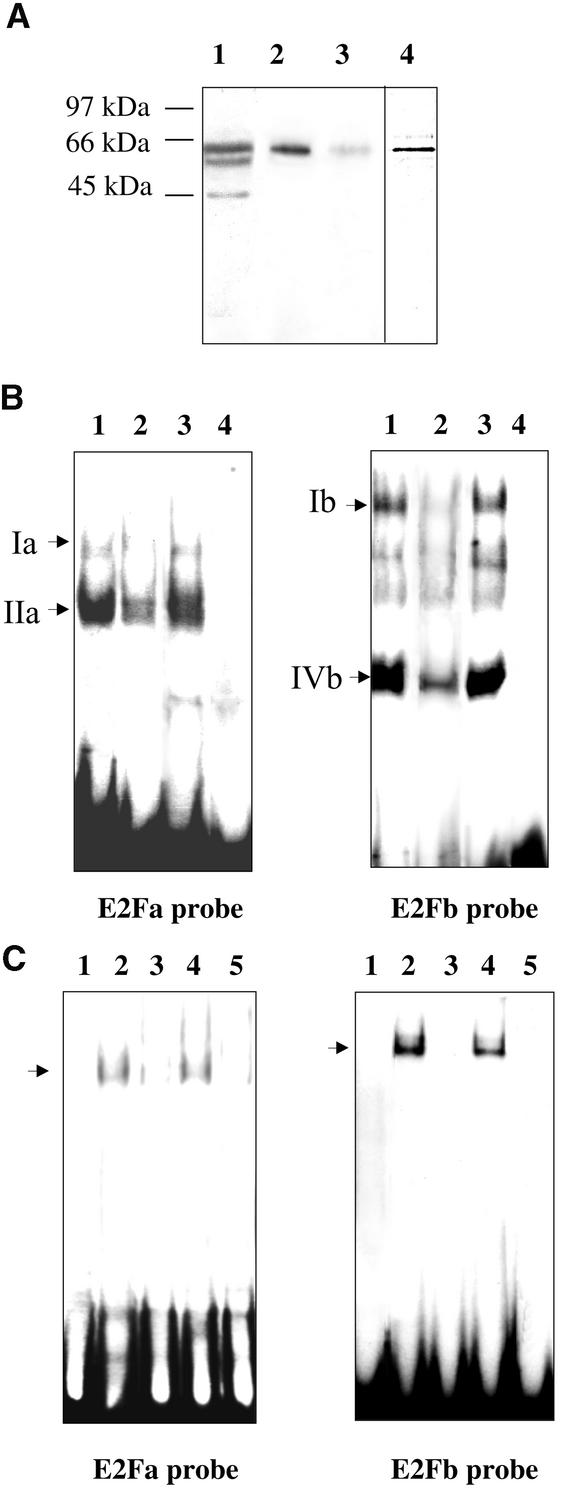
To check the involvement of E2F factors in the binding of nuclear complexes to the E2F probes, we used a rabbit polyclonal antibody raised against the human E2F5 DNA binding domain (amino acids 89 to 200) (Figure 4). The E2F DNA binding domain is well conserved (75% homology) between the various members of the mammalian E2F family (Sardet et al., 1995) and the recently discovered plant E2F proteins (Ramirez-Parra et al., 1999; Sekine et al., 1999). When used on a protein gel blot of BY2 tobacco nuclear extract, the antibody revealed three bands, two corresponding to proteins with apparent molecular masses of ∼60 kD and one corresponding to a 45-kD protein (lane 1). As a control, the antibody was tested on a protein gel blot of a recently cloned tobacco E2F protein purified as described in Methods. When analyzed on a silver-stained gel, the purified protein had the same electrophoretic mobility (lane 4) as the highest-mass band of the nuclear extract. This protein was recognized by the anti-E2F5 antibody (lane 2) in a specific interaction, as shown by the competition with excess purified human E2F5 protein (lane 3).
Figure 4.
Involvement of E2F Factor in Nuclear Complexes Binding to E2F-like Sites of the Tobacco RNR2 Promoter.
(A) Protein gel blots incubated with the antibody directed against the DNA binding domain of human E2F5 factor: 6 μg of nuclear extract from mid-log-phase BY2 cells (lane 1); 15 ng of the purified tobacco E2F protein (lanes 2 and 3); competition with 0.1 μM purified human E2F5 protein (lane 3). Silver-staining of the purified tobacco E2F protein (lane 4).
(B) EMSA of nuclear extracts from mid-log-phase BY2 cells with the 32P-labeled DNA probes E2Fa and E2Fb: free probes (lanes 4); complexes with 6 μg of nuclear extract (lanes 1); competitions with the antibody directed against the DNA binding domain of human E2F5 factor (lanes 2); and competitions with an unrelated antibody directed against α-tubulin (lanes 3). Specific E2F complexes are indicated by arrows.
(C) EMSA of the purified tobacco E2F protein with the 32P-labeled DNA probes E2Fa and E2Fb: free probes (lanes 1); complexes with 300 ng of tobacco E2F protein (lanes 2); competitions with a 200-fold molar excess of the unlabeled probes (lanes 3) or of the E2F site-mutated oligonucleotides (lanes 4); competition with the antibody directed against the DNA binding domain of human E2F5 factor (lanes 5). Specific E2F complexes are indicated by arrows.
In a second step, the antibody was tested in EMSA with nuclear extracts from mid-log-phase cells (Figure 4B). Binding of complexes Ia and Ib to the E2Fa and E2Fb sites, respectively, was markedly impeded by the anti-E2F5 antibody (lanes 2) but not by an unrelated antibody raised against α-tubulin (lanes 3). The bindings of complexes IIa and IVb were partially inhibited, and the CHR binding complex was not affected. Together, these results suggest that one or more E2F-related proteins could belong to the nuclear complexes bound to the E2F elements of the RNR2 promoter.
To confirm the ability of the two E2F sites to bind E2F factors, we tested the purified tobacco E2F protein in EMSAs (Figure 4C). A single strong signal was obtained with the E2Fb probe, and a faint signal was observed with the E2Fa probe (lanes 2). The specificity of the complexes was confirmed by competition experiments, based on the fact that E2F-specific complexes are titrated out by an excess of the unlabeled E2F probe (lanes 3) but not by the mutated version of the E2F sites (lanes 4). As an additional control, the antibody raised against the DNA binding domain of the human E2F5 was used. Prior incubation of the antibody with the tobacco E2F protein prevented subsequent binding of the protein to its specific sites on the E2Fa and E2Fb probes (lanes 5). These results suggest that the purified E2F transcription factor can bind both E2F sites on the tobacco RNR2 promoter, albeit with greater affinity for the E2Fb than for the E2Fa site.
Binding of Nuclear Proteins to E2F Elements Is Cell Cycle Regulated
To detect changes in protein binding to the E2F-like motifs during the cell cycle, nuclear extracts were prepared from synchronized BY2 cells at different steps of the cell cycle (Figure 5A). EMSAs were performed with equal amounts of nuclear extracts (Figure 5B). Protein binding to the E2F-like sites fluctuated during the cell cycle. The two specific complexes Ia and IIa, which bind to the E2Fa probe, displayed maximal binding activity in S phase and minimal binding activity in mitosis. However, complex Ia was also markedly present in G1 phase, whereas the amount of complex IIa was quite high in G2 phase. The binding activity of complex IVb to the E2Fb probe also was cell cycle regulated, with a maximal value in S phase and a substantial amount in G2 phase, whereas complex Ib showed a relatively constitutive binding activity throughout the cell cycle progression. Complex IIIb, specific to the CHR motif, was present mainly in G2 phase.
Figure 5.
Fluctuations in Binding Activities of E2F-like Elements during Cell Cycle Progression.
(A) Tobacco BY2 cells were synchronized by treatment with aphidicolin, and cell cycle progression was monitored by measuring DNA synthesis (diamonds: cpm × 102/μg protein) and mitotic index (crosses). Cells were harvested in G2, M, G1, or S phases (arrowheads) of the cell cycle.
(B) EMSA of G2, M, G1, and S phase nuclear extracts with the 32P-labeled DNA probes E2Fa and E2Fb. Lanes C are free probes. Specific complexes are indicated by arrows.
Effect of E2F Sites on RNR2 Promoter Activity
In investigating the relevance of the two E2F sites in the RNR2 promoter activity, various promoter constructs were used to drive a firefly luciferase (LUC) gene containing a plant intron (Mankin et al., 1997). Site-directed mutageneses were performed to modify the E2F sites in a 550-bp proximal promoter fragment (wild type) containing all the remarkable motifs described above. The E2Fa and E2Fb/CDE sites were each mutated in the E2FaMU and E2FbMU constructs by using the same nucleotide changes as in bandshift assays, and both E2F sites were mutated in the E2Fa + bMU construct. These constructs were introduced into BY2 cells through stable transformation by way of Agrobacterium.
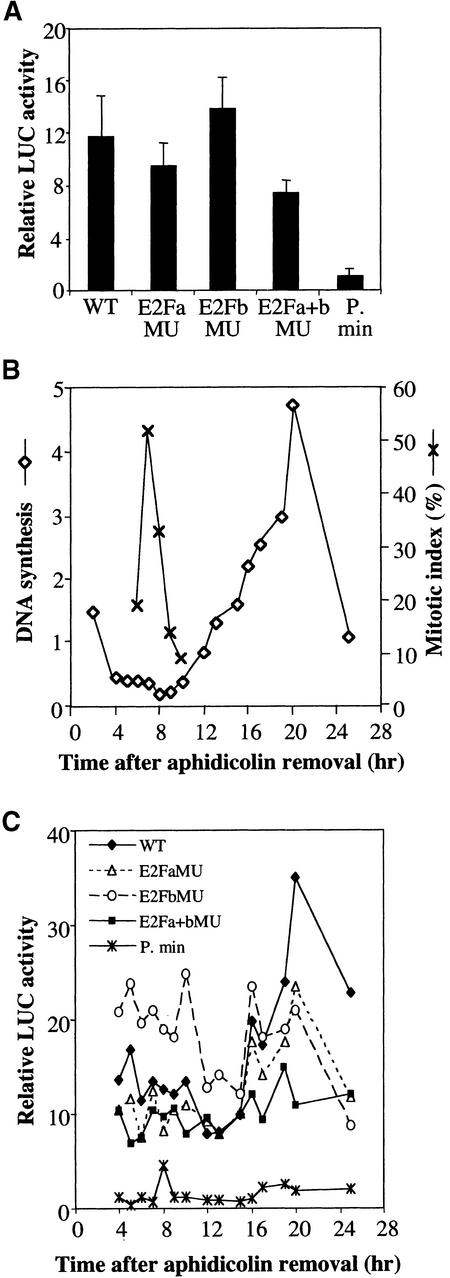
LUC activities were first measured in mid-log-phase transgenic cells (Figure 6A). Activity of the wild-type construct was 12-fold more than in a minimal promoter construct truncated to the TATA box (P. min). The E2FbMU construct showed a slightly higher LUC activity (118%) than the wild-type control construct, whereas the E2FaMU and E2Fa + bMU constructs presented lower LUC activities than the control (80 and 63%, respectively).
Figure 6.
Role of E2F Elements in the Tobacco RNR2 Promoter Activity.
Various RNR2 promoter constructs were fused to the LUC reporter gene: wild-type promoter (WT, −531 to +19), promoters with E2Fa or E2Fb mutated sites (E2FaMU, E2FbMU), promoter with both mutated E2F sites (E2Fa + bMU), and the minimal RNR2 promoter (P. min, −100 to +19). LUC activities were measured in transgenic BY2 cell lines (population of 500 to 1000 independent calli) containing the promoter constructs, as described in Methods.
(A) LUC activities in mid-log-phase cells. Error bars indicate sd.
(B) Transgenic cell lines synchronized by treatment with aphidicolin and monitored for cell cycle progression by measurements of DNA synthesis (diamonds: cpm × 102/μg protein) and mitotic index (crosses). Cell cycle progression was similar for all the transgenic lines. A representative experiment is shown.
(C) LUC activities of cells harvested at different points of the cell cycle.
For monitoring the LUC activity of the constructs during cell cycle progression, the transgenic cells were synchronized (Figure 6B), and the LUC activities were measured at different times after removal of aphidicolin (Figure 6C). As expected, the LUC activity of the wild-type construct was low outside S phase and increased concomitantly with DNA synthesis in S phase, whereas the LUC activity of the minimal promoter construct remained at a basal value throughout the cell cycle. Outside S phase, E2FaMU and E2Fa + bMU constructs had similar LUC activities, which were slightly less than in the wild-type control, similar to that in asynchronous cells. In contrast, the E2FbMu construct presented a twofold greater LUC activity relative to the wild-type construct during G2 and M phases and then decreased during G1 phase. This increased activity, already observed in asynchronous cells, could reflect the loss of a repressor element in the nuclear complex bound to the E2Fb/CDE element outside of S phase. At the G1/S transition (15 to 16 hr), LUC activities of the E2FaMU and E2FbMU constructs increased similarly, but the maximal activities in S phase remained less than for the wild-type construct (67 and 60%, respectively). Interestingly, mutation of both E2F motifs greatly affected the S-phase activity of the promoter, which remained very low. Thus, these results suggest that one E2F site is sufficient to allow G1/S induction of the RNR2 promoter but that both E2F sites are required for a maximal promoter induction in S phase. Additionally, we have revealed a dual function for the E2Fb/CDE element that acts as a repressor outside S phase and as an activator during S phase.
DISCUSSION
Singular Structure of a Plant RNR2 Promoter
In this article, we present the structural and functional analyses of the promoter sequence of a plant gene encoding ribonucleotide reductase, namely, the RNR2 small subunit.
Potential cis elements were found in a 500-bp proximal region of the promoter (Figure 1) that was sufficient to drive LUC gene induction at the G1/S transition of the cell cycle in synchronized BY2 cells (Figure 6C). The tobacco RNR2 promoter contained E2F-like motifs, similar to several animal genes induced from mid-G1 to S phase (Slansky and Farnham, 1996a), but in contrast to the animal RNR promoters, which do not possess E2F cis elements (Filatov and Thelander, 1995). The E2F motifs of the tobacco RNR2 promoter consisted in a simple element called E2Fa and a composite element consisting of a reverse E2Fb motif overlapping a CDE-CHR site. A similar organization of E2F-like motifs also exists in the 5′ flanking region of an RNR2 gene in Arabidopsis (EMBL accession number AB 023,040), and diversely organized E2F-like elements also were found in RNR1 promoters of tobacco (M.-E. Chabouté, B. Clément, G. Philipps, unpublished data) and Arabidopsis (EMBL accession number AC 007,019). Therefore, existence of E2F motifs in RNR promoters could be a general rule in plants. Promoters with two copies of E2F elements, some of which as overlapping inverted repeats, often have been found in animals. The general feature of these arrangements is that the two copies do not have exactly the same functions in promoter regulation but act synergistically to confer the final gene expression pattern (see below).
An AP1-like, an ATF-like, and three Myb-like sequences were identified close to the E2F motifs in the RNR2 promoter. Such an occurrence of general transcription factor binding sites may be a general feature of E2F cycle-regulated genes; it already has been described in animals, and in some cases, functional cooperation between E2F and general transcription factors has been demonstrated (Zwicker et al., 1995; Zwicker and Müller, 1997; Fry and Farnham, 1999).
In Vitro and in Vivo Protection of E2F-like Elements of the RNR2 Promoter during the Cell Cycle
To understand the mechanism by which the tobacco RNR2 gene is transcriptionally regulated, we have focused our in vitro analyses on the putative E2F elements, motifs that had been shown to be essential for the induction of many cell cycle–regulated genes in animals. Specific interaction of nuclear complexes with the two E2F-like elements was demonstrated by EMSA (Figure 3). Moreover, by using a purified tobacco E2F protein produced from a recently isolated E2F cDNA (Sekine et al., 1999), we ascertained that both E2F sites could interact with the E2F protein (Figure 4C), albeit with different affinities, strongly for the E2Fb site and more faintly for the E2Fa site. The stronger signal observed with the E2Fb probe could be explained by the presence on this probe of two overlapping E2F sites, E2Fb and CDE-CHR, which could enhance the stability of E2F binding as a dimer. Such a phenomenon has been described for two overlapping inverted-repeat E2F sites found in the mammalian dihydrofolate reductase (DHFR) promoter (Wade et al., 1995). The faint signal observed with the E2Fa probe suggests that the E2Fa motif–E2F protein interaction might require stabilization by another component, such as the E2F partner protein DP as described in animals, or by another protein. In this respect, antibody competition assays have demonstrated that the mammalian E2F4 factor belongs to the complex bound to a variant E2F site in the mouse cyclin E promoter; however, in vitro–produced E2F4 and DP1 failed to bind the cis element, suggesting that an additional protein was required for binding (Le Cam et al., 1999). Alternatively, the E2Fa binding complex might involve an E2F protein different from the available tobacco protein. Indeed, to date, only a few members of the E2F family have been cloned from plants, one from wheat and one from tobacco (Ramirez-Parra et al., 1999; Sekine et al., 1999). However, analysis of the Arabidopsis expressed sequence tags database reveals that at least three different E2F factors exist in Arabidopsis, and our protein gel blot analysis (Figure 4A) suggests that tobacco also contains at least three E2F proteins. The purified tobacco E2F protein migrated as a 60-kD protein, thereby displaying the same electrophoretic behavior as the wheat E2F (Ramirez-Parra et al., 1999) and the human E2F1 and E2F4 factors (Helin et al., 1992; Beijersbergen et al., 1994). In contrast, the behavior of the 45-kD nuclear tobacco E2F protein in protein gel blot electrophoresis was similar to that of human E2F5 (Hijmans et al., 1995). These observations suggest that plants also may have different families of E2F transcription factors, as animals do.
Antibody competition assays for the E2F DNA binding site (Figure 4B) provided evidence that E2F factors belong to the identified nuclear complexes and are directly involved in the binding of both E2F elements on the RNR2 promoter. The binding capacity of complexes Ia and IIa to the E2Fa site and of IVb to the E2Fb site fluctuated during the cell cycle to reach a maximum in S phase, whereas the binding activity of complex Ib to the E2Fb site was relatively nonvarying (Figure 5B). Thus, two types of complexes seem to be involved in the binding of E2F motifs, some constitutive and some fluctuating throughout the cell cycle, the latter being possibly involved in upregulating the RNR2 expression at the G1/S transition. The presence of complexes IIIb and IVb in G2 phase could well correlate with the binding of a repressor protein on the CDE-CHR motif at this stage, whereas the presence of complex IVb in S phase might involve the binding of an activator, as it was recently demonstrated for the induction mechanism of CDE-CHR–containing mammalian gene promoters (Liu et al., 1998; Philips et al., 1999).
The data obtained from the in vitro analysis correlated quite well with the in vivo footprinting results that also showed cell cycle–modulated protection of E2F sites (Figure 2). Actually, the E2Fa site showed a strong protection in G1 and S phases on the upper strand, which could correspond to binding of complex Ia, mostly in G1 and S phases. In vivo and in vitro binding to E2F element specifically in G1 and S phases already has been described for an E2F site in direct orientation in the G1/S-induced mammalian DHFR promoter (Wells et al., 1996). A more constitutive protection was observed on the lower strand of the E2Fa motif, except for a hypersensitive site in G2 and S phases that could be correlated with the binding of complex IIa; this binding is maximal in S phase and quite marked in G2 phase. The CDE motif overlapping the E2Fb motif on the upper strand displayed some protection in G2 and S phases of the cell cycle, and hypersensitive sites were present mainly in G2. We therefore can postulate the binding of complex IVb to the CDE element. In contrast, the E2Fb motif on the lower strand was strongly protected at all phases of the cell cycle and could be a good candidate for binding complex Ib, which presented a nonvarying binding activity throughout the cell cycle. Constitutive binding of E2F-containing complexes already has been described in animal gene promoters, notably over the E2F site of the human thymidine kinase promoter (Kim and Lee, 1991; Tommasi and Pfeifer, 1997) and over a reverse E2F motif of the DHFR promoter (Wells et al., 1996). In this case, the reverse E2F motif has been shown to bind an E2F protein different from that found on the E2F site occupied at G1/S (Wells et al., 1997).
Comparing the in vivo footprint patterns with the protein/DNA contact points highlighted by the crystal structure of the human E2F4/DP2/DNA complex (Zheng et al., 1999) provides further evidence for in vivo binding of E2F-containing complexes on the corresponding elements of the RNR2 promoter. Within the E2Fa motif, the two protected G residues (−348, −349) on the outermost right of the lower and upper strands, respectively, correspond to contact points between the human E2F4 and DNA, whereas the outermost left hyperreactive G residue (−352) is located in a region subject to local compression, which might well result in hypersensitivity to DMS. The protection of the two internal G residues fits well with the contact points between human DP2 and DNA. Interpretation of the E2F/DP contact points on the E2Fb/CDE motif is somewhat more complex because this motif is composed of two overlapping E2F elements; however, hypersensitive sites are found at similar locations as on the E2Fa motif, and the position of the protected G residues is consistent with E2F or DP binding on the direct or reverse strands.
Both E2F Elements Function Synergistically for Induction of the RNR2 Promoter at the G1/S Transition
Additional evidence for the fundamental role of the two E2F elements in the RNR2 promoter activity resulted from functional studies of various RNR2 promoter–LUC constructs in synchronized transgenic BY2 cells (Figure 6C).
The LUC activity of the wild-type construct paralleled the RNR2 mRNA steady-state level (Chabouté et al., 1998): low outside S phase, and high in S phase. Mutations of both E2F binding sites resulted in a slight decrease of transcription in asynchronous cells and in synchronous cells outside S phase, which suggests that E2F sites are not essential for basal promoter activity. In contrast, this mutation nearly prevented the G1/S-specific induction of RNR2 promoter, a result similar to that reported when both E2F sites in the mammalian DHFR and CDC6 gene promoters were mutated (Means et al., 1992; Hateboer et al., 1998).
Separate mutations of each E2F element indicated that RNR2 promoter induction is achieved by a differential regulation on the two E2F motifs. The E2Fa element acted essentially as an activator, whereas the E2Fb/CDE appeared to behave as a repressor outside S phase and to switch from a negative to a positive regulator during G1 phase. Similarly, the simultaneous occurrence of activating and repressing E2F sites within the same promoter recently was revealed in the promoters of the mammalian cdc25A phosphatase and of a ran binding protein involved in cell cycle regulation (Di Fiore et al., 1999; Vigo et al., 1999). Moreover, dual functions of the same E2F element, first as a repressor and then as an activator through cell cycle progression, have been described in animals, essentially for the CDE-CHR element. Actually, the repressor CDF complexes binding on the CDE and CHR elements compete with activating E2F complexes binding on the CDE element. Depending on the timing of appearance of the various E2F family members and their respective affinities to the CDE elements, the CDF complexes formed with various gene promoters are sequentially displaced by the activating E2F complexes. Such a mechanism mediates the successive inductions of B-myb, cyclinA, cdc2, and cdc25C genes from mid-G1 to late S phases (Liu et al., 1997, 1998; Lucibello et al., 1997).
Our data provide evidence for a similar complexity in the mechanisms of cell cycle gene regulation through the E2F transcription factor in plants. Further analysis of the various components of the RNR2 promoter transcriptional complexes should enlighten the mechanisms involved in G1/S-specific gene induction. In this respect, the recent isolation in plants of E2F cDNAs (Ramirez-Parra et al., 1999; Sekine et al., 1999) as well as cDNAs for the E2F-inhibitory protein, Rb (Xie et al., 1996; Nakagami et al., 1999), will help to elucidate the nature of the various E2F site binding complexes and unravel E2F-mediated regulation pathways in plants.
METHODS
Plant Cell Culture and Synchronization
The tobacco BY2 cell suspension was maintained by weekly subculturing as previously described (Nagata et al., 1992). The synchronization procedure consisted of treating freshly subcultured stationary-phase cells with aphidicolin (3 μg/mL; Sigma) for 24 hr, followed by extensive washes (Nagata et al., 1992; Reichheld et al., 1995). DNA synthesis was evaluated on 1-mL samples of cell suspension (106 cells) taken at different times after the aphidicolin release. DNA was labeled with 1 μCi 3H-dTTP (90 to 120 Ci/mmol; Du Pont–New England Nuclear) for 30 min at 27°C. Cells then were washed twice with culture medium and frozen in liquid nitrogen. 3H-dTTP incorporation in the nascent DNA was measured as described (Lepetit et al., 1992). Mitotic index was estimated by microscopic analysis in UV light of 500 cells stained with 0.25 μg/mL Hoechst No. 33258-bis benzimide (Sigma) in the presence of 0.2% Triton X-100.
Isolation of the Tobacco RNR2 Promoter
The RNR2 gene promoter was amplified by inverse polymerase chain reaction (PCR) on genomic DNA. The complete promoter sequence was obtained by a two-step approach. In the first step, genomic DNA was digested with TspRI, and the restricted DNA fragments were ligated and submitted to a first PCR with a set of primers (a1 and a2; Figure 1) located just downstream of the ATG. A second PCR with a second set of nested primers (a1' and a2'; Figure 1) was performed with 3 μL of the diluted (1:100 volume) PCR product. The 600-bp amplified DNA fragment was cloned directly into the EcoRV site modified with a 5′ T overhang of the pBluescript II KS+ (Stratagene, La Jolla, CA) and sequenced. The result was confirmed by sequencing other DNA fragments amplified from different PCRs. In the second step, a DNA sequence located further upstream was amplified by another inverse PCR performed on a Cac8I genomic digest with two sets of primers located on the promoter sequence already obtained (b and b'; Figure 1). Based on the overall sequence, a 550-bp RNR2 promoter fragment was amplified by using primers P1 and P2, which contain a BamH1 site at their 5′ ends (Figure 1). The amplified fragment was subcloned into the BamHI site of the pKS+ vector and sequenced on both strands.
In Vivo Dimethyl Sulfate Genomic Footprinting
In vivo DMS genomic footprinting was performed as described (Reichheld et al., 1998). Ten milliliters of cell suspension (106 cells/mL) was treated for 2 min at 20°C with 0.5% DMS to modify the guanine residues that are not protected by proteins. DMS then was removed by washing the cells extensively with ice-cold water. DNA was extracted according to Zimmermann and Goldberg (1977). As a reference, naked genomic DNA was treated in the same conditions, and the reaction was stopped by precipitation of DNA with 0.3 M sodium acetate and 3 volumes of ethanol. DNA was cleaved at the level of DMS-modified residues with 1 M piperidine at 95°C for 30 min, lyophilized three times, and precipitated. Two micrograms of cleaved DNA then was amplified by ligation-mediated PCR. Two sets of three overlapping primers were used to analyze the lower and upper strands of the RNR2 promoter separately (see Figure 2C). The asymmetric linkers used for ligation had the canonical sequences 5′-GCGGTGACCCGGGGAGATCTGAATTC-3′ and 5′-GAATTCAGATC-3′. The annealing temperatures in the PCRs were Tm + 1°C for the first primers, Tm + 2°C for the second primers, and Tm + 4°C for the third primers. The third primers were end-labeled with γ32P-ATP by T4 DNA polynucleotide kinase (New England Biolabs, Beverly, MA) to a specific activity of 0.5 × 106 to106 cpm/pmol. After precipitation, the amplified products were analyzed on standard 6% sequencing gels, and the gels were dried and autoradiographed.
Nuclear Extracts and Electrophoretic Mobility-Shift Assays
Tobacco BY2 cells were harvested by centrifugation at 200g for 10 min and frozen in liquid nitrogen. Pelleted cells were resuspended in 4 mL/g of buffer A containing 25 mM Tris-HCl, pH 6.5, 0.45 M sucrose, 5 mM MgCl2, 5 mM β-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride (Sigma), 250 μg/L pepstatin (Sigma), and 500 μg/L leupeptin (Sigma) and were disrupted with a French press (3000 p.s.i.). Nuclear extracts were prepared as described by Shen and Gigot (1997) and stored in aliquots at −80°C.
Electrophoretic mobility-shift assays (EMSAs) were performed with the nuclear extracts, and the double-stranded oligonucleotides were end-labeled by filling in their 5′ overhangs with α-32P dCTP and Klenow fragment (Gibco BRL). The probes were purified on nondenaturing 10% polyacrylamide gels, and 20,000 cpm of the probes (0.1 to 0.5 ng) was added to 6 μg of nuclear proteins in a final volume of 20 μL of binding buffer (25 mM Hepes-KOH, pH 8.0, 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 5 mM β-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride, 250 μg/L pepstatin, and 500 μg/L leupeptin) in the presence of 0.05% Nonidet P-40 and 1 μg of poly (dI-dC)-poly (dI-dC) (Pharmacia) as a nonspecific competitor. For competition experiments or immunoassays, unlabeled double-stranded oligonucleotides or 1 μL of antibodies (anti-E2F5 [Santa Cruz Biotechnology, Santa Cruz, CA]; anti-α-tubulin [Amersham Pharmacia Biotech]) was added to the reaction mix 30 min and 1 hr, respectively, before adding the DNA probe. After 1 hr on ice, the samples were analyzed on nondenaturing 6% polyacrylamide gels in 0.5 × TBE (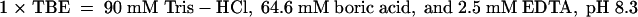 ) at 8 V/cm at 4°C. After electrophoresis, the gels were dried and autoradiographed.
) at 8 V/cm at 4°C. After electrophoresis, the gels were dried and autoradiographed.
Production and Purification of a Tobacco E2F Protein
Spodoptera frugiperda (Sf9) cells were maintained at 27°C in Grace's insect medium (Gibco BRL) containing 10% fetal bovine serum and gentamicin in 100-mL spinner bottles. Viral infection was performed in 60-mm-diameter dishes. The cDNA fragment containing the entire coding region of NtE2F (Sekine et al., 1999) was inserted into the pFastBac HTa plasmid (Gibco BRL). The resulting plasmid, containing a His-tag sequence fused to the N-terminal end of the NtE2F, was transformed into Escherichia coli DH10 Bac, host for baculovirus expression vector (Gibco BRL) for transposition into the bacmid. The recombinant bacmid was isolated and transfected into Sf9 cells with a liposome-mediated transfection kit (Gibco BRL). Recombinant viruses were assayed for expression by protein gel blots and were used to infect Sf9 cells for 72 hr. His-NtE2F was purified from the lysed cells by use of a nickel affinity column according to the manufacturer's instructions (Takara Suzo Co., Shiga, Japan). To further purify His-NtE2F protein, we subjected the eluted fractions to 10% SDS-PAGE, and the fractions corresponding to the NtE2F protein were collected with an electro-elutor (Nihon-Eido Co., Tokyo, Japan).
Protein Gel Blots
For protein gel blot analysis, proteins fractionated by SDS-PAGE were transferred to Immobilon-P membranes (Millipore Corp., Bedford, MA) by using a mini-transblot transfer cell apparatus (Bio-Rad). The blots were incubated with a polyclonal serum raised against the DNA binding domain of the human E2F5 factor (Santa Cruz Biotechnology), and immunodetection was performed with enhanced chemiluminescence detection reagents (Pierce, Rockford, IL).
Promoter–Luciferase Reporter Gene Constructs
The cauliflower mosaic virus 35S promoter was removed from the luciferase (LUC)/intron reporter gene plasmid pLuk07 (Mankin et al., 1997) by digestion with KpnI-NcoI. The E2F-like motifs of the RNR2 promoter were mutated by PCR-based site-directed mutagenesis. The amplified products were relegated, sequenced, and subcloned into the pLuk07 plasmid. The KpnI-XbaI fragments carrying the RNR2 promoter–LUC fusion were subcloned at the KpnI-XbaI sites of the binary vector pCGN 1549 (Calgene, Davis, CA). Plasmids constructs were introduced into Agrobacterium tumefaciens LBA 4404 and used to transform tobacco BY2 cells, as described by An (1987). Approximately 500 to 1000 kanamycin-resistant calli were pooled and reintroduced into suspension cultures. The transgenic cell suspensions were maintained by subculturing 2 mL of stationary-phase cells in 80 mL of fresh medium supplemented with carbenicillin (500 μg/mL) and kanamycin (100 μg/mL). After eight rounds of subculture, carbenicillin was omitted from the medium.
LUC Assay
Two milliliters of cells was washed twice in PBS buffer (140 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, and Na2HPO4, pH 7.4) and lysed by a 10-min incubation at room temperature in 200 μL of lysis buffer (100 mM potassium phosphate buffer, pH 7.8, 1 mM DTT, and 0.2% Triton X-100). After centrifugation at 3000g for 3 min, the supernatant was frozen in liquid nitrogen and stored at −80°C. LUC activity assays were performed with the Luciferase assay kit (Tropix; Perkin-Elmer) in a microplate luminometer (TR 717 Tropix; Perkin-Elmer) according to the manufacturer's instructions.
Acknowledgments
We thank Dr. L. Mankin for supplying the LUC-int vector, Dr. C. Sardet for providing GST (glutathione S-transferase)-human E2F5 purified protein, and Calgene for providing the pCGN vector.
References
- An, G. (1987). Binary Ti vectors for plant transformation and promoter analysis. Methods Enzymol. 153, 292–305. [Google Scholar]
- Beijersbergen, R.L., Kerhoven, R.M., Zhu, L., Carlee, L., Voorhoeve, P.M., and Bernards, R. (1994). E2F–4, a new member of the E2F gene family, has oncogenic activity and associates with p107 in vivo. Genes Dev. 8, 2680–2690. [DOI] [PubMed] [Google Scholar]
- Björklund, S., Skogman, E., and Thelander, L. (1992). An S-phase specific release from a transcriptional block regulates the expression of mouse ribonucleotide reductase R2 subunit. EMBO J. 11, 4953–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabouté, M.-E., Combettes, B., Clément, B., Gigot, C., and Philipps, G. (1998). Molecular characterization of tobacco ribonucleotide reductase RNR1 and RNR2 cDNAs and cell cycle–regulated expression in synchronized plant cells. Plant Mol. Biol. 38, 797–806. [DOI] [PubMed] [Google Scholar]
- Chaubet, N., Flénet, M., Clément, B., Brignon, P., and Gigot, C. (1996). Identification of cis-elements regulating the expression of an Arabidopsis histone H4 gene. Plant J. 10, 425–435. [DOI] [PubMed] [Google Scholar]
- Di Fiore, B., Guarguaglini, G., Palena, A., Kerkhoven, R.M., Bernards, R., and Lavia, P. (1999). Two E2F sites control growth-regulated and cell cycle–regulated transcription of the Htf9-a/RanBP1 gene through functionally distinct mechanisms. J. Biol. Chem. 274, 10339–10348. [DOI] [PubMed] [Google Scholar]
- Dou, Q.-P., and Pardee, A.B. (1996). Transcriptional activation of thymidine kinase, a marker for cell cycle control. Progr. Nucleic Acids Res. Mol. Biol. 53, 197–217. [DOI] [PubMed] [Google Scholar]
- Dyson, N. (1998). The regulation of E2F by pRB-family proteins. Genes Dev. 12, 2245–2262. [DOI] [PubMed] [Google Scholar]
- Elledge, S.J., and Davis, R.W. (1987). Identification and isolation of the gene encoding the small subunit of ribonucleotide reductase from Saccharomyces cerevisiae: DNA damage-inducible gene required for mitotic viability. Mol. Cell. Biol. 7, 2783–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge, S.J., Zhou, Z., and Allen, J.B. (1992). Ribonucleotide reductase: Regulation, regulation, regulation. Trends Biochem. Sci. 17, 119–123. [DOI] [PubMed] [Google Scholar]
- Faisst, S., and Meyer, S. (1992). Compilation of vertebrate-encoded transcription factors. Nucleic Acids Res. 20, 3–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov, D., and Thelander, L. (1995). Role of a proximal NF-Y binding promoter element in S phase–specific expression of mouse ribonucleotide reductase R2 gene. J. Biol. Chem. 270, 25239–25243. [DOI] [PubMed] [Google Scholar]
- Fry, C.J., and Farnham, P.J. (1999). Context-dependent transcriptional regulation. J. Biol. Chem. 274, 29583–29586. [DOI] [PubMed] [Google Scholar]
- Greenberg, G.R., and Hilfinger, J.M. (1996). Regulation of synthesis of ribonucleotide reductase and relationship to DNA replication in various systems. Progr. Nucleic Acids Res. Mol. Biol. 53, 345–392. [DOI] [PubMed] [Google Scholar]
- Hateboer, G., Wobst, A., Petersen, B.O., Le Cam, L., Vigo, E., Sardet, C., and Helin, K. (1998). Cell cycle regulated expression of mammalian CDC6 is dependent on E2F. Mol. Cell. Biol. 18, 6679–6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin, K., Lees, J.A., Vidal, M., Dyson, N., and Fattaey, A. (1992). A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell 70, 337–350. [DOI] [PubMed] [Google Scholar]
- Hijmans, E.M., Voorhoeve, P.M., Beijersbergen, R.L., van't Veer, L.J., and Bernards, R. (1995). E2F–5, a new E2F family member that interacts with p130 in vivo. Mol. Cell. Biol. 15, 3082–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H., and Martin, C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41, 577–585. [DOI] [PubMed] [Google Scholar]
- Johansson, E., Skogman, E., and Thelander, L. (1995). The TATA-less promoter of mouse ribonucleotide reductase R1 gene contains a TFII-I binding initiator element essential for cell cycle–regulated transcription. J. Biol. Chem. 270, 30162–30167. [DOI] [PubMed] [Google Scholar]
- Johnson, L.F. (1992). G1 events and the regulation of genes for S-phase enzymes. Curr. Opin. Cell Biol. 4, 149–154. [DOI] [PubMed] [Google Scholar]
- Kim, Y.K., and Lee, A.S. (1991). Identification of a 70-base-pair cell cycle regulatory unit within the promoter of the human thymidine kinase gene and its interaction with cellular factors. Mol. Cell. Biol. 11, 2296–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi, S., Suzuka, I., and Ohashi, Y. (1995). Two of three promoter elements identified in a rice gene for proliferating cell nuclear antigen are essential for meristematic tissue-specific expression. Plant J. 7, 877–886. [DOI] [PubMed] [Google Scholar]
- Le Cam, L., Polanowska, J., Fabbrizio, E., Olivier, M., Philips, A., Eaton, E.N., Classon, M., Geng, Y., and Sardet, C. (1999). Timing of cyclin E gene expression depends on the regulated association of a bipartite repressor element with a novel E2F complex. EMBO J. 18, 1878–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepetit, M., Ehling, M., Atanassova, R., Chaubet, N., and Gigot, C. (1992). A plant histone promoter can direct both replication-dependent and -independent gene expression in transgenic plants. Mol. Gen. Genet. 231, 276–285. [DOI] [PubMed] [Google Scholar]
- Liu, N., Lucibello, F.C., Körner, K., Wolfraim, L.A., Zwicker, J., and Müller, R. (1997). CDF-1, a novel E2F-unrelated factor, interacts with cell cycle–regulated repressor elements in multiple promoters. Nucleic Acids Res. 25, 4915–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, N., Lucibello, F.C., Engeland, K., and Müller, R. (1998). A new model of cell cycle–regulated transcription: Repression of the cyclin A promoter by CDF-1 and anti-repression by E2F. Oncogene 16, 2957–2963. [DOI] [PubMed] [Google Scholar]
- Lowndes, N.F., Johnson, A.L., Breeden, L., and Johnston, L.H. (1992). SWI6 protein is required for transcription of the periodically expressed DNA synthesis genes in budding yeast. Nature 357, 505–512. [DOI] [PubMed] [Google Scholar]
- Lucibello, F.C., Liu, N., Zwicker, J., Gross, C., and Müller, R. (1997). The differential binding of E2F and CDF repressor complexes contributes to the timing of cell cycle–regulated transcription. Nucleic Acids Res. 25, 4921–4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankin, S.L., Allen, G.C., and Thompson, W.F. (1997). Introduction of a plant intron into the luciferase gene of Photinus pyralis. Plant Mol. Biol. Rep. 15, 186–196. [Google Scholar]
- Means, A.L., Slansky, J.E., McMahon, S.L., Knuth, M.W., and Farnham, P.J. (1992). The HIP1 binding site is required for growth regulation of the dihydrofolate reductase gene promoter. Mol. Cell. Biol. 12, 1054–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, T., Nemoto, Y., and Hazezawa, S. (1992). Tobacco BY2 cell line as the “HeLa” cell in the cell biology of higher plants. Int. Rev. Cytol. 132, 1–30. [Google Scholar]
- Nakagami, H., Sekine, M., Murakami, H., and Shinmyo, A. (1999). Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with cdc2/cyclin D in vitro. Plant J. 18, 243–252. [DOI] [PubMed] [Google Scholar]
- Pearson, B.E., Nasheuer, H.-P., and Wang, T.S.-F. (1991). Human DNA polymerase α gene: Sequences controlling expression in cycling and serum-stimulated cells. Mol. Cell. Biol. 11, 2081–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipps, G., Clément, B., and Gigot, C. (1995). Molecular characterization and cell cycle–regulated expression of a cDNA clone from Arabidopsis thaliana homologous to the small subunit of ribonucleotide reductase. FEBS Lett. 358, 67–70. [DOI] [PubMed] [Google Scholar]
- Philips, A., Chambeyron, S., Lamb, N., Vié, A., and Blanchard, J.M. (1999). CHF: A novel factor binding to cyclin A CHR corepressor element. Oncogene 18, 6222–6232. [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra, E., Xie, Q., Boniotti, M.B., and Gutierrez, C. (1999). The cloning of plant E2F, a retinoblastoma-binding protein, reveals unique and conserved features with animal G1/S regulators. Nucleic Acids Res. 27, 3527–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard, P. (1988). Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 57, 349–374. [DOI] [PubMed] [Google Scholar]
- Reichheld, J.P., Sonobe, S., Clément, B., Chaubet, N., and Gigot, C. (1995). Cell cycle–regulated histone gene expression in synchronized plant cells. Plant J. 7, 245–252. [Google Scholar]
- Reichheld, J.P., Gigot, C., and Chaubet-Gigot, N. (1998). Multilevel regulation of histone gene expression during the cell cycle in tobacco cells. Nucleic Acids Res. 26, 3255–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotheneder, H., Geymayer, S., and Haidweger, E. (1999). Transcription factors of the Sp1 family: Interaction with E2F and regulation of the murine thymidine kinase promoter. J. Mol. Biol. 293, 1005–1015. [DOI] [PubMed] [Google Scholar]
- Sardet, C., Vidal, M., Cobrinik, D., Geng, Y., Onufryk, C., Chen, A., and Weinberg, R.A. (1995). E2F–4 and E2F–5, two members of the E2F family, are expressed in the early phases of the cell cycle. Proc. Natl. Acad. Sci. USA 92, 2403–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler, U., Beckmann, H., and Cashmore, A.R. (1992). TGA1 and G-box binding factors: Two distinct classes of Arabidop-sis leucine zipper proteins compete for the G-box element TGACGTGG. Plant Cell 4, 1309–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine, M., Ito, M., Uemukai, K., Maeda, Y., Nakagami, H., and Shinmyo, A. (1999). Isolation and characterization of the E2F-like gene in plants. FEBS Lett. 460, 117–122. [DOI] [PubMed] [Google Scholar]
- Shen, W.H., and Gigot, C. (1997). Protein complexes binding to cis elements of the plant histone gene promoters: Multiplicity, phosphorylation and cell cycle alteration. Plant Mol. Biol. 33, 367–379. [DOI] [PubMed] [Google Scholar]
- Slansky, J.E., and Farnham, P.J. (1996a). Introduction to the E2F family: Protein structure and gene regulation. In Transcriptional Control of Cell Growth: The E2F Gene Family, P.J. Farnham, ed (Berlin, Heidelberg: Springer-Verlag), pp. 1–30. [DOI] [PubMed]
- Slansky, J.E., and Farnham, P.J. (1996. b). Transcriptional regulation of the dihydrofolate reductase gene. BioEssays 18, 55–62. [DOI] [PubMed] [Google Scholar]
- Slansky, J.E., Li, Y., Kaelin, W.G., and Farnham, P.J. (1993). A protein synthesis-dependent increase in E2F1 mRNA correlates with growth regulation of the dihydrofolate reductase promoter. Mol. Cell. Biol. 13, 1610–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka, K.-I., Kaya, H., Nakayama, T., Araki, T., Meshi, T., and Iwabuchi, M. (1999). Identification of three kinds of mutually related composite elements conferring S phase–specific transcriptional activation. Plant J. 18, 611–623. [DOI] [PubMed] [Google Scholar]
- Terada, R., Nakayama, T., Iwabuchi, M., and Shimamoto, K. (1995). A type I element composed of the hexameric (ACGTCA) and octamer (CGCGGATC) motifs plays a role(s) in meristematic expression of a wheat histone H3 gene in transgenic rice plants. Plant Mol. Biol. 27, 17–26. [DOI] [PubMed] [Google Scholar]
- Thelander, L., Eriksson, L., and Akerman, M. (1980). Ribonucleotide reductase of calf thymus. Separation of the enzyme into two nonidentical subunits, proteins M1 and M2. J. Biol. Chem. 255, 7426–7432. [PubMed] [Google Scholar]
- Tommasi, S., and Pfeifer, G.P. (1997). Constitutive protection of E2F recognition sequences in the human thymidine kinase promoter during cell cycle progression. J. Biol. Chem. 272, 30483–30490. [DOI] [PubMed] [Google Scholar]
- Verma, R., Patapoutian, A., Gordon, C.B., and Campbell, J.L. (1991). Identification and purification of a factor that binds to the Mlu I cell cycle box of yeast DNA replication genes. Proc. Natl. Acad. Sci. USA 88, 7155–7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigo, E., Müller, H., Prosperini, E., Hateboer, G., Cartwright, P., Moroni, M.C., and Helin, K. (1999). CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Mol. Cell. Biol. 19, 6379–6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, M., Blake, M.C., Jambou, R.C., Helin, K., Harlow, E., and Azizkhan, J.C. (1995). An inverted repeat motif stabilizes binding of E2F and enhances transcription of the dihydrofolate reductase gene. J. Biol. Chem. 270, 9783–9791. [DOI] [PubMed] [Google Scholar]
- Wells, J., Held, P., Illenye, S., and Heintz, N.H. (1996). Protein–DNA interactions at the major and minor promoters of the divergently transcribed dhfr and rep3 genes during the Chinese hamster ovary cell cycle. Mol. Cell. Biol. 16, 634–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, J.M., Illenye, S., Magae, J., Wu, C.L., and Heintz, N.H. (1997). Accumulation of E2F–4.DP-1 DNA binding complexes correlates with induction of dhfr gene expression during the G1 to S phase transition. J. Biol. Chem. 272, 4483–4492. [DOI] [PubMed] [Google Scholar]
- Xie, Q., Sanz-Burgos, A.P., Hannon, G.J., and Gutierrez, C. (1996). Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J. 15, 4900–4908. [PMC free article] [PubMed] [Google Scholar]
- Zheng, N., Fraenkel, E., Pabo, C.O., and Pavletich, N.P. (1999). Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev. 13, 666–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, J.I., and Goldberg, R.B. (1977). DNA sequence organization in the genome of Nicotiana tabacum. Chromosoma 59, 227–252. [Google Scholar]
- Zwicker, J., and Müller, R. (1997). Cell-cycle regulation of gene expression by transcriptional repression. Trends Genet. 13, 3–6. [DOI] [PubMed] [Google Scholar]
- Zwicker, J., Lucibello, F.C., Wolfraim, L.A., Gross, C., Truss, M., Engeland, K., and Müller, R. (1995). Cell cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J. 14, 4514–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]