Abstract
Very long chain lipids contribute to the hydrophobic cuticle on the surface of all land plants and are an essential component of the extracellular pollen coat in the Brassicaceae. Mutations in Arabidopsis CER genes eliminate very long chain lipids from the cuticle surface and, in some cases, from the pollen coat. In Arabidopsis, the loss of pollen coat lipids can disrupt interactions with the stigma, inhibiting pollen hydration and causing sterility. We have positionally cloned CER6 and demonstrate that a wild-type copy complements the cer6-2 defect. In addition, we have identified a fertile, intragenic suppressor, cer6-2R, that partially restores pollen coat lipids but does not rescue the stem wax defect, suggesting an intriguing difference in the requirements for CER6 activity on stems and the pollen coat. Importantly, analysis of this suppressor demonstrates that low amounts of very long chain lipids are sufficient for pollen hydration and germination. The predicted CER6 amino acid sequence resembles that of fatty acid–condensing enzymes, consistent with its role in the production of epicuticular and pollen coat lipids >28 carbons long. DNA sequence analysis revealed the nature of the cer6-1, cer6-2, and cer6-2R mutations, and segregation analysis showed that CER6 is identical to CUT1, a cDNA previously mapped to a different chromosome arm. Instead, we have determined that a new gene, CER60, with a high degree of nucleotide and amino acid similarity to CER6, resides at the original CUT1 locus.
INTRODUCTION
The Arabidopsis extracellular pollen coat is essential for pollen recognition by the stigma surface. cer6-2 mutations inhibit pollen hydration by depleting pollen coat lipids and proteins (Preuss et al., 1993), both of which are functionally important (Wolters-Arts et al., 1998; Mayfield and Preuss, 2000). cer6 pollen is viable, and the sterile phenotype is reversed at high humidity, where atmospheric water bypasses the need for hydration by the stigma. In addition, cer6 mutations act specifically at pollen hydration without disrupting pollen adhesion (Zinkl et al., 1999). Here, we report positional cloning of CER6 and analysis of an intragenic suppressor of cer6-2, elucidating the role of CER6 in pollen coat composition and maintenance.
The Arabidopsis CER genes form a large family, originally identified by the glossy, bright-green appearance of cer mutant stems (Koornneef et al., 1989; McNevin et al., 1993). CER proteins, and their homologs in species such as maize and barley, play a role in forming a hydrophobic waxy cuticle that is critical for reducing water loss, averting pathogen attack, and minimizing damage by UV irradiation (reviewed in Post-Beittenmiller, 1996). CER genes also contribute to the synthesis of the lipid-rich pollen coat—several Arabidopsis cer mutants (cer1, cer2, cer3, and cer6) exhibit defective pollen recognition, failed pollen hydration, and consequently, male sterility (Preuss et al., 1993; Hulskamp et al., 1995). The lipids in both the pollen coat and cuticle consist of a mixture of very long chain fatty acids (VLCFAs) and their derivatives (Post-Beittenmiller, 1996). VLCFAs are synthesized by the repeated transfer of two-carbon units from malonyl-acyl carrier protein to C16 and C18 fatty acid precursors. After reaching an appropriate length, VLCFAs are modified to generate alkanes, esters, primary and secondary alcohols, aldehydes, and ketones.
All classes of lipids are detected in cer6 mutants, indicating that this gene is not required for forming fatty acid derivatives. Instead, CER6 is probably involved in lipid elongation, because the quantities of all derivatives >28 carbons long are substantially decreased in cer6 pollen, stems, and leaves (McNevin et al., 1993; Preuss et al., 1993; Jenks et al., 1995). Elongation of fatty acids requires four enzymatic steps: condensation, reduction, dehydration, and a second reduction (reviewed in von Wettstein-Knowles, 1982). The first step, condensation, is thought to be rate-limiting (Millar and Kunst, 1997). Recently, Millar et al. (1999) identified CUT1, an Arabidopsis cDNA encoding a putative condensing enzyme. Plants overexpressing CUT1 have a sense-suppressed phenotype strikingly similar to cer6: they lack stem epicuticular waxes, have substantial decreases in lipids >28 carbons long, and exhibit conditional sterility. However, because mapping CUT1 showed it was not linked to CER6, the two were considered to be distinct (Millar et al., 1999).
As described below, we positionally cloned CER6, sequenced the cer6-1 (Koornneef et al., 1989) and cer6-2 alleles (Preuss et al., 1993), identified and sequenced an intragenic suppressor of cer6-2, placed the CER6 gene on several high-resolution genetic maps, and demonstrated that the cloned gene complements cer6 mutant phenotypes. Unexpectedly, we found that CER6 can exhibit differential activity in pollen and stems; fertility, but not stem wax, was completely restored in the suppressor, as well as in some complemented mutants. Furthermore, we showed that CUT1 and CER6 correspond to the same locus and that the mapping data described by Millar et al. (1999) point to a novel CER6-like gene, CER60.
RESULTS
Mapping CER6
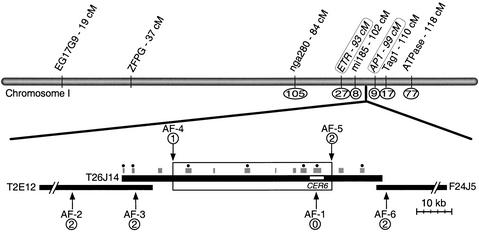
We positionally cloned the CER6 gene by analyzing the segregation of polymerase chain reaction (PCR)–based markers in F2 plants derived from a cross of a Landsberg cer6-2 homozygote to wild-type Columbia (Konieczny and Ausubel, 1993; Bell and Ecker, 1994). After analyzing 490 recombinant chromosomes, we mapped cer6-2 between the molecular markers mi185 and AP1 (see www.Arabidopsis.org/maps.html), consistent with its previously described (Koornneef, 1994) position on chromosome 1 at ∼107 centimorgans (cM) (see Figure 1). We refined the location of CER6 by generating additional PCR-based markers, narrowing the region to 45 kb contained entirely within the sequenced bacterial artificial chromosome (BAC) T26J14 (GenBank accession number AC011915; Figure 1).
Figure 1.
Genetic and Physical Mapping of CER6 on Chromosome 1 and T26J14.
cer6-2 was positionally mapped between mi185 and AP1 on chromosome 1 by scoring PCR-based markers; the indicated positions in centimorgans correspond to the map generated from recombinant inbred lines (http://nasc.nott.ac.uk/new_ri_map.html). New PCR-based markers were generated at ETR and AP1; the centimorgan positions correspond to the phenotypic marker map (Koornneef, 1994) and within the BAC clones T2E12, T26J14, and F24J5 (AF-1 through AF-6). Circled numbers indicate the number of recombination events detected between each marker and CER6. The 45-kb region between AF-4 and AF-5 (open box) defines the boundaries of the region containing CER6; annotated genes (gray boxes), some of which correspond to ESTs (filled circles), are indicated. The white line within the T26J14 BAC indicates the fragment used for complementation experiments.
Annotation of the 45-kb region revealed seven predicted genes (GenBank accession number AC011915), four of which are associated with expressed sequence tags (ESTs) (Figure 1). Moreover, this region included a gene, T26J14.10, for which the predicted exon sequence was entirely identical to that of the CUT1 cDNA, which had been previously mapped to 38 cM on chromosome 1, near the marker ZFPG (Millar et al., 1999). Sense-suppressed CUT1 plants display phenotypes similar to cer6 mutants, and as described below, we found that CUT1 actually maps to the CER6 region. Consistent with the Arabidopsis gene nomenclature standards (Meinke and Koornneef, 1997), we hereafter refer to CUT1 as CER6.
We used PCR to amplify the CER6 sequence, identified a polymorphism within the gene (AF-1), and verified that this polymorphism cosegregated with the cer6-2 mutant phenotype: every homozygous mutant plant from the cer6-2 mapping population contained Landsberg DNA at this locus (Figure 1). In addition, we used tetrad analysis to monitor the segregation of AF-1 in 32 plants derived from eight pollen tetrads (Copenhaver et al., 1998). Because analysis of the segregation of >100 markers in these tetrads has revealed the locations of all the crossover sites, any new marker can be placed precisely on the genetic map. Using this method, we placed CER6 on chromosome 1 between ETR (94 cM) and TAG-1 (110 cM) and excluded the possibility that the gene lies near ZFPG. Finally, by scoring AF-1 segregation on the same recombinant inbred population used by Millar et al. (1999), we placed CER6 uniquely on chromosome 1 at 107 cM, with an LOD score of 3.0 (http://nasc.nott.ac.uk/RI_data/html/chrom1.html).
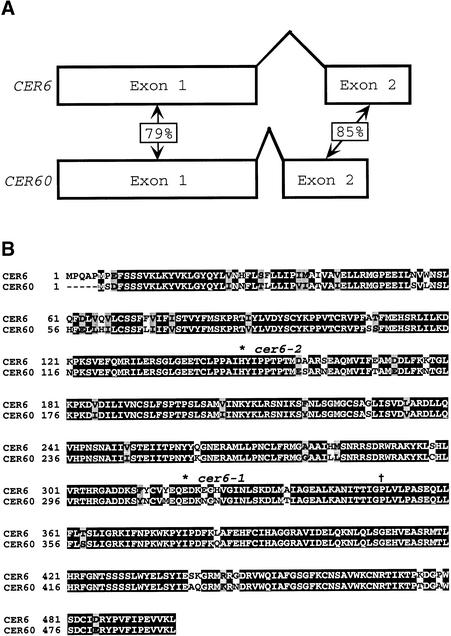
The mapping data we obtained are consistent with the virtually complete genomic sequence generated by the Arabidopsis Genome Initiative. Only one gene identical to CER6 has been found, consistent with the high-stringency DNA gel blot analysis preformed by Millar et al. (1999). In an attempt to account for the previous erroneous placement of CUT1, we investigated the genomic sequence near ZFPG (BAC clone F2J7; GenBank accession number AC079281) and found a novel gene that shares 79 and 85% nucleotide identity with CER6 in exons 1 and 2, respectively, but showed no similarity in the upstream, downstream, or intron sequences (Figure 2A). The similarity in DNA sequence between the 38- and 107-cM regions on chromosome 1 could have confounded the previous mapping experiments with restriction fragment length polymorphisms. Because no previously described cer defect has been mapped to this region, we termed this gene CER60. BLAST searches of the Arabidopsis EST database revealed three high-quality matches for CER60 and defined the splice junction (GenBank accession numbers AV527833, BE527614, and AI997914). At the protein level, CER6 and CER60 share 88% identity and 94% similarity (Figure 2B). Despite this high degree of conservation, however, the presence of wild-type CER60 does not eliminate the phenotype of cer6 mutants. Although ESTs from both genes are found in whole-plant libraries, their tissue specificities could differ. Furthermore, ∼10-fold more ESTs corresponding to CER6 than to CER60 are known, suggesting that the former gene is expressed at higher levels than the latter.
Figure 2.
Sequence Analysis of CER6 and CER60.
(A) Scale drawing of the CER6 and CER60 genes, indicating the percentage of DNA sequence identity for each exon.
(B) Sequence alignment of CER6 and CER60, generated by using ClustalW 1.8 (http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html) and Boxplot (http://www.ch.embnet.org/software/BOX_form.html). (*), residues altered by the mutations; black shading, identical residues; gray shading, similar residues; (†), position of the exon1/exon2 splice junction. Dashes were introduced to optimize alignment.
Analysis and Complementation of cer6 Mutations
We sequenced the cer6-1 and cer6-2 alleles and identified the corresponding lesions. Because both were Landsberg-derived, we sequenced the wild-type Landsberg CER6 gene and showed that it differed by only two silent substitutions from Columbia: F361 (TTC to TTT) and K467 (AAA to AAG). cer6-1 is a three-base deletion eliminating the E319 codon; cer6-2 is an A-to-C transversion that changes H148 to proline (Figure 2B). The cer6-1 deletion lies in a region conserved among other Arabidopsis VLCFA-condensing enzymes (Millar et al., 1999); the H148 altered by cer6-2 is not conserved, but the introduction of a proline residue may alter the protein structure. Interestingly, of the two alleles, cer6-2 has the stronger phenotype, in terms of both stem epicuticular lipids and pollen coat morphology (Preuss et al., 1993).
We introduced a genomic fragment containing T26J14.10 into cer6-2 mutants (Figure 1), identified the transformed plants carrying the gene, and analyzed fertility and stem wax phenotypes (Table 1). All 13 transformants containing CER6 produced at least 30-fold more seed than did the cer6-2 mutants, and most exhibited deposits of epicuticular stem lipids. Transformants containing only the T-DNA vector were identical to the cer6-2 parent. These results confirmed that T26J14.10 complements cer6-2; differences in the extent of complementation may reflect effects of position on gene expression or differences in insertion number.
Table 1.
Complementation of Fertility and Stem Cuticle Phenotype of cer6-2 Mutants with CER6
| Linea | Stem Epicuticular Waxb |
Seeds (n)c | Kan Resistanceb,d |
WT CER6b,e |
|---|---|---|---|---|
| Ler | +++ | 27.2 ± 2.9 (23) | NA | + |
| cer6-2 | − | 0.3 ± 0.6 (14) | NA | − |
| 6-2 FJ | +++ | 29.5 ± 2.0 (12) | + | + |
| 6-2 FG | +++ | 27.4 ± 4.7 (15) | + | + |
| 6-2 FI | +++ | 24.4 ± 6.0 (18) | + | + |
| 6-2 R1 | +++ | 22.2 ± 2.9 (15) | + | + |
| 6-2 FF | +++ | 20.8 ± 2.7 (18) | + | + |
| 6-2 RA | ++ | 28.3 ± 3.3 (19) | + | + |
| 6-2 F4 | ++ | 21.6 ± 4.1 (19) | + | + |
| 6-2 FK | + | 21.8 ± 5.1 (8) | + | + |
| 6-2 F5 | + | 11.5 ± 3.0 (13) | + | + |
| 6-2 FH | −/+ | 26.1 ± 3.3 (19) | + | + |
| 6-2 F3 | −/+ | 23.1 ± 3.1 (16) | + | + |
| 6-2 RB | − | 26.3 ± 2.2 (16) | + | + |
| 6-2 FD | − | 15.9 ± 3.3 (14) | + | + |
| 6-2 FA | − | 0.4 ± 1.0 (10) | + | − |
| 6-2 FB | − | 0.4 ± 0.7 (10) | + | − |
Lines 6-2 FJ through 6-2 FD denote cer6-2 lines transgenic for a wild-type copy of CER6, ranked by their stem cuticle phenotypes. Lines 6-2 FA and 6-2 FB contain vector sequences but lack CER6. Ler, Landsberg erecta.
(+++), wild-type wax levels; (++), (+), (−/+), intermediate wax; (−), no wax.
Mature seeds per half seed pod and standard deviation.
The presence of the kanamycin resistance gene was determined by PCR. NA, not applicable.
The presence of wild-type (WT) CER6 was assessed using two dCAPS markers.
Differential Requirements for CER6 in Transgenic Plants and Suppressors
The cer6 mutations, as with many other cer defects, reduce the abundance of long-chain lipids both in the pollen coat and on the stem surface (Preuss et al., 1993). Interestingly, both defects were not rescued equally by introducing a wild-type CER6 allele into cer6-2 mutants: some transformed plants with restored fertility still exhibited a strong defect in stem wax, whereas the opposite result, sterile transformants with abundant stem lipids, was not observed (Table 1). This unexpected finding suggests that CER6 activity may be greater in reproductive tissues than in stems or that restoring fertility requires less lipids than are required to restore stem wax structures.
Additional support for this differential activity was obtained from the analysis of six intragenic cer6-2 suppressors that were fertile but lacked abundant epicuticular waxes. All six suppressors were dominant: crosses to cer6-2 mutants yielded fertile progeny with glossy stems. Crosses to the wild type confirmed that all six suppressors were tightly linked to cer6-2: one-quarter of the F2 plants had glossy stems, and of those, 782 of 800 were fertile (as opposed to the 600 of 800 expected if the suppressor were not linked). The few sterile glossy plants observed in the F2 population reflected unrelated sterile mutations detected in the mutagenized background. Although the suppressors were isolated as independent M1 mutants, DNA sequencing surprisingly revealed the same reversion event in all six lines. This suppressing mutation, which alters the cer6-2 lesion, is designated cer6-2R. As described above, cer6-2 changes codon 148 from CAT (His) to CCT (Pro); the suppressing mutation is a C-to-T transition that generates a TCT (Ser) codon at position 148.
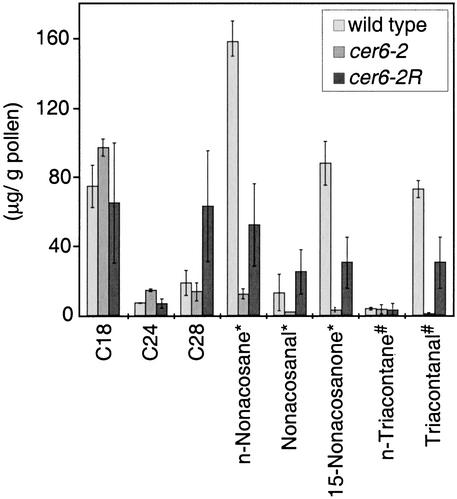
Ultrastructural and chemical analyses of the suppressors confirmed that partial restoration of CER6 was sufficient for a functional pollen coat but not for the development of a normal epicuticular wax layer. Transmission electron microscopy of cer6-2 pollen revealed an unevenly distributed pollen coat with few lipid droplets (Preuss et al., 1993), whereas both wild-type and cer6-2R pollen grains were completely surrounded by an abundant pollen coat containing many lipid droplets (Figure 3). Furthermore, as shown in Figure 4, gas chromatography/mass spectrometry analysis of the pollen coat lipids demonstrated that the C29 and C30 lipids were partially restored in cer6-2R but not to wild-type quantities. Although no epicuticular wax was apparent on the stems of cer6-2R plants, scanning electron microscopy revealed a slight restoration of wax crystals relative to cer6-2 stems (Figure 5). Thus, the suppressing mutation that alters the cer6-2 P148 codon is unable to restore enough CER6 activity to generate a normal stem cuticle, but it is able to generate a morphologically and functionally normal pollen coat, even with C29 and C30 lipids in less than wild-type amounts. Additionally, because cer6-2/cer6-2R heterozygous plants are also fertile, we conclude that one copy of CER6-2R generates sufficient lipid for a functional pollen coat.
Figure 3.
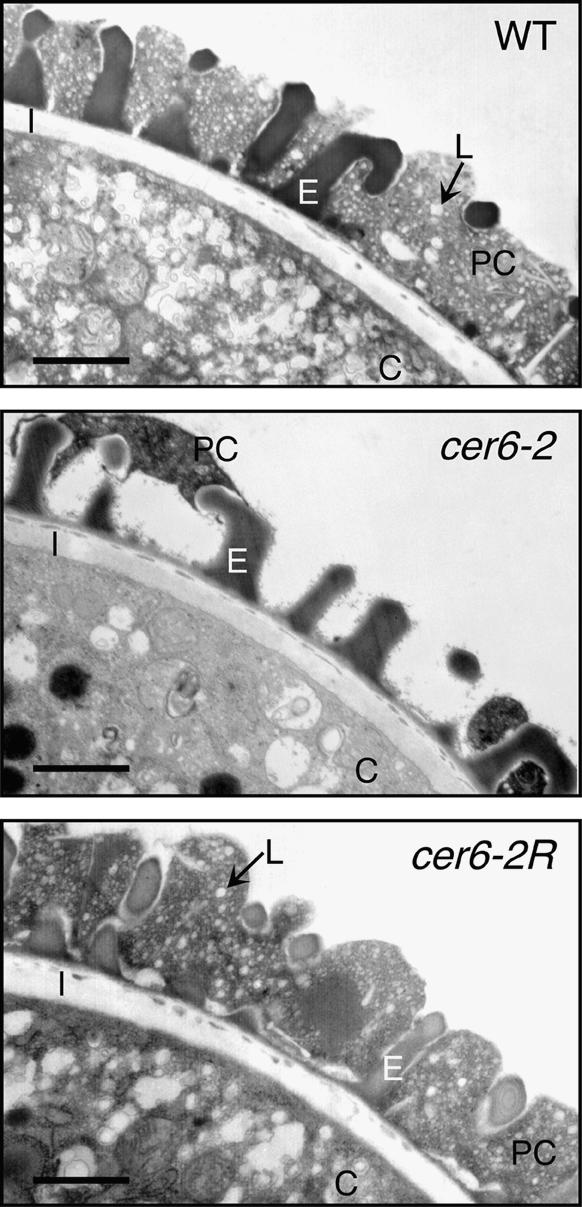
Morphology of the cer6-2R Pollen Coat.
Transmission electron microscopy of wild-type (WT), cer6-2, and cer6-2R pollen grains, showing the resemblance between cer6-2R pollen grains and the wild type. C, cytoplasm; E, exine; I, intine; L, lipid droplet; PC, pollen coat. 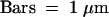 .
.
Figure 4.
Lipid Contents in the Pollen Coats of Wild-Type, cer6-2, and cer6-2R Pollen.
Chloroform-extracted lipids were analyzed by gas chromatography/mass spectrometry. Lipids constituting >1% of wild-type pollen coat lipids are reported. Quantities were normalized to an internal control. All classes of lipids for a given carbon chain length—18, 24, or 28—were pooled for that length. Error bars indicate standard deviation; for the wild type (derived from Landsberg and Columbia), 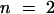 ; for cer6-2,
; for cer6-2, 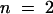 ; and for cer6-2R,
; and for cer6-2R, 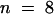 . (*), C29 lipids; (#), C30 lipids.
. (*), C29 lipids; (#), C30 lipids.
Figure 5.
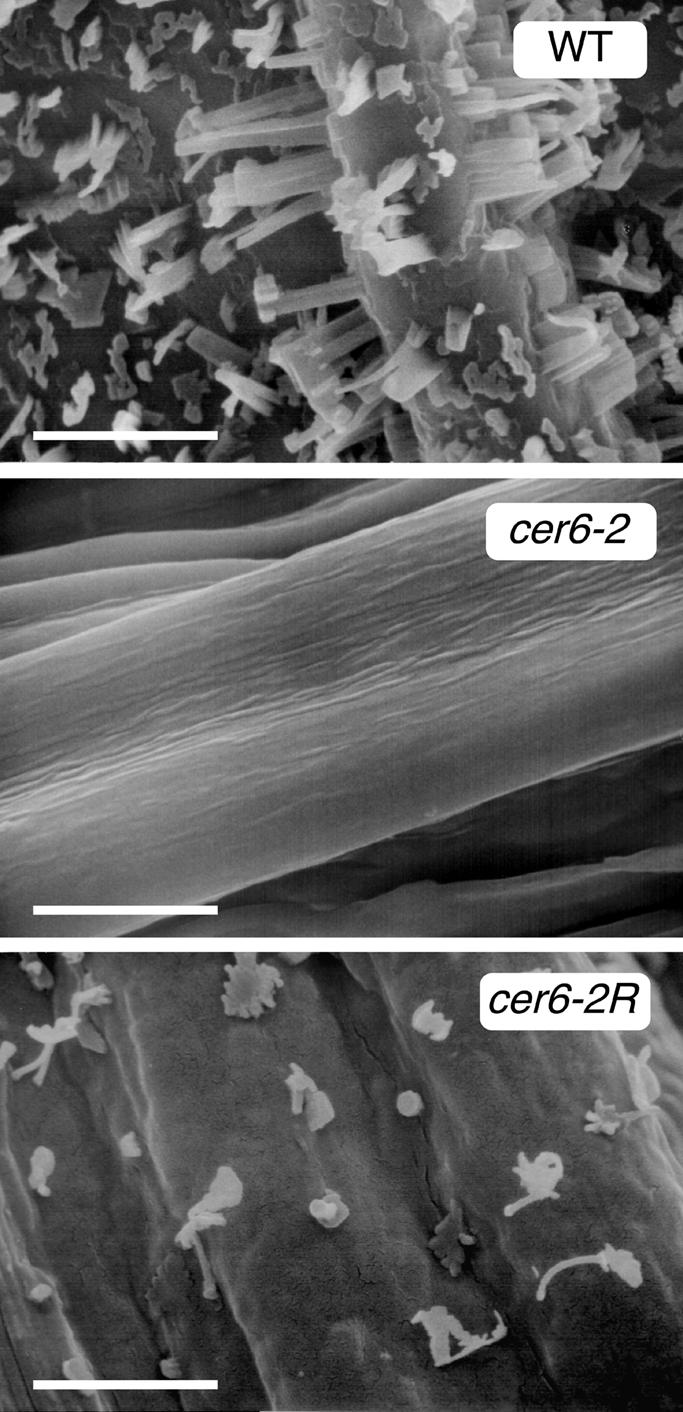
Presence of Lipid Crystals on cer6-2R Stems.
Scanning electron microscopy of wild-type (WT), cer6-2, and cer6-2R stems shows fewer crystals on cer6-2R stems than on those of the wild type.  .
.
DISCUSSION
Here, we describe the cloning of CER6, a lipid biosynthesis gene critical for Arabidopsis pollination, the identification of the cer6-1 and cer6-2 lesions, and the characterization of an intragenic suppressor of cer6-2 sterility. CER6 is identical to CUT1, a cDNA conferring a cer6-like phenotype through sense suppression (Millar et al., 1999). Using a molecular marker and three independent mapping populations, we showed that CER6 maps to chromosome 1 at 107 cM. Furthermore, because CUT1 was previously mapped to 38 cM on chromosome 1, we analyzed the DNA sequence of the surrounding region and identified a related gene, CER60. This gene is the likely source of error in the previous attempt to map a CUT1 restriction fragment length polymorphism.
The stem cuticle and pollen coat of cer6-2 mutants are deficient in all classes of very long chain lipids (Preuss et al., 1993), and the predicted CER6 amino acid sequence is highly similar to that of VLCFA-condensing enzymes and to other condensing enzymes such as chalcone synthase (Millar et al., 1999). Sense-suppressed mutants overexpressing the CER6 message can display even stronger lipid synthesis defects (Millar et al., 1999), raising the possibility that such mutants are deficient in messages from both CER6 and the closely related gene, CER60. Although CER60 probably encodes another VLCFA-condensing enzyme, understanding the role of this gene in plant development will require analysis of mutants that are defective only in CER60.
In Arabidopsis, a family of at least 22 CER genes controls the formation of cuticular lipids (Koornneef et al., 1989; McNevin et al., 1993). To date, the genes corresponding to four cer mutants have been cloned from Arabidopsis: CER1 (Aarts et al., 1995), CER2 (Negruk et al., 1996; Xia et al., 1996), CER3 (Hannoufa et al., 1996), and CER6 reported here. cer1 mutants accumulate C30 aldehyde and are deficient in C29 alkane, secondary alcohols, and ketones (Hannoufa et al., 1993); the CER1 gene shares several characteristics with decarbonylases, indicating a role in the conversion of aldehydes to alkanes. Like cer6 mutants, cer2 plants accumulate shorter lipids and lack longer ones (Hannoufa et al., 1993; Jenks et al., 1995); CER2 shares sequence similarity with GL2, a maize protein having an unknown enzymatic role required for lipid biosynthesis. Neither the sequence of CER3 nor the phenotype of the cer3 mutant suggests a role for this putative nuclear protein in lipid biosynthesis.
In addition to cloning CER6, we identified an intragenic suppressor, cer6-2R, that only partially restores pollen coat lipids but fully restores fertility. These data indicate that pollen hydration and germination can proceed when the coat contains only a small amount of very long chain lipids. Moreover, the epicuticular wax defects seen with the cer6-2R suppressor, and in some cer6-2 mutants transformed with CER6, apparently indicate different requirements for very long chain lipids in the assembly of the pollen coat and in the stem cuticle. Possibly, cer6-2R is the only mutation that can fully restore cer6-2 fertility without producing normal amounts of stem lipids. We independently isolated cer6-2R six times and found no mutations that upregulate cer6-2, alter CER60, or modify any other Arabidopsis lipid-condensing enzyme. Consequently, we conclude that the proline residue at position 148 in CER6 is detrimental to its overall structure and function. Because the abundance of very long chain lipids in cer6-2R plants is much less than in the wild type, this suppressor provides an important opportunity to dissect the requirements for lipids in the assembly of the pollen coating, interaction with pollen coat proteins, and migration of the coating onto the stigma surface. Moreover, this allele will be of use in elucidating the role of different classes and quantities of long-chain lipids in the formation of epicuticular wax structures.
METHODS
Arabidopsis thaliana Strains
cer6-1 was generated by fast neutron mutagenesis (Koornneef et al., 1989); cer6-2 was generated with ethyl nitrosourea (Preuss et al., 1993). Both lines are available from the Arabidopsis Biological Resource Center (Ohio State University; http://aims.cps.msu.edu/aims). Recombinant inbred lines were also provided by the Arabidopsis Biological Resource Center (Lister and Dean, 1993). cer6-2R was generated by mutagenizing cer6-2 seeds with 1% ethyl methanesulfonate; fertile M1 plants were identified as described previously (Ohad et al., 1996).
Molecular Markers and DNA Sequencing
All polymerase chain reaction (PCR)–based genetic markers used in this study are available from the Arabidopsis Information Resource (http://www.arabidopsis.org/aboutcaps.html). The identity of the recombinant inbred lines was verified by scoring the markers AthATPase and ZFPG (chromosome 1, 118 and 37 cM, respectively), nga361 (chromosome 2, 63 cM), and ca72 (chromosome 5, 30 cM). Primers for the AF-1 marker, which corresponds to CER6, were AF-1F (5′-GGCTAGAAGCGAGGCTCAGATGG) and AF-1R (5′-GCCAGCTTGAAA-TCCGGTATGTATG). Digestion of the AF-1 PCR product with MnlI revealed an additional cleavage site in Columbia DNA compared with Landsberg. Two dCAPS markers (Neff et al., 1998) were designed; because each uses the same forward primer (dCER6-F, 5′-TCGTGTCCCCTTCGCAACTTTCAT) with the reverse primer dCER6-2R (5′-GTCCATGGTTGGTGTGGGAGGAATCTAA) or dCER6wt-R (5′-GTCCATGGTTGGTGTGGGAGGAATATCA), detection of the cer6-2 or wild-type CER6 allele, respectively, is unambiguous. Digestion of the dCER6-F/dCER6-2R PCR product with DdeI cleaves the mutant allele, whereas digestion of the dCER6-F/dCER6wt-R PCR product with NlaIII cleaves the wild-type allele.
The 3.6-kb region that includes CER6 was sequenced as six overlapping regions from wild-type Landsberg, cer6-1, cer6-2, and cer6-2R genomic DNA. Fragments were amplified with Pfu Turbo (Stratagene, La Jolla, CA), cloned into the pCR2.1 vector by using the TOPO-TA cloning kit (Invitrogen, Carlsbad, CA), and transformed into TOP10F′ cells (Invitrogen). At least two clones from independent PCR reactions were sequenced.
Mutant Analysis and Complementation
Pollen coat collection, lipid extraction, gas chromatographic/mass spectrometric lipid analysis (Mayfield and Preuss, 2000), and scanning and transmission electron microscopy (Preuss et al., 1993) were performed as previously described.
A 3988-bp fragment containing the entire CER6 coding sequence, 1143 bp 5′, 650 bp 3′, and no other predicted genes, was generated by EcoRV digestion of the bacterial artificial chromosome (BAC) T10H19. This fragment was introduced into the SmaI site of a lacZ-containing version of pBI-121 (Clonetech, Palo Alto, CA), generating pBI-CER6. These CER6-containing clones were electroporated into Escherichia coli DH5α and Agrobacterium tumefaciens GV3101, and Agrobacterium-carrying pBI-CER6 were introduced into cer6-2 plants by vacuum infiltration (Bechtold and Pelletier, 1998). To obtain seeds, infiltrated cer6-2 plants were exposed to humid conditions as described previously (Preuss et al., 1993). Transformed seedlings were selected on agar plates containing 0.5 × Murashige and Skoog (1962) medium, 50 mg/mL kanamycin (Kan), and 25 mg/mL benomyl; Kan-resistant plants were transferred to soil after 4 weeks. The presence of the Kan resistance gene was confirmed by PCR; the presence of a wild-type CER6 allele was monitored by using the dCAPS markers. The stem cuticle of the transformants was assessed visually, and fertility was assessed by counting the mature seeds per half seedpod.
Acknowledgments
We thank Brooke Bevis, Natalie Cullinane, Sarah Kniss, and Annette Weglinski for assistance in the mapping of this gene; Bonnie Bartel for sharing markers; Chris Town and Samir Kaul for sharing DNA sequences; and members of the Preuss, Greenberg, and Malamy laboratories for helpful discussions. This work was supported by Grant No. MCB-9513511 from the National Science Foundation awarded to D.P., Grant No. 97-35304-4941 from the U.S. Department of Agriculture awarded to R.L.F., and a predoctoral fellowship from the National Science Foundation awarded to A.F. Pollen lipid analysis by the Michigan State University Mass Spectrometry Facility was supported by Grant No. DRR-00480 from the National Institutes of Health.
References
- Aarts, M.G., Keijzer, C.J., Stiekema, W.J., and Pereira, A. (1995). Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7, 2115–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–143. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G.P., Browne, W.E., and Preuss, D. (1998). Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads. Proc. Natl. Acad. Sci. USA 95, 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannoufa, A., McNevin, J.P., and Lemieux, B. (1993). Epicuticular wax of Arabidopsis thaliana eceriferum (cer) mutants. Phytochemistry 33, 851–855. [DOI] [PubMed] [Google Scholar]
- Hannoufa, A., Negruk, V., Eisner, G., and Lemieux, B. (1996). The CER3 gene of Arabidopsis thaliana is expressed in leaves, stems, roots, flowers and apical meristems. Plant J. 10, 459–467. [DOI] [PubMed] [Google Scholar]
- Hulskamp, M., Kopczak, S.D., Horejsi, T.F., Kihl, B.K., and Pruitt, R.E. (1995). Identification of genes required for pollen–stigma recognition in Arabidopsis thaliana. Plant J. 8, 703–714. [DOI] [PubMed] [Google Scholar]
- Jenks, M.A., Tuttle, H.A., Eigenbrode, S.D., and Feldmann, K.A. (1995). Leaf epicuticular waxes of the eceriferum mutants in Arabidopsis. Plant Physiol. 108, 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Koornneef, M. (1994). Arabidopsis genetics. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 89–120.
- Koornneef, M., Hanhart, C.J., and Thiel, F. (1989). A genetic and phenotypic description of eceriferum (cer) mutants in Arabidopsis thaliana. J. Hered. 80, 118–122. [Google Scholar]
- Lister, C., and Dean, C. (1993). Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 4, 745–750. [DOI] [PubMed] [Google Scholar]
- Mayfield, J.A., and Preuss, D. (2000). Rapid initiation of Arabidopsis pollination requires the oleosin-domain protein GRP17. Nat. Cell Biol. 2, 128–130. [DOI] [PubMed] [Google Scholar]
- McNevin, J.P., Woodward, W., Hannoura, A., Feldmann, K.A., and Lemieux, B. (1993). Isolation and characterization of eceriferum (CER) mutants induced by T-DNA insertions in Arabidopsis thaliana. Genome 36, 610–618. [DOI] [PubMed] [Google Scholar]
- Meinke, D., and Koornneef, M. (1997). Community standards: A new series of guidelines for plant science. Plant J. 12, 247–253. [Google Scholar]
- Millar, A.A., and Kunst, L. (1997). Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 12, 121–131. [DOI] [PubMed] [Google Scholar]
- Millar, A.A., Clemens, S., Zachgo, S., Giblin, E.M., Taylor, D.C., and Kunst, L. (1999). CUT1, an Arabidopsis gene re-quired for cuticular wax biosynthesis and pollen fertility, en-codes a very-long-chain fatty acid condensing enzyme. Plant Cell 11, 825–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Neff, M.M., Neff, J.D., Chory, J., and Pepper, A.E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 14, 387–392. [DOI] [PubMed] [Google Scholar]
- Negruk, V., Yang, P., Subramanian, M., McNevin, J.P., and Lemieux, B. (1996). Molecular cloning and characterization of the CER2 gene of Arabidopsis thaliana. Plant J. 9, 137–145. [DOI] [PubMed] [Google Scholar]
- Ohad, N., Margossian, L., Hsu, Y.-C., Williams, C., Repetti, P., and Fischer, R.L. (1996). A mutation that allows endosperm development without fertilization. Proc. Natl. Acad. Sci. USA 93, 5319–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post-Beittenmiller, D. (1996). Biochemistry and molecular biology of wax production in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 405–430. [DOI] [PubMed] [Google Scholar]
- Preuss, D., Lemieux, B., Yen, G., and Davis, R.W. (1993). A conditional sterile mutation eliminates surface components from Arabidopsis pollen and disrupts cell signaling during fertilization. Genes Dev. 7, 974–985. [DOI] [PubMed] [Google Scholar]
- von Wettstein-Knowles, P. (1982). Elongases and epicuticular wax biosynthesis. Physiol. Veg. 20, 797–809. [Google Scholar]
- Wolters-Arts, M., Lush, W.M., and Mariani, C. (1998). Lipids are required for directional pollen-tube growth. Nature 392, 818–821. [DOI] [PubMed] [Google Scholar]
- Xia, Y., Nikolau, B.J., and Schnable, P.S. (1996). Cloning and characterization of CER2, an Arabidopsis gene that affects cuticular wax accumulation. Plant Cell 8, 1291–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkl, G.M., Zwiebel, B.I., Grier, D.G., and Preuss, D. (1999). Pollen–stigma adhesion in Arabidopsis: A species-specific interaction mediated by lipophilic molecules in the pollen exine. Development 126, 5431–5440. [DOI] [PubMed] [Google Scholar]